Abstract
The high mobility group 14/17 (HMG-14/ -17) proteins form specific complexes with nucleosome core particles and produce distinct footprints on nucleosomal DNA. Therefore, they could be an integral part of the chromatin fiber. Here we show that during the cell cycle these proteins are transiently dissociated from chromatin. They colocalize with the nuclear DNA in interphase and prophase but not in metaphase and anaphase. They relocate into the nucleus and colocalize again with the DNA in late telophase, concomitantly with the appearance of the nuclear envelope. Thus, these nucleosomal binding proteins are not always associated with chromatin. Using reconstituted nuclei and permeabilized cells, we demonstrate that these two small proteins, with a molecular mass <10 kD, are actively imported into the nucleus. We identify the major elements involved in the nuclear import of these chromosomal proteins: HMG-14/-17 proteins contain an intrinsic bipartite nuclear localization signal, and their entry into the nucleus through nuclear pores requires energy and the participation of importin α. These findings suggest that the cell cycle–related association of HMG-14/-17 with chromatin is dependent on, and perhaps regulated by, nuclear import processes.
Keywords: HMG proteins, nuclear transport, cell cycle, localization, nuclear proteins
Throughout the cell cycle the histones remain associated with the DNA and the chromatin maintains its nucleosomal organization (van Holde, 1988; Wolffe, 1995). In contrast, the pattern of association of nonhistone chromosomal proteins with the chromatin fiber during the cell cycle is not uniform and cannot be predicted. Studies on the high mobility group (HMG)1 proteins provide a good example of the variability in the pattern of nuclear localization of nonhistone proteins during the cell cycle. The HMG proteins are one of the most abundant and well characterized class of nonhistone nuclear proteins which seem to function as architectural elements in chromatin (reviewed in Bustin and Reeves, 1996). Members of this group are found in all of the cells of higher eukaryotes and share certain physical properties: they can be extracted from chromatin or nuclei with 0.35 M NaCl, they have a high content of charged amino acids, they are soluble in 5% perchloric or trichloroacetic acid, and they have a molecular mass <30 kD. Currently, the HMG proteins are grouped into three families: the HMG-1/-2 family, the HMG-I/Y/C family, and the HMG-14/-17 family. Each of these families seems to have a distinct type of function in the cell nucleus (reviewed in Bustin et al., 1990; Bustin and Reeves, 1996). Members of the most abundant class of HMGs, the HMG-1/-2 class, are transiently associated with chromatin, shuttle between the nucleus and cytoplasm, and are not found in mitotic chromatin (Bustin and Neihart, 1979; Isackson et al., 1980; Einck and Bustin, 1985; Falciola et al., 1997; Shirakawa et al., 1997). In contrast, the HMG-I/Y/C group seems to be associated with chromatin throughout the cell cycle and has been detected in metaphase chromosomes (Disney et al., 1989; Saitoh and Laemmli, 1994).
The cell cycle–related cellular location of the third class of HMGs, the HMG-14/-17 class, has not been studied. HMG-14/-17 proteins are the only nuclear proteins known to specifically bind to the 146-bp nucleosome core particle (reviewed in Bustin and Reeves, 1996). They associate with the nucleosome core in a highly specific way and form complexes containing either two molecules of HMG-14 or two molecules of HMG-17 (Postnikov et al., 1995). They generate a distinct footprint on the nucleosomal DNA, suggesting that they occupy specific sites on the nucleosome core, independent of the underlying DNA sequence (Alfonso et al., 1994). In view of these and additional findings (reviewed in Bustin and Reeves, 1996), the proteins could be considered to be an integral part of the chromatin fiber, and would be expected to remain associated with nucleosomes throughout the cell cycle. However, here we demonstrate that HMG-14/-17 proteins are transiently displaced from chromatin and are not detectable in metaphase chromosomes. They reenter the nucleus after the formation of the nuclear envelope.
Our finding that the HMG-14/-17 proteins enter the nucleus only after the reformation of the nuclear membrane suggests a link between their intranuclear location and cell cycle events. Although the HMG proteins are among the most abundant class of nonhistone proteins, little is known about their nuclear transport. A highly unusual, tetrapartite nuclear localization signal (NLS) for HMG-2 has been proposed recently (Shirakawa et al., 1997). The HMG-14/ -17 proteins are significantly smaller than either HMG-2 or any other proteins whose nuclear entry has been studied. Thus, it is not known whether they contain intrinsic NLSs, or whether they enter the nucleus associated with another component (Nigg, 1997) or perhaps even by passive diffusion (Harootunian et al., 1993; Jans et al., 1996; Chatterjee and Stochaj, 1998).
In this article we report that during the cell cycle these nucleosomal nonhistone proteins are released from chromatin, and that their nuclear reentry displays all the hallmarks of facilitated nuclear transport. The results may be pertinent to understanding the mechanism whereby HMG-14 and HMG-17 segregate to separate intranuclear foci and create clusters of nucleosomes complexed with either HMG-14 or HMG-17 (Postnikov et al., 1997).
Materials and Methods
Antibodies, Proteins, and Peptides
Affinity pure antibodies to histone H1 and HMGs were prepared as described (Bustin, 1989) and tested on numerous occasion by ELISA, Western, immunodiffusion, and radioimmunoassay (Bustin, 1989). Monoclonal antibodies to nucleoporin p62 were a gift from M.C. Dabauvalle (Dabauvalle et al., 1988). Recombinant proteins (Bustin et al., 1991), truncated HMG-14/-17 proteins (Trieschmann et al., 1995b ), and the S88C-HMG-14 recombinant mutant (Trieschmann et al., 1998) were prepared as described. The truncation mutants were detected with antibodies directed against the nucleosomal binding domain which is present in all mutants. Peptides were synthesized and HPLC-purified in the laboratory of Dr. Dieter Palm (Department of Physiological Chemistry I, Biocenter, Wurzburg, Germany): PKRKC (HMG-NLS1), PKRKTKGKRGAC (HMG- NLS1+2), AGRKGKTKRKPC (revHMG-NLS1+2), PKKKRKVEDC (SV-40-NLS), and DEVKRKKKPC (revSV-40-NLS).
