Abstract
The fission yeast Schizosaccharomyces pombe divides symmetrically using a medial F-actin– based contractile ring to produce equal-sized daughter cells. Mutants defective in two previously described genes, mid1 and pom1, frequently divide asymmetrically. Here we present the identification of three new temperature-sensitive mutants defective in localization of the division plane. All three mutants have mutations in the polo kinase gene, plo1, and show defects very similar to those of mid1 mutants in both the placement and organization of the medial ring. In both cases, ring formation is frequently initiated near the cell poles, indicating that Mid1p and Plo1p function in recruiting medial ring components to the cell center. It has been reported previously that during mitosis Mid1p becomes hyperphosphorylated and relocates from the nucleus to a medial ring. Here we show that Mid1p first forms a diffuse cortical band during spindle formation and then coalesces into a ring before anaphase. Plo1p is required for Mid1p to exit the nucleus and form a ring, and Pom1p is required for proper placement of the Mid1p ring. Upon overexpression of Plo1p, Mid1p exits the nucleus prematurely and displays a reduced mobility on gels similar to that of the hyperphosphorylated form observed previously in mitotic cells. Genetic and two-hybrid analyses suggest that Plo1p and Mid1p act in a common pathway distinct from that involving Pom1p. Plo1p localizes to the spindle pole bodies and spindles of mitotic cells and also to the medial ring at the time of its formation. Taken together, the data indicate that Plo1p plays a role in the positioning of division sites by regulating Mid1p. Given its previously known functions in mitosis and the timing of cytokinesis, Plo1p is thus implicated as a key molecule in the spatial and temporal coordination of cytokinesis with mitosis.
Keywords: plo1, mid1/dmf1, pom1, cytokinesis, S. pombe
Mitosis and cytokinesis are tightly coordinated both temporally and spatially, which ensures that each daughter cell will receive a full set of chromosomes together with a proper complement of cytoplasm and organelles. Proper placement and orientation of the cleavage plane are also important during development, where programmed changes in the division plane can produce daughter cells with different fates and determine the positions of cells relative to each other.
During mitosis, the whole cytoskeleton reorganizes and the cytokinesis machinery assembles. A central part of this machinery in many cell types is a cortical contractile ring that contains F-actin, myosin II, and numerous other proteins and forms during mitosis at the site where cytokinesis will occur (Satterwhite and Pollard, 1992; Fishkind and Wang, 1995). It was observed very early that the contractile ring typically forms perpendicular to the mitotic spindle, and numerous studies have suggested that the mitotic apparatus plays an active role in determining where and when cytokinesis will occur. Two models have been proposed to explain how the contractile ring is positioned. One model proposes that overlapping astral microtubules at the cell cortex determine the position of the contractile ring, whereas the other model suggests that signals emanating from the spindle midzone are critical for this process (Rappaport, 1986, 1996; Strome, 1993; Oegema and Mitchison, 1997). The models have in common that the position of the mitotic apparatus determines the site of cell division. In organisms such as C. elegans (Guo and Kemphues, 1996; Zwaal et al., 1996), Drosophila melanogaster (Kraut et al., 1996; Doe, 1996), and Saccharomyces cerevisiae (Stearns, 1997), some progress has been made in identifying proteins that are involved in positioning and orienting the mitotic spindle. However, little is known about the proteins involved in signaling to the cell cortex to assemble the contractile ring.
The fission yeast Schizosaccharomyces pombe is an attractive model system to study the temporal and spatial coordination of mitosis and cytokinesis (for review see Gould and Simanis, 1997). The cylindrical S. pombe cells undergo symmetrical division to form two daughter cells of equal size. As in higher eukaryotes, the initiation of cell division is normally dependent on the onset of mitosis (Minet et al., 1979) and is preceded by the formation of an actin-based medial ring at the cell cortex (Marks and Hyams, 1985), which constricts during septation (Jochová et al., 1991; McCollum et al., 1995; Kitayama et al., 1997). In addition to actin, numerous other proteins have recently been shown to be components of the medial ring (reviewed by Gould and Simanis, 1997). The mitotic spindle itself is not required for formation and placement of the medial ring (Chang et al., 1996), but the position of the nucleus may determine the site of ring formation (Chang and Nurse, 1996). The nucleus itself seems to be positioned in the cell center by interactions of cytoplasmic microtubules with the spindle pole body (SPB),1 which is associated with the nuclear membrane (Ding et al., 1997; Hagan and Yanagida, 1997 and references cited therein). The SPB is the centrosome-equivalent in S. pombe; it duplicates just before the onset of mitosis to organize the mitotic spindle (Masuda et al., 1992; Hagan and Yanagida, 1995; Ding et al., 1997).
Several genes involved in positioning and orienting the cell division site in S. pombe have recently been identified; these include mid1/dmf1 (Chang et al., 1996; Sohrmann et al., 1996), the pos genes (Edamatsu and Toyoshima, 1996), and pom1 (Bähler and Pringle, 1998). Mutants defective in any of these genes show a frequent mislocalization and/or misorientation of division septa, although the nuclei occupy a normal position in the cell center before nuclear division. Mid1p is nuclear during interphase and forms a cortical medial ring during mitosis; this relocalization seems to occur shortly before actin ring formation and correlates with increased phosphorylation of Mid1p (Sohrmann et al., 1996). Pom1p is a putative protein kinase that is involved in the spatial control of both polarized growth and cytokinesis; it localizes both to the cell ends and, during cytokinesis, to the cell center (Bähler and Pringle, 1998).
Here we report the isolation of three temperature-sensitive S. pombe mutants that have phenotypes similar to mid1 mutants and harbor mutant alleles of the essential gene plo1 (Ohkura et al., 1995). Plo1p belongs to a conserved family of Ser/Thr protein kinases, the polo-like kinases, that were first identified in Drosophila (Sunkel and Glover, 1988; Llamazares et al., 1991). Polo kinases seem to be involved in centrosome maturation and mitotic spindle formation as well as in late stages of mitosis and cell division (for review, see Glover et al., 1996). In S. pombe, Plo1p is required for the formation of a bipolar spindle, the medial ring, and the division septum, and overexpression of Plo1p can induce septation in interphase cells (Ohkura et al., 1995). These data indicate that Plo1p is important for cytokinesis as well as mitosis and is perhaps involved in the temporal coordination of these two events. Our studies suggest that the Plo1p kinase also functions in the spatial coordination of mitosis and cytokinesis, possibly by phosphorylating and thus regulating the behavior of Mid1p.
Materials and Methods
Strains, Media, and Genetic and Molecular Biology Methods
S. pombe strains used in this study are listed in Table I; all strains are isogenic to 972 (Leupold, 1970). Growth media were as described by Moreno et al. (1991). Except where noted, cells for experiments were grown in EMM minimal medium. Overexpression of Plo1p was carried out by expressing plo1 from the thiamine-repressible nmt1 promoter in plasmid pRep1 (Maundrell, 1993), as described previously (Ohkura et al., 1995). Standard genetic and recombinant-DNA methods (Sambrook et al., 1989; Moreno et al., 1991) were used except where noted. Yeast transformations were performed using either a lithium acetate method (Keeney and Boeke, 1994) or electroporation (Prentice, 1992). DNA was prepared from bacteria and isolated from agarose gels using QIAGEN (Chatsworth, CA) kits and from yeast cells as described by Hoffman and Winston (1987). DNA was sequenced using Sequenase 2.0 (United States Biochemical Co., Cleveland, OH) or by the UNC-CH Automated Sequencing Facility on a Model 373A DNA Sequencer using the Taq DyeDeoxy Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Oligonucleotide primers were obtained from Integrated DNA Technologies or from Operon.
Table I.
S. pombe Strains Used in This Study
| Strain | Genotype | Reference/source | ||
|---|---|---|---|---|
| 975 | wild-type h + | Leupold, 1970 | ||
| 972 | wild-type h − | Leupold, 1970 | ||
| JB13 | ura4-D18 leu1-32 h − | J. Kohli | ||
| YDM105 | ade6-210 leu1-32 ura4-D18 h − | Gould lab collection | ||
| YDM106 | ade6-210 leu1-32 ura4-D18 h + | Gould lab collection | ||
| YDM110 | plo1-1 leu1-32 ura4-D18 h + | See text | ||
| YDM114 | plo1-24C ade6-210 leu1-32 ura4-D18 h + | See text | ||
| JB200 | plo1-25 leu1-32 h − | See text | ||
| JB201 | plo1-25 ura4-D18 h + | See text | ||
| YDM296 | mid1-18* ade6-210 leu1-32 ura4-D18 h − | Sohrmann et al., 1996 | ||
| JB41 | mid1-ΔF ade6-216 leu1-32 ura4-D18 h − | Sohrmann et al., 1996 | ||
| JB20 | cdc25-22 h − | Thuriaux et al., 1980 | ||
| YDM152 | cdc25-22 ura4-D18 h + | Gould lab collection | ||
| JB109 | pom1-Δ1 ade6-216 ura4-D18 h + | Bähler and Pringle, 1998 | ||
| JB110 | pom1-Δ1 ura4-D18 h − | Bähler and Pringle, 1998 | ||
| JB120 | pom1-Δ1 cdc25-22 ura4-D18 h − | Bähler and Pringle, 1998 | ||
| JB43 | mid1-ΔF cdc25-22 ura4-D18 h + | JB41 × YDM152 | ||
| YDM403 | mid1-GFP::ura4 + ade6-210 leu1-32 ura4-D18 h − | See text | ||
| JB250 | mid1-GFP::ura4 + plo1-1 ade6-210 leu1-32 ura4-D18 h − | YDM403 × YDM110 | ||
| YDM603 | mid1-13Myc::ura4 + ade6-210 leu1-32 ura4-D18 h − | See text | ||
| YDM607 | mid1-13Myc::ura4 + plo1-1 ade6-210 leu1-32 ura4-D18 h − | YDM603 × YDM110 | ||
| YDM604 | mid1-13Myc::ura4 + plo1-25 ade6-210 leu1-32 ura4-D18 h − | YDM603 × JB201 | ||
| YDM608 | mid1-13Myc::ura4 + plo1-24C ade6-210 leu1-32 ura4-D18 h − | YDM603 × YDM114 | ||
| YDM609 | mid1-13Myc::ura4 + pom1-Δ1 ade6-210 leu1-32 ura4-D18 h − | YDM603 × JB109 | ||
| JB206 | plo1-GFPS65T h − | See text | ||
| YDM457 | plo1-GFPS65T cdc25-22 h − | JB206 × YDM152 | ||
| JB214 | plo1-GFPS65T mid1-ΔF cdc25-22 h − | JB206 × JB43 |
mid1-18 (Balasubramanian et al., 1998) corresponds to dmf1-6 (Sohrmann et al., 1996).
