Abstract
We isolated two novel actin filament (F-actin)–binding proteins from rat brain and rat 3Y1 fibroblast. They were splicing variants, and we named brain big one b-nexilin and fibroblast small one s-nexilin. b-Nexilin purified from rat brain was a protein of 656 amino acids (aa) with a calculated molecular weight of 78,392, whereas s-nexilin, encoded by the cDNA isolated from rat 3Y1 cells by the reverse transcriptase-PCR method, was a protein of 606 aa with a calculated molecular weight of 71,942. b-Nexilin had two F-actin– binding domains (ABDs) at the NH2-terminal and middle regions, whereas s-nexilin had one ABD at the middle region because 64 aa residues were deleted and 14 aa residues were inserted in the first NH2-terminal ABD of b-nexilin, and thereby the first ABD lost its activity. b- and s-nexilins bound along the sides of F-actin, but only b-nexilin showed F-actin cross-linking activity. b-Nexilin was mainly expressed in brain and testis, whereas s-nexilin was mainly expressed in testis, spleen, and fibroblasts, such as rat 3Y1 and mouse Swiss 3T3 cells, but neither b- nor s-nexilin was detected in liver, kidney, or cultured epithelial cells. An immunofluorescence microscopic study revealed that s-nexilin was colocalized with vinculin, talin, and paxillin at cell– matrix adherens junction (AJ) and focal contacts, but not at cell–cell AJ, in 3Y1 cells. Overexpressed b- and s-nexilins were localized at focal contacts but not at cell–cell AJ. These results indicate that nexilin is a novel F-actin–binding protein localized at cell–matrix AJ.
Keywords: actin-binding protein, focal contact, stress fiber, adherens junction, integrin
The actin filament (F-actin)1–associated adherens junctions (AJs) are subclassed into two types: cell–matrix and cell–cell AJs (Geiger et al., 1985). Cell–matrix AJ includes focal contacts, also referred to as focal adhesions or adhesion plaques, which are the specialized structures at the ends of stress fibers, up to 10 μm in length and 0.5 μm in width (Abercrombie and Dunn, 1975; Izzard and Lochner, 1976; Heath and Dunn, 1978). Originally, focal contacts were described in living or fixed fibroblasts cultured on glass or plastic (Abercrombie et al., 1971). Typical focal contacts are mainly developed in cultured cells and are rarely found in the organism. However, since cultured cells are amenable to microscopic and biochemical analyses and can be experimentally manipulated, the focal contacts in cultured cells have been extensively studied as a model system for cell–matrix AJ. A transmembrane connection between the actin cytoskeleton and the extracellular matrix through the integrin family occurs at these sites of the plasma membrane (Schwartz et al., 1995). At the extracellular surface, integrin interacts with matrix proteins, whereas the cytoplasmic domain interacts directly or indirectly with F-actin–binding proteins, including α-actinin, vinculin, and talin (Jockusch et al., 1995; Burridge and Chrzanowska-Wodnicka, 1996). Moreover, a number of structural and regulatory proteins, such as paxillin (Turner et al., 1990), tensin (Wilkins et al., 1986; Lo et al., 1994; Chuang et al., 1995), zyxin (Beckerle, 1986; Crawford and Beckerle, 1991), profilin (Carlsson et al., 1977), gelsolin (Wang et al., 1984), and FAK (Schaller et al., 1992), have been identified and characterized. Most of these proteins are found not only at cell–matrix AJ but also at cell–cell AJ, but paxillin and talin are restricted to cell–matrix AJ (Geiger et al., 1985; Tidball et al., 1986; Drenkhahn et al., 1988; Turner et al., 1990).
Molecular linkage between the actin cytoskeleton and the plasma membrane at cell–cell AJ has also extensively been investigated (Geiger, 1983, 1989; Geiger and Ginsberg, 1991; Turner and Burridge, 1991; Luna and Hitt, 1992; Tsukita et al., 1992, 1997; Bretscher, 1993). Many linker proteins have been isolated and characterized at cell–cell AJ, where cadherins interact with each other at the extracellular surface (Takeichi, 1988, 1991; Geiger and Ginsberg, 1991; Tsukita et al., 1992). At the cytoplasmic surface, cadherin is associated with α-, β-, and γ-catenins (Ozawa et al., 1989; Nagafuchi et al., 1991; Takeichi, 1991; Tsukita et al., 1992). α-Catenin directly interacts with F-actin and indirectly interacts with F-actin through α-actinin and/ or ZO-1 (Knudsen et al., 1995; Rimm et al., 1995; Itoh et al., 1997). Thus, many F-actin–binding proteins appear to link the actin cytoskeleton to the plasma membrane and have important roles in the dynamic regulation of both cell–matrix and cell–cell AJ.
In contrast to cadherin-based cell–cell AJ in epithelial and endothelial cells, the molecular mechanism of synaptic junction has not fully been understood, although recent studies have revealed that cadherin is concentrated at synaptic junctions (Fannon and Colman, 1996; Uchida et al., 1996). To identify F-actin–binding proteins, which regulate synaptic junctions, we have been attempting to isolate novel F-actin–binding proteins from rat brain using a blot overlay method with 125I-labeled F-actin. We had isolated and reported four novel F-actin–binding proteins: neurabin-I (Nakanishi et al., 1997), neurabin-II (Satoh et al., 1998), afadin (Mandai et al., 1997), and frabin (Obaishi et al., 1998). Neurabin-I is an F-actin–binding protein specifically expressed in neural tissue and is implicated in both neurite formation and maintenance of synaptic junction (Nakanishi et al., 1997). Neurabin-II is an F-actin–binding protein ubiquitously expressed (Satoh et al., 1998). Neurabin-II is enriched at the postsynaptic density fraction in rat brain and the cell–cell AJ fraction in rat liver. Neurabin-II is identical to the recently reported protein phosphatase 1–binding protein, named spinophilin (Allen et al., 1997). Afadin has two splicing variants: l-afadin ubiquitously expressed and s-afadin mainly expressed in neural tissue. s-Afadin lacks the F-actin–binding domain. l-Afadin is localized at cell–cell AJ. Frabin shows a homology to FGD1, which has been identified to be the genetic locus responsible for faciogenital dysplasia (Pasteris et al., 1994). Frabin is capable of inducing filopodium formation and c-Jun NH2-terminal kinase activation as described for FGD1 (Zheng et al., 1996).
During the purification of these F-actin–binding proteins, we purified the fifth F-actin–binding protein from rat brain, which was localized at cell–matrix AJ. It has two splicing variants that have different physical and functional properties. We named big and small ones b- and s-nexilins, respectively, from a Latin word nexilis meaning “bound together.” We describe here these novel F-actin– binding proteins.
Materials and Methods
125I-Labeled F-Actin Blot Overlay
125I-labeled F-actin blot overlay was done as described (Chia et al., 1991; Pestonjamasp et al., 1995). In brief, purified actin monomer (G-actin) was labeled with 125I–Bolton Hunter reagent. 125I-labeled G-actin (1 mg/ml; average specific activity, 63.3 μCi/mg) was polymerized with 18 μg/ml of gelsolin (Sigma Chemical Co., St. Louis, MO) (molar ratio 100:1) by incubation at 4°C for 10 min in a solution containing 20 mM Pipes at pH 7.0, 50 mM KCl, and 2 mM MgCl2. Phalloidin was then added to give a final concentration of 40 μM. The mixture was then incubated at room temperature for another 15 min and stored at 4°C as 125I-labeled F-actin. The sample to be tested was subjected to SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was blocked in TBS containing 5% defatted powder milk. The membrane was then incubated at room temperature for 1 h with 10 μg/ml of 125I-labeled F-actin in TBS containing 5% defatted powder milk and 4 μM phalloidin. After the incubation, the membrane was washed with TBS containing 0.5% Tween 20, followed by autoradiography using an image analyzer (Fujix BAS-2000; Fuji Photo Film Co., Tokyo, Japan).
For competition experiments, 125I-labeled F-actin was prepared as described. 10 μg/ml of 125I-labeled F-actin was incubated at room temperature for 30 min with 0.66 mg/ml of myosin subfragment 1 (S1) (Sigma Chemical Co.) in a solution containing 20 mM Pipes at pH 7.0, 58 mM KCl, 2 mM MgCl2, 130 μM CaCl2, and 0.4 μM phalloidin. Where indicated, 4 mM MgATP was added to the mixture. After the incubation, the mixture was diluted with an equal volume of TBS containing 0.5% Tween 20 and 10% defatted powder milk, and it was added to the blot membrane, followed by incubation at room temperature for 1 h.
Purification of b-Nexilin
The synaptic soluble fraction was prepared from 480 adult rat brains as described (Mizoguchi et al., 1990) and stored at −80°C until use. 1/12 of the fraction (420 ml, 360 mg of protein) was adjusted to 0.2 M NaCl with 4 M NaCl and applied to a Q-Sepharose FF column (2.6 × 10 cm; Amersham Pharmacia Biotech, Piscataway, NJ) equilibrated with buffer A (20 mM Tris-Cl at pH 7.5 and 1 mM DTT) containing 0.2 M NaCl. After the column was washed with 250 ml of buffer A containing 0.2 M NaCl, elution was performed with 350 ml of buffer A containing 0.5 M NaCl at a flow rate of 5 ml/min. Fractions of 10 ml each were collected. b-Nexilin appeared in fractions 5–19. These fractions (150 ml, 152 mg of protein) were collected, and NaCl was added to give a final concentration of 2 M. The sample was applied to a phenyl-Sepharose column (2.6 × 10 cm; Amersham Pharmacia Biotech) equilibrated with buffer A containing 2 M NaCl. After the column was washed with 250 ml of the same buffer, elution was performed with a 360-ml linear gradient of NaCl (2.0–0 M) in buffer A, followed by 180 ml of buffer A, at a flow rate of 3 ml/min. Fractions of 6 ml each were collected. b-Nexilin appeared in fractions 45–49. The active fractions (30 ml, 3.7 mg of protein) were collected. The two samples were combined and applied to a phenyl-5PW RP column (0.75 × 7.5 cm; Tosoh, Tokyo, Japan) equilibrated with 0.05% trifluoroacetic acid. Elution was performed with a 320-ml linear gradient of acetonitrile (10– 80%) in 0.05% trifluoroacetic acid. Fractions of 1.33 ml each were collected. b-Nexilin appeared in fractions 44 and 45. The active fractions of the six phenyl-5PW RP column chromatographies (7.98 ml, 40 μg of protein) were collected, lyophilized, and stored at −80°C.