Immunofluorescence on Cultured Cells
Mouse 3T3 cells and human Hep2 cells were cultured in DME (GIBCO-BRL, Eggenstein, Germany) supplemented with 10% FCS (GIBCO-BRL) at 37°C in a 5% CO2 incubator. Cells grown on coverslips were rinsed in PBS and fixed with 2% formaldehyde in PBS for 10 min at room temperature. After fixation, the coverslips were incubated as follows: washed for 2 × 3 min in PBS, incubated for 10 min in PBS containing 0.1% Triton X-100, rinsed in PBS; washed for 2 × 5 min in PBS, treated with primary antibody (1–2 μg/ml for anti–HMG-14 or anti–HMG-17; 4.5 μg/μl for antibody to peptide 2) for 30–40 min at room temperature in a moist chamber, rinsed in PBS, and washed for 2 × 5 min in PBS. Bound primary antibodies were detected with Tx-red conjugated to goat anti– rabbit, or with FITC-conjugated goat anti–mouse (Dianova) incubated for 30 min at room temperature, and counterstained with 1 μg/ml Hoechst 33258 for 5 min, rinsed in PBS, washed for 2 × 5 min in PBS, and mounted in Mowiol. For double-immunolabeling experiments, both first antibodies were incubated at the same time for 45 min.
Xenopus Egg Extracts and Nuclear Reconstitution
Low speed extract for nuclear reconstitution (Newport, 1987) was prepared by crushing the eggs in egg buffer (EB: 100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, 1 mM DTT, 50 mM sucrose, 5 μg/ml cytochalasin B [Sigma Chemical Co., St. Louis, MO], 10 mM K-Hepes, pH 7.7) and centrifugation at 12,000 g for 10 min at 4°C. The crude interphase extract was clarified by four additional centrifugations at 12,000 g. Aliquots were then frozen in liquid nitrogen and stored up to 1 mo at −80°C. For high speed extract preparation (used as cytosol for import assay with permeabilized cells) the crude extract was centrifuged at 150,000 g for 2 h in an ultracentrifuge (Beckman Instruments, Inc., Fullerton, CA). Formation of nuclei was initiated by adding demembranated Xenopus sperm nuclei (500/μl extract) prepared according to Blow and Laskey (1986) together with an ATP regenerating system (Newport, 1987) and cycloheximide (100 μg/ ml). To facilitate nuclear transport, GTP (0.5 mM final concentration) was added. The mixture was incubated at 22°C. The time required for the nuclear formation was tested, for each batch of extract, by staining of 2 μl of the incubation mixture with Hoechst 33258 and by the presence of prenucleolar bodies (Bell et al., 1997) and an envelope as seen in phase optics.
Isolation of Reconstituted Nuclei and Import Assays with Reconstituted Nuclei
To isolate the reconstituted nuclei, the reaction mixture was diluted with 4 vol of EB, laid over a 1 M sucrose cushion, and centrifuged at 1,000 g, for 30 min, at 4°C. The pelleted nuclei were washed twice in EB and resuspended in a desired solution or extract. Recombinant HMG proteins were added (1 μg/50-μl extract) either directly to the extract after nuclear formation, or to isolated nuclei. The isolated nuclei (prepared as described above) were resuspended in an egg extract diluted with EB to a final protein concentration of 10 μg/ml. The diluted extract was preincubated for 30 min, at 22°C, with an ATP regenerating system and GTP and only then added to the nuclei. Then, the import assay was performed for 15 min or as indicated in the figures. For immunofluorescence, the in vitro assembled nuclei were fixed by adding 4 vol of 2% formaldehyde in PBS, then spun onto microscope slides and processed as described (Dabauvalle et al., 1991). After immunofluorescence, the slides were washed in PBS, the second antibodies were added as described, and the slides were mounted in Vectashield mounting medium containing Dapi (Vector Laboratories, Inc., Burlingame, CA). For WGA inhibition studies, the isolated reconstituted nuclei were resuspended in preincubated extract (see above) containing 50 μg/ml WGA coupled to digoxigenin and incubated for 20 min before the addition of HMG proteins. Then, nuclei were fixed and used for immunofluorescence as described above.
Competition Experiments Using Reconstituted Nuclei
Transport of HMG-17 and HMG-14 into the nucleus was inhibited with a HPLC-purified SV-40 T-antigen NLS homologue (PKKKRKVED-C). Peptides were added to the extract containing the reconstituted nuclei before the addition of HMG proteins. After 3–5 min, the HMG proteins were added and the mixture was incubated for a further 10 min. Nuclei were then isolated by diluting the mixture with 4 vol EB and pelleting them at 3,000 g. Nuclei were washed once with EB and used for SDS-PAGE and Western blot analysis.
SDS-PAGE and Western Blotting
Proteins were separated using polyacrylamide minigels and transferred onto a PVDF membrane (Immobilon; Millipore Corp., Bedford, MA) by tank blotting (45 min, 50 V at 4°C). After transfer, the membranes were blocked with 5% nonfat dry milk in TBS (10 mM Tris/Cl, pH 7.4, 140 mM NaCl). HMG proteins were detected using antibodies recognizing the nucleosomal binding region of both HMG-14 and HMG-17, at a concentration of 0.1 μg/ml. First antibodies were incubated for 1 h at room temperature. After 3 × 15 min washes in TBS, bound primary antibodies were detected with an anti–rabbit IgG coupled to peroxidase (Boehringer Mannheim, Mannheim, Germany; 0.2 U/ml, 1 h at room temperature) by enhanced chemical luminescence (Pierce, Rockford, IL).
Preparation of Fluorescent Conjugates and Fluorescein Conjugation
The phycobiliprotein allophycocyanin (APC; Calbiochem, La Jolla, CA) was activated with a 100-fold molar excess of sulfo-SMCC (Sigma) at 1 h at room temperature (Adam et al., 1990). Excess cross-linker was removed by centrifugation in a spin column (30-kD cutoff; MembraPure). The SMCC-APC was washed several times with 50 mM Hepes (pH 7.0) by spin column centrifugation and the concentration finally was readjusted. Peptides and the S88C-HMG-14 mutant (Trieschmann et al., 1998) were solubilized in 50 mM Hepes, pH 7.0, and incubated at a 50-fold molar excess with the SMCC-activated APC, at 4°C, overnight. Nonbound mutant protein and peptides were removed by centrifugation in a spin column with a 30-kD cutoff. Approximately 15–25 peptides or S88C-HMG-14 were conjugated to each APC molecule as estimated by mobility shift on SDS-polyacrylamide gels. For fluorescein conjugation (Schmidt and Krohne, 1995) of the S88C-HMG-14, at the single, mutated -SH, a 10–20-fold excess of 5-iodoacetamido-fluorescein (5-IAF; Molecular Probes, Eugene, OR) was added to 1 mmol protein solubilized in 10 mM Hepes, pH 6.9, and the mixture was incubated overnight at 22°C. Nonbound 5-IAF was removed with a spin-column with a 10-kD cutoff. Protein labeling efficiency was determined by fractionating the modified protein on SDS-PAGE and UV illumination of nonstained gels.
