Abstract
A feedback control mechanism, or cell cycle checkpoint, delays the onset of anaphase until all the chromosomes are correctly aligned on the mitotic spindle. Previously, we showed that the murine homologue of Bub1 is not only required for checkpoint response to spindle damage, but also restrains progression through a normal mitosis (Taylor, S.S., and F. McKeon. 1997. Cell. 89:727–735). Here, we describe the identification of a human homologue of Bub3, a 37-kD protein with four WD repeats. Like Bub1, Bub3 localizes to kinetochores before chromosome alignment. In addition, Bub3 and Bub1 interact in mammalian cells. Deletion mapping was used to identify the domain of Bub1 required for binding Bub3. Significantly, this same domain is required for kinetochore localization of Bub1, suggesting that the role of Bub3 is to localize Bub1 to the kinetochore, thereby activating the checkpoint in response to unattached kinetochores. The identification of a human Mad3/Bub1-related protein kinase, hBubR1, which can also bind Bub3 in mammalian cells, is described. Ectopically expressed hBubR1 also localizes to kinetochores during prometaphase, but only when hBub3 is overexpressed. We discuss the implications of the common interaction between Bub1 and hBubR1 with hBub3 for checkpoint control.
Keywords: hBub3, mitosis, cell cycle, checkpoint, anaphase
Before cell division, the replicated genome must be accurately segregated to ensure the continued growth and development of the daughter cells. To maintain the fidelity of chromosome segregation, eukaryotes have evolved a feedback control mechanism, often referred to as a cell cycle checkpoint (Hartwell and Weinert, 1989), which delays the onset of anaphase until all the chromosomes are correctly aligned on the microtubule spindle apparatus.
Evidence that the mitotic checkpoint monitors chromosome alignment has been mounting for many years. It has long been appreciated that insect spermatocytes remain blocked at the first meiotic metaphase if one or more chromosomes are not correctly attached to the meiotic spindle (Callan and Jacobs, 1957). In addition, analyses of mitotic newt heart cells suggested that the arrival of the last kinetochore at the metaphase plate may play a critical role in regulating anaphase onset (Zirkle, 1970a ,b).
More recently, it has been shown that anaphase is inhibited until all the chromosomes achieve bipolar attachments (Rieder et al., 1994) and that unattached kinetochores generate the inhibitory signal (Rieder et al., 1995). Micromanipulation experiments with insect spermatocytes suggest that this inhibitory signal is generated by tension-sensitive kinases and/or phosphatases located at the kinetochore (Nicklas, 1997). When a chromosome is not attached to the spindle, or only mono-oriented, the kinetochores are not under tension. In the absence of tension, kinetochores express the 3F3/2 phosphoepitope, suggesting that a kinase is active, and anaphase is delayed (Gorbsky and Ricketts, 1993; Li and Nicklas, 1995, 1997; Nicklas et al., 1995). Upon achieving a bipolar attachment, kinetochores experience tension due to opposing spindle forces. When under tension, kinetochores lack the 3F3/2 phosphoepitope, suggesting a loss of kinase activity, and anaphase ensues. Significantly, microinjection of the 3F3/2 antibody into living cells prevents kinetochore dephosphorylation and delays the onset of anaphase (Campbell and Gorbsky, 1995).
Although the evidence implicating tension as the regulator of anaphase onset in insect spermatocytes is compelling (Nicklas, 1997), it is not known whether tension is the key factor in other cell types. Furthermore, it is not known whether the effect of tension is direct. Tension at the kinetochore is known to stabilize microtubule attachment (Nicklas and Koch, 1969) and hence microtubule attachment may be the event that directly regulates anaphase. Indeed, in mitotic PtK1 cells, laser ablation of the distal, unattached kinetochore of a mono-oriented chromosome resulted in a timely anaphase, suggesting that microtubule attachment, not tension, regulates anaphase (Rieder et al., 1995). However, it has been suggested that, in these irradiated cells, the remaining attached kinetochore is still under tension due to polar ejection forces (Rieder et al., 1995).
Formal demonstration that the onset of anaphase is regulated by a cell cycle checkpoint, as defined by Hartwell and Weinert (1989), came with the identification of several Saccharomyces cerevisiae mutants that failed to undergo mitotic arrest in response to spindle damage (Hoyt et al., 1991; Li and Murray, 1991). Genetic screens identified BUB 1, 2, and 3 (Hoyt et al., 1991) and MAD 1, 2, and 3 (Li and Murray, 1991) as necessary for the preanaphase delay in response to spindle damage. Mad1, a phosphoprotein that can bind Mad2, is hyperphosphorylated in response to spindle damage (Hardwick and Murray, 1995). This phosphorylation appears to be carried out by Mps1 (Hardwick et al., 1996), a protein kinase required for spindle pole body duplication and checkpoint response to spindle damage (Weiss and Winey, 1996). Bub1 is a protein kinase that can bind and phosphorylate Bub3 (Roberts et al., 1994; Farr and Hoyt, 1998), and appears to act upstream of Mad1 (Hardwick and Murray, 1995). Both Mad3, which shares homology with the NH2-terminal, noncatalytic domain of Bub1, and Bub2 appear to act downstream of Mad1 phosphorylation (Hardwick and Murray, 1995).
One of the downstream targets of the mitotic checkpoint is likely to be the anaphase-promoting complex (APC)1 or cyclosome (Irniger et al., 1995; King et al., 1995; Sudakin et al., 1995; Tugendreich et al., 1995). Activation of APC is required to initiate both the onset of anaphase, by degrading inhibitors of sister chromatid separation such as Pds1 and Cut2 (Cohen-Fix et al., 1996; Funabiki et al., 1996), and the exit from mitosis, by degrading mitotic cyclins and the spindle component, Ase1 (Juang et al., 1997). Recently, Mad2 has been shown to interact with APC (Li et al., 1997; He et al., 1997; Hwang et al., 1998; Kim et al., 1998) and repress its activity by inhibiting cdc20, an APC activator (Visintin et al., 1997; Hwang et al., 1998; Kim et al., 1998). Although these observations link the checkpoint pathway to the cell cycle machinery that initiates anaphase, the mechanism by which Mad2 inhibits APC remains unclear. In addition, how spindle events regulate Mad2 activity remains uncertain.
The BUB and MAD genes are also required for the mitotic delay induced by mutations in centromeric DNA sequences and kinetochore proteins (Wang and Burke, 1995; Pangilinan and Spencer, 1996; Wells and Murray, 1996). These observations suggested that the Mad and Bub proteins may play a role in the kinetochore attachment checkpoint described in larger eukaryotes. Additional support for this hypothesis came with the identification of vertebrate homologues of Mad2 (Chen et al., 1996; Li and Benezra, 1996), which localize to kinetochores of chromosomes before alignment on the metaphase spindle. Indeed, XMad2 is required to maintain MPF activity in a Xenopus egg extract system that reconstitutes the mitotic checkpoint (Chen et al., 1996), and electroporation of anti-HsMad2 antibodies into HeLa cells inhibited mitotic arrest in response to spindle disruption (Li and Benezra, 1996).
The murine homologue of Bub1 also localizes to the kinetochores of unaligned chromosomes (Taylor and McKeon, 1997). Expression of a dominant-negative mBub1 mutant in HeLa cells compromised their ability to maintain mitotic arrest in response to spindle damage. In addition, disrupting Bub1 function accelerated passage through a normal mitosis, suggesting that Bub1 monitors the interactions between microtubules and kinetochores during every cell cycle (Taylor and McKeon, 1997). To further characterize how Bub1 activity regulates mitotic progression, we set out to analyze proteins that interact with Bub1. In S. cerevisiae, Bub3 binds to and is a substrate of Bub1 (Roberts et al., 1994). Here we describe the identification of a human homologue of Bub3, which appears to be required for kinetochore localization of Bub1. In addition, we describe a human protein that shares significant homology with the NH2-terminal domain of Mad3 from budding yeast (ScMad3). However, unlike ScMad3, this human protein has a COOH-terminal extension that contains a protein kinase domain, suggesting that it is a Bub1-related protein. This human Mad3/Bub1-related protein, hBubR1, also binds to hBub3 and can localize to kinetochores during prometaphase. The implications of the common interaction between Bub1 and BubR1 with Bub3 are discussed.
