Abstract
Gle2p is implicated in nuclear export of poly(A)+ RNA and nuclear pore complex (NPC) structure and distribution in Saccharomyces cerevisiae. Gle2p is anchored at the nuclear envelope (NE) via a short Gle2p-binding motif within Nup116p called GLEBS. The molecular mechanism by which Gle2p and the Gle2p–Nup116p interaction function in mRNA export is unknown. Here we show that RAE1, the mammalian homologue of Gle2p, binds to a GLEBS-like NUP98 motif at the NPC through multiple domains that include WD-repeats and a COOH-terminal non–WD-repeat extension. This interaction is direct, as evidenced by in vitro binding studies and chemical cross-linking. Microinjection experiments performed in Xenopus laevis oocytes demonstrate that RAE1 shuttles between the nucleus and the cytoplasm and is exported from the nucleus in a temperature-dependent and RanGTP-independent manner. Docking of RAE1 to the NE is highly dependent on new mRNA synthesis. Overexpression of the GLEBS-like motif also inhibits NE binding of RAE1 and induces nuclear accumulation of poly(A)+ RNA. Both effects are abrogated either by the introduction of point mutations in the GLEBS-like motif or by overexpression of RAE1, indicating a direct role for RAE1 and the NUP98–RAE1 interaction in mRNA export. Together, our data suggest that RAE1 is a shuttling transport factor that directly contributes to nuclear export of mRNAs through its ability to anchor to a specific NUP98 motif at the NPC.
Keywords: nuclear pore complex, mRNA export, RNA polymerase II, NUP98, RAE1
Nucleocytoplasmic transport is a signal-mediated process in which soluble carriers bind to a specific cargo in one compartment, guide it to and through the nuclear pore complex (NPC),1 release it in the other compartment and finally recycle to the original compartment (Nigg, 1997; Cole and Hammell, 1998; Ohno et al., 1998; Weis, 1998). This model is based primarily on our knowledge of the protein import machinery (Görlich, 1997). However, there are several lines of evidence to suggest that it holds true for protein and RNA export pathways as well. Kinetic competition studies in Xenopus oocytes have indicated that export of several classes of RNA, including 5S rRNA, U snRNAs, tRNAs, and mRNAs, seems to be mediated by distinct and class-specific transport factors (Jarmolowski et al., 1994; Izaurralde and Mattaj, 1995). The first direct mediator of RNA export identified is the HIV-1 protein REV, which specifically binds to a REV-responsive element in nonspliced viral mRNAs (Fischer et al., 1995). REV contains a leucine-rich–type nuclear export signal (NES) that is recognized by the nuclear export receptor CRM1, which mediates export of the viral RNA (Fornerod et al., 1997a; Fukuda et al., 1997; Stade et al., 1997). CRM1 also mediates export of U snRNAs, probably via direct binding to the U snRNP–associating protein CBC, or via an unknown NES-containing bridging factor (Izaurralde et al., 1995a). Recently, a second export receptor, hLos1p or exportin-t, was shown to bind tRNA molecules and promote their nuclear export (Arts et al., 1998; Kutay et al., 1998).
It is generally believed that mRNA molecules are exported from the nucleus as mRNA–protein (mRNP) particles (Nakielny and Dreyfuss, 1997). The protein composition of these complex structures is unclear, although a series of abundant nuclear RNA-binding proteins has been identified (Dreyfuss et al., 1993; Misteli and Spector, 1998). There is some evidence to suggest that important signals for export of mRNA are provided by its interacting proteins (Izaurralde et al., 1997a; Michael et al., 1995, 1997; Nakielny et al., 1997). However, it is unclear whether multiple proteins provide independent signals for export of mRNP, whether these signals would be additive, cooperative, or redundant, and whether they occur at different stages of the export pathway (Nigg, 1997). Several laboratories have screened for mutations in yeast that result in nuclear accumulation of poly(A)-containing RNAs (outlined by Doye and Hurt, 1997). Accordingly, a variety of NPC proteins and soluble factors (often referred to as mRNA transport factors) have been identified with possible roles in nuclear export of mRNA. The majority of these yeast mutants display additional defects mostly in NPC structure and distribution, but also in nuclear localization signal (NLS)–mediated protein import, RNA processing, or nucleolar organization (summarized in Doye and Hurt, 1997). Therefore, it is very difficult to be sure whether the observed nuclear accumulation of poly(A)+ RNA is the primary defect (Ohno et al., 1998; Weis, 1998). Some NPC mutants might be expected to induce an mRNA defect, because mRNPs are relatively large and their passage through the NPC channel may require a nucleoporin-dependent remodeling process (Mehlin et al., 1995). Mutations in three yeast proteins induce nuclear accumulation of poly(A)+ RNA in the absence of impaired nuclear protein import or gross structural NPC defects: Gle1p, an essential RNA export mediator that interacts with Rip1p (Murphy et al., 1996; Segref et al., 1997); Mex67p, a poly(A)+ RNA binding protein (Segref et al., 1997); and Dbp5p, a cytosolic RNA helicase (Snay-Hodge et al., 1998; Tseng et al., 1998). Human Gle1p (hGlep1) seems to be involved in mRNA export as well, although there is evidence to suggest that yeast and human Gle1p function differently (Watkins et al., 1998). The human homologue of Mex67p, TAP, has recently been shown to function in the nuclear export of retroviral RNAs with a constitutive transport element (Gruter et al., 1998).
Genetic studies in yeast have implicated the Schizosaccharomyces pombe Rae1p (Brown et al., 1995; Whalen et al., 1997) and its Saccharomyces cerevisiae homologue Gle2p (Murphy et al., 1996) in mRNA export. Gle2p is known to bind the nuclear envelope (NE) via Nup116p, which contains a short Gle2p-binding motif called GLEBS that is conserved in the mammalian NUP98 protein (Bailer et al., 1998). However, the precise role of Gle2p and the Gle2p– Nup116p interaction in mRNA export remains a mystery. Here we report the identification and characterization of the interaction between human RAE1 and NUP98, and outline a molecular mechanism by which RAE1 and the RAE1–NUP98 interaction can function in nuclear export of mRNA.
Materials and Methods
Expression Constructs of NUP98 and Mouse RAE1 Fragments
Human NUP98 cDNA was obtained as described (Kasper et al., 1999). A full-length mouse RAE1 cDNA was generated from two partial cDNA clones, 473342 and 465642, obtained from the I.M.A.G.E. consortium. HA1-NUP98, mouse HA1-RAE1, and mutants thereof were generated by PCR using plaque-forming unit DNA polymerase (Stratagene Inc.), and cloned in pUHD10S for expression in HtTA cells (Kasper et al., 1999). All constructs were verified by DNA sequence analysis. HA1-RAE1 cDNA was cloned into vector pSP73 (Promega Corp.) for in vitro translation purposes. [35S]-methionine–labeled HA1-RAE1 protein was produced using the TNT-coupled rabbit reticulocyte lysate system (Promega Corp.) as indicated by the manufacturer. To isolate pure populations of transiently transfected HtTA and baby hamster kidney (BHK)grβ cells, HA1-NUP98(150–224) was cloned into the EcoRI–XhoI sites of vector MSCV-IRES-GFP (Persons et al., 1997).
