Abstract
A set of nuclear mutants of C. reinhardtii were identified that specifically lack translation of the chloroplast-encoded psbA mRNA, which encodes the photosystem II reaction center polypeptide D1. Two of these mutants are deficient in the 47-kD member (RB47) of the psbA RNA-binding complex, which has previously been identified both genetically and biochemically as a putative translational activator of the chloroplast psbA mRNA. RB47 is a member of the poly(A)-binding protein family, and binds with high affinity and specificity to the 5′ untranslated region of the psbA mRNA. The results presented here confirm RB47's role as a message-specific translational activator in the chloroplast, and bring together genetic and biochemical data to form a cohesive model for light- activated translational regulation in the chloroplast.
Keywords: genetic translation, initiation factors, translational regulation, gene expression
Analysis of photosynthetic gene expression in the unicellular green algae Chlamydomonas reinhardtii has shown that for chloroplast mRNAs, translational regulation is a prevalent mechanism used for controlling the synthesis of key photosynthetic proteins. Translation of a number of these chloroplast mRNAs is strongly influenced by light, with high levels of protein synthesis initiated from preexisting pools of mRNA only after transfer of cells into light. Genetic analysis has identified a number of nuclear mutants in which translation of specific chloroplast mRNAs is lacking. In many of these mutants, translation of a single mRNA was affected, whereas other photosynthetic mRNAs were translated at normal levels (Kuchka et al., 1988; Rochaix et al., 1989; Girard-Bascou et al., 1992; Yohn et al., 1996). Mutants affecting mRNA processing (Goldschmidt-Clermont et al., 1990) and mRNA stability (Kuchka et al., 1989) have also been identified, suggesting that these processes also play roles in gene expression within the chloroplast. In addition, protein turnover has been shown to affect accumulation of photosynthetic proteins in the chloroplast (Erickson et al., 1986; Mayfield et al., 1987). Though each of these processes plays some role in regulating protein accumulation, translational regulation appears to be the predominant form of light-mediated gene regulation in the chloroplast (for review see Rochaix, 1992; Gillham et al., 1994; Mayfield et al., 1995).
Analysis of nuclear mutants that affected translation of specific chloroplast mRNAs suggested that nuclear factors act as translational activators. These translational activators interact with the 5′ untranslated region (UTR)1 of chloroplast-encoded mRNAs to facilitate translation of the downstream coding region (Mayfield et al., 1995). This RNA–protein interaction was first suggested by chloroplast mutations that were localized to the 5′ UTR of the mRNA and that affected translation of the downstream encoded message (Rochaix et al., 1989). A chloroplast suppressor was also identified that restored translation to a nuclear mutant lacking translation of the chloroplast psbC mRNA (which encodes P6, a component of photosystem II [PS II]). This suppressor mutation was localized in the 5′ UTR of the psbC mRNA (Rochaix et al., 1989). The 5′ UTR of the psbA mRNA (encoding D1, a PSII subunit) of tobacco was shown to confer light-regulated translation on a reporter coding region in vivo (Staub and Maliga, 1993). Site-directed mutagenesis to the 5′ UTR of psbA mRNA of C. reinhardtii has further identified RNA elements critical for translation of this mRNA. A consensus Shine-Dalgarno sequence located 26 nucleotides 5′ of the initiation codon was required for mRNA/ribosome association and translation, whereas changes to a predicted stem-loop structure located adjacent to this potential ribosome binding sequence dramatically reduced psbA translation in the light (Mayfield et al., 1994).
RNA-binding proteins, which were originally identified based on in vitro binding that correlated with translation rates under a variety of conditions, are part of a complex that binds to the 5′ UTR of the psbA mRNA with high affinity and specificity (Danon and Mayfield, 1991). The biochemical identification of these putative translational activators of psbA, as well as elucidation of mechanisms that control psbA RNA-binding activity (Danon and Mayfield, 1994a ,b), suggested that multiple components are required for light-activated translation of this specific chloroplast-encoded message. Three of the components of this complex, the 38-, 47-, and 60-kD proteins, have been cloned and sequenced. This characterization identified the 47-kD protein (RB47) as a member of the poly(A)-binding protein family (PABP) (Yohn et al., 1998), the 60-kD protein (RB60) as a protein disulfide isomerase (PDI) (Kim and Mayfield, 1997), and the 38-kD protein as a novel protein with no characterized homologue identified in other species. These proteins bind as a complex to the 5′ UTR of the psbA mRNA, along with uncharacterized protein of 55 kD. The light dependence of this RNA binding was shown to be mediated by redox potential (Danon and Mayfield, 1994b ), specifically through the 60-kD PDI modulating redox status of RB47 (Kim and Mayfield, 1997).
Characterization of C. reinhardtii nuclear mutants deficient in psbA translation has shown that members of the psbA RNA-binding complex are affected in these mutants. The nuclear mutant F35 was deficient in translation initiation of the psbA mRNA, implying that initiation may be the stage at which translation is regulated by the translational activator proteins (Girard-Bascou et al., 1992; Yohn et al., 1996). Both the 47- and 55-kD psbA RNA-binding proteins accumulated at reduced levels in this mutant, suggesting that a reduction in accumulation of these two putative translational activators might have caused the loss of translation of psbA mRNA in this strain (Yohn et al., 1996). The nuclear mutant nac1-18 lacks translation of both psbA and psbD mRNAs (Cohen, A., C.B. Yohn, and S.P. Mayfield, manuscript submitted for publication). This mutant was shown to be blocked in translation elongation of the psbD mRNA. This translation arrest led to accumulation of aberrant forms of the RB47 protein (Cohen et al., manuscript submitted for publication), subsequently resulting in a reduction of psbA translation initiation.
The data gathered to date suggest a role for RB47 and the psbA RNA-binding complex in translation initiation of the psbA mRNA. To further identify and characterize proteins required for psbA translation, a library of mutants was created by random DNA insertion mutagenesis (Tam and Lefebvre, 1993), and screened for defects in translation of the chloroplast-encoded psbA mRNA. We have characterized five nuclear mutants of C. reinhardtii that specifically lack translation of the psbA mRNA. One of these mutants, hf149, completely lacks the 47-kD psbA RNA-binding protein (RB47) and any detectable psbA mRNA-binding activity. Another mutant, hf261, contains only 10% of the wild-type (wt) levels of this protein and ∼10% of the psbA mRNA-binding activity. In both of these mutants, the psbA mRNA fails to associate with ribosomes. Other chloroplast mRNAs associate with polyribosomes and are translated at normal levels in these strains. The three additional nonallelic mutations show related phenotypes in terms of reduced mRNA translation and reduced ribosome association of the psbA mRNA. However, all three of these mutants have psbA RNA-binding activity and accumulate RB47 protein. The data shown here provide direct evidence that the psbA RNA-binding complex is required for psbA mRNA translational activation, and identifies these proteins, and the PABP homologue RB47 in particular, as translational activators of the psbA mRNA.