Mutant Screens
The plo1-1 and plo1-24C mutants were isolated by screening cells of strain 972 that had been mutagenized with nitrosoguanidine or N-nitroso-N-ethylurea (both Sigma Chemical Co., St. Louis, MO), respectively. Mutagenized clones were screened for those that grew poorly, or not at all, at 36°C and had morphologies characteristic of S. pombe cytokinesis mutants. These mutants were then stained with Calcofluor, which stains the septum, to identify those with defects in septum formation or placement. The original mutant isolates were backcrossed four times to strains YDM105 and YDM106 to yield strains YDM110 and YDM114. The plo1-25 mutant was isolated in a screen of cells that had been mutagenized with UV. Cells of strain 972 were grown in EMM medium to stationary phase and spread on YE plates to give ∼1,000 cells/plate. The plates were subjected to short-wave UV radiation for 80 s, which resulted in ∼90% killing. Surviving cells were grown at 25°C for 5 d. Individual colonies were then isolated, replica plated to 36°C, stained with Calcofluor, and screened visually as described by Bähler and Pringle (1998). The original mutant isolate was backcrossed four times to wild-type strains 975 and JB13, yielding strains JB200 and JB201.
Demonstration That the Mutants Contained plo1 Mutations
Identification of the gene defined by the plo1-1 and plo1-24C mutations exploited the finding (during allelism tests of the newly isolated mutants) that although plo1-1 and pom1 (Bähler and Pringle, 1998) single mutants were viable at 36°C, a double mutant was inviable at this temperature (see Table II). A plo1-1 pom1 double mutant was transformed with a genomic library (provided by A. Carr, University of Sussex, Falmer, East Sussex, UK) constructed in pUR19 (Barbet et al., 1992), and transformants were grown for 3 d at 25°C and then replica plated to 36°C. Thirteen transformants showed plasmid-dependent rescue at 36°C. These plasmids were isolated and found to contain either identical or overlapping inserts derived from a region of chromosome I that had been sequenced by the Sanger Center as part of the S. pombe genome project. The region common to all of these plasmids contained plo1, and the plasmids were found to rescue the slow growth and morphological phenotypes of both plo1-1 and plo1-24C single mutants but not those of a pom1 single mutant. Integrating a plo1-containing plasmid and crossing to plo1-1 and plo1-24C strains confirmed that the mutations were tightly linked to the plo1 locus. Finally, both the plo1-1 and plo1-24C mutant strains were rescued by a plo1 + cDNA expressed under the control of the nmt1 promoter (provided by C. Albright, Vanderbilt University, Nashville, TN).
Table II.
Genetic Interactions among plo1, mid1, and pom1 Mutations
| Mutant | Growth at the indicated temperature* | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 18°C | 25°C | 30°C | 32°C | 36°C | ||||||
| plo1-1 | +++ | +++ | +++ | +++ | ++ | |||||
| plo1-24C | +++ | +++ | +++ | ++ | + | |||||
| mid1-ΔF | ++ | ++ | ++ | ++ | + | |||||
| pom1-Δ1 | +++ | +++ | +++ | +++ | ++ | |||||
| plo1-1 mid1-ΔF | ++ | ++ | ++ | ++ | + | |||||
| plo1-24C mid1-ΔF | ++ | ++ | ++ | ++ | + | |||||
| mid1-ΔF pom1-Δ1 | − | − | − | − | − | |||||
| plo1-1 pom1-Δ1 | ++ | ++ | + | − | − | |||||
| plo1-24C pom1-Δ1 | +++ | +++ | ++ | + | − | |||||
Double mutants were constructed by crossing appropriate strains (Table I) and dissecting tetrads. To evaluate growth at different temperatures, cells were diluted to single colonies on YE medium containing phloxin B and incubated at the temperatures indicated. (+++) Colony formation similar to wild-type and most cells healthy (phloxin-pink); (++) colonies with significant numbers of dead cells (phloxin-red); (+) very slow growth, small colonies with high numbers of dead cells; (−) no colony formation.
To clone the gene defined by the plo1-25 mutation, strain JB200 was transformed with an S. pombe genomic DNA library (a gift of P. Young, Queens University, Kingston, Ontario, Canada) constructed in plasmid pWH5 (Wright et al., 1986). Transformants were grown for 4 d at 30°C, replica plated onto EMM plates containing 2 μg/ml phloxin B (which accumulates in dead cells, staining them red; Kohli et al., 1977), and grown over night at 37°C. Transformants that did not accumulate phloxin B were tested for plasmid-dependent rescue, and five plasmids were recovered that complemented both the slow growth and morphological defects of the plo1-25 mutation at 37°C upon retransformation. These plasmids had identical or overlapping inserts as judged from restriction digests. Partial sequencing of two plasmids revealed that the inserts contained a region of chromosome I that contained plo1 and three adjacent ORFs, and a plo1 + cDNA plasmid (see above) rescued both the slow growth and morphological defects of plo1-25 strains at 37°C. A cross of plo1-25 (strain JB200) with plo1-24C (strain YDM114) did not yield any wild-type recombinants among 600 spore colonies, and plo1-25 did not complement plo1-24C in a diploid strain, as judged by its slow growth and aberrant morphology at 37°C.
Tagging of Mid1p and Plo1p
Mid1p was tagged at its COOH terminus with GFP(S65T) or 13 tandem copies of the myc epitope (13Myc; Bähler et al., 1998) by first amplifying by PCR the COOH-terminal 900 bp of the mid1 gene. The 5′ primer contained a KpnI site followed by an in frame stop codon, and the 3′ primer contained an NdeI and BamHI site. The PCR product was then cloned into the integrating vector pJK210 (Keeney and Boeke, 1994) by using KpnI and BamHI sites that were included in the primers. GFP and 13Myc-encoding sequences were then cloned into this plasmid as NdeI and BamH1/BglII fragments respectively to generate COOH-terminal fusions with Mid1p. These plasmids were then linearized in the mid1 gene by digestion with PstI and transformed into strain YDM105 to generate strains in which the wild-type mid1 locus had been replaced by the tagged genes. Note that integration of this plasmid results in the full-length tagged gene as well as an untagged COOH-terminal fragment of the mid1 gene which is not expressed due to the in frame stop codon introduced at its NH2-terminus. The GFP and 13Myc tagged strains (YDM403 and YDM603, respectively) were checked for correct integration at the mid1 locus by PCR. These strains were indistinguishable from wild-type with respect to growth rates and cellular morphology.
Plo1p was tagged at its COOH terminus with green fluorescent protein (GFP) carrying the S65T mutation (Heim et al., 1995) by direct chromosomal integration into strain 972 of a fragment generated by PCR using plasmid pFA6a-GFP(S65T)-kanMX6 as template (Bähler et al., 1998). The two primers had 80-bp tails corresponding to the regions just upstream and 150–230 bp downstream of the plo1 stop codon. The resulting strain (JB206) was checked for correct integration by PCR as described by Bähler et al. (1998).
Two-Hybrid Analyses
Two-hybrid assays (Fields and Sternglanz, 1994) were performed as described previously (Gyuris et al., 1993; Ausubel et al., 1995). Sequences to be tested were fused to the LexA DNA-binding domain (DBD) in plasmid pEG202 (Zervos et al., 1993) or to the activation domain (AD) in pJG4-5PL, a modified version of plasmid pJG4-5 (Gyuris et al., 1993) that contains the polylinker region from pEG202 (DeMarini et al., 1998). Full-length plo1, pom1, and mid1 and the NH2-terminal and COOH-terminal halves of plo1 (encoding amino acids 1–318 and 320–683, respectively) were amplified by PCR using the Expand PCR System (Boehringer Mannheim Corp., Indianapolis, IL) and either the cloned genes (plo1, pom1) or genomic DNA (mid1) as templates. Appropriate restriction sites were introduced with the primers. The PCR products were cloned at the NcoI-XhoI sites of pEG202 and pJG4-5PL using the pGEM-T Easy system (Promega Corp., Madison, WI). Constructs containing the S. cerevisiae MSB2 gene (Simon et al., 1995) were used as negative controls. The pEG202 constructs contain nine additional amino acids (EFPGIRRPW) between the LexA DBD and the first amino acid of the fused protein, and all constructs but one have the original stop codons of the fused genes. In the case of the plo1 NH2-terminal fusion, a stop codon was inserted (in the PCR primer) after the codon for amino acid 318. Strain EGY48 (Zervos et al., 1993) containing the LexAop-lacZ reporter plasmid pSH18-34 (Gyuris et al., 1993) was cotransformed with either pEG202 or a pEG202-derived plasmid expressing a LexA DBD-fusion protein and with either pJG4-5PL or a pJG4-5PL–derived plasmid expressing an AD-fusion protein. β-Galactosidase activities were measured in four independent isolates of each strain as described by Ausubel et al. (1995).
