Abstract
La (SS-B) is a highly expressed protein that is able to bind 3′-oligouridylate and other common RNA sequence/structural motifs. By virtue of these interactions, La is present in a myriad of nuclear and cytoplasmic ribonucleoprotein complexes in vivo where it may function as an RNA-folding protein or RNA chaperone. We have recently characterized the nuclear import pathway of the S. cerevisiae La, Lhp1p. The soluble transport factor, or karyopherin, that mediates the import of Lhp1p is Kap108p/Sxm1p. We have now determined a 113-amino acid domain of Lhp1p that is brought to the nucleus by Kap108p. Unexpectedly, this domain does not coincide with the previously identified nuclear localization signal of human La. Furthermore, when expressed in Saccharomyces cerevisiae, the nuclear localization of Schizosaccharomyces pombe, Drosophila, and human La proteins are independent of Kap108p. We have been able to reconstitute the nuclear import of human La into permeabilized HeLa cells using the recombinant human factors karyopherin α2, karyopherin β1, Ran, and p10. As such, the yeast and human La proteins are imported using different sequence motifs and dissimilar karyopherins. Our results are consistent with an intermingling of the nuclear import and evolution of La.
Keywords: nuclear transport, karyopherin, protein evolution, RNA biogenesis, RNA-binding proteins
The La (SS-B) protein was originally characterized as an autoreactive antigen in patients afflicted with systemic lupus erythematosus (Mattioli and Reichlin, 1974; for reviews see Tan, 1989; Van Venrooij and Pruijn, 1995). La is an RNA-binding protein that is found complexed to a wide variety of RNAs including precursors to tRNA and 5S RNA (Hendrick et al., 1981; Rinke and Steitz, 1982, 1985). By virtue of its affinity for 3′ oligouridylate, La is able to bind to the oligo(U)-tail present in all RNA polymerase III transcripts (Stefano, 1984). Since these oligo(U)-tails are often cleaved during RNA maturation, the interaction between La and these RNAs is likely to be transient. Recently, additional modes of RNA recognition by La have been described. These modes include general recognition of 5′ triphosphates of tRNA precursors and specific recognition of AUG within a Kozak consensus translation initiation sequence (McBratney and Sarnow, 1996; Fan et al., 1998). Human La is a multidomain protein that dimerizes via a COOH-terminal domain and contains a putative ATP-binding cassette (Topfer et al., 1993; Craig et al., 1997). La proteins have been identified in Saccharomyces cerevisiae, Schizosaccharomyces pombe, mosquito, Drosophila, Xenopus, mouse, rat, bovine, and human species (Chambers et al., 1988; Chan et al., 1989; Scherly et al., 1993; Semsei et al., 1993; Topfer et al., 1993; Bai et al., 1994; Yoo and Wolin, 1994; Pardigon and Strauss, 1996; Van Horn et al., 1997).2 Evolution has conserved the domain arrangement and biochemical properties of the La proteins. The La proteins share an NH2-terminal “La domain,” central RNA recognition motif (RRM),1 and a highly charged COOH terminus. The region of most divergence among the La proteins is their COOH-terminal half, where apparently unconserved features of the mammalian La proteins, such as an atypical RRM, an ATP-binding motif, and a dimerization domain, are located (Birney et al., 1993; Topfer et al., 1993; Craig et al., 1997).
Defining a unique function of La has been difficult. It participates in both transcriptional and posttranscriptional processes (for review of such proteins see Ladomery, 1997). La has been shown to be an important constituent of RNA polymerase III initiation, termination, and recycling reactions in vitro (Gottlieb and Steitz, 1989a ,b; Maraia, 1996). The highly unusual histone mRNAs, which are devoid of introns and poly(A) tails, were demonstrated to be specifically protected from cell cycle–dependent degradation in vitro by La, possibly through its interaction with a 3′-stem loop (McLaren et al., 1997). La was also shown to be essential for the cap-independent, internal ribosome entry site-mediated (IRES) translation of several viral RNAs, including those of polio, human immunodeficiency virus, and hepatitis C (Meerovitch et al., 1993; Svitkin et al., 1994; Ali and Siddiqui, 1997). A genetic analysis of the S. cerevisiae La, Lhp1p, led to the discovery of a role for Lhp1p in 3′-endonucleolytic cleavage of tRNA precursors in vivo (Yoo and Wolin, 1997). Based on this function, S. cerevisiae and S. pombe each appear to have only one La protein (Yoo and Wolin, 1994; Van Horn et al., 1997). Deletion of the S. pombe La, Sla1, leads to tRNA processing defects that can be complemented by the expression of Sla1 or LHP1 or even human La, indicating functional conservation throughout the La family (Van Horn et al., 1997). Recently, 5′ and 3′ processing of tRNA precursors was shown to be affected by La in human HeLa cell extracts, further confirming the functional conservation of La (Fan et al., 1998). Additionally, human La has been shown to have an ATP hydrolysis activity and to be an ATP-dependent RNA/DNA and RNA/RNA helicase (Bachmann et al., 1990; Huhn et al., 1997).
In an attempt to unify these seemingly disparate observations, it has been suggested that La acts as a general RNA-folding protein, or RNA chaperone (Meerovitch and Sonenberg, 1993; for reviews see Herschlag, 1995; Weeks, 1997). In such a scenario, La, which is highly expressed and binds to many different RNAs, stabilizes unfolded and/or folded RNA domains, possibly decreasing “off-pathway” folding events in a manner analogous to the action of the hsp70 class of chaperones on polypeptides (for reviews see Hartl, 1996; Bukau and Horwich, 1998).
To carry out its myriad of cellular tasks, La is needed in both the nucleus, for its transcription and tRNA-processing activities, and the cytoplasm, for its translation and mRNA protection activities. Indeed, La has been shown to shuttle from its steady-state nuclear localization to the cytoplasm after herpes or polio infection, consistent with its observed functions in viral translation (Bachmann et al., 1989; Meerovitch et al., 1993). A cell cycle–dependent redistribution to the nucleolus has also been observed (Deng et al., 1981). Nucleocytoplasmic transport of macromolecules occurs via several distinct pathways (for reviews see Pemberton et al., 1998; Wozniak et al., 1998). A unique soluble factor, or karyopherin (Kap) governs each individual pathway. (Having been identified nearly coincidentally in several labs, these factors have been given many names, including: importins, exportins, transportins, RanBPs, PTACs, p97, and nuclear localization sequences [NLS] receptor [Pemberton et al., 1998].) Each Kap recognizes cognate transport substrates and transports them through the nuclear pore complex (NPC) in concert with the small GTPase Ran and its cofactors (for review see Moore, 1998). Only Kapα/Kapβ1 (Kap60p/Kap95p in S. cerevisiae) functions as a dimer, all of the other Kaps characterized are monomeric, able to recognize both substrate and the NPC. In the Kapα/Kapβ1 case, α-Kaps recognize NLSs rich in basic residues whereas Kapβ1 binds to the NPC. A 27-amino acid (aa) NLS of the human La has recently been delineated (Simons et al., 1996). Consistent with a need for regulated and specific nucleocytoplasmic transport of La, we have previously demonstrated a requirement for a distinct Kap for the nuclear localization of Lhp1p in S. cerevisiae (Rosenblum et al., 1997). Sxm1p/Kap108p mediates this pathway. In the course of further characterization of the nuclear import of Lhp1p via Kap108p, we have now identified an evolutionary divergence in the pathway of nuclear import of La proteins. This divergence coincides with a significant increase in the complexity of La proteins and suggests interplay between nuclear import and evolution of this class of proteins.
Materials and Methods
Strains and Plasmids
DF5 was used as the parent strain for all new strains constructed (Finley et al., 1987). The Lhp1–PrA strains and the Kap108–PrA strain in the wild-type background were described previously (Rosenblum et al., 1997). The Kap95-PrA strain was the generous gift of M.P. Rout (Rockefeller University, New York, NY) and J.D. Aitchison (University of Alberta, Alberta, Canada) (Aitchison et al., 1996). The Kap108-PrA/Δlhp1 strain was generated by direct integration of a PCR product (as described in Aitchison et al., 1995) directly into a haploid Δlhp1 strain (provided by S. Wolin, Yale University, New Haven, CT). Proper integration was assessed by PCR and Western blotting as described previously (Rosenblum et al., 1997). Yeast strains with and without plasmids were grown at 30°C in dropout and YPD media, respectively (Ausubel et al., 1997).