Materials and Methods
Cell Growth Conditions
C. reinhardtii strains were grown in complete media (Tris-acetate-phosphate [TAP]) (Harris, 1989) to a density of 5 × 106 cells/ml under constant light. Cells were harvested by centrifugation at 4°C for 5 min at 4,000 g. Cells were either used immediately or frozen in liquid N2 for storage at −70°C.
Mutagenesis and Screening
The C. reinhardtii strain Arg7/cw15, an arginine auxotroph and cell wall-deficient mutant, was grown in TAP media with 50 mg/liter l-arginine to a density of ∼106 cells/ml under constant light. Cells were washed with TAP without l-arginine and resuspended at ∼5 × 107 cells/ml in TAP. For each glass bead transformation (Kindle, 1990), 400 ml of cells were placed in a 15-ml sterile conical tube (Falcon Plastics, Cockeysville, MD) with 300 ml of sterile, acid-washed glass beads (0.5-mm average diam). 2 mg of linearized DNA was added and polyethylene glycol (molecular weight of 8,000 D) was added to 5% (wt/vol) final concentration. The DNA was a 7.8-kb genomic fragment containing the argininosuccinate lyase gene of C. reinhardtii (Debuchy et al., 1989) cloned into pBlueScript, and then linearized with XbaI in the polylinker. The cells, with beads DNA and PEG, were vortexed at full speed for 10 s. After allowing the beads to settle, cells were pipetted from the tube and spread on TAP media plates (with 1.5% agar). Transformant colonies were allowed to grow in dim light for 10–14 d.
Transformants with a high fluorescence (hf) phenotype were identified with a video system based on a setup from Bennoun and Delepelaire (1982). Transformants were illuminated with a 300-W quartz-iodine lamp filtered by a blue glass filter (model 51710, peak transmittance 480 nm; Oriel Corp., Stratford, CT). A charge-coupled device camera (model XC-77; Sony Corp., Park Ridge, NJ) with a red filter (Wratten gelatin filter 89B, peak transmittance 720 nm; Eastman-Kodak Co., Rochester, NY) attached to the lens was used to visualize chlorophyll fluorescence in the transformant colonies. The video signal was input into a Macintosh computer running the public domain NIH Image program (developed at the U.S. National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/) via a framegrabber (model LG-3; Scion Corp., Frederick, MD). Integrating 4–8 frames allowed for easy visualization and selection of hf mutants.
Membrane and Soluble Protein Isolation and Immunoblotting
Protein isolation, electrophoresis, and immunoblotting were performed as described by Mayfield et al. (1994), with the following modifications. After extraction, membrane or soluble proteins were separated by electrophoresis and electroblotted to nitrocellulose (Schleicher & Schuell, Keene, NH) in 10 mM cyclohexylamino-1-propanesulfonic acid (CAPS), pH 11, with 10% methanol. Rabbit polyclonal antisera specific for D1, D2, ATPase, or LHC II proteins were added to blots of membrane proteins, whereas antisera specific for OEE1 or the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase were used with blots of soluble proteins.
Pulse Labeling of Proteins with [14C]Acetate
Cells were labeled with [14C]acetate for 10 min in the presence of cycloheximide as described by Mayfield et al. (1994). Membrane proteins were isolated, separated by size on SDS-PAGE, and then visualized by fluorography as described by Malnoë et al. (1988).
Genetic Analysis
Standard protocols were used for crosses, tetrad dissection, and the scoring of the phenotypes of meiotic progeny (Levine and Ebersold, 1960). In most cases, tetrads resulting from backcrosses and intercrosses were incomplete, with fewer than four viable progeny. For one cross (hf859 × F35), random spore analysis was used to generate a larger number of meiotic progeny. In this case, the mating was done according to standard protocols, but zygotes were subsequently collected, transferred to 10 ml of TAP liquid medium supplemented with arginine, and then allowed to germinate for 14–16 h in bright light. After germination, tetrads were collected by centrifugation at 2,500 rpm for 10 min, resuspended in 0.5 ml TAP liquid medium supplemented with arginine, vortexed briefly to disperse meiotic progeny cells, and then plated onto TAP plates supplemented with arginine. Colonies were visible in 7–10 d.
Chromatography
Approximately 5 × 109 cells were resuspended in low salt extraction buffer (10 mM Tris, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 5 mM β-mercaptoethanol). After passage through a cell disruption bomb (Parr Instrument Co., Moline, IL), the soluble cell extract was applied to a 5-ml Econo-Pac heparin cartridge (Bio-Rad Laboratories, Inc., Hercules, CA) as described in Danon and Mayfield (1991). Partial purification of proteins over heparin agarose was used to reduce RNase activity and enrich for mRNA-binding proteins.
T1-Gel Mobility Shift
Gel mobility shift (GMS) coupled with T1 RNase protection assays were performed with heparin agarose fractions as described in Danon and Mayfield (1991). Equal quantities of protein lysate were incubated with 0.065 pmol 32P-labeled transcript, 20 mg tRNA, and 1 mg total RNA from the C. reinhardtii strain Fud7 (a chloroplast mutant with a psbA gene deletion) (Bennoun et al., 1986). After 15 min at room temperature, 0.5 U of T1 RNase (Pharmacia Biotech., Inc., Piscataway, NJ) was added, and then the reaction was incubated for an additional 5 min at room temperature before electrophoresis of the sample on a 1× TBE, 5% native polyacrylamide gel. RNA–protein complexes were detected by autoradiography.