Materials and Methods
cDNA Cloning and Manipulation
Human expressed sequence tags (ESTs) with homology to ScBub3 (EMBL/GenBank/DDBJ accession number H83201; WashU-Merck EST Project, St. Louis, MO), ScMad3 (accession number AA314793; Adams et al., 1995), and mBub1 (accession number R94348; WashU-Merck EST Project) were identified by searching the National Center for Biotechnology Information dEST databases using the BLAST algorithm (Altschul et al., 1990). Based on the EST sequences, the following oligonucleotides were designed and used to screen pools of cDNAs by PCR: hBub3 Fwd 5′ GTAACAGCTGCTGGGATTACACATGGC 3′(This is a vector- derived sequence and was used in a vector-insert PCR screen); hBub3 Rvs 5′ CTCCCTGCGCTGCTGCACGTAACC 3′; hBubR1 Fwd 5′ TAGCTC- CGAGGGCAGGTTGC 3′; hBubR1 Rvs 5′ CATAAACGCCCTAAT- TTAAGCCAGAG 3′; hBub1 Fwd 5′ACGTATGGGGCCAAGTGT- AGG 3′; hBub1 Rvs 5′ GTCTGTTACTGTCTGGGCTTTCAA 3′. Positive subpools were screened by hybridization using either the ESTs (hBub3 and hBub1) or the PCR product (hBubR1) as probes. cDNAs were constructed using the SuperScript plasmid system for cDNA synthesis and plasmid cloning kit (GIBCO BRL, Gaithersburg, MD). mRNA was purified from the U2OS osteosarcoma cell line using the FastTrack Kit (Invitrogen, Carlsbad, CA). PCR reactions and filter hybridizations were done according to standard methods (Sambrook et al., 1989). cDNA inserts were sequenced on both strands using an ABI 373A sequencer (BioPolymers Facility, Howard Hughes Medical Institute, Harvard Medical School, Boston, MA). Sequence alignments were performed using the MegAlign software (DNAStar, Madison, WI). The expression vectors were constructed by directly subcloning cDNA fragments and/or PCR amplification using Pfu polymerase (Stratagene, La Jolla, CA). All the expression vectors are based on pcDNA-3 (Invitrogen), modified to include the 5′ untranslated sequence from the human lamin A cDNA and NH2-terminal Myc, green fluorescent protein, or GST tags. Mutagenesis of hBub3 was performed using the GeneEditor kit (Promega Corp., Madison, WI) according to the manufacturer's instructions. The hBub3ΔVAVE mutant was constructed using the following oligonucleotide: 5′ TCT ATT GAA GGC CGA AGA TCT TAT TTG GAC CCA AGC 3′, which results in a substitution of four amino acids (217 VAVE 220) with two amino acids (217 RS 218), thus introducing a novel BglII restriction site.
Cell Culture and Transfections
BHK cells were cultured in a humidified incubator at 37°C plus 5% CO2 in Dulbecco's modified Eagle's media supplemented with 10% fetal calf serum (HyClone, Logan, UT), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM glutamine. All media reagents were from GIBCO BRL unless stated otherwise. Transient transfections for immunofluorescence analysis were performed on 18-mm coverslips using the calcium phosphate method as described (Heald et al., 1993). Transient transfections for purification of GST-hBub3 complexes were performed using Lipofectamine (GIBCO BRL). Briefly, 1.6 × 105 BHK cells were plated in 60-mm dishes, cultured for 24 h, and then washed three times with serum-free media. 1 μg of DNA, purified by two rounds of cesium chloride/ethidium bromide density centrifugation, was complexed with 16 μg of Lipofectamine in serum-free media at room temperature for 20 min and then added to the cells in a final volume of 1 ml of serum-free media for 16 h. The cells were then fed with media plus serum and cultured for a further 24 h.
Immunofluorescence
24 h after transfection, BHK cells were fixed for 5 min in 1% formaldehyde, freshly diluted from a 37% stock (J.T. Baker, Phillipsburg, NJ) in PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4). After fixation, cells were washed three times in PBST (PBS plus 0.1% Triton X-100) and then blocked in PBST plus 5% nonfat dried milk (NFDM) for 30 min. To detect Myc-tagged proteins, the cells were incubated with the 9E10 monoclonal antibody diluted 1:500 in PBST plus 5% NFDM for 30 min. After three washes in PBST, the cells were incubated with a Cy3-conjugated donkey anti–mouse secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) diluted 1:500 in PBST plus 5% NFDM for 30 min. After three washes in PBST the chromatin was stained for 1 min with Hoechst 33258 at 1 μg/ml in PBST. The coverslips were then inverted on microscope slides using 90% glycerol plus 10% 0.2M Tris-HCl, pH 7.5, as mounting media. Kinetochores were labeled using a CREST autoimmune serum diluted 1:1,000 and a Cy3-conjugated donkey anti–human secondary antibody (Jackson ImmunoResearch Laboratories, Inc.). Cells were examined on an Axioplan 2 microscope (Carl Zeiss, Inc., Thornwood, NY) equipped with fluorescence using an oil immersion 63× objective lens. Images were captured using a Sony color video camera (Park Ridge, NJ) driven by the Northern Exposure software (Empix Imaging, Inc., Mississauga, ON), and processed for printing using Adobe Photoshop (Adobe Systems, Inc., San Jose, CA).
Purification and Analysis of GST–hBub3 Complexes
Soluble protein lysates were prepared by scraping transfected cells in 500 μl of lysis buffer (100 mM NaCl, 50 mM Hepes, pH 7.4, 20 mM β-glycerophosphate, 1 mM sodium vanadate, 10 mM EDTA, 10 mM EGTA, 1 mM DTT, 1 mM PMSF, 1 μg/ml antipain, 1 μg/ml aprotinin, 5 μg/ml bestatin, 5 μg/ml chymostatin, 2 μg/ml leupeptin, and 1 μg/ml pepstatin), followed by centrifugation at 14,000 g for 10 min at 4°C. To purify GST–hBub3 complexes, 20 μl of glutathione Sepharose beads (Pharmacia Biotech, Inc., Piscataway, NJ), equilibrated in lysis buffer and resuspended as a 50% slurry, were added to the lysates and incubated at 4°C for 30 min. After five washes in lysis buffer, proteins were eluted off the beads by boiling in SDS sample buffer, resolved by SDS-PAGE, and electroblotted onto Immobilon-P membranes (Millipore Corp., Waters Chromatography, Milford, MA). GST–hBub3 and Myc-tagged proteins were then labeled using a rabbit polyclonal antiserum containing both anti-GST and anti-Myc antibodies diluted at 1:1,000 in TBST (50 mM Tris, pH 7.6, 150 mM NaCl, 0.1% Tween-20) plus 5% NFDM for 1 h at room temperature. After washing in TBST, bound primary antibodies were labeled with a horseradish peroxidase-conjugated goat anti–rabbit antibody (Zymed Labs, Inc., South San Francisco, CA) diluted 1:500 in TBST plus 5% NFDM. After washing in TBST, bound secondary antibodies were detected using the SuperSignal chemiluminescence system (Pierce Chemical Co., Rockford, IL). To perform in vitro kinase assays, GST–mBub1 and GST– hBubR1 were expressed and purified as described above for GST–hBub3. After washing in lysis buffer, the beads were equilibrated in kinase buffer (50 mM NaCl, 50 mM Hepes, pH 7.4, 20 mM β-glycerophosphate, 10 mM MgCl2, 10 mM MnCl2, 1 mM sodium vanadate, 1 mM DTT, and 1 mM PMSF) and then incubated with 10 μCi 32P γ-ATP (ICN Biomedicals, Inc., Irvine, CA) at 30°C for 30 min. The beads were then washed three times in lysis buffer and bound proteins were eluted by boiling in SDS sample buffer. Eluted proteins were resolved by SDS-PAGE, exposed to a PhosphorImager screen, and analyzed using ImageQuant software (both from Molecular Dynamics, Inc., Sunnyvale, CA).
Results
Structural Comparison of Bub3 Homologues
A screen of EST databases identified a partial human cDNA with homology to the S. cerevisiae protein Bub3 (ScBub3) (Hoyt et al., 1991). We isolated a corresponding full-length human cDNA (hBub3, EMBL/GenBank/ DDBJ accession number AF053304) with a 984-bp open reading frame encoding a 328-amino acid protein with a predicted molecular mass of ∼37 kD. hBub3 shares ∼34% identity and 69% similarity with ScBub3 throughout the entire protein. Both ScBub3 and hBub3 contain four WD repeats, three in the NH2-terminal region and one towards the COOH terminus (Fig. 1). hBub3 also shares significant homology with the Rae1 proteins from human (Bharathi et al., 1997), S. cerevisiae (Murphy et al., 1996) and Schizosaccharomyces pombe (Brown et al., 1995), which have been implicated in nucleocytoplasmic transport. An alignment of the Bub3 and Rae1 homologues (Fig. 1) indicates that the Bub3 and Rae1 proteins represent two separate families.