Microscopy and Protein Methods
Conventional fluorescence microscopy of GFP-containing or stained cells was performed as described previously (Bähler and Pringle, 1998). In some experiments, the medial ring was visualized using a GFP-Cdc4p fusion protein expressed from a plasmid under the control of an attenuated nmt1 promoter (Balasubramanian et al., 1997). Calcofluor staining of GFP-Cdc4p–expressing cells was performed as described by Balasubramanian et al. (1997). Immunofluorescence was performed as described previously (Balasubramanian et al., 1997) except as noted below. Antibodies to Cdc4p (McCollum et al., 1995), Mid1p (Sohrmann et al., 1996), GFP (a gift of Ken Sawin and Paul Nurse, ICRF, London, UK), Myc (monoclonal 9E10 monoclonal; a gift from Hayes McDonald, Vanderbilt University, Nashville, TN), actin (Karpova et al., 1993), and tubulin (monoclonal TAT1; Woods et al., 1989, or tubulin polyclonal serum; Gundersen et al., 1984; provided by Kevin Vaughn and Richard Vallee, UMMC, Worcester, MA) were diluted 1:100, 1:30, 1:200, 1:300, 1:100, and 1:10 or 1:200, respectively, and detected using anti–rabbit IgG tagged with Texas red, TRITC, or Alexa 594; TRITC-tagged anti–goat IgG; or anti–mouse IgG tagged with FITC or Alexa 488 (Molecular Probes, Eugene, OR; Jackson ImmunoResearch Laboratories, West Grove, PA), as appropriate, as described previously (McCollum et al., 1995; Bähler and Pringle, 1998). The TAT1 antibody was used for all tubulin staining experiments except for double staining with Myc monoclonal antibodies, in which case the rabbit polyclonal anti-tubulin antibody was used. For double staining of Mid1p and spindles, cell were fixed with 3.8% formaldehyde for 30 min at growth temperature and prepared for immunofluorescence as described (Bähler and Pringle, 1998). For double staining of actin and spindles, cells were treated also with methanol and acetone at −20°C for 6 min and 30 s, respectively, before incubation with the primary antibodies. For double staining of microtubules and Cdc4p, cells were fixed with a mixture of glutaraldehyde and formaldehyde (Moreno et al., 1991). Double staining of Mid1p and F-actin was performed as described by Sohrmann et al. (1996).
For time-lapse studies, including observations by deconvolution microscopy, exponentially growing cells expressing GFP-Cdc4p were concentrated ∼10-fold, and 10 μl of cell suspension were placed on a microscope slide and overlaid with a 1% YE agar slab. Cells were then viewed on a Zeiss Axiophot inverted fluorescence microscope using a 100× lens and an FITC filter set (Carl Zeiss, Inc., Thornwood, NY). Images were collected at 0.25 or 0.5 μm intervals through the cells, and two-dimensional representations of the three-dimensional images were calculated using a deconvolution algorithm (Wang, 1998). Time-lapse studies of pom1-Δ1 cells were conducted as described above except that all images were collected in a single focal plane.
Proteins were extracted as described by Moreno et al. (1991). SDS-PAGE was performed as described Ausubel et al. (1995). Western blotting was performed using a BioRad Immun-Star kit and a 1:2,000 dilution of 9E10 Myc antibodies.
Results
Identification of plo1 Mutants Defective in Medial Ring Placement and Organization
We identified three temperature-sensitive mutants that displayed defects in septum placement (see Materials and Methods). Complementation and linkage analysis (see Materials and Methods and data not shown) revealed that all three mutants had mutations in the same gene. Molecular cloning, mapping of an integrated plasmid, and rescue with a plo1 + cDNA plasmid showed that the three mutants harbored alleles (plo1-1, plo1-24C, and plo1-25) of the previously described plo1 gene (see Materials and Methods). At permissive temperature (25°C), mutant cells carrying any of the three alleles were wild-type in appearance and divided symmetrically (data not shown). However, at restrictive temperature (36°C), the mutants, although viable, exhibited a variety of defects in septum positioning and structure, with the plo1-1 mutant displaying the most severe defects. In contrast to the septa of wild-type cells (Fig. 1 B), the plo1 mutant septa were often misplaced from the cell center and/or not perpendicular to the long axis of the cell; in extreme cases, the mutant septa ran longitudinally through the cell (Fig. 1 D). Because the medial ring is thought to guide the placement of the septum, we examined the localization of the medial ring in plo1-1 mutant cells using a GFP-Cdc4p fusion protein. Cdc4p encodes a putative myosin light chain which localizes to the medial ring (McCollum et al., 1995; Balasubramanian et al., 1997). At restrictive temperature, the mutant cells displayed misplaced and disorganized medial rings (Fig. 1 C) whose positions corresponded approximately to those of the misplaced septa (Fig. 1 D). Similar results were obtained by staining cells with antibodies against either actin or Cdc4p (data not shown). These data suggest that the underlying plo1 mutant defect is in medial ring formation.
Figure 1.

Medial ring and septum defects in plo1-1 mutant cells and comparison of the plo1-1 and mid1-18 defects. Wild-type strain JB13 (A and B), plo1-1 mutant strain YDM110 (C–E), and mid1-18 strain YDM296 (F), all expressing GFP-Cdc4p fusion protein, were grown at 25°C and shifted to 36°C for 2 h before examination. (A–D) Cells were fixed and stained with Calcofluor to visualize septal material; GFP-Cdc4p fluorescence (A and C) and Calcofluor fluorescence (B and D) of the same cells are shown. (E and F) Living cells were mounted on a microscope slide and overlaid with an agar slab, and GFP-Cdc4p was viewed by fluorescence microscopy at 36°C. Images were collected every 0.25 μm and processed by deconvolution methods to generate a two-dimensional projection of the three-dimensional image (see Materials and Methods).
Previous analysis has shown that plo1 null mutants are defective in forming a bipolar spindle as well as in cytokinesis (Ohkura et al., 1995). To ask if the new plo1 mutations also cause defects in spindle formation, the mutant strains were stained with anti-tubulin and anti-Cdc4p antibodies after 2 h of growth at 36°C. Both plo1-24C and plo1-25 mutants showed spindle defects similar to those described previously for the null mutant; in both cases, a majority of the cells that had entered mitosis (as judged by the breakdown of cytoplasmic microtubules) displayed highly condensed chromosomes and either a small dot of tubulin staining in the nucleus or what appeared to be a short monopolar spindle (Fig. 2, A and B, upper 2 cells). Similar defects were observed in a small percentage of plo1-1 mutant cells. The block in spindle formation appeared to be transient: after 2 h at 36°C, some cells had formed seemingly normal spindles and proceeded through anaphase (Fig. 2, A and B, lower 3 cells), and after 4 h at 36°C, almost all plo1-24C and plo1-25 mutant cells had completed nuclear division at least once and contained two to four nuclei (data not shown). Cells that were blocked in spindle formation did not form medial rings (Fig. 2 C, upper 2 cells), but aberrant medial ring structures were observed after spindle formation (Fig. 2 C, lower 3 cells).
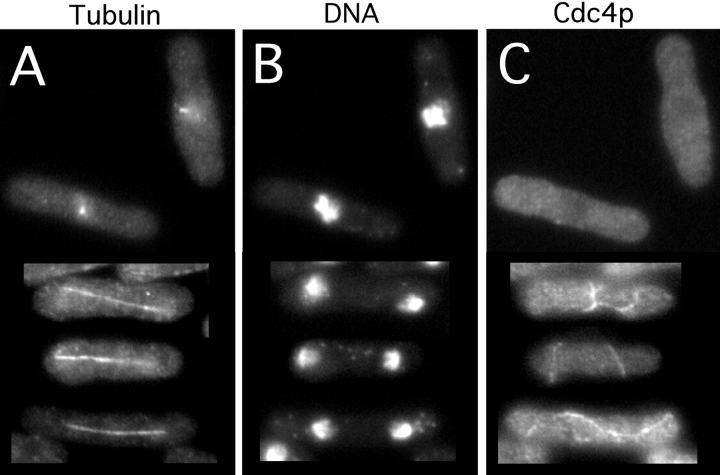
Figure 2.
Spindle formation defects in plo1-24C mutant cells. Cells of strain YDM114 were grown at 25°C to exponential phase, shifted to 36°C for 2 h, and triple stained for tubulin (A), DNA (B), and Cdc4p (C). The upper two cells were blocked at an early stage of spindle formation with highly condensed chromatin and no Cdc4p rings, whereas the lower three cells had leaked past the block and formed spindles and aberrant medial rings.
Comparison of the Medial Ring Defects in plo1, mid1, and pom1 Mutants
In addition to plo1, two other genes, mid1 and pom1, have been shown to be required for proper septum placement. Most pom1 mutant cells form relatively normal-looking medial rings and septa that are displaced towards one end of the cell (Bähler and Pringle, 1998). In contrast, the defect reported for mid1 mutants (Chang et al., 1996; Sohrmann et al., 1996) appeared more similar to that of the plo1 mutants in that the medial rings and septa were typically disorganized as well as misplaced. To characterize better the structures of the medial rings in these two mutants, plo1-1 and mid1-18 cells expressing GFP-Cdc4p were shifted to 36°C for 2 h and examined by deconvolution fluorescence microscopy (see Materials and Methods) to generate two-dimensional images of all the Cdc4p-containing structures in the cells. The two mutants appeared essentially indistinguishable (Fig. 1, E and F). In both mutants, many of the cells contained GFP-Cdc4p not only in rings but also in interconnected networks of cables. A similar analysis of pom1 mutant cells revealed Cdc4p rings of seemingly normal structure that were displaced from the middle of the cell (data not shown).