Determination of Partial Amino Acid Sequence of b-Nexilin
The purified phenyl-5PW RP sample (40 μg of protein) was subjected to SDS-PAGE (8% polyacrylamide gel). A protein band corresponding to b-nexilin was cut out from the gel and digested with a lysyl endopeptidase, and the digested peptides were separated by TSKgel ODS-80Ts (4.6 × 150 mm; Tosoh) reverse-phase high-pressure liquid column chromatography as described (Imazumi et al., 1994). The amino acid (aa) sequences of the eight peptides were determined with a peptide sequencer.
Molecular Cloning of b- and s-Nexilins
Computer homology search revealed that two of the eight peptides were contained within a peptide sequence deduced from the cDNA fragment isolated from a human cDNA library (Elisei et al., 1993). Therefore, a probe was obtained by the PCR method with oligonucleotide primers from the cDNA fragment. To obtain full-length b-nexilin, a rat brain cDNA library in λZAPII (Stratagene, La Jolla, CA) was screened using the probe.
Reverse transcriptase–PCR was performed to amplify a fragment of mRNA encoding s-nexilin expressed in 3Y1 cells. For that purpose, total RNA was prepared from 3Y1 cells using the ISOGEN RNA extraction kit (Nippon Gene, Tokyo, Japan). Reverse transcription of 0.5 μg of total RNA was carried out using Ready-To-Go You-Prime First-Strand Beads and pd(N)6 (Amersham Pharmacia Biotech). For PCR, an aliquot of cDNA was amplified using primers based on the NH2- and COOH-terminal regions of b-nexilin. The primer sequences were 5′-atgaatgacgtgtcacagaaggca-3′ and 5′-ctagtagtcatccatttcaatggt-3′ for the NH2- and COOH-terminal regions, respectively. DNA sequencing was performed by the dideoxy nucleotide termination method using a DNA sequencer (ALF express; Amersham Pharmacia Biotech). For coiled-coil prediction, the PAIRCOIL program was used (Berger et al., 1995).
Expression and Purification of Recombinant b- and s-Nexilins
Prokaryotic and eukaryotic expression vectors were constructed in pGEX (Amersham Pharmacia Biotech), pCMV5 (Takeuchi et al., 1997), pCIneo (Promega Corp., Madison, WI), and pRSET (Invitrogen Corp., Carlsbad, CA) using standard molecular biological methods (Sambrook et al., 1989). Glutathione S-transferase (GST) fusion constructs of b- and s-nexilins contained the following amino acid (aa) residues: pGEX-b-nexilin-1, aa 1–150; pGEX-b-nexilin-1′, aa 1–240; pGEX-b-nexilin-2, aa 77–240; pGEX-b-nexilin-3, aa 151–283; pGEX-b-nexilin-4, aa 211–330; pGEX-b-nexilin-5, aa 284–450; pGEX-b-nexilin-6, aa 493–656; pGEX-s-nexilin-1, aa 1–100; and pGEX-s-nexilin-1′, aa 1–190. The GST fusion proteins were purified by use of glutathione-Sepharose beads according to the manufacturer's protocol (Amersham Pharmacia Biotech). Full-length b- and s-nexilins were constructed in pCMV5 and pRSET vectors. Full-length and various truncated mutants of b- and s-nexilins were constructed in pCIneo-myc vector. pCIneo-myc vector was constructed by ligating the oligonucleotides, 5′-ctagaccccccaacatggagcagaagcttatcagcgaggaggacctgctcgagg-3′/ 5′-aattctcgagcaggtcctcctcgctgataagcttctgctccatgttggggggt-3′, into the NheI and EcoRI sites of pCIneo. The myc-epitope (MEQKLISEEDL) was inserted at the NH2 termini of full-length b- and s-nexilins. Various myc-tagged truncated mutants of b- and s-nexilins contained the following aa residues: pCIneo-myc-s-nexilin-M1, aa 1–100; pCIneo-myc-s-nexilin-M2, aa 1–233; pCIneo-myc-FC, aa 161–430 for b-nexilin, aa 211–480 for b-nexilin; pCIneo-myc-ABD, aa 234–400 for s-nexilin, aa 284-450 for b-nexilin; pCIneo-myc-CC, aa 101–233 for s-nexilin, aa 151–283 for b-nexilin; pCIneo-myc-b-nexilin-ABD, aa 1–150; and pCIneo-myc-b-nexilin-M1, aa 1–283. pRSET was designed to express the proteins with the NH2-terminal six histidine residues (His6). The His6-tagged proteins were purified by use of TALON metal affinity beads according to the manufacturer's protocol (CLONTECH Labs, Palo Alto, CA).
Assay for Cosedimentation of b- and s-Nexilins with F-Actin
G-actin was polymerized by incubation at room temperature for 30 min in a polymerization buffer (20 mM imidazole/Cl at pH 7.0, 2 mM MgCl2, 1 mM ATP, 0.5 mM DTT, and 90 mM KCl). His6-b- or His6-s-nexilin in an indicated amount was incubated at room temperature for 30 min with 0.3 mg/ml of F-actin in a solution containing 25 mM imidazole/Cl at pH 7.0, 2 mM MgCl2, 1 mM ATP, 0.05 mM DTT, 27 mM KCl, and 100 mM NaCl, and the mixture (50 μl) was placed over a 50-μl cushion of 30% sucrose in the polymerization buffer. To estimate K d values, various amounts of His6-b- or His6-s-nexilin were incubated with 0.1 mg/ml of F-actin in a solution containing 32 mM imidazole/Cl at pH 7.0, 2 mM MgCl2, 1 mM ATP, 0.05 mM DTT, 27 mM KCl, and 100 mM NaCl. After the sample was centrifuged at 130,000 g for 20 min, the supernatant was removed from the cushion and the pellet was brought to the original volume in an SDS sample buffer. The comparable amounts of the supernatant and pellet fractions were subjected to SDS-PAGE, followed by protein staining with Coomassie brilliant blue to quantitate recombinant b- and s-nexilins cosedimented with F-actin using a densitometer.
For the cosedimentation assay using the crude samples, the cell lysates of Escherichia coli expressing the GST fusion proteins of nexilin were centrifuged at 100,000 g for 1 h, and the supernatant was used for the assay. In brief, after G-actin was polymerized as described above, the supernatant (5 μl each) of the lysates was incubated at room temperature for 30 min with 0.3 mg/ml of F-actin in a solution containing 25 mM imidazole/Cl at pH 7.0, 2 mM MgCl2, 1 mM ATP, 0.05 mM DTT, 27 mM KCl, and 100 mM NaCl, and the mixture (50 μl) was placed over a 50-μl cushion of 30% sucrose in the polymerization buffer. After the sample was centrifuged at 130,000 g for 20 min, the supernatant and pellet were subjected to SDS-PAGE, followed by Western blot analysis using the anti-GST and antiactin antibodies.
Assay for F-Actin Cross-linking Activity of b- and s-Nexilins
Electron microscopy was performed as described (Endo and Masaki, 1982; Kato et al., 1996). In brief, 0.28 mg/ml of G-actin was incubated at 25°C for 45 min with His6-b- (40 μg/ml of protein) or His6-s-nexilin (37 μg/ml of protein) in a solution containing 25 mM imidazole/Cl at pH 7.0, 140 mM NaCl, 2 mM MgCl2, 0.1 mM ATP, and 1 mM EGTA. The samples were negatively stained with 2% uranyl acetate and viewed with an electron microscope (model H-7100; Hitachi, Tokyo, Japan).
Low shear viscometry was performed as described (Pollard and Cooper, 1982; Kato et al., 1996). In brief, His6-b- or His6-s-nexilin in an indicated amount was mixed with 0.28 mg/ml of G-actin in a solution containing 20 mM imidazole/Cl at pH 7.0, 140 mM NaCl, 0.1 mM ATP, 0.5 mM DTT, and 1 mM EGTA, and the solution was sucked into a 0.1-ml micropipette. After the incubation at 25°C for 35 min, the time for a stainless ball to fall a fixed distance in the pipette was measured.
Cell Culture and Transfection
Rat 3Y1 and mouse mammary tumor MTD-1A cells were kindly supplied by Dr. Sh. Tsukita (Kyoto University, Kyoto, Japan). MDCK cells were supplied by Dr. W. Birchmeier (Max-Delbruck-Center for Molecular Medicine, Berlin, Germany). These cells were maintained in DME containing 10% FCS (GIBCO BRL; Life Technologies, Grand Island, NY), penicillin (100 U/ml), and streptomycin (100 mg/ml). Transfection of pCIneo-myc-b- or pCIneo-myc-s-nexilin into 3Y1 cells was carried out using LipofectAMINE reagent (GIBCO BRL). Transfection of pCMV-b- or pCMV-s-nexilin into COS7 cells was carried out using the DEAE-dextran method (Hata and Südhof, 1995).