Protein Electrophoresis and Immunoblotting of HA Proteins
Equal quantities of heparin agarose fractions were mixed with 2× sample buffer (5% SDS, 5% β-mercaptoethanol, 400 mM Tris-HCl, pH 6.8, 10% sucrose), heated to 60°C for 10 min, and then separated by size using SDS-PAGE. The gel was either stained with Coomassie blue in 40% methanol, 10% acetic acid or electroblotted to nitrocellulose as described above. Blotted nitrocellulose filters were treated as described above, except polyclonal antisera specific for the RB38, RB47, or RB60 protein (Danon and Mayfield, 1991) was used and the incubation continued at 4°C for 16 h. Goat anti–rabbit IgG alkaline phosphatase conjugate (Bio-Rad Laboratories. Inc.) was used as the secondary antibody to visualize RB38, RB47, or RB60.
RNA/Polysome Isolation and Analyses
Preparation of RNA and fractionation of polysomes were performed as described by Barkan (1988) with modifications as described in Yohn et al. (1996).
Results
Generation and Selection of PS II Mutants of C. reinhardtii
A library of nuclear mutants was generated by insertional DNA mutagenesis using the arg7 gene to complement the arg7 nuclear mutant of C. reinhardtii (Purton and Rochaix, 1995). Transformants were screened for potential PS II– deficient mutations by chlorophyll fluorescence, immunoblot, and Northern analyses (refer to Materials and Methods and see Table I). Five mutants were identified (hf149, hf233, hf261, hf859, and hf1085) which failed to accumulate the D1 protein but contained wild-type amounts of other chloroplast proteins. Preparations of membrane proteins were analyzed for the presence of D1, D2, ATPase, the light-harvesting complex (LHC), and photosystem I. Soluble protein fractions were analyzed for the presence of ribulose bis-phosphate oxygenase/carboxylase and a component of the PS II oxygen evolution complex (OEE1). As expected, none of the mutants that lacked accumulation of D1 had detectable accumulation of the D2 protein (Table I). Other chloroplast localized proteins accumulate at near wild-type levels in each of the mutant strains (Table I). A representative Western showing each of the proteins analyzed in the hf149 strain is shown in Fig. 1.
Table I.
Protein and RNA Levels in Nuclear Mutant Strains
| D1 | D2 | LHC | ATPase | CP I | Rubisco | OEE1 | psbA | psbD | rbcL | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| wt | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ||||||||||
| hfl49 | − | − | ++ | ++ | nd | ++ | ++ | + | ++ | ++ | ||||||||||
| hf233 | − | − | ++ | ++ | nd | nd | nd | + | +++ | ++ | ||||||||||
| hf261 | − | − | ++ | ++ | nd | nd | nd | + | ++ | ++ | ||||||||||
| hf859 | − | − | ++ | ++ | ++ | ++ | − | + | ++ | ++ | ||||||||||
| hf1085 | − | − | ++ | ++ | ++ | ++ | + | + | +++ | ++ |
+++, greater than 120% wt levels; ++, wt levels; +, less than 50% wt levels; −, less than 10% wt levels; nd, not determined. The five nuclear mutants were tested for the presence of several different chloroplast-localized proteins and RNAs. Immunoblots of membrane or soluble proteins were assayed for D1, D2, LHC, ATPase, photosystem I components (CP I), ribulose bis-phosphate oxygenase/carboxylase (RBCase), and a component of the PS II OEE1. Northern blot analysis of total RNA from wt and the mutant strains was used to determine the levels of psbA (encoding D1), psbD (encoding D2), and rbcL (encoding RBCase LS) mRNA present.
Figure 1.

Protein accumulation in wt and mutant strains. Equal quantities of membrane or soluble proteins were separated by SDS-PAGE and blotted to nitrocellulose. Identical filters were probed with antiserum against D1, D2, LHCII, ATPase, ribulose bis-phosphate oxygenase/ carboxylase, and a component of the PS II OEE1. Representative immunoblots of proteins from nuclear mutant hf149 compared with wt and Fud7, a chloroplast mutant with a psbA gene deletion (4).
To determine if these mutants have a specific defect in psbA translation, the mutant strains were pulse labeled for 10 min with [14C]acetate to assay protein synthesis rates. Cycloheximide was added to inhibit cytoplasmic, but not chloroplast, translation. Fig. 2 shows the relative levels of expression of the predominant chloroplast-encoded proteins in the hf149 mutant. This strain has undetectable levels of psbA translation with near wt translation of other chloroplast encoded mRNAs. The other four mutants had similar profiles (data not shown). Slight changes in the pattern of other chloroplast proteins were observed in the mutants compared with wt, these changes are likely due to degradation of other PS II proteins (e.g., P5 or P6), which turn over more rapidly in the absence of D1 accumulation.
Figure 2.
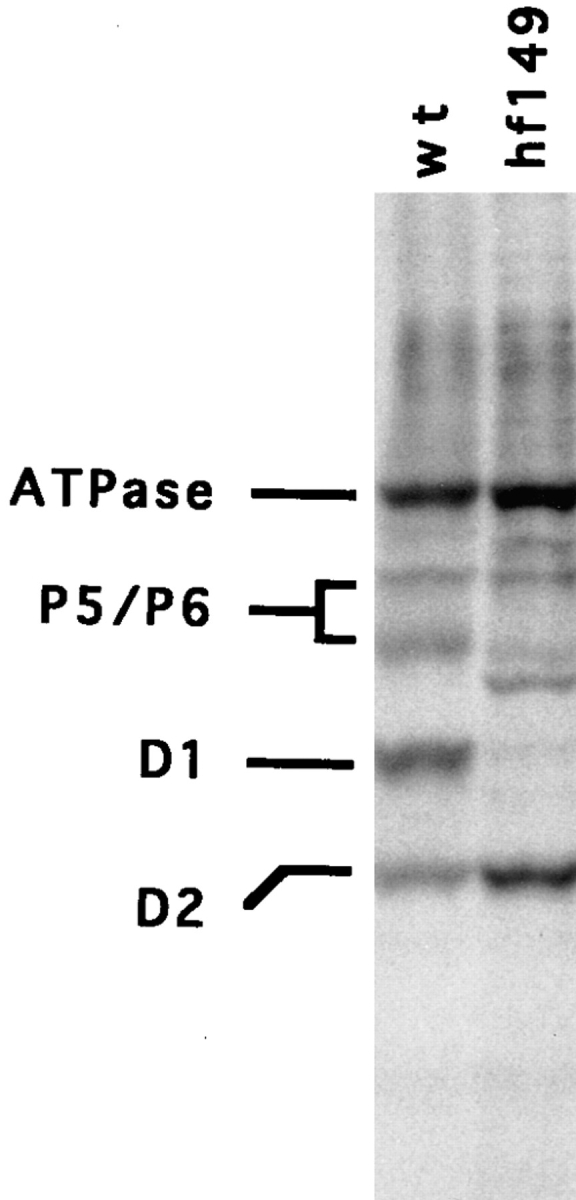
Synthesis of chloroplast-encoded proteins from wt and hf149 mutant strains measured by [14C]acetate pulse labeling. Equal quantities of labeled proteins were loaded in each lane. The positions of the major components of PS II (D1, D2, P5, and P6) as well as ATPase are indicated.
