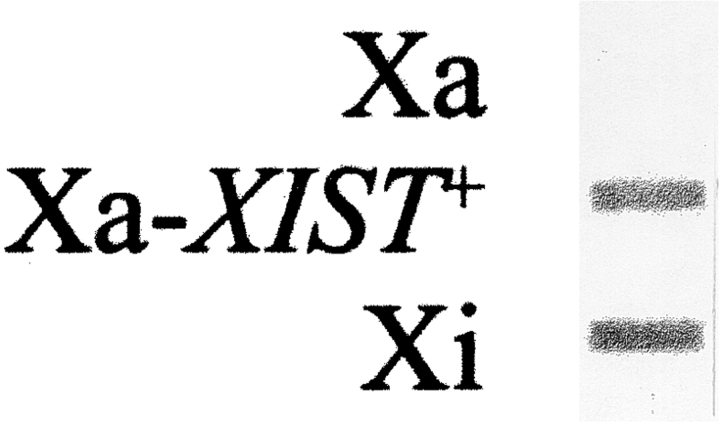Abstract
These studies address whether XIST RNA is properly localized to the X chromosome in somatic cells where human XIST expression is reactivated, but fails to result in X inactivation (Tinker, A.V., and C.J. Brown. 1998. Nucl. Acids Res. 26:2935–2940). Despite a nuclear RNA accumulation of normal abundance and stability, XIST RNA does not localize in reactivants or in naturally inactive human X chromosomes in mouse/ human hybrid cells. The XIST transcripts are fully stabilized despite their inability to localize, and hence XIST RNA localization can be uncoupled from stabilization, indicating that these are separate steps controlled by distinct mechanisms. Mouse Xist RNA tightly localized to an active X chromosome, demonstrating for the first time that the active X chromosome in somatic cells is competent to associate with Xist RNA. These results imply that species-specific factors, present even in mature, somatic cells that do not normally express Xist, are necessary for localization. When Xist RNA is properly localized to an active mouse X chromosome, X inactivation does not result. Therefore, there is not a strict correlation between Xist localization and chromatin inactivation. Moreover, expression, stabilization, and localization of Xist RNA are not sufficient for X inactivation. We hypothesize that chromosomal association of XIST RNA may initiate subsequent developmental events required to enact transcriptional silencing.
Xinactivation, the process whereby an entire X chromosome is transcriptionally silenced in mammalian females (Lyon, 1961), has been studied for several decades, yet the underlying process remains largely undefined. A wealth of developmental evidence has implicated mouse Xist (and its human counterpart XIST) in the process of X inactivation. Recently it has been demonstrated that Xist is necessary for X inactivation during development, as mouse X chromosomes deleted for Xist failed to inactivate (Penny et al., 1996; Marahens et al., 1997). Transfection of ES cells further showed that Xist is sufficient for X inactivation, as an Xist-containing cosmid or YAC introduced onto an autosome causes the chromosome to manifest signs of inactivation (Herzing et al., 1997; Lee and Jaenisch, 1997).
The transcript from XIST is a large untranslated RNA, which we have shown to have a unique and highly specific location in the nucleus (Brown et al., 1992; Clemson et al., 1996). Unlike other nuclear RNAs, the bulk of the signal is fully processed message that is stably maintained in a specific site in the nucleus, and is not detected in the cytoplasm. This abundant stable transcript maintains a tight association with the X chromosome, mirroring its shape and size as if painting the chromosome (Clemson et al., 1996). When Xist is expressed ectopically through ES cell transfection, the RNA shows the same association with autosomal chromatin after differentiation, which subsequently exhibits hallmarks of inactivation such as heterochromatinization, loss of acetylated histone H4 antibody staining, and inactivation of sequences close to the insertion site (Herzing et al., 1997; Lee and Jaenisch, 1997). The apparent structural association of XIST/Xist with the inactive X chromosome (Clemson et al., 1996; Lee et al., 1996; Lee and Jaenisch, 1997) coupled with the evidence revealing Xist's importance in the process of X inactivation (Lee et al., 1996; Penny et al., 1996; Herzing et al., 1997; Lee and Jaenisch, 1997) suggests that this RNA is a functional nuclear RNA that is most likely contributing to, by the act of its association, chromatin inactivation. As such, it is predicted that the precise localization of XIST RNA is critical for its function.
Although a spate of recent evidence has clearly demonstrated the absolute requirement for Xist in X inactivation during mouse development, studies indicate that its human counterpart, XIST, is not essential for X inactivation in cultured somatic cells. Brown and Willard (1994) showed that XIST expression is apparently not necessary for maintaining X inactivation in mouse/human somatic cell hybrids, and Rack et al. (1994) showed that inactive X chromosomes in human leukemic cells that had lost the X-inactivation center maintained their inactivation. These results are surprising in light of the fact that XIST RNA is constitutively expressed in differentiated female cells, suggesting that it is functioning in the maintenance of chromosome inactivation. Two recent papers show that expression of XIST RNA is not sufficient to cause inactivation (Hansen et al., 1998; Tinker and Brown, 1998). In these reports, rodent/human somatic cell hybrids retaining the human active X chromosome were treated with several rounds of 5-azadeoxycytidine (5azadC),1 a demethylating agent, to create clones that reactivated the dormant XIST gene. Using mouse/human hybrids, Tinker and Brown (1998) analyzed eight X-linked genes by reverse transcription (RT)- PCR for expression, with none of the genes showing signs of inactivation as a result of XIST expression, while Hansen et al. (1998) reactivated XIST expression in hamster–human hybrids, and detected no silencing of two X-linked genes. These results contrast with those that show that forced Xist expression caused by mutant methyltransferase results in silencing of X-linked genes in differentiating male ES cells and developing mice embryos (Panning and Jaenisch, 1996).
The requirement for Xist in X inactivation in developing mice embryos, coupled with the precise spatial positioning of the RNA to inactivated chromatin (Clemson et al., 1996; Lee et al., 1996; Panning and Jaenisch, 1996), strongly suggests that there is a correlation between XIST RNA binding and inactivation of chromatin; however, it is not known conclusively whether Xist localization results in X inactivation. Somatic cell hybrids where XIST is apparently neither necessary (Brown and Willard, 1994) nor sufficient (Tinker and Brown, 1998) for X inactivation allow us to investigate rigorously whether association of XIST RNA with the X chromosome corresponds with transcriptional silencing. Interestingly, our results reveal that the human XIST RNA shows aberrant positioning in mouse/ human hybrid cells that express the normally dormant XIST gene by means of demethylation, in that the RNA is more disperse and spread across the entire nucleus. Even in hybrids that contain a naturally inactive human X chromosome, the XIST RNA does not show normal tight localization to the X chromosome. These results suggest that the impotence of XIST RNA in this system may be due to failure of the RNA to localize. Furthermore, these results imply that the ability to localize to the chromosome is not an innate feature of the XIST RNA, but is afforded by specific factors that are not fully substituted by mouse homologues. We also show that when the RNA is not restricted to the chromosome, it is still found as a large and stable entity, indicating that stabilization and localization of XIST RNA are uncoupled. Using demethylation, we then forced expression of Xist RNA from the active murine X chromosome, and examined the relationship of the RNA to the X chromosome. Unlike its human counterpart, the murine Xist RNA strictly localized to the X chromosome in the mouse background. Since the reactivated Xist RNA correctly localized in a somatic cell, we conclude that its association to the X chromosome does not have to occur during development to mark the chromosome for Xist RNA binding. Furthermore, Xist RNA does not discriminate between active and inactive chromatin as it paints the active X chromosome completely. Additionally, this result shows that the factors required to deposit the RNA at the X chromosome are present and available in terminally differentiated cells that do not normally express Xist. Finally, we show that when Xist RNA is properly localized to an active mouse X chromosome, inactivation of the X chromosome does not result. This important result clearly demonstrates a lack of correlation between Xist RNA localization and inactivation, and also indicates that when removed from the normal developmental context, even with proper expression, stabilization, and localization, Xist is not able to cause X inactivation.
