Abstract
Occludin is the only known integral membrane protein localizing at tight junctions (TJ), but recent targeted disruption analysis of the occludin gene indicated the existence of as yet unidentified integral membrane proteins in TJ. We therefore re-examined the isolated junction fraction from chicken liver, from which occludin was first identified. Among numerous components of this fraction, only a broad silver-stained band ∼22 kD was detected with the occludin band through 4 M guanidine-HCl extraction as well as sonication followed by stepwise sucrose density gradient centrifugation. Two distinct peptide sequences were obtained from the lower and upper halves of the broad band, and similarity searches of databases allowed us to isolate two full-length cDNAs encoding related mouse 22-kD proteins consisting of 211 and 230 amino acids, respectively. Hydrophilicity analysis suggested that both bore four transmembrane domains, although they did not show any sequence similarity to occludin. Immunofluorescence and immunoelectron microscopy revealed that both proteins tagged with FLAG or GFP were targeted to and incorporated into the TJ strand itself. We designated them as “claudin-1” and “claudin-2”, respectively. Although the precise structure/function relationship of the claudins to TJ still remains elusive, these findings indicated that multiple integral membrane proteins with four putative transmembrane domains, occludin and claudins, constitute TJ strands.
Occludin is the only known integral membrane protein with four transmembrane domains that is exclusively localized at tight junctions (TJ)1 (Furuse et al., 1993; Ando-Akatsuka et al., 1996). TJ represent one mode of cell-to-cell adhesion in epithelial or endothelial cell sheets. These junctions constitute continuous, circumferential seals around cells that serve as a physical barrier preventing solutes and water from passing freely through the paracellular space (barrier function) (for reviews see Gumbiner, 1987, 1993; Schneeberger and Lynch, 1992; Anderson and Van Itallie, 1995). TJ are also thought to play a role as a boundary between the apical and the basolateral plasma membrane domains to create and maintain cell polarity (fence function) (Rodriguez-Boulan and Nelson, 1989). On ultrathin-section electron microscopy, TJ appear as a series of discrete sites of apparent fusion, involving the outer leaflet of the plasma membrane of adjacent cells (Farquhar and Palade, 1963). In freeze-fracture electron micrographs, TJ appear as a set of continuous, anastomosing intramembranous particle strands (TJ strands) or fibrils in the P-face (the outwardly facing cytoplasmic leaflet) with complementary grooves in the E-face (the inwardly facing extracytoplasmic leaflet) (Staehelin, 1973, 1974).
Occludin has been shown to be directly involved in the formation of TJ strands. In immunoreplica analyses, anti-occludin antibodies specifically labeled the TJ strand itself (Fujimoto, 1995; Furuse et al., 1996; Saitou et al., 1997). When overexpressed in insect Sf9 cells, occludin was highly accumulated in the cytoplasmic vesicular structures to form characteristic multilamellar bodies that bore apparent fusion sites as well as short TJ strand-like structures (Furuse et al., 1996). The overexpression of occludin in cultured MDCK cells increased the number of TJ strands (McCarthy et al., 1996). Furthermore, the serine/threonine phosphorylation level of occludin showed a strong correlation with the TJ formation (Sakakibara et al., 1997). These findings indicated that occludin plays a role in TJ formation and its regulation together with TJ-associated peripheral membrane proteins such as ZO-1 (Stevenson et al., 1986), ZO-2 (Gumbiner et al., 1991), ZO-3 (Haskins et al., 1998), cingulin (Citi et al., 1988), 7H6 antigen (Zhong et al., 1993), and symplekin (Keon et al., 1996).
Occludin has also been shown to be a functional component of TJ. Overexpression of full-length occludin in cultured MDCK cells elevated their trans-epithelial resistance (TER) (McCarthy et al., 1996; Balda et al., 1996), and introduction of COOH-terminally truncated occludin into MDCK cells or Xenopus embryo cells resulted in increased paracellular leakage of small molecular mass tracers (Balda et al., 1996; Chen et al., 1997). The TER of cultured Xenopus epithelial cells was reduced by addition of a synthetic peptide corresponding to the second extracellular loop of occludin into the culture medium (Wong and Gumbiner, 1997). Furthermore, in transfected fibroblasts, occludin was reported to show some cell adhesion activity (Van Itallie and Anderson, 1997). In addition to these findings suggesting the involvement of occludin in the TJ barrier and cell adhesion, the TJ fence function was also shown to be affected, when COOH-terminally truncated occludin was introduced into MDCK cells (Balda et al., 1996).
On the other hand, some recent observations suggested that occludin is not the only integral membrane protein in TJ strands. The introduction of COOH-terminally truncated occludin into MDCK cells caused the reconcentration of endogenous occludin in a dotted manner along the cell–cell border, whereas the continuous network of TJ strands was not affected (Balda et al., 1996). When a synthetic peptide corresponding to the second extracellular loop of occludin was added to the culture medium, it drove out the endogenous occludin from junctional areas of cultured epithelial cells without affecting the gross epithelial cell morphology (Wong and Gumbiner, 1997). Furthermore, endothelial cells in non-neuronal tissue and Sertoli cells in some species bore TJ but expressed only trace amounts of occludin (Hirase et al., 1997; Moroi et al., 1998).
Recently, we succeeded in targeted disruption of both alleles of the occludin gene in embryonic stem (ES) cells (Saitou et al., 1998). Surprisingly, when the occludin-deficient ES cells were differentiated into epithelial cells by embryoid body formation, well-developed TJ were found between adjacent epithelial cells. This finding conclusively indicated that occludin is not necessarily required for TJ formation itself, and that there are as yet unidentified TJ integral membrane protein(s) that can form strand structures without occludin. To understand the function and the molecular architecture of TJ in more detail, it is necessary to identify other integral membrane protein(s) of TJ strands. Identification of integral membrane proteins of TJ was originally regarded as difficult, but it is now possible to use occludin as a probe.
In this study, we examined the integral membrane proteins in the isolated junction fraction from chicken liver, which is highly enriched in TJ, and found that a broad ∼22-kD band was detected with occludin through 4 M guanidine extraction as well as sonication, followed by stepwise sucrose density gradient centrifugation. Through peptide sequencing of this band and homology search, we isolated full-length cDNAs encoding mouse homologues of two chicken proteins found in the broad ∼22-kD band. These proteins showed sequence similarity to each other and bore four putative transmembrane domains, but did not show any sequence similarity to occludin. Furthermore, transfection experiments revealed that both molecules were exclusively localized at TJ, especially at the TJ strand itself together with occludin. The results of this study provide new insight into the molecular architecture of TJ.
Materials and Methods
Antibodies and Cells
Rabbit anti–chicken occludin polyclonal antibody (pAb) (F44), rat anti– chicken occludin mAb (Oc-1), and rat anti–mouse occludin mAb (MOC37) were raised and characterized as described previously (Furuse et al., 1993; Saitou et al., 1997; Sakakibara et al., 1997). Rat anti–mouse E-cadherin mAb (ECCD2) was kindly provided by Dr. M. Takeichi (Kyoto University, Kyoto, Japan). Mouse anti-FLAG mAb (M2) and rabbit anti-GFP (green fluorescent protein) pAb was purchased from Eastman Kodak Co. (New Haven, CT) and Clontech (Palo Alto, CA), respectively. MDCK type II (MDCK II) cells were grown in DME supplemented with 10% FCS.
Isolated Junction Fraction from Chick Liver
The junction fraction was prepared from the livers of 1- or 2-d-old male chicks from the crude membrane and the bile canaliculi fractions according to the method described previously (Tsukita and Tsukita, 1989; Furuse et al., 1993). A total of 320 chicks yielded ∼20 ml of sucrose solution of the junction fraction. For immunoabsorption of occludin, 100 μl of the isolated junction fraction was washed with 10 mM Tris-HCl solution, pH 7.5, by centrifugation, and then the pellet was suspended in 100 μl of SDS buffer (0.1% SDS, 150 mM NaCl, 10 mM Tris-HCl, pH 7.5) by pipetting several times. After a 15-min incubation at room temperature, 100 μl of NP-40 buffer (2% NP-40, 150 mM NaCl, 10 mM Tris-HCl, pH 7.5) was added, followed by centrifugation at 15,000 g for 20 min. A 40-μl bed volume of protein A–Sepharose 4B (Pharmacia Biotech Sevrage, Uppsala, Sweden), which had been pre-incubated with 25 μl of rabbit anti–chicken occludin serum (F44) or control rabbit serum, was added to the supernatant and rotated for 2 h at 4°C. The beads were removed by brief centrifugation, and then the supernatant was processed for SDS-PAGE.
To thoroughly extract peripheral membrane proteins from isolated junctions, 400 μl of the junction fraction was washed twice with 10 mM Tris-HCl solution, pH 7.5, by centrifugation (15,000 g, 5 min). The pellet was resuspended and incubated in 800 μl of a solution containing 4 M guanidine-HCl and 10 mM Tris-HCl, pH 7.5, on ice for 30 min, and then diluted with 700 μl of 10 mM Tris-HCl solution, pH 7.5. Extracted peripheral membrane proteins were removed by centrifugation (15,000 g, 20 min), and the pellet was used for analysis of integral membrane proteins by SDS-PAGE.