Import Assay with Permeabilized Cells
Cell permeabilization of Hep2 cells and the import assay was done as described by Adam et al. (1990). The transport mixture contained high speed Xenopus egg extract diluted in import buffer to give a final concentration of 10 mg/ml protein, 20 mM Hepes, pH 7.3, 110 mM potassium acetate, 2 mM DTT, 1.0 mM EGTA, 2 mM ATP, 20 mM creatine phosphate, 100 μg/ml creatine phosphokinase (Sigma), 0.5 mM GTP, and 100 μg/ml import substrate. For ATP depletion, the extract was preincubated for 20 min with 50 U/ml apyrase (Sigma). For WGA inhibition, permeabilized cells were preincubated with extract containing 50 μg/ml WGA (Sigma). For competition experiments a 100-fold molar excess of peptide over import substrate was added to the mixture. The permeabilized cells were incubated with 25 μl of transport mixture for 1 h at 22°C. Then, the coverslips were blotted, fixed with 2% formaldehyde, and mounted in Mowiol. The preparations were analyzed immediately using a Leica confocal laser scanning microscope with the help of the TCS-NT software.
Results
Displacement of HMG-14/-17 from Mitotic Chromosomes
To examine the cellular distribution of HMG-14 and HMG-17 throughout the cell cycle, we used affinity pure antibodies to visualize the location of the proteins in cultured human Hep2 and mouse 3T3 cells. The immunofluorescent patterns obtained with antibodies to HMG-17 (Fig. 1, A and C) are the same as those obtained with anti–HMG-14 (not shown) and are observed in both Hep2 (Fig. 1) and mouse 3T3 (not shown) cells. In interphase cells, HMG-14/-17 proteins are present in the nucleus and appear to be concentrated in multiple dots which are scattered throughout the nucleus, with the exception of the nucleolus. During prophase, the dotted pattern is less distinct, the staining seems more diffuse, and the intensity of the immunofluorescent signal diminishes. In metaphase, HMG-17 proteins are found in the cytoplasm and are not associated with the chromosomes. In anaphase, the fluorescent signal obtained with the antibodies remains clearly distinct from the DNA signal obtained with Hoechst. The protein is dispersed throughout the cytoplasm and does not colocalize with the DNA, suggesting that it is not associated with chromatin. In late telophase the immunofluorescent and the Hoechst signal colocalize, suggesting that the protein reentered the nucleus. The failure of the HMG-17 antibodies to stain mitotic chromosomes is not due to the inaccessibility of the antigenic sites in chromosomes to antibody binding since our controls with histone H1 antibodies (Fig. 1 B) and many previous experiments (reviewed in Bustin, 1989; Martinez-Balbas et al., 1995; Falciola et al., 1997) demonstrated that these chromosomes stain intensely with antibody to histones. Thus, we conclude that during mitosis HMG-14/-17 proteins are not associated with chromatin.
Figure 1.

HMG proteins are absent from mitotic chromosomes. (A) Immunofluorescence analysis of the distribution of HMG-17 in cultured Hep2 cells with affinity pure antibodies to HMG-17 is shown in the upper panels. The lower panels show staining for DNA with Hoechst. Cell cycle stages are indicated below the panels. Staining for HMG proteins shows a punctuate staining in interphase. In metaphase or anaphase (arrow, early; arrowhead, late) the protein is present in the cytoplasm and not associated with chromatin. Nuclear staining reappears in the late telophase cells. (B) Control with antibodies to histone H1 showing antibody binding in metaphase (a) and corresponding Hoechst (a′). Arrows point to a cell in metaphase. (C) Confocal microscopy depicting immunofluorescence in human Hep2 cells stained with antibodies to HMG-17.
The results suggest that HMG-14/-17 proteins are displaced from the chromatin and then are rapidly reintroduced into postmitotic nuclei. Double immunofluorescence, with antibodies to HMG-17 and monoclonal antibodies to nucleoporin p62, reveals a correlation between the presence of the nuclear pore complexes (NPC) and the nuclear localization of HMG-14/-17 protein (Fig. 2). In metaphase and anaphase, when the nuclear envelope is dissociated, the cells do not stain with either antibodies to nucleoporin p62 or with antibodies to HMG-17 (Fig. 2, a and a′). In late telophase, the nuclear envelope is reassembled with NPC and produces an intense signal with antibodies to nucleoporin p62 (Fig. 2 b′). At this stage the HMG-17 protein concentrates in the newly formed nuclei and the intense anti HMG-17 immunofluorescent signal colocalizes with the DNA (Fig. 2, b and b′′). The results indicate that the proteins accumulate in the nuclei after the end of mitosis and only after the formation of the nuclear membrane.
Figure 2.
Nuclear staining of HMG-17 protein correlates with the presence of the nuclear envelope. Human Hep2 cells were double labeled with affinity pure anti–HMG-17 (a and b) and with the monoclonal antibody PI1, specific for nucleoporin p62 (a′ and b′), and counterstained for DNA with Hoechst (a′′ and b′′). Bound anti–HMG-17 and PI1 were detected with an Tx-red anti– rabbit and a FITC-coupled anti–mouse, respectively. The arrows in a, a′, and a′′ point to a cell in mitosis. The arrows in b, b′, and b′′ point to cells in late telophase. Bar represents 10 μm.
Nuclear Accumulation of HMG-14/-17 In Vitro
Since nuclei of higher eukaryotes contain HMG-14/-17, we initially studied the nuclear transport of these proteins with in vitro reconstituted nuclei which we formed by adding demembranated Xenopus sperm to Xenopus egg extracts (Newport, 1987). These nuclei are fully functional in nuclear protein import (Newmeyer et al., 1986). By Western analysis we demonstrated that the egg extracts do not contain HMG-14/-17 proteins (Crippa et al., 1993), therefore it is possible to follow the distribution of exogenously added recombinant HMG proteins, by immunofluorescence microscopy.
Within 60–90 min after the addition of Xenopus chromatin to the extract, nuclei were reconstituted as determined by Hoechst staining and phase-contrast microscopy (Fig. 3 A). Recombinant HMG-14 or HMG-17 were added to these reconstituted nuclei, and after 15 min of incubation the nuclei were fixed and analyzed by immunofluorescence. In parallel, another aliquot of the nuclei was isolated by centrifugation through a sucrose cushion, and the protein content of the nuclei was examined by electrophoresis in SDS-containing polyacrylamide gels. Both assays revealed that the HMG proteins accumulated in the reconstituted nuclei. By immunofluorescence, the nuclei were positive only when recombinant protein was added to the extract (Fig. 3 A). The presence of HMG-17 in these nuclei was verified by SDS-polyacrylamide gel analysis and subsequent staining with Commassie blue (Fig. 3 B), and by Western analysis (for example see Fig. 5).