Materials and Methods
Cells, Cell Culture, and Treatment with 5-Azadeoxycytidine
Derivatives of the AHA-11aB1 active X chromosome–containing human/ mouse somatic cell hybrid that expressed human XIST RNA were isolated after treatment with successive rounds of the demethylating agents 5-azadeoxycytidine or 5-azacytidine (Tinker and Brown, 1998). To create additional hybrid subclones that expressed the murine Xist gene, AHA-A5-2b cells (that stably expressed human XIST from the active X chromosome) or subclones that had undergone one round of 5-azadeoxycytidine treatment, but did not stably express human XIST, were plated at 104 cells/60-mm culture dish and allowed to attach for 4–6 h before being treated with 5-azadeoxycytidine (0.2 μg/ml) or 5-azacytidine (4 μM) for 24 h. Individual colonies were isolated by trypsinization in cloning cylinders, and were plated in 60-mm dishes. Cells were grown to confluence and subcloned, and RNA was isolated for RT-PCR examination of Xist expression using primers MX23b and MIX20 (Kay et al., 1993). RT was as described previously (Brown et al., 1990). Xist-positive clones were grown for in situ hybridization (see below), and were subjected to one to four additional rounds of treatment with demethylating agents. Cells were maintained in alpha-MEM with 7.5% FCS supplemented with penicillin/streptomycin (GIBCO BRL, Gaithersburg, MD).
Cell Preparation for In Situ Hybridization
Our standard cell fixation has been described previously (Lawrence et al., 1989), and will be summarized briefly here. Monolayer cells grown on glass coverslips were extracted with 0.5% Triton X-100, 5% vanadyl ribonucleoside complex to preserve RNA (GIBCO BRL) in CSK buffer (Fey et al., 1986) for 2 min on ice. Cells were then fixed in 4% paraformaldehyde for 10 min at room temperature, and were stored in 70% ethanol at 4°C.
DNA Probes
For fluorescence in situ RNA hybridization, the following probes were used: human PGK1, a genomic lambda clone for PGK1 isolated from the American Type Culture Collection (Rockville, MD) X chromosome library LAOXNL01; XIST (human XIST probes generously provided by Hunt Willard, Case Western Reserve University): XIST G1A, an ∼10-kb genomic plasmid spanning from the fourth intron to the 3′ end of the human XIST gene; pXISTHbC1A, a 1.6-kb cDNA clone wholly contained within exon 1 of XIST; mouse Xist, a genomic clone spanning exons 5 and 6 (generously provided by Barbara Panning, Massachusetts Institute of Technology); mouse Zfx: PDP1115, a 6.8-kb cDNA obtained from American Type Culture Collection (cat. 63069); and mouse Pgk1, a 1.8-kb cDNA clone from American Type Culture Collection (cat. 57222).
In Situ Hybridization and Detection
Hybridization and detection was performed as described previously (Lawrence et al., 1988; Johnson et al., 1991; Xing et al., 1993), so will be detailed briefly here. DNA probes were nick-translated using biotin-11-dUTP or digoxigenin-16-dUTP (Boehringer Mannheim Corp., Indianapolis, IN). For exclusive RNA hybridization, cells were hybridized under nondenaturing conditions (such that cellular DNA was not accessible) overnight at 37°C in 50% formamide, 2× SSC using a probe concentration of 5 μg/ml. Human and mouse Cot-1 was included in the hybridization buffer for the human and murine RNA hybridizations, respectively. Posthybridization washes were performed as follows: 50% formamide, 2× SSC for 30 min at 37°C; 2× SSC for 30 min at 37°C; 1× SSC for 30 min at room temperature (rt) with agitation; and 4× SSC for 30 min at rt with agitation. Hybridization was detected with either antidigoxigenin antibody (Boehringer Mannheim Corp.) coupled with rhodamine or fluorescein at 200 μg/ml in 1% BSA, 4× SSC; or with fluorescein-conjugated avidin (Boehringer Mannheim) at 2.5 mg/ml in 1% BSA, 4× SSC for 1 h at 37°C. Postdetection washes were performed as follows: 4× SSC for 20 min at rt with agitation, 4× SSC, 0.1% Triton for 20 min at rt with agitation, and then 4× SSC for 20 min at rt with agitation. Simultaneous detection of whole X chromosome library and RNA was done as described previously (Clemson et al., 1996) with minor modifications. In brief, cells were extracted and fixed as described above. The cells were then denatured at 80°C for 5 min in 70% formamide, 2× SSC, and were immediately dehydrated through an ice-cold ethanol series. The Total Chromosome Library Coatasome X-digoxigenylated™ (Oncor Inc., Gaithersburg, MD) was used for human chromosome detection, while biotinylated Mouse Chromosome X Paint Probe™ (Oncor Inc.) was used for mouse X detection. 10 μl of the whole chromosome probe mixture was denatured for 10 min at 75°C, and was allowed to preanneal for 30 min at 37°C. Samples were then hybridized with the probes overnight at 37°C. Posthybridization washes, detection, and postdetection washes were performed as described above.
XIST Stability
To determine the stability of the XIST RNA in hybrid and lymphoblast cells, recently expanded cultures were treated with 2 μg/ml actinomycin in DMSO. Control cultures received only the DMSO. Whole-cell RNA was harvested by acid guanidinium thiocyanate-phenol-chloroform extraction (Chomczynski and Sacchi, 1987), and was reverse-transcribed as above. cDNA was diluted 1/100, and RT-PCR was performed with primers for XIST (Duncan et al., 1993) and actin (Chariot and Castronovo, 1996). PCR was demonstrated to be in a linear range of amplification, and then products were quantitated with NIH Image (National Institutes of Health, Bethesda, MD) on scanned images of ethidium bromide–stained agarose gels. To determine the half-life of the XIST message, the ratio of XIST product to actin product was divided by the average ratio of XIST to actin in the DMSO-treated samples. The half-life of actin cytoplasmic mRNA has previously been estimated at 25 h (Sympson and Geoghan, 1990), and therefore should not have varied substantially over the 8-h time period used here. Stability of XIST RNA in hybrids was also determined by hybridization to slot blots, and yielded a similar half-life of 5 h (data not shown).
