Abstract
In the Drosophila embryo, the correct association of muscles with their specific tendon cells is achieved through reciprocal interactions between these two distinct cell types. Tendon cell differentiation is initiated by activation of the EGF-receptor signaling pathway within these cells by Vein, a neuregulin-like factor secreted by the approaching myotube. Here, we describe the cloning and the molecular and genetic analyses of kakapo, a Drosophila gene, expressed in the tendons, that is essential for muscle-dependent tendon cell differentiation. Kakapo is a large intracellular protein and contains structural domains also found in cytoskeletal-related vertebrate proteins (including plakin, dystrophin, and Gas2 family members). kakapo mutant embryos exhibit abnormal muscle-dependent tendon cell differentiation. A major defect in the kakapo mutant tendon cells is the failure of Vein to be localized at the muscle–tendon junctional site; instead, Vein is dispersed and its levels are reduced. This may lead to aberrant differentiation of tendon cells and consequently to the kakapo mutant deranged somatic muscle phenotype.
Keywords: muscles, Drosophila, tendons, Egfr, neuregulin, cytoskeleton
Differentiation of the somatic musculature of the Drosophila embryo is a complex, multistep process, resulting in a segmentally reiterated pattern of muscles that govern larval locomotion via muscle connections to discrete attachment sites in the epidermis. After initial periods of independent differentiation, the mesodermally derived muscle cells and the epidermal attachment cells use a complex signaling mechanism, through which connections between the two cell types and final differentiation are achieved.
During the second half of Drosophila embryogenesis, each of the specific somatic myotubes extends its leading edge towards a specific location, at which a group of epidermal muscle attachment (EMA)1 cells is located. At the end of this extension process, each myotube forms a physical contact with a specific EMA cell, which is then induced to develop into a mature tendon cell (Bate, 1993; Becker et al., 1997).
The larval tendon cells develop in the embryo in two sequential steps: initially, a subset of ectodermally derived competent EMA cells is defined along the A-P and D-V axes. In a second step, the portion of these competent cells that are bound to muscles differentiate into mature tendon cells (Becker et al., 1997). The expression of the regulatory protein Stripe, a transcription factor of the early growth response (EGR) family, determines the fate of the EMA competent cells at the first phase of tendon cell development (Lee et al., 1995; Frommer et al., 1996). Stripe expression leads to the expression of an array of EMA-specific genes that contribute to the correct guidance of the myotubes (Becker et al., 1997; Vorbrüggen et al., 1997).
The second phase of tendon cell differentiation depends on inductive interactions between the myotube and the EMA cell. These interactions lead to terminal differentiation of the EMA competent cells into tendon cells, in which high protein levels of Stripe, Groovin (Volk and VijayRaghavan, 1994), and Alien (Goubeaud et al., 1996) are maintained, and the transcription of the genes delilah (Armand et al., 1994) and β1 tubulin (Buttgereit et al., 1991) is induced.
The inductive signal responsible for triggering the muscle-dependent differentiation of the tendon cells is provided by Vein, a secreted protein that is homologous to vertebrate neuregulins (Schnepp et al., 1996). Vein is necessary and sufficient to induce the expression of tendon-specific genes, including stripe, groovin, delilah, and β1 tubulin (Yarnitzky et al., 1997). Vein activity is mediated through its activation of the Drosophila EGF receptor homologue, DER, expressed on the EMA cells (Yarnitzky et al., 1997; Schnepp et al., 1998). Thus, Vein acts as a secreted differentiation factor that mediates the muscle-dependent differentiation of the EMA cells into tendon cells.
Although vein mRNA is produced in the muscle cells, Vein protein is highly concentrated in the intercellular space between the muscles and the tendon cells, where intense adherens type junctions are formed (Yarnitzky et al., 1997). This junctional space contains electron-dense material, which presumably represents protein aggregates of various extracellular matrix components (Tepass and Hartenstein, 1994). Since the primary sequence of Vein includes a signal peptide but no transmembrane domain, it is assumed that Vein protein is secreted from the myotube and accumulates at the muscle–tendon junctional space. The molecular mechanism that is responsible for Vein localization at this site is yet to be elucidated. The Vein ligand is a relatively weak activator of the EGF receptor pathway (Schnepp et al., 1998; Yarnitzky et al., 1998); therefore, a mechanism regulating Vein accumulation at the site of activity may be essential for a proper activation of the pathway.
This paper describes the molecular cloning and functional analysis of the EMA-specific gene groovin (grv), which is identical to kakapo (Prout et al., 1997). kakapo (kak) has been previously identified in a genetic screen for mutations that cause wing blisters. This phenotype results from impaired adhesion between the dorsal and ventral wing epithelia (Prout et al., 1997). Kak is a large intracellular protein, expressed mainly along the EMA cell plasma membrane. The primary sequence of Kak exhibits sequence motifs that indicate its possible association with the cytoskeleton of the EMA cell. Analysis of kak mutant embryos shows that kak is essential for proper completion of the muscle-dependent tendon cell differentiation program. Our results suggest that the primary role of Kak is to mediate the restricted localization and accumulation of Vein protein at the muscle–tendon junctional site.
Materials and Methods
Fly Strains
Fly stocks used were y w (used as a “wild-type” strain). Df(2R)MK1 = Df (2R) 50B3-5;50D1-4 was provided by V. Harternstein (University of California, Los Angeles, CA). b pr cn bw was provided by J. Campos-Ortega (University of Köln, Germany) and was isogenized in our lab. P{ry +17.2 = neo-FRT} 42D; ry 605 (referred to as FRT42D in this paper) was provided by the Bloomington stock center (Bloomington, IN). FRT42DV104/SM5 (referred to as V104 or kakV104 in this paper) was provided by N. Brown (Cambridge University, UK; Walsh and Brown, 1998). Seven alleles of kak (Prout et al., 1997) were obtained from J. Fristrom and M. Prout (University of California, Berkeley, CA). kakA405 was generated in our lab by repetitive EMS mutagenesis (screen for lethal mutations over Df(2R)MK1; see below).
EMS mutagenesis was performed essentially as described in Girgliatti (1986). Isogenized b pr cn bw males were EMS mutagenized and their F1 male progeny were tested for lethality over Df(2R)MK1. kakA405 was generated in this screen.
Immunochemical Reagents and DNA Probes for Whole Mount In Situ Hybridization
Primary antibodies: anti-Groovin monoclonal antibody (mAb#19) was raised in mice immunized with Drosophila embryonic membrane proteins (>100 kD) (Volk and VijayRaghavan, 1994). Hybridomas were screened by immunostaining and selected according to their staining pattern. Although mAb#19 reacts specifically with the Kak COOH terminus protein sequence (as shown in Results), we found that it cross-reacts with additional, Kak-unrelated epitope(s) present along the embryonic segmental grooves. Polyclonal anti-Kak antibodies were generated in guinea pigs by immunizing with a Kak–glutathione-S-transferase (GST) fusion protein (containing the Kak sequence encoded by a 4.6-kbp fragment of kak cDNA [position 6394–10993, encoding amino acids 2136–3668]). Polyclonal anti-Stripe (Becker et al., 1997), anti-Vein, and anti-Delilah antibodies were prepared in our lab (Yarnitzky et al., 1997).
Rabbit anti-myosin heavy chain antibodies were obtained from P. Fisher (SUNY, Stony Brook, NY). Secondary antibodies used included: Cy3-conjugated anti–guinea pig and anti–rat fluorescein-conjugated anti– rabbit, and HRP-conjugated anti–rabbit and anti–mouse IgM antibodies (Jackson ImmunoResearch, West Grove, PA).
Nonradioactive digoxygenin-labeled DNA probes for various kak cDNA fragments and a β1 tubulin fragment (D. Buttgereit, University of Marburg, Germany) were used for whole embryo in situ hybridization.
Histochemical Staining
For sectioning, embryos were first stained with anti-Kak polyclonal antibody and then embedded in JB-4 embedding media (Polysciences, Inc., Warrington, PA) as described (Volk, 1992). Sections (3–4-μm width) were examined under a Zeiss Axioscope microscope (Thornwood, NY).
Whole Mount Embryonic Staining
Antibody staining was performed essentially as described previously (Ashburner, 1989), except that the embryos were fixed with 3% paraformaldehyde. In situ hybridization was performed by the method of Tautz and Pfeifle (1989) using a digoxygenin-labeled DNA probe.
Western Analysis
Western analysis was performed according to standard procedures and is described in Volk (1992). SuperSignal chemiluminescent substrate (Pierce Chemical Co., Rockford, IL) was used for signal detection.
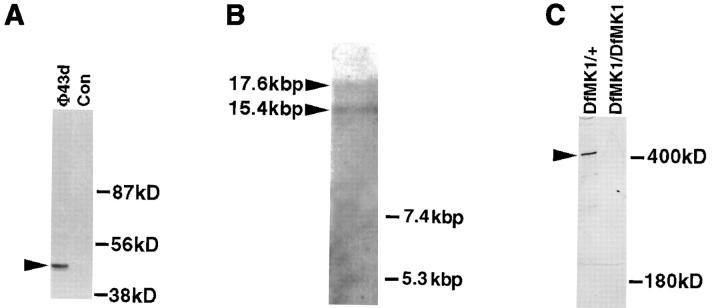
For Western blot analysis of single embryos, kakV104/+ flies were produced and crossed with each other. The embryos of this cross were incubated for 25 h in 25°C, and each of the nonhatched embryos was boiled in sample buffer and subjected to SDS-PAGE in a single lane. The truncated band (Fig. 4 D) is representative of seven individual embryos.