Figure 3.
Recombinant HMG-14/-17 proteins accumulate in reconstituted nuclei. (A) Immunofluorescence analysis of reconstituted nuclei after addition of recombinant HMG-17 (a) or HMG-14 (b). Corresponding DNA stain with Hoechst and phase are shown in a′ and b′, and a′′ and b′′, respectively. Bar represents 10 μm. (B) Coomassie stained SDS-gel depicting proteins isolated from purified, reconstituted nuclei (105 nuclei/lane) which were either incubated with (lanes 1 and 2) or without (lane 3) recombinant HMG-17 (arrow). As a marker, 200 ng recombinant HMG-17 protein was loaded in lane 4. The nuclei were incubated in the presence of HMG-17 for either 30 min (lane 1) or 15 min (lane 2).
Figure 5.
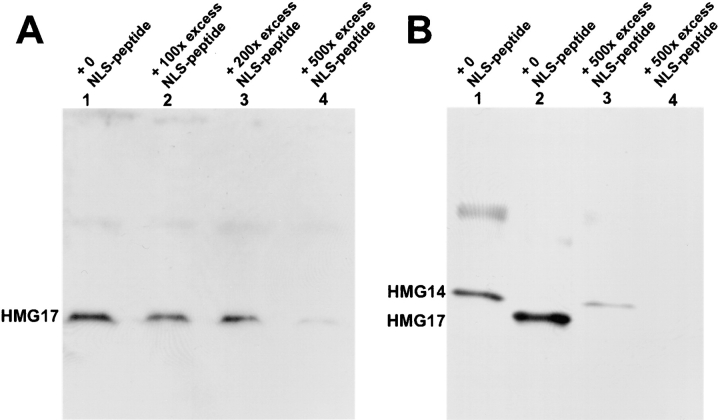
Nuclear transport of HMG-17 and HMG-14 can be competed with the SV-40 T-antigen NLS. Isolated reconstituted nuclei were resuspended in extracts containing various amounts of the NLS peptide (indicated on the top of the lanes as molar excess of peptide over protein) for 3–5 min before the addition of HMG-14/-17 protein. After a 10-min incubation with HMG-14/-17, the nuclei were isolated and the amount of HMG-14/-17 in the nuclei was analyzed by Western blotting. 30,000 nuclei were loaded per lane. Increasing amounts of NLS peptide lead to decreased nuclear HMG-14/-17. (A) Dose-dependent inhibition of HMG-17 transport by the NLS peptide. (B) The transport of HMG-14 (lanes 1 and 3) or HMG-17 (lanes 2 and 4) in the presence of the indicated amounts of NLS peptide.
To test whether the accumulation of HMG-14/-17 into the reconstituted nuclei involved active transport, we isolated the nuclei by centrifugation through a sucrose cushion and resuspended them either in the Xenopus egg extract or in the buffer used to prepare the extract. The nuclei resuspended in the Xenopus egg extract accumulated exogenously added recombinant HMG-17 or HMG-14, whereas the nuclei resuspended in the control buffer did not (not shown). Therefore, we conclude that the accumulation of HMG-14/-17 into the nuclei depends on the activity of factors present in the extract and is not diffusion driven.
Identification of the HMG-14/-17 Nuclear Translocation Signal
We used NH2-terminal and COOH-terminal truncation mutants of HMG-17 protein to search for putative NLS in this protein. In these experiments, the protein was localized with an antibody elicited against the conserved nucleosome binding domain of HMG-17 (domain B in the diagrams, Fig. 4). Although the nuclei were not fully rounded, they contain a functional nuclear membrane as demonstrated before (Bell et al., 1997), and as evident from our experiments, indicating that HMG-14/-17 import requires extract, is dependent on a NLS, and is inhibited by WGA (see below). Truncation mutants lacking either the first 16, or even only the first 4, NH2-terminal amino acids did not accumulate into the nuclei, suggesting that the NH2-terminal region of the protein is necessary for nuclear transport (Fig. 4, b and c). In contrast, a deletion mutant lacking the 22 COOH-terminal amino acids did accumulate in the reconstituted nuclei (Fig. 4 d). However, an even smaller truncation mutant, lacking the 37 COOH-terminal amino acids, but containing an intact NH2-terminal, was not transported into the nuclei (Fig. 4 e).
Figure 4.
Identification of the HMG-17 NLS. a–e show staining with antibodies directed against the nucleosomal binding region (domain B) which is present in all the truncation mutants diagrammed on the right. a′–e′ show the corresponding Hoechst images. Bar is 10 μm. The following mutants are used: ΔN4, lacking the first four NH2-terminal amino acids; ΔN16, lacking the first 16 NH2-terminal amino acids; ΔC37, lacking the last 37 COOH-terminal amino acids; ΔC22, lacking the last 22 COOH-terminal amino acids.
Therefore, we conclude that the first four NH2-terminal amino acids (PKRK), which are absolutely conserved in every member of the HMG-14/-17 protein family (Bustin and Reeves, 1996), are necessary but not sufficient for nuclear localization. Likewise, in the COOH-terminal region of the protein there is an additional region which is necessary, but not sufficient, for nuclear translocation. In the COOH-terminal part of the HMG-14/-17 we have identified previously a pentapeptide with the sequence KGK (K/R)G which is highly conserved among all the HMG-14/ -17 proteins (Bustin and Reeves, 1996). Both regions, the NH2-terminal four amino acids and the conserved pentapeptide, contain basic amino acids which are characteristic of elements in the NLS (for reviews see Gorlich, 1997; Nigg, 1997). Therefore, we conclude that the HMG-14/-17 protein family has an intrinsic, bipartite NLS. The presence of a bipartite NLS in HMG-14/-17 proteins was further verified in experiments with permeabilized Hep2 cells (see below).