For quantitation of relative amounts of XIST RNA, hybridization of slot blots was performed as described previously (Brown et al., 1991). In brief, total cellular RNA was prepared using the guanidinium thiocyanate method. 5 μg of RNA from an inactive X–containing somatic cell hybrid (Willard et al., 1993) and the AHA-11aB1 active X–containing hybrid as well as its demethylated XIST+ derivative was denatured in formaldehyde/formamide and transferred to a nitrocellulose membrane. The blots were hybridized with 32P-labeled XIST14A cDNA probe (Brown et al., 1991). To control for loading equal amounts of the RNA, the blots were stripped and reprobed for the MIC2 gene.
Imaging
Large numbers of cells were evaluated by direct examination using an Axiophot microscope equipped with an 100× 1.4 NA Plan ApoChromat™ objective (Carl Zeiss Inc., Thornwood, NY) with a 2.5× photo eyepiece and multibandpass epifluorescence filters (Chroma, Brattleboro, VT). Representative images were recorded with a CCD camera (200 series; Photometric, Tucson, AZ) with a pixel size of 19 μM and a 14-bit A/D converter (data acquisition system by G.W. Hannaway and Associates, Boulder, CO).
Results
Human XIST Transcripts in Hybrid Cells Show Abnormal Nuclear Localization
In other recent work (Tinker and Brown, 1998), subclones of AHA-11aB1 mouse/human somatic cells (AHA hybrids) were treated with three rounds of demethylation, each of which involved culturing cells for 24 h with 5azadC. After removing the 5azadC and culturing cells in normal media, many clones were isolated that stably expressed RNA from the normally dormant XIST gene on the active human X chromosome. Subsequent RT-PCR analysis showed that none of eight X-linked genes examined were inactivated as a result of XIST expression from the active X chromosome. As it was possible that some of the cells in the population examined might have lost XIST expression, and that the gene expression detected by RT-PCR could have been derived from this subset of cells, here we used in situ hybridization to confirm the lack of inactivation at a single cell level. Using genomic probes to the human genes, we examined expression of PGK-1 and XIST RNA in the AHA-A5-2b mouse/human hybrid cells that contain a human X chromosome expressing RNA from the hypomethylated XIST gene. PGK-1 RNA was detected in the majority of the cells that also expressed XIST RNA, confirming the PCR data from Tinker and Brown (1998) that the X chromosome is not inactivated as a result of reactivated XIST expression.
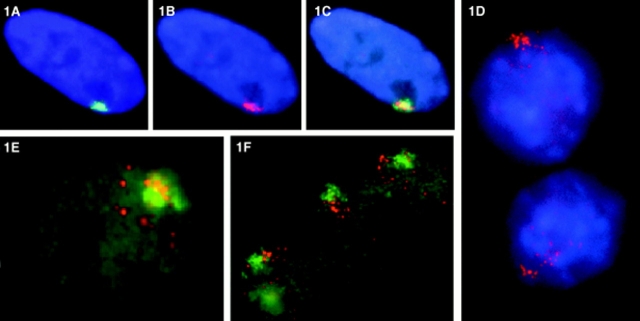
To begin our analysis of the correlation between XIST localization and inactivation of chromatin, we examined the distribution of the XIST RNA in AHA mouse/human hybrids that had previously been identified as stable XIST expressers by RT-PCR. Using in situ hybridization, we analyzed a total of six different hybrid clones, and found that almost every cell in each population was expressing XIST RNA. Furthermore, the hybrids continue to show stable XIST expression for over 2 yr after the original demethylation, confirming that transcriptional reactivation of the human XIST locus is quite stable (Tinker and Brown, 1998). The XIST RNA signal in the reactivated hybrid cells was similar in brightness and apparent abundance to normal diploid fibroblasts (Fig. 1, A and D), and the particulate nature of XIST RNA, which we have reported previously in normal fibroblasts (Clemson et al., 1996), is highly apparent in the AHA hybrids (Fig. 1, D–F). However, while the distribution of the XIST RNA in normal diploid fibroblasts is discrete and highly contained in a small area in the nucleus (Fig. 1 A), the reactivated XIST RNA in the hybrids was distinctly different (Fig. 1, D–F) in that it was more spread out, occupying a much larger area of the nucleus. This less-contained pattern revealed numerous tiny distinct particles that spread over a significant portion of the nucleus. In normal diploid fibroblasts, the XIST RNA associates closely with the inactive X chromosome in a pattern that is similar in both size and shape (Fig. 1, A–C; Clemson et al., 1996). While the dispersed pattern of XIST RNA in the hybrid cells suggested that it was not closely associating with the X chromosome, it is possible that the human X chromosome in a mouse background also occupies a broad, more disperse territory. To examine the specific relationship of the XIST RNA to the X chromosome in the AHA hybrid cells, the RNA signal was directly compared with that from a whole X chromosome library hybridization in 100 cells in three separate experiments. In >90% of the AHA reactivants, the RNA signal was obviously not strictly associated with the X chromosome (Fig. 1 E). While there generally was some density of XIST RNA signal over the chromosome, a large quantity of signal was usually seen spreading out away from the X chromosome.
Figure 1.
Fluorescence in situ hybridization detection of XIST RNA in normal human and mouse/human hybrid cells. Digoxigenin and biotinylated probes were hybridized in situ and detected with fluorochrome-conjugated avidin or antidigoxigenin antibody. Fluorochromes used were FITC (green), rhodamine (red), and DAPI (blue). (A) In normal human diploid fibroblasts (46, XX WI-38 cells), the XIST RNA (green) occupies a discrete location in the nucleus. (B) The XIST RNA has a similar shape to the X chromosome territory defined by the whole X chromosome library signal (red; the weak red signal in the middle of this cell is the active X chromosome which is out of the plane of focus). (C) The XIST RNA essentially paints the X chromosome in these normal nuclei (overlap is yellow). (D) In mouse/human hybrids containing a single active human X chromosome (AHA-A5) that expresses XIST through treatment with the demethylating agent 5azadC, the XIST RNA (red) is disperse, and spreads throughout the nucleus. (E) The XIST RNA (red) does not associate or strictly localize to the human whole X chromosome library signal (green) in the hybrid cells. (F) In mouse/human hybrid cells (t86-B1maz1b-3a) that contain a single human inactive X that expresses XIST endogenously (no 5azadC treatment), the human XIST RNA (red) also shows aberrant localization to the whole X chromosome library signal (green).
To determine if the lack of localization is due to artifacts created by reactivating XIST expression from the active X chromosome (e.g., the active X chromosome may be refractory to XIST binding), we examined the distribution of XIST RNA in t86-B1maz1b-3a mouse–human hybrid cells that retain a normal human inactive X chromosome (Willard et al., 1993). In over 100 cells examined in four separate experiments, the distribution of the XIST RNA in the hybrid cells was identical to that in the AHA-A5 hybrids; the XIST RNA was disperse, and spread into the nucleoplasm with no strict limitation to the X chromosome library signal (Fig. 1 F). This result indicates that the lack of localization is not an artifact of global demethlyation, as these cells were not treated with demethylating agents, and still show no detectable localization of the XIST RNA to the X chromosome. It also strongly suggests that the aberrant localization is not due solely to inherent differences between active and inactive X chromosomes, as the human X chromosome in this cell line is an inactive X chromosome known to remain transcriptionally inert (Brown et al., 1997).
