Abstract
We have investigated the role of membrane proteins and lipids during early phases of the cotranslational insertion of secretory proteins into the translocation channel of the endoplasmic reticulum (ER) membrane. We demonstrate that all steps, including the one during which signal sequence recognition occurs, can be reproduced with purified translocation components in detergent solution, in the absence of bulk lipids or a bilayer. Photocross-linking experiments with native membranes show that upon complete insertion into the channel signal sequences are both precisely positioned with respect to the protein components of the channel and contact lipids. Together, these results indicate that signal sequences are bound to a specific binding site at the interface between the channel and the surrounding lipids, and are recognized ultimately by protein–protein interactions. Our data also suggest that at least some signal sequences reach the binding site by transfer through the interior of the channel.
Keywords: signal sequence, endoplasmic reticulum, translocation, cross-linking, Sec61p
Cotranslational protein transport across the membrane of the endoplasmic reticulum (ER)1 is initiated in the cytosol when the signal sequence of a growing polypeptide chain has emerged from the translating ribosome and is recognized by the signal recognition particle (SRP) through its 54-kD subunit (for review, see Walter and Johnson, 1994; Rapoport et al., 1996). The complex consisting of the ribosome, the nascent polypeptide chain, and SRP is then targeted to the membrane via two interactions. One occurs between the SRP and the SRP receptor (docking protein; Gilmore et al., 1982; Meyer et al., 1982), and the other between the ribosome and the Sec61p complex (Kalies et al., 1994), a membrane component forming the channel through which polypeptides traverse the membrane (Deshaies and Schekman, 1987; Görlich et al., 1992b ; Mothes et al., 1994; Hanein et al., 1996). The initial interaction between the ribosome– nascent chain complex and the Sec61p complex is weak; it is sensitive to high salt concentrations and the nascent chain remains accessible to added proteases (Jungnickel and Rapoport, 1995). Once the nascent polypeptide chain has reached a critical length, a second stage is attained in which the ribosome is bound more tightly to the Sec61p complex; it is no longer extractable with high salt and the nascent chain is inaccessible to proteolysis, reflecting its insertion into the Sec61p channel (Jungnickel and Rapoport, 1995). Cross-linking experiments show that at this stage the signal sequence contacts another translocation component, the translocating chain associating membrane (TRAM) protein (Görlich et al., 1992a ; Jungnickel and Rapoport, 1995). Although additional steps may occur during initiation of translocation (Nicchitta et al., 1991; Nicchitta and Zheng, 1997), once inserted into the channel, the nascent chain appears to be committed to translocation.
Recent experiments have provided evidence that the transition between the two modes of interaction of the ribosome–nascent chain complex with the membrane is a decisive step during protein translocation (Jungnickel and Rapoport, 1995). The transition involves signal sequence recognition and occurs at the same nascent chain length as the opening of the channel towards the lumen, as indicated by fluorescence quenching experiments (Crowley et al., 1994). Thus, tightening of the ribosome–Sec61p interaction, signal sequence recognition inside the membrane, and gating of the channel all seem to be simultaneous events. The molecular mechanisms by which these important steps take place are unknown.
A critical problem is how the signal sequence is recognized inside the membrane. According to one model, a signal sequence would partition into the bilayer by virtue of its hydrophobicity, and would never interact in a specific manner with the channel proteins. Nonfunctional signal sequences would be unable to enter the lipid phase. Experiments with synthetic signal peptides have provided evidence that such a simple partitioning process could explain the discrimination between functional and nonfunctional, i.e., less hydrophobic, signal sequences (Briggs et al., 1985). However, in these experiments, the lipid phase could have mimicked the hydrophobic pocket of a receptor protein. Other models have indeed proposed that the signal sequence must bind to a particular site of a channel protein in order to initiate translocation (Blobel and Dobberstein, 1975; Prehn et al., 1980; Robinson et al., 1987). In this case, the binding strength between the signal sequence and the receptor protein would distinguish functional and nonfunctional signal sequences. The Sec61p complex and the TRAM protein would be the best candidates for signal sequence receptors: they can be cross-linked to the signal sequences of inserted nascent chains and are essential and sufficient for the translocation of all (Sec61p complex) or most (TRAM) substrates tested to date (Görlich et al., 1992b ; Görlich and Rapoport, 1993; Jungnickel and Rapoport, 1995; Voigt et al., 1996). Since all experiments so far have been performed with these proteins in a phospholipid bilayer, discrimination between the two types of models has not been possible. The demonstrated requirement of the Sec61p complex for signal sequence recognition (Jungnickel and Rapoport, 1995) may reflect its participation as a ribosome receptor (Kalies et al., 1994) rather than as a binding site for the signal sequence.
We now report that all early steps of cotranslational protein translocation, including the decisive step during which signal sequence recognition occurs, can be reproduced with purified translocation components in detergent solution, in the absence of a lipid bilayer. Photocross-linking experiments with native membranes indicate that upon complete insertion into the channel, signal sequences are in proximity to both lipids and channel components, contacting the latter in a specific manner. These results suggest that signal sequences are recognized by a specific binding site at the interface between channel and lipids, and are not simply partitioned into the lipid phase. Our data also suggest that at least some signal sequences reach the binding site by transfer through the interior of the channel.
Materials and Methods
Generation of Ribosome–Nascent Chain Complexes
Truncated mRNAs coding for 59 or 86 amino acids of wild-type preprolactin as well as mRNA coding for a signal sequence deletion mutant (pPLΔ13-15) were generated as described (Jungnickel and Rapoport, 1995). Truncated mRNAs coding for 50 or 86 amino acids of prepro-α-factor were produced by transcription of PCR-amplified fragments of the gene. The 3′ end primer introduced two or three additional methionines to allow detection of the polypeptide chains by incorporation of [35S]methionine. Ribosome–nascent chain complexes (RNCs) were generated in a wheat germ translation system and either used directly or isolated by centrifugation as described (Jungnickel and Rapoport, 1995).
Experiments with Detergent Extracts of Microsomes
To produce a detergent extract, canine pancreatic microsomes stripped of ribosomes by puromycin and high salt treatment (PK-RM) were resuspended at a concentration of 1–2 eq/μl (eq; for definition, see Walter et al., 1981) in a buffer containing 50 mM Hepes, pH 7.5, 400 mM KoAc, 5 mM MgAc, 2 mM β-mercaptoethanol, 10% glycerol, 1 μg/ml pepstatin, 3 μg/ml elastatinal, 5 μg/ml chymostatin, 10 μg/ml leupeptin. They were subsequently solubilized with 1% digitonin (final concentration). After incubation for 10 min on ice, the sample was centrifuged for 10 min at 100,000 rpm in a Beckman TL100 rotor. Sec61p immunodepletion of the detergent extract (Kutay et al., 1995) and purification of the Sec61p complex (Görlich and Rapoport, 1993) were as described. 1–2 eq detergent extract of PK-RM were mixed with 2 μl of a suspension of RNCs (ribosome concentration of about 150 nM in a buffer containing 50 mM Hepes, pH 7.5, 150 mM potassium acetate, and 2 mM magnesium acetate). The final volume was 10 μl and the concentrations of potassium acetate and magnesium acetate were adjusted to 150 mM and 2 mM, respectively.
Nascent Chain Insertion with Purified Translocation Components
Sec61p complex and TRAM were purified as before (Görlich and Rapoport, 1993) and the protein concentrations estimated to be 0.5 mg/ml by Coomassie blue staining and comparison with bovine serum albumin as standard. For insertion assays, RNCs containing 0.2 pmol ribosomes were incubated in a final volume of 10 μl with different proteins and detergents as indicated in the figures. All samples were adjusted to final concentrations of 50 mM Hepes, pH 7.5, 150 mM potassium acetate, 8 mM magnesium acetate, and 150 mM sucrose. The lipid mixture was prepared as described (Görlich and Rapoport, 1993) and consists of phosphatidyl- choline, -ethanolamine, -inositol, and -serine (Sigma Chemical Co., St. Louis, MO) in a ratio of 63:18:9:2. In nascent chain insertion assays, lipid was used at a final concentration of 2 mg/ml.