The green fluorescent protein (GFP) constructs were assembled in pYX242 (Novagen, Madison, WI). pYX242 is a 2-μ plasmid with the triose phosphate isomerase promoter and LEU2 marker. The GFP used, a yeast codon-optimized, fluorescence optimized mutant (yEGFP3), was the kind gift of B. Cormack (Stanford University, Stanford, CA) (Cormack et al., 1997). All restriction enzymes were purchased from New England Biolabs (Beverly, MA), Pfu was purchased from Stratagene (La Jolla, CA). yEGFP3 was cloned into pYX242 using the enzymes HindIII and SalI to generate the plasmid pYX242–GFP. LHP1 was amplified from S. cerevisiae genomic DNA (Promega, Madison, WI) and cloned into pYX242–GFP using primer-encoded BamHI and ApaI sites to generate pYXLhp1–GFP. Restriction of pYXLhp1–GFP with HindIII, filling in the overhangs with T4 DNA polymerase and religating generated the fragment 1 construct, pYXfrag1–GFP. Fragments 2 and 3 were cloned with primer-encoded EcoRI/BamHI and BamHI/HindIII sites, respectively to generate pYXfrag2–GFP and pYXfrag3–GFP. The human La was amplified from a human fetal bone marrow cDNA library (Clontech, Palo Alto, CA) using primer-encoded BamHI and HindIII sites, generating pYXhsLa–GFP. Drosophila and S. pombe Las were amplified from cDNAs kindly provided by S. Wolin and cloned into pYX242–GFP using primer-encoded BamHI and HindIII sites to generate pYXdmLa–GFP and pYXspLa–GFP. pYXspLa35–GFP and pYXspLa67–GFP, which contain the last 35 and 67 aa of Sla1, respectively, fused to GFP, and were cloned from Sla1 cDNA using primer-encoded EcoRI and HindIII sites. The construct encoding the last 35 aa of Drosophila La fused to GFP, pYXdmLa35–GFP, was generated by restriction of pYXdmLa–GFP with EcoRI and religating. Yeast strains were transformed with plasmids via electroporation (Ausubel et al., 1997).
To express human La in bacteria for in vitro import assays, the hsLa– GFP cassette from pYXhsLa–GFP was amplified and cloned into pET21b (Novagen) using primer-encoded BamHI and SalI sites. The Escherichia coli strains XL1-blue and BL21(DE3) (Stratagene) were used for general cloning and protein expression, respectively. Bacterial strains were transformed by heat shock (Ausubel et al., 1997).
Western Blotting
Total yeast extracts were isolated as previously described (Rosenblum et al., 1997). After electrophoresis through a 7.5% polyacrylamide gel, proteins were transferred to nitrocellulose (0.4 μ, Schleicher & Schuell, Keene, NH). Blots were visualized with amido black (0.1% amido black in 40% methanol, 10% acetic acid) before blocking in 5% milk in TBS (10 mM Tris, 150 mM NaCl, pH 7.5) for 15 min. Blots were incubated with polyclonal anti-GFP antibodies (Clontech) at 1:1,500 dilution in 5% nonfat dry milk in TBS for 2 h. After incubation, blots were rinsed with water, washed twice with 0.05% NP-40 in TBS, and then once with TBS. The blots were incubated with horseradish peroxidase-coupled donkey anti– rabbit antibodies (Amersham, Arlington Heights, IL) at 1:2,000 dilution in 5% milk in TBS for 1 h. After this incubation, blots were washed in a similar manner to before. Enzyme-linked antibodies were detected using luminol-based chemiluminescence (Schneppenheim et al., 1991).
Protein Purification
For the purification of protein A fusion proteins from yeast, cytosol was prepared from strains essentially as described (Rout and Blobel, 1993). For a typical purification, cytosol from ∼2 g (wet weight) of cells in a final volume of 50 ml was incubated overnight at 4°C with 25 μl of IgG Sepharose (made from coupling CNBr-activated Sepharose 4B [Pharmacia Biotech, Piscataway, NJ] to rabbit IgG [Cappel, Malvern, PA] following standard protocols). The Sepharose was isolated and washed at least six times with 1 ml TB (20 mM Hepes, pH 7.5, 110 mM KOAC, 2 mM MgCl2, 0.1% Tween 20), before elution with acid (0.5% HOAc, 150 mM NaCl) or MgCl2 (various concentrations in 20 mM Hepes, pH 7.5, 0.05% Tween 20). Protein samples were concentrated by the method of Wessel and Flugge (1984). Proteins were identified using a combination of MALDI-TOF mass spectrometry and Edman sequencing (Fernandez et al., 1994; Gharahdaghi et al., 1996). Peptide masses were compared with several databases (http://prospector.ucsf.edu or http://prowl.rockefeller.edu/cgi-bin/ProFound).
For the purification of hsLa–GFP from E. coli, BL21(DE3) cells harboring pEThLa–GFP were grown in 30 ml 2× YT at 37°C to an OD600 of 0.7. This starter culture was then diluted 1:20 in fresh 2× YT and grown to an OD600 of 0.7 at 37°C. Recombinant protein expression was induced by addition of isopropyl B-p-thiogalactopyranoside to 0.5 mM. Cells were allowed to grow for an additional 3 h at 30°C. Cells were harvested, resuspended in 30 ml of 50 mM KH2PO4, 300 mM NaCl, 30 mM beta-mercaptoethanol, pH 8.0, and lysed by passing twice through a French pressure cell at 900 pounds per square inch. After sedimentation of cell debris, the fusion protein was purified by virtue of the plasmid-derived hexahistidine sequence using 1.5 ml Talon resin (Clontech) following the protocols supplied by the manufacturer.
Sequence Analysis
Sequences were aligned using ClustalW 1.6 (Megalign Program; DNAStar, Inc., Madison, WI). Phylogenetic analysis was performed using Phylip (PHYLIP v 3.5c; distributed by J. Felsenstein, University of Washington, Seattle, WA). Both programs were used with standard settings.
Fluorescence Microscopy
GFP was visualized in live yeast after overnight growth in selective medium or on selective agar. 4 μl of culture or one colony resuspended in 4 μl of water was placed on a slide. All micrographs were taken using a 63× oil objective on an Axiophot microscope (Carl Zeiss, Thornwood, NY) and Zeiss filter set number 9 (excitation, 450–490 nm; beamsplitter, 510 nm; emission, long-pass 520 nm) with coincident differential interference contrast (Nomarski) optics. Images were transferred directly to Adobe Photoshop 3.0.5m (Adobe Systems, San Jose, CA) using a Sony DKS-5000 digital photo camera (Tokyo, Japan).
Nuclear Import Assay
Import assays were performed in digitonin-permeabilized HeLa cells essentially as described (Moore and Blobel, 1992). Each assay contained 1 mM GTP and a combination of the following factors: 0.5 μg of Kapα2, 0.5 μg of Kapβ1, 4 μg of Ran, 60 ng of p10, 1 μg of human La-GFP, and 500 μg of HeLa S-100 cytosol.
Results
Kap108p Directly Binds both Lhp1p and rpL11p
Previously, we demonstrated that Kap108p could be isolated from yeast cytosol in a complex whose main components were Lhp1p and rpL11p (Rosenblum et al., 1997) (rpL11p was previously known as Rpl16p, for new S. cerevisiae ribosomal protein nomenclature see Mager et al., 1997). At the time we could not differentiate between two coexisting, dimeric complexes of Kap108p and a single trimeric complex of all three members. To address this issue, we generated a yeast strain that contained a genomically expressed fusion between protein A and Kap108p (Kap108– PrA) but was deleted for LHP1. We then isolated Kap108–PrA by virtue of the affinity for IgG-Sepharose of protein A. The results are shown in Fig. 1 a. In the first lane, for comparison, cytosolic Kap108–PrA was purified from cells wild type for LHP1 and the bound proteins were eluted with acid. The second panel of Fig. 1 a shows the result of a similar experiment done in parallel with cytosol from the LHP1-deletion strain. rpL11p eluted from Kap108–PrA in the LHP1 deletion strain at the same concentrations of MgCl2 that dissociate this complex isolated from a strain containing Lhp1p. As such, it is evident that Lhp1p does not mediate the rpL11p/Kap108p interaction. We have also detected other, smaller ribosomal proteins in complex with Kap108p (Rosenblum et al., 1997). In Fig. 1 a, right, these smaller ribosomal proteins, between ∼11 and 17 kD, appear to elute from the column earlier than rpL11p, peaking in the 100 mM MgCl2 fraction. Therefore, we cannot exclude the possibility that these ribosomal proteins bind to Kap108p via rpL11p.
Figure 1.