Importin α Is Involved in the Nuclear Transport of HMG-14/-17
Most proteins containing an intrinsic mono- or bipartite NLS bind to the nuclear pore by a protein complex containing the cytoplasmic receptor importin α; however, additional cytoplasmic receptors (Rout et al., 1997) have been identified recently (for reviews see Gorlich, 1997; Nigg, 1997; Weis, 1998). To identify the main cytoplasmic receptor involved in the nuclear import of HMG-14/-17 proteins, we tested whether a peptide homologous to the SV-40 T-antigen NLS, which is imported into the nuclei via the importin α pathway, inhibits the accumulation of the HMGs into the reconstituted nuclei. The peptide and either HMG-14 or HMG-17 protein were added to reconstituted nuclei and after 15 min of incubation the nuclei were isolated by centrifugation through a sucrose cushion. Western analysis of the protein content in these nuclei indicated that the NLS peptide inhibited the nuclear accumulation of both the HMG-17 (Fig. 5 A) and HMG-14 (Fig. 5 B) whereas a positively charged control peptide (RAKPAKLPKAAPSPKADKERSRPKPQPKEP), did not (not shown). In the presence of a 500-fold molar excess of NLS peptide, the nuclear content of HMG-17 and HMG-14 was reduced by >95 and 70%, respectively. Nuclear import of HMG-14/-17 could be competed also by the recombinant chicken thyroid receptor protein which contains a bipartite NLS. In addition, by affinity chromatography of Xenopus egg extracts, we found that whereas HMG-17 affinity columns bound importin α, BSA affinity columns did not (not shown). These results suggest that HMG-14/ -17 proteins are transported into the nucleus by a complex containing importin α, the major cytoplasmic receptor of most karyophilic proteins studied. In addition, the results indicate that the transport of HMG-14 is mediated by the same cytoplasmic factors as HMG-17. Indeed, excess of HMG-14 protein inhibited the import of HMG-17 protein (not shown).
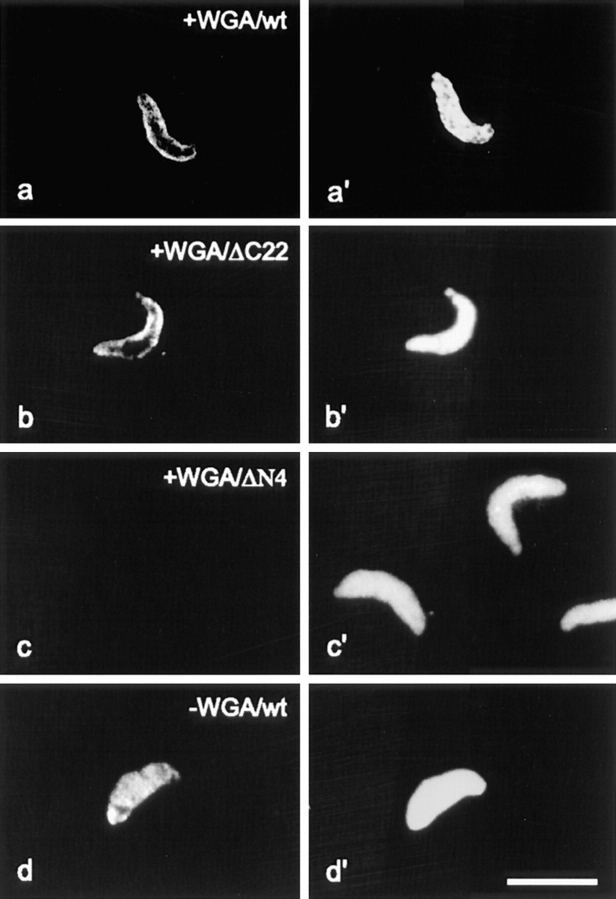
In an additional test, we examined whether the nuclear accumulation of these proteins could be blocked by WGA. WGA binds to GlucNAc residues in the nuclear pore complex and blocks the facilitated passage of proteins through the pores (Michaud and Goldfarb, 1993). In these experiments, we first reconstituted the nuclei and then added WGA, before the addition of various recombinant HMG-17 proteins. In the presence of WGA, the immunofluorescence signal outlines the periphery of the reconstituted nuclei. Thus, in the presence of WGA, the intact HMG-17 (Fig. 6 a) or the mutant lacking the 22 COOH-terminal amino acids (Fig. 6 b) is docked onto the nuclear membrane but cannot enter the nucleus. In contrast, a mutant lacking the first four amino acids, i.e., one of the NLS elements necessary for transport to the nuclei, did not accumulate at the periphery of the nuclei (Fig. 6 c). In the absence of WGA, the protein is transported through the nuclear pores and accumulates inside the nucleus (Fig. 6 d). We conclude that HMG-17 bind to the nuclear membrane and enter the nucleus through nuclear pores through a facilitated process.
Figure 6.
The NH2-terminal, first element of HMG-NLS, is essential for docking of HMGs to the nuclear periphery. Wild-type (wt) recombinant HMG-17 or truncated versions of HMG-17 were added to Xenopus extract preincubated with WGA (see Materials and Methods). The localization of the added proteins was visualized by immunofluorescence (a–d). In a′–d′ corresponding DNA staining is shown. In the presence of WGA, wtHMG-17 and a truncation mutant containing both elements of the NLS dock to the pores as visualized by the ring-like staining around the nucleus (a and b). In contrast, a truncation mutant lacking the first four amino acids (i.e., the fist NLS element) fails to bind to the nuclear pores (c). In the absence of WGA, HMG17 enters the nucleus (d). Bar represents 10 μm.
Nuclear Import in Permeabilized Tissue Culture Cells
We next tested whether the cell cycle–related reentry of HMG-14/-17 into the nucleus of tissue culture cells (Figs. 1 and 2) is indeed dependent on active import. In these experiments we followed the entry of various modified proteins into digitonin-permeabilized (Adam et al., 1990) Hep2 cells. We used either a mutant HMG-14 fluorescently labeled with 5-IAF, or various conjugates of the autofluorescent protein APC. When injected into the cytoplasm of live Hep2 cells, the 5-IAF–modified HMG-14 rapidly accumulated in the nucleus (not shown). Thus, the modification does not interfere with the entry of HMG-14 into the nucleus.
The results clearly showed that in permeabilized cells nuclear accumulation of modified HMG-14 protein depends on the addition of Xenopus egg extracts (Fig. 7, a and b). Extracts treated with apyrase (Fig. 7 c) or containing WGA (Fig. 7 d) did not facilitate the entry of the fluorescent HMG-14 into the nucleus. The nuclear import of HMG-14 was prevented with the SV-40 nuclear localization sequence (Fig. 7 f), but not with the reverse SV-40 NLS, which is known to be nonfunctional (Adam et al., 1989; Gorlich et al., 1994). Similar experiments demonstrate that both elements of the bipartite HMG-14/-17 NLS are necessary for nuclear import (Fig. 8). Thus, while the APC conjugated with the intact HMG-14 accumulates in the nucleus (Fig. 8 a), the protein modified with only the first NLS element (PKRK) did not (Fig. 8 b). When APC was modified with a peptide whose sequence consisted of both HMG-14 NLS elements, the protein accumulated in the nucleus (Fig. 8 d). In contrast, when APC was modified with the same peptide but in a reverse sequence it did not accumulate in the nucleus (Fig. 8 c). Control experiments verified that APC by itself does not contain an intrinsic NLS (Adam et al., 1989). The protein accumulates in the nucleus when modified by the SV-40 NLS in the correct sequence (Fig. 8 f) but not when modified with the reverse SV-40 NLS (Fig. 8 e).