Figure 4.
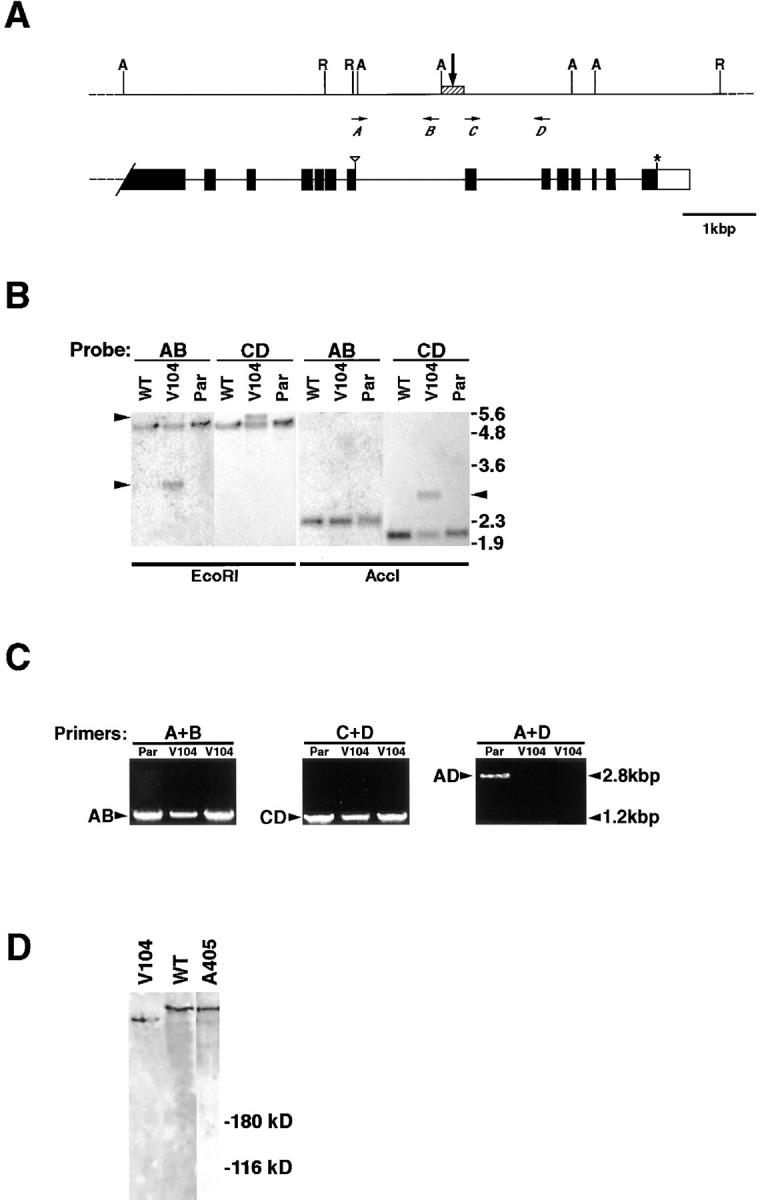
The V104 mutation falls within the kak locus. (A) A scheme describing the COOH-terminal genomic region of the kakapo gene and the rearrangement break point deduced from Southern and PCR analysis. In the upper scheme, the restriction sites of EcoRI (R) and AccI (A) are indicated. Horizontal arrows A–D represent the relative location of the primers used for amplifying the probes AB, CD, and AD described below. The vertical arrow points to the rearrangement region (hatched box). In the lower scheme exons (black boxes), stop codon (asterisk) and noncoding sequences (open box) are indicated. The triangle marks amino acid position 3871 in Kak protein sequence. (B) Southern analysis of EcoRI- or AccI-digested genomic DNA isolated from an isogenized b pr cn bw strain (WT), kakV104/b pr cn bw strain (V104), or the parental FRT42D strain (Par), reacted with either AB or CD probes. The AB and CD probes flank the rearrangement point in kakV104 chromosome and were generated by PCR amplification using primers A and B, or C and D (see scheme in A). Note that while both AB and CD probes react with a single 5-kbp EcoRI band in all chromosomes, in V104 the AB probe recognizes an extra band of 3 kbp, while CD probe recognizes a distinct 5.5-kbp band (left arrowheads). Likewise, probe AB recognizes a 2.3-kbp AccI band common to all chromosomes, while probe CD recognizes, in addition to a 2-kbp common band, an extra 2.8-kbp band present uniquely in the V104 chromosome (right arrowhead). Notice that because of the presence of an AccI site between the rearrangement point and the AB sequence (see A), the AB probe is not expected to recognize the chromosomal rearrangement in V104. (C) PCR analysis on individual homozygous kakV104 (V104) embryos selected as described in Materials and Methods, with either A and B (A+B) or C and D (C+D) primers flanking the rearrangement point, or with A and D primers (A+D) located 3′ and 5′ to the rearrangement point (see scheme in A). Note that both AB and CD DNA fragments are amplified in the PCR reaction on template DNA taken from two individual kakV104mutant embryos, or from the parental FRT42D genomic DNA. However, the 2.8-kbp AD PCR fragment is amplified only from the FRT42D genomic template DNA and not from the template DNA taken from the kakV104mutant embryos. The sequence of all PCR fragments was verified by sequence analysis. (D) Western analysis of kakV104or kakA405 mutant embryos. Single kakV104or kakA405 homozygous mutant embryos were selected (see Materials and Methods), boiled in protein sample buffer, and analyzed in Western using anti-Kak polyclonal antibody. A representative lane of the Kak-reactive band from the kakV104mutant embryos (V104) runs slightly faster relative to the wild-type (WT) band. The size of Kak protein in kakA405 mutant embryos (A405) is similar to that of the wild-type band.
Expression of FLAG-tagged Kakapo
The kak cDNA insert was amplified by PCR from phage Φ43d and subcloned in pECE-FLAG (SmaI-XbaI). 293 cultured human cells were transfected with 20 μg of the above construct by the standard calcium phosphate transfection method. Transfected and nontransfected cells were harvested 48 h after transfection and lysed, and the resulting protein extracts were immunoprecipitated with anti-FLAG antibodies coupled to agarose beads (anti-FLAG M2 affinity gel; Kodak No. IB13021). Immunoprecipitated proteins were eluted in sample buffer and resolved on 7.5% SDS-PAGE, followed by Western analysis using mAb#19.
Preparation of Fly Genomic DNA and Southern Analysis
FRT42DV104/SM5 flies were crossed to b pr cn bw flies to out-cross SM5. Genomic fly DNA was prepared from FRT42DV104/b pr cn bw progeny of the above cross and from the FRT42D and b pr cn bw lines, according to standard procedure. The genomic DNA was subjected to Southern analysis using genomic probes. The probes were amplified by PCR using Taq DNA polymerase (D1806; Sigma Chemical Co., St. Louis, MO) on genomic DNA of homozygous FRT42D flies. The following primers were used (see also Fig. 4 A): primer A: 5′-CTGTTATGGTGCGCGTGG-3′; primer B: 5′-CGGGTTCGGTTTATCAAG-3′; primer C: 5′-CACTAACATAGAGCTACG-3′; and primer D: 5′-CAGTTGTTGTTGGATGAC-3′.
PCR Analysis on Individual Mutant Embryos
Genomic DNA from individual embryos was prepared as described by Rastelli et al. (1993). kakV104/SM6B, Cy, Roi, al, dp, cn, sp, P[ry +, eve-lacZ] embryos were collected, aged, permeabilized in heptane/PBS mixture, rinsed, and stained for X-gal for 1.5 h. White embryos were selected, squashed in Gloor and Engel's buffer (10 mM Tris, pH 8.2, 1 mM EDTA, 25 mM NaCl, 200 μg/ml proteinase K), and incubated at 37°C for 30 min and at 95°C for 2 min. For each PCR reaction, 1–2 μl of this lysate was used.
Cloning of groovin/kakapo cDNA and Molecular Analysis
Anti-Grv monoclonal antibody (mAb#19) was used to screen a Drosophila embryonic (9–12 h) expression library cloned in λgt11 (Zinn et al., 1988). A single clone (Φ43d) was isolated, and it exhibited a similar pattern of expression, by in situ hybridization, as that revealed by the mAb#19 staining. A total of 12,989 bp of kak cDNA sequence was isolated by successive screens with 5′-end fragments from isolated clones (Φ9-1-1, Φ4, Φ6, Φ6P, and Φ3). The most 5′ cDNA sequences (1–3, 13, 700PCR, and 800V) were isolated by PCR amplification from the phage expression library. All clones and amplified fragments were tested by in situ hybridization, and they exhibited an EMA cell–specific expression pattern.