Cell Culture, Transfections, and Electron Microscopy
HtTA-1 and BHKgrβ cells (Bastos et al., 1996) were grown in DMEM containing 10% FBS. Cells were transfected with Superfect transfection reagent (QIAGEN Inc.) according to the manufacturer's instructions. Metabolic labeling of HtTA cells was as described (Kasper et al., 1999). For indirect immunofluorescence studies, HtTA cells were transiently transfected in 24-well dishes, seeded on microscope slides 6-h after transfection, and stained 16–18 h later. To study the effect of the RNA–polymerase II inhibition on the RAE1 distribution in HtTA cells, the culture medium was supplemented with 0.04 or 5.0 μg/ml actinomycin D (AMD; Boehringer Mannheim Biochemicals), or 50 μg/ml DRB (Fluka Chemical Corp.). To obtain pure populations of transiently transfected HtTA or BHKgrβ cells, green fluorescent protein (GFP)–positive cells were isolated by fluorescence-activated cell sorting (Persons et al., 1997). Electron microscopy was as previously described (van Deursen et al., 1996).
Antibody Production
To generate RAE1-specific antibodies, we cloned cDNA sequences encoding mouse RAE1 amino acids 188–347 into pQE30 (QIAGEN Inc.). HIS-tagged recombinant mouse RAE1 protein was produced in Escherichia coli DH12S cells, purified with Ni-NTA (nitrilotriacetic acid) agarose beads (QIAGEN Inc.) according to the manufacturer's instructions, and injected into rabbits. NUP98- and CRM1-specific antisera were generated as described (Kasper et al., 1999). Antibodies were affinity purified using recombinant antigen bound to ProBlott (Perkin-Elmer Corp.) as previously reported (van Deursen et al., 1996).
Immunofluorescence, Coimmunoprecipitations, and Western Blotting
Indirect immunofluorescence, coimmunoprecipitations, and Western blot analyses were carried out as previously described in detail (Kasper et al., 1999).
Generation of Recombinant Proteins
A cDNA fragment encoding NUP98(150–224) was cloned in pQE31 and pGEX-5X-1 for expression of HIS- and glutathione-S-transferase (GST)– tagged recombinant NUP98(150–224). Recombinant protein expression in E. coli strain DH12S was induced by addition of 1 mM IPTG (isopropyl-β-d-thigalactopyranoside), followed by incubation at 25°C for 4–5 h. Harvested bacteria were suspended in 50 mM Tris-HCl, pH 7.5, 1 M NaCl, 2 mM PMSF, 1 mM leupeptin, 2 mM aprotinin, and 1 mM pepstatin. After 30 min of lysozyme treatment at 4°C, bacteria were lysed by sonication. HIS-NUP98(150–224) and GST-NUP98(150–224) were then purified from bacterial lysates using Ni-NTA agarose and glutathione beads (Sepharose 4B; Pharmacia LKB Biotechnology Inc.), respectively (according to standard procedures), and used in pull-down assays. A cDNA fragment encoding RAE1(1–368) was cloned in pGEX-5X-1 to express GST-RAE1 in E. coli. Bacterial pellets were suspended in PBS with 2 mM PMSF, 1 mM leupeptin, 2 mM aprotinin, and 1 mM pepstatin. Lysis was performed by sonification and GST-RAE1 was purified with glutathione beads using standard procedures. Purified RAE1 was separated from the GST-affinity tag using Factor Xa protease (Pharmacia LKB Biotechnology Inc.).
Pull-Down Assays and Chemical Cross-Linking
Glutathione beads with 100 ng purified GST-NUP98(150–224) were washed three times with binding buffer (20 mM Hepes, pH 7.9, 100 mM KCl, 5 mM MgCl2, 0.1% Tween-20, 20% glycerol, 0.01% bovine serum albumin, 1 mM dithiothreitol, 1 mM PMSF, 1 mM leupeptin, 2 mM aprotinin and 1 mM pepstatin), preblocked for 10 min with rabbit serum, washed with binding buffer, and resuspended in 60 μl binding buffer. Then, 10 μl in vitro–transcribed and –translated [35S]-methionine–labeled HA-RAE1 was added to the beads and the mixture was incubated at 4°C for 1 h (vortexed every 5 min). Beads were washed six times with binding buffer and boiled in 15 μl SDS sample buffer. Samples were analyzed by SDS-PAGE (10% polyacrylamide), followed by autoradiography. Pull-down assays with Ni-NTA agarose aliquots containing 100 ng of HIS-NUP98(150–224) were performed in the same way. For chemical cross-linking, pellets were resuspended in 10 μl PBS with 1 mM disuccinimidyl suberate (DSS; Pierce) after the last wash in our pull-down protocol. Cross-linking was at 4°C for 20 min. Finally, 10 μl SDS sample buffer was added and boiled samples were analyzed by SDS-PAGE (9% polyacrylamide) followed by autoradiography.
Tryptic Digestion and RNA Degradation
Trypsin digestion of in vitro–translated or recombinant RAE1 was as previously described (Audigier, 1994). The reaction was stopped with 2 μl of 100 mM benzamidine. For enzymatic RNA degradation, 10 μl in vitro– transcribed and –translated [35S]-methionine–labeled HA1-RAE1 was incubated with either 0.1 unit Micrococcal nuclease (Sigma Chemical Co.) or 10 μg RNase A. Both incubations were at 37°C for 30 min in the presence of 1 mM PMSF, 1 mM leupeptin, 2 mM aprotinin, and 1 mM pepstatin.
Detection of Poly(A)+ RNA
HtTA or BHKgrβ cells were stained for specific proteins and poly(A)+ RNA by use of the following combined immunostaining/in situ hybridization procedure. At 6 h after transfection, HtTA cells were seeded on microscope slides, and ∼16 h later they were fixed in PBS/3% formaldehyde for 15 min at 4°C. After five washes in PBS, cells were permeabilized in PBS/0.5% Triton X-100 for 5 min at 4°C. They were then washed in PBS (5×) and incubated with either 12CA5 antibodies (9 μg/ml) alone or 12CA5 and anti–RAE1 antibodies in PBS/2% BSA/0.2% Triton X-100/ 200 U/ml RNasin for 30 min at room temperature (RT). Cells were then washed in PBS (5×) and incubated with fluorochrome-coupled secondary antibodies for 30 min at RT: RAE1 antibodies were detected with Texas red–conjugated goat anti–rabbit antibodies (5 μg/ml), and 12CA5 mouse monoclonal antibodies were detected with R-phycoerythrin–conjugated goat anti–mouse antibodies (Caltag Laboratories). Cells were washed in PBS (5×), and then fixed in PBS/3% formaldehyde for 5 min at RT. After five rinses in PBS, the cells were equilibrated in 2× SSC for the in situ hybridization. Hybridization was performed by using an FITC-coupled oligo-(dT) 50 mer probe as detailed by Amberg et al. (1992). At the end of the procedure, the cells were mounted on the slides in Vectashield (Vector Laboratories, Inc.) and analyzed by using laser scanning confocal microscopy. The above procedure is very well suited for detection of alterations in nuclear poly(A)+ RNA levels. However, due to the relatively short fixation time of 15 min, a fraction of the cytoplasmic poly(A)+ RNA pool is lost. Therefore, if we wanted to study the cytoplasmic poly(A)+ RNA levels in detail, we extended the 3% formaldehyde fixation step by 15 min and fixed at RT.
Oocyte Injections
[35S]-methionine–labeled protein for microinjection into Xenopus oocytes was synthesized in a rabbit reticulocyte lysate as indicated by the manufacturer (Promega Corp.). Templates were pT7-CBP80 (Izaurralde et al., 1995a), pSP73-HA1-RAE1 (mouse), and pT7-GST-NES, encoding a GST fusion with the HIV-1 Rev nuclear export signal (Fischer et al., 1995). E. coli–expressed Rna1p was coinjected at a concentration of 80 μM in the injection mixture as described (Izaurralde et al., 1997b). Microinjections, incubations, and protein extraction and analysis were performed as described (Jarmolowski et al., 1994). Nuclear export of RAE1 was quantified by measuring the nuclear and cytoplasmic fractions of RAE1 with a phosphoimager (we corrected for leakage of RAE1 into the cytoplasm by quantifying the percentage of cytoplasmic CBP80).