From the observations described above, it was not clear how the medial ring defects in the three mutants arose. For example, it seemed possible either that the mutants initiated medial ring formation at the wrong place or that they initiated medial ring formation at the right place but were unable to anchor the ring properly, so that it could drift away. In addition, in the cases of plo1 and mid1, it seemed possible either that the aberrant Cdc4p-containing structures formed de novo or that they formed by the breakdown of normally structured rings that had formed initially. To explore these possibilities, we used time-lapse video microscopy of living cells expressing the GFP-Cdc4p fusion protein at restrictive temperature. The medial rings formed in the pom1 mutant were observed readily in a single focal plane. However, the aberrant structures formed in the plo1 and mid1 mutants were difficult to observe in a single focal plane, so we used compiled images derived from serial optical sections (see above and Materials and Methods). Images were collected at 5-min intervals (the shortest convenient interval for the serial sectioning). For each strain, time courses were done on at least six cells, and representative series are shown in Fig. 3. In the pom1 mutant, medial rings first appeared at incorrect locations and subsequently did not move; except for their locations, the rings appeared normal. In contrast, in the plo1 and mid1 mutants, the structure as well as the placement of the medial rings was clearly aberrant from the beginning. Most cells began by forming Cdc4p-containing cables that ran along the long axis of the cell. These structures often appeared first at the cell ends (Fig. 3, asterisks) and then grew and moved into different regions of the cell. Occasionally, some cells managed to assemble a relatively normal looking medial ring (Fig. 3, arrowhead). It seemed possible that the deconvolution processing methods could have caused some structures present in the original images to be lost or could even have caused the appearance of structures that were not present in the original images. However, examination of the individual sets of raw images showed that all structures found in the deconvoluted images were present in the raw data, and all structures found in the raw images were represented in the deconvoluted images (data not shown). It also seemed possible that a crucial early stage of medial ring formation had been missed in these experiments because of the 5-min interval between images. However, capturing images of plo1-1 cells in a single focal plane every 30 s gave very similar results: of 22 cells examined, 18 had Cdc4p-containing cables appear first at their tips (data not shown). In addition, examination of medial ring formation in asynchronous populations of the plo1-1 and mid1-18 mutants 1 h after shift to restrictive temperature also showed that a majority of these cells (66 of 99 for mid1-18 and 79 of 110 for plo1-1) formed Cdc4p-containing cables at their ends (data not shown).
Figure 3.
Time-lapse analysis of medial ring formation in mutant cells. Wild-type (JB13), pom1-Δ1 (JB110), plo1-1 (YDM110), and mid1-18 (YDM296) cells, all expressing GFP-Cdc4p fusion protein, were grown at 25°C, then shifted to 36°C for 1 h in liquid culture. The living cells were then mounted on a microscope slide overlaid with an agar slab and viewed by fluorescence microscopy at 36°C. For wild-type, plo1-1, and mid1-18 cells, images were collected every 0.5 μm at 5-min intervals and processed by deconvolution methods to generate two-dimensional projections of the three-dimensional images (see Materials and Methods). Images of pom1-Δ1 cells were taken in a single focal plane. Asterisks and the arrowhead indicate structures described in the text.
If actin is not recruited efficiently to the middle of the cell in the plo1 and mid1 mutants, it might then begin to form filaments at the cell ends, where it is most concentrated in interphase cells. This might explain the frequent origination of the Cdc4p-containing structures at the cell ends in these mutants (see above). In this case, the effect might be more dramatic in highly elongated cells. Therefore, we examined medial ring formation in cdc25-22 mutant cells, which become highly elongated at the restrictive temperature due to a block in late G2 (Thuriaux et al., 1980; Mitchison and Nurse, 1985) and undergo cytokinesis synchronously upon return to permissive temperature. When cdc25-22 single-mutant cells were blocked at restrictive temperature for 4 h and then returned to permissive temperature, 66 of 66 cells examined developed a Cdc4p-containing ring at the cell center (Fig. 4, wt). Similarly, in cdc25-22 pom1-Δ1 double mutants, 69 of 69 cells examined formed rings at or near the cell center (Fig. 4, pom1-Δ; see Discussion). In contrast, 82 of 91 cdc25-22 mid1-ΔF double-mutant cells examined formed rings and filaments close to the very ends of the cells (Fig. 4, mid1-Δ), consistent with the hypothesis that Mid1p (and presumably Plo1p) is required for recruitment of actin from the cell ends to the cell center at the time of ring formation.
Figure 4.
Medial ring formation in elongated cells. cdc25-22 (strain YDM152; wt), cdc25-22 pom1-Δ1 (strain JB120), and cdc25-22 mid1-ΔF (strain JB43) cells, all expressing GFP-Cdc4p fusion protein and growing exponentially at 25°C, were shifted to 36°C for 4 hr, returned to 25°C, and observed 45 min later.
Early Events in Medial Ring Formation
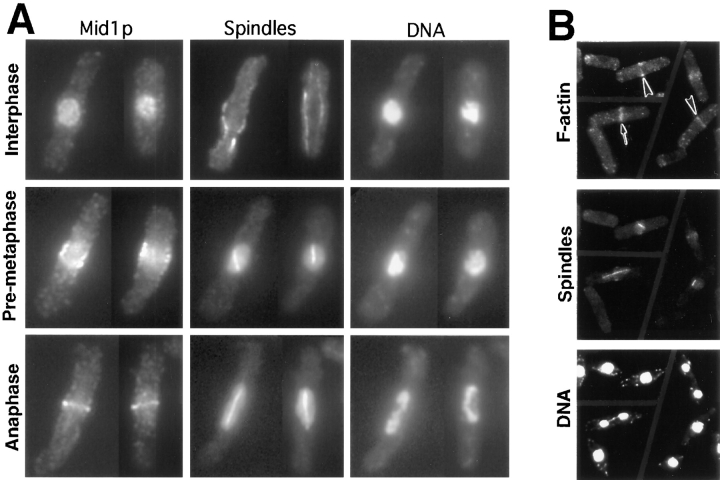
The formation of a medial ring during mitosis is the earliest known step of cytokinesis and appears to determine the site at which division will occur (see Introduction). However, the precise timing of the appearance of various medial ring components relative to each other and to the stages of mitosis has not been well characterized. To study this timing in more detail, we first stained cells simultaneously for Mid1p, mitotic spindles, and DNA. As reported previously (Sohrmann et al., 1996), Mid1p was nuclear in all cells without mitotic spindles (interphase cells; Fig. 5 A, Interphase) and was visible as a discrete medial ring in metaphase or anaphase cells with well separated spindle poles (Fig. 5 A, Anaphase). However, in 46 of 50 cells with short spindles (pre-metaphase cells), although some Mid1p typically remained in the nucleus, most Mid1p was found in a diffuse band at the cell cortex in the nuclear region (Fig. 5 A, Pre-metaphase). Thus, Mid1p leaves the nucleus at an early stage of mitosis but does not organize into a tight ring until the spindle is fully formed at metaphase or anaphase onset.
Figure 5.
Timing of Mid1p ring formation and F-actin ring formation in wild-type cells. (A) Mid1p-GFP expressing cells (strain YDM403) growing exponentially at 32°C were triple stained for Mid1p (using GFP-specific antibodies), spindles, and DNA. Representative interphase, pre-metaphase, and anaphase cells are shown. Very similar results were obtained when strain 972 (wild-type) cells were stained with Mid1p-specific antibodies or when Mid1p-13Myc-expressing cells (strain YDM603) were stained with antibodies to the Myc epitope. (B) Strain 972 cells growing exponentially at 36°C were triple stained for F-actin, spindles, and DNA. The fixation protocols used visualize Mid1p, Mid1p-fusion proteins, F-actin, and spindle microtubules well, but preserve cytoplasmic microtubules poorly. Large arrowheads, pre-metaphase cells with short spindles; arrow, cell with an anaphase spindle.
We also stained cells simultaneously for actin and mitotic spindles. Surprisingly, actin was already detectable as a discrete ring in 16 of 20 pre-metaphase cells with short spindles (Fig. 5 B, arrowheads), although these early rings often seemed to stain less intensely or to be less well centered and/or less well organized (e.g., split into two strands) than the actin rings seen at later stages (Fig. 5 B, arrow, and data not shown). Taken together, the data indicate that a discrete ring of actin appears during spindle formation, at a time when Mid1p is present as a diffuse cortical band. However, the actin ring may become more coherent and better centered later in mitosis as other components (including Mid1p) join the medial ring structure.
Roles of Plo1p and Pom1p in Formation and Localization of the Mid1p Ring
It was shown previously that the relocalization of Mid1p from the nucleus to the medial ring is correlated with an increase in Mid1p phosphorylation (Sohrmann et al., 1996). Since both plo1 and pom1 encode putative protein kinases that are involved in medial ring formation, we asked if these kinases were required for Mid1p localization to the medial ring. At permissive temperature, the localization of Mid1p in both interphase and mitotic cells of the plo1-1 mutant was indistinguishable from that in wild-type cells (Fig. 6 A, and data not shown). However, at restrictive temperature, Mid1p appeared to remain in the nucleus throughout mitosis and failed to form a discrete ring at the cell cortex (Fig. 6 B). In plo1-25 and plo1-24C strains, at least some Mid1p appeared to leave the nucleus during mitosis, but Mid1p rings were never observed (data not shown). Control experiments showed that Mid1p rings formed normally at 36°C in wild-type cells (data not shown).
Figure 6.