DNA fragments subcloned in Bluescript (Stratagene, La Jolla, CA), λgt11 DNA, or PCR products were sequenced by an automated sequencer (Applied Biosystems, Foster City, CA). The sequences were analyzed by AutoAssembler DNA sequence assembly software package (version 1.0.3; Applied Biosystems).
For Northern analysis, a kak cDNA 4.6-kbp (position 6394–10993) probe was used.
Sequence and Computer Analysis
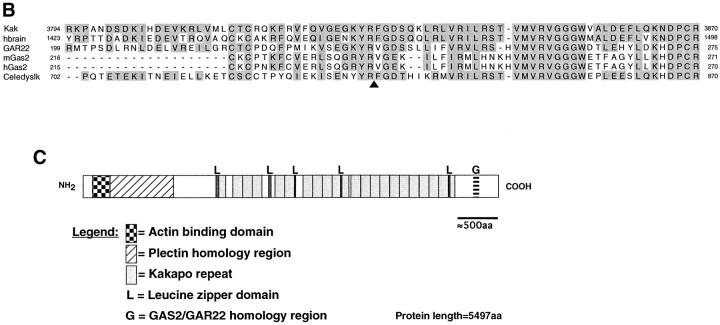
The predicted amino acid sequence was analyzed by the Wisconsin Package Version 9.1 (Genetics Computer Group [GCG], Madison, WI). Nonredundant databases were searched against the Kak amino acid sequence. The results of BLASTP 1.4.11 (Altschul et al., 1990), which are representative of other results obtained, have indicated homology to the dystrophin protein family. The region of Kak internal repeats was identified using COMPARE and DOTPLOT, with the parameters window length = 100, stringency = 39–45 points (not shown). The repeats were aligned using PILEUP, and the output file was further processed using the SeqVu 1.0.1 program (The Garavan Institute, Sydney, Australia). The repeat border definition is in line with the dystrophin consensus CS1 (Koenig and Kunkel, 1990). The Kak consensus repeat contains a similar amino acid content and order as the dystrophin consensus CS1 (46% identity and 51% similarity). The putative Leucine-zipper domains were identified by the MOTIFS program of GCG. The results of BLASTP 1.4.11 also indicated that Kak COOH terminus shares a homology domain with additional proteins in the database (Fig. 2 B). The proteins that share this homology domain with Kak COOH terminus are human brain mRNA (KIAA0465 protein; sequence data available from GenBank/EMBL/ DDBJ under accession number AB007934), human GAR22 (accession number Y07846), mouse Gas2 (accession number M21828), human Gas2 (accession number U95032), and a putative open reading frame (ORF) from Caenorhabditis elegans (Wilson et al., 1994; accession number U40800; gi1065956).
Figure 2.
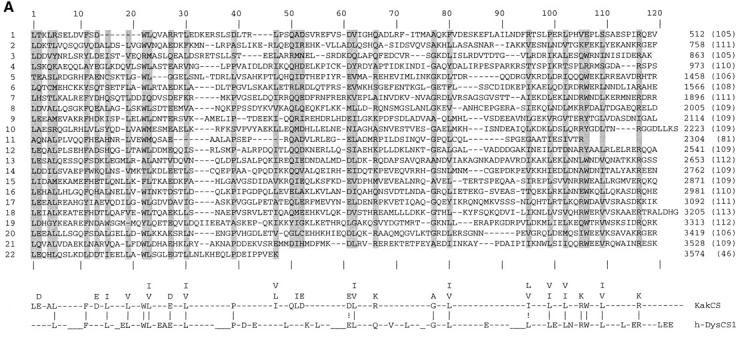
kakapo is homologous to cytoskeletal-associated proteins. (A) Sequence alignment of the Kak repeats. The amino acid sequence of the 22 repeats marked in Fig. 1 D are aligned. Gaps (−) were added to optimize alignments. The numerical order of the repeats is shown on the left and the sequence position of the residue that ends each repeat is given on the right. The length of each repeat is indicated in parenthesis. The repeat consensus sequence (KakCS) indicates amino acids that are conserved in a given position in 11 or more repeats and/or share this position with a related amino acid(s) (designated above KakCS sequence) and are shaded in the repeat alignment. The alignment between Kak repeat consensus and human dystrophin repeat consensus CS1 (h-DysCS1; Koenig and Kunkel, 1990) is presented at the bottom of this figure. Gaps (−) in h-DysCS1 were added manually to optimize alignment with KakCS. Identical residues are marked with “|” and related amino acids are marked by “!”. KakCS shares 46% identity (51% similarity) with h-DysCS1. (B) Kak shares a homology domain with proteins of the Gas2/GAR22 family of intracellular proteins and with a partially cloned cDNA KIAA0465 from human brain. Alignment of Kak, human brain cDNA (hbrain), GAR22, mouse Gas2 (mGas2), human Gas2 (hGas2), and a putative ORF from C. elegans (Celedyslk) homology domains is shown. The amino acid sequence of the homology domain in each of the aligned proteins was compared with that of Kak. The degree of identity is as follows: 72%, hbrain; 50%, GAR22; 44%, mGas2 and hGas2; and of the C. elegans protein 27% identity over the first 36 aligned amino acids and 71% identity over the last 39 amino acids. Shaded positions represent residues that are identical or similar to the corresponding residue in Kak. The start and end amino acid positions are indicated. In “Celedyslk,” a stretch of 96 amino acids was removed to optimize the alignment (the position is marked by an arrowhead under the sequence). (C) A schematic linear representation of the Kak protein, showing the relative positions of the putative actin binding site (checkered box) and plectin homology region (hatched box) (described in Gregory and Brown, 1998), putative Leucine-zipper domains (L), Kakapo repeats (gray boxes), and the Gas2/GAR22 homology region (G).
Results
Cloning of groovin/kakapo
We have previously identified a monoclonal antibody (mAb#19), which stained the EMA cells in a specific manner, and according to its prominent staining along the embryonic segmental grooves, we named the corresponding antigen Groovin (Volk and VijayRaghavan, 1994). To gain an insight into the molecular nature of Groovin, we used mAb#19 to screen an embryonic cDNA expression library. Based on this screen, we isolated a single reactive phage clone (Φ43d). In situ hybridization with the isolated 43d cDNA insert exhibits embryonic expression in the EMA cells (similar to the expression pattern obtained with the monoclonal antibody). The specific reactivity of mAb#19 with the expressed protein sequence of 43d cDNA was verified by the following experiment. The cDNA insert from Φ43d was subcloned into a FLAG-tag– containing vector to enable transfection of human cultured 293 cells. After transfection with the 43d-containing construct, cells were lysed, immunoprecipitated with anti-FLAG antibody, and subjected to Western analysis with mAb#19. A specific reactive band of the expected 43-kD size was detected only in the transfected cells (Fig. 1 A). The selected cDNA clone was subsequently used to screen and clone continuous cDNA sequences. As shown below, we have demonstrated that groovin is the structural gene for the kakapo (Prout et al., 1997) locus. We will refer to this gene as kakapo (kak).
Figure 1.
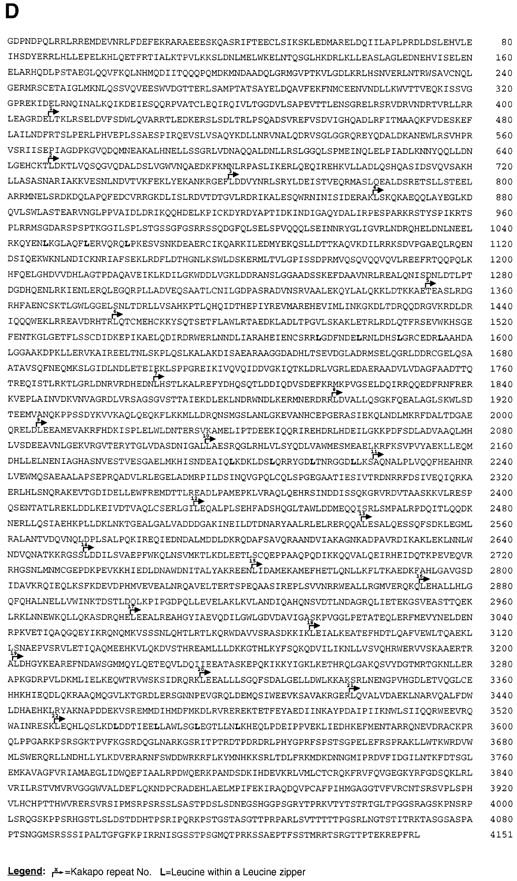
Structural and molecular analysis of kakapo. (A) The specific reactivity of mAb#19 with the expressed protein sequence of the cDNA clone isolated from phage Φ43d (representing the COOH-terminal end of Kak) is demonstrated. The 43d cDNA insert was subcloned in frame to FLAG-tagged–containing vector. This construct was used to transfect 293 human cultured cells, and the extract from the transfected cells was first immunoprecipitated with anti-FLAG antibody and then subjected to Western analysis with mAb#19. While mAb#19 reacts with the expected 43-kD polypeptide in the transfected cells (arrowhead in lane Φ43d), it did not react with any band in the control nontransfected cells (Con). (B) Northern blot analysis of poly (A+) RNA prepared from 12–20-h-old embryos. The kak mRNA was detected using a kak cDNA fragment of 4.6 kbp (nucleotides 6394–10993 of the cDNA). RNA molecular marker sizes are shown on the right. kak cDNA probe hybridizes specifically with two transcripts of ∼15.4 and ∼17.6 kbp (arrowheads). (C) Western blot analysis of wild-type (Df(2R)MK1/+) or mutant (Df(2R)- MK1/Df(2R)MK1) embryos with anti-Kak polyclonal antibody. Each lane represents extract made from 30 embryos (stages 16– 17). Mutant embryos were selected according to their phenotype. The anti-Kak antibody recognizes a specific band of ∼400 kD. We used as markers (shown on the right) the Drosophila laminin A chain (∼400 kD) and standard molecular mass markers (only the ∼180 kD size is shown). The specificity of the polyclonal antibody is indicated by its lack of reactivity with extracts from embryos homozygous for Df MK1. (D) The deduced amino acid sequence of Kak. The 4,151–amino acid sequence was derived from the single ORF in the 12,989-nucleotide cDNA sequence. The sequence presented lacks an initiator methionine. kak ORF terminates at position 12454 and is followed by a putative polyadenylation signal at position 12979. The amino acid residue number is indicated to the right of each line. Leucine residues in potential Leucine-zipper domains are in bold text. Kakapo repeats are delineated with bent arrows and are numbered. The presented sequence data are available from EMBL nucleotide sequence database under accession number Y09430. (Note: All the amino acid positions indicated in this paper refer to the amino acid sequence presented in Fig. 1 D, which does not include the two alternative NH2-terminal spliced forms described in Gregory and Brown, 1998.)