Site-specific Photocross-linking
cDNAs coding for preprolactin and prepro-α-factor were cloned into pAlter (Promega Corp., Madison, WI) and stop codons (TAG) were introduced at various positions in the regions coding for the hydrophobic portions of the signal sequences. mRNAs were translated for 20 min in the wheat germ system in the presence of [35S]methionine, SRP, and suppressor tRNA carrying a modified phenylalanine (TmdPhe) as described (High et al., 1993; Martoglio et al., 1995; Mothes et al., 1997). Translation was stopped by addition of 5 mM cycloheximide. To a 10-μl reaction, microsomal membranes (1 eq) were added for 5 min on ice and 10 min at 26°C, and the samples were irradiated.
Site-directed photocross-linking with the benzophenone photophore was accomplished after translation in the presence of suppressor-tRNA carrying l-2-amino-5-(p-benzoylphenyl)pentanoic acid. This amino acid was prepared by asymmetric synthesis following the general method of Belokon et al. (1988). In brief, the Ni(II)-complex of the Schiff base derived from (S)-2-[(N-benzoyl)amino]benzophenone was alkylated with p-(3-bromopropyl)benzophenone (Kanamori et al., 1997) following method C (Belokon et al., 1988). The diastereomeric complexes were separated by silica gel chromatography and the main isomer (>90%) was decomposed in refluxing HCl and methanol to give l-2-amino-5-(p-benzoylphenyl)pentanoic acid (Bap) after cation exchange chromatography. The amino acid was purified by crystallization from methanol water (found: C, 72.60; H, 6.36; N, 4.8%; for C18H19NO3 expected: C, 72.71; H, 6.44; N, 4.71%; [a]D (c = 1.073, DMF- 5 N HCl) +28.32). Bap was converted into the N-ter.-butyloxycarbonyl (Boc)-protected form and the carboxyl function of the latter activated by formation of the cyanomethyl ester using established protocols (Moroder et al., 1976; Robertson et al., 1991). The activated ester was condensed to the dinucleotide pdCpA to give 5′-phospho-2′-deoxycitydyl(3′-5′)-2′ (3′)-O-[N-Boc-l-2-amino-5-(p-benzoylphenyl)-pentanoyl)]adenosine (Boc-Bap-pdCpA; Robertson et al., 1991). Before ligation to abbreviated suppressor tRNA, Boc-Bap-pdCpA was treated with trifluoroacetic acid to remove the Boc protecting group. Ligation of Bap-pdCpA to suppressor tRNA was done as described earlier for the preparation of (Tmd)phenylalanyl-suppressor-tRNA (Martoglio et al., 1995; Mothes et al., 1997).
Product Analysis
Immunoprecipitations of cross-linked products with antibodies against Sec61α or TRAM (Görlich et al., 1992b ) were performed as described. Protease treatment of samples was carried out as described by Jungnickel and Rapoport (1995), except that 0.5 μl of a saturated solution of phenylmethylsulfonyl fluoride in isopropanol was added to the samples to stop proteolysis. In time course experiments (Figs. 3 and 4), aliquots of the reaction mixture were taken starting immediately after warming to 26°C. The samples were placed on ice, and after the last aliquot was taken all samples were treated with proteinase K. Precipitation with trichloroacetic acid was carried out in the presence of a precipitation aid, either an equal volume of 20% Triton X-100 or 2 μl of wheat germ extract. Extraction of membranes with alkali and cleavage of lipid cross-links with phospholipase A2 were done as described (Mothes et al., 1997).
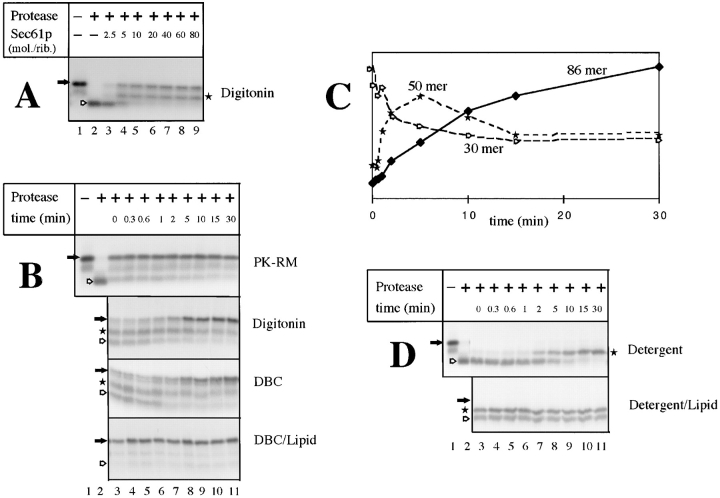
Figure 3.
Insertion of nascent preprolactin chains into the purified, soluble Sec61p channel. (A) Isolated RNCs (∼0.2 pmol ribosomes) carrying preprolactin chains of 86 amino acids were incubated for 15 min at 26°C with different amounts of purified Sec61p complex in 1% digitonin. The samples were then treated with proteinase K and analyzed by SDS-PAGE. The positions of the nascent chains (filled arrow) and of the proteolytic fragments of 30 (hollow arrow) and of 50 (star) amino acids are indicated. (B) Isolated RNCs were incubated at 26°C for different time periods with either ribosome-stripped canine pancreatic microsomes (PK-RM), or with purified Sec61p complex in either 1% digitonin, 0.3% deoxyBigCHAP (DBC), or a mixture of 0.6% deoxyBigCHAP and 2 mg/ ml phospholipids (DBC/Lipid; approximate molar ratio of 80 mol of Sec61p complex per mole of ribosomes). The samples were treated with proteinase K and analyzed by SDS-PAGE. Filled arrows, hollow arrows, and stars indicate the positions of the nondegraded nascent chains, and of the fragments of 30 and 50 amino acids, respectively. (C) An experiment in digitonin, similar to that in the second panel of B, was performed with a Sec61p/ribosome molar ratio of 10 and quantitated to demonstrate that the fragment of 50 amino acids corresponds to a kinetic intermediate. (D) RNCs containing 83 amino acids of a signal sequence mutant of preprolactin (pPLΔ13-15) were incubated for different time periods at 26°C with purified Sec61p in either digitonin or a deoxyBigCHAP–lipid mixture (DBC/Lipid; molar ratio of Sec61p complex to ribosomes of 10), and treated with proteinase K. The positions of the nascent chains (filled arrow) and of their proteolytic fragments (hollow arrow and star) are indicated.
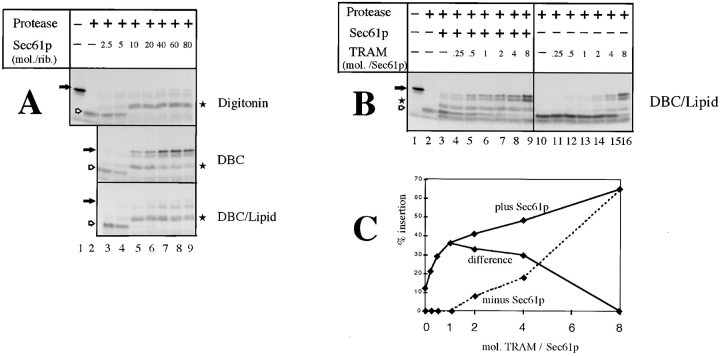
Figure 4.
Insertion of nascent prepro-α-factor chains into the soluble translocation channel. (A) RNCs containing prepro-α-factor chains of 86 amino acids (0.2 pmol ribosomes) were incubated with increasing amounts of purified Sec61p complex in either 1% digitonin, 0.3% deoxyBigCHAP (DBC), or a mixture of 0.6% DBC and 2 mg/ml phospholipids (DBC/Lipid). Insertion of the nascent chains was tested by protection against digestion with proteinase K. The filled arrows indicate the positions of the nondegraded nascent chains, the hollow arrows and stars those of proteolytic fragments of 30 and 50 amino acids, respectively. (B) RNCs containing prepro-α-factor chains (0.2 pmol ribosomes) were incubated with purified Sec61p complex (2 pmol) and increasing amounts of TRAM in a DBC–lipid mixture. Insertion of the nascent chains was analyzed by incubation with proteinase K. (C) The amounts of fully protected prepro-α-factor chains in the experiment shown in B were quantitated. The difference curve indicates that the maximum stimulatory effect of TRAM is seen at an approximate 1:1 molar ratio.