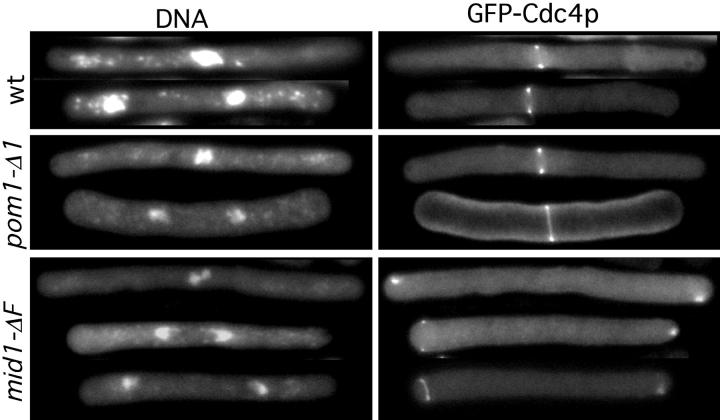
Failure of Mid1p nuclear exit and ring formation in the plo1-1 mutant. (A) mid1p-GFP plo1-1 cells (strain JB250) growing exponentially at 25°C were fixed and triple stained for Mid1p, spindles, and DNA as in Fig. 5. Very similar results were obtained when plo1-1 cells (strain YDM110) were stained with Mid1p-specific antibodies or when mid1-13Myc plo1-1 cells (strain YDM607) were stained with Myc-specific antibodies. (B) plo1-1 cells expressing Mid1p-GFP (strain JB250) or Mid1p-13Myc (strain YDM607) cells growing exponentially at 25°C were shifted to 36°C for 2.5 h before fixation. The fixed cells were triple stained for Mid1p-GFP (top) or Mid1p-13Myc (bottom) using GFP-specific or Myc-specific antibodies as appropriate, for spindles, and for DNA as in Fig. 5. Very similar results were obtained when plo1-1 cells (strain YDM110) were stained with Mid1p-specific antibodies. Note that the anti-tubulin antibodies used to stain the Mid1p-13Myc cells (see Materials and Methods) do not give the background nuclear staining like the TAT-1 anti-tubulin monoclonal antibody which is used above.
The behavior of Mid1p differed in a pom1 deletion strain. In this case, interphase cells displayed nuclear Mid1p staining that was reproducibly weaker and more diffuse than that in wild-type cells (Fig. 7, A and B). During mitosis, the pom1 mutant cells did form Mid1p rings (Fig. 7, A and B), but these rings were frequently misplaced (Fig. 7, A and C, arrows) like the medial rings and septa seen in pom1 mutants (Bähler and Pringle, 1998; see also above). Double staining for Mid1p and actin showed that the mislocalized Mid1p and actin rings coincided, as expected (Fig. 7, C and D, arrows). The weaker nuclear staining of interphase cells might have resulted from a reduction in Mid1p levels in the pom1 mutant strain. However, when we determined Mid1p levels in wild-type, plo1, and pom1 mutant backgrounds using strains that expressed Mid1p-13Myc from the normal chromosomal mid1 locus (see Materials and Methods), all strains displayed similar levels of protein after incubation at 36°C (Fig. 8 A). Thus, Pom1p appears to be necessary both for normal nuclear localization of Mid1p in interphase cells and for normal placement of the Mid1p ring in mitotic cells, but not for the actual formation of the Mid1p ring.
Figure 7.

Mislocalization of Mid1p rings in a pom1 mutant. Cells of pom1-Δ1 strain JB110 growing exponentially at 30°C were double stained for Mid1p using Mid1p-specific antibodies (A) and DNA (B) or for Mid1p (C) and F-actin (D). Arrows indicate the positions of mislocalized Mid1p and actin rings.
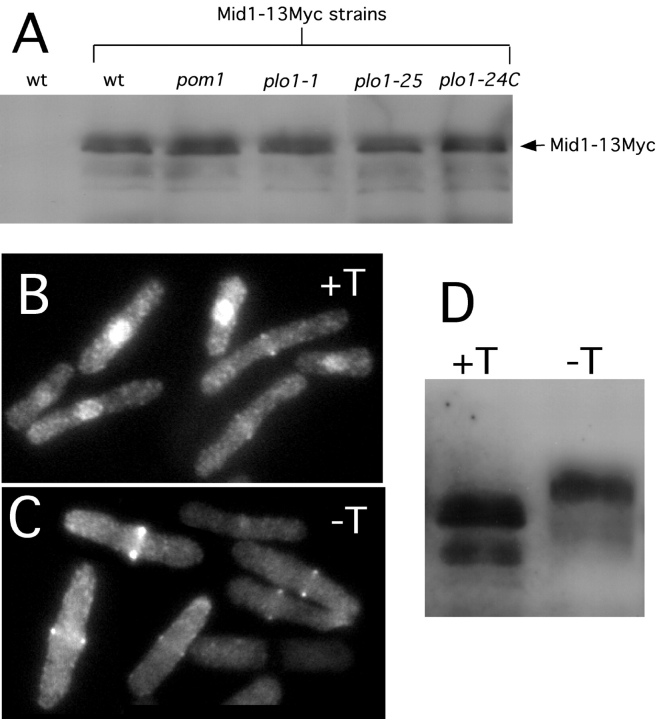
Figure 8.
Mid1p levels in plo1 and pom1 mutants, and the effect of Plo1p overproduction on Mid1p. (A) Strains expressing either normal Mid1p (no Myc) (YDM105) or Mid1p-13Myc (wt, YDM603; pom1, YDM609; plo1-1, YDM607; plo1-25, YDM604; and plo1-24C, YDM608) were grown at 25°C, shifted to 36°C for 2.5 h, and harvested. Protein lysates were prepared from each strain and equal amounts of protein were analyzed by Western blotting using anti-Myc antibodies. (B-D) Mid1p-13Myc-expressing cells (YDM603) containing a plasmid (pREP1Plo1) for expression of plo1 from the thiamine-repressible nmt1 promoter, were grown at 30°C in the presence of thiamine. This culture was then used to inoculate two new cultures either with (promoter off) or without (promoter on) thiamine, and growth was continued for 16 h. Portions of each culture were then either fixed and stained for Mid1p-13Myc using Myc-specific antibodies or used to prepare lysates for analysis by western blotting. (B and C) Mid1p-13Myc staining in cells either not overexpressing (B) or overexpressing (C) Plo1p. (D) Western blot analysis of Mid1p-13Myc in cells either not overexpressing (+T) or overexpressing (−T) Plo1p.
Because Mid1p becomes hyperphosphorylated in mitosis and displays a mobility shift on gels (Sohrmann et al., 1996), we might have expected to observe some difference in mobility between Mid1p in plo1 (or pom1) mutant and wild-type cells. However, the Mid1p mobility shift was only observed using cells that had been highly synchronized in mitosis using a cdc25 block and release, and not in asynchronous cells like those examined here (Sohrmann et al., 1996). Another way to examine the effect of Plo1p activity on Mid1p is to examine Mid1p in cells overexpressing Plo1p. Thus, we examined the localization and electrophoretic mobility of Mid1p-13Myc in cells expressing plo1 under control of the thiamine-repressible nmt1 promoter. Cells in which the promoter was repressed grew normally and displayed a Mid1p localization pattern similar to that seen in wild-type cells (Fig. 8 B). Among 124 cells examined, 106 displayed Mid1p localization to the nucleus and 12 displayed Mid1p rings. However, when the promoter was induced, among 107 cells examined, none displayed nuclear staining and 83 had unequivocal Mid1p rings (Fig. 8 C). Furthermore, overexpression of Plo1p caused a very noticeable retardation in Mid1p mobility (Fig. 8 D), presumably due to phosphorylation, like that previously observed in mitotic cells (Sohrmann et al., 1996).
Genetic and Two-Hybrid Interactions among plo1, mid1, and pom1
To gain further insights into the functional relationships among Plo1p, Mid1p, and Pom1p, we examined double mutants to uncover possible genetic interactions (synthetic lethality, suppression, or epistasis) between mutations in the corresponding genes. Double mutants containing any of the three plo1 alleles together with mid1-ΔF were viable and phenotypically indistinguishable from the single mutants (Table II and data not shown). In contrast, the mid1-18 allele showed a temperature-sensitive synthetic lethality with pom1-Δ1 (Bähler and Pringle, 1998). Moreover, mid1-ΔF pom1-Δ1 double-mutant spores were inviable at all temperatures tested (Table II); they germinated but did not divide (data not shown). Double mutants containing pom1-Δ1 together with any of the three plo1 alleles also showed a temperature-sensitive synthetic lethality (Table II and data not shown). Interestingly, this effect was most pronounced with the plo1-1 allele, which has the most severe defects in septum placement and organization (Fig. 1). The plo1-1 single mutant grows better at high temperatures than does the plo1-24C single mutant (Table II), which has the most severe defect in spindle formation (Fig. 2). Taken together, these data suggest that Plo1p and Mid1p may function in a common pathway, whereas Pom1p has a distinct and complementary role in septation. Plo1p/ Mid1p and Pom1p together define an essential function for the cell.
We also analyzed interactions among Plo1p, Mid1p, and Pom1p using the yeast two-hybrid system (Table III). Although backgrounds were high, full-length Mid1p appeared to interact significantly with both full-length Plo1p and the COOH-terminal half of Plo1p, although not with the NH2-terminal half of Plo1p, which contains the protein kinase catalytic domain (Ohkura et al., 1995). In contrast, neither Mid1p nor Plo1p interacted detectably with full-length Pom1p. These data support the hypothesis that Plo1p and Mid1p function together in a pathway that is distinct from the Pom1p pathway.
Table III.
Two-hybrid Interaction between Mid1p and Plo1p
| AD fusion in pJG4-5PL | LexA-DBD fusion in pEG202 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| plo1 | plo1 (NH2-terminal) | plo1 (COOH-terminal) | pom1 | MSB2 * | ||||||
| mid1 | 1,042 | 23 | 5,119 | 3 | 3 | |||||
| plo1 | 6 | |||||||||
| MSB2 * | 50 | 21 | 760 | 4 | ||||||
| No insert | 61 | 14 | 589 | 4 | ||||||
β-Galactosidase activities were determined in Materials and Methods. The reported values are the averages of at least four determinations.