Genetic Analysis of Mutants
Intercrosses between mutant strains showed that mutants hf261, hf859, and hf1085 fall into different complementation groups (Table II). Additionally, all three of these mutants identify genes that are distinct from the gene mutated in F35, another nuclear mutant affected in translation initiation of the psbA mRNA (Yohn et al., 1996). Strains hf149 and hf233 were not included in this analysis because these strains failed to mate with the other mutants, or produced too few viable progeny for a reliable analysis. These data show that at least four nuclear loci are specifically required for the expression of the psbA mRNA in the chloroplast.
Table II.
Complementation of Nuclear Mutant Strains
| Intercross | Total number of progeny | ac secregants | AC segregants | |||
|---|---|---|---|---|---|---|
| hf1085 × hf261 | 46 | 19 | 27 | |||
| hf1085 × hf859 | 6 | 0 | 6 | |||
| hf1085 × F35 | 10 | 6 | 4 | |||
| hf859 × hf261 | 8 | 2 | 6 | |||
| hf859 × F35 | 91 | 88 | 3 | |||
| hf261 × F35 | 15 | 13 | 2 |
hf, high fluorescent; ac, acetate requiring; AC, nonacetate requiring. The presence of nonacetate-requiring (AC, i.e., wt) segregants suggests that the genes affected within these strains are in different complementation groups.
Accumulation and Polysomal Association of psbA mRNA in Mutants
The absence of D1 protein synthesis in the nuclear mutants could be due to a variety of blocks in psbA gene expression. To determine if the psbA gene is transcribed and if psbA mRNA accumulates in the mutant strains, Northern blot analysis of psbA mRNA, encoding the D1 protein, was undertaken. As shown in Table I and Fig. 3, psbA and other chloroplast mRNAs accumulate in each of the mutants, although the psbA mRNA is at a somewhat reduced level relative to wt. The fact that measurable amounts of psbA mRNA accumulate, whereas no D1 protein synthesis is observed, suggests that translation is the defective step for D1 protein accumulation in each of these mutants.
Figure 3.
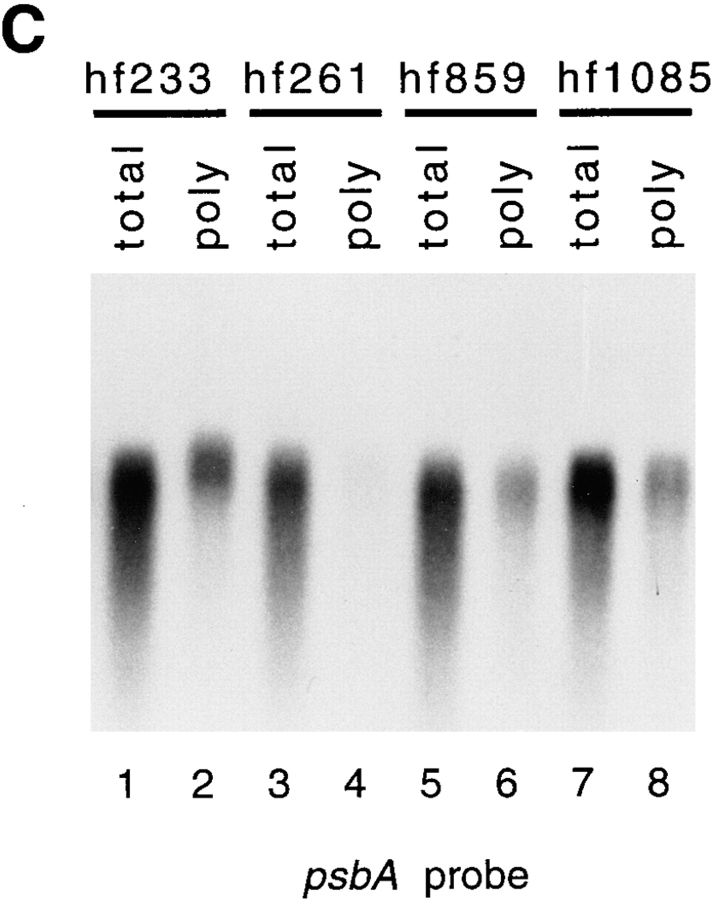
Northern blot analysis of psbA (A and C) and psbD (B) mRNA accumulation in wt and mutant strains. Total RNA and RNA associated with polysomes was examined for the presence of these RNAs. 2 μg of RNA was loaded in each lane. (A and B) Total and polysomal RNA from the hf149 mutant strain probed with labeled psbA (A) or psbD (B) cDNAs. Dilutions of wt total RNA are shown for comparison to levels of RNA in the hf149 mutant strain. (C) Levels of total and polysomal psbA RNA are shown for the hf233, hf261, hf859, and hf1085 mutant strains.
To identify the point at which translation is blocked, RNA/ribosome association was assayed for each of the mutant strains. This analysis should determine if the mRNA is able to associate with ribosomes and suggest whether the mutations affect translation at initiation or in events after initiation (i.e., elongation or termination). Polyribosomes were separated from mRNA not associated with ribosomes by pelleting through a sucrose cushion. psbA mRNA associated with polysomes and from whole cells was assayed in the hf149 mutant by hybridization of a psbA-specific probe. Reduced levels of psbA mRNA are associated with polysomal RNA relative to wt (Fig. 3 A), suggesting that the hf149 mutant is affected in its ability to initiate translation of the psbA mRNA. psbD mRNA shows essentially wt accumulation and polysome association in hf149 (Fig. 3 B), indicating that the inability of mRNA to associate with ribosomes in the hf149 strain is specific to the psbA mRNA. Mutants hf261 and hf859 also show greatly reduced psbA/ribosome association, whereas hf1085 has slightly reduced psbA polysome association (Fig. 3 C). Mutant hf233 appears to have fairly normal levels of psbA mRNA associated with polysomes. Each of these mutants have normal levels of psbD mRNA associated with ribosomes as expected (data not shown).