Since the signal intensities in the different cell lines seemed equivalent (Fig. 1, A, D, and F), it appeared that hybrid cells that ectopically express XIST produce similar amounts of RNA, as do diploid cells that express XIST endogenously. However, we wished to confirm that aberrations in expression from the reactivated XIST gene do not cause the mislocalization. For example, demethylation could produce an overabundance of XIST RNA, causing the transcripts to spill from the site of transcription into the surrounding nuclear space. Alternatively, reactivation could produce quantities of RNA that are insufficient to paint the chromosome. This possibility was addressed through a slot blot hybridization assay (Fig. 2). XIST RNA from a hybrid cell line containing an active X (Xa), reactivated X (Xa-XIST+), and normally expressing inactive X chromosome (Xi) were compared. No XIST RNA was detected from the active X chromosome, while a similar abundance of RNA was observed in the Xa-XIST+ and Xi cell lines. This result confirms the in situ hybridization data that suggested similar amounts in the different somatic cells examined, and clearly shows that XIST reactivation by demethylation produces essentially the same levels of XIST RNA as the XIST gene on the naturally inactivated X chromosome. This result further suggests that lack of localization is not due to artifacts created by demethylation.
Figure 2.
Reactivated and endogenous XIST expression is similar in hybrid cells. Total cellular RNA from the following cells were transferred to nitrocellulose and hybridized with an XIST cDNA probe: AHA-11aB1, the active X-containing hybrid cells (Xa); AHA-2C-5C-9, the demethylated derivative that ectopically expresses XIST (Xa-XIST +); and t11-4Aaz5 cells containing a human inactive X chromosome that normally expresses XIST (Xi). The blots were scanned, and the density of signal from the Xi and Xa-XIST+ clones were essentially identical, demonstrating that the RNA is produced in similar quantities from endogenously expressing and demethylated XIST genes.
Stability and Localization of XIST RNA are Uncoupled
Next we examined the stability of the XIST RNA in hybrid cells, as previous results indicate that this nuclear RNA is highly stable (Clemson et al., 1996), and that nuclear stability is an important developmentally regulated feature of this RNA (Panning et al., 1997; Sheardown et al., 1997). In XX somatic cells, only the inactive X chromosome expresses XIST (Brown et al., 1992; Clemson et al., 1996; Lee et al., 1996), and the transcripts that coat the chromosome form a nuclear accumulation much larger than other mRNAs examined (Clemson et al., 1996). This large XIST RNA accumulation was shown to be composed of two populations: a small focus of short-lived nascent transcripts found at the inactive X chromosome simply because it is transcribed there, and a large spliced and stable population that represents the mature, likely functional RNA (Clemson et al., 1996). Before X inactivation in female cells or undifferentiated ES cells, XIST RNA is observed as tiny dots on both X chromosomes, or as a single dot in male cells (Lee et al., 1996; Panning and Jaenisch, 1996; Panning et al., 1997; Sheardown et al., 1997). Very recently it has been shown that during early development, XIST RNA is initially expressed in an unstable form from both active and inactive X chromosomes, and becomes stabilized from the inactive X as development proceeds. The transition from single dot to large accumulation is not regulated by increased transcription, but by stabilization of the XIST transcripts in cis from the inactive X chromosome (Panning et al., 1997; Sheardown et al., 1997).
It is not known whether deposition and stabilization of XIST RNA arise from a single mechanism, or if separate mechanisms are involved in stabilizing and positioning the RNA. Analysis of XIST RNA in hybrids provides important insights in this regard. In all six hybrid lines examined, it was clear that the XIST RNA comprised a large accumulation, with the quantity of signal indistinguishable from that observed in normal cells (Figs. 1 and 2), suggesting that the XIST RNA represents a unique XIST intermediate, one in which the transcripts are expressed and accumulating, but not localizing to the chromosome. These results are the first to indicate that association of XIST RNA with the X chromosome does not automatically arise from the increased stability that occurs after differentiation.
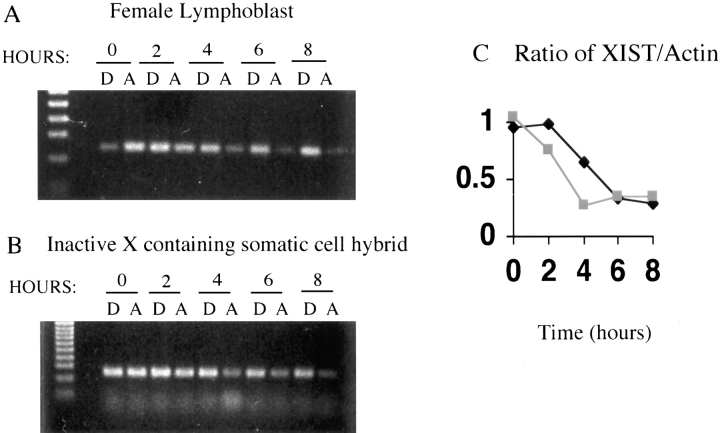
To examine stability by another approach, we compared the half-life of XIST RNA after actinomycin D treatment in unsynchronized human female lymphoblasts (GM07350), where the transcripts are expected to be stable, and in the mouse/human hybrid cell line t75-2maz-34-1B that expresses XIST from the inactive human X chromosome. Fig. 3 shows the results of RT-PCR with primers for human XIST from RNA isolated from cells after up to 8 h of actinomycin treatment. By normalizing to RT-PCR products for actin mRNA (which has a cytoplasmic half-life of ∼25 h [Sympson and Geoghan, 1990]) the half-life of the RNA from the two cell lines could be determined. Both the hybrids and the lymphoblasts showed a half-life for XIST RNA of ∼5 h (Fig. 3 B), which is similar to the half-life reported for stable XIST transcripts in diploid fibroblasts (Sheardown et al., 1997). The similarity of the half-life in the two cell lines confirms that the transcripts in the hybrids are stable. The similar half-lives, but very different patterns of XIST RNA localization in the nucleus, strongly suggest that stabilization of XIST RNA does not arise from chromosomal localization, and vice versa. Therefore, some additional mechanism other than stability is responsible for localizing the XIST transcripts to the X chromosome.
Figure 3.
Stability of the XIST RNA after treatment of cells with actinomycin. Cells were harvested after 0–8 h of actinomycin treatment (A) or control DMSO treatment (D) as the actinomycin is dissolved in DMSO. RT-PCR was performed on the RNA isolated from these cells with primers for XIST (A and B) and actin (not shown). The RT-PCR products for a female lymphoblast (GM07350; A) and an inactive X-containing human/ mouse somatic cell hybrid (t75-2maz34-1a; B) are shown, and the ratio of XIST product to actin product (normalized to the ratio of XIST to actin for the control DMSO treatments) is plotted in C. The black line shows the ratio for the female cells, while the grey line shows the ratio for the hybrid cell line.