Figure 7.
Active nuclear import of fluorescein-labeled HMG-14 in permeabilized cells. Nuclear import of the protein is facilitated by incubation with egg extract (b) but not by buffer (a). The nuclear import is energy dependent (c) and is inhibited by WGA (d) and by a peptide corresponding to the SV-40-NLS (f). It is not inhibited by the reverse SV-40-NLS (e). The pictures were taken on a Leica confocal laser scanning microscope. Bar represents 10 μm.
Figure 8.
Both elements of HMG-14 are required for nuclear import. The import of various APC constructs (see Materials and Methods) in permeabilized cells was analyzed by fluorescence microscopy. APC conjugated with the entire HMG-14 (a), with a peptide containing both elements of the HMG NLS (b), or with the SV-40 NLS (f), enters the nucleus. In contrast, APC conjugated with only the first HMG-14 NLS element (b), with a peptide whose sequence is the reverse of the two HMG-14 NLS elements (c), or with the nonfunctional, reverse SV-40 NLS, does not enter the nucleus of the permeabilized cells. Bars represent 10 μm.
Therefore, we conclude that both in tissue culture cells and in reconstituted nuclei, the small molecular weight HMG-14/-17 proteins are actively transported into nucleus, through nuclear pores by a complex containing importin α.
Discussion
In this article we report that HMG-14 and HMG-17, the only nonhistone proteins known to bind specifically to the 146-bp nucleosome core particle, are absent from mitotic chromatin and reenter the nucleus only after the reformation of the nuclear envelope. Thus, these proteins are not always associated with nucleosomes; their entry into the nucleus correlates with cell cycle events. We find that these small basic nuclear proteins, with a molecular mass <10 kD, are actively transported into nuclei, and identify the four major elements involved in this process: the intrinsic NLS of the HMG-14/-17 family of proteins, a cytoplasmic receptor, an energy-dependent step necessary for their entry into the nucleus, and the NPC as their gateway into the nucleus. These findings suggest that the cell cycle–related interactions of HMG-14/-17 with chromatin may be dependent on, and perhaps regulated by, nuclear import processes.
Displacement of HMG-14/-17 Proteins from Mitotic Chromatin
Various experiments indicate that chromosomal proteins HMG-14/-17 bind to nucleosome cores in a precise way (Alfonso et al., 1994; Trieschmann et al., 1998) to form complexes containing either two molecules of HMG-14 or two molecules of HMG-17 (Postnikov et al., 1995). These, and numerous other results on the organization of these proteins in the nucleus (reviewed in Bustin and Reeves, 1996), suggest that HMG-14/-17 may be an integral part of the chromatin fiber and would remain associated with chromatin throughout the cell cycle. Thus, our present results indicating that these HMGs are not present in mitotic chromosomes were unexpected and suggest that the behavior of these proteins is similar to that of certain transcription factors (Martinez-Balbas et al., 1995; Segil et al., 1996) and HMG-1 (Isackson et al., 1980; Falciola et al., 1997) which are also displaced from mitotic chromatin.
The correlation between loss of transcription factors and the condensation of chromatin during mitosis suggests a linkage between the two events (Martinez-Balbas et al., 1995). A similar linkage may exists between the loss of HMG-14/-17 and the state of chromatin condensation, especially in light of the recent findings that these two proteins unfold the higher order chromatin fiber (Trieschmann et al., 1995a ; Ding et al., 1997). Thus, displacement of HMG-14/-17 from chromatin would be compatible with a folded structure, whereas their reentry into the nucleus would promote unfolding which in turn may increase the accessibility of the underlying DNA sequence to transcription or other regulatory factors. An interesting implication of these findings is that the temporary displacement of the HMGs from chromosomes may be used for resetting the higher order chromatin structure in anticipation of transcriptional events occurring during the G1 phase. An alternative possibility is that the proteins accumulate in G1 in anticipation of incorporation into transcribing, or early replicating, chromatin (Bustin et al., 1995).
It has been suggested that HMG-14/-17 modulate the chromatin structure of transcriptionally active genes and confer DNase I sensitivity to active chromatin (Weisbrod and Weintraub, 1979). In some cases the DNase I sensitivity is also observed in mitotic chromosomes (Kerem et al., 1984). An attractive hypothesis was that the HMG-14/-17 remain associated with mitotic chromatin and mark the chromatin regions destined for transcription in the daughter cells. It is still possible that a small fraction of the HMG-14/-17 proteins undetectable by our techniques remains in chromatin and marks certain loci; however, our results clearly demonstrate that the bulk of the proteins leave the mitotic chromatin. Thus, these HMG proteins are not involved in maintaining the DNase I sensitivity of active genes throughout the cell cycle.
Our results also suggest that the cell contains an active mechanism for the displacement of the proteins from chromatin and their reentry into nuclei. The molecular mechanism involved in their displacement from chromosomes may be similar to those used for displacement of transcription factors, which have been discussed in detail elsewhere (Martinez-Balbas et al., 1995). Here we demonstrate that their cell cycle–related reentry into the nucleus is facilitated by active transport through the NPC.
Facilitated Nuclear Import of HMG-14/-17 Proteins
The accumulation of HMG-14/-17 into reconstituted nuclei displays all the hallmarks of facilitated nuclear import. The process requires distinct NLSs, occurs through NPC in an energy-dependent step, and involves cytoplasmic carriers.
Our experiments clearly demonstrate that the HMG-14/ -17 proteins contain an intrinsic, bipartite NLS. The first element of the signal, PKRK, constitutes the first four NH2-terminal amino acids and is absolutely conserved in every member of the HMG-14/-17 protein family, from fish to humans. The second element differs slightly between the HMG-14 and the HMG-17 proteins. In all HMG-17 proteins the sequence of this element is KGKKG, whereas in the HMG-14 proteins the fourth amino acid is R rather then K. The high concentration of basic residues in the NLS is similar to that found in other bi-, or even tetrapartite, NLSs. The two elements of the signal are separated by a stretch of ∼40 amino acids. The separation between the two NLS elements is bigger than usually found in bipartite NLS. However, the peptide sequence intervening between the two elements of the NLS does not contain information that is crucial for the nuclear import function, since a peptide corresponding to only the fused sequence of the two elements still functions as an NLS (Fig. 8 d). Each of the two elements is necessary, but not sufficient, for nuclear import since truncation mutants lacking either element are not accumulated in the nucleus (Fig. 4). Likewise, when only one NLS element was attached to APC, the protein did not enter the nucleus, whereas when a peptide containing both elements was attached the protein did enter the nucleus (Fig. 8). We have also found that phycoerythrin conjugated with a peptide containing the first 10 NH2-terminal amino acids of HMG-17 did not accumulate into nuclei whereas controls in which the protein was conjugated with the NLS of SV-40 T-antigen did (Bustin, M., and J. Hanover, unpublished observations).