Status of psbA RNA-binding Complex and Accumulation of psbA RNA-binding Proteins
Binding of proteins to the 5′ UTR of the psbA mRNA has been shown to correlate with the level of D1 synthesis under varying physiological (Danon and Mayfield, 1991) and biochemical (Danon and Mayfield, 1994a ,b) conditions, and in different genetic backgrounds (Yohn et al., 1996). The psbA RNA-binding activity present in lysates from these translation deficient mutants was analyzed by GMS coupled with RNase T1 protection (Fig. 4, T1-GMS). In all cases, in vitro binding to the 5′ UTR of psbA RNA is less than that observed in wt lysates. Mutant hf149 shows no detectable binding to psbA mRNA in vitro (Fig. 4), whereas mutant hf261 shows binding activity at <10% of wt levels. Mutants hf859 and hf1085 both have ∼50% of the binding activity of wt, whereas mutant hf233 has close to wt levels of psbA RNA binding, although this mutant contains an additional RNA–protein complex that is not seen in the wt or other mutant samples (Fig. 4).
Figure 4.
psbA RNA binding activity in wt and mutant strains. Binding activity with 6 μg of protein was measured by T1-GMS assays. The first lane in each panel contains labeled RNA but no proteins (no lysate) for comparison.
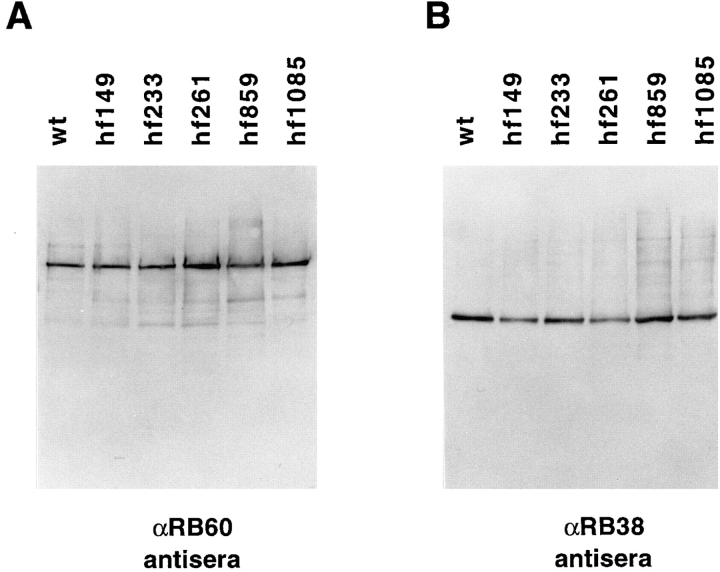
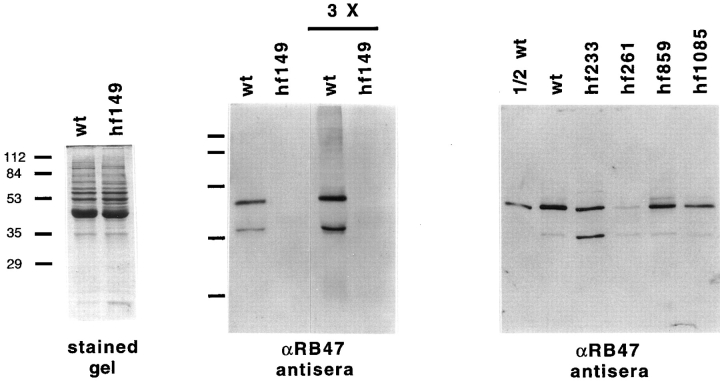
The in vitro binding described above is primarily due to the binding of RB47 of the RNA-binding complex (Yohn et al., 1998). This protein is a member of the PABP family and is in close contact with the psbA mRNA (Danon and Mayfield, 1991). Immunoblot analysis was performed using antisera against RB47, as well as antisera against the RB38 and RB60 components of the complex. Heparin agarose-purified proteins from wt and each of the mutants was assayed. This assay should indicate the amount of these proteins in wt and mutant cells independent of their ability to bind the psbA RNA, since binding of psbA RNA-binding proteins to heparin agarose is unrelated to their ability to bind psbA RNA (Yohn, C.B., and S.P. Mayfield, unpublished data). Both the RB38 and RB60 proteins are present at levels similar to wt in all mutants assayed (Fig. 5). When equal amounts of protein from wt and hf149 are analyzed, no RB47 is found to accumulate in hf149 cells (Fig. 6, middle panel), and RB47 cannot be detected even when a threefold excess of protein (the upper limit for total protein loading in this system) is used. RB47 accumulation in hf261 was found to be <10% of wt levels (Fig. 6, right panel), whereas RB47 accumulation in the other three mutants was found to be at least 50% of wt levels. Accumulation of RB47 appears to correlate closely to the level of in vitro binding detected by T1-GMS (refer to Fig. 4). The loss of psbA binding activity in hf149 and hf261 (and reduction in other mutants) as determined by T1-GMS could therefore be accounted for by the loss of the RB47 protein in these strains. Note that the hf233 strain, which contains an additional band in the RNA-binding assay, has an increased accumulation of a lower molecular weight species of the RB47 protein (Fig. 6, right panel).
Figure 5.
Protein accumulation of psbA-specific RNA-binding proteins RB60 and RB38 in wt and mutant strains. Antisera against RB60 (A) and RB38 (B) were used for immunoblots of two identical filters with 3 μg of heparin agarose-purified proteins loaded in each lane. Proteins were separated by SDS-PAGE and blotted to nitrocellulose filters before decoration with specific antisera.
Figure 6.
Protein accumulation of psbA specific RNA-binding protein RB47 in wt and mutant strains. Left panel, Coomassie-stained gel of equal amounts of wt and hf149 heparin agarose- purified proteins. Molecular weight standards are indicated at the left. Middle panel, antiserum against RB47 was used for an immunoblot of a gel with lanes identical to the left panel as well as additional lanes with threefold excess (3×) protein loaded for both wt and hf149. Right panel, immunoblot using antiserum against RB47 on heparin agarose-purified proteins from wt and hf233, hf261, hf859, and hf1085 mutant strains. A dilution of wt is shown for comparison to the mutant strains.