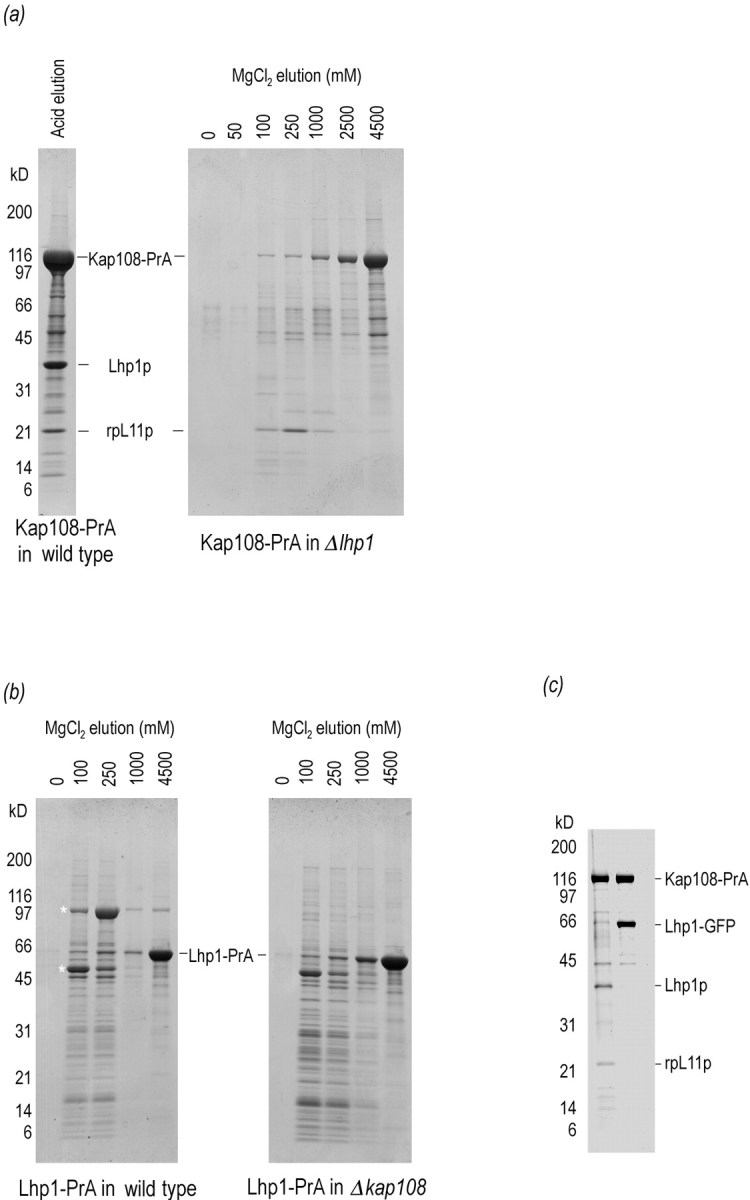
Kap108p binds directly to both Lhp1p and rpL11p. The binding sites of Lhp1p and rpL11p on Kap108p overlap. (a) Genomically expressed Kap108– PrA was isolated from cytosol of a wild-type strain (left) and an lhp1-deletion strain (right). After incubation with IgG-Sepharose, unbound proteins were removed and bound proteins were eluted with acid (left) and a MgCl2 gradient (right). After separation on 4–20% SDS-PAGE gels, proteins were visualized with Coomassie blue R. (b) Genomically expressed Lhp1–PrA was isolated from cytosol of a wild-type strain (left) and a KAP108-deletion strain (right). Purified cytosol from each strain was incubated with IgG-Sepharose, washed and eluted with a MgCl2 step gradient. After separation on 4–20% SDS-PAGE gels, proteins were visualized with Coomassie blue R. Asterisk, Lhp1–PrA-associated proteins. (c) Cytosolic Kap108– PrA was isolated from cytosol of a strain without (left lane), and with (right lane), a plasmid expressing an Lhp1– GFP fusion protein. After isolation of Kap108–PrA, bound proteins were eluted with acid. After separation on a 10–20% SDS-PAGE gel, proteins were visualized with Coomassie blue R.
In a similar fashion, we determined whether the Kap108p/ Lhp1p interaction might be mediated by rpL11p. To do so, we purified Lhp1p, by virtue of an Lhp1p–protein A fusion protein, from strains with and without Kap108p. As shown in Fig. 1 b, the profile of proteins associated with Lhp1p is quite different from that of proteins associated with Kap108p. The profiles of proteins bound to Lhp1– PrA are identical between strains with (Fig. 1 b, left) and without (Fig. 1 b, right) Kap108p with the exception of a prominent band of ∼105 kD. As expected, more Lhp1–PrA and associated factors are present in the cytosol of the KAP108-deficient strain. We identified two Lhp1p-associated proteins. The 50-kD protein eluting mostly at 100 mM MgCl2 was identified by sequencing three peptides, xxxANLIAAGAxxL, xLLCGAAIGTIDAD, and xxxAQLEGxV (Fernandez et al., 1994). The peptides uniquely identified Pur5p, inosine monophosphate dehydrogenase, the enzyme that catalyzes the first dedicated step of GMP biosynthesis. The 105-kD protein eluting mostly in the 250 mM MgCl2 fraction and present only in strains containing Kap108p was analyzed by mass spectrometry (Gharahdaghi et al., 1996). A database search was performed using the observed masses of eleven tryptic peptides generated from this protein (from 964.1 to 2,132 D), and a mass tolerance of 0.5 D. This search uniquely identified this protein as Kap108p, with the peptides covering 14% of the Kap108p sequence. That the interaction between Lhp1–PrA and Kap108p is direct is strongly suggested by the final two lanes of Fig. 1 b, left. In these two lanes, the only predominant proteins remaining on the column were Lhp1–PrA and Kap108p. Further evidence that the Lhp1–PrA/Kap108p interaction is not bridged by rpL11p is suggested by the absence of a distinct band at 22 kD eluting with or after Kap108p from Lhp1–PrA.
As both Lhp1p and rpL11p could bind directly to Kap108p, we determined whether these two substrates bind to independent sites on Kap108p or if their binding sites functionally overlap. This experiment was done in the cellular milieu to avoid nonspecific interactions between these three highly charged proteins. To do so, we constructed a plasmid to overexpress an Lhp1–GFP fusion protein in S. cerevisiae. When Kap108–PrA was purified from a strain harboring this plasmid, a large quantity of Lhp1–GFP copurified, but no endogenous Lhp1p or rpL11p could be seen by Coomassie blue staining (Fig. 1 c). Since Lhp1–GFP could compete with both Lhp1p and rpL11p for binding to Kap108–PrA, it is evident that the binding sites for Lhp1p and rpL11p on Kap108p overlap functionally. This functional overlap could be due to identical or overlapping binding sites or to steric interactions between the ligands.
The Lhp1p NLS Includes an RNP Consensus Domain and Does Not Correspond to the NLS of the Human La
To further characterize the nuclear import of Lhp1p via Kap108p we next delimited a subdomain of Lhp1p that was capable of conferring nuclear localization via Kap108p. To do so, we constructed plasmids for the in vivo expression of Lhp1p and three fragments of Lhp1p, each fused to GFP. A schematic of these constructs is shown in Fig. 2 a. These fragments overlap by at least 25 amino acids to minimize the likelihood that an NLS would be missed because it had been split between two fragments. That these fragments were properly expressed as full-length products was confirmed by Western blotting with anti-GFP antibodies (Fig. 2 b). Consistent with our previous observations with Lhp1–PrA, in wild-type cells, Lhp1–GFP is restricted to the nucleus, whereas in the KAP108 deletion, Lhp1–GFP is unable to target the nucleus (Fig. 2 c, first row) (Rosenblum et al., 1997). As such, the GFP tag, like the PrA tag we have previously used, did not affect the nuclear targeting of Lhp1p. Fragment 1, aa 1–136, contains the La domain which is highly conserved throughout the La family (Van Horn et al., 1997). As a GFP fusion, fragment 1 was unable to accumulate in nuclei of wild-type cells (Fig. 2 c, second row). Fragment 2 spans aa 112–224, whereas the RRM spans aa 124–209. This fragment, which was not as highly expressed as the other fragments (Fig. 2 b), did accumulate in nuclei of wild-type cells (Fig. 2 c, third row). In contrast, when fragment 2 was expressed in cells deleted for KAP108, no accumulation of GFP was seen in nuclei (Fig. 2 c, third row). Fragment 3 is composed of the COOH terminus of Lhp1p, aa 188–275. This fragment is composed of just under 50% of charged residues, and encompasses a region that corresponds to the NLS of human La that has recently been defined by microinjection of in vitro translated hsLa mutants into Xenopus laevis oocytes followed by dissection (Simons et al., 1996). Interestingly, this region was also able to direct nuclear accumulation, yet this accumulation was insensitive to the presence of Kap108p (Fig. 2 c, fourth row). That fragments 2 and 3 contained discrete NLSs was suggested by their different dependence on Kap108p for nuclear localization. To demonstrate that each fragment had its own NLS, a construct was made that encoded the portion of fragment 3 that did not overlap with fragment 2. This shorter fragment demonstrated localization identical to that of fragment 3, in both wild-type and KAP108-deleted cells (data not shown). As full-length Lhp1p is unable to accumulate in the nucleus in the absence of Kap108p (Fig. 2 c, first row), it appears that fragment 3 contains not the endogenous NLS of Lhp1p, but a secondary NLS unmasked by its expression out of context of Lhp1p.