Samples were separated by SDS-PAGE in 10–20% linear acrylamide gels, except in the case of protease protection experiments, in which 12% Tris-Tricine gels were used. Quantitations were done with a PhosphoImager (BAS1000; Fuji Photo Film Co., Tokyo, Japan).
Results
Early Steps of Preprolactin Translocation Reproduced in Detergent Solution
Previous experiments with native microsomes have demonstrated that a nascent 86–amino acid fragment of the secretory protein preprolactin can be targeted to the membrane, triggers tight ribosome binding, and reaches a protease-protected state corresponding to nascent chain insertion into the translocation channel (Jungnickel and Rapoport, 1995). These reactions can be performed independently of SRP and SRP receptor if membrane binding sites are offered in excess over ribosomes (Jungnickel and Rapoport, 1995, Lauring et al., 1995; Neuhof et al., 1998; Raden and Gilmore, 1998). We first used photocross-linking to test whether the same translocation steps can occur with a crude detergent extract of microsomes. Ribosome– nascent chain complexes (RNCs) were produced by in vitro translation of a truncated mRNA in a wheat germ system. Translation was carried out in the presence of modified lysyl-tRNA, leading to the incorporation of photoreactive lysine derivatives at positions where lysines normally occur (Wiedmann et al., 1987). In the fragment of 86 amino acids (86mer), two lysines preceding the hydrophobic core of the signal sequence have emerged from the ribosome and can give rise to cross-links. RNCs containing the 86mer were isolated by sedimentation and incubated with either intact dog pancreatic microsomes or with a digitonin extract prepared from them. After irradiation, several cross-linked products were visible (Fig. 1 A, lanes 2 and 3 vs. lane 1). The major bands could be immunoprecipitated with antibodies against Sec61α (α subunit of the Sec61p complex) or TRAM (lanes 4–7). These data demonstrate that cross-linking to Sec61α occurs with equal efficiency in intact membranes and in detergent solution. Cross-links to TRAM were significantly weaker in detergent solution, perhaps because TRAM does not show strong interactions with either the Sec61p complex (Görlich and Rapoport, 1993) or the ribosome–nascent chain complex (Kalies et al., 1994) after solubilization in digitonin.
Figure 1.
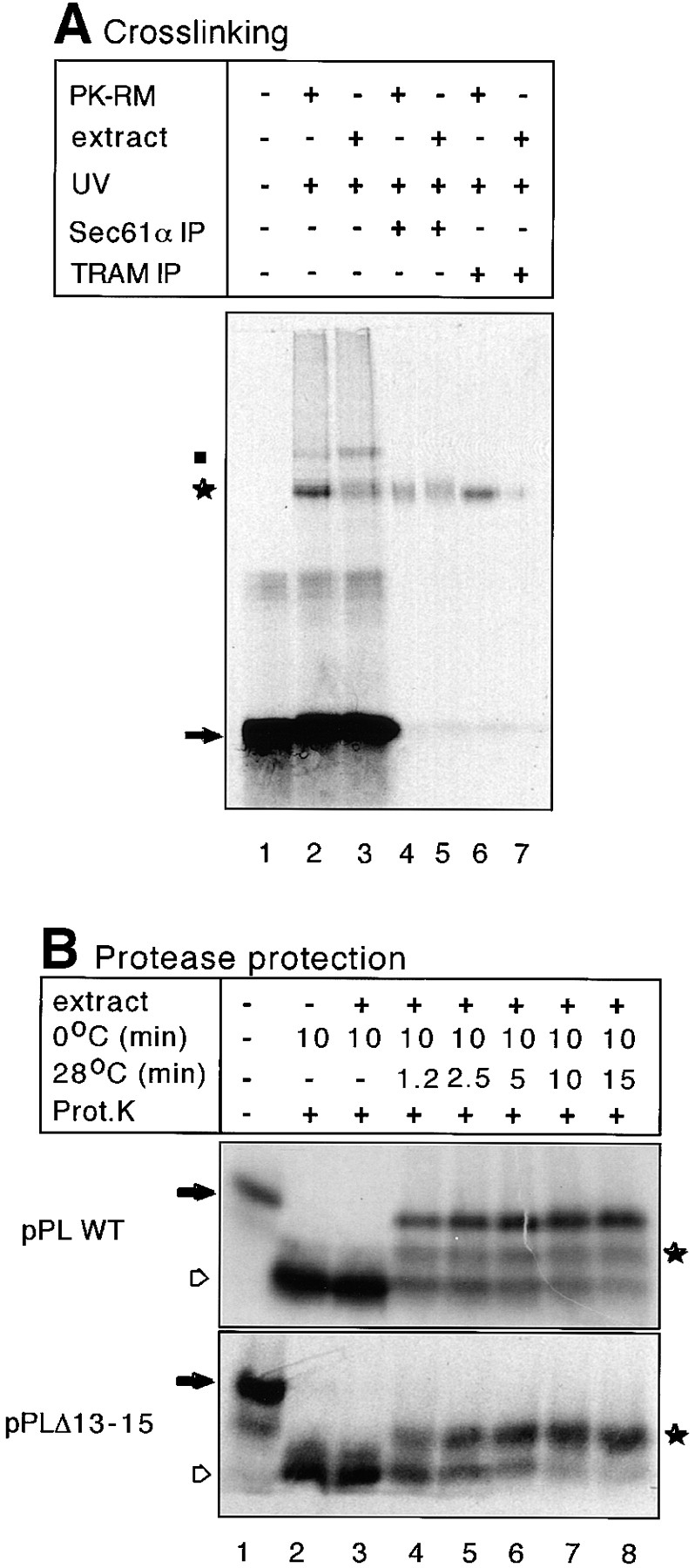
Early steps of preprolactin translocation reproduced with a detergent extract of microsomes. (A) A preprolactin fragment of 86 amino acids containing photoreactive lysine derivatives at positions 4 and 9 of the signal sequence was synthesized in vitro and RNCs were isolated by centrifugation through a sucrose cushion. After resuspension, they were incubated with either intact microsomal membranes (PK-RM) or a digitonin extract (extract) prepared from them. The samples were irradiated with UV light, as indcated, and immunoprecipitated (IP) with antibodies to Sec61α or TRAM. The samples were subjected to SDS-PAGE and analyzed by autoradiography. The arrow and star indicate the noncross-linked nascent chain and its cross-links to membrane proteins, respectively. Weak cross-linking to the 54-kD subunit of SRP (dot) is caused by some residual SRP in the membranes. (B) RNCs containing nascent chains of 86 amino acids of wild-type preprolactin (pPLWT) or of 83 amino acids of a signal sequence mutant (pPLΔ13-15) were incubated with a digitonin extract of microsomes for 10 min on ice followed by incubation for different time periods at 28°C. The samples were analyzed by treatment with proteinase K (Prot.K). The filled arrow indicates the position of the nascent chains. The hollow arrow indicates the fragment of ∼30 amino acids that is protected against proteolysis by the ribosome alone and the star indicates a fragment of ∼50 amino acids.
To determine whether in the soluble system the nascent chain is inserted into the translocation channel, we used a protease-protection assay (Connolly et al., 1989; Jungnickel and Rapoport, 1995). In the absence of membrane extract, the addition of proteinase K led to the degradation of the nascent preprolactin chains to small fragments of ∼30 amino acids (Fig. 1 B, top, lane 2 vs. lane 1, marked by a hollow arrow), which correspond to the COOH-terminal parts of the nascent chains within the ribosomes. In the presence of membrane extract this fragment persisted if the incubation was carried out at 0°C (lane 3). At 28°C, however, an increasing percentage of the 86mer became resistant to proteolysis (lanes 4–8). These chains are thus protected by both the ribosome and associated membrane proteins. The results are similar to those with native membranes (Jungnickel and Rapoport, 1995). In the soluble system a minor fraction of the nascent chains is degraded to fragments of ∼50 amino acids (marked by a star). These chains are likely inserted with the NH2-terminal portion not completely protected by membrane proteins since the proteolytic fragments can be precipitated with cetyltrimethylammonium bromide and are thus still associated with the tRNA at their COOH termini (not shown).