Figure 1.

Sequence comparison of Bub3 homologues. Schematic representation of hBub3 showing the location of the four WD repeats and an alignment of the Bub3 and Rae1 homologues from human (h), S. cerevisiae (Sc) and S. pombe (Sp). The sequences are arranged such that the WD repeats are aligned with each other and with the WD repeat consensus (Dalrymple et al., 1989). The positions of WD repeat consensus residues are indicated with a +. Note the conserved glycine in the Rae1 homologues, marked with an *, that is not conserved in the Bub3 homologues. The rae1-1 loss of function allele in S. pombe results from a substitution of this glycine with a glutamic acid (Brown et al., 1995). The four amino acids deleted to generate the hBub3ΔVAVE mutant are identified with a solid bar.
hBub3 Localizes to Kinetochores before Chromosome Alignment
To determine the subcellular localization of hBub3, the DNA corresponding to the open reading frame was cloned into a GFP-tagged expression vector and transiently transfected into BHK cells. After fixation, chromatin was stained with Hoechst dye and kinetochores labeled with CREST autoantibodies. During interphase, the GFP fluorescence was diffusely localized in the nucleus (Fig. 2 A). During prophase and prometaphase, GFP fluorescence was concentrated in pairs of foci that colocalized with the CREST antigens (Fig. 2, B and C). During metaphase and anaphase, the GFP fluorescence was distributed throughout the cell, whereas the CREST antigens remained associated with the chromatin (Fig. 2, D and E). Based on these observations, we conclude that hBub3 localizes to kinetochores during prophase and prometaphase, but not during and after metaphase. This kinetochore localization pattern is very similar to that of murine Bub1 (Taylor and McKeon, 1997).
Figure 2.
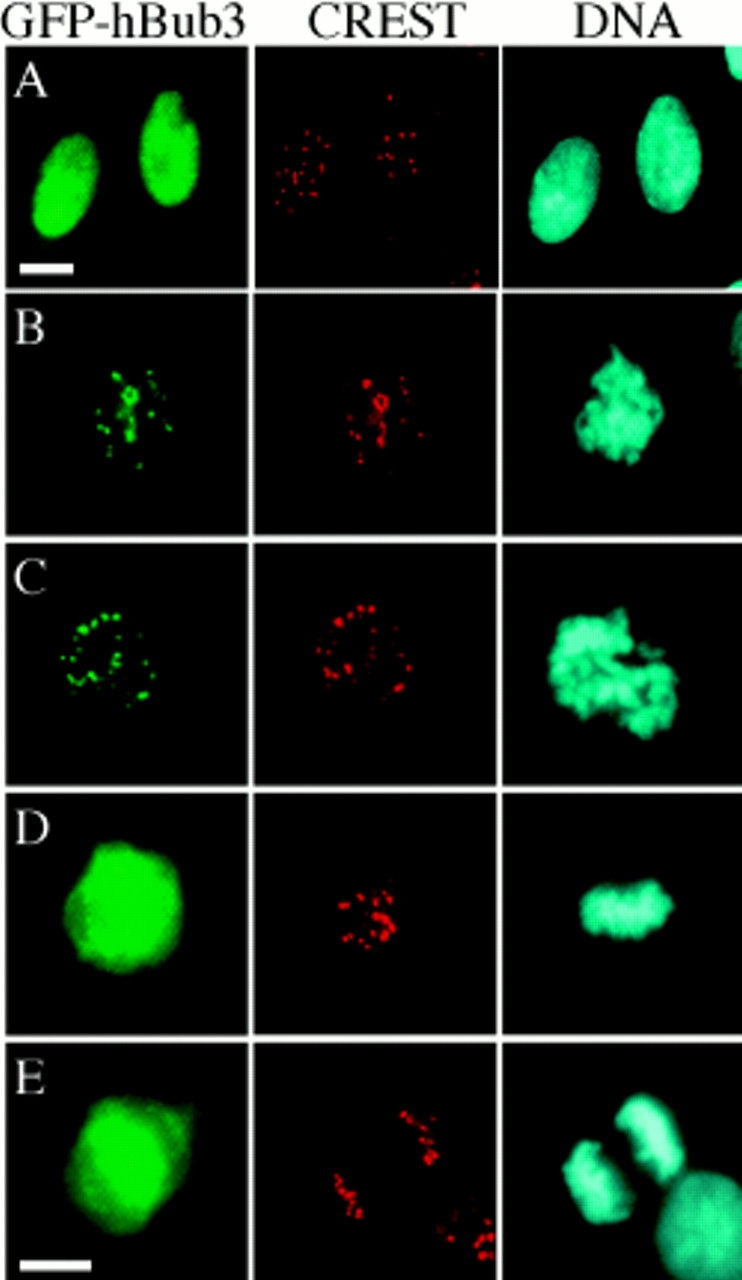
hBub3 localizes to kinetochores during prometaphase. BHK cells were transfected with a GFP–hBub3 expression construct, fixed, and then stained with a CREST antiserum to identify the kinetochores (red) and Hoechst dye to visualize the chromatin (blue). GFP fluorescence is shown in green. (A) Transfected cells showing that hBub3 is diffusely nuclear during interphase. (B) Transfected prophase and (C) prometaphase cells showing colocalization of GFP–hBub3 with kinetochores. (D) Transfected metaphase and (E) anaphase cells showing GFP–hBub3 diffusely distributed throughout the cell. Bars, 10 μm. B–D are to the same scale as E.
A Domain in the NH2 Terminus of mBub1 Binds hBub3
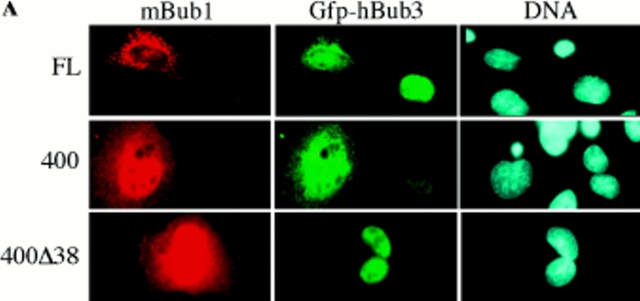
In interphase cells, ectopically expressed mBub1 is predominately cytoplasmic (Taylor and McKeon, 1997) whereas hBub3 is nuclear (Fig. 2 A). These different localization patterns provided an opportunity to test whether Bub3 and Bub1 associate in mammalian cells. We reasoned that if these two proteins interacted, the localization pattern of one or the other may be altered when coexpressed. The top row in Fig. 3 A shows two cells expressing hBub3, only one of which is expressing mBub1. In the cell not expressing mBub1, hBub3 is exclusively nuclear. However, in the cell expressing mBub1, hBub3 localizes to both the cytoplasm and nucleus. This cytoplasmic sequestration of hBub3 was dependent on the amount of mBub1 cotransfected, but was not observed when a control plasmid was cotransfected (Fig. 3 B).
Figure 3.