Results
Human RAE1 Interacts with a GLEBS-like NUP98 Motif
We overexpressed an HA1-tagged NUP98 cDNA (Fig. 1 A) in HtTA cells and immunoprecipitated the cell lysates with 12CA5 monoclonal antibody against the tag. A protein of ∼40 kD specifically coprecipitated with HA1-NUP98 (Fig. 1 B). We wanted to test whether this interacting protein was the human homologue of the S. cerevisiae Gle2p because of (a) the molecular weight similarity and (b) the presence of a motif within NUP98 that is similar to the Gle2p-binding sequence of Nup116p (Bailer et al., 1998). We obtained a murine cDNA clone with similarity to yeast gle2/rae1 cDNA (designated RAE1) and generated polyclonal antisera in rabbits against the carboxy-terminal half of RAE1. We overexpressed HA1-NUP98 in HtTA cells, immunoprecipitated the cell lysates with 12CA5 monoclonal antibody against the HA1 tag, and performed a Western blot analysis using affinity-purified antibodies raised against mouse RAE1. As shown in Fig. 1 C, the RAE1 antibodies recognized the HA1-NUP98 coprecipitating protein, demonstrating that NUP98 indeed interacts with the human homologue of the yeast Gle2p. To ensure that the interaction of NUP98 with RAE1 was not an overexpression artifact, we precipitated NUP98 from nontransfected HtTA cells with affinity-purified NUP98 antibodies and determined whether RAE1 was coisolated by Western blot analysis. RAE1 indeed coprecipitated with NUP98 from HtTA cells (Fig. 1 D, lanes 3 and 4). When affinity-purified RAE1 antibodies were used in the immunoprecipitation step, NUP98 was coisolated with RAE1 (Fig. 1 D, lanes 1 and 2), thereby confirming that NUP98 and RAE1 are in a complex in human cells. It should be emphasized that our data do not rule out the possibility that the NUP98–RAE1 complex is part of a larger protein assembly.
Figure 1.
A GLEBS-like motif within NUP98 is necessary and sufficient for interaction with human RAE1. (A) Schematic of the NUP98 structure. Vertical bars indicate FG (phenylalanine-glycine) repeats; HA1, hemagglutinin tag; NRM, nucleoporin RNA-binding motif (shaded box); Nup116p homology region (gray box). (B) [35S]-methionine–labeled proteins immunoprecipitated with monoclonal antibody 12CA5 from HtTA cells transiently expressing an HA1-tagged version of NUP98, separated by SDS-PAGE (8% polyacrylamide) and visualized by autoradiography. A molecular weight standard is indicated at right. (C) Western blot analysis (8% polyacrylamide) of the 40-kD protein coimmunoprecipitated with HA1-NUP98 transiently expressed in HtTA cells (lanes 2 and 4). Nontransfected cells served as a negative control (lanes 1 and 3). The blots were first incubated with 12CA5 antibody (lanes 1 and 2), and then with affinity-purified anti–mouse RAE1 antibodies (lanes 3 and 4). The position of human RAE1 is indicated with an arrow. Molecular weight standards are indicated at left. (D) Western blot analysis (8% polyacrylamide) of proteins coimmunoprecipitated from HtTA lysates with anti–RAE1 (left) or anti–NUP98 antibodies (right). The antibodies used to visualize NUP98 or RAE1 proteins are indicated above the lanes. Molecular weight standards are indicated to the left. (E) Structure of NUP98 mutants used to define the GLEBS-like motif. The various NUP98 motifs are as indicated in A. (F) Western blot analysis (7% polyacrylamide) of proteins precipitated with 12CA5 antibody from HtTA cells transiently transfected with HA1-NUP98 or HA1-NUP98Δ(192–221). Molecular weight standards are indicated at left. (G) As in F for a set of HA1-tagged GLEBS-like motif mutants. Immunoprecipitated proteins were split in two equal portions, half was run through a 15% polyacrylamide gel to verify proper expression of HA1-tagged mutant peptides (top), and half on an 8% polyacrylamide gel to determine coimmunoprecipitation of RAE1. Because HA1-NUP98(150–186) protein (lane 3, top) was expressed at a lower level than the other HA1-tagged mutants, we also collected longer exposures of the RAE1 immunoblot (bottom); however, we were still unable to detect a RAE1-specific signal in lane 3. With the transfection protocol applied, levels of HA1-NUP98(150–224) were consistently higher than those of HA1-NUP98(181–224), which causes the difference in intensity of RAE1 signals in lanes 2 and 4. Molecular weight standards for each gel are indicated at left.
To test the role of the GLEBS-like motif of NUP98 in RAE1 binding, an HA1-NUP98 mutant lacking amino acids 192–221 was generated (Fig. 1 E). This mutant, designated as HA1-NUP98Δ(192–221), failed to coimmunoprecipitate RAE1 (Fig. 1 F, lanes 2 and 5), although, like full-length NUP98, it localized at the NE (data not shown). Hence, the GLEBS-like motif of NUP98 was necessary for binding RAE1. We then asked whether this region of NUP98 was sufficient for RAE1 binding. We expressed amino acids 150–224 of NUP98 as an HA1-tagged fusion protein in HtTA cells and performed a co-IP Western analysis. As shown in Fig. 1 G (lane 2), NUP98(150–224) indeed coimmunoprecipitated RAE1. Additional mutagenesis studies revealed that the actual NUP98 interaction motif is located within residues 181–224 (Fig. 1 G, lane 4). Computer analysis (using the GCG program PEPTIDESTRUCTURE) identified a potential alpha-helical region from amino acids 187–212. Three helix-breaking proline mutations introduced in this region abrogated the NUP98(181–224) interaction with RAE1 (Fig. 1 G, lane 5), further confirming that the NUP98(181–224) segment contains the GLEBS-like motif.
The RAE1–NUP98 Interaction Is Direct and mRNA Independent
To further characterize the RAE1–NUP98 interaction, we produced RAE1 and the GLEBS-like motif in vitro, and analyzed whether they bind directly or indirectly via an adaptor protein or a molecule of mRNA. Presumably, members of the WD-repeat superfamily all form compact globular propeller structures that are resistant to proteolysis (Garcia-Higuera et al., 1996a,b). It has been shown previously that most WD-repeat proteins fold into their native globular structure when synthesized in vitro in a rabbit reticulocyte lysate system, but not when synthesized in E. coli (Garcia-Higuera et al., 1996a; Neer et al., 1994a). We synthesized RAE1 protein both in a rabbit reticulocyte lysate system (see Fig. 2 C, lane 1) and in E. coli (not shown) and performed pull-down assays with HIS-NUP98(150–224) or GST-NUP98(150–224) purified from E. coli (Fig. 2, A and B). Both HIS- and GST-NUP98 (150–224) bind to RAE1 (hemagglutinin [HA]-tagged) generated in a rabbit reticulocyte lysate system (Fig. 2 C, lanes 4 and 6), but failed to bind recombinant RAE1 synthesized in E. coli (not shown). To test the folding of RAE1 purified from E. coli and in a rabbit reticulocyte lysate, we analyzed their sensitivity to tryptic cleavage (Neer et al., 1994a; Garcia-Higuera et al., 1996a, 1998). We found that tryptic cleavage of in vitro–translated [35S]-methionine–labeled HA1-RAE1 removes ∼3 kD and leaves a large stable fragment of 42 kD, despite the presence of many potential cleavage sites (data not shown). By contrast, RAE1 from E. coli was extensively degraded because of cleavage at multiple tryptic sites (data not shown). This result implies that in E. coli RAE1 cannot fold into a compact, globular structure capable of interacting with NUP98.