Nascent chain insertion in the soluble system also requires a functional signal sequence in the translocation substrate. With a fragment of a preprolactin mutant carrying a nonfunctional signal sequence (pPLΔ13-15; Jungnickel and Rapoport, 1995), addition of a detergent extract of microsomes did not give rise to completely protected chains even after prolonged incubation (Fig. 1 B, bottom, lanes 4–8). Instead, as with native membranes, a protected fragment of 50 amino acids appeared (star; Jungnickel and Rapoport, 1995). Taken together, these data indicate that a lipid bilayer is not required for signal sequence recognition and insertion of a nascent chain into the translocation channel.
The Sec61p Complex Is Necessary and Sufficient in the Soluble System
Previous experiments with reconstituted proteoliposomes demonstrated that the Sec61p complex is necessary and sufficient for early steps of preprolactin translocation (Görlich and Rapoport, 1993; Jungnickel and Rapoport, 1995). Therefore, we asked whether this was also the case in the solubilized system.
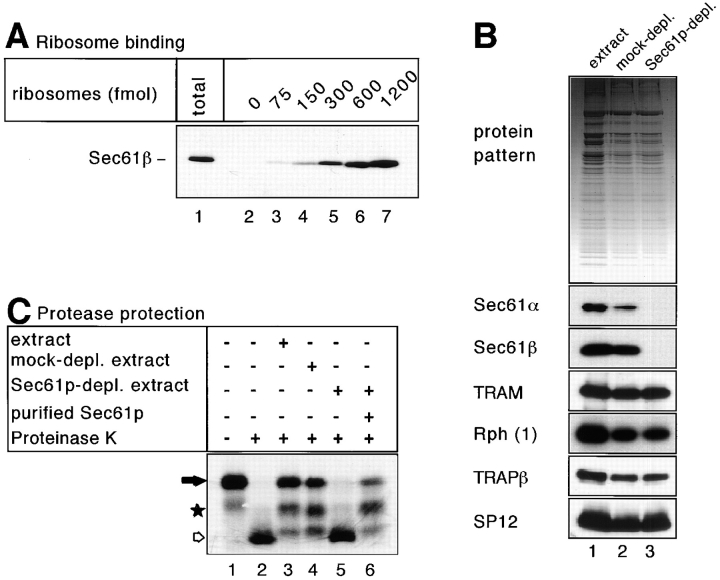
We first tested whether the Sec61p complex binds to ribosomes in solution. A detergent extract of microsomes was mixed with defined amounts of isolated ribosomes lacking nascent chains. Upon sedimentation, the amounts of Sec61p complex cosedimenting with the ribosomes were determined by immunoblotting with antibodies against Sec61β (Fig. 2 A). With high amounts of ribosomes (1.2 pmol), nearly all Sec61p complex was recovered in the pellet (lane 7 vs. 1). Since ∼5–7 pmol of Sec61p complex were present in the assay, several molecules must be bound per ribosome under these conditions, consistent with the fact that ribosomes are bound to oligomers of the Sec61p complex in membranes (Hanein et al., 1996). The results also agree with those of Beckmann et al. (1997) on the binding of yeast Sec61p complex to yeast ribosomes in detergent solution.
Figure 2.
Sec61p is essential for protease protection of nascent preprolactin chains in the soluble system. (A) Nontranslating ribosomes in the indicated amounts were incubated with a detergent extract of microsomes (corresponding to ∼5–7 pmol Sec61p). After sedimentation of the ribosomes through a sucrose cushion, the amount of cosedimenting Sec61p complex was determined by immunoblotting with antibodies against the β-subunit of the Sec61p complex (Sec61β). Lane 1 shows the total amount of Sec61p complex added to the ribosomes. (B) The upper part shows the protein pattern of an untreated detergent extract, of a mock-immunodepleted extract after incubation with protein A/G–Sepharose (mock-depl.), and of an extract immunodepleted with antibodies to Sec61β that had been coupled to protein A/G– Sepharose (Sec61p-depl.). The lower part shows immunoblots of the same samples using antibodies against Sec61α, Sec61β, TRAM, ribophorin 1 (Rph 1), the β subunit of the translocon- associated protein complex (TRAPβ), and the 12-kD subunit of signal peptidase (SP12). (C) RNCs containing 86 amino acids of preprolactin were incubated with the extracts shown in (B) and treated with proteinase K. In lane 6, purified Sec61p complex was added in amounts corresponding to those in the original sample. The positions of the nascent chains (filled arrow) and of their proteolytic fragments (star and hollow arrow) are indicated.
To investigate whether in the solubilized system the Sec61p complex is essential for nascent chain insertion into the translocation channel, we immunodepleted the Sec61p complex from a detergent extract of canine microsomes. About 97–98% of the α and β subunits of the Sec61p complex were removed with a resin containing immobilized antibodies against Sec61β, whereas the overall protein pattern did not change significantly (Fig. 2 B, lane 3 vs. 1). Membrane proteins presumed to be located at the translocation site but not associated with the Sec61p complex (signal peptidase, TRAP, ribophorin I, and TRAM) were not depleted (Fig. 2 B), demonstrating the specificity of immunodepletion. A general dilution of all membrane proteins occurred during the immunodepletion but was identical to that in an extract mock-depleted with protein A/G–Sepharose alone (lane 2 vs. 3). The Sec61p depleted extract did not give rise to fully protected nascent preprolactin chains (Fig. 2 C, lane 5), in contrast to the mock- depleted extract that behaved identically to the nontreated extract (lanes 3 and 4). Upon readdition of purified Sec61p complex to the depleted extract, fully protected forms of the nascent chain as well as fragments of ∼50 amino acids reappeared (lane 6). We conclude that in the solubilized system the Sec61p complex is essential for the insertion of the nascent chain into a protease-protected environment.
Next, we tested whether the purified, soluble Sec61p complex alone is sufficient for insertion of nascent preprolactin chains into the translocation site. When isolated RNCs were incubated with increasing amounts of purified Sec61p complex in digitonin, both fully protected 86mers and fragments of 50 amino acids appeared and the fragments of 30 amino acids gradually disappeared (Fig. 3 A, lanes 4–9). 5–10 molecules Sec61p per ribosome were needed for maximum insertion. Similar results were obtained with RNCs that were used directly after translation without prior isolation by sedimentation (not shown). The insertion reaction in digitonin was significantly slower than with intact membranes (Fig. 3 B, compare second panel and top panels). The absence of lipids was found to be a decisive factor for the slower kinetics, as shown by experiments in which the reaction in deoxyBigCHAP (DBC) was compared with that in a mixture of DBC and lipids (third and fourth panels; the experiment could not be performed with digitonin because the lipid mixture was insoluble in this detergent). The time course of insertion into the purified, soluble Sec61p channel indicates that the protected fragment of 50 amino acids is a kinetic intermediate. This was particularly obvious at low concentrations of Sec61p complex at which the intensity of this fragment first increased and then decreased, concomitantly with the appearance of the fully protected form (see quantitation in Fig. 3 C).
With a nascent chain of a signal sequence mutant, only protected fragments of 50 amino acids were generated (Fig. 3 D). Thus, the insertion process is halted at an intermediate stage and the transition to the fully inserted stage requires a functional signal sequence. The appearance of the protected fragment of 50 amino acids was slower in detergent than in a detergent and lipid mixture, suggesting that the effect of lipids on the insertion of the wild-type protein (see Fig. 3 B) is largely on steps preceding signal sequence recognition. Taken together, these results show that the soluble Sec61p complex alone is sufficient for ribosome binding, protection of the nascent chain against protease, and signal sequence recognition. Lipids are not required for these reactions but accelerate at least the early steps of the insertion process.