Figure 2.
The Kap108p-dependent NLS of Lhp1p overlaps with the RNA recognition motif of Lhp1p. (a) A diagram of the constructs localized by virtue of fusion with GFP. Three relevant features of Lhp1p, the location of the La domain, RRM, and corresponding region of the human La NLS are noted above the bar representing full-length Lhp1p. (b) Approximately equal amounts of total protein from cells expressing the reporter constructs were separated on a 7.5% SDS-PAGE gel and transferred to nitrocellulose. Polyclonal anti-GFP antibodies were used to visualize the fusion proteins. (c) GFP fusion proteins were visualized in live wild-type and Δkap108 cells. Coincident fluorescence and Nomarski optics were used to localize GFP relative to the cell periphery. Fusion proteins were considered nuclear (Nuc) if they were able to concentrate in the nucleus, otherwise they were considered cytoplasmic (Cyt).
In an attempt to further analyze the fragment 2-type NLS, we first selected a 20-aa portion of this fragment that appeared similar to a stretch of rpL11p. This portion, from lysine 152 to phenylalanine 171 of Lhp1p was highly charged, with 10 charged amino acids, eight of them basic. However, this portion, when fused to GFP, was unable to target the nucleus (data not shown). Next, two proteins were identified that appeared to have significant homology to fragment 2, Snp1p, and the unknown open reading frame YOL041c. Although both of these proteins were nuclear as GFP fusions in wild-type cells, neither lost this localization when expressed in Δkap108 cells, suggesting that they are not substrates of Kap108p (data not shown).
It should be noted that although fragment 2 was the only construct examined that was able to confer Kap108p-dependent nuclear import, it did not contain as strong of a signal for nuclear import as full-length Lhp1p. Whereas no Lhp1–GFP could be seen in the cytoplasm of wild-type cells, fragment 2–GFP was observable in the cytoplasm of wild-type cells. This is the case even though fragment 2 overlapped the Kap108p-independent fragments by 25 NH2-terminal and 37 COOH-terminal amino acids. The most likely explanation for the inefficient import of fragment 2–GFP is that fragment 2, although 113-aa long, does not contain complete structural information for interaction with Kap108p. Another possibility is that fragment 2 is actively exported from the nucleus, perhaps by virtue of binding to RNA.
Heterologous La Proteins, When Expressed in S. cerevisiae, Are Nuclear Even in the Absence of Kap108p
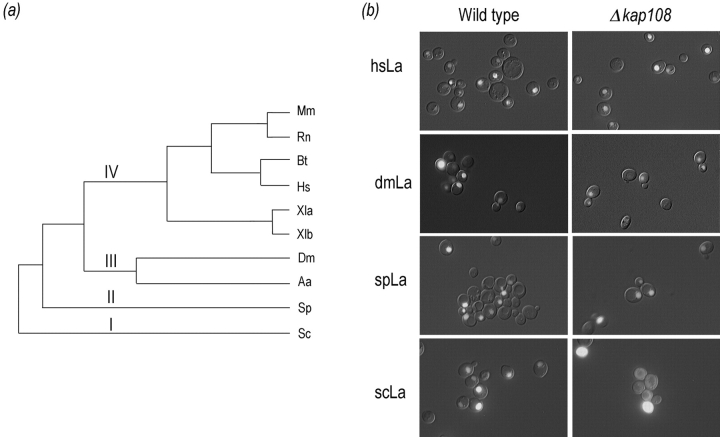
As the Kap108p-dependent NLS of Lhp1p did not coincide with the NLS of human La mapped using the Xenopus assay, we undertook an analysis of intracellular targeting of heterologous La proteins in S. cerevisiae. In the course of evolution, four major branches of La proteins have arisen (Fig. 3 a). The clearest diversion in primary structure among these branches occurs between branches I and II and branches III and IV. In diverging from branches I and II, the branch III and IV proteins grew in primary sequence by ∼30%. For example, spLa is 298-aa long whereas dmLa is 390-aa long. The added sequence is most apparent in the COOH-terminal half of the larger proteins. scLa and spLa are 36% identical, spLa and dmLa are 28% identical, and dmLa and hsLa are 34% identical. GFP fusion constructs were generated for each of these proteins. Each construct was transformed into wild-type cells and cells deleted for KAP108. As shown in Fig. 3 b (left column), human, Drosophila, and S. pombe La proteins were targeted to the nucleus when expressed in S. cerevisiae as fusions with GFP. Significantly, all of the fusion proteins were entirely restricted to the nucleus in wild-type cells, indicating that they were substrates of an S. cerevisiae Kap. Surprisingly, this Kap is Kap108p only for Lhp1p, the endogenous S. cerevisiae La protein (Fig. 3 b, right column). Even Sla1, the S. pombe La protein that is 36% identical to Lhp1p, is targeted to the nucleus in the absence of Kap108p. This targeting of Sla1 occurs even though it, like Lhp1p, diverges markedly from the COOH-terminal domain shared by the La proteins of higher eukaryotes (Van Horn et al., 1997). The NLS of the human La resides in precisely this COOH-terminal domain (Simons et al., 1996).
Figure 3.
La proteins localize to the nucleus in S. cerevisiae, but only Lhp1p, the endogenous S. cerevisiae La, needs Kap108p to do so. (a) A phylogenetic tree of the ten cloned La proteins is shown, branches I–IV are indicated. The phylogenetic tree was generated by aligning the NH2-terminal 250 aa of each La protein using ClustalW 1.6 followed by phylogenetic analysis using Phylip (the aligned sequences are available at http://129.85.13.212/LAS.aln). This region contains the La domain and an RRM for each protein. Species abbreviations and references for cDNA sequencing are as follows: Mm, Mus musculus (Topfer et al., 1993); Rn, Rattus norvegicus (Semsei et al., 1993); Bs, Bos taurus (Chan et al., 1989); Hs, Homo sapiens (Chambers et al., 1988); Xla and Xlb, Xenopus laevis (Scherly et al., 1993); Dm, Drosophila melanogaster (Bai et al., 1994); Aa, Aedes albopictus (Pardigon and Strauss, 1996); Sp, Schizosaccharomyces pombe (Van Horn et al., 1997); Sc, Saccharomyces cerevisiae (Yoo and Wolin, 1994). (b) La proteins from the four main branches of the phylogenetic tree in Fig. 3 a were expressed as GFP fusions in S. cerevisiae. Each protein (rows) was expressed in both wild-type (left column) and Δkap108 (right column) cells. Fusion proteins were visualized as described above.