Figure 2.
Chemical cross-linking of in vitro–translated HA1-RAE1 to E. coli–purified GLEBS-like motifs. (A) Purified recombinant HIS-NUP98(150–224) (∼11-kD) protein separated by SDS-PAGE (15% polyacrylamide gel) and detected by Coomassie brilliant blue (CBB; Bio-Rad Laboratories) staining. (B) Purified recombinant GST (29 kD) and GST-NUP98(150–224) (∼38 kD) protein run on a 10% polyacrylamide gel and stained with CBB. (C) Pull-down assays performed with [35S]-methionine–labeled HA1-RAE1 synthesized in vitro (45 kD), and GST- or HIS-NUP98(150–224) fusion proteins purified from E. coli. 5% input shows 5% of the labeled HA1-RAE1 protein used in each pull-down assay. Typically, the in vitro–translated RAE1 appears as a doublet, representing fragments with and without a HA1 tag (the RAE1 cDNA was cloned in pSP73 as a HA1 fusion gene that retained the endogenous RAE1 translation initiation codon). GST beads and Ni-NTA agarose acted as negative control for binding in GST-NUP98(150–224) and HIS-NUP98(150–224) pull-down assays, respectively. Comparable amounts of GST, GST-NUP98(150–224), and HIS-NUP98(150–224) proteins were used in each pull-down assay. The experiment shown is representative for two independent experiments. A cross-linked GST-NUP98(150–224)/RAE1 product of ∼84 kD and a cross-linked HIS-NUP98(150–224)/RAE1 product of ∼56 kD were obtained specifically in DSS-treated samples. Note that cross-linking of RAE1 to HIS-NUP98(150–224) was more efficient than to GST-NUP98(150–224). Molecular weight standards are indicated at right.
To investigate whether RAE1 and NUP98 establish direct contact, we used chemical cross-linkers (Yi et al., 1991; Neer et al., 1994a; Garcia-Higuera et al., 1996b). Cross-linking of residues from in vitro–translated [35S]- methionine–labeled HA1-RAE1 and recombinant HIS-NUP98(150–224) or GST-NUP98(150–224) should yield specific cross-linked products of ∼56 and ∼83 kD, respectively. The predicted cross-linked products were indeed obtained with DSS (Fig. 2 C, lanes 3 and 5), a reagent that cross-links mainly lysine residues, but not with BMH, a sulfhydryl-reactive cross-linker (data not shown). DSS-mediated coupling of RAE1 to the GLEBS-like motif of NUP98 revealed that the interaction is direct and not mediated though another protein.
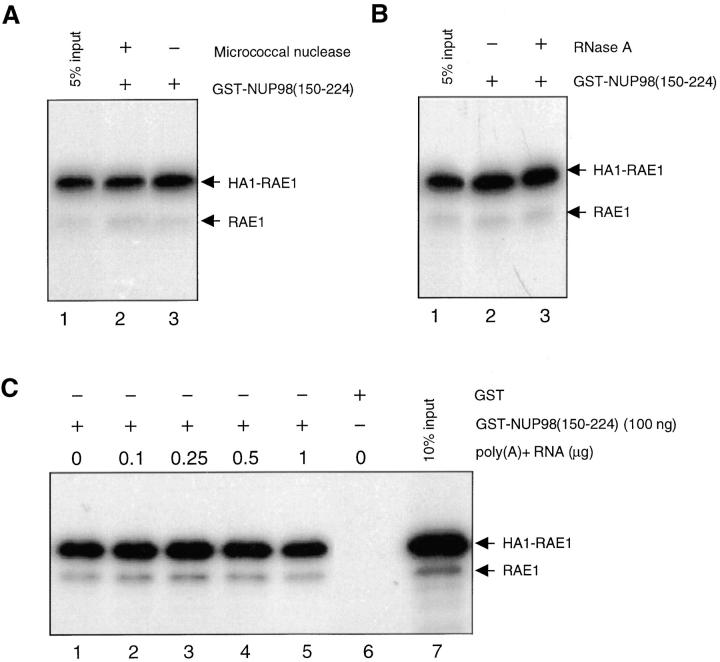
It has been reported that RAE1 can be UV cross-linked to poly(A)+ RNA (Kraemer and Blobel, 1997), and theoretically, binding between RAE1 and GLEBS-like motif may be established via mRNA. To investigate this possibility, we synthesized [35S]-methionine–labeled HA1-RAE1 protein, removed the mRNA from the reticulocyte lysate with either Micrococcal nuclease or RNase A, and performed pull-down assays with GST-NUP98(150–224) beads. Neither the nuclease (Fig. 3 A) nor the RNase A treatment (Fig. 3 B) had any effects on the binding ability of RAE1 to NUP98 in vitro. Pull-down assays were also performed after addition of various amounts of poly(A)+ mRNA isolated from HtTA cells. However, the binding efficiency of HA1-RAE1 to the GLEBS-like motif was similar irrespective of the mRNA amount present during the binding reaction (Fig. 3 C). In summary, our in vitro-binding studies indicate that the interaction between RAE1 and the GLEBS-like motif of NUP98 is direct and mRNA independent.
Figure 3.
Poly(A)+ RNA is not a cofactor in the binding reaction between RAE1 and the GLEBS-like motif of NUP98 in vitro. (A) Pull-down assay performed with GST-NUP98(150– 224) fusion protein purified from E. coli and [35S]-methionine– labeled in vitro–translated HA1-RAE1 pretreated with or without Micrococcal nuclease. GST alone did not pull-down HA1-RAE1 (not shown). (B) As in A, but pretreated with or without RNase A. GST alone did not pull-down HA1-RAE1 (not shown). (C) Pull-down assay with GST-NUP98(150–224) and in vitro–translated HA1-RAE1 in the presence of various concentrations of poly(A)+ RNA isolated from HtTA cells. GST alone does not pull-down HA1-RAE1, as indicated in lane 6.
The COOH-Terminal Non–WD-repeat Extension of RAE1 Is Essential for NUP98 Binding
To start investigating how RAE1 binds to the GLEBS-like motif of NUP98 at the NPC, we generated a series of mutants with deletions in the NH2- or COOH-terminal non– WD-repeat extensions and tested them for their ability to interact with NUP98 by using a coimmunoprecipitation approach (Fig. 4 A). Mutant HA1-RAE1(33–368), which lacks the entire non–WD-repeat NH2-terminal extension, was able to coprecipitate NUP98, although with reduced efficiency compared with wild-type RAE1 (Fig. 4 B, lanes 1 and 3; n = 3 independent experiments). Extension of this deletion into the first WD repeat [HA1-RAE1(66– 368)] abolished interaction with NUP98 (Fig. 4 B, lane 6). HA1-RAE1(1–329), which lacks the COOH-terminal 39 amino acids, again failed to coprecipitate NUP98 (Fig. 4 B, lane 5). By contrast, mutant HA1-RAE1(1–359), which lacks the COOH-terminal nine residues containing a highly conserved basic motif that has been shown to be essential for Rae1p function in S. pombe (Whalen et al., 1997), could coprecipitate NUP98 (Fig. 4 B, lane 4). Hence, the basic motif of RAE1 may attribute a critical cellular function of RAE1 other than binding to NUP98.
Figure 4.