For fragmentation of isolated junctions, 1.8 ml of the junction fraction was washed twice and suspended in 600 μl of 10 mM Hepes solution, pH 7.5, and then sonicated for 10 s on ice four times using a Sonifier 250 (Branson Ultrasonics Co., Danbury, CT). The sonicated fraction was mixed with 50% (wt/wt) sucrose solution to make 38% (wt/wt) sucrose solution. This solution was then centrifuged through a discontinuous gradient consisting of 50%, 46%, 42%, 38% (containing sample), 34%, 30%, 25%, 20%, and 0% sucrose solutions (wt/wt) in an SW41 rotor (Beckman Instruments, Inc., Palo Alto, CA) at 40,000 rpm for 1.8 h at 4°C. Each interface was carefully collected. 0%:20% and 42%:46% interfaces; were mixed with 20%:25% and 42%:46% interfaces; these were called 0%:25% and 42%:50% interface fractions, respectively. 200-μl aliquots of each fraction were diluted with 800 μl of 10 mM Hepes solution, pH 7.5, and centrifuged in a TLA100.2 rotor (Beckman Instruments, Inc.) at 80,000 rpm for 1 h at 4°C. Pellets were processed for SDS-PAGE.
SDS-PAGE and Immunoblotting
One-dimensional SDS-PAGE (12.5%) was performed according to the method of Laemmli (1970), and gels were stained with a silver-staining kit (Wako Pure Chemicals, Osaka, Japan). For immunoblotting, proteins were electrophoretically transferred from gels onto nitrocellulose membranes, which were then incubated with the first antibody. Bound antibodies were detected with biotinylated second antibodies and streptavidin-conjugated alkaline phosphatase (Amersham Corp., Arlington Heights, IL). Nitroblue tetrazolium and bromochloroindolyl phosphate were used as substrates for detection of alkaline phosphatase.
Peptide Sequencing
The 4 M guanidine-insoluble fractions of isolated junctions were electrophoresed on a 12.5% polyacrylamide gels. The lower and upper halves of the broad 22-kD band (see Fig. 1, band 9) was then subjected to direct peptide sequencing as follows. For amino acid sequencing of the lower half, proteins separated by SDS-PAGE were electrophoretically transferred onto polyvinylidene difluoride (PVDF) membrane (Bio-Rad Laboratories, Hercules, CA). The PVDF membrane was stained with Coomassie brilliant blue R-250 (CBB), and the lower half of the transferred 22-kD band was excised and subjected to NH2-terminal amino acid sequence analysis (HP G1005A Protein Sequencing Systems; Hewlett-Packard Co., Palo Alto, CA). For amino acid sequencing of the upper half, gels were stained with CBB, the upper half of the broad 22-kD band was excised, and amino acid sequence analysis was performed by the in-gel digestion method described by Rosenfeld et al. (1992). Two distinct peptide sequences (peptide sequence-1 and -2; see Fig. 3) were determined from the lower and upper halves of the 22-kD band, respectively.
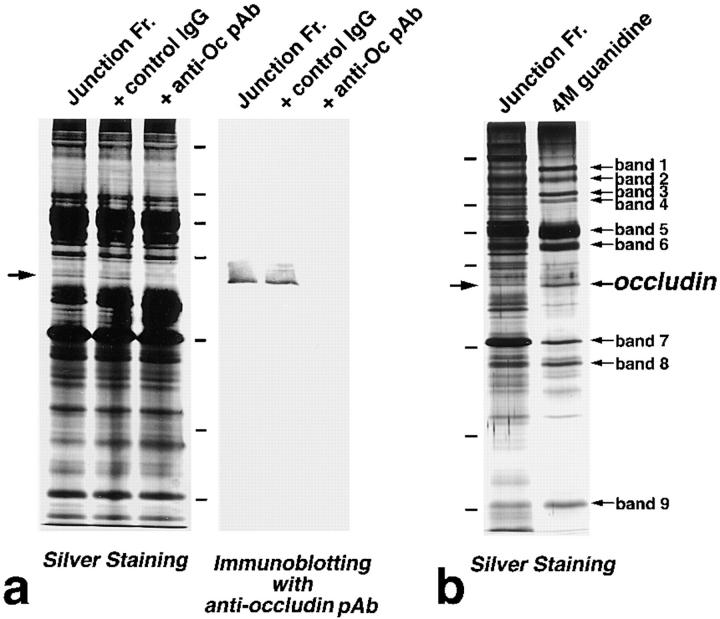
Figure 1.
Identification of the occludin band and nine guanidine-insoluble bands in the isolated junction fraction from chick liver. (a) Immunoabsorption of occludin from the isolated junction fraction. The isolated junction fraction was solubilized with 0.1% SDS. After dilution, immunoprecipitation was carried out with rabbit anti–chicken occludin pAb (F44) or control rabbit serum, and then the supernatant was processed for SDS-PAGE. In silver-stained gels, a 60-kD band (arrow) that was detected in the non-treated (Junction Fr.) as well as control serum-treated (+control IgG) fraction, disappeared in the F44-treated fraction (+anti-Oc pAb). Thus, occludin was identified as a 60-kD band in the junction fraction by silver staining. Accompanying immunoblots with anti-occludin pAb (F44) confirmed this notion. (b) Guanidine extraction of isolated junction fraction. To thoroughly extract peripheral membrane proteins from isolated junctions, the fraction was treated with 4 M guanidine-HCl. The silver-stained banding pattern was compared between non-treated (Junction Fr.) and guanidine-treated (4 M guanidine) samples. Occludin in the non-treated fraction (arrow) was concentrated by guanidine treatment (occludin). In addition to the occludin band, nine guanidine-insoluble bands were identified (bands 1–9), the amounts of which were similar to or greater than that of occludin. Bars indicate molecular masses of 200, 116, 97, 66, 45, 31, and 21 kD, respectively, from the top.
Figure 3.
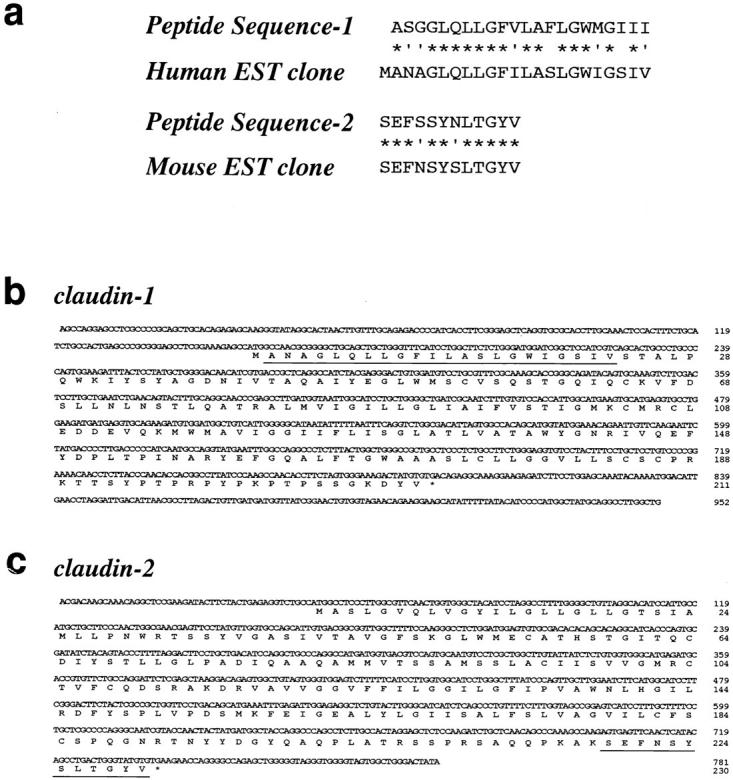
Cloning and sequencing of cDNAs encoding claudin-1 and -2. (a) Peptide sequencing. The lower and upper halves of band 9 (see Fig. 1 b) were separately subjected to the direct peptide sequencing, and yielded peptide sequence-1 and -2, respectively. Database searching identified human and mouse EST clones (these sequence data are available from GenBank/ EMBL/DDBJ under accession numbers AA305424 and AA116709, respectively), which encoded sequences showing significant similarity to peptide sequence-1 and -2, respectively. Identity and homology are indicated by asterisks and dots, respectively. (b and c) Nucleotide and deduced amino acid sequences of mouse claudin-1 and -2. Based on the above peptide sequences, two distinct cDNAs were isolated from a mouse liver cDNA library (for details, see Materials and Methods). Products encoded by these cDNAs were designated as claudin-1 and -2. Sequences homologous to peptide sequence-1 and -2 (a) are underlined. Since peptide sequence-1 was determined as the NH2-terminal sequence (for details, see Materials and Methods), the first ATG codon was conclusively determined in claudin-1 cDNA. Sequence similarity between claudin-1 and -2 (see Fig. 4 a) then enabled us to estimate the first ATG codon in claudin-2 cDNA. Claudin-1 and -2 cDNAs encoded 211– and 230– amino acid polypeptides with predicted molecular masses of 22.9 kD and 24.5 kD, respectively.
cDNA Cloning and Sequencing
Mouse liver total RNA was isolated according to the method described by Chomczynski and Sacchi (1987). Poly A(+) was prepared from the total RNA using oligo dT-cellulose (New England Biolabs, Inc., Bevery, MA). First strand cDNA was prepared from this polyA(+) RNA with Superscript™ II reverse transcriptase (GIBCO BRL, Gaithersburg, MD) and used for PCR. A λgt10 mouse liver cDNA library was constructed using a Time Saver TM cDNA Synthesis Kit (Pharmacia Biotech Sevrage) and used for cDNA screening.