We speculate that localization of this RNA to the X chromatin is brought about by specific autosomal factors that allow deposition of the human XIST transcripts along the chromosome. As there is ∼30% sequence divergence between mouse and human Xist/XIST sequences (Brockdorff et al., 1992; Brown et al., 1992), it is not surprising if the homologues required for localization of Xist RNA are unable to substitute completely for the human factors. In the hopes of finding a human autosome that would allow for XIST localization, and ultimately to identify potential localization factors, we examined the distribution of XIST RNA in six different hybrids containing a human inactive X chromosome. As these hybrids contained various other human chromosomes, we hoped to identify a hybrid that contained the necessary human factor to allow localization of human XIST. None of the hybrids showed localized XIST RNA, although all exhibited large apparently stable XIST RNA accumulations. The consistent failure of hybrid cells to localize the RNA correctly suggests that either the necessary locus was missing from these hybrids, or that more than one factor is required concurrently, and that these hybrids did not have the right combination of human chromosomes. We favor the latter possibility, as these hybrids have been shown by PCR or karyotyping to include collectively most of the human chromosome complement.
Reactivated Mouse Xist RNA Localizes to the Active X Chromosome
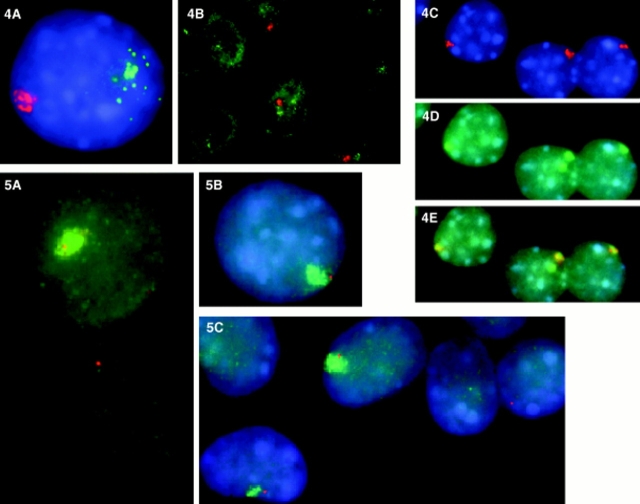
If the failure of the human XIST RNA to localize is the result of mouse/human species differences, then it follows that if murine Xist were reactivated, the RNA would be properly localized as the full complement of murine factors would be present. To test this hypothesis, we isolated clones that expressed not only human XIST RNA, but also mouse Xist RNA after additional rounds of demethylation. We studied the nuclear Xist/XIST RNA expression pattern in AHA-A5-2b-g cells, AHA hybrids expressing human XIST that had been treated with additional rounds of demethylating agents to induce mouse Xist expression, providing an opportunity to compare directly mouse and human XIST RNA in the same cell. The mouse cells used in the generation of these hybrids (A9) do not express Xist RNA without treatment; and these cells maintained ectopic murine Xist expression less consistently than human XIST expression (see below). In our analysis of 80 cells expressing both human and mouse Xist, >70% showed the following pattern of expression: the human XIST RNA showed a small focal concentration, with particles apparently drifting away from the site of transcription into the rest of the nucleoplasm; in contrast, the murine Xist RNA signal was a large but discrete pattern that was much more confined in the nucleus than its human counterpart (Fig. 4, A and B). We directly compared the distribution of the murine Xist signal to the mouse chromosome library signal in 50 different cells. Like the human XIST RNA in its normal human diploid environment, the murine transcripts formed a punctate accumulation very similar in size and shape to the mouse X chromosome paint, revealing a close association with the active murine X chromosome (Fig. 4, C–E). Clearly, murine Xist RNA can properly localize in the mouse background, while in the same cell the human XIST RNA cannot.
Figure 4.
In situ RNA and murine X whole chromosome library detection in AS-2B mouse/human hybrid cells that contain both an active human and mouse X chromosome. Both human and murine Xist RNA is expressed from these chromosomes as a result of 5azadC treatment. (A and B) The difference between the reactivated murine Xist RNA and human XIST RNA is illustrated in these cells, with the murine RNA appearing highly localized (red) and the human XIST RNA (green) showing a focus of expression (most likely representing the site of transcription) with particles that drift away into the nucleoplasm. Reactivated murine Xist RNA localizes to the active X chromosome (C–E). (C) The murine Xist RNA (red) is discretely contained in the hybrid cell nucleus (D) in a pattern similar to the murine X chromosome signal (green; the mouse X chromosome library cross-reacts slightly with the human X chromosome, giving a second green signal apparent in the middle cell in D); (E) the murine Xist RNA colocalizes with the active murine X chromosome (overlap is yellow).Figure 5. The mouse X chromosome painted by the Xist RNA does not inactivate. Codetection of murine Xist RNA (green) and Zfx RNA or Pgk1 RNA (red) in undenatured 1B-5C-3B cells. (A and B) Zfx RNA (red) is expressed from cells that also express mouse Xist RNA (green); (C) Pgk1 RNA expression (red) is also expressed from chromosomes that express Xist RNA (green), suggesting that the active X chromosome is not inactivated as a result of Xist localization. Expression of mouse genes appears to be unaffected in cells that express mouse Xist. (A) There is a similar pattern and intensity of expression of Zfx RNA in cells that express mouse Xist (top), and those that do not (bottom). (C). Similarly, there is comparable Pgk1 expression in cells that express mouse Xist (left) and cells that do not (far right).
These results support the hypothesis that the aberrant human XIST RNA location is due to a lack of species-specific factors necessary for localization. Interestingly, it is apparent that the factors required to localize the Xist RNA are present in terminally differentiated cells that normally do not express Xist RNA. This important result is informative on several other levels. First, it shows that the lack of localization of the human XIST was not due to 5azadC treatment, as the localization of the murine Xist was observed after 5azadC treatment; hence, demethylation does not disallow localization of the transcripts. Second, the mislocalization is not due to general disruption of nuclear structure in hybrid cells. Perhaps most importantly, it shows that the XIST RNA does not discriminate between active and inactive X chromosomes. In these mouse cells the murine X chromosome has already been selected in early embryogenesis to remain active, but is able to stably express and bind the Xist RNA. Hence, active X chromosomes are not recalcitrant to Xist RNA binding. The fact that the reactivated murine Xist localizes unequivocally shows that the proper deposition of this RNA at the X chromosome can occur outside the realm of early development, and therefore, that no developmental factors are required to set up a pattern of Xist chromosome binding. Apparently all the requirements for expression, stability, and localization of Xist RNA are available in somatic cells.
The Mouse X Chromosome Painted by Xist RNA Does Not Inactivate
Studies in transgenic mice have definitively demonstrated that Xist is not only necessary (Penny et al., 1996; Marahens et al., 1997), but also sufficient (Herzing et al., 1997; Lee and Jaenisch, 1997) to initiate X inactivation in developing mouse embryos. In our analysis, we have circumvented development by recreating, in terminally differentiated cells, several of the steps known to correlate with X inactivation in embryonic cells: expression of Xist RNA (Kay et al., 1993; Kay et al., 1994); transition from a single dot of expression to stabilization of a large nuclear RNA accumulation (Panning et al., 1997; Sheardown et al., 1997); and deposition of these stable Xist transcripts along the entire length of the chromosome (Clemson et al., 1996). The obvious next question is, does inactivation of the chromatin result from properly expressed, stabilized, and localized Xist RNA?