The S. cerevisiae MSB2 gene (Bender and Pringle, 1992) was used as a negative control (see also Cvrcková et al., 1995; Simon et al., 1995).
Localization of Plo1p
To determine the localization of Plo1p, a strain was constructed that expressed a Plo1p-GFP fusion protein from the normal chromosomal plo1 locus (see Materials and Methods). Examination of asynchronous cultures of living cells showed that shorter cells displayed no detectable signal (Fig 9 A, cell 1). However, in longer cells, which were presumably in late G2 or early mitosis, a faint nuclear staining was observed along with a brighter spot at the periphery of the nucleus that probably corresponded to the SPB (Fig. 9 A, cell 2), as judged by observations on cells with spindles. During spindle formation, there was a strong Plo1-GFP signal at both spindle poles and a faint signal along the length of the spindle (Fig. 9 A, cells 3–6). At the onset of anaphase, there was a strong diminution in the signal, and detectable signal was lost before cells divided. The loss of Plo1p-GFP signal during anaphase was also observed in living cells using real-time video microscopy and by examining cells synchronized by cdc25-22 block and release (data not shown). In some cells, a faint cortical ring could be observed in the middle of the cell (data not shown). This ring was only observed just before anaphase, as judged from spindle length, and was hard to document.
Figure 9.
Localization of Plo1p-GFP in wild-type, cdc25-22, and cdc25-22 mid1-ΔF cells. (A) plo1-GFP strain JB206 was grown at 30°C, and cells were photographed at various stages in the cell cycle as described in the text. (B and C) plo1-GFP cdc25-22 cells (strain YDM457; B) and plo1-GFP cdc25-22 mid1-ΔF cells (strain JB214; C) were blocked at 36°C for 4 h and then shifted to 25°C for 45 min. The arrowheads indicate medial Plo1p rings.
We then examined Plo1p-GFP localization in cdc25-22 mutant cells, for two reasons. First, we have found that some faint structures are more easily visualized in large cells such as those of cdc25-22 mutants. Second, we thought that the faint cortical ring structure might be more readily visualized in cells that were proceeding synchronously through division (see above). We first examined Plo1p-GFP localization in living cdc25-22 cells at 25°C. Even at this temperature, the cells are somewhat elongated compared with wild type because of a delay in G2. The Plo1p signal in these cells seemed more intense than that in wild-type cells, but the localization observed was essentially the same, except that the medial ring in metaphase cells was more clearly visible in the cdc25-22 cells (data not shown). cdc25-22 cells that had been blocked at 36°C and then shifted down to 25°C showed an even more intense Plo1p-GFP signal, and the ring was clearly visualized in 44 of 48 metaphase cells examined (Fig. 8 B, arrowheads). Such rings were not observed in cells at earlier or later stages of the cell cycle. Given the timing and appearance of the Plo1p-GFP ring, it seemed likely that Plo1p localized to the medial ring at about the time that the Mid1p ring was observed. However, we were unable to demonstrate colocalization of the Plo1p-GFP ring with medial ring components using immunofluorescence because the Plo1p-GFP ring staining did not survive any of a variety of different fixation methods. To determine whether Plo1p localization depended on the presence of Mid1p, cdc25-22 mid1-ΔF cells expressing Plo1p-GFP were blocked at 36°C and then shifted down to 25°C as described above. These cells showed Plo1p-GFP signal along the spindle and at the spindle poles similar to that seen in wild-type or cdc25-22 cells, but no Plo1p-GFP rings were observed in 45 metaphase cells examined (Fig. 8 C). Thus, Mid1p seems to be required for Plo1p to localize to the medial ring but not to the spindle or spindle poles.
Discussion
Proper segregation of chromosomes to each daughter cell requires precise temporal and spatial coordination between the mitotic apparatus and the cleavage furrow. Polo family kinases have been shown to be essential for spindle formation in S. pombe (Ohkura et al., 1995), Drosophila (Sunkel and Glover, 1988; Llamazares et al., 1991), and human cells (Lane and Nigg, 1996). In this report, we identify cleavage plane positioning as an additional function of the Plo1p kinase in S. pombe, suggesting that Plo1p may function in the spatial and temporal coupling of spindle formation with cytokinesis.
Function of Plo1p and Mid1p in a Common Pathway for Medial Ring Positioning
At the onset of mitosis, Mid1p becomes hyperphosphorylated, leaves the nucleus, and forms a medial ring (Sohrmann et al., 1996). It seems likely that mutants defective in phosphorylating Mid1p and/or in redistributing it from the nucleus to the medial ring would have phenotypes similar to those of mid1 loss-of-function mutants. Mutations in the pom1 (Bähler and Pringle, 1998) and plo1 (this study) protein kinase genes also cause defects in positioning the cell division site, suggesting that one or both of these putative kinases might regulate the behavior of Mid1p. In fact, several lines of evidence suggest that Plo1p and Mid1p function in a common pathway for medial ring placement and that Pom1p functions in a distinct pathway. First, comparison of pom1, plo1, and mid1 mutants showed that the plo1 and mid1 mutants have the most similar phenotypes. Second, mid1 pom1 and plo1 pom1 double mutants both display strong negative interactions (unconditional or temperature-sensitive synthetic lethality), whereas plo1 mid1 double mutants resemble the single mutants. Third, Mid1p and Plo1p were found to interact in the yeast two-hybrid system, whereas Pom1p did not interact detectably with either Plo1p or Mid1p. Fourth, Plo1p overexpression leads to a Mid1p mobility shift consistent with its hyperphosphorylation and causes Mid1p to leave the nucleus and form a ring even in interphase cells. Finally, Mid1p ring formation does not occur in cells carrying any of the three plo1 mutations described here. Interestingly, the phenotypes of the three plo1 mutants differ somewhat with respect to Mid1p localization. The plo1-1 mutant, which has the most severe defect in medial ring positioning, retains Mid1p in the nucleus at mitosis, whereas in the plo1-24C and plo1-25 mutants, some Mid1p does leave the nucleus although it does not localize to a ring. These allelic differences could be explained if Plo1p were required both for nuclear export of Mid1p and for Mid1p ring formation. In contrast, Pom1p is not required for Mid1p ring formation, since Mid1p localizes to the misplaced rings formed in pom1 mutants. In addition to its role in the proper positioning of the medial ring, Pom1p may be involved in the nuclear import of Mid1p after mitosis, because pom1 mutant cells in interphase show fainter nuclear staining for Mid1p. The evidence that Plo1p and Mid1p function in a common pathway suggests the possibility that Mid1p may be a substrate of the Plo1p kinase. The function of Pom1p in division site selection remains more obscure and will be an important subject for future study.
Role of Mid1p in Placement of the Medial Ring
It has been reported that the Mid1p ring forms before the actin ring and thus that Mid1p may mark the site for actin ring formation (Sohrmann et al., 1996). However, it remained unclear how Mid1p could go from the nucleus to a well defined cortical ring. It seemed possible either that Mid1p localized to an existing ring structure at the cortex or that there was an intermediate stage in Mid1p ring formation that had not been recognized previously. We found the latter to be the case: in pre-metaphase cells with incomplete or short mitotic spindles, Mid1p was present in a diffuse cortical band in the medial region of the cell, and discrete Mid1p rings were not observed until cells were in metaphase or early anaphase. Surprisingly, actin rings were observed at the medial cortex in the pre-metaphase cells. Although these rings were faint and sometimes branched or slightly misplaced in comparison to the actin rings in metaphase or anaphase cells, their formation clearly preceded the formation of the discrete Mid1p ring. It is not clear why our results differ from those of the previous study. One possibility is that the early actin rings were not preserved well under the fixation and staining conditions employed by Sohrmann et al. (1996), which differed from those used here (see Materials and Methods).
How, then, does Mid1p define the position at which the actin ring forms? Before the onset of mitosis, actin is present primarily at the growing ends of the cells. When Mid1p leaves the nucleus early in mitosis to form the broad band at the cortex overlying the nucleus, it may recruit actin and/or actin-binding proteins to this zone from the ends of the cell. Actin filaments that form in this zone may then coalesce into the well-defined ring structure through a self-focusing mechanism that involves actin-bundling proteins and perhaps tension provided by myosin. Mid1p may also play a role in this maturation process, since mid1 mutants form rings that are not only misplaced but often branched and disorganized as well. However, Mid1p is clearly not required for actin ring formation per se, since mid1 temperature-sensitive and null mutants are both capable of forming rings (Chang et al., 1996; Sohrmann et al., 1996; this study).
One prediction of this model for Mid1p function is that in the absence of Mid1p, actin and/or actin-binding proteins would not be recruited efficiently to the cell middle, and actin ring formation might then occur at the cell ends, where the concentration of actin is highest. A previous study examining the localization of the medial ring component Cdc12p in a mid1 mutant observed that a majority of cells displayed a dot of Cdc12p near the cell center with a cable going from this dot towards the cell tip, suggesting that the ring originated in the center of the cell and extended towards the tip (Chang et al., 1997). In contrast we found that many cells first formed medial ring-like structures at the cell ends in both mid1 and plo1 mutants. It seemed possible that this effect would be exaggerated in elongated cells, because the actin at the cell ends would be even further from the middle of the cell when ring formation began, and this was indeed observed in elongated mid1-ΔF cdc25 mutant cells, which typically formed rings or cables at their extreme tips. In contrast, pom1 cdc25 mutant cells, typically formed the medial ring close to the cell center, suggesting that pom1 mutants may misplace the ring with respect to the nucleus but not the growing ends of the cell. Taken together, these findings support the hypothesis that Mid1p, but not Pom1p, is required to recruit actin and/or actin-binding proteins to the cell center. It is not clear why Chang et al. (1997) reached a different conclusion. The discrepancy may result from the use of different mid1 alleles or of different markers of medial ring formation in the two studies. Alternatively, as the previous studies did not include real-time observations, the structures observed might actually have originated at the cell tips and then moved toward the cell center.