The contiguous nucleotide sequence of partially overlapping cDNA clones spans 12,989 bp and contains a single continuous ORF (positions 1–12453) encoding 4,151 amino acids (Fig. 1 D). The most 5′ sequences of the cDNA, encoding the putative initiator methionine of the ORF, were cloned by Gregory and Brown (1998). Thus, the complete cDNA of kak spans 17,420 bp and encodes a novel protein of 5,497 amino acids of a putative size of 627 kD. All the amino acid positions indicated in this paper refer to the amino acid sequence presented in Fig. 1 D, which does not include the two alternative NH2-terminal spliced forms described in Gregory and Brown (1998).
In situ hybridization to polytene chromosomes using a 1.4-kbp fragment of kak cDNA (position 11588–12989) indicates that kak maps to a single location on the second chromosome, at position 50C3-6 of the cytological map (not shown). Northern blot analysis using a kak cDNA fragment of 4.6 kbp (position 6394–10993) revealed two transcripts of ∼17.6 and ∼15.4 kbp (Fig. 1 B). Western blot analysis of protein extracts from embryos (12–20 h) shows a major protein band in the ∼400-kD size range (determined by comparing the Kak band to that of laminin A chain, Fig. 1 C). This major band is seen when using polyclonal antibodies raised against the protein product of a 4.6-kbp fragment of kak cDNA (encoding for amino acids 2136–3668) fused to GST. The specificity of the antibody is demonstrated by its failure to react with extracts derived from embryos homozygous for a deficiency (Df(2R)MK1; see Materials and Methods) that uncovers the kak locus (Fig. 1 C). The discrepancy between the calculated protein molecular mass (627 kD) and the 400-kD band obtained in Western analysis may be indicative of posttranslational modifications of the translated sequences.
Kakapo Is a Cytoskeletal-associated Protein
The primary amino acid sequence of Kak reveals several domains and motifs that show high degrees of similarity to three distinct vertebrate cytoskeletal-related protein families, namely plakin (Ruhrberg and Watt, 1997), dystrophin (Koenig et al., 1988), and Gas2/GAR22 (Schneider et al., 1988; Zucman-Rossi et al., 1996) (Fig. 2, A–C).
The NH2-terminal region of Kak is homologous to the NH2-terminal domain of members of the plakin family of cytoskeletal cross-linker proteins, comprising plectin, BPAG1, and ACF7 (Ruhrberg and Watt, 1997). These large proteins link actin microfilaments and intermediate filaments to the plasma membrane at specialized attachment sites, called hemidesmosomes. Abnormal function of various plakin family members leads to skin (e.g., bullous pemphigous) as well as neurological (e.g., dystonia musculorum) disorders (for reviews see Ruhrberg and Watt, 1997; Fuchs and Cleveland, 1998). The region of similarity between Kak and plakin family members includes the actin binding region but does not exhibit similarity to the intermediate filament–associated domain; homologies with plakin family members are described in more detail in Gregory and Brown (1998).
The central region of Kak (amino acids 408–3574) consists of 22 repeats, 105–113 amino acids long (Figs. 1 D and 2 A). A computerized search has indicated that the central region of Kak shares sequence similarity (∼20% identity) with spectrin-like repeats present in an array of cytoskeletal-associated proteins, including dystrophin, α-actinin, and spectrin (Davison and Critchley, 1988). These repeats are predicted to adopt a triple-helical conformation (Speicher and Marchesi, 1984). In dystrophin, the multiple repeat domain functions as a spacer between the NH2-terminal actin-binding domain and the COOH-terminal domain associated with a group of membrane proteins (Ervasti and Campbell, 1991). The consensus sequence deduced from the alignment of the spectrin-like repeats in Kak shares 46% identity (51% similarity) with the human dystrophin repeat consensus, CS1 (Koenig and Kunkel, 1990) (Fig. 2 A). This similarity suggests the presence of a similar domain containing multiple spectrin-like, triple-helical repeats in the central region of Kak protein. This region also contains five Leucine-zipper motifs (Figs. 1 D and 2 C).
A somewhat lower degree of similarity between the Kak COOH-terminal domain (sequence 3725–3793) and the region in dystrophin containing the two EF-hand motifs is also observed (not shown). The COOH-terminal domain of dystrophin-related proteins is highly conserved (Roberts and Bobrow, 1998) and includes a WW domain, implicated in mediating interactions with the transmembrane protein, β-dystroglycan (Jung et al., 1995). This domain also includes two putative Ca2+-binding EF-hands (Kawasaki and Kretsinger, 1995) and a region involved in the binding to members of the syntrophin family of PDZ domain–containing proteins (Ahn and Kunkel, 1995; Suzuki et al., 1995). The similarity between Kak and dystrophin in the COOH-terminal domain is detected only along the EF-hand motifs (∼30% identity along a sequence of 70 amino acids). This limited similarity may suggest that although Kak does not appear to be a dystrophin family member, these genes may share a common ancestor.
The COOH-terminal region of Kak shows sequence conservation with yet another family of cytoskeletal-related proteins representative by the Gas2/GAR22 proteins (Schneider et al., 1988; Zucman-Rossi et al., 1996) (Fig. 2 B). Mouse Gas2, a member of this protein family, belongs to a set of proteins that was shown to be selectively expressed in growth-arrested cells in culture (Schneider et al., 1988). It is a highly regulated protein (Brancolini et al., 1992; Manzow et al., 1996) that interacts with the microfilament system (Brancolini et al., 1992; Brancolini and Schneider, 1994). Deletion analysis of the Gas2 protein suggests that the region in Gas2 that is homologous to Kak has a significant function in cytoskeletal organization (Brancolini et al., 1995). During apoptosis, Gas2 is cleaved by ICE proteases, presumably leading to microfilament derangement (Brancolini et al., 1995). An additional partially cloned cDNA species of unknown function from human brain (Seki et al., 1997) exhibits a high level of identity to the COOH-terminal domain of Kak protein (Fig. 2 B). This similarity extends beyond the Gas2 homology domain and exhibits 40% overall identity along the entire 1,658–amino acid sequence available in the data base. It is not clear whether this partially cloned cDNA represents a human Kak homologue. In addition, a putative protein from C. elegans (Wilson et al., 1994) shares domain contents with Kak, including plectin, dystrophin, and Gas2/ GAR22-like domains mentioned above (see also Gregory and Brown, 1998). The function of this putative protein is yet to be elucidated.
Taken together, the deduced amino acid sequence of Kak (summarized in Fig. 2 C) predicts a novel large intracellular protein that carries two distinct cytoskeletal-associated domains separated by a spacer consisting of elongated triple-helical spectrin-like repeats. At the NH2-terminal domain, Kak may interact with actin microfilaments, while at its COOH terminus, it may be associated with membrane structures or with additional cytoskeletal components. The similarities between Kak and its C. elegans and putative human homologues suggests that Kak structure is conserved through evolution.
Kakapo Marks the Epidermal Muscle Attachment Cells in the Embryo
The expression pattern of kak mRNA and protein during various stages of embryonic development were characterized (Fig. 3). kak mRNA expression is restricted to ectodermally derived cells, and is initially observed in extended germ band embryos during late stage 11. During germ band retraction, the mRNA is expressed by cells along the future segmental grooves and in a small cluster of ectodermal cells at the middle of each hemi-segment (Fig. 3, A–D). Since at these stages the somatic myotubes have not yet extended their leading edge into their final position, we assume that the initial activation of kak transcription is independent of muscle-dependent cues.