Multiple domains of RAE1 are necessary but not sufficient for interaction with NUP98. (A) Schematic representation of HA1-RAE1 deletion mutants. The four WD-repeat motifs are indicated as gray boxes. All mutants expressed an NH2-terminal HA1 tag. (B) Western blot analysis of proteins precipitated with 12CA5 antibody from lysates of HtTA cells transiently transfected with HA1-RAE1 or various deletion mutants. Precipitated proteins were visualized with 12CA5 (top) and anti–NUP98 antibodies (bottom). The HA1-RAE1 mutants used are indicated above the lanes. Molecular weight standards are indicated to the left. Results shown are representative for three independent experiments. (C–G) Confocal images detailing the subcellular distribution HA1-RAE1 deletion mutants moderately expressed in HtTA cells and stained with 12CA5 antibody. Note that the ability to coprecipitate NUP98 correlates with prominent NE staining.
In nontransfected HtTA cells, RAE1 was localized prominently at the NE, but substantial amounts of RAE1 were also found in the nucleus and cytoplasm (see Fig. 6 B). A very similar distribution pattern was observed in HtTA cells that moderately overexpress HA1-RAE1 (Fig. 4 C); however, more robust overexpression resulted in a disproportionate increase of RAE1 levels in the nucleus (not shown). As expected, all deletion mutants but the ones that failed to coprecipitate NUP98 displayed overt NE localization when transiently expressed in HtTA cells (Fig. 4, D–F). The above experiments suggest that both the WD repeat propeller and the COOH-terminal non– WD-repeat extension of RAE1 contribute to NUP98 binding.
Figure 6.
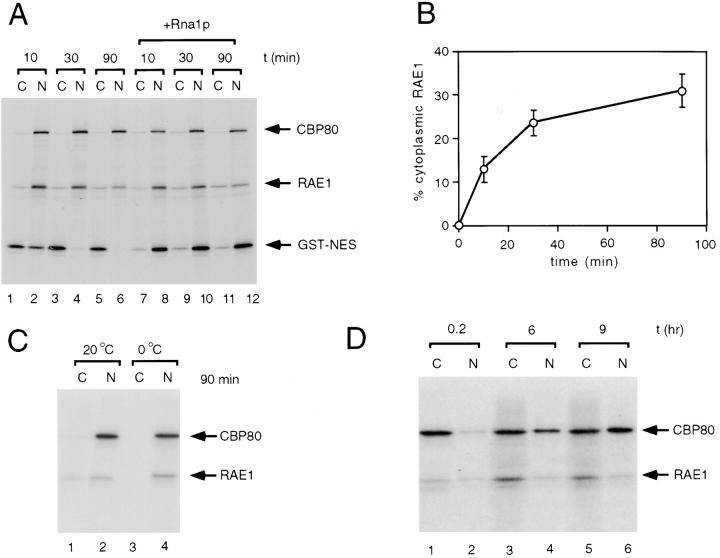
RAE1 shuttles between the nucleus and the cytoplasm. (A) A mixture of in vitro–translated [35S]-methionine–labeled CBP80, HA1-RAE1 (seen as a doublet that represents HA1-RAE1 and RAE1), and GTS-NES was injected into Xenopus laevis oocytes nuclei, either in the absence (lanes 1–6) or presence (lanes 7–12) of 80 μM Rna1p. After injection, oocytes were incubated at 20°C and protein samples from nuclear (N) and cytoplasmic (C) fractions were collected at 10, 30, or 90 min. Proteins were separated by SDS-PAGE and detected by fluorography. (B) Kinetics of RAE1 distribution after nuclear injection. Values were obtained from three experiments; error bars represent standard deviations. (C) Comparison of nuclear export of RAE1 at 20° (lanes 1 and 2) and 0°C (lanes 3 and 4). (D) Nuclear uptake of RAE1 after cytoplasmic injection after 0.2 (lanes 1 and 2), 6 (lanes 3 and 4), or 9 h (lanes 5 and 6).
Point Mutation's Individual WD-repeats Differentially Affect NUP98 Binding
The HA1-RAE1(66–368) mutant suggested that the WD-repeat propeller of RAE1 is implicated in NUP98 binding. To further define the role of the WD-repeats in the RAE1–NUP98 interaction, we mutated single WD-repeats at their highly conserved aspartic acid residue positioned in the turn connecting the β strands b and c of a propeller blade (Fig. 5, A and B). We targeted these particular residues because their conservation in 85% of all known WD-repeats indicates that they perform an important role within the propeller structure (Neer et al., 1994b; Neer and Smith, 1996; Garcia-Higuera et al., 1998). Moreover, it has been reported that point mutations at the conserved aspartic acid residues of the WD-repeat proteins Gβ and sec13 can cause local distortions in the structure of individual propeller blades without affecting the overall structure of the propeller (Garcia-Higuera et al., 1998). We expressed the point mutants in HtTA cells and determined their ability to interact with NUP98. We found that the point mutation in propeller blade 4 (Fig. 5 C, lane 3), but not the point mutation in blade 2 or 3 (Fig. 5 C, lanes 1 and 2, respectively), abolished RAE1's ability to bind to NUP98. Accordingly, HA1-RAE1-D2 and -D3 (Fig. 5, E and F, respectively) displayed a prominent NE localization when transiently expressed in HtTA cells. However, HA1-RAE1-D4 (Fig. 5 G) was undetectable at the NE. Together, the above studies underscore that the WD-repeat propeller is implicated in the binding of RAE1 to NUP98. Moreover, they suggest that individual WD-repeats differentially support the RAE1–NUP98 interaction.
Figure 5.
A single point mutation in a highly conserved WD-repeat residue abrogates RAE1–NUP98 interaction at the NPC. (A) A scale drawing of HA1-RAE1 depicting the positions of the highly conserved D residues within the last three WD-repeats that we individually mutated to A. (B) Schematic representation of a blade from a WD-repeat propeller structure (Garcia-Higuera et al., 1998). (a–d) The four β strands within the blade are indicated. Note that the position of the conserved D residue in the hairpin turn between b and c is highlighted (star). (C) Western blot analysis of proteins precipitated with 12CA5 antibody from lysates of HtTA cells transiently transfected with HA1-RAE1 or the point mutants. Precipitated proteins were visualized with 12CA5 (top) and anti–NUP98 antibodies (bottom). The HA1-RAE1 mutants used are indicated above the lanes. Molecular weight standards are indicated at left. Results shown are representative for three independent experiments. (D–G) Representative confocal images detailing the subcellular distribution of HA1-RAE1 mutants in HtTA cells (12CA5 antibody staining).
RAE1 Shuttles between the Nucleus and the Cytoplasm
RAE1 could be permanently or transiently bound to NUP98 at the NPC, or both. As a first step to investigate whether RAE1 has dynamic properties, we microinjected in vitro–translated [35S]-methionine–labeled mouse RAE1 into Xenopus oocytes and analyzed its ability to shuttle between the nucleus and the cytoplasm. The NLS-containing protein CBP80 was coinjected with RAE1 to serve as a control for nuclear protein import, and proper injection and dissection of the oocytes (Izaurralde et al., 1995b). We injected RAE1 into the oocyte nucleus and quantified the fraction of RAE1 appearing in the cytoplasm at various time points after injection by phosphoimager analysis. Fig. 6, A (lanes 1–6) and B, demonstrates that ∼24% (SD ± 4%, n = 3 independent experiments) of the RAE1 molecules injected into the nucleus is present in the cytoplasm within 30 min after injection and maximal cytoplasmic levels of 31% (SD ± 4%, n = 3) are achieved within 90 min after injection. These data suggest (a) that an equilibrium between export and import has been established around 90 min after injection or (b) that the majority of the microinjected RAE1 molecules seems to be export incompetent. In addition, export of microinjected RAE1 was completely inhibited at 0°C (Fig. 6 C, lanes 3 and 4), indicating that RAE1 export is temperature dependent and not driven by “simple” diffusion. We also investigated whether RanGTP mediates nuclear export of RAE1 by reducing the level of RanGTP via nuclear injection of Rna1p, a normally cytoplasmic GTPase-activating protein for Ran (Izaurralde et al., 1997b). As shown in Fig. 6 A (compare lanes 1–6 with 7–12), coinjection of 80 μM Rna1p (1–10 μM usually induces a significant inhibition in RanGTP-dependent export) did not significantly inhibit nuclear export of RAE1, while, in accordance with previous data (Izaurralde et al., 1997b; Richards et al., 1997) nuclear export of NES-tagged GST substrates was dramatically reduced. As expected from its nuclear localization, RAE1 protein injected into the oocyte cytoplasm was able to migrate rapidly into the nucleus (Fig. 6 D, lanes 1–6).