Antibodies and Immunofluorescence Staining
A rabbit polyclonal antibody against b-nexilin was raised against GST-b-nexilin-4 (aa 211–330, the antinexilin antibody). The antiserum was affinity-purified with the GST fusion protein covalently coupled to NHS-activated Sepharose (Amersham Pharmacia Biotech). The specificity of this antibody was confirmed by Western blot analysis of the pCMV-b-nexilin– transfected COS7 cells versus mock cells. Hybridoma cells expressing the mouse monoclonal anti-myc antibody (9E10) were purchased from American Type Culture Collection (Rockville, MD). The rabbit monoclonal antiactin antibody was purchased from Amersham Pharmacia Biotech. The mouse monoclonal antipaxillin antibody was purchased from Transduction Laboratories (Lexington, KY). The mouse monoclonal antivinculin and antitalin antibodies were purchased from Sigma Chemical Co. The mouse monoclonal anti-GST antibody was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Rhodamine-phalloidin was purchased from Molecular Probes, Inc. (Eugene, OR). The rat monoclonal anti–E-cadherin and anti–P-cadherin antibodies were purchased from TAKARA shuzo, Inc. (Shiga, Japan). Second antibodies for immunofluorescence microscopy were obtained from Chemicon International, Inc. (Temecula, CA).
For localization of s-nexilin and other marker proteins in 3Y1 cells, immunofluorescence microscopy was performed as described (Kotani et al., 1997). In brief, the cells were fixed in 3.7% formaldehyde in PBS for 20 min. The fixed cells were incubated with 50 mM NH4Cl in PBS for 10 min and permeabilized with PBS containing 0.2% Triton X-100 for 10 min. After being soaked in 10% FCS/PBS for 30 min, the cells were treated with the first antibody in 10% FCS/PBS for 1 h. The cells were then washed with PBS three times, followed by incubation with the second antibody in 10% FCS/PBS for 1 h. For the detection of F-actin, rhodamine-phalloidin was mixed with the second antibody solution. After being washed with PBS three times, the cells were examined using a confocal laser scanning microscope (model LSM 410; Carl Zeiss, Oberkochen, Germany).
Other Procedures
G-actin was purified from rabbit skeletal muscle as described (Pardee and Spudich, 1982). Protein concentrations were determined with BSA as a reference protein (Bradford, 1976). SDS-PAGE was performed as described (Laemmli, 1970). Protein markers used were either myosin (200 kD), β-galactosidase (116 kD), phosphorylase b (97 kD), BSA (66 kD), and ovalbumin (45 kD), or myosin (206 kD), β-galactosidase (117 kD), BSA (89 kD), and ovalbumin (47 kD).
Results
Purification of b-Nexilin and Molecular Cloning of Its cDNA
During the purification of neurabin-I, -II, afadin, and frabin from rat brain using a blot overlay method with 125I-labeled F-actin (Mandai et al., 1997; Nakanishi et al., 1997; Obaishi et al., 1998; Satoh et al., 1998), we identified a band of 125I-labeled F-actin–binding activity with a molecular mass of about 97 kD (p97). p97 was highly purified by column chromatographies, including Q-Sepharose, phenyl-Sepharose, and phenyl-5PW RP column chromatographies. On the final phenyl-5PW RP column chromatography, the F-actin–binding protein band closely coincided with a protein with a molecular mass of ∼97 kD, which was identified by protein staining with Coomassie brilliant blue (Fig. 1).
Figure 1.
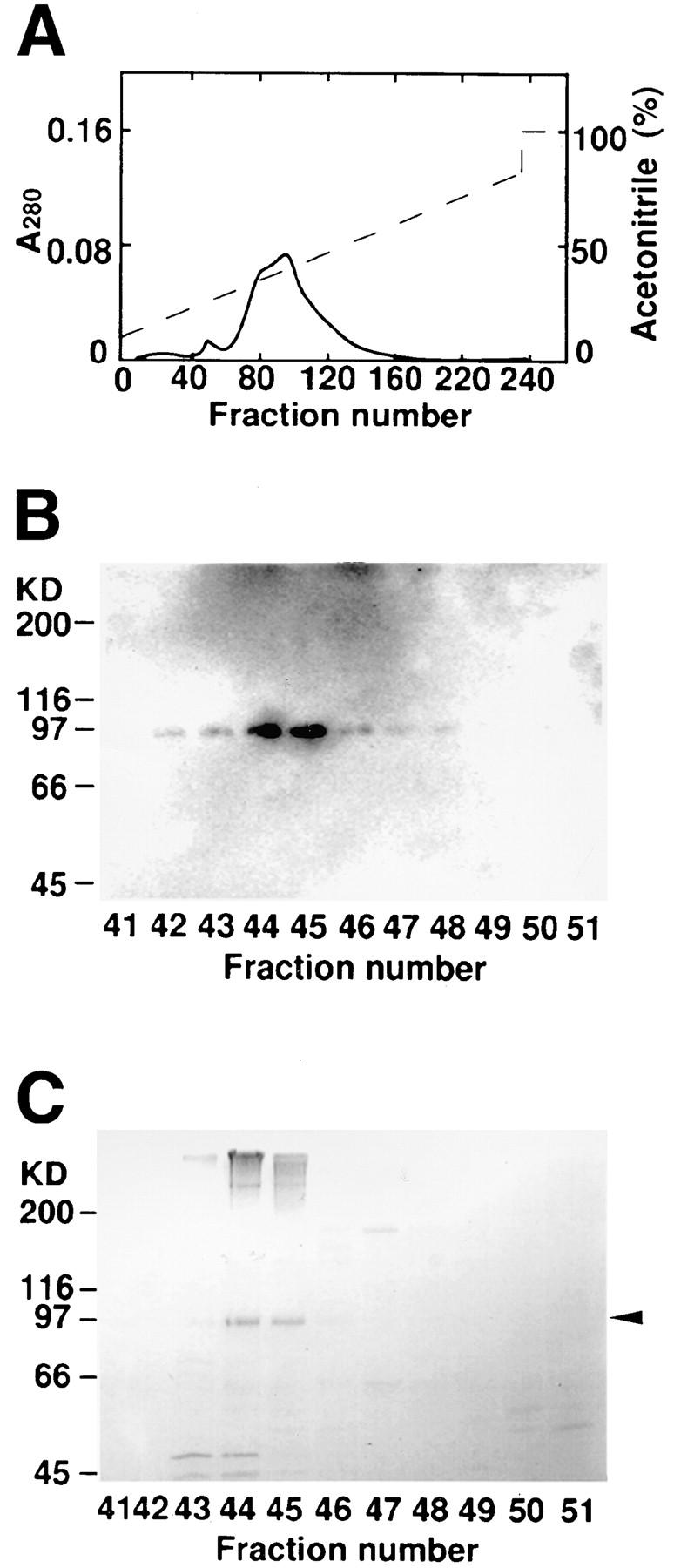
Phenyl-5PW RP column chromatography. (A) Absorbance at 280 nm (A 280). (B) 125I-labeled F-actin blot overlay. An aliquot (33 μl) of each fraction was subjected to SDS-PAGE (8% polyacrylamide gel), followed by 125I-labeled F-actin blot overlay. (C) Protein staining with Coomassie brilliant blue. An aliquot (200 μl) of each fraction was subjected to SDS-PAGE (8% polyacrylamide gel), followed by protein staining with Coomassie brilliant blue. Arrowhead indicates p97 (b-nexilin).
When the peptide mapping of the phenyl-5PW RP sample of p97 was performed, over 20 peptides were observed. Of these peptides, the aa sequences of the eight peptides were determined. Computer homology search revealed that two of the eight peptides were contained within a peptide sequence deduced from the cDNA fragment (sequence data available from GenBank/EMBL/DDBJ under accession number S67069) isolated from a human cDNA library (Elisei et al., 1993). Therefore, using a probe obtained by the PCR method with oligonucleotide primers from the cDNA fragment, we isolated several clones from a rat brain cDNA library and obtained a full-length clone by connecting two clones among them. The encoded protein consisted of 656 aa and showed a calculated molecular weight of 78,392 (Fig. 2 A). It included all the aa sequences of the eight peptides. The molecular mass value calculated from the predicted aa sequence was slightly less than that estimated by SDS-PAGE. To confirm whether this clone encoded a full-length cDNA of p97, we constructed the eukaryotic expression vector with this cDNA and expressed the protein in COS7 cells. Western blot analysis indicated that the expressed protein showed a mobility similar to that of native p97 on SDS-PAGE (Fig. 3 Aa). Although the reason for the slight difference between the molecular mass value calculated from the predicted aa sequence and that estimated by SDS-PAGE was not known, we concluded that this clone encoded the full-length cDNA of p97.
Figure 2.

Deduced amino acid sequences of b- and s-nexilins. (A) Deduced aa sequence of b-nexilin. The aa sequences of the eight peptide peaks derived from the purified sample of b-nexilin are indicated by underlining. (B) Aligned sequences of b- and s-nexilins. The deletion and insertion are indicated by a box and black shading, respectively. These sequence data are available from GenBank/EMBL/DDBJ under accession numbers AF056034 for b-nexilin and AF056035 for s-nexilin.
Figure 3.