The cDNA encoding claudin-1 was isolated as follows (see Fig. 3). Similarity search using the GenBank/EMBL/DDBJ databases with the MPsrch TPN program identified a human EST clone (these data are available under accession number AA305424) encoding a polypeptide showing significant similarity to peptide sequence-1 (see Fig. 3 a). Mouse liver first strand cDNA was thus subjected to PCR using two primers, 5′-ATGGCCAACGCGGGGCTGCAGCTGTTGGG-3′ and 5′-CAGATTCAGCAAGGAGTCAAGACTTT-3′, designed from the human EST sequence, and the expected 216-base DNA fragment was amplified. Sequencing confirmed that it encoded the mouse homologue of human EST product. Using this cDNA fragment as a template, a DIG-labeled probe was prepared using a DIG High Prime Labeling Kit (Boehringer Mannheim GmbH, Mannheim, Germany), and used to screen the λgt10 mouse liver cDNA library. One positive cDNA clone (IN01) with a 0.9-kb insert was obtained, and then subcloned into pBluescript SK(−). As peptide sequence–1 was obtained as the NH2-terminal sequence, this insert was interpreted to contain the 3′ end of the open reading frame (ORF) and 3′ noncoding region, but not the 5′ end of the ORF. Repeated searches of updated databases identified a mouse EST clone (these data are available from GenBank/ EMBL/DDBJ under accession number AA562761). This clone contained the 5′ end sequence of IN01 at its 3′ end, and encoded a polypeptide that showed significant similarity to peptide sequence–1, indicating that this clone contained the 5′ noncoding region and 5′ end of ORF. Two primers, 5′-AGCCAGGAGCCTCGCCCCGCAGCTGCA-3′ and 5′-CAGCCAAGGCCTGCATAGCCATGG-3′, were designed from the EST clone and IN01, respectively, and using them cDNA encoding the whole ORF was amplified by PCR from the mouse liver first strand cDNA. The amplified cDNA was subcloned into the pGEM-T Easy Vector (Promega Corp., Madison, WI), and the resulting construct was designated as pGTCL-1. To ensure no sequence errors occurred during PCR, the nucleic acid sequence was determined from three independently amplified fragments.
The cDNA encoding claudin-2 was isolated as follows (see Fig. 3). Similarity search using the Genbank/EMBL/DDBJ database with the MPsrch TPN program identified another mouse EST clone (these data are available under accession number AA116709) encoding a polypeptide showing significant similarity to the peptide sequence–2 (see Fig. 3 a). This clone appeared to contain the 3′ end of the ORF and 3′ noncoding region but not the 5′ end of the ORF. A mouse liver cDNA library was then prepared using a MarathonTM cDNA Amplification Kit (Clontech), and from this cDNA library the remaining 5′ part of the cDNA was amplified by PCR using two primers, 5′-GCTGTTGAGCAGATCTTGGAGAGCTCC-3′ (designed from the EST sequence) and AP-1 (provided with the Kit). Sequencing confirmed that the amplified cDNA contained the 5′ noncoding region and 5′ end of the ORF. Two primers, 5′-ACGACAAGCAAACAGGCTCCGAAG-3′ and 5′-TATAGTCCCAGCCACTAC-3′, were designed from the EST sequence and the amplified cDNA, and then the cDNA encoding the whole ORF was amplified by PCR. This cDNA was subcloned into the pGEM-T Easy vector (Promega), and the resulting construct was designated as pGTCL-2. To ensure no sequence errors occurred during PCR, the nucleic acid sequence was determined from three independently amplified fragments. DNA sequencing was performed using a Dye Terminator Cycle Sequence Kit (Applied Biosystems, Inc., Foster City, CA). For further plasmid construction, the insert was excised from pGTCL-2 by EcoRI digestion, and then subcloned into pBluescript SK(−) to make pSKCL-2.
Mammalian Expression Vectors and Transfection
Claudin-1 and -2 were tagged with FLAG-peptide or GFP at their COOH termini. To construct a FLAG-tagged claudin-1 expression vector (pCCL-1F), BamHI site was introduced at the stop codon of claudin-1 cDNA by PCR with primers 5′-GGCGACATTAGTGGCCACAFCATG-3′ and 5′-CGCGGATCCCACATAGTCTTTCCCACT-3′. The EcoRI–BamHI fragment of this PCR product and SacI–EcoRI fragment excised from pGTCL-1 were simultaneously ligated into SacI–BamHI-digested pBluescript SK(−) to construct pSKCL-1CB. An adapter DNA encoding a FLAG peptide was produced by annealing two oligonucleotides, 5′-GATCCGACTACAAGGACGACGATGACAAGTAGATCT-3′ and 5′-AGATCTACTTGTCATCGTCGTCCTTGTAGTCG-3′, followed by phosphorylation with T4 polynucleotide kinase. The plasmid pSKCL-1F containing an insert encoding FLAG-tagged claudin-1 was produced by ligating BamHI-digested FLAG adapter and BamHI–EcoRV-digested pSKCL-1CB. The insert was excised from pSKCL-1F by SacI–KpnI digestion followed by blunting with T4 polymerase, and then introduced into pCAGGSneodelEcoRI (Niwa et al., 1991), provided by Dr. J. Myazaki (Osaka University). The final expression vector for FLAG-tagged claudin-1 was designated as pCCL-1F. FLAG-tagged claudin-2 expression vector (pCCL-2F) was constructed in basically the same manner. A BamHI site was introduced at the stop codon of claudin-2 cDNA by PCR with primers 5′-ACGACAAGCAAACAGGCTCCGAAG-3′ and 5′-GCGGGATCCCACATACCCAGTCAGG-3′. The SacI–XhoI fragment of pSKCL-2 and XhoI–BamHI fragment of the PCR fragment were ligated and subcloned into SacI–BamHI-digested pSKCL-2F to be tagged with FLAG peptide. The FLAG-tagged claudin-2 cDNA was subcloned into pCAGGSneodelEcoRI to make the expression vector pCCL-2F.
To construct the GFP-tagged claudin-1 expression vector pBCL-1G, GFP cDNA was excised from pQBI25 (Quantum Biotechnologies, Inc., Quebec, Canada) and an XbaI site and GGGGGGGGGGGG (encoding four glysine residues) were introduced upstream of GFP in pBluescript (pGGLG4). The XbaI site was introduced at the stop codon of claudin-1 cDNA by PCR, and then ligated into the XbaI site of pGGLG4. The claudin-1–GFP fusion protein cDNA was introduced in the expression vector pBATEM2, from which E-cadherin cDNA was excised (Nose et al., 1988), to make the expression vector pBCL-1G.
MDCK II cells were used for transfection. To improve the transfection efficiency, cells on 35-mm dishes were cultured overnight in DME containing 50 μM Ca2+ (LCM) supplemented with 5% Ca2+-depleted FCS, and then washed twice with serum-free LCM. 1-μg aliquots of pCCL-1F, pCCL-2F, or pBCL-1G with 0.1 μg of pSTneoB were introduced into such pretreated cells in 1 ml LCM medium using Lipofectamine Plus (GIBCO BRL). After a 5-h incubation, 2 ml of DME containing 10% FCS was added. Cells were then cultured for 1 d, replated, and then cultured on two 9-cm dishes in DME containing 10% FCS and 300 μg of Geneticine (GIBCO BRL) to select stable transfectants. Geneticine-resistant colonies were isolated after 10–14-d selection. The clones expressing tagged proteins were screened by fluorescence microscopy with anti-FLAG antibody or GFP fluorescence.
Immunofluorescence Microscopy
Stable transfectants plated on glass coverslips were fixed with 1% formaldehyde in PBS for 10 min at room temperature. The fixed cells were treated with 0.2% Triton X-100 in PBS for 10 min and washed three times with PBS. After soaking in PBS containing 1% BSA, the samples were treated with primary antibodies for 1 h in a moist chamber. They were then washed three times with PBS, followed by a 30-min incubation with secondary antibodies. FITC-conjugated goat anti–rat IgG antibody (Biosource), rhodamine-conjugated goat anti–mouse IgG antibody (Chemicon, Temecula, CA), or Cy3-conjugated goat anti–rat IgG antibody (Amersham Pharmacia Biotech, Bucks, UK) were used as secondary antibodies. Cells were washed three times and then mounted in 90% glycerol-PBS containing 0.1% para-phenylendiamine and 1% n-propylgalate. Specimens were observed using a fluorescence microscope, Axiophot photomicroscope (Carl Zeiss, Inc., Thornwood, NY), or an MRC 1024 confocal fluorescence microscope (Bio-Rad Laboratories) equipped with a Zeiss Axiophot photomicroscope.