Human La, Expressed in Yeast, Interacts with Kap95p/Kap60p
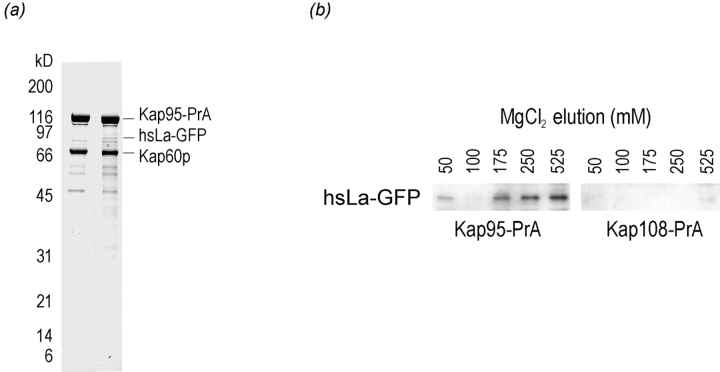
Two sequences similar to the consensus bipartite NLS recognized by Kapα/Kapβ1 have been identified in hsLa and termed box A and box B (Simons et al., 1996). Box A perfectly corresponds to the consensus, with 2 NH2-terminal basic residues, a 10-aa spacer, and a COOH-terminal cluster of three basic residues, whereas box B does not, having a 12-aa spacer and only two basic residues on the COOH-terminal side. Interestingly, box A was shown to be unable to confer nuclear localization on hsLa whereas box B was shown to be the hsLa NLS (Simons et al., 1996). Although we were initially drawn to the differences between box B and the consensus, the ability of human La to localize to the nucleus in the absence of Kap108p led us to consider that human La is brought to the nucleus in an α-Kap– dependent manner. We tested for this biochemically in yeast by introducing our hsLa–GFP plasmid into a strain that carried a genomic protein A fusion with the S. cerevisiae Kapβ1, Kap95p (Kap95–PrA). We then purified cytosolic Kap95p–PrA from this strain by immunoaffinity chromatography. As shown in Fig. 4 a, Kap60p, the S. cerevisiae Kapα, copurifies with Kap95–PrA from an untransformed strain. As Kap95p/Kap60p appears to be the major route to the nucleus, transporting perhaps hundreds of proteins, individual import substrates cannot generally be visualized by Coomassie blue staining (Aitchison et al., 1996; Makkerh et al., 1996). This inability to clearly see substrates most likely results from the stoichiometry of the interaction—each substrate is probably two to three orders of magnitude less abundant than the isolated Kap60p/ Kap95p dimer. Surprisingly, when Kap95–PrA is isolated from cytosol of a strain carrying the human La–GFP fusion, a clear band corresponding to the fusion can be seen by Coomassie blue staining. The ability to visualize hsLa– GFP by Coomassie staining most likely results from the high expression of this construct, making the fusion protein one of the main substrates of this pathway. That the fusion product is observable is even more remarkable when the trimeric nature of the isolated complex is taken into account; purified Kap95–PrA is bound to Kap60p, which in turn is bound to human La–GFP. The identity of the hsLa–GFP band was confirmed by Western blot analysis with anti-GFP antibodies (Fig. 4 b). Significantly, in a separate experiment performed in parallel, no hsLa–GFP copurified with Kap108–PrA (Fig. 4 b).
Figure 4.
Human La interacts with yeast Kap60p/ Kap95p. (a) Proteins associated with Kap95–PrA were isolated from cytosol of strains without (left) and with (right) a plasmid expressing human La as a GFP fusion (hsLa–GFP). Extracts were passed over an IgG-Sepharose column, which was then washed. Bound proteins were eluted with acid and subjected to SDS-PAGE on a 10–20% gel. Proteins were stained with Coomassie blue R. (b) Proteins associated with Kap95–PrA, (left) and Kap108–PrA, (right), were isolated from cytosol. In both cases, the strains harbored a plasmid expressing hsLa–GFP. In this case, bound proteins were eluted with a MgCl2 gradient, separated by SDS-PAGE, and then transferred to nitrocellulose. The GFP moiety was detected by immunoblotting with anti-GFP antibodies. Only the relevant part of the blot is shown.
Human Kapα2, Kapβ1, p10, and Ran Reconstitute Import of Human La
The foregoing experiments strongly suggested that human La is targeted to the S. cerevisiae nucleus via the Kap60p/ Kap95p pathway. To address the targeting of La in human cells, we subjected recombinant hsLa–GFP to the permeabilized HeLa cell assay in the presence of recombinant human factors. As shown in Fig. 5 (first panel), the fusion protein, when added alone to permeabilized HeLa cells, did not localize to the nucleus. In the presence of HeLa cytosol, however, hsLa–GFP was rapidly and efficiently targeted to the nuclei of HeLa cells (Fig. 5, second panel). In the presence of Kapα2 and Kapβ1, a portion of hsLa–GFP could be seen to accumulate around the nuclear rim (Fig. 5, third panel). Furthermore, with the addition of Kapα2, Kapβ1, Ran and p10, the efficient nuclear import of HeLa cytosol was completely reconstituted (Fig. 5, bottom panel). As is the case with other Kapα/Kapβ1 substrates, hsLa–GFP, in the presence of either Kapα2 or Kapβ1, with or without Ran and p10, did not localize to the nucleus (data not shown). The ability to reconstitute hsLa– GFP nuclear import with human Kapα/Kapβ1 and the similarity of the previously mapped hsLa NLS to the consensus bipartite NLS, combined with our experiments in S. cerevisiae (Figs. 2 and 3) provide firm evidence that La is targeted to the nucleus via the Kapα/Kapβ1 pathway in human cells.
Figure 5.

Recombinant human Kapα2, Kapβ1, Ran, and p10 reconstitute nuclear import of human La. A bacterially expressed, human La–GFP fusion protein was subjected to the nuclear import assay in permeabilized HeLa cells in the presence of: (first panel) no added factors, (second panel) whole HeLa cytosol, (third panel) Kapα2 and Kapβ1, or (fourth panel) Kapα2, Kapβ1, Ran, and p10.
Appearance of a Kapα/Kapβ1 NLS in the La Family
As we had detected a division in the route to the nucleus taken by La proteins of different species, we sought a possible evolutionary explanation. We had mapped the Lhp1p NLS to the central region, overlapping its RRM, whereas the human La NLS resides at the COOH terminus. In Fig. 6 a, we have listed the COOH-terminal 35 amino acids of each of the La proteins cloned to date, highlighting basic residues which are critical for the interaction with Kapα (Conti et al., 1998). Although this stretch from each protein contains several basic residues, scLa and spLa only have one basic residue on the COOH-terminal end of this region, whereas bipartite NLSs generally have three basic residues in their COOH-terminal cluster (Makkerh et al., 1996; Conti et al., 1998). The La proteins from the multicellular eukaryotes have clear bipartite NLSs at their COOH termini and are ∼100 amino acids larger than those from the yeasts (Fig. 6 a, right column). To asses the ability of this region to act as an NLS in the branch II and branch III proteins, we constructed reporter constructs of the last 35 aa of both spLa and dmLa. When expressed in S. cerevisiae, the last 35 aa of dmLa concentrated in the nucleus, with no observable GFP signal in the cytoplasm (Fig. 6 b). This suggests that the last 35 aa of dmLa can mediate its nuclear import, presumably by Kap60p/ Kap95p in S. cerevisiae as a bipartite consensus region begins at Lys-358, 35 aa from the COOH terminus of dmLa. In contrast, the last 35 aa of spLa did not concentrate in the nucleus, possibly due to the insufficiency of the single COOH-terminal basic residue (Fig. 6 b). However, a larger COOH-terminal construct of Sla1, beginning with Met-232 could concentrate in the nucleus (data not shown). This region of Sla1 contains a putative bipartite NLS beginning at Lys-255, which is 44 aa from the COOH terminus.
Figure 6.
The extreme COOH termini of La proteins are key elements of their evolution. (a) The final 35 amino acids of ten La proteins (for abbreviations see Fig. 3 legend). White on black background, basic residues; gray on shaded background, acidic residues; gray, other residues. (b) The final 35 amino acids of S. pombe and D. melanogaster were each expressed as fusions with GFP in S. cerevisiae. The fusion constructs were visualized as previously described.
Discussion
A Karyopherin That Interacts Directly with a Multifunctional, Conserved RNA-binding Protein and at Least One Ribosomal Protein
We have previously described Kap108p as a protein essential for the nuclear localization of Lhp1p (Rosenblum et al., 1997). By the experiments described above (Fig. 1) we have now extended this finding to indicate that Kap108p imports Lhp1p without an adapter protein. It appears that of the entire karyopherin superfamily, only Kapβ1, or Kap95p in S. cerevisiae, needs an adapter. Significantly, rpL11p and Lhp1p seem to compete for binding to Kap108p in vivo.
We have looked for mislocalization of rpL11p–GFP fusions in KAP108-deletion strains, but such fusions were only mislocalized in KAP123-deletion strains (data not shown). As nuclear import of ribosomal proteins appears to be a distributed, overlapping cellular task, we cannot rule out the possibility that Kap108p plays a role in the import of rpL11p (Rout et al., 1997). In addition, our observations, particularly with regard to the in vivo competition between Lhp1p and rpL11p, are also consistent with a regulatory role for the interaction between rpL11p and Kap108p. Not only is RPL11 one of the most highly transcribed genes in S. cerevisiae, its expression is also highly regulated (Neuman-Silberberg et al., 1995; Velculescu et al., 1997). There could, therefore, be feedback between expression of rpL11p and nuclear import of Lhp1p.