The bipartite NLS is similar to that found in several nuclear proteins (Gorlich and Mattaj, 1996; Nigg, 1997). This signal is different from tetrapartite NLS found in HMG-2 protein in which the various elements are separated by long stretches of amino acids. Presumably, the tertiary structure of the HMG-2 protein juxtaposes the various elements of the NLS into a conformation which can be recognized by the cytoplasmic carrier. Thus, the nuclear transport of HMG-2 may be determined by the higher order structure rather than the primary sequence of the protein (Shirakawa et al., 1997). A similar situation could exist in HMG-14/-17 whose nuclear localization elements are separated by a stretch of ∼40 amino acids, which is somewhat longer than that generally found in proteins with bipartite NLS. Indeed, a peptide in which the two HMG-17 NLS elements were juxtaposed enabled the nuclear entry of APC.
The NLS of the HMG-14/-17 proteins is recognized by a cytoplasmic component present in the egg extract. The accumulation is not diffusion driven since nuclei suspended in buffer, rather than egg extract, did not accumulate HMG proteins. The cytoplasmic factors facilitating the nuclear import of HMG-14/-17 seem to be those commonly used by other karyophilic proteins since the T-antigen NLS and proteins carrying a bipartite NLS (not shown) fully inhibited the nuclear import of these HMGs. Additional evidence that the nuclear import of HMG-14/-17 proteins proceeds by the same pathway as that of most karyophilic proteins is indicated by our finding that WGA, which binds to N-acetyl glucosamine residues of pore complex proteins, inhibits their accumulation in the nucleus (Gorlich, 1997; Nigg, 1997; Weis, 1998).
In summary, all the data are consistent with the notion that chromosomal proteins HMG-14/-17 are imported into the nuclei by a facilitated process and that the key components of this process are the same as those generally used for nuclear import of karyophilic proteins. Most probably these findings apply to nonpermeabilized tissue culture cells since we observed that the microinjected 5-IAF– labeled HMG-14 rapidly enters into the nucleus, and that the HMG-14/-17 proteins reenter the nucleus in late telophase, only after the formation of the nuclear envelope. HMG-14/-17 proteins are the smallest proteins known to be transported into the nucleus by an active mechanism. A regulated nuclear import, perhaps during a special time window in the cell cycle, may affect the deposition of these proteins to their target places in chromatin.
Acknowledgments
We thank Dr. M.C. Dabauvalle for anti p62. We thank Drs. M.C. Dabauvalle and G. Krohne for critically reviewing this manuscript.
Abbreviations used in this paper
- APC
allophycocyanin
- EB
egg buffer
- HMG
high mobility group
- 5-IAF
5-iodoacetamido-fluorescein
- NLS
nuclear localization signal
- NPC
nuclear pore complexes
Footnotes
This work was partially supported by grant HO1804/1-1 from the Deutsche Forschungsgemeinschaft to Robert Hock, and by an award from the Humboldt Foundation.
References
- Adam S, Lobi T, Mitchell M, Gerace L. Identification of specific binding proteins for a nuclear localization sequence. Nature. 1989;337:276–279. doi: 10.1038/337276a0. [DOI] [PubMed] [Google Scholar]
- Adam S, Sterne-Marr R, Gerace L. Nuclear protein import in permeabilized mammalian cells requires solubilized cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso PJ, Crippa MP, Hayes JJ, Bustin M. The footprint of chromosomal proteins HMG-14 and HMG-17 on chromatin subunits. J Mol Biol. 1994;236:189–198. doi: 10.1006/jmbi.1994.1128. [DOI] [PubMed] [Google Scholar]
- Bell P, Mais C, McStay B, Scheer U. Association of the nucleolar transcription factor UBF with the transcriptionally inactive rRNA genes of pronuclei and early Xenopusembryos. J Cell Sci. 1997;110:2053–2063. doi: 10.1242/jcs.110.17.2053. [DOI] [PubMed] [Google Scholar]
- Blow J, Laskey R. Initiation of replication in nuclei and purified DNA by a cell free extract of Xenopuseggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Bustin, M. 1989. Preparation and application of immunological probes for nucleosomes. In Methods in Enzymology. Vol. 170. P.M. Wassarman and R.D. Kornberg, editors. Academic Press, New York. 214–251. [DOI] [PubMed]
- Bustin M, Neihart N. Antibodies against chromosomal HMG protein stain the cytoplasm of mammalian cells. Cell. 1979;16:181–189. doi: 10.1016/0092-8674(79)90199-5. [DOI] [PubMed] [Google Scholar]
- Bustin, M., and R. Reeves. 1996. High mobility group chromosomal proteins: architectural components that facilitate chromatin function. In Progress in Nucleic Acid Research and Molecular Biology. Vol. 54. W.E. Cohn and K. Moldave, editors. Academic Press, San Diego. 35–100. [DOI] [PubMed]
- Bustin M, Crippa MP, Pash JM. Immunochemical analysis of the exposure of high mobility group protein 14 and 17 surfaces in chromatin. J Biol Chem. 1990;265:20077–20080. [PubMed] [Google Scholar]
- Bustin M, Becerra PS, Crippa MP, Lehn DA, Pash JM, Shiloach J. Recombinant human chromosomal proteins HMG-14 and HMG-17. Nucleic Acids Res. 1991;19:3115–3121. doi: 10.1093/nar/19.11.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M, Trieschmann L, Postnikov YV. The HMG-14/-17 chromosomal protein family: architectural elements that enhance transcription from chromatin templates. Semin Cell Biol. 1995;6:247–255. doi: 10.1006/scel.1995.0033. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Stochaj U. Diffusion of proteins across the nuclear envelope of HeLa cells. Biotechniques. 1998;24:668–674. doi: 10.2144/98244rr04. [DOI] [PubMed] [Google Scholar]
- Crippa MP, Trieschmann L, Alfonso PJ, Wolffe AP, Bustin M. Deposition of chromosomal protein HMG-17 during replication affects the nucleosomal ladder and transcriptional potential of nascent chromatin. EMBO (Eur Mol Biol Organ) J. 1993;12:3855–3864. doi: 10.1002/j.1460-2075.1993.tb06064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabauvalle M, Benavente R, Chaly N. Monoclonal antibodies to a Mr 68,000 pore complex glycoprotein interfere with nuclear protein uptake in Xenopusoocytes. Chromosoma. 1988;97:193–197. doi: 10.1007/BF00292960. [DOI] [PubMed] [Google Scholar]