Figure 3.
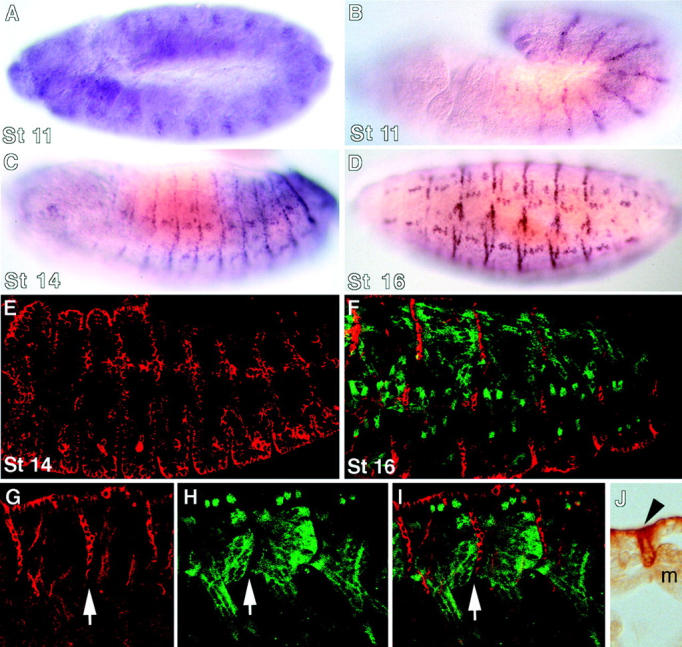
kakapo mRNA and protein expression pattern. In situ hybridization with kak cDNA fragments (A–D) and antibody staining using anti-Kak polyclonal antibody (E–J) show the dynamics of Kak expression during distinct developmental stages. kak mRNA expression starts at the extended germ-band (stage 11) as segmented patches in the ectoderm (A). In late stage 11, the mRNA is detected along the segment borders (B). After germ-band retraction (stage 14), kak mRNA expression is also seen within the segments (C). At stage 16, the expression pattern becomes localized at all the EMA cells (D). Kak protein expression follows that of the mRNA expression at stages 14 and 16 (E and F, respectively). The complementary pattern of the somatic muscle pattern with that of EMA cells is shown by double labeling the embryos with anti–myosin heavy chain (green) and anti-Kak (red) in F–I. G–I represent single (G and H) and overlap (I) images of the same embryo. Arrows in G–I indicate the same row of EMA cells bound to an array of ventral longitudinal muscles. Notice the association of Kak protein with the circumference of the EMA cells in G–I, and in the cross section of an embryo labeled with anti-Kak antibody at stage 16 (J; EMA cell marked by an arrowhead; m, myotube).
The expression of Kak protein was studied using a polyclonal antibody raised against a bacterially expressed cDNA fragment from the COOH-terminal region. The protein expression pattern of Kak follows its mRNA expression and is apparent from stage 14 of embryonic development. During the final stages of muscle development (e.g., stage 16), high levels of Kak protein are detected in muscle-bound tendon cells, as demonstrated in embryos double-labeled for Kak and myosin (Fig. 3, G–I). Kak protein is concentrated at the circumference of the tendon cell, presumably along the tendon cell plasma membrane (Fig. 3 J). Low levels of Kak protein are observed in other tissues, including epidermal cells and chordotonal organs (not shown).
The Isolation of Mutations in the kakapo Locus
To gain a functional insight as to the role of Kak during embryonic development, we screened for EMS-induced mutations that are lethal when crossed with Df(2R)MK1, a deletion that uncovers the kak locus. We recovered 15 lethal mutations that fall into several complementation groups. One allele designated A405 failed to complement a collection of seven alleles of the kak gene (Prout et al., 1997). A genetic screen (Walsh and Brown, 1998), similar to that of Prout et al. (1997), identified a complementation group that proved to be allelic to kak. A representative allele from this complementation group, V104, failed to complement the A405 allele. We concluded that A405 is allelic to kak.
We performed molecular analysis of kakV104 genomic DNA and identified a chromosomal rearrangement in the 3′ end region of the kak gene (see Fig. 4). We carried out Southern analysis with an array of probes covering the entire kak cDNA sequence and identified a region in the COOH terminus of the gene that exhibited a distinct pattern of restriction fragments. The corresponding genomic region was sequenced from the parental (FRT42D) genomic DNA. To further narrow down the region containing the chromosomal rearrangement, we used smaller genomic sequences as probes and tested their potential to recognize the rearrangement. We identified two sequential genomic sequences (AB and CD; see map in Fig. 4 A) that detect a rearrangement in kakV104 mutant chromosome in Southern analysis but react with distinct restriction fragments in the rearranged chromosome (Fig. 4 B). Thus, the rearrangement in kakV104 mutant chromosome is likely to be introduced in the genomic region that includes both fragments (see map in Fig. 4 A).
To further characterize the rearrangement in kakV104, we performed PCR, followed by sequence analysis of individual homozygous kakV104 embryos in the suspected rearranged genomic region (see Materials and Methods for details). Although the size and sequence of the AB and CD genomic fragments was not altered in the mutant embryos, we were unable to amplify the 2.8-kbp AD fragment, which includes both fragments and an additional 400-nucleotide fragment in between (Fig. 4 C). This result together with the Southern analysis (described above) strongly argues that the x ray–induced rearrangement falls into the 400-nucleotide stretch between the AB and CD fragments (Fig. 4 A, hatched box).
The rearrangement at the kak 3′ end region in kakV104chromosome predicts an alteration within the last 280 amino acids of the mutant Kak protein. Indeed, Western analysis of individual homozygous kakV104 embryos with the anti-Kak polyclonal antibody reveals a truncated protein that runs slightly faster compared with the wild-type protein (Fig. 4 D). Thus, it is suggestive that the alteration in kak 3′ end region leads to premature translational termination.
Taken together, the Southern, PCR, and Western analyses are consistent with the notion that a DNA rearrangement in the kak gene was introduced in the kakV104 chromosome. Molecular analysis described in the accompanying paper (Gregory and Brown, 1998) shows that in kak P-element mutant allele (l(2)k03010), the P-element is inserted at the 5′ intronic sequences of the kak gene. Our molecular analyses and the indication that kakV104 is allelic to kakl(2)k03010 mutant strongly argue that these mutations affect the kak gene.
The EMA Cells in kak Mutant Embryos Fail to Differentiate Properly
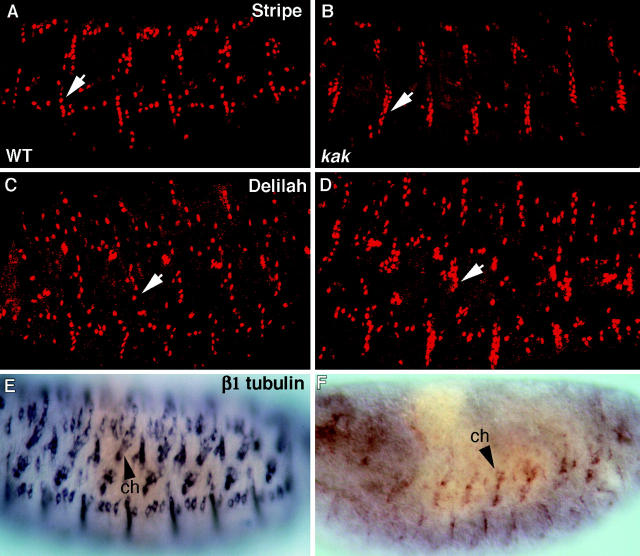
To elucidate the function of kak in EMA cell differentiation, we analyzed embryos transheterozygous for kakV104/ kakA405 alleles for expression of various markers characteristic of tendon cell terminal differentiation, including Stripe, Delilah, and β1 tubulin mRNA. This analysis shows that an excess of EMA cells, marked by the expression of Stripe and Delilah, is observed at a number of sites in the epidermis (Fig. 5). This phenotype is particularly notable in domains in which a group of muscles extend together towards neighboring epidermal attachment cells, such as along the ventral segmental border cells, to which the four ventral longitudinal muscles bind (Fig. 5 B, arrow). The excess in EMA cells was also observed in additional kak allelic combinations, e.g., embryos transheterozygous for kakA405/DfMK1. The EMS-induced kakA405 allele appears to represent a severe kakapo allele. Although the size of Kak protein is not altered, as concluded from Western analysis of individual kakA405 homozygous mutant embryos (see Fig. 4 D), their mutant phenotype is quite similar to that of kakA405/DfMK1 mutant embryos (not shown).
Figure 5.