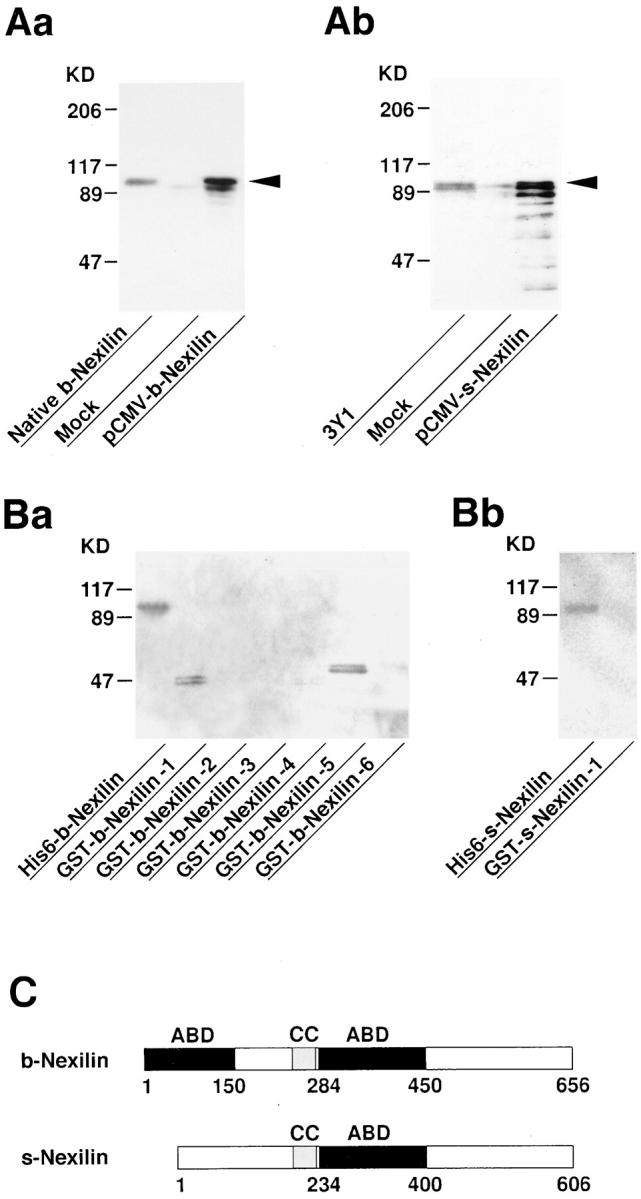
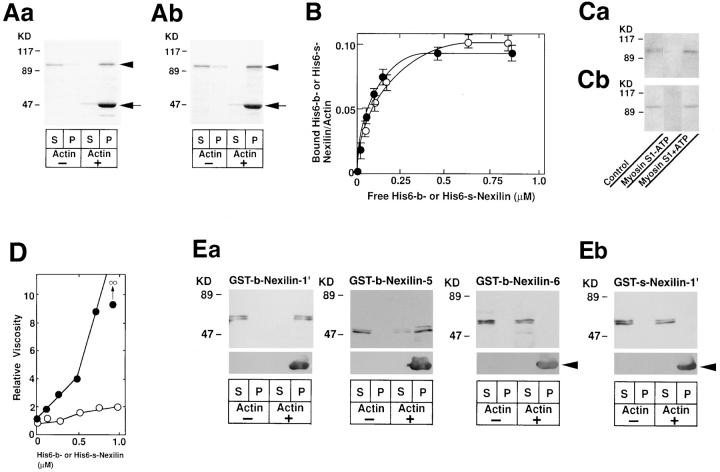
Molecular characterization of b- and s-nexilins. (A) Western blot analysis of recombinant b- and s-nexilins. The pCMV-b- or pCMV-s-nexilin was transfected into COS7 cells, and the cell lysates were subjected to SDS-PAGE (8% polyacrylamide gel), followed by Western blot analysis using the antinexilin antibody. Arrowheads indicate b- and s-nexilins. (Aa) b-Nexilin and (Ab) s-nexilin. (B) 125I-labeled F-actin–binding activity of various truncated forms of b- and s-nexilins. The purified proteins (0.2 μg of protein each), except for GST-b-nexilin-1, were subjected to SDS-PAGE (10% polyacrylamide gel), followed by 125I-labeled F-actin blot overlay. GST-b-nexilin-1 was recovered in the inclusion bodies during the preparation of the supernatant of the cell lysates, and thus an aliquot of the cell lysates was used for the assay. GST-b-Nexilin-1, aa 1–150; GST-b-Nexilin-2, aa 77– 240; GST-b-Nexilin-3, aa 151–283; GST-b-Nexilin-4, aa 211–330; GST-b-Nexilin-5, aa 284–450; GST-b-Nexilin-6, aa 493–656; and GST-s-Nexilin-1, aa 1–100. (Ba) b-Nexilin and (Bb) s-nexilin. (C) Schematic drawing of the structures of b- and s-nexilins. ABD, F-actin–binding domain; CC, coiled-coil region.
We named p97 b-nexilin since p97 was the bigger of the two splicing variants described below. To determine the F-actin–binding domain of b-nexilin, we prepared the fusion proteins of several truncated forms of b-nexilin with GST and examined the binding of 125I-labeled F-actin to these fusion proteins. Under the conditions where full-length His6-b-nexilin showed 125I-labeled F-actin–binding activity, GST-b-nexilin-1 (aa 1–150) and GST-b-nexilin-5 (aa 284–450) showed activity, whereas the other truncated forms of b-nexilin did not show activity (Fig. 3 Ba). This result suggests that b-nexilin has two F-actin–binding domains at the NH2-terminal (aa 1–150) and middle (aa 284– 450) regions, and that b-nexilin interacts with F-actin through these domains (Fig. 3 C). This conclusion was supported by two other lines of evidence described below (see Figs. 4 Ea and 7). The two F-actin–binding domains showed no significant homology to any known F-actin– binding protein in current protein database. In addition to the two F-actin–binding domains, b-nexilin had a sequence stretch that had high probability of forming a predicted coiled-coil structure (Fig. 3 C). The coiled-coil probability of the sequence stretch at the middle region (aa 244–281) was calculated to be 0.9–1.0 (data not shown). The NH2- and COOH-terminal regions of b-nexilin did not show any tendency to form a coiled-coil structure.
Figure 4.
Biochemical properties of b- and s-nexilins. (A) Cosedimentation of His6-b- and His6-s-nexilins with F-actin. His6-b- (6.5 μg of protein) or His6-s-nexilin (6.0 μg of protein) was mixed with F-actin, followed by ultracentrifugation. S, supernatant; P, pellet. Arrows indicate actin. Arrowheads indicate His6-b- and His6-s-nexilins. (Aa) His6-b-nexilin and (Ab) His6-s-nexilin. (B) Binding of His6-b- and His6-s-nexilins to F-actin. Various amounts of His6-b- or His6-s-nexilin were mixed with F-actin, followed by ultracentrifugation. Amounts of the free and bound His6-b- or His6-s-nexilin were calculated by determining the protein amounts from the supernatant and pellet fractions with a densitometer. The values are means ± SEM of three independent experiments. (Filled circles) His6-b-nexilin and (open circles) His6-s-nexilin. (C) Inhibition by myosin S1 of the binding of His6-b- and His6-s-nexilins to 125I-labeled F-actin. His6-b- or His6-s-nexilin (50 ng of protein each) was subjected to SDS-PAGE (8% polyacrylamide gel), followed by the blot overlay with 125I-labeled F-actin pretreated with myosin S1 in the presence or absence of ATP. (Ca) His6-b-nexilin and (Cb) His6-s-nexilin. (D) F-actin cross-linking activity of His6-b- and His6-s-nexilins estimated by low shear viscometry. (Filled circles) His6-b-nexilin and (open circles) His6-s-nexilin. (E) Cosedimentation of various truncated forms of b- and s-nexilins having or lacking the F-actin–binding domains. The supernatant (5 μl each) of the lysates of E. coli expressing the GST fusion proteins was mixed with F-actin. After the sample was centrifuged, the supernatant and pellet were subjected to SDS-PAGE (10% polyacrylamide gel), followed by Western blot analysis using the anti-GST and antiactin antibodies. GST-b-Nexilin-1′, aa 1–240; GST-b-Nexilin-5, aa 284–450; GST-b-Nexilin-6, aa 493–656; and GST-s-Nexilin-1′, aa 1–190. S, supernatant; P, pellet. Arrowheads indicate actin. (Ea) b-Nexilin and (Eb) s-nexilin.
Biochemical Properties of b-Nexilin
In addition to 125I-labeled F-actin blot overlay, the binding of b-nexilin to F-actin was examined by cosedimentation of b-nexilin with F-actin. When full-length His6-b-nexilin was incubated with F-actin, followed by ultracentrifugation, His6-b-nexilin was recovered with F-actin in the pellet (Fig. 4 Aa). The stoichiometry of the binding of His6-b-nexilin to actin was calculated to be one His6-b-nexilin molecule per about nine actin molecules (Fig. 4 B). The K d value was calculated to be 8.0 ± 0.1 × 10−7 M. It was examined by competition experiments using myosin S1, a well-characterized protein that binds along the sides of F-actin (Rayment et al., 1993; Schröder et al., 1993), whether b-nexilin bound along the sides of F-actin or at the ends. The binding of His6-b-nexilin to 125I-labeled F-actin was inhibited by an excessive amount of myosin S1 (Fig. 4 Ca). This inhibition was reversed by the addition of MgATP because MgATP dissociates the actin–myosin complex (Fraser et al., 1975). b-Nexilin increased the viscosity in a dose-dependent manner, and at 1.0 μM, the viscosity became unmeasurably high as estimated by the falling ball method for low shear viscometry (Fig. 4 D). Preliminary results from electron microscopy confirmed the cross-linking activity of b-nexilin as demonstrated by low shear viscometry (data not shown). All of these results suggest that b-nexilin binds along the sides of F-actin and shows cross-linking activity, presumably because of the two F-actin– binding domains. We confirmed the F-actin–binding domains of b-nexilin shown in Fig. 3 C by the cosedimentation assay using the GST fusion proteins. The purified samples of the GST fusion proteins having the F-actin– binding domains were insoluble under several conditions where full-length b- or s-nexilin was not (data not shown). Therefore, we used the crude samples of E. coli (the supernatant of the cell lysates) expressing the GST fusion proteins. As to GST-b-nexilin-1 (aa 1–150) in Fig. 3 Ba, because it was recovered in the inclusion bodies during the preparation of the supernatant of the cell lysates, we used another GST fusion protein, GST-b-nexilin-1′ (aa 1–240) for the assay. GST-b-nexilin-1′ and GST-b-nexilin-5 (aa 284–450) were cosedimented with F-actin, whereas GST-b-nexilin-6 (aa 493–656) was not (Fig. 4 Ea). GST and GST-b-nexilin-3 (aa 151–283) were not cosedimented with F-actin (data not shown). These reuslts are consistent with those of Fig. 3 Ba and support the conclusion shown in Fig. 3 C that b-nexilin has two F-actin–binding domains at the NH2-terminal and middle regions, and that b-nexilin interacts with F-actin through these domains.