It is unlikely that the nascent chain insertion observed in detergent solution involves residual lipids that remained tightly bound to the Sec61p complex throughout the purification procedure. Lipid cross-links were not seen in experiments in which the environment of the signal sequence of preprolactin was tested after its incubation with the purified Sec61p complex (not shown). However, cross-links to lipids were observed with digitonin-solubilized microsomes (not shown) or native membranes (see Fig. 6). We were also unable to detect phospholipid in the purified Sec61p complex using a phosphate assay. With a detection limit of 5 nmol phosphate, no lipid was found in 0.5–1 nmol of the Sec61p complex. Each Sec61p molecule could therefore be associated with a maximum of 5–10 phospholipid molecules. Therefore, these results suggest that the insertion of the nascent chain into the channel and signal sequence recognition do not require lipids. In addition, the stimulating effect of lipids on the kinetics of the insertion process required >300 lipid molecules per Sec61p (data not shown), suggesting that lipids are a particularly good solvent for the Sec61p complex but do not serve as specific ligands.
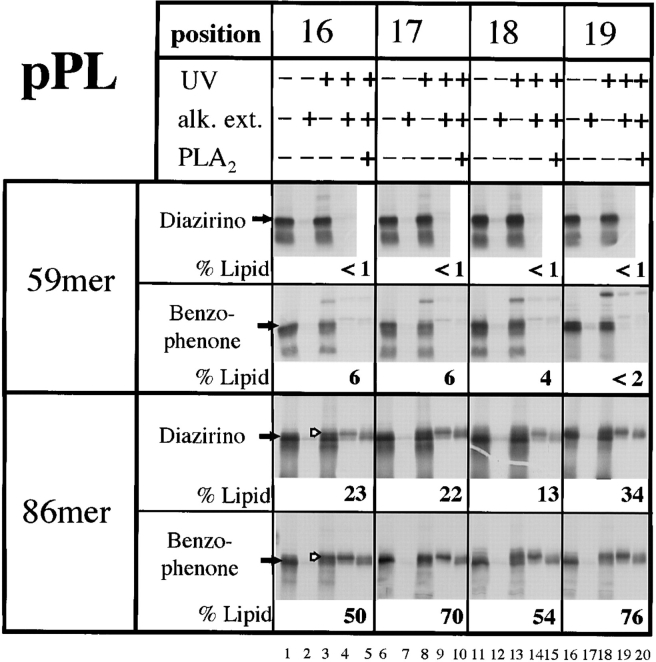
Figure 6.
Site-specific photocross-linking of the signal sequence of nascent preprolactin chains to phospholipids. RNCs containing 59 or 86 amino acids of preprolactin (59mer or 86mer) were synthesized with photoreactive diazirino or benzophenone probes at various positions of the signal sequence. The RNCs were incubated with canine microsomes and subjected to UV irradiation, as indicated. To enrich for lipid cross-links, the alkali-extracted membrane pellet (alk. ext.) was also analyzed. To verify cross-linking to lipids, some of the samples were treated with phospholipase A2 (PLA2). The percentage of radioactivity in the nascent chain that is cross-linked to lipids was determined with a phosphoimager. The filled arrows indicate the positions of the noncross-linked nascent chains, the hollow arrows indicate the positions of the lipid cross-links. The upper band seen with the benzophenone cross-linker is probably a ribosomal protein since it is not enriched in the alkali-extracted membrane pellet.
Insertion of Nascent Prepro-α-Factor Chains into the Solubilized Channel
Next we tested whether nascent chain insertion in solution can also be reproduced for the TRAM-dependent translocation substrate prepro-α-factor. We again used nascent chains of 86 amino acid length in protease protection experiments. With the purified Sec61p complex alone in digitonin, fragments of ∼50 amino acids, but no full-length 86mers, were protected against proteolysis (Fig. 4 A, top). These results agree with those obtained with reconstituted proteoliposomes (Voigt et al., 1996) and indicate that partial insertion of prepro-α-factor chains by the Sec61p complex can occur in the absence of lipids. Although in proteoliposomes the additional presence of TRAM leads to full protection of the nascent chains, in digitonin solution it had no effect (data not shown), perhaps because TRAM only poorly functions in this detergent (note also the reduced cross-linking to TRAM in Fig. 1 A). In the detergent DBC, on the other hand, protection of 86mers was observed in the absence of TRAM addition. Whereas these results are somewhat surprising because in proteoliposomes TRAM is required for full insertion, they do demonstrate that, as for preprolactin, insertion of prepro-α-factor chains can occur in the absence of lipids. The apparent lack of a TRAM requirement may be explained by contamination of the Sec61p preparation with TRAM (1:30 molar ratio according to immunoblots; not shown). Indeed, upon further removal of TRAM from the Sec61p complex by concanavalin A–Sepharose, insertion was reduced and could be restored by readdition of TRAM (not shown). Interestingly, when lipids were added to the Sec61p complex in DBC, no protection of full-length 86mers was observed (third panel); protection of the 86mers was now entirely dependent upon the addition of TRAM (Fig. 4 B). At high concentrations, TRAM alone showed insertion activity (lanes 14–16), which again is best explained by cross-contamination with Sec61p complex (1:30 molar ratio according to immunoblots; not shown). The synergism of Sec61p complex and TRAM was most pronounced at an equimolar ratio (Fig. 4 C), which corresponds to the approximate ratio in native microsomes (Görlich and Rapoport, 1993). Thus, lipids are required in the soluble system to best mimic the physiological situation in intact membranes. It is unclear why in DBC, in the absence of lipids, minor contaminations of the Sec61p complex with TRAM suffice for full insertion (Fig. 4 A, second panel).
Specific Contact of Signal Sequences with the Sec61p Complex and TRAM
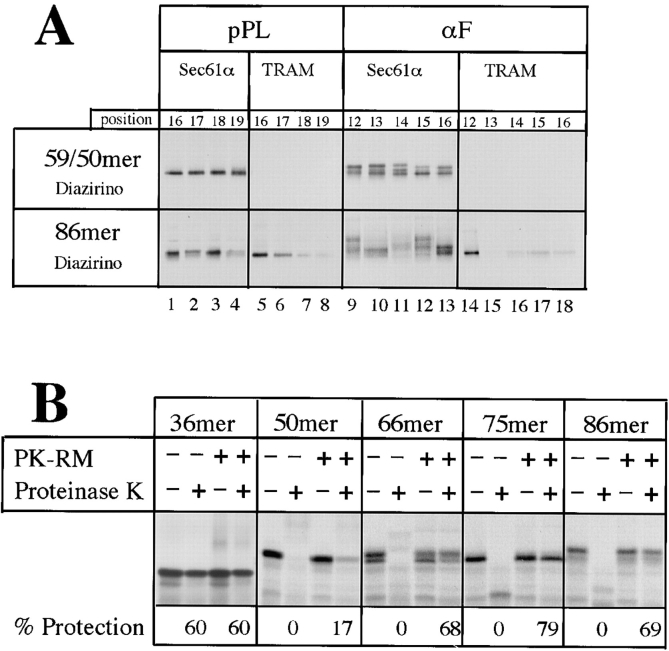
The absence of a lipid requirement for nascent chain insertion suggested that the signal sequence is recognized through a protein interaction with the Sec61p complex and TRAM. To investigate this possibility, we characterized the molecular environment of the signal sequence in native membranes, using a site-specific photocross-linking approach. Stop codons at different positions within the signal sequence-coding region were suppressed in vitro by translation in the presence of a modified suppressor Phe-tRNA containing a diazirino group in the side chain of the amino acid. Cross-linking experiments were first performed with short nascent preprolactin chains of 59 amino acids (59mer) corresponding to an early stage of translocation preceding signal sequence recognition. At this stage the ribosome–membrane junction is not yet tight and the nascent chain is still accessible to proteolysis (Jungnickel and Rapoport, 1995). The signal sequence of these short chains could be cross-linked to Sec61α but not to TRAM (Fig. 5 A, top, lanes 1–8). Four neighboring positions within the signal sequence gave about equal efficiencies of cross-linking to Sec61α. With preprolactin chains of 86 amino acids (86mer), corresponding to a stage beyond signal sequence recognition at which the ribosome–membrane junction is tight and the nascent chain is completely protected against proteolysis (Connolly et al., 1989; Jungnickel and Rapoport, 1995), the signal sequence could be cross-linked to both Sec61α and TRAM (Fig. 5 A, lower panel, lanes 1–8). Interestingly, neighboring positions within the signal sequence behaved reproducibly differently. For example, positions 16 and 18 gave stronger Sec61α cross-links than position 19, and positions 16 and 17 much stronger TRAM cross-links than positions 18 and 19. These data indicate that after nascent chain insertion into the translocation channel, the signal sequence of preprolactin contacts both Sec61α and TRAM in a specific, nonrandom manner.