Tendon cell differentiation is abnormal in kak mutant embryos. Staining with anti-Stripe (A and B), anti-Delilah (C and D), and in situ hybridization with β1 tubulin probe (E and F) is analyzed in kakV104/kakA405 mutant embryos (B, D, and F), and compared with that of wild-type (A, C, and E) embryos. An excess in Stripe- or Delilah-expressing EMA cells is observed in the kak mutant embryos (compare arrows in A and B, and in C and D). β1 tubulin expression in all the EMA cells is significantly reduced (F), as compared with its expression in the chordotonal organs (ch, arrowhead).
To further study the state of differentiation of the EMA cells in the kak mutant embryos, we analyzed the expression of the β1 tubulin gene. In wild-type embryos, the expression of the β1 tubulin gene is significantly elevated towards the end of tendon cell differentiation. In contrast to the expression of Stripe and Delilah, the mRNA expression of β1 tubulin in kak mutant embryos is significantly reduced, suggesting that transcription of the latter gene requires different levels of signaling (Fig. 5 B). We suspected that Vein signaling, which is required for terminal differentiation of tendon cells (Yarnitzky et al., 1997), may be reduced in the mutant embryos; while there is enough signal to trigger Delilah and Stripe expression, it is not capable of inducing β1 tubulin transcription.
Vein Localization Is Abnormal in kak Mutant Embryos
Previously, we showed that the expression of delilah, stripe, and β1 tubulin is induced in the epidermal attachment cells as a result of the EGF-receptor pathway activation by the neuregulin-like growth factor, Vein (Yarnitzky et al., 1997). Vein protein localization is restricted to the muscle–tendon junctional site in wild-type embryos. However, in kakV104/kakA405 mutant embryos, Vein protein is not localized and appears rather diffuse (Fig. 6). This altered pattern of Vein may explain the multiple number of cells expressing delilah and stripe; since Vein is not strictly localized at a given muscle–tendon junction site, it apparently weakly activates the EGF-receptor pathway in neighboring cells as well. We presume that the only cells that can respond to the ectopic Vein protein are the competent population of EMA cells, defined by the early expression of stripe (see introduction). These cells express stripe during early developmental stages in a muscle-independent manner and normally lose their stripe expression by stage 16 of embryonic development. When these competent EMA cells receive the muscle-derived Vein signal, the expression of stripe and delilah is reactivated. It appears that only this population of cells is capable of responding to Vein, since the pattern of the ectopic Stripe- or Delilah-expressing cells in the kak mutant embryos resembles that of the early population of Stripe-expressing cells (not shown).
Figure 6.
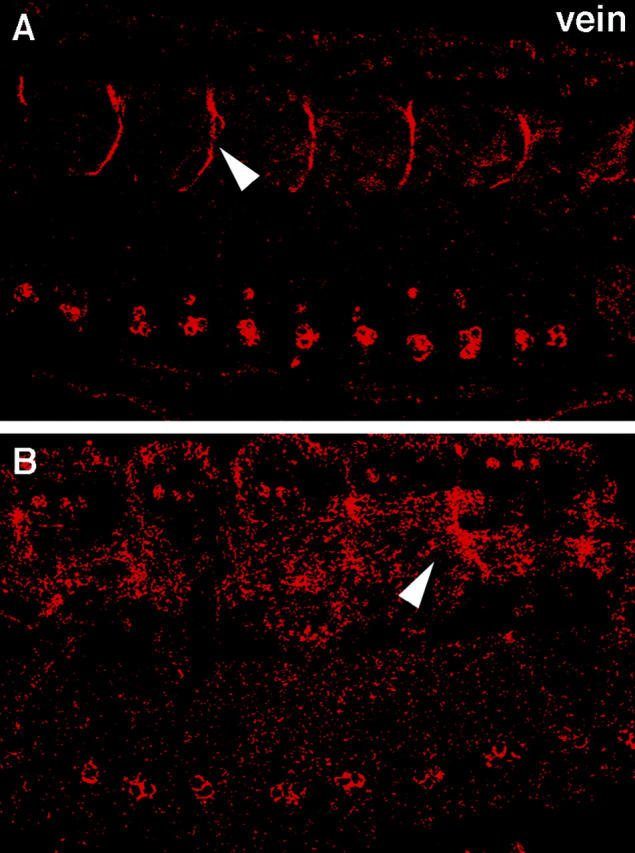
Abnormal localization of Vein protein in the muscle– tendon junction site of kak mutant embryos. Wild-type and kakV104/kakA405 mutant embryos were stained with anti-Vein antibody. The localization of Vein protein in wild-type embryos is prominent at the muscle–tendon junction site (arrowhead in A). In kak mutant embryos, the localization of Vein is diffuse in the region of muscle–tendon junction (arrowhead in B).
The reduced levels of β1 tubulin mRNA in the mutant tendon cells may also result from the abnormal pattern of Vein localization, since lower levels of Vein may not be sufficient to induce maximal β1 tubulin expression. It therefore appears that the primary defect in kak mutant embryos stems from the lack of Vein accumulation at the muscle–tendon junctional site.
The Somatic Muscle Pattern Is Disrupted in kakapo Mutant Embryos
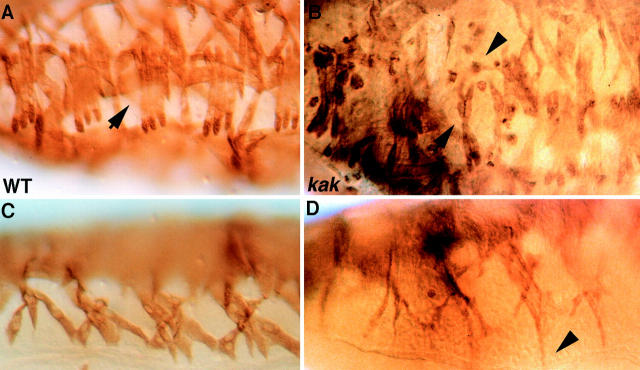
Finally, we wished to examine whether the abnormal differentiation of the EMA cells in kak mutant embryos is reflected by the pattern of the somatic musculature. kakV104/ kakA405 mutant embryos at stage 16 of embryonic development were labeled with anti–myosin heavy chain antibody to visualize the somatic muscles, and the muscle pattern was compared with that of wild-type embryos. A significant disruption of the somatic muscle pattern is observed in kak mutant embryos (Fig. 7). In many cases, individual myotubes are not oriented correctly, and in some cases the myotube rounds up. Since Kak cannot be detected in myotubes using our antibodies, it is assumed that the somatic muscle derangement is secondary to the abnormal differentiation of the EMA cells. A similar phenotype is also observed in stripe mutant embryos, in which the EMA cells do not differentiate correctly (Frommer et al., 1996). The similarity between the stripe and kak muscle phenotype and the reduced β1 tubulin mRNA expression are consistent with the conclusion that EMA cell differentiation is defective in kak mutants. The correct recognition between the muscle and the tendon cell is essential for arresting the extension of the myotube and establishment of the final pattern of somatic musculature (Yarnitzky et al., 1997). It appears that the muscle development in kakV104/ kakA405 embryos does not represent a complete loss of function phenotype since a more severe muscle defect is observed in kakV104/DfMK1 embryos (not shown).
Figure 7.
The somatic muscle pattern is deranged in kak mutant embryos. Wild-type embryos (A and C) and kakV104/kakA405 mutant embryos (B and D) were stained with anti–myosin heavy chain antibody. Lateral (A and B) and ventral (C and D) views of the embryos are shown. The pattern of the somatic muscles is disrupted in kak mutant embryos. Notice that the muscles continue to extend their leading edge ignoring the attachment sites (arrowhead in D). In some cases, muscles are missing (e.g., compare arrow in B to A), and some rounded muscles are also observed (arrowhead in B).
Discussion
Intercellular inductive interactions take place continuously during development and underlie proper organogenesis. In this paper, we describe a novel protein, Kakapo, that is essential for the inductive interactions between muscles and their epidermal attachment cells during Drosophila embryogenesis. The accompanying papers describe additional functions for this protein in epidermal integrity and cytoskeletal arrangement (Gregory and Brown, 1998) and in proper formation of the neuromuscular junction (Prokop et al., 1998).
Our results are consistent with a model in which Kak is essential for the concentration of Vein at the muscle–tendon junctional site. This localization is important for the accumulation of Vein protein at high levels; such levels are critical to ensure that a single EMA cell responds to the approaching muscle by turning on its differentiation program towards a fully developed tendon cell.
Possible Mechanisms by which Kakapo Exerts Its Function
Kak is an intracellular protein that exhibits two structural domains associated with cytoskeletal elements: the actin binding domain at its NH2-terminal and the Gas2 domain at its COOH-terminal end, spaced by an elongated, multiple spectrin-like repeat domain. How could this intracellular protein affect the localization of Vein at the extracellular matrix surrounding the EMA cell? We consider at least two possibilities, which are not mutually exclusive. The first is the association of Kak with the unique cytoskeletal network of the EMA cell, which is critical for its polarized organization. Tendon cell polarity may be essential for maintaining the characteristic junctional complexes formed between the basal surfaces of the EMA cell and the muscle cells. The space between these junctional complexes contains many extracellular matrix proteins, some of which may posses a Vein binding function. Impaired tendon cell polarity may lead to the loss of the putative Vein-binding component(s). Alternatively, Kak may be associated with a transmembrane protein(s) responsible for Vein localization either by direct binding or by association with additional extracellular matrix components that may directly bind Vein. Immunoprecipitation experiments with anti-Kak antibody (Strumpf, D., and T. Volk, unpublished) indicated that Kakapo forms protein complexes containing the extracellular protein Tiggrin (Fogerty et al., 1994). These results favor the latter possibility that Kak is directly associated with protein complexes that may be important for Vein binding. The reduced amount of electron-dense material observed at the muscle–tendon junction site in the kak mutant embryos described in Prokop et al. (1998) is in line with both mechanisms mentioned above.