The molecular mass value of b-nexilin was estimated by sucrose density gradient ultracentrifugation. On sucrose density gradient ultracentrifugation, His6-b-nexilin appeared at a position corresponding to a molecular mass of about 66 kD (S value, ∼4.4) (data not shown). Because the molecular mass value of b-nexilin estimated by SDS-PAGE is ∼97 kD, this result suggests that b-nexilin exists as a monomer.
Tissue Distribution of b-Nexilin
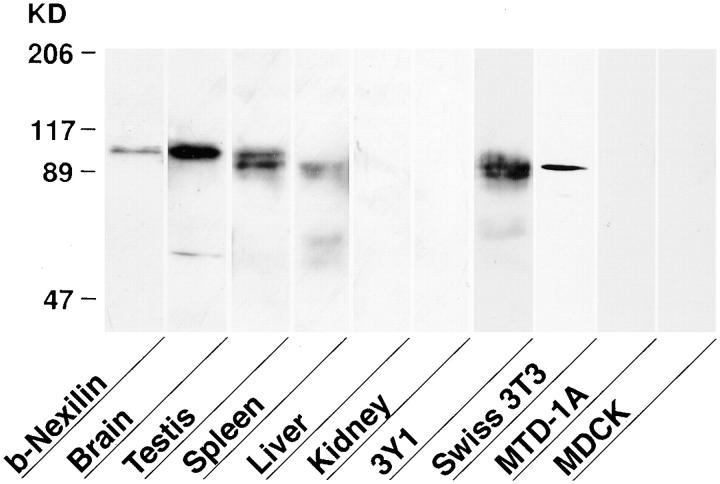
Western blot analysis showed that the antinexilin antibody recognized a protein band with a molecular mass of ∼97 kD in brain and testis and a protein band with a slightly smaller size (∼95 kD) in testis, spleen, and fibroblasts such as rat 3Y1 and mouse Swiss 3T3 cells (Fig. 5). Neither a big nor small protein was detected in liver, kidney, or epithelial cells, such as MDCK and MTD-1A cells. Because liver and kidney contain abundant epithelial cells, these results suggest that nexilin is expressed in nonepithelial cells but not in epithelial cells.
Figure 5.
Tissue distribution of b-nexilin. For preparation of cell lysates, cells were harvested with a cell scraper and suspended in PBS. After sonication, samples were treated with SDS–sample buffer. The homogenates of various rat tissues (25 μg of protein each) and whole-cell lysates of various cell lines (50 μg of protein each) were subjected to SDS-PAGE (8% polyacrylamide gel), followed by Western blot analysis using the antinexilin antibody.
Molecular Cloning and Physical Properties of s-Nexilin
The results described above led us to speculate about the existence of a smaller isoform or a smaller splicing variant of b-nexilin. Therefore, we attempted to isolate it and cloned a small splicing variant from 3Y1 cells by use of the reverse transcriptase-PCR method. The encoded protein had 606 aa and showed a calculated molecular weight of 71,942. The protein showed the aa sequence identical to that of b-nexilin except that it had 64 aa deletion and 14 aa insertion in the first F-actin–binding domain of b-nexilin (Fig. 2 B). We therefore named this variant s-nexilin (a small variant of nexilin). To confirm whether this clone encoded a full-length cDNA, we constructed the eukaryotic expression vector with this cDNA and expressed the protein in COS7 cells. Western blot analysis indicated that the expressed protein with a molecular mass of ∼95 kD showed two bands with mobilities similar to those of the two bands endogenously detected in 3Y1 cells (Fig. 3 Ab). Although the reason for the slight difference between the molecular mass value calculated from the predicted aa sequence and that estimated by SDS-PAGE was not known, we concluded that this clone encoded the full-length cDNA of s-nexilin. The reason for the presence of the two bands on SDS-PAGE is not known, but the two bands may be caused by posttranslational modifications, such as phosphorylation.
To determine whether the NH2-terminal region of s-nexilin still has its F-actin–binding activity, we prepared a fusion protein of the truncated form of s-nexilin with GST and examined the binding of 125I-labeled F-actin to this protein. Under the conditions where full-length His6-s-nexilin showed 125I-labeled F-actin–binding activity, GST-s-nexilin-1 (aa 1–100) did not show activity (Fig. 3 Bb). This result suggests that s-nexilin has only one F-actin–binding domain at the middle region (aa 234–400), and that s-nexilin interacts with F-actin through this domain (Fig. 3 C). This conclusion was supported by two other lines of evidence described below (see Figs. 4 Eb and 7).
The binding of s-nexilin to F-actin was examined by cosedimentation of s-nexilin with F-actin. When full-length His6-s-nexilin was incubated with F-actin, followed by ultracentrifugation, His6-s-nexilin was recovered with F-actin in the pellet (Fig. 4 Ab). The stoichiometry of the binding of His6-s-nexilin to actin was calculated to be one His6-s-nexilin molecule per about ten actin molecules (Fig. 4 B). The K d value was calculated to be 9.0 ± 0.2 × 10−7 M. s-Nexilin also bound along the sides of F-actin (Fig. 4 Cb). However, s-nexilin did not increase the viscosity estimated by the falling ball method (Fig. 4 D). Consistently, preliminary results from electron microscopy indicated that s-nexilin had no cross-linking activity (data not shown).
We confirmed the F-actin–binding domain of s-nexilin shown in Fig. 3 C. GST-s-nexilin-1′ (aa 1–190) was not cosedimented with F-actin (Fig. 4 Eb). This result is consistent with that of Fig. 3 Bb and supports the conclusion shown in Fig. 3 C that s-nexilin has only one F-actin–binding domain at the middle region, and that s-nexilin interacts with F-actin through this domain. Sucrose density gradient ultracentrifugation analysis revealed that His6-s-nexilin appeared at a position corresponding to a molecular mass of ∼66 kD (S value, ∼4.4) (data not shown), suggesting that s-nexilin exists as a monomer. All of these results suggest that s-nexilin binds along the sides of F-actin but lacks cross-linking activity, presumably because of one F-actin– binding domain.
Localization of s-Nexilin in 3Y1 Cells
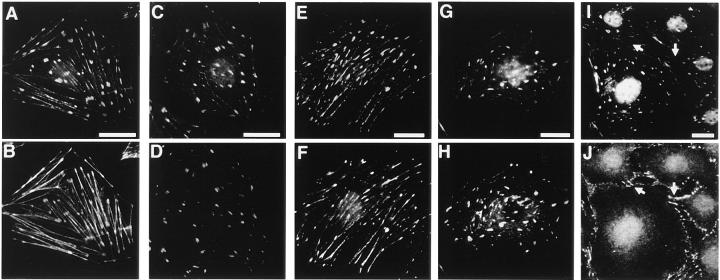
Since s-nexilin was expressed in 3Y1 cells, we examined the localization of s-nexilin in 3Y1 cells. An immunofluorescence microscopic study revealed that s-nexilin was localized at the ends of stress fibers, which are known to be focal contacts (Fig. 6, A and B). To confirm localization of s-nexilin at focal contacts, we compared the localization of s-nexilin with those of vinculin, talin, and paxillin, which are typical focal contact proteins and thereby known to be good markers for cell–matrix AJ (Geiger, 1979; Burridge and Connell, 1983; Turner et al., 1990). s-Nexilin was colocalized with vinculin, talin, and paxillin (Fig. 6, C–H). However, s-nexilin was not colocalized with P-cadherin, which is known to be a marker for cell–cell AJ (Itoh et al., 1991), in 3Y1 cells that were densely plated (Fig. 6, I and J). These results indicate that s-nexilin is specifically localized at focal contacts but not at cell–cell AJ.
Figure 6.
Colocalization of s-nexilin with vinculin, talin, and paxillin in 3Y1 cells. 3Y1 cells were doubly stained with the antinexilin antibody and phalloidin for F-actin, the antinexilin and antivinculin antibodies, the antinexilin and antitalin antibodies, the antinexilin and antipaxillin antibodies, or the antinexilin and anti–P-cadherin antibodies. Arrows indicate P-cadherin–positive cell–cell AJ. (A, C, E, G, and I) s-Nexilin, (B) actin, (D) vinculin, (F) paxillin, (H) talin, and (J) P-cadherin. Bars, 50 μm.
Localization of Overexpressed Full-Length and Truncated Mutants of b- and s-Nexilins in 3Y1 Cells
We examined whether exogenously expressed b- or s-nexilin was localized at cell–matrix AJ in 3Y1 cells. s-Nexilin was localized at focal contacts (Fig. 7, A, a and B, a). b-Nexilin was also localized at focal contacts (data not shown). Similar results were obtained in other cells, such as mouse 3T3, MTD-1A, and MDCK cells (data not shown). Neither b- nor s-nexilin was localized at cell–cell junctions in 3Y1 cells (data not shown). Even in the case of epithelial cells with prominent cell–cell junctions, such as MTD-1A and MDCK cells, overexpressed b- and s-nexilins were consistently localized at focal contacts but not at cell–cell junctions (data not shown). These results indicate that both b- and s-nexilins have potential to be localized at focal contacts.
Figure 7.