While we were able to isolate stable long-term human XIST reactivants after three rounds of treatment with 5azadC, we were not able to isolate a clone that expressed mouse Xist long-term, despite multiple serial treatments with demethylating agents. After selection and culture of single cell isolates, the level of Xist RNA declined over a period of several weeks as detected by RT-PCR. By in situ techniques we saw that initially almost every cell in the population expressed Xist. Gradually, the number of cells that expressed Xist declined (Fig. 4 B), until eventually no more cells expressed Xist RNA.
While we initially thought that this reduction was due to some selective disadvantage conferred by mouse Xist expression (for example, the cells could be dying because the mouse Xist RNA inactivated the sole murine X chromosome), several considerations make this possibility less likely. First, we detected no change in cell growth rate or evidence of large scale cell death in the reactivated hybrids, both of which would be expected if Xist expression was causing large-scale toxicity. Second, we have isolated clones that continued to express mouse Xist in >30% of the population for as long as 2 mo after the initial demethylation. Additionally, we were able to subclone pure populations of Xist RNA expressers from clones that had reduced expression of Xist to only a few cells, which would not be expected if the cells were gradually dying. While it is possible that the reactivants were selected against, we consider it more likely that Xist was being remethylated gradually, much like what was seen for the unstable human reactivants (Tinker and Brown, 1998). Our failure to obtain a clone that expressed mouse Xist longer than several months, in contrast to the human gene, may reflect differences between the methylation of the two genes; for example, the murine gene may have more methylation which must be completely removed before remethylation occurs, causing loss of Xist expression. Ultimately, the difficulty we had in creating long-term mouse Xist reactivants did not cause a problem for our experiments, as any cells that showed detectable mouse Xist always showed the same large intense pattern, allowing us to perform analysis of individual cells, even when as little as 10% of the population expressed Xist RNA.
The definitive way to assess whether the mouse X chromosome painted by reactivated Xist RNA is inactivated is to examine expression of individual genes in single cells. Because of the heterogeneity of Xist expression in our cell population, RT-PCR analysis could not address the critical question of whether individual genes were still expressed. We therefore used dual in situ detection of murine Xist with X-linked genes to examine directly X inactivation in individual cells. Pgk1 is subject to X inactivation in humans and mice (Disteche, 1995), while Zfx is an X-linked gene that escapes inactivation in humans, but is subject to X inactivation in the mouse (Adler et al., 1991). Zfx and Pgk1 nuclear RNA expression were examined in single cells that expressed Xist RNA (Fig. 5, A–C), using cDNA clones. In multiple experiments encompassing over 200 hybrid cells, these two genes were clearly transcribed in cells that were also expressing and localizing Xist RNA. While we were also able to detect the human RNA with the Pgk-1 probe, we included in our analysis only those signals that were closely associated with the mouse Xist RNA signal, confirming that it was transcribed from the mouse X.
To evaluate whether the Xist RNA localization resulted in a more subtle transcriptional downregulation of the genes vs. a complete silencing of the X chromosome, we compared the distribution of both Pgk1 and Zfx RNA from cells either expressing or not expressing murine Xist RNA. As exemplified in Fig. 5, A and C, the Pgk1 and Zfx RNA signals were not discernibly different in cells devoid of Xist expression vs. those in which the mouse X is painted by Xist RNA, suggesting that the murine X chromosome has not undergone detectable transcriptional downregulation as a result of Xist expression and localization. While we cannot rule out the possibility that, over the long term, progressive inactivation will occur, expression of Zfx and Pgk-1 was analyzed in subclones that expressed and localized murine Xist RNA for over 2 mo. Therefore, our results show that Xist RNA can localize to the mouse X chromosome without inactivation of specific genes for many generations. We conclude that the association of Xist RNA with chromatin is not strictly correlated with transcriptional silencing. Furthermore, these results indicate that expression, stabilization, and proper localization of Xist RNA in somatic cells is not sufficient to cause inactivation.
Discussion
Understanding the relationship of XIST RNA localization to X inactivation and the mechanisms governing localization to the chromosome is likely key to understanding the process of X inactivation itself. Results presented here show that despite a nuclear RNA accumulation of normal abundance and stability, XIST RNA does not localize to reactivated or inactive human X chromosomes in mouse/ human hybrid cells. To determine if the improper location of RNA in these hybrids explained the lack of subsequent inactivation, we created clones that stably expressed not only the human XIST, but mouse Xist as well. Distribution of the mouse Xist RNA in these hybrids clearly showed a tight association with the active murine X chromosome, in marked contrast to the nonlocalized human XIST RNA in the same cells. This important result also allows several conclusions: (a) demethylation with 5azadC does not affect the ability of Xist RNA to localize, since the murine Xist RNA does localize properly in the same cells; (b) expression of this RNA does not have to begin in early development to allow localization; and (c) Xist RNA does not discriminate between the developmentally selected active and the inactive X chromosome. Collectively, these results indicate that the most likely reason for the lack of localization to the human X chromosome is mouse/human species differences. Since completion of this work, it was shown that inactive human X chromosomes, when inserted into mouse embryonal carcinoma cells, activate expression of many genes, yet fail to stop expressing XIST RNA (Yoshida et al., 1997). Results presented here indicate that the XIST RNA may not closely associate with the chromosome in the mouse environment.
As many pre-mRNAs are most concentrated near their site of transcription (Xing and Lawrence, 1993; Xing et al., 1995), it is not clear to what extent limited localization of the XIST RNA with the X chromosome in the hybrids is simply a consequence of its transcription, or if it reflects a weak affinity for the chromosome. For example, the XIST RNA may be transcribed but unable to localize to the chromosome at all, causing the transcripts to drift away gradually into the nucleoplasm. Alternatively, the murine factors may be able to localize the human transcripts, but because of either reduced affinity or availability, allow only a small fraction of the transcripts to associate with the X chromosome.
Localization of XIST RNA Does Not Arise from Stability
Recent results showed the transition of Xist RNA from a small dot to an abundant accumulation in early development, and suggested that the increased stability allowed the transcripts to localize to the inactive X chromosome (Panning et al., 1997; Sheardown et al., 1997). Our examination of the stability of the human RNA in human/rodent hybrids indicates that the transcripts are fully stabilized despite the inability to localize to the X chromosome. Hence, localization of XIST RNA does not result from increased stability. Our results make it clear that the process required for transcript stabilization can occur after development as the half-life of XIST RNA in these hybrids is similar to that recently reported in normal diploid fibroblasts (Sheardown et al., 1997).
Differing Impact of Xist Expression Throughout Development
The observation that the chromosome is completely painted by the Xist RNA, but still shows normal expression of both Zfx and Pgk1 RNA in the same cell shows that the chromosome is still transcriptionally active, and that there is no strict correlation between Xist RNA chromosomal localization and inactivation of chromatin. We conclude that expression, stabilization, and localization of Xist RNA is not sufficient for X inactivation. Importantly, these results also suggest that there is a difference between the role of Xist/XIST in immature vs. fully differentiated cell types. In creating a system where a single copy of the Xist gene produces RNA that is stably localized in fully differentiated cells, we are able to compare our results with those that show the absolute requirement for Xist in X inactivation (Penny et al., 1996; Marahens et al., 1997) and the sufficiency of Xist with limited surrounding sequences to cause silencing (Herzing et al., 1997; Lee and Jaenisch, 1997). The critical difference is that in these previous studies, Xist is ectopically expressed in embryonic cells before differentiation, while in experiments reported here, Xist expression is forced in fully differentiated somatic cells. The contrasting results suggest that XIST RNA is fully competent to enact chromatin inactivation during embryogenesis, but later in development loses this ability either through lack of contributing factors or a change in the RNA itself.