Electron Microscopy
Isolated junction fraction or sonicated samples were fixed overnight with a fixative containing 2.5% glutaraldehyde, 2% formaldehyde, 0.1% tannic acid, and 0.1 M cacodylate buffer, pH 7.2. After washing with 0.1 M cacodylate buffer, pH 7.2, samples were then postfixed with 1% OsO4 in 0.1 M cacodylate buffer, pH 7.2, for 2 h, followed by en bloc staining with 1% uranyl acetate for 1 h. Samples were dehydrated and then embedded in Epon 812. Ultrathin sections were cut with a diamond knife, and stained doubly with uranyl acetate and lead citrate. Samples were examined with a JEOL 1200EX electron microscope at an accelerating voltage of 100 kV.
Immunoelectron microscopy to examine freeze-fracture replicas was performed as described (Fujimoto, 1995). Cultured transfectant cells were scraped with a rubber policeman followed by centrifugation, and the pellet was quickly frozen by being slammed against a pure copper block cooled by liquid helium gas (Heuser et al., 1979). The frozen samples were fractured at −10°C and platinum-shadowed unidirectionally at an angle of 45° in Balzers Freeze Etching System (BAF 400T; Balzers Corp., Hudson, NH). The samples were immersed in a sample lysis buffer containing 2.5% SDS, 10 mM Tris-HCl, and 0.6 M sucrose, pH 8.2, for 12 h at room temperature, and then replicas floating off the samples were washed with PBS. Under these conditions, integral membrane proteins were captured by replicas, and their cytoplasmic domain was accessible to antibodies. The replicas were incubated with the anti-FLAG mAb or anti-GFP pAb for 60 min, and then washed with PBS several times. They were then incubated with goat anti–mouse or anti–rabbit IgG coupled to 10-nm gold (Amersham Pharmacia Biotech). The samples were washed with PBS, picked up on Formvar-filmed grids, and examined in a JEOL 1200EX electron microscope at an accelerating voltage of 80 kV.
Northern Blotting
The expression of claudin-1 and -2 in various mouse tissues was examined by Northern blotting using Mouse Multiple Tissue Northern Blot (Clontech). DNA fragments of claudin-1 (IN01) and claudin-2 (227–669) were radiolabeled with [32P]dCTP and used as probes for Northern blotting. Hybridization was performed in ExpressHyb™ Hybridization Solution (Clontech) at 68°C for 12 h. The membranes were washed with 2× SSC containing 0.1% SDS at room temperature for 30 min, and then with 0.1× SSC containing 0.1% SDS at 50°C for 30 min. The membranes were exposed to imaging plates for 12 h, and the signals were visualized using a Bio-Imaging Analyzer BAS2000 (Fuji Photo Film Co. Ltd., Tokyo, Japan).
Results
Identification of Occludin and ∼22-kD Band in SDS-PAGE of Isolated Junction Fractions from Chicken Liver
Occludin was first identified by mAb production using the isolated junction fraction from chicken liver as antigens (Furuse et al., 1993). However, it has not been determined whether occludin is present in the fraction in amount sufficient to be detected by silver staining. The isolated junction fraction was then solubilized with a solution containing 0.1% SDS, and occludin was immunoabsorbed by anti–chicken occludin pAb (F44). When compared with the silver-stained banding patterns of non-treated and control IgG-treated isolated junction fractions, that of F44-treated junction fraction specifically lacked a band of ∼60 kD (Fig. 1 a). We thus concluded that occludin can be detected as a single 60-kD band in silver-stained gels of the isolated junction fraction. This conclusion was confirmed by accompanying immunoblotting analysis with anti–chicken occludin pAb (Fig. 1 a), although occludin was detected as multiple bands in immunoblots as reported previously (Sakakibara et al., 1997).
Occludin knockout analyses suggested the existence of as yet unidentified TJ integral membrane protein(s) (Saitou et al., 1998). As occludin-deficient epithelial cells still bore well-developed TJ strands, we expected that the content of the as yet unidentified TJ integral membrane protein(s) in native TJ strands would not be less than that of occludin. If this was the case, the as yet unidentified TJ integral membrane protein(s) should also be detectable as a silver-stained band or bands in the isolated junction fraction. We attempted to identify a band or bands in the isolated junction fraction, which could be co-isolated with the 60-kD occludin band during the following treatments.
First, to thoroughly remove peripheral proteins and leave only integral membrane proteins, we treated the isolated junction fraction with 4 M guanidine-HCl, pH 7.5. This treatment has been used to remove all peripheral membrane proteins from isolated desmosomes (Blaschuk et al., 1986). As expected, the 60-kD occludin band resisted guanidine extraction (Fig. 1 b). In addition to the occludin band, nine other bands were detected in the guanidine-insoluble fraction in amounts similar to or greater than occludin. These bands were tentatively named bands 1–9 from the top of the gel.
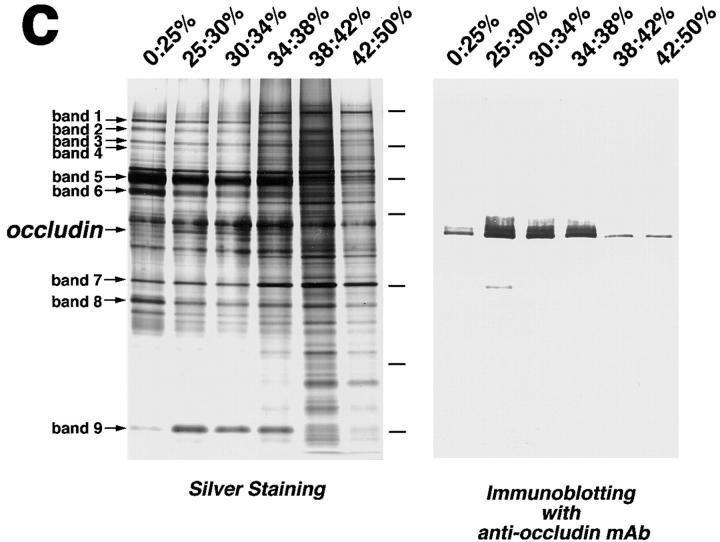
We next sonicated the isolated junction fraction, and fragmented structures were recovered as a pellet by centrifugation. Electron microscopy revealed that in the pellet actin filaments became undetectable and that plasma membranes were fragmented into small vesicles, some of which appeared to contain fragmented TJ (Fig. 2, a and b). The sonicated fraction was then fractionated by centrifugation in a stepwise discontinuous sucrose density gradient (Fig. 2 c). Each interface was resolved by SDS-PAGE followed by silver staining, and the distribution of the occludin band was compared with those of the other guanidine-insoluble bands (bands 1–9). Occludin was abundantly recovered at the 25:30%, 30:34%, and 34:38% interfaces, but in only trace amounts at 0:25%, 38:42%, and 42:50% interfaces, which was confirmed by immunoblotting with anti-occludin mAb. Among bands 1–9, only band 9 was recovered with the same distribution pattern as occludin. In contrast, the other guanidine-insoluble bands 1–8 behaved differently from occludin; bands 1–6 and 8, and band 7 were abundant at the 0:25% interface and 38:42%/42:50% interfaces, respectively.
Figure 2.
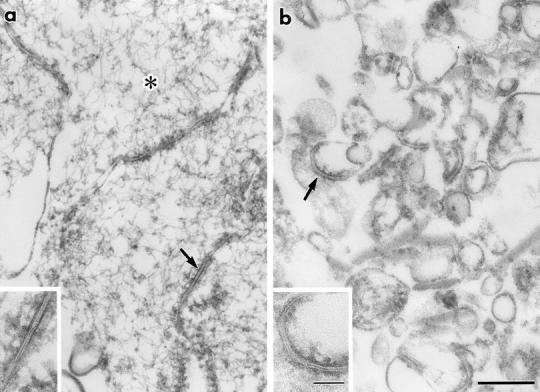
Behavior of the occludin and nine guanidine- insoluble bands on sonication followed by sucrose density gradient centrifugation. (a and b) Ultrathin sectional electron microscopic images of non-treated (a) and sonicated (b) junction fraction fixed in the presence of 0.1% tannic acid. The non-treated fraction was characterized by numerous actin filaments (*) and isolated junctions (arrow), which contained typical tight junctions (inset). After sonication followed by centrifugation, no actin filaments were detected, and isolated junctions were fragmented into small vesicular structures (arrow), some of which still contained typical tight junctions (inset). (c) Fractionation of sonicated junction fraction. After sonication, the isolated junctions were fractionated by stepwise sucrose density gradient centrifugation. 0:25%, 25:30%, 30:34%, 34:38%, 38:42%, and 42:50% interfaces were collected, and subjected to SDS-PAGE followed by silver staining. The distribution of the occludin band (occludin) was compared with those of nine guanidine-insoluble bands (band 1–9), and only band 9 was copartitioned with occludin. Silver staining and accompanying immunoblots with anti-occludin mAb (Oc-1) revealed that occludin was mainly recovered at 25:30%, 30:34%, and 34:38% interfaces, where band 9 was also characteristically accumulated. Bars in c indicate molecular masses of 200, 116, 97, 66, 45, 31, and 21 kD, respectively, from the top. Bar, 0.2 μm.
We concluded that among the numerous bands in isolated junction fractions, only band 9 behaved similarly to occludin, not only after guanidine treatment, but also sonication followed by sucrose density gradient centrifugation. We speculated that the 22-kD polypeptide in the band 9 is a good candidate as an integral membrane protein comprising TJ strands together with occludin.