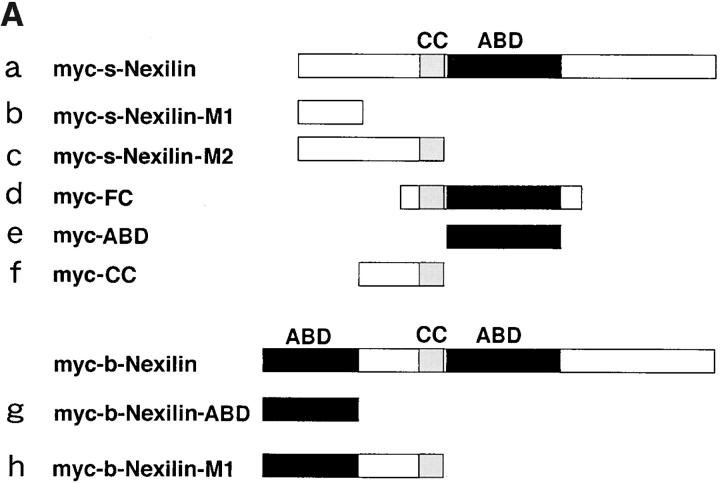
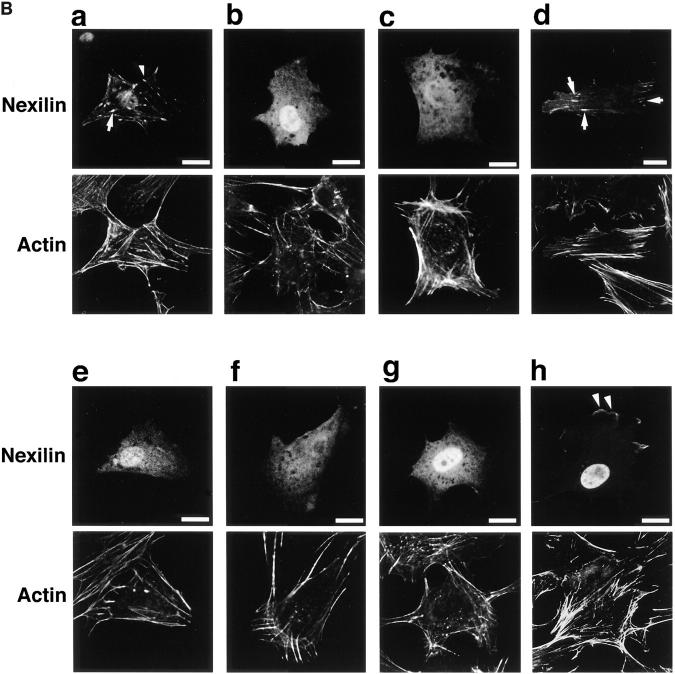
Localization of overexpressed full-length and various truncated mutants of b- and s-nexilins in 3Y1 cells. (A) Constructions of truncated mutants of b- and s-nexilins. Myc-s-Nexilin-M1, aa 1–100; myc-s-Nexilin-M2, aa 1–233; myc-FC, aa 161–430 for s-nexilin, aa 211–480 for b-nexilin; myc-ABD, aa 234– 400 for s-nexilin, aa 284–450 for b-nexilin; myc-CC, aa 101–233 for s-nexilin, aa 151– 283 for b-nexilin; myc-b-Nexilin-ABD, aa 1–150; and myc-b-Nexilin-M1, aa 1–283. a–h for each mutant correspond to a–h in B. (B) Localization of full-length and various truncated mutants of b- and s-nexilins. 3Y1 cells transiently expressing myc-tagged truncated mutants of b- or s-nexilin were doubly stained with the anti-myc antibody and phalloidin for F-actin. Arrows indicate focal contacts. Arrowheads indicate the region at the cell periphery where myc-b-nexilin-M1 is localized. Bars, 50 μm.
To clarify focal contact-targeting and F-actin–binding domains in intact cells, we examined which overexpressed truncated mutant of nexilin was localized at focal contacts or colocalized with endogenous F-actin in 3Y1 cells. Myc-ABD and myc-b-nexilin-ABD, having only the second and first F-actin–binding domains, respectively, were diffusely distributed in the cytoplasm and were not colocalized with F-actin (Fig. 7, A, e and g, and B, e and g). Myc-b-nexilin-M1, having the first F-actin–binding domain plus the following predicted coiled-coil region, was colocalized with F-actin at the cell periphery (Fig. 7, A, h and B, h). Myc-FC, having the predicted coiled-coil region and the second F-actin–binding domain, was colocalized with F-actin at focal contacts (Fig. 7, A, d and B, d). Myc-CC, having only the predicted coiled-coil region that was overlapped in myc-b-nexilin-M1 and myc-FC, was diffusely distributed in the cytoplasm (Fig. 7, A, f and B, f). Moreover, myc-s-nexilin-M1, which lacked the first F-actin–binding domain, and myc-s-nexilin-M2, which lacked the first F-actin–binding domain but had the predicted coiled-coil region, were also diffusely distributed in the cytoplasm (Fig. 7, A, b and c, and B, b and c). These reuslts are consistent with those of Fig. 3, Ba and Bb, and support the conclusion shown in Fig. 3 C that b-nexilin has two F-actin–binding domains at the NH2-terminal and middle regions, and that s-nexilin has only one F-actin–binding domain at the middle region. At present, we do not know why the myc-tagged mutants, having only the F-actin–binding domains, are not colocalized with F-actin in intact cells, but the mutants, having the F-actin–binding domains plus the predicted coiled-coil region, are colocalized with F-actin. Moreover, these results suggest that a focal contact-targeting domain is located at the region containing both the predicted coiled-coil region and the second F-actin–binding domain.
Discussion
We isolated and characterized here two novel F-actin– binding proteins, which were likely splicing variants derived from the same gene. We named the big and small ones b- and s-nexilins, respectively. We have concluded that both the proteins are real F-actin–binding proteins on the basis of the following observations: (a) full-length b- and s-nexilins showed 125I-labeled F-actin–binding activity; (b) both full-length b- and s-nexilins were cosedimented with F-actin; (c) full-length b-nexilin showed F-actin cross-linking activity; and (d) when full-length b- or s-nexilin was expressed in COS7 cells, both the proteins were colocalized with F-actin (data not shown).
Structural analysis of b- and s-nexilins showed that the nucleotide sequence of the s-nexilin cDNA was identical to that of the b-nexilin cDNA except for the two splicing regions that were located in the first F-actin–binding domain of b-nexilin. Consequently, the NH2-terminal region of s-nexilin lost F-actin–binding activity. Thus, the big variant of nexilin has two F-actin–binding domains, whereas the small one has one F-actin–binding domain (Fig. 3 C). Among many F-actin–binding proteins so far identified, some variants lose the F-actin–binding domain. For instance, BPAG1n, the neuronal splicing form, contains the functional F-actin–binding domain at the NH2-terminal region but BPAG1e, which is mainly expressed in epidermis, lacks the domain (Yang et al., 1996). l-Afadin also has the F-actin–binding domain at the COOH-terminal region, but s-afadin lacks this domain (Mandai et al., 1997).
Among many F-actin–binding proteins having F-actin cross-linking activity, the members of the α-actinin/spectrin superfamily have most extensively been studied (Hartwig and Kwiatkowski, 1991; Matsudaira, 1991). Most of them usually form oligomers by association at rod domains and hence show F-actin cross-linking activity, but the members of the fimbrin (plastin) subfamily show F-actin cross-linking activity without oligomerization since they have two F-actin–binding domains in a single subunit (Bretscher and Weber, 1980; De Arruda et al., 1990). We have shown here that b-nexilin, but not s-nexilin, has F-actin cross-linking activity, although neither b- nor s-nexilin forms a dimer or an oligomer as estimated by sucrose density gradient ultracentrifugation. Therefore, the structural property of b-nexilin is apparently similar to that of the fimbrin subfamily, although there is no significant homology between the primary structures of the F-actin–binding domains of b- and s-nexilins and those of the α-actinin/spectrin superfamily.
In addition to the F-actin–binding domains, both b- and s-nexilins have predicted coiled-coil structures that have been identified in a variety of cytoskeleton proteins (Lupas et al., 1991; Lupas, 1996). They typically form rodlike, homodimeric, or heterodimeric structures. Moreover, we have identified a focal contact-targeting domain that contained the predicted coiled-coil region and the F-actin– binding domain at the middle region. Although it is unknown why these two regions are necessary for the targeting of b- and s-nexilins to focal contacts, it could be speculated that there is a focal contact-specific protein(s) that interacts with the predicted coiled-coil region of nexilin and that the interactions of nexilin with both this protein and F-actin are essential for its correct localization.
Furthermore, b- and s-nexilins show different tissue expression patterns. b-Nexilin is mainly expressed in brain and testis, whereas s-nexilin is expressed in testis, spleen, and fibroblasts. It may be noted that neither b- nor s-nexilin is expressed in epithelial cells such as MTD-1A and MDCK cells. Consistently, neither b- nor s-nexilin is expressed in rat tissues such as liver and kidney, where there are abundant epithelial cells. At present, it is not known why nexilin is not expressed in epithelial cells, but there may be another isoform of nexilin that is specifically expressed in epithelial cells.
The subcellular distribution analysis of s-nexilin in 3Y1 cells indicates that it is localized at cell–matrix AJ, but not at cell–cell AJ. Consistently, overexpressed s-nexilin was localized at the same area. Overexpressed b-nexilin was also localized at cell–matrix AJ, suggesting that it is localized there in brain and testis. Many F-actin–binding proteins localized at cell–matrix AJ have been isolated and characterized (Jockusch et al., 1995; Burridge and Chrzanowska-Wodnicka, 1996). However, only talin has been shown to be specifically localized at cell–matrix AJ but not at cell–cell AJ (Geiger et al., 1985; Tidball et al., 1986; Drenckhahn et al., 1988). Talin is expressed not only in nonepithelial cells but also in epithelial cells (Philp et al., 1990). Nexilin is the second F-actin–binding protein that is specifically localized at cell–matrix AJ, but this protein is expressed only in nonepithelial cells. Like other F-actin– binding proteins, nexilin may also be involved in the linkage between the actin cytoskeleton and cell–matrix AJ in nonepithelial cells, but its precise role in this linkage remains unknown.