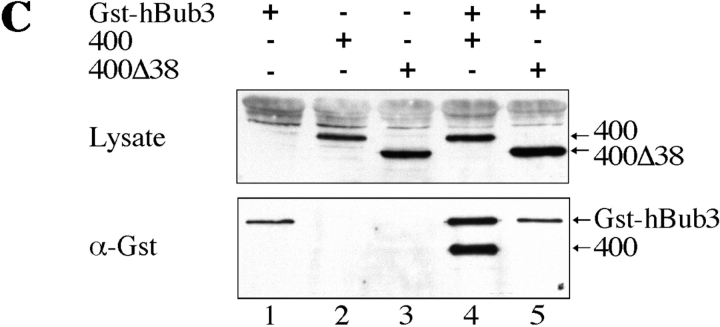
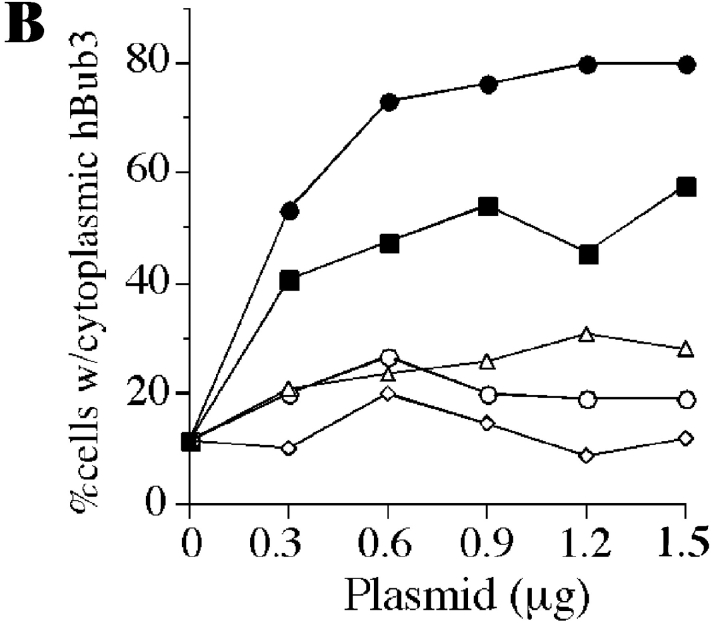
A domain in the NH2-terminus of mBub1 binds hBub3. (A) BHK cells were cotransfected with GFP–hBub3 and Myc-tagged mBub1 expression constructs. After fixation, cells were stained with the 9E10 monoclonal antibody (red) to localize mBub1 and Hoechst dye to visualize the chromatin (blue). GFP fluorescence is shown in green. Coexpression of both full-length mBub1 (top row) and the NH2-terminal domain (NH2-mBub1[400], middle row) results in a cytoplasmic sequestration of GFP–hBub3. In contrast, NH2-mBub1(400Δ38) does not sequester GFP–hBub3 in the cytoplasm (bottom row). (B) BHK cells were cotransfected with 0.5 μg of GFP–hBub3 and increasing amounts of full-length mBub1 (solid squares), the NH2-terminal domain (amino acids 1–331) (solid circles), the central domain (amino acids 332–731) (open triangles), the COOH-terminal kinase domain (amino acids 732–1,058) (open circles) or a control plasmid (open diamonds). The total amount of DNA used in each transfection was maintained at 2.0 μg by addition of carrier DNA where necessary. The percentage of cells with cytoplasmic GFP–hBub3 was scored and plotted against the amount of cotransfected mBub1. Only coexpression of full-length mBub1 and the NH2-terminal domain resulted in a dose-dependent, cytoplasmic sequestration of GFP–hBub3. (C) BHK cells were transfected with GST–hBub3 (lanes 1, 4, and 5) and/or Myc-tagged NH2-mBub1(400) (lanes 2 and 4) and/or Myc-tagged NH2-mBub1(400Δ38) (lanes 3 and 5). Proteins from cell lysates (top) and purified GST–hBub3 complexes (bottom) were resolved by SDS-PAGE, transferred to a membrane, and then probed with anti-GST and anti-Myc antibodies. In the absence of GST–hBub3, neither NH2-mBub1(400) nor NH2-mBub1(400Δ38) purify with the glutathione beads (lanes 2 and 3). In the presence of GST–hBub3, NH2-mBub1(400) does purify with the glutathione beads (lane 4) but NH2-mBub1(400Δ38) does not (lane 5). Note that GST–hBub3 cannot be identified in the cell lysates due to several comigrating background bands.
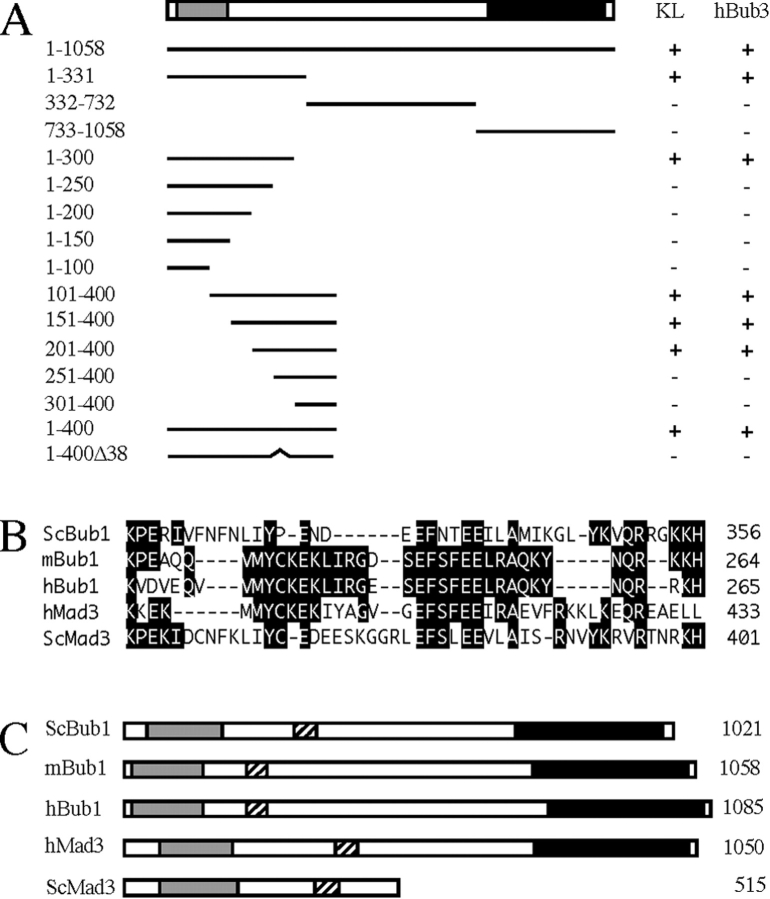
To determine which domain of mBub1 was responsible for the cytoplasmic sequestration of hBub3, a series of mBub1 deletion mutants were cotransfected with hBub3. When the three major subdomains of mBub1 were assayed (Fig. 4 A), only the NH2-terminal domain (amino acids 1–331) was capable of sequestering hBub3 in the cytoplasm in a dose-dependent manner (Fig. 3 B). Further deletion mapping of the NH2-terminal domain of mBub1 identified a region within amino acids 201–300 as being required for localizing hBub3 in the cytoplasm (Fig. 4 A). Closer inspection of the amino acid sequence in this region identified a domain that shows significant homology between the Bub1 proteins from mouse, human, and S. cerevisiae. (Fig. 4 B). This 37-amino acid region of mBub1 shares ∼36% identity with the corresponding domain in ScBub1. To test whether this domain is required for the Bub1/Bub3 interaction, we constructed a mutant lacking this region (NH2-mBub1[400Δ38]). This deletion abolished the ability of the NH2 terminus of mBub1 to sequester hBub3 in the cytoplasm (Fig. 3 A).
Figure 4.
The hBub3-binding domain and kinetochore localization domain of mBub1 overlap. (A) A schematic representation of mBub1 and the Myc-tagged deletion mutants tested for kinetochore localization and GFP–hBub3 interaction after transient transfection of BHK cells. The lightly shaded box represents the NH2-terminal homology domain defined previously (Taylor and McKeon, 1997), whereas the darkly shaded box represents the COOH-terminal kinase domain. A + under KL indicates that kinetochore localization of the Myc epitope was observed during prometaphase. A + under hBub3 indicates that cytoplasmic sequestration of GFP–hBub3 was observed in interphase cells. The numbers on the right correspond to the amino acids expressed by each construct. This analysis shows that amino acids 201–300 are required for both kinetochore localization and hBub3 binding. (B) A sequence alignment of the Bub1 and Mad3-related proteins within the Bub3-binding domain. Deletion of this domain from mBub1 and hBubR1 abolishes hBub3 binding (refer to Figs. 3 and 7). (C) Schematic representations of the Bub1 and Mad3-related proteins showing the relative positions of the NH2-terminal homology domains (lightly shaded boxes), the kinase domains (darkly shaded boxes), and the Bub3-binding domains (hatched boxes).
To test whether the cytoplasmic sequestration of hBub3 reflects a physical interaction between the two proteins, hBub3 was expressed as a GST fusion protein in BHK cells along with the NH2-terminal 400 amino acids of mBub1 (NH2-mBub1[400]). GST-hBub3 was then purified from cell lysates by affinity chromatography using glutathione–Sepharose beads and the affinity complexes were analyzed by Western blotting. NH2-mBub1(400) clearly purifies with the glutathione beads in a GST–hBub3-dependent manner (Fig. 3 C, lane 4). In contrast, NH2-mBub1(400Δ38), which does not sequester hBub3 in the cytoplasm, does not copurify with GST–hBub3 (Fig. 3 C, lane 5). Based on these observations, we conclude that Bub1 and Bub3 can bind in mammalian cells, and that a 38-amino acid domain in the NH2 terminus, which is conserved among the Bub1 and Mad3-related proteins, is required for this interaction.