An S. cerevisiae-specific Nuclear Import Pathway
The beta karyopherin superfamily in S. cerevisiae likely contains as many as 14 members. Since S. cerevisiae is the only eukaryote whose genome has been sequenced so far, the number of karyopherins in other organisms is currently unknown. Direct homologues of Kap95p, Cse1p, Kap104p, Kap121p, Crm1p, and Los1p have been identified in higher eukaryotes and expressed sequence tags for homologues of other yeast Kaps have been detected (Pemberton et al., 1998; Wozniak et al., 1998). In addition, splicing variants of the human homologues of Kap104p have been identified (Siomi et al., 1997) (Bonifaci, N., and G. Blobel, unpublished data). As such, it is possible that humans have many more than 14 Kaps. No direct homologues of Kap108p have been identified, and our finding that Kapα/Kapβ1 can import hsLa into human cells suggests that no new Kaps for the import of La proteins will be needed. However, two Kaps, Xenopus RanBP7 and human RanBP8, appear to be equally related to both Kap108p and Nmd5p (Kap119p), a protein that we have recently shown to be a Kap (Albertini et al., 1998). The functional relevance of this homology between RanBP7/ RanBP8 and Kap108p/Nmd5p (Kap119p) is not known, since import substrates for RanBP7 and RanBP8 have not been determined. Also, Kap108p and Nmd5p (Kap119p), although the most similar pair of all the yeast Kaps (26% identical) appear to have no obvious functional overlap (Albertini et al., 1998).
The maintenance of the Kap108p import pathway in S. cerevisiae may have more to do with the early stage of evolution of Lhp1p than with a need for a specific pathway for this protein. Lhp1p is clearly a structural and functional homologue of human La (Yoo and Wolin, 1994; Van Horn et al., 1997; Fan et al., 1998). Even so, we have shown here that the nuclear import of La proteins in yeast and humans are not mediated by directly homologous Kaps. Import of spLa, dmLa, and hsLa in S. cerevisiae takes place even in the absence of Kap108p. In addition, we have been unable to observe an interaction between hsLa and Kap108p (Fig. 4 b). As such it appears that S. pombe, Drosophila, and humans should not need a Kap108p homologue for La import. Although we have been able to directly assess the Kap requirement in human cells, we have not yet done so in S. pombe or Drosophila, so it remains possible that additional Kaps mediate La import in these organisms. Although La seems to use different Kaps for import in S. cerevisiae and S. pombe, it is possible that other proteins will be identified that diverge differently. For this reason, although we believe our results indicate that spLa and dmLa are transported to the nucleus by an α-Kap–dependent pathway, it remains possible that other mechanisms exist for the transport of these proteins in S. pombe and Drosophila. It is clear that this is not the case for humans, as the hsLa NLS has been thoroughly characterized and shown to be similar to the consensus bipartite NLS (Simons et al., 1996). Three scenarios exist for the conservation of nuclear import pathways: (a) directly homologous Kaps can transport directly homologous substrates; (b) directly homologous Kaps can transport nonhomologous substrates, as is the case for the yeast Kap104p and the human Kapβ2 (Siomi et al., 1998); and (c) nonhomologous Kaps can transport directly homologous substrates, as we have described here. It remains to be seen how many examples of each of these scenarios will be found.
Divergence of a Nuclear Import Pathway and Evolution of La
La proteins have been cloned from nine divergent species and are highly conserved (Fig. 3 a). In addition to their sequence conservation, functional conservation of La's role in tRNA processing has been rigorously demonstrated (Van Horn et al., 1997; Fan et al., 1998). However, La proteins in higher eukaryotes have been shown to have many functions other than facilitating tRNA processing. These functions occur in both the nucleoplasm and cytoplasm. Other nucleoplasmic functions of La proteins include facilitating initiation, termination, and recycling of RNA polymerase III transcripts (Gottlieb and Steitz, 1989a ,b; Fan et al., 1997; Lin-Marq and Clarkson, 1998). Although human La, like homologous La proteins, is nuclear (or nucleolar depending on cell cycle stage [Deng et al., 1981]) at steady state, potentially cytoplasmic functions have been described for this protein. These include: stimulating cap-independent IRES-dependent translation for a variety of viruses (Svitkin et al., 1994; Ali and Siddiqui, 1997); unwinding double-stranded RNA and RNA/DNA duplexes (Bachmann et al., 1990; Huhn et al., 1997) and protecting the highly regulated and structurally unusual histone mRNA from cell cycle–dependent degradation (McLaren et al., 1997). It remains to be seen whether the S. cerevisiae La protein has any of these functions. It has been shown that the inability to translate certain IRES-dependent viral transcripts in S. cerevisiae extracts is not due to inactivity of Lhp1p. Instead, a small RNA inhibitor of this function of La has been identified in S. cerevisiae (Das et al., 1994, 1996).
The evolution of La, a multidomain, multifunctional protein most likely occurred through a number of discrete steps. It is possible that evolution of a Kapα/Kapβ1-dependent NLS allowed La to attain additional functionality, perhaps by decreasing the mass of La used as a signal for nuclear localization (see Fig. 7 for a schematic representation of the four La proteins studied here). As shown above, even a large, 113-aa domain encompassing a Kap108p-dependent NLS only weakly concentrated fragment 2 in the nucleus (Fig. 2). NLSs for α-Kap–independent β-Kaps appear to be larger than NLSs recognized by Kapα. The NLS of Nab2p for Kap104p-mediated import has been mapped to a region 50- or 110-aa long, and the hnRNP A1 NLS for Kapβ2-mediated import is at least 38-aa long (Siomi and Dreyfuss, 1995; Siomi et al., 1998; Truant et al., 1998). In addition, Kap111p/Mtr10p, which imports Npl3p, uses an NLS on the order of 130-aa long (Pemberton et al., 1997; Senger et al., 1998). In contrast, a 27-aa domain of human La (Simons et al., 1996) and a 35-aa domain of Drosophila La (Fig. 6 b) strongly concentrate fusion proteins to the nucleus via α-Kap–dependent pathways. In fact, α-Kap–dependent NLSs are often shorter than 12 aa and can even be as short as 5 aa (Dingwall and Laskey, 1991). In addition, the Kap108p-dependent NLS of Lhp1p includes the RNA-binding domain, and it is therefore possible that RNA binding and Kap108p binding of Lhp1p are mutually exclusive (many NLSs of DNA- or RNA-binding proteins have been mapped to sites overlapping or adjacent to DNA- or RNA-binding domains; LaCasse and Lefebvre, 1995). An example of this exclusivity of binding La has been described for the human protein. A monoclonal antibody, mAb SW3, which binds within the RRM of hsLa, is unable to bind to RNA-associated La (Pruijn et al., 1995). It could be envisioned that directionality for Lhp1p import could be conveyed in this manner. Newly synthesized, cytoplasmic Lhp1p could be recognized in the cytoplasm by Kap108p. Kap108p could then direct Lhp1p through the NPC by virtue of its interaction with nucleoporins (Rosenblum et al., 1997). Once in the nucleus, the Kap108p/Lhp1p complex could be dissembled in the presence of a high concentration of oligouridylate-containing RNA polymerase III transcripts in the nucleus (Senger et al., 1998). The overlap between the Lhp1p NLS and its RNA recognition motif may also affect cytoplasmic functions of La proteins. As shown in Fig. 1 b, a large proportion of cytoplasmic Lhp1p is stably bound to Kap108p. Given that this binding appears to be mediated by a large region that includes the RNA-binding domain of Lhp1p, it appears unlikely that this domain would be accessible for cytoplasmic functions in S. cerevisiae. As such, there may have been an evolutionary advantage to have the NLS of the more complex La proteins in a region not involved in direct RNA binding.
Figure 7.
A representation of important domains for four evolutionarily distinct La proteins. Bars proportional to the length of the human, Drosophila, S. pombe, and S. cerevisiae La proteins are shown. Black lines, RRMs; two-headed arrows, proteins NLSs. The ATP-binding domain of human La is indicated.
It is possible that using different domains for an NLS would require different modes of regulation for nuclear import/export. One possible mode for regulation of hsLa is by phosphorylation. Phosphorylation of the major casein kinase II site of La, serine 366, has recently been shown to impair the RNA pol III transcription factor activity of La in vitro (Fan et al., 1997). As a corollary to this observation, it would be expected that the unphosphorylated form of La would be present in the nucleus where it performs its transcription-related functions. Our observation that bacterially expressed, and therefore unphosphorylated, hsLa–GFP is actively transported into HeLa cell nuclei is consistent with this expectation. Further, the proximity of Ser-366 to the NLS of La (aa 382–408) raises the possibility that La activity may be coupled to its localization. Such feedback would appear to be particularly important for proteins like La that have nuclear and cytoplasmic functions.