Cloning of Full-Length cDNAs Encoding Mouse Claudin-1 and -2
The broad band 9, ∼22 kD in the guanidine-insoluble fraction, was tentatively divided into lower and upper halves, which were then subjected to direct peptide sequencing. As shown in Fig. 3 a, one peptide sequence was obtained from each half of the band (peptide sequence 1 and 2, respectively). A similarity search identified two EST clones with homology to these peptide sequences, human EST (these sequence data are available from GenBank/EMBL/ DDBJ under accession number AA305424) and mouse EST (available under accession number AA116709), respectively. Using these EST sequences, two distinct but related cDNAs were isolated from mouse liver cDNA.
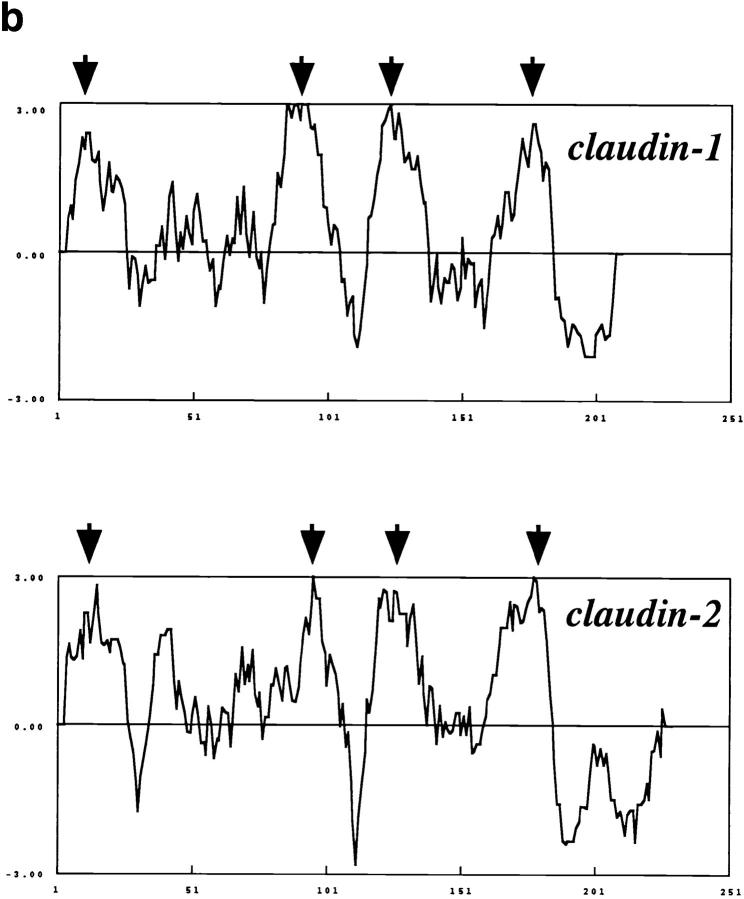
The complete nucleotide sequences of these cDNAs and their deduced amino acid sequences are shown in Fig. 3, b and c. These cDNAs contained open reading frames encoding 211– and 230–amino acid polypeptides with predicted molecular masses of 22.9 and 24.5 kD. As shown in Fig. 4 a, these polypeptides showed sequence similarity to each other (38% identity at the amino acid sequence level) but not to occludin. We called these proteins claudin-1 and -2, respectively (the reason for this nomenclature will be explained in the Discussion session). Furthermore, hydrophilicity analysis predicted that, like occludin, both claudin-1 and -2 contained four transmembrane domains (Fig. 4 b). If this prediction is the case, their putative first extracellular loop appears to be rather hydrophobic, especially in claudin-2. Occludin was reported to bear glycine/tyrosine-enriched extracellular loops, but neither claudin-1 nor -2 showed such characteristics.
Figure 4.
Structures of mouse claudin-1 and -2. (a) Comparison of amino acid sequences of mouse claudin-1 and -2 by the GENETYX program. Identity and homology are indicated by asterisks and dots, respectively. They showed 38% identity at the amino acid sequence level. (b) Hydrophilicity plots for claudin-1 and -2 prepared using the Kyte and Doolittle algorithm. The plot records the average hydrophilicity along the sequence over a window of eight residues. Hydrophilic and hydrophobic residues are in the lower and upper parts of the frames, respectively. The axes are numbered in amino acid residues. Both claudin-1 and -2 appeared to contain four major hydrophobic, potentially membrane-spanning regions (arrows), although the possibility was not excluded that five transmembrane domains were included in these molecules only from these plots as discussed in the text.
Expression and Distribution of Mouse Claudin-1 and -2
To examine the expression and distribution patterns of claudin-1 and -2, we attempted to produce specific mAbs or pAbs. As antigens, we used GST fusion proteins with the COOH-terminal cytoplasmic domains of claudin-1 and -2 (24 and 42 amino acids, respectively) produced in Escherichia coli, or synthetic peptides corresponding to their cytoplasmic domains conjugated to hemocyanin. However, it proved very difficult to obtain specific mAbs or pAbs.
We then introduced FLAG- or GFP-tagged claudin-1 and -2 into cultured MDCK cells, and MDCK transfectants stably expressing FLAG–claudin-1, FLAG–claudin-2, and GFP–claudin-1 with expected molecular masses were obtained. The subcellular distributions of these tagged proteins were then examined by confocal microscopy in comparison with those of occludin and E-cadherin. As shown in Fig. 5, both introduced FLAG–claudin-1 and FLAG–claudin-2 were precisely colocalized with occludin at the level of TJ. Computer-generated cross-sectional images revealed that FLAG–claudin-1 and FLAG–claudin-2 were precisely colocalized with occludin at the most apical part of the lateral membranes of confluent MDCK transfectants, whereas E-cadherin appeared to be localized more basally than the distribution patterns of claudin-1 or claudin-2 (Fig. 6). The same results were also obtained from GFP-tagged claudin-1–expressing MDCK cells, excluding the possibility that tag peptides affected the subcellular distribution of introduced proteins. These findings indicated that both claudin-1 and -2 were concentrated at TJ.
Figure 5.
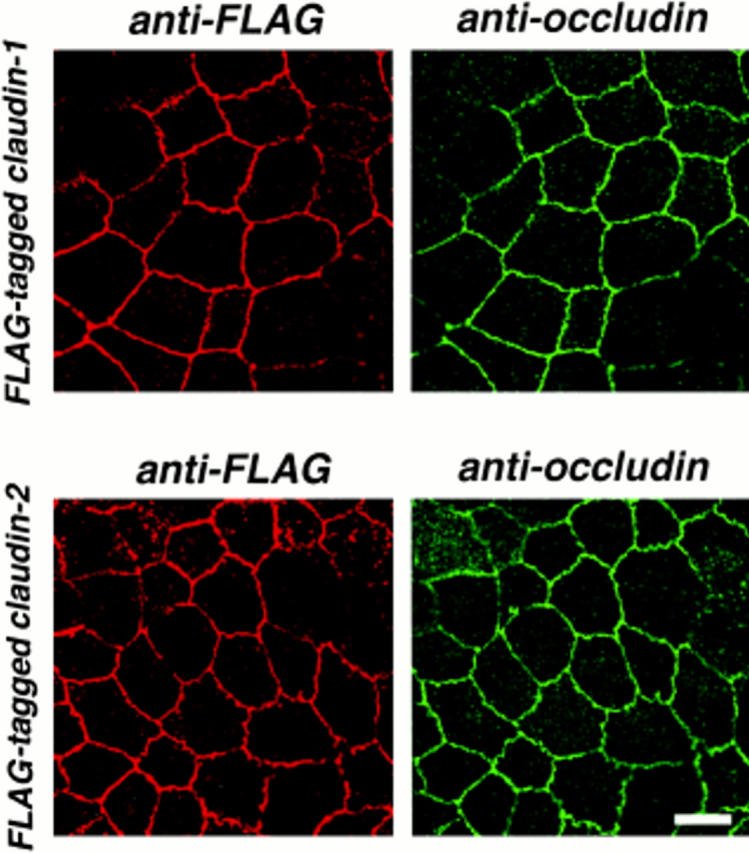
Colocalization of FLAG-tagged claudin-1 and -2 with occludin in MDCK transfectants. Confluent cultures of MDCK transfectants expressing FLAG–claudin-1 or FLAG–claudin-2 (FLAG-tagged claudin-1 or FLAG-tagged claudin-2) were doubly stained with mouse anti-FLAG mAb (anti-FLAG) and rat anti-occludin mAb MOC37 (anti-occludin). Images were obtained at the focal plane of the most apical region of lateral membranes by confocal microscopy. Both FLAG–claudin-1 and FLAG– claudin-2 were precisely colocalized with occludin at tight junction regions. Bar, 10 μm.
Figure 6.