The Bub3-binding Domain of Bub1 Is Also Required for Kinetochore Localization
We previously showed that the kinetochore localization domain of mBub1 resides within the NH2-terminal 331 amino acids (Taylor and McKeon, 1997). To further define the kinetochore localization domain, we assayed the deletion mutants shown in Fig. 4 A for kinetochore localization during mitosis by transient transfection into BHK cells. This analysis defined a region within amino acids 201–300 as being required for kinetochore localization, suggesting that the hBub3-binding domain and the kinetochore localization domain of mBub1 may overlap (Fig. 4 A). Significantly, the 38-amino acid deletion that abolished the ability of mBub1 to interact with hBub3 also abolished its ability to localize to the kinetochore. These observations suggest that the Bub1/Bub3 interaction is required for localizing Bub1 to the kinetochore in mitosis.
The Bub3-binding Domain Is Conserved in hBubR1
The NH2 terminus of Mad3 from S. cerevisiae (ScMad3, EMBL/GenBank/DDBJ accession number Z49288) shares significant homology with the NH2-terminal domain of Bub1 (Hardwick, K.G., and A.W. Murray, personal communication) (refer to Fig. 4 C and see Fig. 5 A). Interestingly, ScMad3 also shares homology with Bub1 within the Bub3-binding domain (Fig. 4 B). This observation tempted us to speculate that perhaps Mad3 may also bind Bub3. To test this possibility, we set out to clone a mammalian homologue of ScMad3. A screen of EST databases identified a partial human cDNA with homology to the ScMad3. Using this EST, we isolated a corresponding full-length cDNA (EMBL/GenBank/DDBJ accession number AF053306) with an open reading frame of 3,153 bp, encoding a 1,050-amino acid protein with a predicted molecular weight of ∼119 kD.
Figure 5.

Sequence comparison of Bub1 and Mad3-related proteins. (A) Sequence alignment of the NH2-terminal domains of the Bub1 and Mad3-related proteins from S. cerevisiae (Sc), mouse (m), and human (h). (B) Sequence alignment of the COOH-terminal kinase domains of the Bub1-related proteins. The 12 subdomains conserved among protein kinases (Hanks and Hunter, 1995) are underlined.
The NH2-terminal domain of this human Mad3-like protein (Fig. 4 C, lightly shaded box) shares ∼26% identity with ScBub1 and 35% identity with ScMad3 (Fig. 5 A). However, unlike ScMad3, this human protein has a COOH-terminal kinase domain (Fig. 4 C). Although this kinase domain shares homology with the Bub1 kinase domains, it is clearly distinct (Fig. 5 B). For example, whereas ScBub1 and mBub1 share 33% identity in the kinase domain, ScBub1 and the human Mad3/Bub1-related protein share only 20% identity. Furthermore, mBub1 is more closely related to ScBub1 (33% identity) than it is to the human Mad3/Bub1-related protein (26% identity) in the kinase domain. These observations suggest that this human protein is perhaps not an additional member of the Bub1 family, but more likely a Mad3-related protein. This human Mad3/Bub1-related kinase is identical to a Bub1-related protein recently described by Cahill et al. (1998). Therefore, in an effort to maintain a consistent nomenclature, we will refer to this Mad3/Bub1-related protein as hBubR1.
Exogenous hBubR1 Localizes to the Kinetochore Only When Bub3 Is Overexpressed
To test whether hBubR1 localizes to kinetochores, the open reading frame of its cDNA was cloned into an Myc-tagged mammalian expression vector and transiently transfected into BHK cells. During interphase, hBubR1 was predominantly cytoplasmic (Fig. 6 A). During mitosis, hBubR1 was diffusely distributed throughout the cell (Fig. 6 C), although occasionally, very faint kinetochore staining was observed during prometaphase (data not shown). Note that mBub1 is also cytoplasmic when ectopically expressed in interphase BHK cells, but exhibits clear kinetochore staining in transfected prometaphase cells (Taylor and McKeon, 1997).
Figure 6.
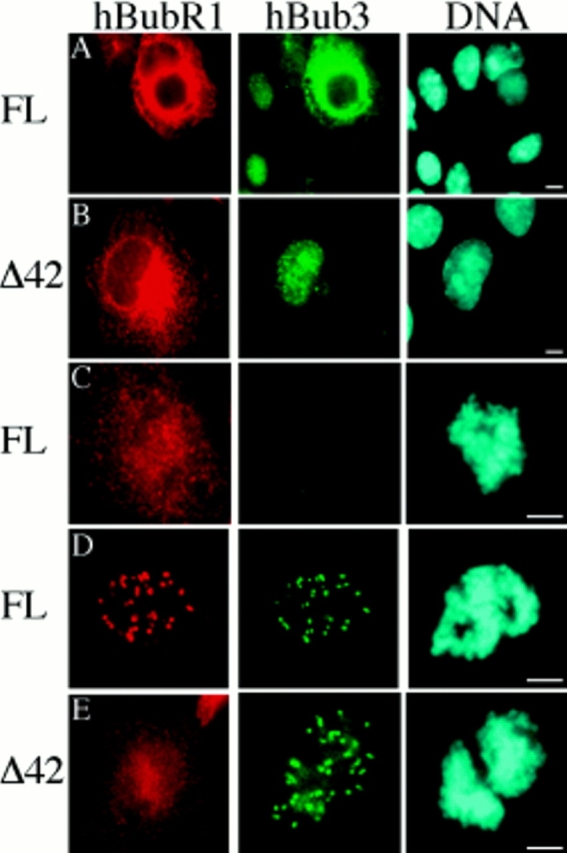
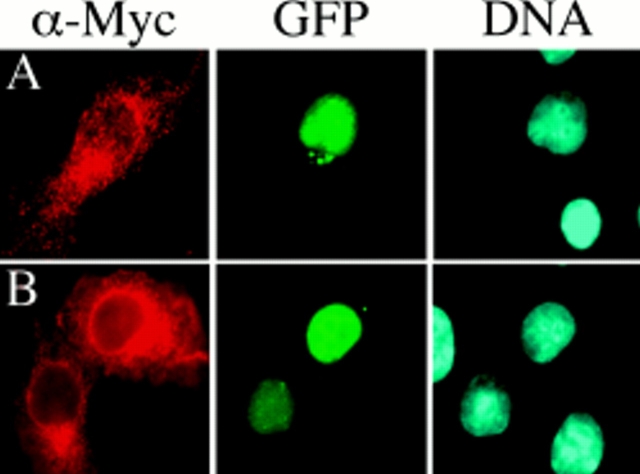
hBubR1 localizes to kinetochores when hBub3 is coexpressed. BHK cells were transiently transfected with either a full-length Myc-tagged hBubR1 expression construct (FL) or an hBubR1 mutant lacking the domain shown in Fig. 4 B (Δ42), fixed, and then stained with the 9E10 monoclonal antibody to determine the subcellular localization of hBubR1 (red). Cells were also stained with Hoechst to visualize the chromatin (blue). In A, B, D, and E, cells were cotransfected with GFP–hBub3 (green). (A) Transfected cells showing that ectopically expressed hBubR1 is predominantly cytoplasmic during interphase. In addition, coexpression of hBubR1 results in cytoplasmic sequestration of GFP–hBub3. (B) Expression of hBubR1Δ42 does not result in cytoplasmic sequestration of GFP–hBub3. (C) Transfected prometaphase cell showing ectopically expressed hBubR1 diffusely localized throughout the cell. (D) A prometaphase cell showing localization of hBubR1 at kinetochores when GFP– hBub3 is coexpressed. (E) Two transfected prometaphase cells showing diffuse localization of hBubR1Δ42 despite coexpression of GFP–hBub3. Bars, 5 μm.
The different localizations of ectopically expressed hBubR1 and hBub3 during interphase allowed us to test whether these two proteins interact by performing cotransfections as described above to test the Bub1–Bub3 interaction. Fig. 6 A shows several cells that have been cotransfected with hBubR1 and hBub3. In the cells not expressing hBubR1, hBub3 is predominantly nuclear. In the cell coexpressing hBubR1, hBub3 is clearly localized to the cytoplasm, suggesting that hBubR1 and hBub3 can interact in mammalian cells. Significantly, in prometaphase cells coexpressing hBub3 and hBubR1, localization of hBubR1 at kinetochores was observed (Fig. 6 D).