A scheme for the evolution of La can be generated, taking into account known domains of La and our results regarding the divergence of its nuclear targeting (see Fig. 7 for a schematic of relevant domains of representative La proteins). In such a scheme, the common ancestor of the modern La proteins (Fig. 3 a) was imported via an α-Kap– independent β-Kap. The progenitor Kap recognized the progenitor La via a large, complex domain. Branch II diverged from branch I in part by acquiring a strong α-Kap– dependent NLS in the COOH-terminal domain of the protein. In the case of the S. pombe La, the NLS probably begins 44 aa from the COOH terminus. The second major evolutionary divergence (between branch II and branch III) occurred with the acquisition of an even more COOH-terminal NLS, the dmLa NLS, for example, begins within 35 aa of the COOH terminus. This second divergence coincided with a 30% increase in the size of La. Further specialization of the central domain, which was no longer required for nuclear import, led to the addition of an atypical RRM and an ATP-binding motif in the proteins of branch IV (Birney et al., 1993). In the case of human La, the NLS is contained within 27 aa of its COOH terminus (Simons et al., 1996).
Lhp1p also appears to have a weak Kap108p-independent NLS that can be unmasked by removing the first 187 amino acids of the protein. This NLS does not appear to function in the context of full-length Lhp1p (Fig. 2 c, top right panel), but is likely to be relevant evolutionarily, as a corresponding domain of Sla1 does appear to be responsible for nuclear import.
Evolution of the Karyopherin Superfamily
Recently a molecular phylogenetic analysis of the karyopherin superfamily was used to suggest an evolutionary framework for this class of proteins (Malik et al., 1997). This analysis demonstrated that the Arm motifs of α-Kaps and the HEAT motifs of β-Kaps are related. As a result, it is likely that an α-Kap–independent progenitor gave rise to both the α-Kaps and the β-Kaps. The β-Kaps that resulted consisted of both α-Kap–dependent and –independent types (Malik et al., 1997). If this were indeed the case, it might be predicted that such an evolutionary divide might be observed in comparing nuclear transport pathways used for a conserved protein in divergent organisms. It is possible that we have uncovered such a division. An additional possibility is that the evolution of nuclear transport mechanisms of La proteins was intimately related to the specialization of α-Kaps. S. cerevisiae only has one α-Kap whereas humans have at least six α-Kaps, each with a unique substrate specificity and expression pattern (Pemberton et al., 1998). In this light it is possible that the development of additional α-Kaps, which occurred after the divergence of S. cerevisiae from humans (Malik et al., 1997), was a key element in the development of α-Kap– dependent NLSs in La proteins. Given the difference in size between NLSs of α-Kap–dependent and –independent pathways, it would be surprising if other classes of proteins did not recapitulate similar evolutionary strategies. Whether Kap108p-like proteins evolved to specialize a role involving ribosomal proteins will be determined with the continued characterization of this function in S. cerevisiae and the cloning and characterization of Kaps in higher eukaryotes.
Acknowledgments
We thank E. Ellison for valuable technical assistance and members of the laboratory for numerous discussions. In addition, we would like to thank M. Rout and B. Cravatt for insightful comments that improved the manuscript. We are grateful to B. Cormack (yEGFP3 cDNA), M. Matunis (human Kapα2), N. Yaseen (human Ran), S. Wolin (scLa and dmLa cDNA), M. Rout, and J. Aitchison (Kap95-PrA strain and KAP123-deletion strain) for providing valuable reagents, and J. Fernandez and F. Gharahdaghi (Rockefeller University Protein/DNA Technology Center) for protein sequencing and mass spectral analysis.
Abbreviations used in this paper
- aa
amino acid
- GFP
green fluorescent protein
- IRES
internal ribosome entry site
- Kap
karyopherin
- NLS
nuclear localization sequence
- NPC
nuclear pore complex
- RRM
RNA recognition motif
Footnotes
J.S. Rosenblum was supported by a National Institutes of Health postdoctoral fellowship.
2. Throughout the text, depending on context, nonstandard nomenclature of La proteins will be used to facilitate the discussion. Specifically, the S. cerevisiae La, Lhp1p, will sometimes be referred to as scLa and the S. pombe La, Sla1, will be referred to as spLa. Drosophila and human La proteins will be referred to as dmLa and hsLa, respectively.
References
- Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Albertini, M., L.F. Pemberton, J.S. Rosenblum, and G. Blobel. 1998. Nuclear import and the evolution of a multifunctional RNA-binding protein. J. Cell Biol. In press. [DOI] [PMC free article] [PubMed]
- Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.A. Smith, J.G. Seidman, and K. Struhl. 1997. Current Protocols in Molecular Biology. Vol. 2. John Wiley & Sons, Inc., New York. 13.0.1–13.13.9.
- Bachmann M, Falke D, Schröder H-C, Müller WEG. Intracellular distribution of the La antigen in CV-1 cells after herpes simplex virus type 1 infection compared with the localization of U small nuclear ribonucleoprotein particles. J Gen Virol. 1989;70:881–891. doi: 10.1099/0022-1317-70-4-881. [DOI] [PubMed] [Google Scholar]
- Bachmann M, Pfeifer K, Schroder HC, Müller WE. Characterization of the autoantigen La as a nucleic acid-dependent ATPase/dATPase with melting properties. Cell. 1990;60:85–93. doi: 10.1016/0092-8674(90)90718-t. [DOI] [PubMed] [Google Scholar]
- Bai C, Li Z, Tolias PP. Developmental characterization of a DrosophilaRNA-binding protein homologous to the human systemic lupus erythematosus-associated La/SS-B autoantigen. Mol Cell Biol. 1994;14:5123–5129. doi: 10.1128/mcb.14.8.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The hsp70 and hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Chambers JC, Kenan D, Martin BJ, Keene JD. Genomic structure and amino acid sequence domains of the human La autoantigen. J Biol Chem. 1988;263:18043–18051. [PubMed] [Google Scholar]
- Chan EK, Sullivan KF, Tan EM. Ribonucleoprotein SS-B/La belongs to a protein family with consensus sequences for RNA-binding. Nucleic Acids Res. 1989;17:2233–2244. doi: 10.1093/nar/17.6.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- Cormack BP, Bertram G, Egerton M, Gow NAR, Falkow S, Brown AJP. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. . Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- Craig AW, Svitkin YV, Lee HS, Belsham GJ, Sonenberg N. The La autoantigen contains a dimerization domain that is essential for enhancing translation. Mol Cell Biol. 1997;17:163–169. doi: 10.1128/mcb.17.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Coward P, Dasgupta A. A small yeast RNA selectively blocks internal initiation of translation programmed by poliovirus RNA: specific interaction with cellular proteins that bind to the viral 5′-untranslated region. J Virol. 1994;68:7200–7211. doi: 10.1128/jvi.68.11.7200-7211.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Kenan DJ, Bocskai D, Keene JD, Dasgupta A. Sequences within a small yeast RNA required for inhibition of internal initiation of translation: interaction with La and other cellular proteins influences its inhibitory activity. J Virol. 1996;70:1624–1632. doi: 10.1128/jvi.70.3.1624-1632.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JS, Takasaki Y, Tan EM. Nonhistone nuclear antigens reactive with autoantibodies. Immunofluorescent studies of distribution in synchronized cells. J Cell Biol. 1981;91:654–660. doi: 10.1083/jcb.91.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus? . Trends Biol Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Fan H, Goodier JL, Chamberlain JR, Engelke DR, Maraia RJ. 5′ processing of tRNA precursors can be modulated by the human La antigen phosphoprotein. Mol Cell Biol. 1998;18:3201–3211. doi: 10.1128/mcb.18.6.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Sakulich AL, Goodier JL, Zhang X, Qin J, Maraia RJ. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell. 1997;88:707–715. doi: 10.1016/s0092-8674(00)81913-3. [DOI] [PubMed] [Google Scholar]
- Fernandez J, Andrews L, Mische SM. An improved procedure for enzymatic digestion of polyvinylidene difluoride-bound proteins for internal sequence analysis. Anal Biochem. 1994;218:112–117. doi: 10.1006/abio.1994.1148. [DOI] [PubMed] [Google Scholar]
- Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Gharahdaghi F, Kirchner M, Fernandez J, Mische SM. Peptide-mass profiles of polyvinylidene difluoride-bound proteins by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in the presence of nonionic detergents. Anal Biochem. 1996;233:94–99. doi: 10.1006/abio.1996.0012. [DOI] [PubMed] [Google Scholar]