Figure 5.
Site-specific photocross-linking of the signal sequence of nascent polypeptides to translocation components. (A) To analyze a preinsertion stage of translocation, RNCs containing 59 amino acids of preprolactin (pPL) or 50 amino acids of prepro-α-factor (αF) were synthesized with photoreactive probes at various positions of the signal sequence. To this end, truncated mRNAs with stop codons at different positions in the signal sequence-coding region were translated in vitro in the presence of a modified suppressor Phe-tRNA containing a diazirino group in the side chain of the amino acid. The RNCs were incubated with canine microsomes and subjected to irradiation. Cross-linked products were analyzed by immunoprecipitation with antibodies against Sec61α or TRAM, followed by SDS-PAGE and autoradiography. To analyze stages at which the nascent chains were inserted into the channel, RNCs with chains of 86 amino acids containing photoreactive probes at the various positions were employed in similar cross-linking experiments. (B) RNCs carrying prepro-α-factor chains of different length were incubated with or without microsomal membranes and treated with proteinase K. The percentage of nascent chains protected is given.
To perform similar cross-linking experiments with prepro-α-factor, we confirmed the existence of two distinct stages of nascent chain insertion for this substrate. Prepro-α-factor chains of different lengths were incubated with or without microsomes and treated with proteinase K (Fig. 5 B). Chains of 36 amino acids were largely protected against proteolysis even in the absence of membranes, indicating that they were still located inside the ribosome. Chains of 50 amino acids were fully degraded in the absence of membranes and only slightly protected in their presence. Chains of 66, 75, or 86 amino acids were mostly protected in the presence of membranes. These data demonstrate that, as for preprolactin (Jungnickel and Rapoport, 1995), a transition occurs from an early stage in which the polypeptide is largely accessible to proteolysis to one in which the chain is fully inserted.
Cross-linking with the 50mer, containing probes at different positions of the signal sequence, indicated proximity to Sec61α, but not TRAM (Fig. 5 A, top, lanes 9–18). With the longer chains, cross-links to both proteins were seen (lower panel, lanes 9–18). The Sec61α cross-links varied not only in intensity but also in pattern, probably because the same position of the nascent chain can be cross-linked to different regions of the Sec61α molecule. Strong cross-links to TRAM were only seen with position 12. Thus, as for preprolactin, nascent chain insertion of prepro-α-factor proceeds in distinct stages and the signal sequence ultimately contacts the translocation components in a nonrandom manner.
Contact of Signal Sequences with Lipids
To further analyze the molecular environment of signal sequences during the insertion of nascent chains into the translocation site, we tested cross-linking to lipids. With the 59mer of preprolactin containing diazirino probes, none of the four tested positions of the signal sequence gave significant cross-links to phospholipids (Fig. 6, top). In contrast, the signal sequence of the 86mer contacted lipids (Fig. 6, lower panel; quantitation given below the gel lanes), in agreement with previous results (Martoglio et al., 1995). All four positions analyzed gave about equal intensities of lipid cross-links that could be enriched in the alkali-extracted membrane pellet and cleaved with phospholipase A2 (Fig. 6). All lipid cross-links were resistant to proteolysis (data not shown), indicating that they are generated by nascent chains inserted into the channel.
To exclude the possibility that the absence of lipid cross-links with short chains is caused by the specific chemistry of the photoreactive probe, we repeated the experiments with a cross-linker that contains a benzophenone group. In contrast to the diazirino group which irreversibly generates a short-lived carbene that is efficiently quenched by water, benzophenone is reversibly converted into a triplet state that reacts preferentially with C-H bonds, even in the presence of solvent water and bulk nucleophiles (Dorman and Prestwich, 1994). Compared with the diazirino group, the benzophenone group gave higher yields of lipid cross-links with the 86mer (Fig. 6, lower panel). Most importantly, despite higher lipid reactivity, the benzophenone group did not give significant cross-links to lipids with the short chains of 59 amino acids (Fig. 6, top panel). Together with the results of cross-linking to proteins (Fig. 5), it therefore appears that with short preprolactin chains the signal sequence is entirely in a protein environment formed by Sec61α; it is transferred to a specific binding site at the interface between membrane protein components and lipids upon chain elongation and insertion into the channel.
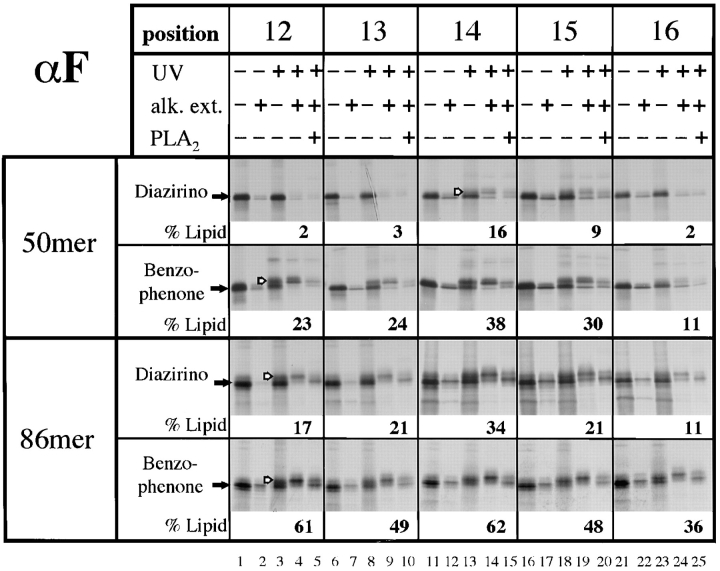
Lipid cross-linking was also tested with nascent chains of the TRAM-dependent substrate prepro-α-factor. With the carbene-generating diazirino cross-linker some cross-linking of short nascent chains to lipids was observed with positions 14, 15 (Fig. 7, top), as well as positions 8, 9, and 10 (data not shown); other positions did not give lipid cross-links. With the chains of 86 amino acids all positions gave cross-links to lipids that were significantly stronger than with the shorter chains (Fig. 7, lower panels). With the benzophenone cross-linker lipid cross-links could be observed from all positions of short chains, but again there was more cross-linking to lipids with the longer chains (Fig. 7). These results indicate that, in contrast to preprolactin, some prepro-α-factor chains have contact with lipids even at early stages of translocation. However, for both substrates the signal sequence contacts more lipids after nascent chain insertion into the translocation site.
Figure 7.
Site-specific photocross-linking of the signal sequence of nascent prepro-α-factor to phospholipids. An experiment similar to that in Fig. 6 was performed with RNCs containing 50 or 86 amino acids of prepro-α-factor (50mer or 86mer).
Discussion
This study shows that early phases of cotranslational protein translocation across the ER membrane, leading to the insertion of a nascent polypeptide chain into the translocation channel, can be reproduced in detergent solution in the absence of bulk lipids or a lipid bilayer. Thus, all steps leading to nascent chain insertion, including signal sequence recognition in the membrane are likely based primarily, if not exclusively, on protein–protein interactions. These data are inconsistent with models that invoke an intact lipid bilayer or inverted micelles as an essential feature for early phases of the translocation process. The demonstration that early steps of cotranslational translocation can occur in detergent solution extends previous results that reproduced posttranslational translocation in the absence of a lipid bilayer (Matlack et al., 1997). Furthermore, our data are in agreement with recent electronmicroscopy results that showed that in detergent solution the yeast Sec61p complex specifically binds to the polypeptide exit site of yeast ribosomes (Beckmann et al., 1997).