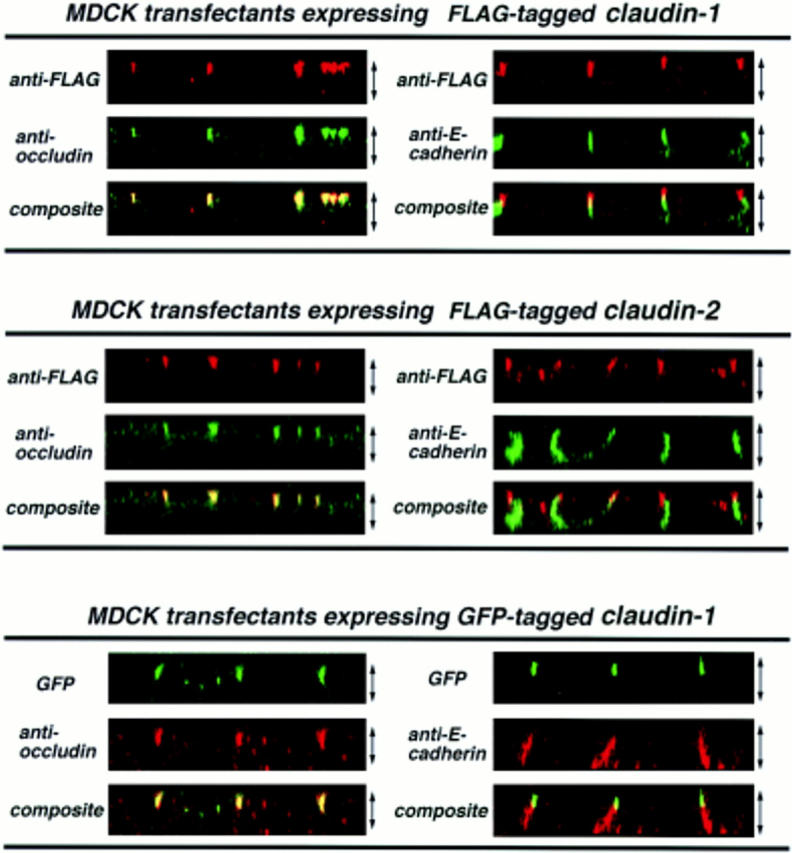
Comparison of the subcellular distributions of claudin-1 and -2 with those of occludin and E-cadherin in MDCK transfectants. Confluent sheets of MDCK transfectants expressing FLAG-tagged claudin-1 or FLAG-tagged claudin-2 were doubly stained with mouse anti-FLAG mAb (M2)/rat anti-occludin mAb (MOC37) or mouse anti-FLAG mAb (M2)/rat anti– E-cadherin mAb (ECCD2). For each sample, 22 optical sections were recorded with 0.5-μm intervals by confocal microscopy, and cross-sectional views were generated. The thickness of each cellular sheet is indicated by arrows. In computer-generated cross-sectional views, FLAG–claudin-1, FLAG– claudin-2 (anti-FLAG), and occludin (anti-occludin) were highly concentrated at the most apical portion of lateral membranes of MDCK cells, and overlaid images showed that FLAG–claudin-1 and FLAG–claudin-2 were precisely colocalized with occludin (composite). In contrast, E-cadherin was distributed along lateral membranes (anti-E-cadherin), and its distribution was more basally than those of FLAG–claudin-1 or FLAG–claudin-2 (composite). The same spatial relationships of claudin-1/occludin and claudin-1/E-cadherin were observed in MDCK transfectants expressing GFP-tagged claudin-1, indicating that tag-peptides did not affect the subcellular distributions of claudins.
To examine whether these proteins were localized on the TJ strands themselves, MDCK transfectants expressing FLAG–claudin-1 (Fig. 7, a and b), GFP–claudin-1 (Fig. 7 c) and FLAG–claudin-2 (Fig. 8) were analyzed by the immuno-freeze fracture replica method using anti-FLAG mAb or anti-GFP pAb. In all cases, TJ regions were specifically labeled. Taking the intrinsic resolution of immunogold labeling (20–25 nm) into consideration, most of the immunogold particles were regarded to directly label the TJ strands themselves, indicating that both claudin-1 and -2 were directly incorporated into TJ strands.
Figure 7.

Immunolabeling of freeze-fracture replicas of MDCK transfectants expressing FLAG- (a and b) or GFP-tagged claudin-1 (c) with anti-FLAG mAb or anti-GFP pAb. TJ regions (arrows), but not other membrane domains such as desmosomes (arrowheads in a; asterisks in b and c), were specifically labeled with immunogold particles. Taking the intrinsic resolution of immunogold labeling (20–25 nm) into consideration, most of the immunogold particles appeared to directly label the TJ strands themselves, indicating that claudin-1 is incorporated into TJ strands. Bars, 0.1 μm.
Figure 8.
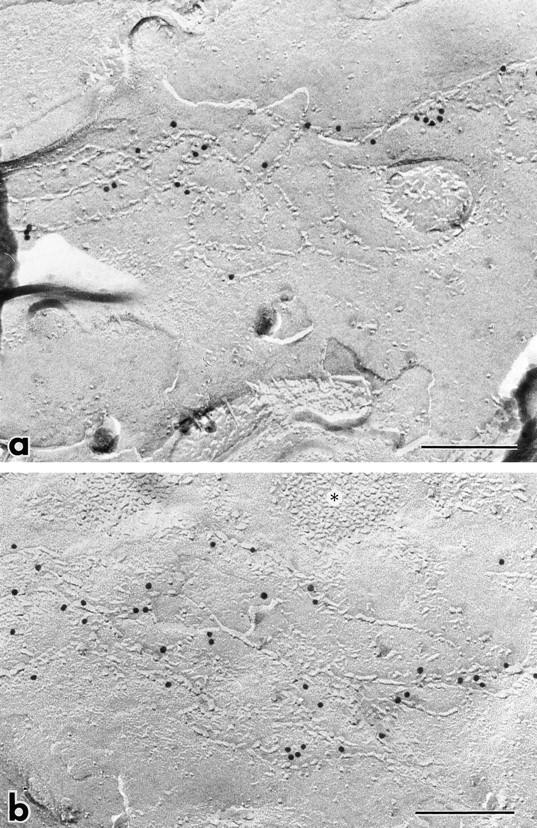
Immunolabeling of freeze-fracture replicas of MDCK transfectants expressing FLAG-tagged claudin-2 with anti-FLAG mAb. TJ regions, but not other membrane domains such as desmosomes (asterisks in b), were specifically labeled with immunogold particles. Taking the intrinsic resolution of immunogold labeling (20–25 nm) into consideration, most of the immunogold particles appeared to directly label the TJ strands themselves, indicating that claudin-2 is incorporated into TJ strands. Bars, 0.1 μm.
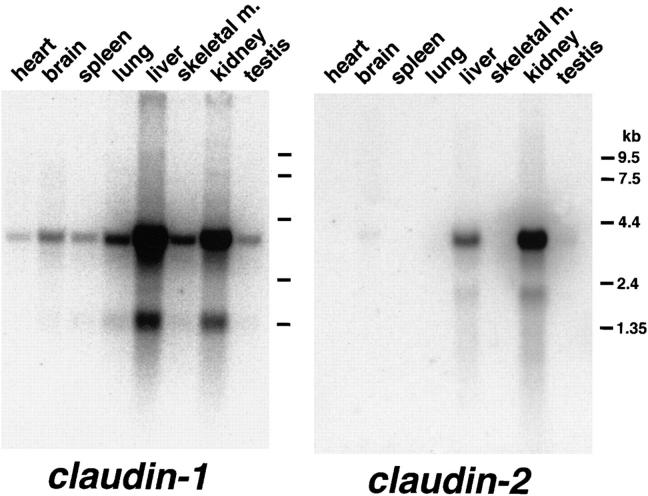
Finally, we examined the expression of claudin-1 and -2 in various tissues by Northern blotting (Fig. 9). Claudin-1 mRNAs were detected as major 4.0-kb and minor 1.3-kb bands in all the tissues examined, and were seen in especially large amounts in liver and kidney. In contrast, the expression of claudin-2 (major 4.0-kb and minor 2.0-kb mRNAs) was restricted to the liver and kidney (with small amounts in the brain).
Figure 9.
Northern blots of mouse claudin-1 and -2 expression. Mouse Multiple Tissue Northern Blot (Clontech) was probed with claudin-1 or claudin-2 cDNA fragments. Claudin-1 mRNAs were detected as major 4.0 kb and minor 1.3 kb bands in all the tissues examined, and were seen in especially large amounts in the liver and kidney. Claudin-2 expression (major 4.0 kb and minor 2.0 kb mRNAs) was restricted to the liver and kidney (with small amounts in the brain).
Discussion
Occludin was the only integral membrane protein known to be localized at TJ strands (Furuse et al., 1993; Ando-Akatsuka et al., 1996; Saitou et al., 1997). However, occludin-deficient epithelial cells, differentiated from occludin gene double knockout ES cells, bore well-developed TJ (Saitou et al., 1998). To interpret this unexpected finding, we searched for as yet unidentified TJ integral membrane proteins. Initially, we expected the occurrence of isotypes of occludin, and performed RT-PCR using various combinations of primers and similarity searches of databases. However, this approach failed to find sequences that showed similarity to occludin. To identify the occludin-binding integral membrane proteins, yeast two-hybrid analysis as well as immunoprecipitation with anti-occludin antibodies were carried out, but again no good candidates were identified.