Association of RAE1 with the NE Requires RNA Polymerase II Activity
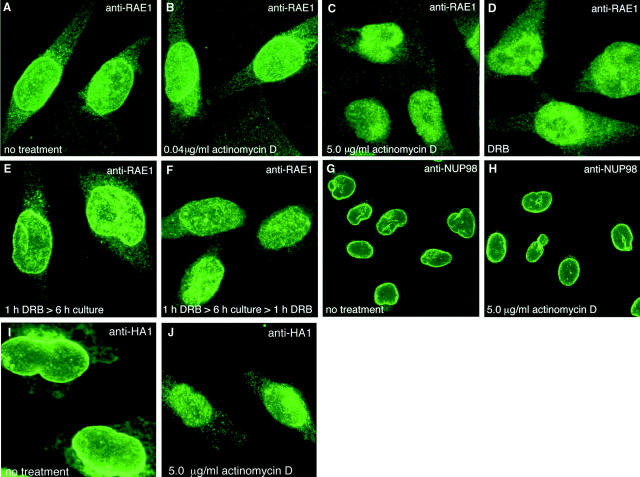
The combination of shuttling and poly(A)+ RNA-binding properties (Kraemer and Blobel, 1997) led us to hypothesize that RAE1 travels between cellular compartments as an mRNA export factor. To investigate this possibility, we stopped mRNA synthesis in HtTA cells by adding RNA polymerase II inhibitors and asked whether the subcellular distribution of RAE1 changed. After 1 h of treatment with 5 μg/ml AMD, a dose that inhibits both RNA polymerase I and II activity, the prominent RAE1 staining at the NE normally seen in HtTA cells was no longer detectable (compare Fig. 7, A and C). In contrast, normal levels of RAE1 at the NE were observed in cells exposed to a dose of AMD that only inhibited RNA polymerase I activity (0.04 μg/ml AMD; Fig. 7 B). When we shortened the treatment with 5 μg/ml AMD from 1 h to 15 min, the drop in RAE1 levels at the NE was still detectable, suggesting that the observed effect can be an immediate early response to RNA polymerase II inhibition. HtTA cells exposed to 50 μg/ml of the RNA polymerase II inhibitor DRB (5,6-dichloro-β-d-ribofuranosylbenzimidazole) showed a RAE1-staining pattern very similar to that observed in cells exposed to a high dose of AMD (Fig. 7 D). The inhibitory effect of DRB is reversible and we analyzed whether normal RAE1 levels at the NE would be restored upon reactivation of RNA polymerase II–mediated transcription. As shown in Fig. 7 E, a prominent NE staining was observed in cells cultured for 6 h in the absence of DRB after a 1-h exposure to this component. As expected, the RAE1 levels at the NE were substantially reduced after a second DRB treatment (Fig. 7 F). In both AMD- and DRB-treated cells, the decrease for NE-associated RAE1 did not coincide with significant alterations in the nuclear and cytoplasmic RAE1 levels. Inhibition of RNA polymerase II activity did not affect NUP98 association with the NE (compare Fig. 7, G and H), demonstrating that the absence of substantial amounts of RAE1 at the NE was not the result of NUP98 relocation. As expected, the nuclear export receptor hCRM1, which mediates export of certain viral RNAs and U snRNP, but not cellular mRNA, retained its NE localization in the presence of AMD (5 μg/ ml) or DRB (data not shown). Together, the above experiments confirm that the NE association of RAE1 is transient rather than stable. Furthermore, they support the hypothesis that RAE1 associates with the NE as part of an mRNP complex.
Figure 7.
RAE1's association with the NE requires RNA polymerase II activity. (A–H) Images of HtTA cells stained for RAE1 or NUP98 after treatment with RNA polymerase inhibitors. RAE1-stained cells treated with (A) 0 μg/ml AMD for 1 h, (B) 0.04 μg/ml AMD for 1 h, (C) 5.0 μg/ml AMD for 1 h, (D) 50 μg/ml DRB for 1 h, (E) 50 μg/ml DRB for 1 h and cultured 6 h in the absence of DRB, and (F) 50 μg/ml DRB for 1 h, cultured 6 h without DRB, and finally treated again with 50 μg/ml DRB for 1 h. Images shown are representative for results obtained from three independent experiments. (G and H) Respective nontreated and 5.0 μg/ml AMD–treated HtTA cells stained with NUP98-specific antibodies. Note that NUP98 levels at the NE are not dependent on RNA polymerase II activity. (I and J) Effect of AMD treatment on HtTA cells that moderately express HA1-RAE1.
It has been demonstrated that an AU1-tagged version of human RAE1 localizes at high levels to the nucleus and at considerably lower levels to the cytoplasm of HeLa cells (Bharathi et al., 1997). When AU1-RAE1–expressing cells were treated with RNA polymerase II inhibitors, no major changes in the distribution of AU1-RAE1 were detected. This result may seem to contradict the data presented in this report; however, it should be stressed that the effects of RNA polymerase II activity on AU1-RAE1 levels at the NE could not be evaluated because the robust nuclear staining masked the NE staining (Bharathi et al., 1997). To reevaluate the effect of RNA polymerase II inhibitors on the distribution of overexpressed RAE1, we transiently expressed HA1-RAE1 in HtTA cells. Typically, when overexpressed at low to moderate levels, HA1-RAE1 prominently localized to the NE, but significant amounts of RAE1 were also found in the nucleus and the cytoplasm (Fig. 7 I). More robust overexpression of HA1-RAE1 resulted mostly in a disproportionate increase of RAE1 levels in the nucleus, which concealed the NE staining (data not shown). We determined the effect of RNA polymerase II inhibition on the distribution of HA1-RAE1 by focusing on cells with a low to moderate level of expression. As shown in Fig. 7 J, HA-RAE1 levels at the NE decreased significantly when cells were treated with 5 μg/ml AMD for 1 h, which is consistent with the results that we obtained by using nontransfected HtTA cells.
Overexpression of the GLEBS-like Motif Inhibits Nuclear Export of Poly(A)+
When HtTA cells transiently expressing HA1-NUP98 (150–224) or HA1-NUP98(181–224) were immunostained with 12CA5 and RAE1 antibodies 24 h after transfection, we noticed a considerable reduction of RAE1 levels at the NE. As shown in Fig. 8, A, A′, B, and B′, NE staining of RAE1 is easily detectable in nontransfected HtTA cells (nt), but not in HA1-NUP98(150–224)– or HA1-NUP98 (181–224)–expressing cells (t). By contrast, NE staining of RAE1 remained intact when HA1-NUP98(181–224)M (Fig. 8, C and C′), a mutated GLEBS-like motif that does not interact with RAE1 (see also Fig. 1 G, lane 5), or full-length HA1-NUP98 was overexpressed (Fig. 8, D and D′). In all cases, NUP98 localization at the pores appeared unchanged (data not shown). The above results suggest that overexpressed GLEBS-like motif of NUP98 acts as a dominant negative inhibitor of RAE1-NPC association by titrating RAE1 from the NPC and/or interfering with RAE1 docking to NUP98 at the NPC.