Our soluble system recapitulates the two-stage nascent chain insertion into the translocation channel observed previously with intact membranes (Jungnickel and Rapoport, 1995; Voigt et al., 1996). In time course experiments in which the insertion of nascent preprolactin chains of 86 amino acids was followed in the soluble system, a protected COOH-terminal fragment of 50 amino acids was transiently generated. With a signal sequence mutant this stage represented the end point of insertion. In the case of prepro-α-factor insertion halted at a similar intermediate in the absence of TRAM. Thus, the transition between the intermediate and fully inserted stages involves signal sequence recognition that, depending on the translocation substrate, requires either the Sec61p complex alone or both the Sec61p complex and TRAM.
The existence of two stages of nascent chain insertion was further supported by photocross-linking experiments using nascent chains of different lengths in conjunction with intact microsomes. Membrane-targeted preprolactin chains of 59 amino acids remain accessible to proteolysis (Jungnickel and Rapoport, 1995) and all tested positions within the signal sequence were found in contact with Sec61α, but not with TRAM or lipids (this paper). Therefore, it appears that at this early stage of translocation the signal sequence is in a proteinacious environment formed exclusively from the Sec61p complex. With preprolactin chains of 86 amino acids, which are inserted into the channel and therefore fully protected against proteolysis (Connolly et al., 1989; Jungnickel and Rapoport, 1995), the signal sequence could be cross-linked to Sec61α, TRAM, and lipids. TRAM not only contacts the NH2 terminus of the signal sequence (High et al., 1993; Mothes et al., 1994) but also certain residues of the hydrophobic core region (this paper). Since even neighboring amino acid positions gave different cross-linking yields with Sec61α and TRAM, the signal sequence must be precisely positioned, rather than randomly oriented, indicating its binding to a specific site. Thus, in agreement with the results from the soluble system, the signal sequence seems to be mostly recognized by protein–protein interactions. Each tested position of the signal sequence is also in contact with lipids, suggesting that the binding site is located at the interface between the channel and surrounding lipids. One possible arrangement is that the signal sequence is bound at the outside of the channel, thus simultaneously contacting both protein and lipid. Alternatively, an equilibrium of different populations may exist with some of the signal sequences located inside the channel with contact to protein only, and others being totally in the lipid phase. Since short preprolactin chains do not give lipid cross-links and appear to be entirely in an environment formed from the Sec61p complex, it seems likely that the signal sequence reaches its binding site at the interface between channel and lipids by transfer through the interior of the Sec61p channel.
Two stages of nascent chain insertion were also observed for the TRAM-dependent translocation substrate prepro-α-factor. Nascent chains of 50 amino acids remained largely accessible to proteolysis and the signal sequence could be cross-linked to Sec61α but not to TRAM, whereas chains of 86 amino acids were resistant to proteolysis and their signal sequences could be cross-linked to Sec61α, TRAM, and lipids. As for preprolactin, with the chains inserted into the channel the cross-linking pattern differed among neighboring positions, indicating again that the signal sequence is bound to a specific site. In contrast to the results with preprolactin, some lipid cross-linking was observed with short prepro-α-factor chains. It is possible that these represent a more advanced stage of translocation than the short preprolactin chains, although in both cases the chains were accessible to proteolysis and did not cross-link to TRAM. Even with prepro-α-factor more cross-linking to lipids was seen with longer chains, suggesting that at least some molecules may follow the route through the interior of the channel. Another possibility is that the population of prepro-α-factor chains that contacts lipids early may be unproductively partitioned into the lipid phase. Some signal sequences, like that of preprolactin, may have a low intrinsic affinity for lipids whereas others, like that of prepro-α-factor, may have a high affinity; the TRAM requirement for translocation of prepro-α-factor might be explained if TRAM prevented partitioning into the lipid phase. Such a hypothesis would be consistent with the observation that in the presence of lipids much higher concentrations of TRAM were required for the insertion of prepro-α-factor into the soluble channel. Yet another possibility is that the prepro-α-factor molecules contacting lipids are in fact on a productive pathway; the signal sequence could take the route through the lipid to reach the binding site at the interface between channel and lipids, a process that might be facilitated by the TRAM protein. The two pathways through the lipid exterior and the proteinaceous interior of the channel may coexist, similarly to the two routes by which substrates of ABC transporters are believed to reach the interior of the channel (Higgins and Gottesman, 1992).
Although lipids were not needed for nascent chain insertion into the soluble channel they do accelerate the process in the case of preprolactin and were required to best mimic the physiological cooperation between the Sec61p complex and the TRAM protein in the insertion of prepro-α-factor chains. The effect of lipids on the kinetics varies with the phospholipid class and depends on the lipid/ detergent ratio, but always requires a molar excess over Sec61p complex (data not shown). Since different detergents also have greatly varying effects, it seems that phospholipids are a particularly good solvent for the functional Sec61p complex, possibly stabilizing an active conformation, but do not bind to it in a specific manner. Indeed, all early steps of preprolactin translocation can occur with the soluble, purified Sec61p complex in which no lipids could be detected. Even though each molecule of the purified Sec61p complex may contain up to 5–10 residual phospholipid molecules, we consider it unlikely that these play a role in signal sequence recognition: whereas the hydrophobic portion of the signal sequence can be cross-linked to phospholipids in native microsomes and crude detergent extracts, no such cross-links appear in the purified, soluble system, indicating that all lipids normally contacting the signal sequence must have been removed during purification of the Sec61p complex. Although it is possible that in these experiments the detergent replaces the lipid as a hydrophobic partitioning phase, our cross-linking data indicate a specific interaction of the signal sequence with protein components. A limited number of binding sites for signal sequences, characteristic for protein–protein interactions, is also suggested by competition experiments with a synthetic signal peptide (our unpublished results). Although rather high concentrations of the peptide were required for efficient competition, specificity is indicated by the fact that a photoreactive derivative of the same peptide gives cross-links to a membrane protein that, according to its size and isoelectric point, is Sec61α (Robinson et al., 1987). Taken together, these data show that signal sequences are ultimately recognized by a proteinacious binding site and exclude partitioning into the lipid bilayer as an exclusive mechanism of their recognition.
The mechanism by which signal sequences are recognized by the Sec61p complex and TRAM is unclear. One may speculate that they interact with, or intercalate between, transmembrane segments, as has been proposed for the homolog of Sec61α, SecYp, in E. coli (Osborne and Silhavy, 1993). Hydrophobic interactions must be most important, explaining why discrimination between functional and nonfunctional synthetic signal peptides can occur in simple model systems in which their partitioning in hydrophobic phases has been studied (Briggs et al., 1985). Hydrophobic interactions must also be decisive in signal recognition by SRP, in which binding occurs through a hydrophobic pocket in the 54-kD subunit (Bernstein et al., 1989).
The present evidence suggests that the translocation channel is gated by the signal sequence. Signal sequence recognition inside the ER membrane occurs at the same nascent chain length as the opening of the channel towards the lumen, determined by fluorescent quenching experiments (Crowley et al., 1994; Jungnickel and Rapoport, 1995). In addition, electrophysiological data demonstrate that synthetic signal peptides can open large ion conducting channels in the cytoplasmic membrane of E. coli (Simon and Blobel, 1992). Together with the present data, it now appears that the signal sequence interacts directly with the Sec61p–SecYp complex and thus opens the channel for the polypeptide chain.
Acknowledgments
We thank S. Heinrich and S. Voigt for providing purified membrane proteins, B. Misselwitz for help with cross-linking experiments, S. Furlong for performing the lipid phosphate analysis, and A. Neuhof, K. Matlack and M. Rolls for critical reading of the manuscript.
Abbreviations used in this paper
- DBC
deoxyBigCHAP
- ER
endoplasmic reticulum
- RNC
ribosome–nascent chain complex
- SRP
signal recognition particle
- TRAM
translocating chain-associating membrane protein
Footnotes
T.A. Rapoport is a Howard Hughes Medical Institute Investigator. The work was further supported by a grant from the National Institutes of Health (GM52586) to T.A. Rapoport and by the Swiss National Science Foundation to J. Brunner.
Drs. Mothes and Jungnickel contributed equally to this work.
Dr. Jungnickel's present address is Institut für Genetik, Universität Köln, Weyertal 121, 50931-Cologne, Germany.