It has been suggested that cell–matrix and cell–cell AJs represent signaling hot spots on the cell surface. For instance, the initial interaction between transmembrane proteins, such as integrins, and extracellular ligands has been shown to turn on a cascade of several signal transduction (Keely et al., 1998). However, little is known about the mechanism by which signals at the cell surface are relayed to the nucleus and vice versa. Recently, several proteins, including c-abl (Lewis et al., 1996), ZO-1 (Gottardi et al., 1996), zyxin (Nix and Beckerle, 1997), and β-catenin (Behrens et al., 1996; Molennar et al., 1996), have been demonstrated to reside both at AJ and in the nucleus. Interestingly, the PSORT protein localization prediction program (Nakai and Kanehisa, 1992) indicates that b- and s-nexilins also partition in the nucleus since these two proteins have the consensus sequences for nucleoplasmin-like nuclear localization signals (Robbins et al., 1991) (data not shown). In fact, we observed weak nuclear stainings in fibroblasts as shown in Fig. 6. However, we have not yet succeeded in demonstrating that transfected full-length b- or s-nexilin is translocated into the nucleus (data not shown). The significance of these nuclear localization signals in a variety of proteins that are localized at cell–matrix and/or cell–cell AJs, including nexilin, is currently unknown.
Acknowledgments
We thank Dr. W. Birchmeier for providing MDCK cells, Dr. Sh. Tsukita for providing rat 3Y1 and MTD-1A cells, Dr. D.W. Russell (University of Texas Southwestern Medical Center, Dallas, TX) for providing the pCMV5 vector, and Dr. Y. Hata (Takai Biotimer Project, ERATO, Kobe, Japan) for providing the pCIneo-myc vector. We also thank Drs. Sh. Tsukita and M. Itoh (Kyoto University, Kyoto, Japan) and Drs. Y. Yoneda and T. Tachibana (Osaka University, Osaka, Japan) for helpful discussions. We are grateful to M. Tokunaga, M. Kinoshita, T. Inoue, Y. Hayashi, and H. Nohara for their technical assistance.
Abbreviations used in this paper
- aa
amino acid(s)
- ABD
F-actin–binding domain(s)
- AJ
adherens junction(s)
- F-actin
actin filament
- G-actin
actin monomer
- GST
glutathione S-transferase
- His6
six histidine residues
- S1
subfragment 1
Footnotes
The work performed at Osaka University Medical School was supported by grants-in-aid for Scientific Research and for Cancer Research from the Ministry of Education, Science, Sports, and Culture, Japan (1997), by grants-in-aid for Abnormalities in Hormone Receptor Mechanisms and for Aging and Health from the Ministry of Health and Welfare, Japan (1997), and by grants from the Human Frontier Science Program (1997).
Address all correspondence to Yoshimi Takai, Department of Molecular Biology and Biochemistry, Osaka University Medical School, Suita 565-0871, Osaka, Japan. Tel.: 81-6-879-3410. Fax: 81-6-879-3419. E-mail: ytakai@molbio.med.osaka-u.ac.jp
References
- Abercrombie M, Dunn GA. Adhesions of fibroblasts to substratum during contact inhibition observed by interference reflection microscopy. Exp Cell Res. 1975;92:57–62. doi: 10.1016/0014-4827(75)90636-9. [DOI] [PubMed] [Google Scholar]
- Abercrombie M, Heaysman JE, Pegram SM. The locomotion of fibroblasts in culture. Exp Cell Res. 1971;67:359–367. doi: 10.1016/0014-4827(71)90420-4. [DOI] [PubMed] [Google Scholar]
- Allen PB, Quimet CC, Greengard P. Spinophilin: a novel protein phosphatase 1 binding protein localized to dendritic spines. Proc Natl Acad Sci USA. 1997;94:9956–9961. doi: 10.1073/pnas.94.18.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckerle MC. Identification of a new protein localized at sites of cell-substrate adhesion. J Cell Biol. 1986;103:1679–1687. doi: 10.1083/jcb.103.5.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Bretscher A, Weber K. Fimbrin, a new microfilament-associated protein present in microvilli and other cell surface structures. J Cell Biol. 1980;86:335–340. doi: 10.1083/jcb.86.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. Microfilaments and membranes. Curr Opin Cell Biol. 1993;5:653–660. doi: 10.1016/0955-0674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- Burridge K, Connell L. A new protein of adhesion plaques and ruffling membranes. J Cell Biol. 1983;97:359–367. doi: 10.1083/jcb.97.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K, Chrzanowska-Wodnicka M. Focal adhesions, contractility, and signaling. Annu Rev Cell Dev Biol. 1996;12:463–518. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]
- Carlsson L, Nystrom LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115:465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- Chia CP, Hitt AL, Luna EJ. Direct binding of F-actin to ponticulin, an integral plasma membrane glycoprotein. Cell Motil Cytoskel. 1991;18:164–179. doi: 10.1002/cm.970180303. [DOI] [PubMed] [Google Scholar]
- Chuang J-Z, Lin DC, Lin S. Molecular cloning, expression, and mapping of the high affinity actin-capping domain of chicken cardiac tensin. J Cell Biol. 1995;128:1095–1109. doi: 10.1083/jcb.128.6.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford AW, Beckerle MC. Purification and characterization of zyxin, an 82,000-dalton component of adherens junctions. J Biol Chem. 1991;266:5847–5853. [PubMed] [Google Scholar]
- De Arruda MV, Watson S, Lin C-S, Leavitt J, Matsudaira P. Fimbrin is a homologue of the cytoplasmic phosphoprotein plasmin and has domains homologous with calmodulin and actin gelatin protein. J Cell Biol. 1990;111:1064–1079. doi: 10.1083/jcb.111.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenckhahn D, Beckerle MC, Burridge K, Otto J. Identification and subcellular location of talin in various cell types and tissues by means of (125I)vinculin overlay, immunoblotting, and immunocytochemistry. Eur J Cell Biol. 1988;46:513–522. [PubMed] [Google Scholar]
- Elisei R, Weightman D, Kendall-Taylor P, Vassart G, Ludgate M. Muscle autoantigens in thyroid-associated ophthalmopathy: the limits of molecular genetics. J Endocrinol Invest. 1993;16:533–540. doi: 10.1007/BF03348900. [DOI] [PubMed] [Google Scholar]
- Endo T, Masaki T. Molecular properties and functions in vitro of chicken smooth-muscle α-actinin in comparison with those of striated-muscle α-actinins. J Biochem (Tokyo) 1982;92:1457–1468. doi: 10.1093/oxfordjournals.jbchem.a134070. [DOI] [PubMed] [Google Scholar]
- Fannon AM, Colman DR. A model for central synaptic junctional complex formation based on the differential adhesive specificities of the cadherins. Neuron. 1996;17:423–434. doi: 10.1016/s0896-6273(00)80175-0. [DOI] [PubMed] [Google Scholar]
- Fraser AB, Eisenberg E, Kielley WW, Carlson FD. The interaction of heavy meromyosin and subfragment 1 with actin. Physical measurements in the presence and absence of adenosine triphosphate. Biochemistry. 1975;14:2207–2214. doi: 10.1021/bi00681a025. [DOI] [PubMed] [Google Scholar]
- Geiger B. A 130K protein from chicken gizzard: its localization at the termini of microfilament bundles in cultured chicken cells. Cell. 1979;18:193–205. doi: 10.1016/0092-8674(79)90368-4. [DOI] [PubMed] [Google Scholar]
- Geiger B. Membrane-cytoskeleton interaction. Biochim Biophys Acta. 1983;737:305–341. doi: 10.1016/0304-4157(83)90005-9. [DOI] [PubMed] [Google Scholar]
- Geiger B. Cytoskeleton-associated cell contacts. Curr Opin Cell Biol. 1989;1:103–109. doi: 10.1016/s0955-0674(89)80045-6. [DOI] [PubMed] [Google Scholar]
- Geiger B, Ginsberg D. The cytoplasmic domain of adherens-type junctions. Cell Motil Cytoskel. 1991;20:1–6. doi: 10.1002/cm.970200102. [DOI] [PubMed] [Google Scholar]
- Geiger B, Volk T, Volberg T. Molecular heterogeneity of adherens junctions. J Cell Biol. 1985;101:1523–1531. doi: 10.1083/jcb.101.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardi CJ, Arpin M, Fanning AS, Louvard D. The junction-associated protein, zonula occludens-1, localizes to the nucleus before the maturation and during the remodeling of cell-cell contacts. Proc Natl Acad Sci USA. 1996;93:10779–10784. doi: 10.1073/pnas.93.20.10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig JH, Kwiatkowski DJ. Actin-binding proteins. Curr Opin Cell Biol. 1991;3:87–97. doi: 10.1016/0955-0674(91)90170-4. [DOI] [PubMed] [Google Scholar]
- Hata Y, Südhof TC. A novel ubiquitous form of Munc-18 interacts with multiple syntaxins. Use of the yeast two-hybrid system to study interactions between proteins involved in membrane traffic. J Biol Chem. 1995;270:13022–13028. doi: 10.1074/jbc.270.22.13022. [DOI] [PubMed] [Google Scholar]
- Heath JP, Dunn GA. Cell to substratum contacts of chick fibroblasts and their relation to the microfilament system: a correlated interference-reflexion and high-voltage electron-microscope study. J Cell Sci. 1978;29:197–212. doi: 10.1242/jcs.29.1.197. [DOI] [PubMed] [Google Scholar]
- Imazumi K, Sasaki T, Takahashi K, Takai Y. Identification of a rabphilin-3A-interacting protein as GTP cyclohydrolase I in PC12 cells. Biochem Biophys Res Commun. 1994;205:1409–1416. doi: 10.1006/bbrc.1994.2822. [DOI] [PubMed] [Google Scholar]