A Domain Conserved between the Bub1 and Mad3-related Proteins Is Required for the hBubR1–hBub3 Interaction
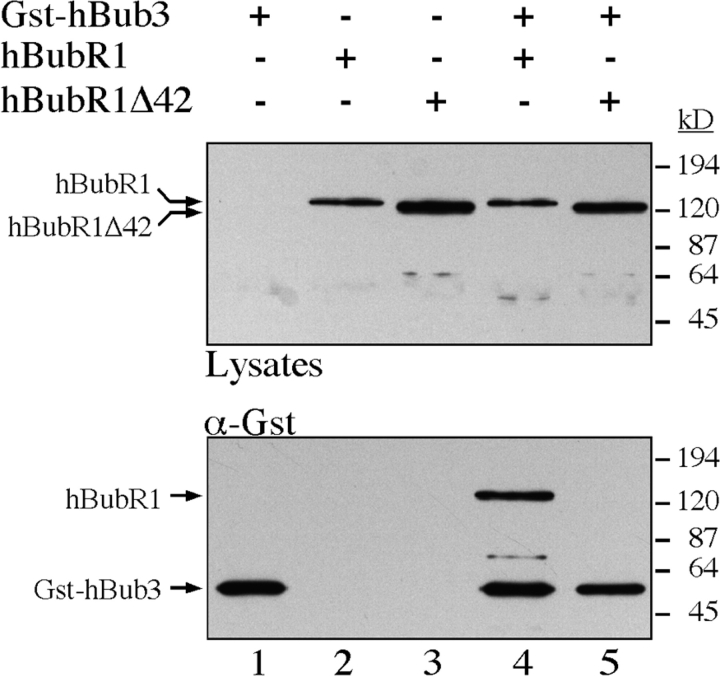
To test whether the homology domain defined in Fig. 4 B is required for the hBubR1/hBub3 interaction, this region was deleted to generate hBubR1Δ42. GST–hBub3 was cotransfected with either hBubR1 or hBubR1Δ42 into BHK cells, followed by affinity purification from cell lysates using glutathione Sepharose beads. The affinity complexes were then analyzed by Western blotting. hBubR1 clearly purifies with the glutathione beads in a GST– hBub3-dependent manner (Fig. 7, lane 4). In contrast, hBubR1Δ42 does not copurify with GST–hBub3 (Fig. 7, lane 5). In addition, hBubR1Δ42 did not sequester coexpressed hBub3 in the cytoplasm (Fig. 6 B). Furthermore, hBubR1Δ42 did not localize to kinetochores when coexpressed with hBub3 (Fig. 6 E). These observations show that hBubR1 can physically interact with hBub3 in mammalian cells, and that a domain conserved between the Bub1 and Mad3-related proteins is required for this binding.
Figure 7.
The Bub3 binding domain of mBub1 and hBubR1 is conserved. BHK cells were transfected with GST–hBub3 (lanes 1, 4, and 5) and/or Myc-tagged hBubR1 (lanes 2 and 4), and/or Myc-tagged hBubR1Δ42 (lanes 3 and 5). Proteins from cell lysates (top) and purified GST– hBub3 complexes (bottom) were resolved by SDS-PAGE, transferred to a membrane, and then probed with anti-GST and anti-Myc antibodies. In the absence of GST–hBub3, neither hBubR1 nor hBubR1Δ42 purify with the glutathione beads (lanes 2 and 3). In the presence of GST–hBub3, hBubR1 purifies with the glutathione beads (lane 4) but hBubR1Δ42 does not (lane 5). Note that GST-hBub3 cannot be identified in the cell lysates.
hBub3ΔVAVE Does Not Interact with mBub1 or hBubR1
To define the domain of Bub3 required for binding to Bub1 and BubR1, several hBub3 mutants were generated and scored for their ability to be sequestered in the cytoplasm when coexpressed with either mBub1 or hBubR1. One mutant, hBub3ΔVAVE, which contains a four-amino acid deletion (amino acids 217–220) from within the central domain (refer to Fig. 1), was not sequestered in the cytoplasm when coexpressed with either mBub1 or hBubR1, but rather remained exclusively nuclear (Fig. 8). These observations suggest that Bub3 binds to both Bub1 and BubR1 in a similar manner. Kinetochore localization of hBub3ΔVAVE was not observed in transfected prometaphase cells (data not shown), suggesting that the ability to bind Bub1 or BubR1 is required for kinetochore localization of Bub3.
Figure 8.
hBub3ΔVAVE does not bind mBub1 or hBubR1. BHK cells were cotransfected with GFP–hBub3ΔVAVE and either (A) Myc-tagged mBub1 or (B) Myc-tagged hBubR1. After transfection, the cells were fixed and then stained with the 9E10 monoclonal antibody to determine the subcellular localization of the Myc-tagged fusion proteins (red). The cells were also stained with Hoechst to visualize the chromatin (blue). GFP fluorescence is shown in green. Despite coexpression of mBub1 and hBubR1, hBub3ΔVAVE remains in the nucleus, suggesting that this hBub3 mutant does not interact with mBub1 or hBubR1.
In Vitro Phosphorylation of mBub1 and hBubR1
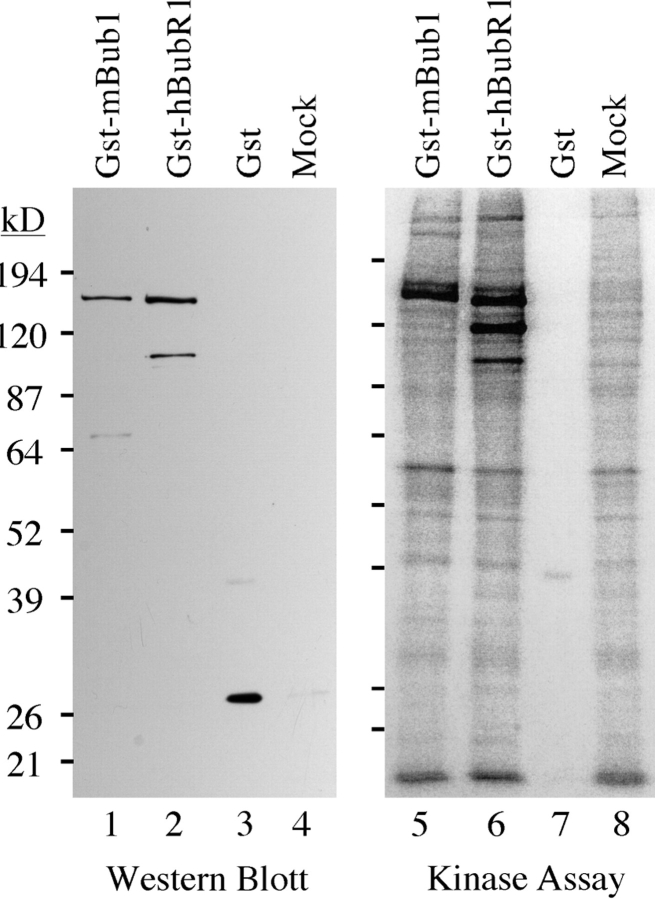
The sequence alignment shown in Fig. 5 B indicates that the kinase domain of hBubR1 differs significantly from the mBub1 kinase domain. In addition, hBubR1 lacks several of the residues that are usually highly conserved among most protein kinases (Hanks and Hunter, 1995). To test whether hBubR1 is indeed a protein kinase, GST– hBubR1 was expressed in BHK cells, affinity purified from cell lysates using glutathione Sepharose beads, and then assayed for in vitro protein kinase activity. Bound proteins were first analyzed by Western blotting to normalize the amount of GST protein used in the subsequent kinase assays (Fig. 9). The kinase assay reveals several phosphorylated proteins associated with the glutathione beads incubated in lysates from cells expressing either GST-mBub1 (Fig. 9, lane 5) and GST–hBubR1 (Fig. 9, lane 6). These phosphorylated proteins were not detected when either equivalent amounts of GST alone (Fig. 9, lane 7) or equivalent amounts of glutathione beads incubated with lysates from mock transfected cells (Fig. 9, lane 8) were compared. The major phosphorylated proteins detected in Fig. 9, lanes 5 and 6 are of the molecular weights expected of GST–mBub1 and GST–hBubR1 respectively, (as judged by Western blotting, see Fig. 9, lanes 1 and 2). This indicates that the phosphorylated bands are GST– mBub1 and GST–hBubR1, suggesting that the observed kinase activity is probably autophosphorylation. Although we cannot rule out the possibility that the observed activities are due to another copurifying protein kinase, these data are consistent with the notion that, like ScBub1 (Roberts et al., 1994), mBub1 and hBubR1 exhibit autophosphorylation activity in vitro.
Figure 9.