Although we have demonstrated in this paper that association of Xist RNA with the X chromosome is not sufficient for transcriptional silencing, the supposition that localization is required for X inactivation is most likely still correct. The process of X inactivation likely involves several steps, including the expression, stabilization, and localization of Xist RNA. Our results not only demonstrate that the localization of XIST RNA with the chromosome does not necessarily culminate in transcriptional silencing, but also suggest that an additional step or mechanism is required to complete the process of X inactivation.
Potential Role of XIST RNA Localization During Development
Based on our observations, we postulate a model in which the XIST RNA is involved in the process of X inactivation, but is not directly responsible for transcriptional downregulation. In this model, we speculate that the Xist RNA serves to mark or potentiate the chromosome for a subsequent step or process. For example, the RNA, by virtue of its binding along the length of the chromosome and its affinity for certain factors, could recruit processes that ultimately result in transcriptional silencing. While there are many examples of processes that could act in concert with Xist RNA, we consider two for discussion: methylation and histone deacetylation. Both are established mechanisms of epigenetic imprinting strongly linked with inactive X chromatin; they are stably maintained and transferred to daughter cells, and are involved with sites along the entire inactive X chromosome. Furthermore, methylation (Lock et al., 1987; Keohane et al., 1996) and overall deacetylation of the inactive X (Keohane et al., 1996) occur after Xist expression early in embryogenesis. Additionally, this idea may explain why Xist is not necessary for maintaining X inactivation in somatic cells, while other results show that it is required during development. XIST RNA could be absolutely required during embryogenesis to establish the pattern of methylation or histone deacetylation by recruiting factors like methyltransferases or deactelyases to the inactive X chromosome; however, once such a pattern was established on the inactive X, then the XIST RNA would no longer be rigorously required. The constitutive expression and localization of XIST RNA (Clemson et al., 1996) in adult cells may serve as a backup mechanism to ensure that epigenetic silencing is rigorously maintained. Ultimately, this model could explain why the XIST gene could be deleted in vitro from somatic cells with no obvious effect (Brown and Willard, 1994); i.e., if the primary source of inactivation maintenance was a process prone to little error, (such as maintenance methyltransferase which has an estimated efficiency of 99.9% [Pfeifer et al., 1990]), then as a backup mechanism XIST RNA could be deleted without obvious phenotype in cultured cells. In contrast, in vivo, where even a slight failure of X inactivation would be deleterious, XIST RNA would be needed as a constant fail-safe mechanism.
Of course, if XIST/Xist initiates subsequent events that ultimately silence the chromosome, then why didn't our reactivated mouse/human hybrid system succumb to transcriptional silencing? It is possible that cells that did inactivate were selected against, allowing us only to detect cells that managed to circumvent X inactivation. This possibility appears less likely because we did not observe any evidence of cell death or change in growth rate, combined with the fact that these cells retained an active human X chromosome, which could compensate for the loss of the single mouse X. Rather, we favor the possibility that the resistance to X inactivation is due to the fact that the Xist RNA was not expressed before differentiation of these cells; e.g., it may be very difficult or impossible to establish de novo inactivation in cells after development as the required enzymes may not be available. For example, the levels and location of de novo and maintenance methyltransferases differ in early embryonic vs. terminally differentiated cells (Razin and Shemer, 1995; Turker and Bestor, 1997).
To reactivate expression of XIST/Xist, the cells were treated with a demethylating agent that can impact cell viability as well as replication timing and condensation of the inactive X chromosome (Gregory et al., 1985; Haaf et al., 1988). Additionally, the new methylation patterns may be slowly incorporated into DNA (Toth et al., 1989). Although we were able to produce clones that expressed human XIST indefinitely, we could not isolate clones that expressed murine Xist longer than several months. Hence, we cannot rule out the possibility that this was not enough time to set up adequate patterns of genomic imprinting that result in silencing of the previously active X-linked genes like Zfx and Pgk1. However, the cells were only cultured in 5azadC for a short period of time (24 h) to cause expression of the mouse or human Xist, and were subsequently cultured without additional 5azadC for as long as several months with a significant fraction (>30%) still expressing mouse Xist. Expression of the X-linked genes Zfx and Pgk1 was analyzed as much as 8 wk after 5azadC treatment, making it less likely that their continued expression is the result of demethylation. Reactivation of normally inactivated genes by 5azadC is extremely rare in hybrids (∼10−4) or normal (10−7) cells (Gartler and Goldman, 1994). It is interesting to speculate that higher susceptibility to reactivation in hybrids is related to mislocalization of XIST RNA. Since X inactivation is well-maintained in hybrids, it is unlikely that the lack of de novo X inactivation is due merely to repression of X inactivation in the mouse–human hybrid system studied here. It is clear that our reactivated hybrid system reconstitutes many steps known to be involved in normal X inactivation; namely expression, stabilization, and localization of Xist RNA. Therefore, it seems that we have created an intermediate to chromosomal inactivation that would not have been seen otherwise; one that makes it possible to analyze and dissect the separate steps involved in X inactivation.
Acknowledgments
The authors wish to thank John McNeil for his expert computer assistance, Meg Byron and Sarah Baldry for their technical assistance, and Patrick Brown for his help on several hybrid in situs.
This work was supported by National Institutes of Health grants GM49254 and GM53234 to J.B. Lawrence and Medical Research Council MT13690 (Canada) to C.J. Brown.