- Itoh M, Yonehara S, Nagafuchi A, Tsukita Sa, Tsukita Sh. A 220-kD undercoat-constitutive protein: its specific localization at cadherin-based cell–cell adhesion sites. J Cell Biol. 1991;115:1449–1462. doi: 10.1083/jcb.115.5.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Nagafuchi A, Moroi S, Tsukita Sh. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to α-catenin and actin filaments. J Cell Biol. 1997;138:181–192. doi: 10.1083/jcb.138.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzard CS, Lochner LR. Cell-to-substrate contacts in living fibroblasts: an interference reflexion study with an evaluation of the technique. J Cell Sci. 1976;21:129–159. doi: 10.1242/jcs.21.1.129. [DOI] [PubMed] [Google Scholar]
- Jockusch BM, Bubeck P, Giehl K, Kroemker M, Moschner J, Rothkegel M, Rüdiger M, Schlüter K, Stanke G, Winkler J. The molecular architecture of focal adhesions. Annu Rev Cell Dev Biol. 1995;11:379–416. doi: 10.1146/annurev.cb.11.110195.002115. [DOI] [PubMed] [Google Scholar]
- Kato M, Sasaki T, Ohya T, Nakanishi H, Nishioka H, Imamura M, Takai Y. Physical and functional interaction of rabphilin-3A with α-actinin. J Biol Chem. 1996;271:31775–31778. doi: 10.1074/jbc.271.50.31775. [DOI] [PubMed] [Google Scholar]
- Keely P, Parise L, Juliano R. Integrins and GTPases in tumour cell growth, motility, and invasion. Trends Cell Biol. 1998;8:101–107. doi: 10.1016/s0962-8924(97)01219-1. [DOI] [PubMed] [Google Scholar]
- Knudsen KA, Soler AP, Johnson KR, Wheelock MJ. Interaction of α-actinin with the cadherin/catenin cell–cell adhesion complex via α-catenin. J Cell Biol. 1995;130:67–77. doi: 10.1083/jcb.130.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani H, Takaishi K, Sasaki T, Takai Y. Rho regulates association of both the ERM family and vinculin with the plasma membranes in MDCK cells. Oncogene. 1997;14:1705–1713. doi: 10.1038/sj.onc.1200998. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lewis JM, Baskaran R, Taagepera S, Schwartz MA, Wang JY. Integrin regulation of c-Abltyrosine kinase activity and cytoplasmic-nuclear transport. Proc Natl Acad Sci USA. 1996;93:15174–15179. doi: 10.1073/pnas.93.26.15174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SH, An Q, Bao S, Wong W-K, Liu Y, Janmey PA, Hartwig JH, Chen LB. Molecular cloning of chick cardiac muscle tensin. J Biol Chem. 1994;269:22310–22319. [PubMed] [Google Scholar]
- Luna EJ, Hitt AL. Cytoskeleton-plasma membrane interactions. Science. 1992;258:955–964. doi: 10.1126/science.1439807. [DOI] [PubMed] [Google Scholar]
- Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mandai K, Nakanishi H, Satoh A, Obaishi H, Wada M, Nishioka H, Itoh M, Mizoguchi A, Aoki T, Fujimoto T, et al. Afadin: a novel actin filament-binding protein with one PDZ domain localized at cadherin-based cell-to-cell adherens junction. J Cell Biol. 1997;139:517–528. doi: 10.1083/jcb.139.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P. Modular organization of actin crosslinking proteins. Trends Biochem Sci. 1991;16:87–92. doi: 10.1016/0968-0004(91)90039-x. [DOI] [PubMed] [Google Scholar]
- Mizoguchi A, Kim S, Ueda T, Kikuchi A, Yorifuji H, Hirooka N, Takai Y. Localization and subcellular distribution of smg p25A, a ras p21-like GTP-binding protein, in rat brain. J Biol Chem. 1990;91:257–269. [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Nagafuchi A, Takeichi M, Tsukita Sh. The 102 kd cadherin-associated protein: similarity to vinculin and posttranscriptional regulation of expression. Cell. 1991;65:849–857. doi: 10.1016/0092-8674(91)90392-c. [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Obaishi H, Satoh A, Wada M, Mandai K, Satoh K, Nishioka H, Matsuura Y, Mizoguchi A, Takai Y. Neurabin: a novel neural tissue–specific actin filament–binding protein involved in neurite formation. J Cell Biol. 1997;139:951–961. doi: 10.1083/jcb.139.4.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix DA, Beckerle MC. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obaishi H, Nakanishi H, Mandai K, Satoh K, Satoh A, Takahashi K, Miyahara M, Nishioka H, Takaishi K, Takai Y. Frabin, a novel FGD1-related actin filament-binding protein capable of changing cell shape and activating c-Jun N-terminal kinase. J Biol Chem. 1998;273:18697–18700. doi: 10.1074/jbc.273.30.18697. [DOI] [PubMed] [Google Scholar]
- Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO (Eur Mol Biol Organ) J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- Pasteris NG, Cadle A, Losie LJ, Porteous MEM, Shwarts CE, Stevenson RE, Glover TW, Wilroy RS, Gorski JL. Isolation and characterization of the faciogenital dysplasia (Aarskog-Scott syndrome) gene: a putative Rho/Rac guanine nucleotide exchange factor. Cell. 1994;79:669–678. doi: 10.1016/0092-8674(94)90552-5. [DOI] [PubMed] [Google Scholar]
- Pestonjamasp K, Amieva MR, Strassel CP, Nauseef WM, Furthmayr H, Luna EJ. Moesin, ezrin, and p205 are actin-binding proteins associated with neutrophil plasma membranes. Mol Biol Cell. 1995;6:247–259. doi: 10.1091/mbc.6.3.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philp NJ, Yoon MY, Hock RS. Identification and localization of talin in chick retinal pigment epithelial cells. Exp Eye Res. 1990;51:191–198. doi: 10.1016/0014-4835(90)90072-3. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Methods to characterize actin filament networks. Methods Enzymol. 1982;85:211–233. doi: 10.1016/0076-6879(82)85022-2. [DOI] [PubMed] [Google Scholar]
- Rayment I, Holden HM, Whittaker M, Yohn CB, Lorenz M, Holmes KC, Milligan RA. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. α1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci USA. 1995;92:8813–8817. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. 2nd edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Satoh A, Nakanishi H, Obaishi H, Wada M, Takahashi K, Satoh K, Hirao K, Nishioka H, Hata Y, Mizoguchi A, Takai Y. Neurabin-II/ spinophilin: an actin filament-binding protein with one PDZ domain localized at cadherin-based cell-cell adhesion sites. J Biol Chem. 1998;273:3470–3475. doi: 10.1074/jbc.273.6.3470. [DOI] [PubMed] [Google Scholar]
- Schaller MD, Borgman CA, Cobbs BS, Vines RR, Reynolds AB, Parsons JT. pp125FAK: a structurally distinctive protein-tyrosine kinase associated with focal adhesions. Proc Natl Acad Sci USA. 1992;89:5192–5196. doi: 10.1073/pnas.89.11.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder RR, Manstein DJ, Jahn W, Holden H, Rayment I, Holmes KC, Spudich JA. Three-dimensional atomic model of F-actin decorated with Dictyosteliummyosin S1. Nature. 1993;364:171–174. doi: 10.1038/364171a0. [DOI] [PubMed] [Google Scholar]
- Schwartz MA, Schaller MD, Ginsberg MH. Integrins: emerging paradigms of signal transduction. Annu Rev Cell Dev Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- Takeichi M. The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development (Camb) 1988;102:639–655. doi: 10.1242/dev.102.4.639. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Hata Y, Hirao K, Toyoda A, Irie M, Takai Y. SAPAPs: a family of PSD-95/SAP90-associated proteins localized at postsynaptic density. J Biol Chem. 1997;272:11943–11951. doi: 10.1074/jbc.272.18.11943. [DOI] [PubMed] [Google Scholar]
- Tidball JG, O'Halloran T, Burridge K. Talin at myotendinous junctions. J Cell Biol. 1986;103:1465–1472. doi: 10.1083/jcb.103.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita Sa, Yonemura S, Tsukita Sh. ERM proteins: head-to-tail regulation of actin-plasma membrane interaction. Trends Biochem Sci. 1997;22:53–58. doi: 10.1016/s0968-0004(96)10071-2. [DOI] [PubMed] [Google Scholar]
- Tsukita Sh, Tsukita Sa, Nagafuchi A, Yonemura S. Molecular linkage between cadherins and actin filaments in cell-cell adherens junctions. Curr Opin Cell Biol. 1992;4:834–839. doi: 10.1016/0955-0674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- Turner CE, Burridge K. Transmembrane molecular assemblies in cell-extracellular matrix interactions. Curr Opin Cell Biol. 1991;3:849–853. doi: 10.1016/0955-0674(91)90059-8. [DOI] [PubMed] [Google Scholar]
- Turner CE, Glenney J, Jr, Burridge K. Paxillin: a new vinculin-binding protein present in focal adhesions. J Cell Biol. 1990;111:1059–1068. doi: 10.1083/jcb.111.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Honjo Y, Johnson KR, Wheelock MJ, Takeichi M. The catenin/cadherin adhesion system is localized in synaptic junctions bordering transmitter release zones. J Cell Biol. 1996;135:767–779. doi: 10.1083/jcb.135.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Yin HL, Krueger JG, Caliguiri LA, Tamm I. Unphosphorylated gelsolin is localized in regions of cell-substratum contact or attachment in Rous sarcoma virus-transformed rat cells. J Cell Biol. 1984;98:761–771. doi: 10.1083/jcb.98.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JA, Risinger MA, Lin S. Studies on proteins that copurify with smooth muscle vinculin: identification of immunologically related species in focal adhesions of nonmuscle and Z-lines of muscle cells. J Cell Biol. 1986;103:1483–1494. doi: 10.1083/jcb.103.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Dowling J, Yu QC, Kouklis P, Cleveland DW, Fuchs E. An essential cytoskeletal linker protein connecting actin microfilaments to intermediate filaments. Cell. 1996;86:655–665. doi: 10.1016/s0092-8674(00)80138-5. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Fisher DJ, Santos MF, Tigyi G, Pasteris NG, Gorski JL, Xu Y. The faciogenital dysplasia gene product FGD1 functions as a Cdc42Hs-specific guanine nucleotide exchange factor. J Biol Chem. 1996;271:33169–33172. doi: 10.1074/jbc.271.52.33169. [DOI] [PubMed] [Google Scholar]