In vitro phosphorylation of mBub1 and hBubR1. BHK cells were transiently transfected with GST–mBub1, GST–hBubR1, and GST alone or mock-transfected as indicated. After transfection, cell lysates were prepared and incubated with glutathione Sepharose beads. Bound proteins were then analyzed by Western blotting to normalize the amounts of GST-fusion proteins used in the subsequent kinase assay. To assay for associated kinase activity, the beads were incubated with 32[P] γ-ATP. Bound proteins were then resolved by SDS-PAGE and analyzed by autoradiography. Lanes 5 and 6 show that there are several phosphorylated proteins associated with the beads incubated in lysates from cells expressing either GST–mBub1 or GST–hBubR1. The Western blot shows that the major phosphorylated proteins correspond to GST–mBub1 (lane 1) and GST– hBubR1 (lane 2), suggesting that the observed kinase activity is autophosphorylation. The major phosphorylated proteins observed in lanes 5 and 6 are not associated with beads incubated in cell lysates expressing GST alone (lane 7) or from mock-transfected cells (lane 8).
Discussion
We describe the cloning of a human homologue of Bub3, which, like murine Bub1, localizes to kinetochores during prometaphase (Taylor and McKeon, 1997). In addition, we show that mBub1 and hBub3 can interact in mammalian cells. We have also identified the Bub3-binding site in Bub1. This same domain is required for kinetochore localization of Bub1, which we have previously shown is required for checkpoint function (Taylor and McKeon, 1997). Therefore, these observations suggest that the role of Bub3 is to facilitate kinetochore localization of Bub1, thereby activating the checkpoint in response to unattached kinetochores. ScBub3 was identified as a high copy suppressor of the bub1-1 allele, suggesting that ScBub1 and ScBub3 interact in yeast (Hoyt et al., 1991). Further support for this interaction came from genetic experiments, which showed that bub3Δ strains have a similar phenotype to bub1Δ strains (Roberts et al., 1994). In addition, bub1Δ/bub3Δ double mutants are phenotypically similar to strains with deletions of either gene, and Bub1p and Bub3p have been shown to coimmunoprecipitate when overexpressed in S. cerevisiae (Roberts et al., 1994). Our observations with Bub1 and Bub3 in mammalian cells are consistent with those in S. cerevisiae and suggest the following model: in the absence of Bub3, Bub1 cannot localize to the kinetochore and hence the checkpoint is not activated in response to unattached kinetochores.
hBub3 also appears to be required for kinetochore localization of hBubR1. Whether Bub3 plays any roles in addition to kinetochore localization of Bub1 and BubR1 remains to be seen. Interestingly, hBub3 contains four WD repeats, a 40-amino acid motif found in many proteins involved in a diverse array of cellular processes, including G protein–linked receptor signaling, RNA processing and export, and cell cycle control (Neer et al., 1994). The exact function of WD repeats is not clear, but it seems likely that they play a role in mediating protein–protein interactions. Indeed, WD repeat proteins are often part of multiprotein complexes (Neer et al., 1994). Whether Bub3 is part of a large multiprotein complex remains to be seen. Thus far, Bub3 has been shown to interact with Bub1 and the human Mad3/Bub1-related protein, hBubR1 (Roberts et al., 1994; this work). In addition, Bub3 and Mad3 have been shown to interact in S. cerevisiae (Hardwick, K.G., and A.W. Murray, personal communication). Recently, Mad1, Mad2, and Mad3 have been shown to interact with cdc20 (Hwang et al., 1998; Kim et al., 1998), which in turn has been shown to interact with the APC (Visintin et al., 1997). Significantly, the vertebrate homologues of Mad2, Bub1, Bub3 and the Mad3-related kinase, BubR1, have been shown to localize to kinetochores before chromosome alignment (Chen et al., 1996; Li and Benezra, 1996; Taylor and McKeon, 1997; this work). It is therefore tempting to speculate that the checkpoint components, including Bub1, BubR1, Bub3, Mad1, Mad2, and Mad3, may be part of a large protein complex that is recruited to unattached kinetochores. This kinetochore-bound form of the checkpoint complex may bind and inhibit cdc20, thereby preventing activation of APC, and hence delaying the onset of anaphase. Based on the observations that Mad2, Bub1, Bub3, and BubR1 are not present at the kinetochores of metaphase chromosomes (Chen et al., 1996; Li and Benezra, 1996; Taylor and McKeon, 1997; this work), it appears likely that the checkpoint complex dissociates from kinetochores upon achieving correct bipolar attachments. Upon dissociation, perhaps the composition or activity of the checkpoint complex is altered, rendering cdc20 active and hence allowing the onset of anaphase. However, although there is evidence that many of these checkpoint components can interact with each other (see above), the existence of such a complex has not yet been demonstrated to exist in vivo. It is therefore possible that some of these proteins interact only transiently as cells progress through mitosis.
hBub3 shares significant homology with the Rae1 family of proteins, both within the WD repeats and in the central, non-WD repeat region (refer to Fig. 1). The S. pombe rae1 gene was identified as a poly(A)+ RNA export mutant (Brown et al., 1995). SpRae1 has recently been shown to interact with the S. pombe homologue of the Nic96 nucleoporin (Yoon et al., 1997). In addition, the S. cerevisiae homologue of Rae1, gle2p, interacts with the Nup100p nucleoporin and other proteins involved in nucleocytoplasmic transport (Murphy et al., 1996). Furthermore, gle2 strains also show defects in poly(A)+ RNA export and have grossly perturbed nuclear pores. These observations suggest that Rae1 is required for nucleocytoplasmic transport and probably does not play a direct role in cell cycle control.
Despite the significant similarity between the Rae1 proteins and the human Bub3 homologue described here (39% identity), several lines of evidence suggest that hBub3 is not a member of the Rae1 family. First, a human cDNA encoding a protein with 49% identity to SpRae1 has recently been identified (Bharathi et al., 1997). This cDNA complements the temperature sensitivity of the rae1-1 allele and is therefore likely to be the human Rae1 homologue. Second, an alignment of the Rae1 and Bub3 proteins identifies several amino acid sequences in the central domain, including the VATAER sequence (amino acids 174–179 in SpRae1), which are conserved in all the Rae1 proteins, but not in the Bub3 homologues. Conversely, there are several sequences, including the SSI(E/ D)GRVAVE sequence (amino acids 211–220 in hBub3), which are conserved in the Bub3 proteins, but are not conserved in the Rae1 homologues. Significantly, deletion of the VAVE sequence abolishes the ability of hBub3 to interact with mBub1 and hBubR1. Note that the S. pombe rae1-1 loss of function allele results in a substitution of the glycine at position 219 with a glutamic acid (Brown et al., 1995). Significantly, this glycine is conserved in the Rae1 homologues, but is replaced by a serine in the Bub3 homologues.
In addition to the sequence analysis, two observations indicate that hBub3 is indeed the homologue of ScBub3. First, hBub3 interacts with mBub1, as do Bub3 and Bub1 in S. cerevisiae (Roberts et al., 1994). Second, hBub3 localizes to kinetochores during prometaphase, a property that one might predict based on the localizations of mBub1 (Taylor and McKeon, 1997) and the vertebrate homologues of Mad2 (Chen et al., 1996; Li and Benezra, 1996). The significance, if any, of the similarity between the Bub3 and Rae1 proteins remains to be determined.
This work also describes the identification of a novel human Mad3/Bub1-related protein, hBubR1. hBubR1 shares homology with Mad3 from S. cerevisiae and yet it is significantly larger (1,050 amino acids) than ScMad3 (515 amino acids), due to a COOH-terminal extension which contains a kinase domain. This suggests that perhaps the human Mad3/Bub1-related kinase is a second Bub1 homologue. Indeed, within the NH2-terminal domains, hBubR1 is more similar to ScBub1 (26% identity) than mBub1 is to ScBub1 (23% identity). In addition, a BLAST search of the yeast genome shows that the kinase most closely related to hBubR1 is ScBub1. However, an alignment of the COOH-terminal kinase domains shows that hBubR1 is clearly distinct from the Bub1 family (refer to Fig. 5 B). Within the kinase domains, there are 85 amino acids conserved among the Bub1 homologues from S. cerevisiae, mouse and human. However, 54 of these are not conserved in hBubR1. Indeed, within the kinase domain, mBub1 is more closely related to ScBub1 (33% identity) than it is to hBubR1 (26% identity). Within the NH2-terminal domain, hBubR1 is significantly more similar to ScMad3 (35% identity) than it is to ScBub1 (26% identity). These observations suggest that perhaps hBubR1 is a Mad3-related protein kinase. Whether hBubR1 and ScMad3 have indeed evolved from a common ancestor will remain uncertain until Mad3-related proteins from other organisms have been identified, or until we have a better understanding of the functions of these two proteins.