References
- Beckmann R, Bubeck D, Grassuci R, Penczek P, Verschoor A, Blobel G, Frank J. Alignment of conduits for the nascent polypeptide chain in the ribosome-sec61 complex. Science. 1997;278:2123–2130. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- Belokon, Y.N., V.I. Bakhmutov, N.I. Chernoglazova, K.A. Kochetkov, S.V. Vitt, N.S. Garbalinkaya, and V.M. Belikov. 1988. General method for the asymmetric synthesis of α-amino acids via alkylation of the chiral nickel(II) Schiff base complexes of glycine and alanine. J. Chem. Soc. Perkin Trans. I: 305–312.
- Bernstein H, Poritz M, Strub K, Hoben P, Brenner S, Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B. Transfer of proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J Cell Biol. 1975;67:852–862. doi: 10.1083/jcb.67.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs MS, Gierasch LM, Zlotnick A, Lear JD, DeGrado WF. In vivo function and membrane binding properties are correlated for Escherichia colilamB signal peptides. Science. 1985;228:1096–1099. doi: 10.1126/science.3158076. [DOI] [PubMed] [Google Scholar]
- Connolly T, Collins P, Gilmore R. Access of proteinase K to partially translocated nascent polypeptides in intact and detergent-solubilized membranes. J Cell Biol. 1989;108:299–307. doi: 10.1083/jcb.108.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley KS, Liao S, Worrell VE, Reinhart GD, Johnson AE. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R. A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J Cell Biol. 1987;105:633–645. doi: 10.1083/jcb.105.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman G, Prestwich GD. Benzophenone photophores in biochemistry. Biochemistry. 1994;33:5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- Gilmore R, Walter P, Blobel G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J Cell Biol. 1982;95:470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- Görlich D, Hartmann E, Prehn S, Rapoport TA. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature. 1992a;357:47–52. doi: 10.1038/357047a0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Hartmann E, Kalies KU, Rapoport TA. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 1992b;71:489–503. doi: 10.1016/0092-8674(92)90517-g. [DOI] [PubMed] [Google Scholar]
- Hanein D, Matlack KES, Jungnickel B, Plath K, Kalies KU, Miller KR, Rapoport TA, Akey CW. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- Higgins CF, Gottesman MM. Is the multidrug transporter a flippase? . Trends Biochem Sci. 1992;17:18–21. doi: 10.1016/0968-0004(92)90419-a. [DOI] [PubMed] [Google Scholar]
- High S, Martoglio B, Görlich D, Andersen SSL, Ashford AJ, Giner A, Hartmann E, Prehn S, Rapoport TA, Dobberstein B, Brunner J. Site-specific photocross-linking reveals that Sec61p and TRAM contact different regions of a membrane-inserted signal sequence. J Biol Chem. 1993;268:26745–26751. [PubMed] [Google Scholar]
- Jungnickel B, Rapoport TA. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell. 1995;82:261–270. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- Kalies KU, Görlich D, Rapoport TA. Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p complex. J Cell Biol. 1994;126:925–934. doi: 10.1083/jcb.126.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori T, Nishikawa SI, Shin I, Schultz PG, Endo T. Probing the environment along the protein import pathways in yeast mitochondria by site-specific photocrosslinking. Proc Natl Acad Sci USA. 1997;94:485–490. doi: 10.1073/pnas.94.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport TA. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO (Eur Mol Biol Organ) J. 1995;14:217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring B, Kreibich G, Wiedmann M. The intrinsic ability of ribosomes to bind to endoplasmic reticulum membranes is regulated by signal recognition particle and nascent-polypeptide-associated complex. Proc Natl Acad Sci USA. 1995;92:9435–9439. doi: 10.1073/pnas.92.21.9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martoglio B, Hofmann MW, Brunner J, Dobberstein B. The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell. 1995;81:207–214. doi: 10.1016/0092-8674(95)90330-5. [DOI] [PubMed] [Google Scholar]
- Matlack KES, Plath K, Misselwitz B, Rapoport TA. Protein transport by purified yeast s complex and Kar2p without membranes. Science. 1997;277:938–941. doi: 10.1126/science.277.5328.938. [DOI] [PubMed] [Google Scholar]
- Meyer DI, Krause E, Dobberstein B. Secretory protein translocation across membranes—the role of the docking protein. Nature. 1982;297:647–650. doi: 10.1038/297647a0. [DOI] [PubMed] [Google Scholar]
- Moroder L, Hallett A, Wünsch E, Keller O, Wersin G. Di-tert.-butyldicarbonat—ein vorteilhaftes Reagenz zur Einführung der tert.-Butyloxycarbonyl-Schutzgruppe. Hoppe-Seyler's Z Physiol Chem. 1976;357:1651–1653. [PubMed] [Google Scholar]
- Mothes W, Prehn S, Rapoport TA. Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. EMBO (Eur Mol Biol Organ) J. 1994;13:3937–3982. doi: 10.1002/j.1460-2075.1994.tb06713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothes W, Heinrich SU, Graf R, Nilsson I, von Heijne G, Brunner J, Rapoport TA. Molecular mechanism of membrane protein integration into the endoplasmic reticulum. Cell. 1997;89:523–533. doi: 10.1016/s0092-8674(00)80234-2. [DOI] [PubMed] [Google Scholar]
- Neuhof A, Rolls MM, Jungnickel B, Kalies KU, Rapoport TA. Binding of signal recognition particle gives ribosome/nascent chain complexes a competitive advantage in endoplasmic reticulum membrane interaction. Mol Biol Cell. 1998;9:103–115. doi: 10.1091/mbc.9.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicchitta CV, Zheng T. Regulation of the ribosome-membrane junction at early stages of presecretory protein translocation in the mammalian endoplasmic reticulum. J Cell Biol. 1997;139:1697–1708. doi: 10.1083/jcb.139.7.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicchitta CV, Migliaccio G, Blobel G. Biochemical fractionation and assembly of the membrane components that mediate nascent chain targeting and translocation. Cell. 1991;65:587–598. doi: 10.1016/0092-8674(91)90091-c. [DOI] [PubMed] [Google Scholar]
- Osborne RS, Silhavy TJ. PrlA suppressor mutations cluster in regions corresponding to three distinct topological domains. EMBO (Eur Mol Biol Organ) J. 1993;12:3391–3398. doi: 10.1002/j.1460-2075.1993.tb06013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prehn S, Tsamaloukas A, Rapoport TA. Demonstration of specific receptors of the rough endoplasmic membrane for the signal sequence of carp preproinsulin. Eur J Biochem. 1980;107:185–195. doi: 10.1111/j.1432-1033.1980.tb04639.x. [DOI] [PubMed] [Google Scholar]
- Raden D, Gilmore R. Signal recognition particle-dependent targeting of ribosomes to the rough endoplasmic reticulum in the absence and presence of the nascent polypeptide-associated complex. Mol Biol Cell. 1998;9:117–130. doi: 10.1091/mbc.9.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- Robertson SA, Ellman JA, Schultz PG. A general and efficient route for chemical aminoacylation of transfer RNAs. J Am Chem Soc. 1991;113:2722–2729. [Google Scholar]
- Robinson A, Kaderbhai MA, Austen BM. Identification of signal sequence binding proteins integrated into the rough endoplasmic reticulum membrane. Biochem J. 1987;242:767–777. doi: 10.1042/bj2420767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon SM, Blobel G. Signal peptides open protein-conducting channels in E-coli. . Cell. 1992;69:677–684. doi: 10.1016/0092-8674(92)90231-z. [DOI] [PubMed] [Google Scholar]
- Voigt S, Jungnickel B, Hartmann E, Rapoport TA. Signal-sequence-dependent function of the TRAM protein during early phases of protein transport across the ER membrane. J Cell Biol. 1996;134:25–35. doi: 10.1083/jcb.134.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- Walter P, Ibrahimi I, Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in vitro assembled polysomes synthesizing secretory protein. J Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann M, Kurzchalia TV, Bielka H, Rapoport TA. Direct probing of the interaction between the signal sequence of nascent preprolactin and the signal recognition particle by specific cross-linking. J Cell Biol. 1987;104:201–208. doi: 10.1083/jcb.104.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]