The excess number of Stripe- and Delilah-expressing cells in the kak mutant embryos may be attributed to the dispersed levels of Vein, which could induce partial activation of the EGF-receptor signaling pathway in neighboring cells. An alternative explanation is that muscle-dependent differentiation of tendon cells may be accompanied by lateral inhibition of neighboring cells. The differentiated tendon cell may activate the Notch-signaling pathway in the surrounding cells. Aberrant contacts between tendon cells and their neighboring EMA competent cells in the kak mutant embryos may prevent efficient lateral inhibition, resulting in an excess of Stripe- and Delilah-expressing cells. An observation that supports this possibility is that an excess in β1 tubulin–expressing cells is detected in Delta mutant embryos (Volk, T., unpublished). Delta, a well-characterized Notch ligand, mediates lateral inhibition in a large array of tissues during embryonic and adult development (Artavanis-Tsakonas et al., 1995). The lack of Delta may prevent lateral inhibition of the competent EMA cells, leading to their differentiation into β1 tubulin– expressing cells. The impaired integrity of the epidermis described by Gregory and Brown (1998) is consistent with this explanation.
Kak May Be Part of a Regulatory Mechanism That Controls Vein Activity
The possibility that Kak directly mediates a Vein-induced signaling cascade was excluded since ectopic expression of Vein in kak mutants can activate delilah gene expression in epidermal cells, as in wild-type embryos (not shown). However, in this experiment the embryos expressing ectopic Vein are exposed to an excess of Vein protein compared with the physiological conditions. High levels of Vein may compensate for a mechanism responsible for accumulation of Vein at specific sites. Recent findings (Schnepp et al., 1998; Yarnitzky et al., 1998) suggest that Vein is a relatively weak activator of the EGF-receptor signaling pathway, compared with Spitz, the TGFα Drosophila homologue. Thus, Vein-mediated signaling may be susceptible to an as-of-yet-uncharacterized regulatory mechanism that controls its localization and accumulation essential for proper receptor activation. Kak may be part of such a regulatory mechanism.
The Function of Kak in the Wing May Be Comparable with Its Function in Muscle-dependent Tendon Cell Differentiation
The kak alleles were initially isolated on the basis of their ability to induce a wing blistering phenotype (Prout et al., 1997). The blistering phenotype is explained by defects in adhesion between the dorsal and ventral aspects of the wing epithelia (Prout et al., 1997; Fristrom and Fristrom, 1993). The failure of kak mutant cells to adhere to wild-type counterpart cells in the wing may result from the impaired differentiation of the mutant cells. Interestingly, Vein mRNA is expressed at high levels in the wing intervein domain after puparium formation (Simcox et al., 1996). This domain includes the cells that adhere to each other at the pupae stage to form the adult wing. The role of Vein in this process is yet to be elucidated. By analogy to its involvement in mediating Vein localization in the embryo during the process of muscle-dependent differentiation, Kak may contribute to the correct localization of Vein protein at the junction formed between the dorsal and ventral intervein epithelia. This localization may be essential for the correct activation of the EGF-receptor pathway and for the proper differentiation of the wing epithelial cells.
Possible Kak-like Activity in Vertebrate Neuromuscular Junction Formation
Several lines of evidence suggest that the process of Vein-mediated tendon cell differentiation shares molecular similarities with neuromuscular synapse formation. The secretion of Vein by the approaching muscle cell, and its restricted localization at the muscle–tendon junctional site, is reminiscent of the specific localization of its mammalian homologue Neuregulin (NRG or ARIA), which is secreted from the motor nerve endings and is localized at postsynaptic sites. NRG is essential for the activation of synapse-specific gene transcription in the postsynaptic muscle nuclei (Falls et al., 1993; Fischbach and Rosen, 1997; Burden, 1998). In a similar manner, Vein is required to positively regulate the transcription of tendon-specific genes, such as delilah and β1 tubulin (Yarnitzky et al., 1997).
The aberrant formation of the Drosophila neuromuscular junction in kak mutant embryos (described by Prokop et al., 1998) is consistent with an important role for this gene in both Vein-mediated tendon cell differentiation and neuromuscular junction formation, supporting a molecular correlation between these processes.
The mechanism that is responsible for the restricted localization of NRG together with its erbB receptors is yet to be elucidated. However, when this localization is defective, the structure and proper function of neuromuscular synapses is disrupted (Sandrock et al., 1997). Thus, vertebrate Kak-like proteins may be part of an intracellular mechanism that is essential for restricting synaptic sites to specific muscle–subcellular domains.
Acknowledgments
We would like to thank P. Fisher and D. Buttgereit for antibody and cDNA reagents; N. Brown, J. Fristrom, M. Prout, V. Hartenstein, and A. Brand for fly strains; N. Brown, S. Gregory, and A. Prokop for sharing unpublished data; E. Schejter, B. Shilo, and S. Schwarzbaum for critical reading of the manuscript and helpful comments; Li Min, Michal Renert, and Inbal Ben-Zvi Zahor for technical assistance; and Yael Rosenberg-Hasson, Naomi Levy-Strumpf, Tali Yarnitzky, and Shirly Becker for their comments and suggestions.
Abbreviations used in this paper
- EMA
epidermal muscle attachment
- kak
kakapo
- ORF
open reading frame
Footnotes
This research was supported by a grant from the Israel Science Foundation (to T. Volk).
Address all correspondence to Talila Volk, Department of Molecular Genetics, The Weizmann Institute of Science, 76100 Rehovot, Israel. Tel.: 972-8-9342426. Fax: 972-8-9344108. E-mail: lgvolk@wiccmail.weizmann.ac.il
References
- Ahn AH, Kunkel LM. Syntrophin binds to an alternatively spliced exon of dystrophin. J Cell Biol. 1995;128:363–371. doi: 10.1083/jcb.128.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Armand P, Knapp AC, Hirsch AJ, Wieschaus EF, Cole MD. A novel basic helix-loop-helix protein is expressed in muscle attachment sites of the Drosophilaepidermis. Mol Cell Biol. 1994;14:4145–4154. doi: 10.1128/mcb.14.6.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- Ashburner, M. 1989. HRP staining of embryos. In Drosophila: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 217–219.
- Bate, M. 1993. The Mesoderm and its derivatives. In The Development of Drosophila melanogaster. M. Bate, and A. Martines Arias, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 1013–1090.
- Becker S, Pasca G, Strumpf D, Min L, Volk T. Reciprocal signaling between Drosophilaepidermal muscle attachment cells and their corresponding muscles. Development (Camb) 1997;124:2615–2622. doi: 10.1242/dev.124.13.2615. [DOI] [PubMed] [Google Scholar]
- Brancolini C, Schneider C. Phosphorylation of the growth arrest-specific protein Gas2 is coupled to actin rearrangements during G0 → G1 transition in NIH 3T3 cells. J Cell Biol. 1994;124:743–756. doi: 10.1083/jcb.124.5.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancolini C, Bottega S, Schneider C. Gas2, a growth arrest-specific protein, is a component of the microfilament network system. J Cell Biol. 1992;117:1251–1261. doi: 10.1083/jcb.117.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancolini C, Benedetti M, Schneider C. Microfilament reorganization during apoptosis: the role of Gas2, a possible substrate for ICE-like proteases. EMBO (Eur Mol Biol Organ) J. 1995;14:5179–5190. doi: 10.1002/j.1460-2075.1995.tb00202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden SJ. The formation of neuromuscular synapses. Genes Dev. 1998;12:133–148. doi: 10.1101/gad.12.2.133. [DOI] [PubMed] [Google Scholar]
- Buttgereit D, Leiss D, Michiels F, Renkawitz-Pohl R. During Drosophilaembryogenesis the β1 tubulin gene is specifically expressed in the nervous system and the apodemes. Mech Dev. 1991;33:107–118. doi: 10.1016/0925-4773(91)90077-j. [DOI] [PubMed] [Google Scholar]
- Davison MD, Critchley DR. α-Actinins and the DMD protein contain spectrin-like repeats [letter] Cell. 1988;52:159–160. doi: 10.1016/0092-8674(88)90503-x. [DOI] [PubMed] [Google Scholar]
- Ervasti JM, Campbell KP. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1991;66:1121–1131. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- Falls DL, Rosen KM, Corfas G, Lane WS, Fischbach GD. ARIA, a protein that stimulates acetylcholine receptor synthesis, is a member of the neu ligand family. Cell. 1993;72:801–815. doi: 10.1016/0092-8674(93)90407-h. [DOI] [PubMed] [Google Scholar]
- Fischbach GD, Rosen KM. ARIA: a neuromuscular junction neuregulin. Annu Rev Neurosci. 1997;20:429–458. doi: 10.1146/annurev.neuro.20.1.429. [DOI] [PubMed] [Google Scholar]
- Fogerty FJ, Fessler LI, Bunch TA, Yaron Y, Parker CG, Nelson RE, Brower DL, Gullberg D, Fessler JH. Tiggrin, a novel Drosophila extracellular matrix protein that functions as a ligand for DrosophilaαPS2β PS integrins. Development (Camb) 1994;120:1747–1758. doi: 10.1242/dev.120.7.1747. [DOI] [PubMed] [Google Scholar]
- Fristrom, D., and J.W. Fristrom. 1993. The metamorphic development of the adult epidermis. In The Development of Drosophila melanogaster. M. Bate and A. Martines Arias, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 843–897.