The functional analysis presented here illustrates similarities between mBub1 and hBubR1, as well as a significant difference. Like mBub1, hBubR1 can bind hBub3 in mammalian cells, and this binding requires a domain that is conserved between the Bub1 and Mad3-related proteins. In addition, hBubR1 can localize to kinetochores during prometaphase and the ability to bind Bub3 is required for this localization. However, unlike mBub1, ectopically expressed hBubR1 only localizes to kinetochores when hBub3 is overexpressed. In contrast, ectopically expressed mBub1 localizes to the kinetochore without coexpression of hBub3 (Taylor and McKeon, 1997). One possible explanation for this observation is that hBubR1 cannot bind hamster Bub3, and hence kinetochore localization is not observed in transfected BHK cells unless exogenous human Bub3 is present. However, significant kinetochore staining was also not observed when hBubR1 was transfected into human cells (data not shown). Until the localization of endogenous hBubR1 has been determined, these observations have to be treated with caution. However, it does suggest that the amount of endogenous Bub3 is limiting with respect to ectopically expressed hBubR1. One possibility is that the endogenous Bub3 is complexed with Bub1 and hence there is no Bub3 available for binding to, and hence kinetochore localization of, transfected hBubR1. This suggests that perhaps the majority of BubR1 may not play a role at the kinetochore. Alternatively, perhaps hBubR1 does play a role at the kinetochore, but that overexpression of hBubR1 somehow prevents the formation of complexes capable of kinetochore localization. The generation of anti-hBubR1 antibodies will hopefully resolve this issue.
The role of hBubR1 remains to be determined. It is possible that Bub1 and BubR1 have similar or partially overlapping roles. Although there is only a single Bub1 kinase encoded in the S. cerevisiae genome, functional redundancy may be beneficial in mammalian cells. Recently, mutant BUB1 alleles have been identified in colorectal tumors displaying a chromosome instability phenotype (Cahill et al., 1998), suggesting that mitotic checkpoint defects may contribute to tumorigenesis. Therefore, overlapping or partially redundant roles for Bub1 and BubR1 may provide a selective advantage to multicellular organisms: increasing the fidelity of chromosome segregation may reduce the possibility of generating potentially tumorigenic aneuploid cells.
Redundancy may allow multiple spindle events to be monitored by the mitotic checkpoint. At present, there is evidence implicating both tension and microtubule attachment as the events which regulate anaphase onset (Li and Nicklas, 1995; Rieder et al., 1995). Perhaps in the quest for enhanced genome stability, both tension and attachment are monitored by the checkpoint. Perhaps, therefore, Bub1 and BubR1 respond to different types of spindle events. Tension and microtubule attachment may also be differentially monitored in mitosis relative to meiosis (Nicklas, 1997). If Bub1 and BubR1 respond differentially to tension and microtubule attachment, perhaps they play differential roles in mitosis and meiosis.
The EMBL/GenBank/DDBJ accession numbers for the cDNA sequences reported here are AF053304 (hBub3); AF053305 (hBub1); and AF053306 (hBubR1).
Acknowledgments
We would like to thank K. Hardwick (University of Edinburgh, Edinburgh, Scotland) and A. Murray (University of California, San Francisco, CA) for helpful discussion. We are grateful to J. Zhu for technical advice and reading the manuscript. We also thank P. Stein and M. Rolls (all three from Harvard Medical School, Boston, MA) for comments on the manuscript.
This work was supported by a National Institutes of Health grant (GM52027) to F. McKeon and a Traveling Research Fellowship from the Wellcome Trust to S.S. Taylor.
Abbreviations used in this paper
- APC
anaphase-promoting complex
- EST
expressed sequence tag
- NFDM
nonfat dry milk
Footnotes
Address all correspondence to Frank McKeon, Department of Cell Biology, Harvard University Medical School, 240 Longwood Avenue, Boston, MA 02115. Tel.: (617) 432-0327. Fax: (617) 432-6655. E-mail: mckeon@warren.med.harvard.edu
References
- Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, White O, et al. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature. 1995;377(Suppl.):3–174. [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bharathi A, Ghosh A, Whalen WA, Yoon JH, Pu R, Dasso M, Dhar R. The human RAE1 gene is a functional homologue of Schizosaccharomyces pombe rae1gene involved in nuclear export of Poly(A)+ RNA. Gene. 1997;198:251–258. doi: 10.1016/s0378-1119(97)00322-3. [DOI] [PubMed] [Google Scholar]
- Brown JA, Bharathi A, Ghosh A, Whalen W, Fitzgerald E, Dhar R. A mutation in the Schizosaccharomyces pombe rae1gene causes defects in poly(A)+ RNA export and in the cytoskeleton. J Biol Chem. 1995;270:7411–7419. doi: 10.1074/jbc.270.13.7411. [DOI] [PubMed] [Google Scholar]
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- Callan HG, Jacobs PA. The meiotic process in Mantis ReligiosaL. males. J Genet. 1957;55:200–217. [Google Scholar]
- Campbell MS, Gorbsky GJ. Microinjection of mitotic cells with the 3F3/2 anti-phosphoepitope antibody delays the onset of anaphase. J Cell Biol. 1995;129:1195–1204. doi: 10.1083/jcb.129.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R-H, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiaeis controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Dalrymple MA, Petersen-Bjorn S, Friesen JD, Beggs JD. The product of the PRP4 gene of S. cerevisiaeshows homology to beta subunits of G proteins. Cell. 1989;58:811–812. doi: 10.1016/0092-8674(89)90930-6. [DOI] [PubMed] [Google Scholar]
- Farr KA, Hoyt MA. Bub1p kinase activates the Saccharomyces cerevisiaespindle assembly checkpoint. Mol Cell Biol. 1998;18:2738–2747. doi: 10.1128/mcb.18.5.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Gorbsky GJ, Ricketts WA. Differential expression of a phosphoepitope at the kinetochores of moving chromosomes. J Cell Biol. 1993;122:1311–1321. doi: 10.1083/jcb.122.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB (Fed Am Soc Exp Biol) J. 1995;9:576–596. [PubMed] [Google Scholar]
- Hardwick KG, Murray AW. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- He X, Patterson TE, Sazer S. The Schizosaccharomyces pombespindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl Acad Sci USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, McLoughlin M, McKeon F. Human wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated Cdc2 kinase. Cell. 1993;74:463–474. doi: 10.1016/0092-8674(93)80048-j. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiaegenes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Juang YL, Huang J, Peters JM, McLaughlin ME, Tai CY, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC26 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Li X, Nicklas RB. Tension-sensitive kinetochore phosphorylation and the chromosome distribution checkpoint in praying mantid spermatocytes. J Cell Sci. 1997;110:537–545. doi: 10.1242/jcs.110.5.537. [DOI] [PubMed] [Google Scholar]
- Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Li Y, Gorbea C, Mahaffey D, Rechsteiner M, Benezra R. MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc Natl Acad Sci USA. 1997;94:12431–12436. doi: 10.1073/pnas.94.23.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R, Watkins JL, Wente SR. GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombeexport factor RAE1, is required for nuclear pore complex structure and function. Mol Biol Cell. 1996;7:1921–1937. doi: 10.1091/mbc.7.12.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Nicklas RB, Koch CA. Chromosome manipulation III: induced reorientation and the experimental control of segregation in meiosis. J Cell Biol. 1969;43:40–50. [Google Scholar]
- Nicklas RB, Ward SC, Gorbsky GJ. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J Cell Biol. 1995;130:929–939. doi: 10.1083/jcb.130.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangilinan F, Spencer F. Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol Cell Biol. 1996;7:1195–1208. doi: 10.1091/mbc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Schultz A, Cole R, Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J Cell Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts BT, Farr KA, Hoyt MA. The Saccharomyces cerevisiae checkpoint gene BUB1encodes a novel protein kinase. Mol Cell Biol. 1994;14:8282–8291. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 545 pp.
- Sudakin V, Ganoth D, Dahan A, Heller H, Hersko J, Luca F, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitination ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–198. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Wang Y, Burke DJ. Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. . Mol Cell Biol. 1995;15:6838–6844. doi: 10.1128/mcb.15.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells WAE, Murray AW. Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J Cell Biol. 1996;133:75–84. doi: 10.1083/jcb.133.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Whalen WA, Bharathi A, Shen R, Dhar R. Npp106p, a Schizosaccharomyces pombe nucleoporin similar to Saccharomyces cerevisiaeNic96p, functionally interacts with Rae1p in mRNA export. Mol Cell Biol. 1997;17:7047–7060. doi: 10.1128/mcb.17.12.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkle RE. Ultraviolet-microbeam irradiation of newt-cell cytoplasm: spindle destruction, false anaphase and delay of true anaphase. Radiat Res. 1970a;41:516–537. [PubMed] [Google Scholar]
- Zirkle RE. Involvement of the prometaphase kinetochore in prevention of precocious anaphase. J Cell Biol. 1970b;47:235a. [Google Scholar]