Localization and Functions of Plo1p
The Plo1p kinase appears to have multiple functions in cell division. To determine whether the localization of Plo1p is consistent with the multiple roles ascribed to it, we examined the localization of a Plo1p-GFP fusion. A distinct signal was observed only in late G2 and/or mitotic cells. Just before spindle formation, a faint nuclear signal was observed along with a stronger signal that appeared to correspond to the SPB. Plo1p then remained at the spindle poles and (more faintly) along the length of the spindle until the onset of anaphase. In addition, in metaphase or early anaphase cells, a faint Plo1p-GFP signal was also observed at the medial ring. As anaphase proceeded, the Plo1p-GFP signal diminished at all locations in the cell and then disappeared, suggesting that the protein was either degraded or dispersed at this time.
These localization data are consistent with the roles ascribed to Plo1p. In particular, the Plo1p associated with the spindle pole bodies and spindle appears to play a role in the formation of a bipolar spindle, as two of the three plo1 mutant strains (plo1-24C and plo1-25) displayed a transient defect in formation of a bipolar spindle similar to that reported previously (Ohkura et al., 1995) for plo1 null mutants. However, once past this block, the plo1-24C and plo1-25 mutants formed spindles and proceeded normally through anaphase, suggesting that Plo1p may not be required for anaphase progression once it is initiated. In addition, the localization of Plo1p is consistent with the other evidence that it functions together with Mid1p in medial ring formation and positioning. The Plo1p-GFP signal appears in the nucleus just at the time of Mid1p exit from the nucleus. In addition, Mid1p fails to exit the nucleus in the plo1-1 mutant, and cells that are strongly overexpressing Plo1p do not display Mid1p nuclear staining. These data suggest that Plo1p may be required for the exit of Mid1p from the nucleus. In addition, the localization of Plo1p to the medial ring raised the possibility that Plo1p might actually mark the site at which the medial ring will form and then recruit Mid1p to that site. However, localization of Plo1p to the medial ring appears to depend on the presence of Mid1p. One model to explain these results is that Plo1p binds Mid1p early in mitosis and triggers its exit from the nucleus, perhaps by phosphorylation. Plo1p might then be transported along with Mid1p to the cortex and the medial ring. A prediction of this model is that in early mitotic cells, Plo1p, like Mid1p, should be present in a diffuse cortical band. This was not seen, possibly because the Plo1p cortical staining was very faint, so that a diffuse band probably would not have been detectable.
Ohkura et al. (1995) found that no medial rings were formed in the absence of Plo1p. In contrast, the plo1 mutants described in this study are capable of making rings, albeit misplaced ones. Thus, the mutant proteins presumably retain some function. Moreover, the differences in mutant phenotypes suggest that the different mutant proteins are selectively defective in particular aspects of Plo1p function. It has also been suggested that Plo1p is required for initiation of medial ring contraction and septation (Ohkura et al., 1995). Although we do not address this function of Plo1p directly in this study, it is worth noting that some plo1-25 and plo1-24C cells proceed through mitosis but do not deposit septum material (Bähler, J., and D. McCollum, unpublished observations), suggesting that they may also be defective in this later stage of cell division.
Are Spindle and Medial Ring Formation Separable Functions of Plo1p?
In mammalian cells and echinoderm eggs, it has been shown that manipulation of the mitotic spindle can cause alterations in placement of the cleavage furrow (for review see Rappaport, 1986, 1996; Oegema and Mitchison, 1997). Thus, it could be argued that the ring placement defects in the plo1 mutants are a secondary consequence of earlier spindle defects. However, this seems unlikely for several reasons. First, the plo1-1 mutant has the strongest defect in medial ring positioning, yet it has virtually no spindle defect. Second, Plo1p appears to act in conjunction with Mid1p to promote medial ring positioning and formation, yet Mid1p ring formation does not depend on the presence of a functional mitotic spindle (Sohrmann et al., 1996). Similarly, other mutants that are either completely defective in spindle formation (such as β-tubulin mutants) or that form monopolar spindles are still capable of forming normal actin rings and septa (Hagan and Yanagida, 1990; Chang et al., 1996). On the other hand, ring formation may depend on execution of the Plo1p function in spindle formation, because plo1 mutant cells blocked transiently in spindle formation do not make medial rings until they escape from this block (Fig. 2).
Evolutionary Conservation of Plo1p Functions
An important question is how conserved the functions of Polo kinase are between S. pombe and multicellular eukaryotes. The spindle defects caused by loss of Polo function are quite similar in S. pombe (Ohkura et al., 1995; this study), Drosophila (Llamazares et al., 1991), and human cells (Lane and Nigg, 1996), suggesting that this function is highly conserved. Although not conclusive, recent evidence suggests that Polo kinase is also involved in cytokinesis in animal cells. In mammalian cells, signaling from the spindle midzone during anaphase is thought to induce the cell cortex to initiate cleavage furrow formation (Oegema and Mitchison, 1997). Interestingly, mammalian Plk1 localizes to the spindle poles up to metaphase but relocalizes to the spindle midzone upon anaphase onset, and kinase activity peaks during mitosis (Golsteyn et al., 1995; Lee et al., 1995). Recent experiments have also demonstrated that overproduction of either wild-type or a kinase-dead version of Plk1 in mammalian cells results in the accumulation of multiple nuclei due to failures in cytokinesis, possibly because a protein required for cytokinesis is titrated away (Mundt et al., 1997; Smith et al., 1997). If Polo kinases do function in cytokinesis in animal cells, it will be interesting to determine if they act through proteins homologous to Mid1p. In this regard, it is intriguing that the Drosophila protein anillin (Field and Alberts, 1995) has some similarities to Mid1p. Both proteins cycle from the nucleus in interphase to the contractile ring in mitosis. In addition, the two proteins have some structural similarities that include a proline-rich domain and a COOH-terminal PH domain (Field and Alberts, 1995; Sohrmann et al., 1996; McCollum, D., unpublished observations).
The S. pombe Polo kinase Plo1p was previously shown to have a role in the formation of a bipolar spindle (Ohkura et al., 1995). In this study, we have confirmed this role and also identified a new role for Plo1p in the positioning and formation of the medial ring that is responsible for cytokinesis. Moreover, we have identified Mid1p as a likely target of Plo1p during medial ring formation, and we have provided evidence that these proteins effect medial ring positioning through a pathway distinct from that involving Pom1p. Targets of Plo1p other than Mid1p are not known in fission yeast, and identification of such targets will be crucial for understanding how a single molecule such as Plo1p is able to promote multiple aspects of mitosis and cytokinesis. The methods of genetic analysis available in S. pombe should greatly facilitate the search for targets of Plo1p and hence perhaps also of other Polo kinases.
Acknowledgments
We are grateful to Charlie Albright, Tony Carr, John Cooper, Keith Gull, Jürg Kohli, Paul Nurse, Ken Sawin, Richard Vallee, Kevin Vaughn, Cindy Sparks, Viesturs Simanis, Marc Sohrmann, and Paul Young for the gifts of plasmids, antibodies, genomic DNA libraries, and strains. We thank Farid Irshad for technical assistance, and Mohan Balasubramanian and Damian Brunner for critical reading of the manuscript.
This work was supported by National Institutes of Health grants GM58406 to D. McCollum and GM31006 to J.R. Pringle and by the Howard Hughes Medical Institute, of which K.L. Gould is an Assistant Investigator. J. Bähler was supported by fellowships from the Swiss National Science Foundation and the Ciba-Geigy-Jubiläums-Stiftung.
Abbreviations used in this paper
- AD
activation domain
- DBD
DNA-binding domain
- GFP
green fluorescent protein
- SPB
spindle pole body
Footnotes
Dr. Bähler's present address is Imperial Cancer Research Fund, Cell Cycle Laboratory, 44 Lincoln's Inn Fields, London WC2A 3PX, UK.
References
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1995. Current Protocols in Molecular Biology. John Wiley and Sons Ltd., New York.