Figure 8.

Overexpression of the GLEBS-like motif of NUP98 results in decreased levels of RAE1 at the NE. (A–D′ inclusive) Paired confocal images from HtTA cells that transiently express various forms of the GLEBS-like motif of NUP98. Cells were double-stained for HA1-tagged protein using the 12CA5 monoclonal antibody (left) and for RAE1 using affinity-purified RAE1 polyclonal rabbit antibodies. Cells shown in the confocal images are representative for results obtained in three to four independent experiments. High magnification images are given to illustrate detail. (A, A′, B, and B′) Overexpression of the GLEBS-like motif leads to a subtle but significant decrease in NE localization of RAE1. Compare NE staining of cells that do not overexpress the RAE1-binding (nt) and cells that do (t). (C, C′, D, and D′) Cells expressing a GLEBS-like motif mutant that is unable to interact with RAE1 or full-length NUP98 (D and D′) display an RAE1-distribution pattern comparable with that of wild-type HtTA cells.
To assess whether this effect is associated with changes in mRNA export, we examined the poly(A)+ RNA distribution in HtTA cells expressing the NUP98 GLEBS-like motif by in situ hybridization with an FITC-labeled oligo- (dT) 50-mer probe (Amberg et al., 1992; van Deursen et al., 1996). With this in situ hybridization protocol, alterations in nuclear poly(A)+ levels can easily be detected. Hybridized cells were examined by confocal microscopy. As shown in detail in Fig. 9 F′, poly(A)+ RNA was detected in both the nucleus and the cytoplasm of nontransfected HtTA cells. In situ hybridization of transiently transfected HtTA cells expressing HA1-NUP98(150–224) (Fig. 9, B and B′) or HA1-NUP98(181–224) (C and C′) revealed a dramatic increase in nuclear labeling (for detailed images, see Fig. 9, G, G′, H, and H′). To confirm that the strong nuclear labeling was indeed due to RNA accumulation, HtTA cells expressing HA1-NUP98(150–224) or HA1-NUP98(181–224) were incubated with RNase for 30 min before in situ hybridization. As expected, no labeling was detectable after such treatment (Fig. 9, J and J′). As an additional control that the signal detected in the nucleus is indeed mRNA, we incubated HA1-NUP98(150–224)– expressing cells for 1 h with DRB before in situ hybridization with the oligo-(dT) 50-mer probe. We quantified the nuclear signal of 20–25 cells by using confocal microscopy and the software program QUANTIFY. We compared the levels obtained with those measured in the same amount of nontreated HA1-NUP98(150–224)–positive cells. In three independent experiments, the nuclear poly(A)+ signal detected in DRB-treated cells was 29, 30, and 36% lower than in nontreated cells. Thus, a proportion of the nuclear poly(A)+ RNA is either exported to the cytoplasm or rapidly degraded (or both), suggesting that at least a proportion of the signal obtained with the oligo-(dT) probe represents nuclear mRNA and not just stable nuclear poly(A)+ RNA (Huang et al., 1994).
Figure 9.

Overexpression of the GLEBS-like motif of NUP98 results in accumulation of poly(A)+ RNA in the nucleus. (A–J′, inclusive) Paired confocal images from HtTA cells that are transiently expressing HA1-NUP98 mutants. These cells were double stained for HA1-tagged protein by immunohistochemistry using the 12CA5 monoclonal antibody (left) and for poly(A)+ RNA by in situ hybridization using a FITC-labeled oligo-(dT) 50 mer (right). The identity of the HA1-tagged NUP98 mutants is indicated as the left of each pair. The poly(A)+ RNA accumulation in B′ appears more robust than that in C′, which represents a photographic rather than a real difference. The arrow in F′ points to one of the sites of preferred poly(A)+ RNA localization (also reported by Huang et al., 1994). K and L Show poly(A)+ distribution in HtTA cells overexpressing mouse RAE1 protein and, at the same time, HA1-NUP98(150–224) or HA1-NUP98(181–224). Each row of three images shows the same field of cells stained for either HA1-tagged protein (left), ectopically expressed mouse RAE1 and native human RAE1 (middle), and poly(A)+ RNA (right). Note that mouse RAE1 overexpression restores proper mRNA export in cells expressing various forms of the GLEBS-like motif. The combined anti–HA1/poly(A)+ staining procedure does not allow extensive blocking of nonspecific 12CA5 antibody binding. Therefore, nontransfected cells display higher levels of background staining than those shown in Fig. 7.
Nuclear accumulation of poly(A)+ RNA induced by HA1-NUP98(150–224) or HA1-NUP98(181–224) expression typically coincided with a decrease in cytoplasmic poly(A)+ RNA levels; however, considerable amounts of polyadenylated RNA were still found in the cytoplasm (Fig. 9, B, B′, C, C′, G, G′, H, and H′). This was further corroborated using an in situ hybridization procedure optimized for detection of cytoplasmic polyadenylated RNA (for details, see Materials and Methods), as is illustrated in Fig. 9, I and I′. No nuclear build up of poly(A)+ RNA was found in cells expressing the GLEBS-like motif mutant HA1-NUP98(181–224)M (Fig. 9, D and D′), which confirms that binding of endogenous RAE1 to overexpressed GLEBS-like motif is essential poly(A)+ RNA accumulation. In HtTA cells overexpressing mouse RAE1 in addition to HA1-NUP98(150–224), or HA1-NUP98(181–224) nuclear accumulation of poly(A)+ RNA was either not seen or hardly detectable (see representative images in Fig. 9, K and L). Thus, proper poly(A)+ RNA export can take place in the presence of transiently expressed GLEBS-like motif if RAE1 levels are increased above normal. RAE1 overexpression probably restores the cellular pool of “free” RAE1 to a level required for proper nuclear mRNA export. We verified that overexpression of HA1-tagged mouse RAE1 (Fig. 9, E and E′) or nontagged RAE1 (not shown) alone did not induce any measurable alterations in poly(A)+ RNA distribution compared with nontransfected HtTA cells.
To verify that the poly(A)+ defect was not the result of a nuclear import defect, we used BHKgrβ cells. These cells express a glucocorticoid receptor–β-galactosidase fusion protein that is strictly localized to the cytoplasm. When exposed to dexamethasone, the fusion protein translocates within 30 min to the nucleus in a quantitative fashion (Bastos et al., 1996). When we transfected BHKgrβ cells with an HA1-NUP98(150–224) construct, they accumulated poly(A)+ RNA in the nucleus (compare Fig. 10, A and A′ with B and B′) with no detectable effect on the import of the glucocorticoid receptor–β-galactosidase fusion protein (Fig. 10, C, C′, D, and D′). Although only one protein was tested, these results show that the NLS-mediated nuclear import pathway remained intact, and that the mRNA export phenotype is not likely to be the result of a general trafficking defect.
Figure 10.
Overexpression of the GLEBS-like motif of NUP98 does not affect nuclear protein import. (A and B) Representative images of normal and HA1-NUP98(150–224)–expressing BHK cells stained with monoclonal antibody 12CA5 and the FITC-labeled oligo-(dT) 50 mer. Note that HA1-NUP98(150–224)–expressing BHK cells accumulate poly(A)+ RNA in their nuclei. (C and D) Immunofluorescence detection of the glucocorticoid receptor–β-galactosidase fusion protein in a representative BHKgrβ cell expressing HA1-NUP98(150–224) before and 30 min after the addition of 10 mg/ml dexamethasone (DMS).