We then re-examined the isolated junction fraction from chick liver, from which occludin was first identified (Tsukita and Tsukita, 1989; Furuse et al., 1993). Among the numerous bands observed on SDS-PAGE of this fraction, we screened for bands that were co-partitioned with the occludin band through guanidine extraction and sonication, followed by sucrose density gradient centrifugation; we identified a broad 22-kD band as a candidate. The broadness of this band was partially explained by the occurrence of multiple (at least two) similar but distinct ∼22-kD polypeptides. Two peptides were obtained from the chicken 22-kD band, and cDNAs encoding their mouse homologues were cloned. These two proteins were structurally related (38% identical at the amino acid sequence level), but showed no sequence similarity to occludin. We did not succeed in obtaining mAbs or pAbs specific for these proteins, probably because of the small size and/or low antigenicity of extramembrane portions of these molecules. Instead, we introduced cDNAs encoding FLAG- or GFP-tagged proteins into MDCK cells, and found that they exclusively targeted to TJ strands at both light and electron microscopic levels. We then designated these newly identified, related 22-kD proteins as “claudin-1” and “claudin-2” from the Latin word “claudere” (to close). This nomenclature is similar to that of TJ-associated MAGUK family members (Woods and Bryant, 1993; Kim, 1995; Anderson et al., 1995) such as ZO-1, ZO-2 and ZO-3 (Haskins et al., 1998).
No sequences similar to occludin were found in databases except for the occludin sequence itself. In contrast, several sequences similar to claudin-1 and -2 have been previously reported as ∼22-kD four transmembrane domain proteins (Briehl and Miesfeld, 1991; Katahira et al., 1997) and also found in databases, although their physiological functions have not been elucidated. These findings point to the existence of a new multiple gene family which could be called the “claudin family” (Morita, K., M. Furuse, and Sh. Tsukita, manuscript in preparation). As reported previously, the levels of occludin expression in various types of cells are generally correlated well with the number of TJ strands (Furuse et al., 1993; Saitou et al., 1997). In contrast, as shown on Northern blots (see Fig. 9), no such correlations were found for the expression of claudin-1 or -2. For example, in the lung which shows well- developed TJ, occludin was abundant, whereas the levels of expression of claudin-1 and -2 were rather low. As occludin-deficient mice were born with normal TJ in various tissues (Saitou, M., K. Fujimoto, Y. Doi, M. Itoh, T. Fujimoto, M. Furuse, H. Takano, Sh. Tsukita, and T. Noda, manuscript in preparation), it is likely that members of the claudin family other than claudin-1 and -2 are also involved in the formation of TJ strands in various tissues.
Hydrophilicity analysis predicted that both claudin-1 and -2 bore four transmembrane domains (see Fig. 4 b). Since the putative first extracellular loop is rather hydrophobic in the four transmembrane domain folding model of claudins, the possibility cannot be excluded that these molecules contain five transmembrane domains. However, considering that claudin-1 and -2 show sequence similarity to several proteins that were reported to have four transmembrane domains as discussed above (Briehl and Miesfeld, 1991; Katahira et al., 1997), and that other newly identified members of claudin family were also predicted to bear four transmembrane domains (Morita, K., M. Furuse, and Sh. Tsukita, manuscript in preparation), it appears to be reasonable to conclude that claudin-1 and -2 belong to the so called “four transmembrane domain proteins.” If this interpretation is the case, the hydrophobic character of the first extracellular loop of claudin-1 and -2 may be implicated in the barrier function of tight junctions.
In chicken liver, at least three proteins, occludin, claudin-1, and claudin-2, comprise TJ strands. Since among the guanidine-insoluble bands in isolated junction fraction, only the band ∼22 kD behaved similarly to the occludin band, integral membrane proteins other than claudin family members or occludin do not appear to be major components of TJ strands. As epithelial cells derived from occludin-deficient ES cells still bore well-developed TJ strands (Saitou et al., 1998), we speculated that under specialized conditions the TJ strand itself can be formed solely from claudin family members without occludin. On the other hand, overexpression of occludin in insect Sf9 cells (Furuse et al., 1996) suggested that occludin alone can form TJ strand-like structures, although these were very short. These observations are similar to the polymerization of intermediate-sized filaments (for review see Fuchs and Weber, 1994; Heins and Aebi, 1994). The constituents of the different types of intermediate-sized filaments differ in amino acid sequence and molecular mass, but they all contain a structurally homologous central rod domain that forms an extended coiled-coil structure when hetero- or homodimers are formed, which allows them to be polymerized into filamentous structures collectively categorized as an intermediate-sized filament. Similarly, the integral membrane proteins with four transmembrane domains such as claudin family members and occludin may be polymerized into TJ strands as both hetero- and homo-polymers, although the efficiency of polymerization may differ depending on their combination. At present, it is still premature to discuss in what manner these integral membrane proteins with four transmembrane domains are integrated into TJ strands. Any model, however, must explain the thickness of TJ strands (∼10 nm), which is similar to the diameter of the gap junction channel (connexon) consisting of six connexin molecules that also bear four transmembrane domains (for reviews see Staehelin, 1973, 1974; Jiang and Goodenough, 1996; Kumar and Gilula, 1996).
ZO-1 has been thought to be associated with TJ strands through its direct interaction with occludin (Furuse et al., 1994). However, in epithelial cells differentiated from occludin-deficient ES cells, ZO-1 was still exclusively localized at TJ (Saitou et al., 1998). As ZO-1 is a multidomain protein containing one SH3, one GUK, and three PDZ domains (Itoh et al., 1993; Tsukita et al., 1993; Willott et al., 1993), it is possible that ZO-1 is associated not only with occludin but also with claudin family members directly or indirectly. Further analyses of the interactions of the cytoplasmic domains of claudin family members with TJ-associated peripheral membrane proteins are required to determine the molecular architecture of TJ in detail.
This study revealed that the molecular architecture of TJ strands is more complex than expected. We concluded that multiple integral membrane proteins with four transmembrane domains, occludin, and claudins, constitute TJ strands. Of course, the most significant limitation of this study is that it has not been definitely proven that claudins (alone or in combination with occludin) actually form TJ strands, but we recently found that the introduction of claudin-1 (and also -2) into cultured L fibroblasts resulted in the formation of a well-developed network of TJ strands at cell–cell borders (Furuse, M., H. Sasaki, K. Fujimoto, and Sh. Tsukita, manuscript in preparation). Molecular manipulation of occludin and claudin functions including their overexpression and targeted gene disruption will lead to a better understanding of how TJ strands are formed as a multi-component complex in vivo.
Acknowledgments
We thank Dr. E. Majima (Apro Science Company, Tokushima, Japan) for his excellent technical assistance and helpful advice in peptide sequencing, and Dr. Y. Ando-Akatsuka (Department of Cell Biology, Faculty of Medicine, Kyoto University, Kyoto, Japan) for her help in constructing GFP– claudin-1. Thanks are due to all the members of our laboratory (Department of Cell Biology, Faculty of Medicine, Kyoto University) for helpful discussions. M. Furuse is grateful to Kanae, Yuhsuke, and Kyoko for their encouragement throughout this study.
This study was supported in part by a Grant-in-Aid for Cancer Research and a Grant-in-Aid for Scientific Research (A) from the Ministry of Education, Science and Culture of Japan.