- Bähler J, Pringle JR. Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev. 1998;12:1356–1370. doi: 10.1101/gad.12.9.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu J-Q, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. . Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, McCollum D, Gould KL. Cytokinesis in the fission yeast Schizosaccharomyces pombe. . Methods Enzymol. 1997;283:494–506. doi: 10.1016/s0076-6879(97)83039-x. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, McCollum D, Chang LC, Wong KCY, Naqvi NI, He X, Sazer S, Gould KL. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 1998;149:1265–1275. doi: 10.1093/genetics/149.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet N, Muriel WJ, Carr AM. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. . Gene. 1992;114:59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- Bender A, Pringle JR. A Ser/Thr-rich multicopy suppressor of a cdc24bud emergence defect. Yeast. 1992;8:315–323. doi: 10.1002/yea.320080409. [DOI] [PubMed] [Google Scholar]
- Chang F, Nurse P. How fission yeast fission in the middle. Cell. 1996;84:191–194. doi: 10.1016/s0092-8674(00)80973-3. [DOI] [PubMed] [Google Scholar]
- Chang F, Woollard A, Nurse P. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J Cell Sci. 1996;109:131–142. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. Cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrcková F, De Virgilio C, Manser E, Pringle JR, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- DeMarini DJ, Pringle JR. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiaecell wall. J Cell Biol. 1998;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, West RR, Morphew M, Oakley BR, McIntosh JR. The spindle pole body of Schizosaccharomyces pombeenters and leaves the nuclear envelope as the cell cycle proceeds. Mol Biol Cell. 1997;8:1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ. Spindle orientation and asymmetric localization in Drosophila: both inscuteable? . Cell. 1996;86:695–697. doi: 10.1016/s0092-8674(00)80142-7. [DOI] [PubMed] [Google Scholar]
- Edamatsu M, Toyoshima YY. Isolation and characterization of pos mutants defective in correct positioning of septum in Schizosaccharomyces pombe. . Zool Sci. 1996;13:235–239. doi: 10.2108/zsj.13.235. [DOI] [PubMed] [Google Scholar]
- Field CM, Alberts BM. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J Cell Biol. 1995;131:165–178. doi: 10.1083/jcb.131.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- Fishkind DJ, Wang Y-l. New horizons for cytokinesis. Curr Opin Cell Biol. 1995;7:23–31. doi: 10.1016/0955-0674(95)80041-7. [DOI] [PubMed] [Google Scholar]
- Glover DM, Ohkura H, Tavares A. Polo kinase: the choreographer of the mitotic stage? . J Cell Biol. 1996;135:1681–1684. doi: 10.1083/jcb.135.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golsteyn RM, Mundt KE, Fry AM, Nigg EA. Cell cycle regulation of the activity and subcellular localization of PLK1, a human protein kinase implicated in mitotic spindle function. J Cell Biol. 1995;129:1617–1628. doi: 10.1083/jcb.129.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KL, Simanis V. The control of septum formation in fission yeast. Genes Dev. 1997;11:2939–2951. doi: 10.1101/gad.11.22.2939. [DOI] [PubMed] [Google Scholar]
- Gundersen GG, Kalnoski MH, Bulinski JC. Distinct populations of microtubules: tyrosinated and nontyrosinated alpha tubulin are distributed differently in vivo. . Cell. 1984;38:779–789. doi: 10.1016/0092-8674(84)90273-3. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. Molecular genetics of asymmetric cleavage in the early Caenorhabditis elegansembryo. Curr Opin Genet Dev. 1996;6:408–415. doi: 10.1016/s0959-437x(96)80061-x. [DOI] [PubMed] [Google Scholar]
- Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Novel potential mitotic motor protein encoded by the fission yeast cut7 +gene. Nature. 1990;347:563–566. doi: 10.1038/347563a0. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene sad1 +associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Evidence for cell cycle-specific, spindle pole body-mediated, nuclear positioning in the fission yeast Schizosaccharomyces pombe. . J Cell Sci. 1997;110:1851–1866. doi: 10.1242/jcs.110.16.1851. [DOI] [PubMed] [Google Scholar]
- Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. . Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Jochová J, Rupes I, Streiblová E. F-actin contractile rings in protoplasts of the yeast Schizosaccharomyces. . Cell Biol Int Rep. 1991;15:607–610. doi: 10.1016/0309-1651(91)90007-6. [DOI] [PubMed] [Google Scholar]
- Karpova TS, Lepetit MM, Cooper JA. Mutations that enhance the cap2 null mutant phenotype in Saccharomyces cerevisiaeaffect the actin cytoskeleton, morphogenesis and pattern of growth. Genetics. 1993;135:693–709. doi: 10.1093/genetics/135.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney JB, Boeke JD. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. . Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama C, Sugimoto A, Yamamoto M. Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. . J Cell Biol. 1997;137:1309–1319. doi: 10.1083/jcb.137.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli J, Hottinger H, Munz P, Strauss A, Thuriaux P. Genetic mapping in Schizosaccharomyces pombeby mitotic and meiotic analysis and induced haploidization. Genetics. 1977;87:471–489. doi: 10.1093/genetics/87.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. . Nature. 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- Lane HA, Nigg EA. Antibody microinjection reveals an essential role for human Polo-like kinase 1 (Plk1) in the functional maturation of mitotic chromosomes. J Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Yuan Y-LO, Kuriyama R, Erikson RL. Plk is an M-phase-specific protein kinase and interacts with a kinesin-like protein, CHO1/MKLP-1. Mol Cell Biol. 1995;15:7143–7151. doi: 10.1128/mcb.15.12.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leupold U. Genetical methods for Schizosaccharomyces pombe. . Methods Cell Physiol. 1970;4:169–177. [Google Scholar]
- Llamazares S, Moreira A, Tavares A, Girdham C, Spruce BA, Gonzalez C, Karess RE, Glover DM, Sunkel CE. polo encodes a protein kinase homolog required for mitosis in Drosophila. . Genes Dev. 1991;5:2153–2165. doi: 10.1101/gad.5.12a.2153. [DOI] [PubMed] [Google Scholar]
- Marks J, Hyams JS. Localization of F-actin through the cell division cycle of S. pombe. . Eur J Cell Biol. 1985;39:27–32. [Google Scholar]
- Masuda H, Sevic M, Cande WZ. In vitro microtubule-nucleating activity of spindle pole bodies in fission yeast Schizosaccharomyces pombe: cell cycle-dependent activation in Xenopuscell-free extracts. J Cell Biol. 1992;117:1055–1066. doi: 10.1083/jcb.117.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- McCollum D, Balasubramanian MK, Pelcher LE, Hemmingsen SM, Gould KL. Schizosaccharomyces pombe cdc4 + gene encodes a novel EF-hand protein essential for cytokinesis. J Cell Biol. 1995;130:651–660. doi: 10.1083/jcb.130.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet M, Nurse P, Thuriaux P, Mitchison JM. Uncontrolled septation in a cell division cycle mutant of the fission yeast Schizosaccharomyces pombe. . J Bacteriol. 1979;137:440–446. doi: 10.1128/jb.137.1.440-446.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison JM, Nurse P. Growth in cell length in the fission yeast Schizosaccharomyces pombe. . J Cell Sci. 1985;75:357–376. doi: 10.1242/jcs.75.1.357. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. . Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Mundt KE, Golsteyn RM, Lane HA, Nigg EA. On the regulation and function of human polo-like kinase 1 (PLK1): effects of overexpression on cell cycle progression. Biochem Biophys Res Commun. 1997;239:377–385. doi: 10.1006/bbrc.1997.7378. [DOI] [PubMed] [Google Scholar]
- Oegema K, Mitchison TJ. Rappaport rules: cleavage furrow induction in animal cells. Proc Natl Acad Sci USA. 1997;94:4817–4820. doi: 10.1073/pnas.94.10.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H, Hagan IM, Glover DM. The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2cells. Genes Dev. 1995;9:1059–1073. doi: 10.1101/gad.9.9.1059. [DOI] [PubMed] [Google Scholar]
- Prentice HL. High efficiency transformation of Schizosaccharomyces pombeby electroporation. Nucleic Acids Res. 1992;20:621. doi: 10.1093/nar/20.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport R. Establishment of the mechanism of cytokinesis in animal cells. Int Rev Cytol. 1986;105:245–281. doi: 10.1016/s0074-7696(08)61065-7. [DOI] [PubMed] [Google Scholar]
- Rappaport, R. 1996. Cytokinesis in Animal Cells. Cambridge University Press, New York. 386 pp.
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Satterwhite LL, Pollard TD. Cytokinesis. Curr Opin Cell Biol. 1992;4:43–52. doi: 10.1016/0955-0674(92)90057-j. [DOI] [PubMed] [Google Scholar]
- Simon M-N, De Virgilio C, Souza B, Pringle JR, Abo A, Reed SI. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature. 1995;376:702–705. doi: 10.1038/376702a0. [DOI] [PubMed] [Google Scholar]
- Smith MR, Wilson ML, Hamanaka R, Chase D, Kung H, Longo DL, Ferris DK. Malignant transformation of mammalian cells initiated by constitutive expression of the Polo-like kinase. Biochem Biophys Res Commun. 1997;234:397–405. doi: 10.1006/bbrc.1997.6633. [DOI] [PubMed] [Google Scholar]
- Sohrmann M, Fankhauser C, Brodbeck C, Simanis V. The dmf1/ mid1gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 1996;10:2707–2719. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- Stearns T. Motoring to the finish: kinesin and dynein work together to orient the yeast mitotic spindle. J Cell Biol. 1997;138:957–960. doi: 10.1083/jcb.138.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S. Determination of cleavage planes. Cell. 1993;72:3–6. doi: 10.1016/0092-8674(93)90041-n. [DOI] [PubMed] [Google Scholar]
- Sunkel CE, Glover DM. polo, a mitotic mutant of Drosophiladisplaying abnormal spindle poles. J Cell Sci. 1988;89:25–38. doi: 10.1242/jcs.89.1.25. [DOI] [PubMed] [Google Scholar]
- Thuriaux P, Sipiczki M, Fantes PA. Genetical analysis of a sterile mutant by protoplast fusion in the fission yeast Schizosaccharomyces pombe. . J Gen Microbiol. 1980;116:525–528. doi: 10.1099/00221287-116-2-525. [DOI] [PubMed] [Google Scholar]
- Wang Y-l. Digital deconvolution of fluorescence images for biologists. Methods Cell Biol. 1998;56:305–315. doi: 10.1016/s0091-679x(08)60432-x. [DOI] [PubMed] [Google Scholar]
- Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K. Definition of individual components within the cytoskeleton of Trypanosoma bruceiby a library of monoclonal antibodies. J Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Wright A, Maundrell K, Heyer W-D, Beach D, Nurse P. Vectors for the construction of gene banks and the integration of cloned genes in Schizosaccharomyces pombe and Saccharomyces cerevisiae. . Plasmid. 1986;15:156–158. doi: 10.1016/0147-619x(86)90051-x. [DOI] [PubMed] [Google Scholar]
- Zervos AS, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- Zwaal RR, Ahringer J, van Luenen HGAM, Rushforth A, Anderson P, Plasterk RHA. G proteins are required for spatial orientation of early cell cleavages in C. elegansembryos. Cell. 1996;86:619–629. doi: 10.1016/s0092-8674(00)80135-x. [DOI] [PubMed] [Google Scholar]