Abbreviations used in this paper
- 5azadC
5-azadeoxycytidine
- rt
room temperature
- RT
reverse transcription
- Xa
active X chromosome
- Xa-XIST+
reactivated X
- Xi
inactive X chromosome
- XIST/Xist
human/ mouse Xi-specific transcript
References
- Adler DA, Bressler SL, Chapman VM, Page DC, Disteche CM. Inactivation of the Zfx gene on the mouse X chromosome. Proc Natl Acad Sci USA. 1991;88:4592–4595. doi: 10.1073/pnas.88.11.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Carrel L, Willard HF. Expression of genes from the human active and inactive X chromosomes. Am J Hum Genet. 1997;60:1333–1343. doi: 10.1086/515488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Flenniken AM, Williams B, Willard HF. X chromosome inactivation of the human TIMP gene. Nucl Acids Res. 1990;18:4191–4195. doi: 10.1093/nar/18.14.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence JB, Willard HF. The Human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Willard HF. The human X inactivation center is not required for maintenance of X chromosome inactivation. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- Chariot A, Castronovo V. Detection of HOXA1 expression in human breast cancer. Biochem Biophys Res Comm. 1996;222:292–297. doi: 10.1006/bbrc.1996.0737. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche CM. Escape from X inactivation in human and mouse. Trends Genet. 1995;11(1):17–22. doi: 10.1016/s0168-9525(00)88981-7. [DOI] [PubMed] [Google Scholar]
- Duncan AM, Macdonald A, Brown CJ, Wolff D, Willard HF, Sutton B. Characterization of a small supernumerary ring X chromosome by fluorescence in situ hybridization. Am J Med Genet. 1993;47:1153–1156. doi: 10.1002/ajmg.1320470804. [DOI] [PubMed] [Google Scholar]
- Fey EG, Krochmalnic G, Penman S. The non-chromatin substructures of the nucleus: the ribonucleoprotein RNP-containing and RNP- depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. JCell Biol. 1986;102:1654–1665. doi: 10.1083/jcb.102.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartler SM, Goldman MA. Reactivation of inactive X-linked genes. Dev Genet. 1994;15:504–514. doi: 10.1002/dvg.1020150609. [DOI] [PubMed] [Google Scholar]
- Gregory P, Greene C, Shapira E, Wang E. Alterations in the time of X chromosome replication induced by 5-azacytidine in a patient with 48,XXXY/47,XXY. Cytogenet Cell Genet. 1985;39:234–236. doi: 10.1159/000132142. [DOI] [PubMed] [Google Scholar]
- Haaf T, Ott G, Schmid M. Inhibition of condensation in the late-replicating X chromosome induced by 5-azadeoxycytidine in human lymphocyte cultures. Hum Genet. 1988;79:18–23. doi: 10.1007/BF00291703. [DOI] [PubMed] [Google Scholar]
- Hansen RS, Canfield T, Stanek A, Keitges E, Gartler S. Reactivation of XIST in normal fibroblasts and a somatic cell hybrid: Abnormal localization of XIST RNA in hybrid cells. Proc Natl Acad Sci USA. 1998;95:5133–5138. doi: 10.1073/pnas.95.9.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzing LB, Romer JT, Horn JM, Ashworth A. Xist has properties of the X-chromosome inactivation centre. Nature. 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- Johnson CV, Singer RH, Lawrence JB. Fluorescent detection of nuclear RNA and DNA: Implication for genome organization. Methods Cell Biol. 1991;35:73–99. [PubMed] [Google Scholar]
- Kay GF, Barton SC, Surani MA, Rastan S. Imprinting and X chromosome counting mechanisms determine Xist expression in early mouse development. Cell. 1994;77:639–650. doi: 10.1016/0092-8674(94)90049-3. [DOI] [PubMed] [Google Scholar]
- Kay GF, Penny GD, Patel D, Ashworth A, Brockdorff N, Rastan S. Expression of Xist during mouse development suggests a role in the initiation of X chromosome inactivation. Cell. 1993;72:171–182. doi: 10.1016/0092-8674(93)90658-d. [DOI] [PubMed] [Google Scholar]
- Keohane AM, O'Neill LP, Belyaev ND, Lavender JS, Turner BM. X-inactivation and histone H4 acetylation in embryonic stem cells. Dev Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH, Marselle LM. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell. 1989;57:493–502. doi: 10.1016/0092-8674(89)90924-0. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Villnave CA, Singer RH. Sensitive high-resolution chromatin and chromosome mapping in situ: presence and orientation of two closely integrated copies of EBV in a lymphoma line. Cell. 1988;52:51–61. doi: 10.1016/0092-8674(88)90530-2. [DOI] [PubMed] [Google Scholar]
- Lee JT, Jaenisch R. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature. 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- Lee JT, Strauss WM, Dausman JA, Jaenisch R. A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell. 1996;86:83–94. doi: 10.1016/s0092-8674(00)80079-3. [DOI] [PubMed] [Google Scholar]
- Lock LF, Takagi N, Martin GR. Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell. 1987;48:39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- Lyon M. Gene action in the X-chromosome of the mouse (Mus musculus L.) . Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Marahens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Panning B, Dausman J, Jaenisch R. X chromosome inactivation is mediated by XistRNA stabilization. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
- Panning B, Jaenisch R. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 1996;10:1991–2002. doi: 10.1101/gad.10.16.1991. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for XIST in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Steigerwald SD, Hansen RS, Gartler SM, Riggs AD. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc Natl Acad Sci USA. 1990;87:8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rack KA, Chelly J, Gibbons RJ, Rider S, Benjamin D, Lafreniere RG, Oscier D, Hendriks RW, Craig IW, Willard HF, et al. Absence of the XIST gene from late-replicating isodicentric X chromosomes in leukaemia. Hum Mol Genet. 1994;3:1053–1059. doi: 10.1093/hmg/3.7.1053. [DOI] [PubMed] [Google Scholar]
- Razin A, Shemer R. DNA methylation patterns in the mammalian genome. Hum Mol Genet. 1995;4:1751–1755. doi: 10.1093/hmg/4.suppl_1.1751. [DOI] [PubMed] [Google Scholar]
- Sheardown SA, Duthie SM, Johnston CM, Newall AET, Formstone EJ, Arkell RM, Nesterova TB, Alghisi GC, Rastan S, Brockdorff N. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell. 1997;91:99–107. doi: 10.1016/s0092-8674(01)80012-x. [DOI] [PubMed] [Google Scholar]
- Sympson CJ, Geoghan TE. Actin gene expression in murine erythroleukemia cells treated with cytochalasin D. Exp Cell Res. 1990;189:28–32. doi: 10.1016/0014-4827(90)90252-6. [DOI] [PubMed] [Google Scholar]
- Tinker AV, Brown CJ. Induction of XIST expression from the human active X chromosome in mouse/human somatic cell hybrids by disruption of methylation. Nucl Acids Res. 1998;26:2935–2940. doi: 10.1093/nar/26.12.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth M, Lichtenberg U, Doerfler W. Genomic sequencing reveals a 5-methylcytosine-free domain in active promoters and the spreading of preimposed methylation patterns. Proc Natl Acad Sci USA. 1989;86:3728–3732. doi: 10.1073/pnas.86.10.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turker MS, Bestor TH. Formation of methylation patterns in the mammalian genome. Mutat Res. 1997;386:119–130. doi: 10.1016/s1383-5742(96)00048-8. [DOI] [PubMed] [Google Scholar]
- Willard HF, Brown CJ, Carrel L, Hendrich B, Miller AP. Epigenetic and chromosomal control of gene expression: molecular and genetic analysis of X chromosome inactivation. Cold Spring Harbor Symp Quant Biol. 1993;58:315–322. doi: 10.1101/sqb.1993.058.01.037. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Dobner PR, Lawrence JB. Higher level organization of individual gene transcription and RNA splicing. Science. 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Moen PT, McNeil JA, Lawrence JB. Nonrandom gene organization: Structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Lawrence JB. Nuclear RNA tracks: structural basis for transcription and splicing? . Trends Cell Biol. 1993;3:346–353. doi: 10.1016/0962-8924(93)90105-a. [DOI] [PubMed] [Google Scholar]
- Yoshida I, Nishita Y, Mohandas TK, Takagi N. Reactivation of an inactive human X chromosome introduced into mouse embryonal carcinoma cells by microcell fusion with persistent expression of XIST. Exp Cell Res. 1997;230:208–219. doi: 10.1006/excr.1996.3393. [DOI] [PubMed] [Google Scholar]