- Frommer G, Vorbrüggen G, Pasca G, Jäckle H, Volk T. Epidermal egr-like zinc finger protein of Drosophilaparticipates in myotube guidance. EMBO (Eur Mol Biol Organ) J. 1996;15:1642–1649. [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Cleveland DW. A structural scaffolding of intermediate filaments in health and disease. Science. 1998;279:514–519. doi: 10.1126/science.279.5350.514. [DOI] [PubMed] [Google Scholar]
- Girgliatti, T. 1986. Mutagenesis. In Drosophila: A Practical Approach. D.B. Roberts, editor. IRL Press, Oxford. 39–58.
- Goubeaud A, Knirr S, Renkawitz-Pohl R, Paululat A. The Drosophila gene alien is expressed in the muscle attachment sites during embryogenesis and encodes a protein highly conserved between plants, Drosophila, and vertebrates. Mech Dev. 1996;57:59–68. doi: 10.1016/0925-4773(96)00532-1. [DOI] [PubMed] [Google Scholar]
- Gregory SL, Brown NH. kakapo, a gene required for adhesion between and within cell layers in Drosophila, encodes a large cytoskeletal linker protein related to plectin and dystrophin. J Cell Biol. 1998;143:1271–1282. doi: 10.1083/jcb.143.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Yang B, Meyer J, Chamberlain JS, Campbell KP. Identification and characterization of the dystrophin anchoring site on β-dystroglycan. J Biol Chem. 1995;270:27305–27310. doi: 10.1074/jbc.270.45.27305. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Kretsinger RH. Calcium-binding proteins 1: EF-hands. Protein Profile. 1995;2:297–490. [PubMed] [Google Scholar]
- Koenig M, Kunkel LM. Detailed analysis of the repeat domain of dystrophin reveals four potential hinge segments that may confer flexibility. J Biol Chem. 1990;265:4560–4566. [PubMed] [Google Scholar]
- Koenig M, Monaco AP, Kunkel LM. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988;53:219–226. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- Lee JC, VijayRaghavan K, Celniker SE, Tanouye MA. Identification of a Drosophilamuscle development gene with structural homology to mammalian early growth response transcription factors. Proc Natl Acad Sci USA. 1995;92:10344–10348. doi: 10.1073/pnas.92.22.10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzow S, Brancolini C, Marks F, Richter KH. Expression of growth arrest-specific (Gas) genes in murine keratinocytes: Gas2 is specifically regulated. Exp Cell Res. 1996;224:200–203. doi: 10.1006/excr.1996.0128. [DOI] [PubMed] [Google Scholar]
- Prokop A, Uhler J, Roote J, Bate M. The kakapo mutation affects terminal arborization and central dendritic sprouting of Drosophilamotorneurons. J Cell Biol. 1998;143:1283–1294. doi: 10.1083/jcb.143.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prout M, Damania Z, Soong J, Fristorm D, Fristrom JW. Autosomal mutations affecting adhesion between wing surfaces in Drosophila melanogaster. . Genetics. 1997;146:275–285. doi: 10.1093/genetics/146.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastelli L, Chan CS, Pirrotta V. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophilaand their dependence on Enhancer of zeste function. EMBO (Eur Mol Biol Organ) J. 1993;12:1513–1522. doi: 10.1002/j.1460-2075.1993.tb05795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RG, Bobrow M. Dystrophins in vertebrates and invertebrates. Hum Mol Genet. 1998;7:589–595. doi: 10.1093/hmg/7.4.589. [DOI] [PubMed] [Google Scholar]
- Ruhrberg C, Watt FM. The plakin family: versatile organizers of cytoskeletal architecture. Curr Opin Genet Dev. 1997;7:392–397. doi: 10.1016/s0959-437x(97)80154-2. [DOI] [PubMed] [Google Scholar]
- Sandrock AW, Jr, Dryer SE, Rosen KM, Gozani SN, Kramer R, Theill LE, Fischbach GD. Maintenance of acetylcholine receptor number by neuregulins at the neuromuscular junction in vivo. Science. 1997;276:599–603. doi: 10.1126/science.276.5312.599. [DOI] [PubMed] [Google Scholar]
- Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- Schnepp B, Grumbling G, Donaldson T, Simcox A. Vein is a novel component in the Drosophilaepidermal growth factor receptor pathway with similarity to the neuregulins. Genes Dev. 1996;10:2302–2313. doi: 10.1101/gad.10.18.2302. [DOI] [PubMed] [Google Scholar]
- Schnepp B, Donaldson T, Grumbling G, Ostrowski S, Schweitzer R, Shilo B-Z, Simcox A. EGF domain swap converts a DrosophilaEGF-receptor activator into an inhibitor. Genes Dev. 1998;12:908–913. doi: 10.1101/gad.12.7.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki N, Ohira M, Nagase T, Ishikawa K, Miyajima N, Nakajima D, Nomura N, Ohara O. Characterization of cDNA clones in size-fractionated cDNA libraries from human brain. DNA Res. 1997;4:345–349. doi: 10.1093/dnares/4.5.345. [DOI] [PubMed] [Google Scholar]
- Simcox AA, Grumbling G, Schnepp B, Bennington C, Mathias, Hersperger E, Shearn A. Molecular, phenotypic, and expression analysis of vein, a gene required for growth of the Drosophilawing disc. Dev Biol. 1996;177:475–489. doi: 10.1006/dbio.1996.0179. [DOI] [PubMed] [Google Scholar]
- Speicher DW, Marchesi VT. Erythrocyte spectrin is comprised of many homologous triple helical segments. Nature. 1984;311:177–180. doi: 10.1038/311177a0. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Yoshida M, Ozawa E. Mammalian α1- and β1-syntrophin bind to the alternative splice-prone region of the dystrophin COOH terminus. J Cell Biol. 1995;128:373–381. doi: 10.1083/jcb.128.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophilaembryos reveals translational control of the segmentation gene hunchback. Chromosoma (Berl) 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Tepass U, Hartenstein V. The development of cellular junctions in the Drosophilaembryo. Dev Biol. 1994;161:563–596. doi: 10.1006/dbio.1994.1054. [DOI] [PubMed] [Google Scholar]
- Volk T. A new member of the spectrin superfamily may participate in the formation of muscle attachments in the Drosophilaembryo. Development (Camb) 1992;116:721–730. doi: 10.1242/dev.116.3.721. [DOI] [PubMed] [Google Scholar]
- Volk T, VijayRaghavan K. A central role for epidermal segment border cells in the induction of muscle patterning in the Drosophilaembryo. Development (Camb) 1994;120:59–70. doi: 10.1242/dev.120.1.59. [DOI] [PubMed] [Google Scholar]
- Vorbrüggen G, Jäckle H. Epidermal muscle attachment site-specific target gene expression and interference with myotube guidance in response to ectopic stripe expression in the developing Drosophilaepidermis. Proc Natl Acad Sci USA. 1997;94:8606–8611. doi: 10.1073/pnas.94.16.8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh EP, Brown NH. A screen to identify Drosophilagenes required for integrin mediated adhesion. Genetics. 1998;150:791–805. doi: 10.1093/genetics/150.2.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans[see comments] Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- Yarnitzky T, Min L, Volk T. The Drosophilaneuregulin homolog Vein mediates inductive interactions between the myotubes and their epidermal attachment cells. Genes Dev. 1997;11:2691–2700. doi: 10.1101/gad.11.20.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnitzky, T., L. Min, and T. Volk. 1998. An interplay between two EGF-receptor ligands, Vein and Spitz, is required for the formation of a subset of muscle precursors in Drosophila. Mech. Dev. In press. [DOI] [PubMed]
- Zinn K, McAllister L, Goodman CS. Sequence analysis and neuronal expression of fasciclin I in grasshopper and Drosophila. . Cell. 1988;53:577–587. doi: 10.1016/0092-8674(88)90574-0. [DOI] [PubMed] [Google Scholar]
- Zucman-Rossi J, Legoix P, Thomas G. Identification of new members of the Gas2 and Ras families in the 22q12 chromosome region. Genomics. 1996;38:247–254. doi: 10.1006/geno.1996.0625. [DOI] [PubMed] [Google Scholar]