- Gottlieb E, Steitz JA. Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO (Eur Mol Biol Organ) J. 1989a;8:851–861. doi: 10.1002/j.1460-2075.1989.tb03446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E, Steitz JA. The RNA binding protein La influences both the accuracy and the efficiency of RNA polymerase III transcription in vitro. EMBO (Eur Mol Biol Organ) J. 1989b;8:841–850. doi: 10.1002/j.1460-2075.1989.tb03445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Wolin SL, Rinke J, Lerner MR, Steitz JA. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol Cell Biol. 1981;1:1138–1149. doi: 10.1128/mcb.1.12.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- Huhn P, Pruijn GJ, van Venrooij WJ, Bachmann M. Characterization of the autoantigen La (SS-B) as a dsRNA unwinding enzyme. Nucleic Acids Res. 1997;25:410–416. doi: 10.1093/nar/25.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCasse EC, Lefebvre YA. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res. 1995;23:1647–1656. doi: 10.1093/nar/23.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladomery M. Multifunctional proteins suggest connections between transcriptional and post-transcriptional processes. Bioessays. 1997;19:903–909. doi: 10.1002/bies.950191010. [DOI] [PubMed] [Google Scholar]
- Lin-Marq N, Clarkson SG. Efficient synthesis, termination and release of RNA polymerase III transcripts in Xenopusextracts depleted of La protein. EMBO (Eur Mol Biol Organ) J. 1998;17:2033–2041. doi: 10.1093/emboj/17.7.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager WH, Planta RJ, Ballesta JG, Lee JC, Mizuta K, Suzuki K, Warner JR, Woolford J. A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. . Nucleic Acids Res. 1997;25:4872–4875. doi: 10.1093/nar/25.24.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makkerh JPS, Dingwall C, Laskey RA. Comparative mutagenesis of nuclear localization signals reveals the importance of neutral and acidic amino acids. Curr Biol. 1996;6:1025–1027. doi: 10.1016/s0960-9822(02)00648-6. [DOI] [PubMed] [Google Scholar]
- Malik HS, Eickbush TH, Goldfarb DS. Evolutionary specialization of the nuclear targeting apparatus. Proc Natl Acad Sci USA. 1997;94:13738–13742. doi: 10.1073/pnas.94.25.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia RJ. Transcription termination factor La is also an initiation factor for RNA polymerase III. Proc Natl Acad Sci USA. 1996;93:3383–3387. doi: 10.1073/pnas.93.8.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli M, Reichlin M. Heterogeneity of RNA protein antigens reactive with sera of patients with systemic lupus erythematosus. Description of a cytoplasmic nonribosomal antigen. Arthritis Rheum. 1974;17:421–429. doi: 10.1002/art.1780170413. [DOI] [PubMed] [Google Scholar]
- McBratney S, Sarnow P. Evidence for involvement of trans-acting factors in selection of the AUG start codon during eukaryotic translational initiation. Mol Cell Biol. 1996;16:3523–3534. doi: 10.1128/mcb.16.7.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren RS, Caruccio N, Ross J. Human La protein: a stabilizer of histone mRNA. Mol Cell Biol. 1997;17:3028–3036. doi: 10.1128/mcb.17.6.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerovitch K, Sonenberg N. Internal initiation of picornavirus RNA translation. Semin Virol. 1993;4:217–227. [Google Scholar]
- Meerovitch K, Svitkin YV, Lee HS, Lejbkowicz F, Kenan DJ, Chan EK, Agol VI, Keene JD, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS. Ran and nuclear transport. J Biol Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- Moore MS, Blobel G. The two steps of nuclear import, targeting to the nuclear envelope and translocation through the nuclear pore, require different cytosolic factors. Cell. 1992;69:939–950. doi: 10.1016/0092-8674(92)90613-h. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Bhattacharya S, Broach JR. Nutrient availability and the RAS/cyclic AMP pathway both induce expression of ribosomal protein genes in Saccharomyces cerevisiaebut by different mechanisms. Mol Cell Biol. 1995;15:3187–3196. doi: 10.1128/mcb.15.6.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardigon N, Strauss JH. Mosquito homolog of the La autoantigen binds to Sindbis virus RNA. J Virol. 1996;70:1173–1181. doi: 10.1128/jvi.70.2.1173-1181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton LF, Blobel G, Rosenblum JS. Transport routes through the nuclear pore complex. Curr Opin Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Rosenblum JS, Blobel G. A distinct and parallel pathway for the nuclear import of an mRNA-binding protein. J Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijn GJ, Thijssen JP, Smith PR, Williams DG, Van Venrooij WJ. Anti-La monoclonal antibodies recognizing epitopes within the RNA-binding domain of the La protein show differential capacities to immunoprecipitate RNA-associated La protein. Eur J Biochem. 1995;232:611–619. doi: 10.1111/j.1432-1033.1995.611zz.x. [DOI] [PubMed] [Google Scholar]
- Rinke J, Steitz JA. Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell. 1982;29:149–159. doi: 10.1016/0092-8674(82)90099-x. [DOI] [PubMed] [Google Scholar]
- Rinke J, Steitz JA. Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucleic Acids Res. 1985;13:2617–2629. doi: 10.1093/nar/13.7.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum JS, Pemberton LF, Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. J Cell Biol. 1997;139:1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Blobel G. Isolation of the yeast nuclear pore complex. J Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Blobel G, Aitchison JD. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Scherly D, Stutz F, Lin-Marq N, Clarkson SG. La proteins from Xenopus laevis. cDNA cloning and developmental expression. J Mol Biol. 1993;231:196–204. doi: 10.1006/jmbi.1993.1275. [DOI] [PubMed] [Google Scholar]
- Schneppenheim R, Budde U, Dahlmann N, Rautenberg P. Luminography—a new, highly sensitive visualization method for electrophoresis. Electrophoresis. 1991;12:367–372. doi: 10.1002/elps.1150120508. [DOI] [PubMed] [Google Scholar]
- Semsei I, Troster H, Bartsch H, Schwemmle M, Igloi GL, Bachmann M. Isolation of rat cDNA clones coding for the autoantigen SS-B/ La: detection of species-specific variations. Gene. 1993;126:265–268. doi: 10.1016/0378-1119(93)90378-g. [DOI] [PubMed] [Google Scholar]
- Senger B, Simos G, Bischoff FR, Podtelejnikov A, Mann M, Hurt E. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO (Eur Mol Biol Organ) J. 1998;17:2196–2207. doi: 10.1093/emboj/17.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons FH, Broers FJ, Van Venrooij WJ, Pruijn GJ. Characterization of cis-acting signals for nuclear import and retention of the La (SS-B) autoantigen. Exp Cell Res. 1996;224:224–236. doi: 10.1006/excr.1996.0132. [DOI] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–559. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Eder PS, Kataoka N, Wan L, Liu Q, Dreyfuss G. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J Cell Biol. 1997;138:1181–1192. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Fromont M, Rain J-C, Wan L, Wang F, Legrain P, Dreyfuss G. Functional conservation of the transportin nuclear import pathway in divergent organisms. Mol Cell Biol. 1998;18:4141–4148. doi: 10.1128/mcb.18.7.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano JE. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell. 1984;36:145–154. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- Svitkin YV, Pause A, Sonenberg N. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J Virol. 1994;68:7001–7007. doi: 10.1128/jvi.68.11.7001-7007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EM. Antinuclear antibodies: diagnostic markers for autoimmune diseases and probes for cell biology. Adv Immunol. 1989;44:93–151. doi: 10.1016/s0065-2776(08)60641-0. [DOI] [PubMed] [Google Scholar]
- Topfer F, Gordon T, McCluskey J. Characterization of the mouse autoantigen La (SS-B). Identification of conserved RNA-binding motifs, a putative ATP binding site and reactivity of recombinant protein with poly(U) and human autoantibodies. J Immunol. 1993;150:3091–3100. [PubMed] [Google Scholar]
- Truant R, Fridell RA, Benson RE, Bogerd H, Cullen BR. Identification and functional characterization of a novel nuclear localization signal present in the yeast Nab2 poly(A)+binding protein. Mol Cell Biol. 1998;18:1449–1458. doi: 10.1128/mcb.18.3.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn DJ, Yoo CJ, Xue D, Shi H, Wolin SL. The La protein in Schizosaccharomyces pombe: a conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA. 1997;3:1434–1443. [PMC free article] [PubMed] [Google Scholar]
- Van Venrooij WJ, Pruijn GJM. Ribonucleoprotein complexes as autoantigens. Curr Opin Immunol. 1995;7:819–824. doi: 10.1016/0952-7915(95)80054-9. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett JDE, Hieter P, Vogelstein B, Kinzler KW. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- Weeks KM. Protein-facilitated RNA folding. Curr Opin Struct Biol. 1997;7:336–342. doi: 10.1016/s0959-440x(97)80048-6. [DOI] [PubMed] [Google Scholar]
- Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Wozniak RW, Rout MP, Aitchison JD. Karyopherins and kissing cousins. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: a yeast homolog of the La autoantigen is dispensable for growth. Mol Cell Biol. 1994;14:5412–5424. doi: 10.1128/mcb.14.8.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell. 1997;89:393–402. doi: 10.1016/s0092-8674(00)80220-2. [DOI] [PubMed] [Google Scholar]