Discussion
Message-specific RNA-binding proteins have previously been implicated in light-regulated translational activation of the chloroplast encoded psbA mRNA (Danon and Mayfield, 1991, 1994a ,b, Yohn et al., 1996). The role of one of these RNA-binding proteins, RB47, is now clearer, since evidence for its function comes from several independent sources. First, biochemical analysis has shown that this protein (along with other members of the psbA RNA-binding complex) bind with high affinity and specificity to the psbA 5′ UTR in a manner consistent with a role in translational regulation; high levels of binding are observed in the light when psbA translation is high, and low levels of binding are observed in the dark when translation is low (Danon and Mayfield, 1991). Second, the predicted amino acid sequence of RB47 suggests the role this protein plays in translation. RB47 belongs to a family of proteins known as poly(A)-binding proteins that bind RNA and have been shown to play a role in translation initiation (Sachs and Davis, 1989; Proweller and Butler, 1996; Tarun and Sachs, 1996; Yohn et al., 1998). Finally, genetic analysis has predicted translational activators of chloroplast mRNAs (Kuchka et al., 1988; Rochaix et al., 1989; Girard-Bascou et al., 1992; Yohn et al., 1996), and here we show by characterization of psbA translation deficient mutants that the absence of RB47 corresponds directly to the loss of translational initiation of the psbA mRNA, thus defining RB47 as a translational activator of the psbA mRNA.
Although proteins that bind to the 5′ UTR of chloroplast mRNAs seem likely candidates for translational activators, no direct link had been made to the body of genetic data before the characterization of these mutants. The hf261 mutant is not likely to be a mutation directly in the RB47 gene, as this mutant is tagged with the arg7 gene and Southern and Northern analysis indicates that the RB47 gene is intact and produces normal amounts of RB47 mRNA in this strain (data not shown). The hf149 mutation is not tagged by the arg7 gene, so this mutation could potentially contain point mutations that disrupt RB47 accumulation while leaving the RB47 gene and RB47 mRNA accumulation intact (data not shown). These genetic data leave open the possibility that the absence of RB47 protein in these strains is the result of a loss of psbA translation, rather than the cause of it. Although this is a formal possibility, it is highly unlikely given the fact that the RB47 protein accumulates in the other psbA translation-deficient mutants described here (hf233, hf859, and hf1085), and that the other psbA RNA-binding proteins RB38 and RB60 accumulate to normal levels in all of the mutant strains. Thus, the hf149 and hf261 mutations provide strong evidence that the RB47 protein is directly involved in translational regulation of the chloroplast-encoded psbA mRNA. Other nuclear mutants determined to affect both RB47 and psbA translation further suggests the importance of this protein in psbA expression (Yohn, et al., 1996; Cohen et al., manuscript submitted for publication). Identification of the specific defect in the hf149 and hf261 mutant, as well as characterization of the other described mutants, should yield further insights into the process of psbA mRNA binding and the mechanism of translational regulation.
The dramatic reduction in the amount of psbA mRNA associated with ribosomes in both the hf149 and hf261 mutants suggests that RB47 is specifically required for ribosomes to initiate translation of the psbA mRNA. Although the identification of message-specific translational activators in the chloroplast has not previously been shown, other organellar systems have been characterized in which similar mechanisms for controlling and coordinating gene expression may be used, most notably the mitochondria of yeast. The COX3 mRNA of Saccharomyces cerevisiae mitochondria is translationally regulated by a complex of at least three proteins which have been shown genetically (Wiesenberger et al., 1995) and biochemically (Brown et al., 1994) to interact with each other and with the COX3 mRNA. One of these proteins (PET122) also interacts with the mitochondrial ribosome (Haffter et al., 1990, 1991; McMullin et al., 1990), suggesting a model for translational activation in which these proteins facilitate the initial interaction between the mRNA and the ribosome. A similar mechanism may be involved with RB47, the psbA mRNA, and chloroplast ribosomes.
The involvement of RB47 in translational initiation of psbA mRNA in the chloroplast is now clear, though the exact role of this PABP is yet to be elucidated. At least four genetic loci are implicated in RB47 accumulation and psbA translation initiation. Some of these genes may encode proteins, other than RB47, that bind to the psbA mRNA and are required for translation initiation of this mRNA. The hf859 and hf1085 mutants may be this type, since these mutations affect the ability of the psbA mRNA to associate with ribosomes, but not to the same degree as the hf149 and hf261 mutants. The psbA mRNA is associated with ribosomes in the hf233 mutant, so this mutation appears to be acting after initiation and thus probably downstream of the other mutations. The hf149 and hf261 mutants appear to act through RB47, either as mutations in the RB47 gene itself, or by specifically affecting accumulation of the RB47 protein. Thus the accumulation of the RB47 protein might also be a regulated event. In fact, cytoplasmic PABPs from other organisms have been shown to be translationally regulated (de Melo Neto et al., 1995; Bag and Wu, 1996). RB47 appears to share this characteristic in that the 5′ UTR of the RB47 mRNA affects its expression in heterologous systems (Mayfield, S.P., unpublished data). This regulated expression of RB47 may be a mechanism for developmental regulation of photosystem accumulation in chloroplasts.