Abbreviations used in this paper
- GFP
green fluorescence protein
- ORF
open reading frame
- pAb
polyclonal antibody
- TJ
tight junctions
Footnotes
Address all correspondence to Shoichiro Tsukita, M.D. and Ph.D., Department of Cell Biology, Faculty of Medicine, Kyoto University, Sakyo-ku, Kyoto 606-01, Japan. Tel.: 81-75-753-4372. Fax: 81-75-753-4660. E-mail: htsukita@mfour.med.kyoto-u.ac.jp
References
- Anderson JM, Fanning AS, Lapierre L, Van Itallie CM. Zonula occludens (ZO)-1 and ZO-2: membrane-associated guanylate kinase homologues (MAGuKs) of the tight junction. Biochem Soc Trans. 1995;23:470–475. doi: 10.1042/bst0230470. [DOI] [PubMed] [Google Scholar]
- Anderson JM, Van Itallie CM. Tight junctions and the molecular basis for regulation of paracellular permeability. Am J Physiol. 1995;269:467–475. doi: 10.1152/ajpgi.1995.269.4.G467. [DOI] [PubMed] [Google Scholar]
- Ando-Akatsuka Y, Saitou M, Hirase T, Kishi M, Sakakibara A, Itoh M, Yonemura S, Furuse M, Tsukita Sh. Interspecies diversity of the occludin sequence: cDNA cloning of human, mouse, dog, and rat-kangaroo homologues. J Cell Biol. 1996;133:43–47. doi: 10.1083/jcb.133.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda MS, Whitney JA, Flores C, González S, Cereijido M, Matter K. Functional dissociation of paracellular permeability and transepithelial electrical resistance and disruption of the apical-basolateral intramembrane diffusion barrier by expression of a mutant tight junction membrane protein. J Cell Biol. 1996;134:1031–1049. doi: 10.1083/jcb.134.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blashcuk OW, Manteuffel RL, Steinberg MS. Purification of desmoglein II: a method for the preparation and fractionation of desmosomal components. Biochim Biophys Acta. 1986;833:426–431. doi: 10.1016/0304-4165(86)90280-1. [DOI] [PubMed] [Google Scholar]
- Briehl MM, Miesfeld RL. Isolation and characterization of transcripts induced by androgen withdrawal and apoptotic cell death in the rat ventral prostate. Mol Endocrinol. 1991;5:1381–1388. doi: 10.1210/mend-5-10-1381. [DOI] [PubMed] [Google Scholar]
- Chen Y-H, Merzdorf C, Paul DL, Goodenough DA. COOH terminus of occludin is required for tight junction barrier function in early Xenopusembryos. J Cell Biol. 1997;138:891–899. doi: 10.1083/jcb.138.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Citi S, Sabanay H, Jakes R, Geiger B, Kendrick-Jones J. Cingulin, a new peripheral component of tight junctions. Nature. 1988;333:272–276. doi: 10.1038/333272a0. [DOI] [PubMed] [Google Scholar]
- Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Weber K. Intermediate filaments: structure, dynamics, function, and disease. Annu Rev Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- Fujimoto K. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. J Cell Sci. 1995;108:3443–3449. doi: 10.1242/jcs.108.11.3443. [DOI] [PubMed] [Google Scholar]
- Furuse M, Fujimoto K, Sato N, Hirase T, Tsukita Sa, Tsukita Sh. Overexpression of occludin, a tight junction-associated integral membrane protein, induces the formation of intracellular multilamellar bodies bearing tight junction-like structures. J Cell Sci. 1996;109:429–435. doi: 10.1242/jcs.109.2.429. [DOI] [PubMed] [Google Scholar]
- Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita Sa, Tsukita Sh. Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol. 1993;123:1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Itoh M, Hirase T, Nagafuchi A, Yonemura S, Tsukita Sa, Tsukita Sh. Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J Cell Biol. 1994;127:1617–1626. doi: 10.1083/jcb.127.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner B. Structure, biochemistry, and assembly of epithelial tight junctions. Am J Physiol. 1987;253:749–758. doi: 10.1152/ajpcell.1987.253.6.C749. [DOI] [PubMed] [Google Scholar]
- Gumbiner B. Breaking through the tight junction barrier. J Cell Biol. 1993;123:1631–1633. doi: 10.1083/jcb.123.6.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner B, Lowenkopf T, Apatira D. Identification of a 160kDa polypeptide that binds to the tight junction protein ZO-1. Proc Natl Acad Sci USA. 1991;88:3460–3464. doi: 10.1073/pnas.88.8.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskins J, Gu L, Wittchen ES, Hibbard J, Stevenson BR. ZO-3, a novel member of the MAGUK protein family found at the tight junction, interacts with ZO-1 and occludin. J Cell Biol. 1998;141:199–208. doi: 10.1083/jcb.141.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins S, Aebi U. Making heads and tails of intermediate filament assembly, dynamics and networks. Curr Opin Cell Biol. 1994;6:25–33. doi: 10.1016/0955-0674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- Heuser JE, Reese TS, Dennis MJ, Jan Y, Yan L, Evans L. Synaptic vesicle exocytosis captured by quick freezing and correlated with quantal transmitter release. J Cell Biol. 1979;81:275–300. doi: 10.1083/jcb.81.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirase T, Staddon JM, Saitou M, Ando-Akatsuka Y, Itoh M, Furuse M, Fujimoto K, Tsukita Sh, Rubin LL. Occludin as a possible determinant of tight junction permeability in endothelial cells. J Cell Sci. 1997;110:1603–1613. doi: 10.1242/jcs.110.14.1603. [DOI] [PubMed] [Google Scholar]
- Itoh M, Nagafuchi A, Yonemura S, Kitani-Yasuda T, Tsukita Sa, Tsukita Sh. The 220-kD protein colocalizing with cadherins in non-epithelial cells is identical to ZO-1, a tight junction-associated protein in epithelial cells: cDNA cloning and immunoelectron microscopy. J Cell Biol. 1993;121:491–502. doi: 10.1083/jcb.121.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JX, Goodenough DA. Heteromeric connexons in lens gap junction channels. Proc Natl Acad Sci USA. 1996;93:1287–1291. doi: 10.1073/pnas.93.3.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Inoue N, Horiguchi Y, Matsuda M, Sugimoto N. Molecular cloning and functional characterization of the receptor for Clostridium perfringensenterotoxin. J Cell Biol. 1997;136:1239–1247. doi: 10.1083/jcb.136.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keon BH, Schäfer S, Kuhn C, Grund C, Franke WW. Symplekin, a novel type of tight junction plaque protein. J Cell Biol. 1996;134:1003–1018. doi: 10.1083/jcb.134.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK. Tight junctions, membrane-associated guanylate kinases and cell signaling. Curr Opin Cell Biol. 1995;7:641–649. doi: 10.1016/0955-0674(95)80105-7. [DOI] [PubMed] [Google Scholar]
- Kumar NM, Gilula NB. The gap junction communication channel. Cell. 1996;84:381–388. doi: 10.1016/s0092-8674(00)81282-9. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McCarthy KM, Skare IB, Stankewich MC, Furuse M, Tsukita Sh, Rogers RA, Lynch RD, Schneeberger EE. Occludin is a functional component of the tight junction. J Cell Sci. 1996;109:2287–2298. doi: 10.1242/jcs.109.9.2287. [DOI] [PubMed] [Google Scholar]
- Moroi, S., M. Saitou, K. Fujimoto, A. Sakakibara, M. Furuse, O. Yoshida, and Sh. Tsukita. 1998. Occludin is concentrated at tight junctions of mouse/rat but not human/guinea pig Sertoli cells in testes. Am. J. Physiol. In press. [DOI] [PubMed]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high- expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Nose A, Nagafuchi A, Takeichi M. Expressed recombinant cadherins mediate cell sorting in model system. Cell. 1988;54:993–1001. doi: 10.1016/0092-8674(88)90114-6. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E, Nelson WJ. Morphogenesis of the polarized epithelial cell phenotype. Science. 1989;245:718–725. doi: 10.1126/science.2672330. [DOI] [PubMed] [Google Scholar]
- Rosenfeld J, Capdevielle J, Guillemot JC, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- Saitou M, Ando-Akatsuka Y, Itoh M, Furuse M, Inazawa J, Fujimoto K, Tsukita Sh. Mammalian occludin in epithelial cells: its expression and subcellular distribution. Eur J Cell Biol. 1997;73:222–231. [PubMed] [Google Scholar]
- Saitou M, Fujimoto K, Doi Y, Itoh M, Fujimoto T, Furuse M, Takano H, Noda T, Tsukita Sh. Occludin-deficient embryonic stem cells can differentiate into polarized epithelial cells bearing tight junctions. J Cell Biol. 1998;141:397–408. doi: 10.1083/jcb.141.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara A, Furuse M, Saitou M, Ando-Akatsuka Y, Tsukita Sh. Possible involvement of phosphorylation of occludin in tight junction formation. J Cell Biol. 1997;137:1393–1401. doi: 10.1083/jcb.137.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger EE, Lynch RD. Structure, function, and regulation of cellular tight junctions. Am J Physiol. 1992;262:647–661. doi: 10.1152/ajplung.1992.262.6.L647. [DOI] [PubMed] [Google Scholar]
- Staehelin LA. Further observations on the fine structure of freeze-cleaved tight junctions. J Cell Sci. 1973;13:763–786. doi: 10.1242/jcs.13.3.763. [DOI] [PubMed] [Google Scholar]
- Staehelin LA. Structure and function of intercellular junctions. Int Rev Cytol. 1974;39:191–283. doi: 10.1016/s0074-7696(08)60940-7. [DOI] [PubMed] [Google Scholar]
- Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol. 1986;103:755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita Sh, Tsukita Sa. Isolation of cell-to-cell adherens junctions from rat liver. JCell Biol. 1989;108:31–41. doi: 10.1083/jcb.108.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita Sh, Itoh M, Nagahchi A, Yonemura S, Tsukita Sa. Submembranous junctional plaque proteins include potential tumor suppressor molecules. J Cell Biol. 1993;123:l049–1053. doi: 10.1083/jcb.123.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie CM, Anderson JM. Occludin confers adhesiveness when expressed in fibroblasts. JCell Sci. 1997;110:1113–1121. doi: 10.1242/jcs.110.9.1113. [DOI] [PubMed] [Google Scholar]
- Willott E, Balda MS, Fanning AS, Jameson B, Van Itallie C, Anderson JM. The tight junction protein ZO-1 is homologous to the Drosophila discs-large tumor suppresser protein of septate junctions. Proc Natl Acad Sci USA. 1993;90:7834–7838. doi: 10.1073/pnas.90.16.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong V, Gumbiner BM. A synthetic peptide corresponding to the extracellular domain of occludin perturbs the tight junction permeability barrier. J Cell Biol. 1997;136:399–409. doi: 10.1083/jcb.136.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DA, Bryant PJ. ZO-1, DlgA and PSD95/SAP90: homologous proteins in tight, septate and synaptic cell junctions. Mech Dev. 1993;44:85–89. doi: 10.1016/0925-4773(93)90059-7. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Saitoh T, Minase T, Sawada N, Enomoto K, Mori M. Monoclonal antibody 7H6 reacts with a novel tight junction-associated protein distinct from ZO-1, cingulin, and ZO-2. J Cell Biol. 1993;120:477–483. doi: 10.1083/jcb.120.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]