- Dabauvalle M, Looks K, Mekert H, Scheer U. Spontaneous assembly of pore complex-containing membranes in Xenopusegg extracts in the absence of chromatin. J Cell Biol. 1991;112:1073–1082. doi: 10.1083/jcb.112.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding HF, Bustin M, Hansen U. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region of chromosomal protein HMG-14. Mol Cell Biol. 1997;17:5843–5855. doi: 10.1128/mcb.17.10.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disney JE, Johnson KR, Magnuson NS, Sylvester SR, Reeves R. High-mobility group protein HMG-I localizes to G/Q- and C-bands of human and mouse chromosomes. J Cell Biol. 1989;109:1975–1982. doi: 10.1083/jcb.109.5.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einck L, Bustin M. The intracellular distribution and function of the high mobility group chromosomal proteins. Exp Cell Res. 1985;156:295–310. doi: 10.1016/0014-4827(85)90539-7. [DOI] [PubMed] [Google Scholar]
- Falciola L, Spada F, Calogero S, Langst G, Viot R, Grummt I, Bianchi M. High mobility group 1 protein is not stably associated with the chromosomes of somatic cells. J Cell Biol. 1997;137:19–26. doi: 10.1083/jcb.137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D. Nuclear protein import. Curr Opin Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Mattaj I. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Prehn S, Laskey R, Hartmann E. Isolation of a protein that is essential for the first step of nuclear import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Harootunian A, Admas S, Wen W, Meinkoth J, Taylor S, Tsien R. Movement of the free catalytic subunit of cAMP-dependent protein kinase in and out of the nucleus can be explained by diffusion. Mol Biol Cell. 1993;1993:993–1002. doi: 10.1091/mbc.4.10.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isackson P, Bidney J, Reeck G, Neihart N, Bustin M. High mobility group chromosomal protein isolated from nuclei and cytosol of cultured hepatoma cells are similar. Biochemistry. 1980;19:4466–4471. doi: 10.1021/bi00560a013. [DOI] [PubMed] [Google Scholar]
- Jans D, Jans P, Briggs L, Sutton V, Trapani J. Nuclear transport of granzyme B. Dependence of perforin in vivo and cytosolic factors in vitro. J Biol Chem. 1996;271:30781–30789. doi: 10.1074/jbc.271.48.30781. [DOI] [PubMed] [Google Scholar]
- Kerem B, Goitein R, Diamond G, Cedar H, Marcus M. Mapping of DNAse I sensitive regions on mitotic chromosomes. Cell. 1984;38:493–499. doi: 10.1016/0092-8674(84)90504-x. [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- Michaud N, Goldfarb D. Most nuclear proteins are imported by a single pathway. Exp Cell Res. 1993;208:128–136. doi: 10.1006/excr.1993.1230. [DOI] [PubMed] [Google Scholar]
- Newmeyer D, Finaly D, Forbes D. In vitro transport of a fluorescent nuclear protein and exclusion of nonnuclear proteins. J Cell Biol. 1986;103:2091–2102. doi: 10.1083/jcb.103.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J. Nuclear reconstitution in vivo: stages of assembly around protein free DNA. Cell. 1987;48:205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- Nigg E. Nucleocytoplasmic transport: signals, mechanism and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Postnikov YV, Trieschmann L, Rickers A, Bustin M. Homodimers of chromosomal proteins HMG-14 and HMG-17 in nucleosome cores. J Mol Biol. 1995;252:423–432. doi: 10.1006/jmbi.1995.0508. [DOI] [PubMed] [Google Scholar]
- Postnikov YV, Herrera JE, Hock R, Scheer U, Bustin M. Clusters of nucleosomes containing chromosomal protein HMG-17 in chromatin. J Mol Biol. 1997;274:454–465. doi: 10.1006/jmbi.1997.1391. [DOI] [PubMed] [Google Scholar]
- Rout M, Blobel G, Atchinson J. A distinct nuclear import pathway used by the ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Saitoh Y, Laemmli U. Methaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell. 1994;76:609–622. doi: 10.1016/0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Krohne G. In vivo assembly kinetics of fluorescently labeled Xenopuslamin A mutants. Eur J Cell Biol. 1995;68:345–354. [PubMed] [Google Scholar]
- Segil N, Geurmah M, Hoffman A, Roeder R, Heintz N. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- Shirakawa H, Tanigawa T, Sugyama S, Kobayashi M, Terashima T, Yoshida K, Arai T, Yoshida M. Nuclear accumulation of HMG2 is mediated by a basic region interspaced with a long DNA-binding sequence, and retention within the nucleus requires the acidic carboxyl terminus. Biochemistry. 1997;36:5992–5999. doi: 10.1021/bi962487n. [DOI] [PubMed] [Google Scholar]
- Trieschmann L, Alfonso PJ, Crippa MP, Wolffe AP, Bustin M. Incorporation of chromosomal proteins HMG-14/-17 into nascent nucleosomes induces an extended chromatin conformation and enhances the utilization of active transcription complexes. EMBO (Eur Mol Biol Organ) J. 1995a;14:1478–1489. doi: 10.1002/j.1460-2075.1995.tb07134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieschmann L, Postnikov Y, Rickers A, Bustin M. Modular structure of chromosomal proteins HMG-14/-17: definition of a transcriptional activation domain distinct from the nucleosomal binding domain. Mol Cell Biol. 1995b;15:6663–6669. doi: 10.1128/mcb.15.12.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieschmann L, Martin B, Bustin M. The chromatin unfolding domain of chromosomal protein HMG-14 targets the N-terminal tail of histone H3 in nucleosomes. Proc Natl Acad Sci USA. 1998;95:5468–5473. doi: 10.1073/pnas.95.10.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Holde, K.E. 1988. Chromatin. Springer-Verlag, New York.
- Weis K. Importin and exportin: how to get in and out of the nucleus. Trends Biochem Sci. 1998;23:185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- Weisbrod S, Weintraub H. Isolation of a subclass of nuclear proteins responsible for conferring a DNase I-sensitive structure of globin chromatin. Proc Natl Acad Sci USA. 1979;76:630–635. doi: 10.1073/pnas.76.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe, A.P. 1995. Chromatin Structure and Function. Academic Press, London.