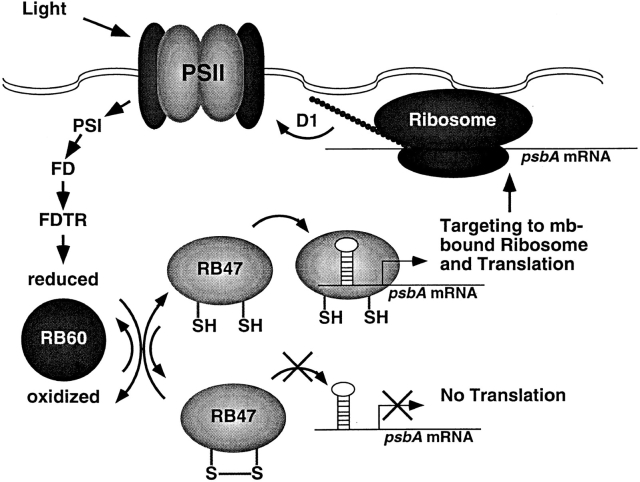
A model for the temporal regulation of psbA expression in the chloroplast is described in Fig. 7. In this model, light acts through PS II and PS I to produce a reducing environment in the chloroplast. Reducing equivalents are passed to the chloroplast PDI (RB60) through ferrodoxin (FD) and ferrodoxin-thioredoxin reductase. RB47's binding activity is then modulated by RB60 in a redox dependent manner through reversible oxidation of sulfhydryl groups within RB47 (Kim and Mayfield, 1997). Binding of RB47 to the psbA mRNA is required for RNA/ribosome association, where this protein acts as a message-specific translation initiation factor, bringing the mRNA and ribosome together. Recently RB47 was shown to be associated with the chloroplast membrane system (Zerges and Rochaix, 1998), suggesting that RB47, along with other members of the psbA RNA-binding complex, could be acting in both translational activation and as a targeting mechanism. Under this type of mechanism, the soluble RB47 protein would bind the psbA mRNA and then direct this mRNA to ribosomes specifically associated with the thylakoid membranes, the site at which the hydrophobic D1 protein is synthesized.
Figure 7.
A model for light-activated translational regulation of the psbA mRNA in the chloroplast. A reducing environment is generated by PSII during the light reactions of photosynthesis. These reducing equivalents are passed through photosystem I (PSI), ferrodoxin (FD), and ferrodoxin-thioredoxin reductase (FDTR) to the chloroplast PDI (RB60). RB60 modulates the binding of RB47 to the psbA mRNA in a redox dependent manner through reversible oxidation of sulfhydryl groups in RB47. Binding of RB47 to the structured 5′ UTR of the psbA mRNA is required for translation of the mRNA. The translational activator RB47 acts concurrently to target the psbA mRNA to the membrane, most likely by facilitating translation initiation on membrane (mb−) bound ribosomes. Once the D1 protein is translated, it can be readily inserted into the membrane to assemble into PSII complexes.
Acknowledgments
We thank J. Allen (The Scripps Research Institute, La Jolla, CA) and J. Akman (California State University, Fullerton, CA) for technical assistance and D. Chagnovich (Skirball Institute, New York) and D. Sommers (The Scripps Research Institute) for critical reading of the manuscript.
This work was supported by funds from the National Science Foundation (MCB-9514698) to S.P. Mayfield, and the U.S. Department of Energy (90ER20009) and U.S. Department of Agriculture (93-37301-9428) to M.R. K Kuchka. A. Cohen was supported by a California State University Research, Scholarship, and Creative Activity Grant (1997–1998). C.B. Yohn was supported by an NSF predoctoral fellowship and an NSF graduate research traineeship.
Abbreviations used in this paper
- FD
ferrodoxin
- hf
high fluorescence
- LHC
light-harvesting complex
- OEE1
oxygen evolution complex
- PABP
poly(A)-binding protein
- PDI
protein disulfide isomerase
- PS II
photosystem II
- T1-GMS
T1 RNase protection-gel mobility shift
- TAP
Tris-acetate-phosphate
- UTR
untranslated region
- wt
wild type
Footnotes
Address all correspondence to Stephen P. Mayfield, Department of Cell Biology, and The Skaggs Institute of Chemical Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037. Tel.: (619) 784-9848. Fax: (619) 784-9840. E-mail: mayfield@scripps.edu
References
- Bag J, Wu J. Translational control of poly(A)-binding protein expression. Eur J Biochem. 1996;237:143–152. doi: 10.1111/j.1432-1033.1996.0143n.x. [DOI] [PubMed] [Google Scholar]
- Barkan A. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO (Eur Mol Biol Organ) J. 1988;7:2637–2644. doi: 10.1002/j.1460-2075.1988.tb03116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennoun, P., and P. Delepelaire. 1982. Isolation of photosynthesis mutants in Chlamydomonas. In Methods in Chloroplast Molecular Biology. M. Edelman, R.B. Hallick, and N.-H. Chua, editors. Elsevier Biomedical Press, Amsterdam, The Netherlands. 25–38.
- Bennoun P, Spierer-Herz M, Erickson J, Girard-Bascou J, Pierre Y, Delsome M, Rochaix J-D. Characterization of photosystem II mutants of Chlamydomonas reinhardtii lacking the psbAgene. Plant Mol Biol. 1986;6:151–160. doi: 10.1007/BF00021484. [DOI] [PubMed] [Google Scholar]
- Brown NG, Costanzo MC, Fox TD. Interactions among three proteins that specifically activate translation of the mitochondrial COX3 mRNA in Saccharomyces cerevisiae. . Mol Cell Biol. 1994;14:1045–1053. doi: 10.1128/mcb.14.2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. Light regulated translational activators: identification of chloroplast gene specific mRNA binding proteins. EMBO (Eur Mol Biol Organ) J. 1991;10:3993–4001. doi: 10.1002/j.1460-2075.1991.tb04974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. ADP-dependent phosphorylation regulates RNA-binding in vitro: implications in light-modulated translation. EMBO (Eur Mol Biol Organ) J. 1994a;13:2227–2235. doi: 10.1002/j.1460-2075.1994.tb06500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. Light-regulated translation of chloroplast messenger RNAs through redox potential. Science. 1994b;266:1717–1719. doi: 10.1126/science.7992056. [DOI] [PubMed] [Google Scholar]
- de Melo Neto, O.P., N. Standart, and C. Martins de Sa. Autoregulation of poly(A)-binding protein synthesis in vitro. Nucleic Acids Res. 1995;23:2198–2205. doi: 10.1093/nar/23.12.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy R, Purton S, Rochaix JD. The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO (Eur Mol Biol Organ) J. 1989;8:2803–2809. doi: 10.1002/j.1460-2075.1989.tb08426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JM, Rahire M, Malnoë P, Girard-Bascou J, Pierre Y, Bennoun P, Rochaix J-D. Lack of the D2 protein in a Chlamydomonas reinhardtii psbDmutant affects photosystem II stability and D1 expression. EMBO (Eur Mol Biol Organ) J. 1986;5:1745–1754. doi: 10.1002/j.1460-2075.1986.tb04422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillham NW, Boynton JE, Hauser CR. Translational regulation of gene expression in chloroplasts and mitochondria. Annu Rev Genet. 1994;28:71–93. doi: 10.1146/annurev.ge.28.120194.000443. [DOI] [PubMed] [Google Scholar]
- Girard-Bascou J, Pierre Y, Drapier D. A nuclear mutation affects the synthesis of the chloroplast psbA gene production Chlamydomonas reinhardtii. . Curr Genet. 1992;22:47–52. doi: 10.1007/BF00351741. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M, Girard-Bascou J, Choquet Y, Rochaix JD. Trans-splicing mutants of Chlamydomonas reinhardtii. . Mol Gen Genet. 1990;223:417–425. doi: 10.1007/BF00264448. [DOI] [PubMed] [Google Scholar]
- Haffter P, McMullin TW, Fox TD. A genetic link between an mRNA-specific translational activator and the translation system in yeast mitochondria. Genetics. 1990;125:495–503. doi: 10.1093/genetics/125.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, McMullin TW, Fox TD. Functional interactions among two yeast mitochondrial ribosomal proteins and an mRNA-specific translational activator. Genetics. 1991;127:319–326. doi: 10.1093/genetics/127.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E.H. 1989. The Chlamydomonas Sourcebook. Academic Press, Inc., San Diego, CA. 25–40.
- Kim J, Mayfield SP. Protein disulfide isomerase as a regulator of chloroplast translational activation. Science. 1997;278:1954–1957. doi: 10.1126/science.278.5345.1954. [DOI] [PubMed] [Google Scholar]
- Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. . Proc Natl Acad Sci USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchka MR, Goldschmidt-Clermont M, van Dillewijn J, Rochaix JD. Mutation at the Chlamydomonas nuclear NAC2 locus specifically affects stability of the chloroplast psbDtranscript encoding polypeptide D2 of PS II. Cell. 1989;58:869–876. doi: 10.1016/0092-8674(89)90939-2. [DOI] [PubMed] [Google Scholar]
- Kuchka MR, Mayfield SP, Rochaix J-D. Nuclear mutations specifically affect the synthesis and/or degradation of the chloroplast-encoded D2 polypeptide of photosystem II in Chlamydomonas reihardtii. . EMBO (Eur Mol Biol Organ) J. 1988;7:319–324. doi: 10.1002/j.1460-2075.1988.tb02815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RP, Ebersold P. The genetics and cytology of Chlamydomonas. . Annu Rev Microbiol. 1960;14:197–216. doi: 10.1146/annurev.mi.14.100160.001213. [DOI] [PubMed] [Google Scholar]
- Malnoë P, Mayfield SP, Rochaix JD. Comparative analysis of the biogenesis of photosystem II in the wild-type and Y-1 mutant of Chlamydomonas reinhardtii. . J Cell Biol. 1988;106:609–616. doi: 10.1083/jcb.106.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield SP, Bennoun P, Rochaix JD. Expression of the nuclear encoded OEE1 protein is required for oxygen evolution and stability of photosystem II particles in Chlamydomonas reinhardtii. . EMBO (Eur Mol Biol Organ) J. 1987;6:313–618. doi: 10.1002/j.1460-2075.1987.tb04756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield SP, Cohen A, Danon A, Yohn CB. Translation of the psbA mRNA of Chlamydomonas reinhardtiirequires a structured RNA element contained within the 5′ untranslated region. J Cell Biol. 1994;127:1537–1545. doi: 10.1083/jcb.127.6.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield SP, Yohn CB, Cohen A, Danon A. Regulation of chloroplast gene expression. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:147–166. [Google Scholar]
- McMullin TW, Haffter P, Fox TD. A novel small-subunit ribosomal protein of yeast mitochondria that interacts functionally with an mRNA-specific translational activator. Mol Cell Biol. 1990;10:4590–4595. doi: 10.1128/mcb.10.9.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proweller A, Butler JS. Ribosomal association of poly(A)-binding protein in poly(A)-deficient Saccharomyces cerevisiae. . J Biol Chem. 1996;271:10859–10865. doi: 10.1074/jbc.271.18.10859. [DOI] [PubMed] [Google Scholar]
- Purton S, Rochaix J-D. Characterization of the ARG7 gene of Chlamydomonas reinhardtiiand its application to nuclear transformation. Eur J Phycol. 1995;30:141–148. [Google Scholar]
- Rochaix JD. Post-transcriptional steps in the expression of chloroplast genes. Annu Rev Cell Biol. 1992;8:1–28. doi: 10.1146/annurev.cb.08.110192.000245. [DOI] [PubMed] [Google Scholar]
- Rochaix JD, Kuchka M, Mayfield S, Schirmer-Rahire M, Girard-Bascou J, Bennoun P. Nuclear and chloroplast mutations affect the synthesis or stability of the chloroplast psbC gene product in Chlamydomonas reinhardtii. . EMBO (Eur Mol Biol Organ) J. 1989;8:1013–1021. doi: 10.1002/j.1460-2075.1989.tb03468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB, Davis RW. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Staub JM, Maliga P. Accumulation of D1 polypeptide in tobacco plastids is regulated via the untranslated region of the psbAmRNA. EMBO (Eur Mol Biol Organ) J. 1993;12:601–606. doi: 10.1002/j.1460-2075.1993.tb05692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam L-W, Lefebvre PA. Cloning of flagellar genes in Chlamydomonas reinhardtiiby DNA insertional mutagenesis. Genetics. 1993;135:375–384. doi: 10.1093/genetics/135.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ, Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO (Eur Mol Biol Organ) J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Wiesenberger G, Costanzo MC, Fox TD. Analysis of the Saccharomyces cerevisiae mitochondrial COX3mRNA 5′ untranslated leader: translational activation and mRNA processing. Mol Cell Biol. 1995;15:3291–3300. doi: 10.1128/mcb.15.6.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn CB, Cohen A, Danon A, Mayfield SP. Altered mRNA binding activity and decreased translational initiation in a nuclear mutant lacking translation of the chloroplast psbAmRNA. Mol Cell Biol. 1996;16:3560–3566. doi: 10.1128/mcb.16.7.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn CB, Cohen A, Danon A, Mayfield SP. A poly(A) binding protein functions in the chloroplast as a message-specific translation factor. Proc Natl Acad Sci USA. 1998;95:2238–2243. doi: 10.1073/pnas.95.5.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges W, Rochaix JD. Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. . J Cell Biol. 1998;140:101–110. doi: 10.1083/jcb.140.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]