Finally, we wanted to exclude that overexpression of the GLEBS-like motif induced NPC herniations similar to those seen in yeast nup116 and gle2 knockouts (Wente and Blobel, 1993; Murphy et al., 1996) and nup116p(ΔGLEBS) mutant cells (Bailer et al., 1998). To this end, we purified transiently transfected HtTA cells expressing both HA1-NUP98(150–224) and green fluorescent protein by FACS® (Becton Dickinson & Co.) sorting and studied their NPC integrity using an electron microscope. None of the HA1-NUP98(150–224)–expressing cells examined displayed any herniated or clustered NPCs (data not shown). Thus, the mRNA export defect is not likely to be secondary to abnormalities in NPC structure and distribution. Taken together, our results suggest a model in which a dominant negative GLEBS-like motif directly interferes with export of poly(A)+ RNA from the nucleus by targeting RAE1 the NUP98-RAE1 interaction, or both.
Discussion
Details about the mechanism by which mRNA is exported from the nucleus remain a mystery. Here we identified and characterized an interaction between human RAE1 and NUP98, and studied its significance in mRNA export. Our studies support a model in which RAE1 is a shuttling transport factor that permits efficient export of mRNA through its ability to anchor to a GLEBS-like NUP98 motif at the NPC. Specifically, we show that: (a) RAE1 binds to the GLEBS-like motif of NUP98 through multiple domains, including the WD propeller and part of the carboxy-terminal non–WD-repeat extension; (b) the RAE1– NUP98 interaction is direct and not via another protein or RNA; (c) RAE1 has the ability to shuttle from the nucleus to the cytoplasm and that its interaction with the NE seems to be transient rather than stable; and (d) the GLEBS-like motif, when overexpressed, binds to RAE1 and inhibits poly(A)+ RNA export from the nucleus, but not NLS-mediated import and NPC structure or distribution.
Shuttling and Dynamic Properties of RAE1
Analysis of the dynamic properties of RAE1 by microinjection of in vitro–translated protein into Xenopus laevis oocytes revealed that RAE1 has the ability to shuttle between the nucleus and the cytoplasm in a rapid manner. Furthermore, nuclear export of RAE1 appears to be established by a temperature-sensitive, RanGTP-independent mechanism. Several studies have demonstrated that GTP-bound Ran has an essential role in nuclear RNA export. However, different RNA classes seem to depend differently on RanGTP for their export from the nucleus. Both U snRNA and tRNA export are highly sensitive towards RanGTP depletion. On the other hand, some mRNAs, such as H4 and DHFR mRNA (Izaurralde et al., 1997b), apparently use RanGTP-dependent as well as RanGTP-independent mechanisms for their nuclear export, whereas export of adenovirus major late transcripts (Izaurralde et al., 1997b) and heat shock mRNAs (Saavedra et al., 1997; Stutz et al., 1997) is unaffected by the absence of nuclear RanGTP. Therefore, the RanGTP insensitivity of RAE1 export does not argue against the idea that RAE1 may be a shuttling nuclear export factor for mRNAs. Indeed, no RanGTP-binding exportin has yet been identified that is directly involved in mRNA export (see also Stutz and Rosbash, 1998), and it remains possible that the effects of RanGTP depletion on mRNA export are indirect.
Two additional findings reported here support RAE1's dynamic properties. First, overexpression of the GLEBS-like motif of NUP98 causes a reduction in the level of RAE1 associated with the NE. If RAE1 is permanently associated with NUP98 at the NE, overexpression of the GLEBS-like motif is expected to have no major effects on RAE1 levels at the NE. On the other hand, if RAE1's association with the NE would be transient rather than permanent, RAE1 molecules released from the NE may form a complex with the overexpressed GLEBS-like motif of NUP98. Once established, such complexes may be defective in docking to NUP98 at the NE and induce a general decline in RAE1 levels at the NE. The second finding that emphasizes the dynamic properties of RAE1 is that the level of RAE1 at the NE appears to be dependent on RNA polymerase II activity. Specifically, we observed that the amount of RAE1 at the NE dropped considerably if cells were exposed to RNA polymerase II inhibitors. When RNA polymerase II activity was restored by removal of the inhibitor, RAE1 levels at the NE returned to normal. Thus, RAE1's translocation from the nucleus to the NE may be dependent on the availability of gene transcripts generated by RNA polymerase II. It can be argued that the effect of RNA polymerase II inhibitors on the NE association of RAE1 is secondary to the depletion or relocation of one or more other cellular factors of unknown identity. Although we cannot exclude this possibility, three of our findings argue against an indirect effect: (a) the RAE1 levels at the NE drop shortly after initiation of AMD treatment (≤15 min), (b) the reversibility of the DRB-induced effect on RAE1, and (c) the observation that the inhibitors do not alter the distribution of the transport factors NUP98 and hCRM1 (Fornerod et al., 1997b). While more detailed analyses need to be done, the RNA polymerase II inhibition experiments are consistent with the notion that RAE1 is a dynamic mRNA export mediator.
On the surface, the observation that the association of RAE1 with the NE depends on RNA polymerase II activity seems to contradict the data from our in vitro binding studies, which indicated that mRNA is not a cofactor in the interaction between RAE1 and the GLEBS-like motif of NUP98. This apparent paradox can be explained if one assumes that docking of a transport substrate from the nucleus to the NPC follows a two-step process. In the first step, the substrate translocates from its site of assembly in the nucleus to the nuclear periphery. In the second step, the substrate anchors to the NPC. In this light, the negative effect of RNA polymerase II inhibitors on the association of RAE1 with the NE may reflect a defect not in the ability to anchor to NUP98 at the nuclear face of the NPC but rather in RAE1's translocation from the nucleus to the NPC. Guidance of RAE1 to the NPC may be a signal-mediated process relying on external export signals, perhaps provided by proteins within the mRNP transport substrate. Because so many proteins are associated with the mRNA export substrate, multiple associated proteins may provide independent signals for export of mRNP.
RAE1 and mRNA Export
Our most powerful evidence for a direct role of RAE1 and the RAE1–NUP98 interaction in the mRNA export pathway comes from GLEBS-like motif overexpression studies. Typically, overexpression of the GLEBS-like motif of NUP98 resulted in reduced levels of RAE1 at the NE and nuclear accumulation of poly(A)+ RNA. Presuming that RAE1 is a component of mRNP (Kraemer and Blobel, 1997), two possibilities could explain these data. It seems fair to argue that RAE1 associates with mRNP since RAE1 and poly(A)+ RNA can be cross-linked by UV irradiation in vivo (Kraemer and Blobel, 1997). Thus, at least part of the cellular RAE1 pool is in close proximity to mRNA, although it remains to be determined whether RAE1 and mRNA interact directly (RAE1 sequences lack similarity to any known mRNA-binding motifs), or indirectly via one of the mRNA-associated proteins. In the first possibility, one might propose that the NUP98 GLEBS-like motif binds to an RAE1 molecule that is in a complex with mRNP. The presence of the GLEBS-like motif in the resulting complex would then prevent its anchoring to full-length NUP98 at the NPC. In the second possibility, one might argue that the GLEBS-like motif associates with free RAE1 protein to affect its binding to mRNP. Lack of RAE1 within the mRNP particle would then affect its proper anchoring to the NPC.
We found that cells that co-overexpress the GLEBS-like motif together with full-length RAE1 displayed neither reduced levels of RAE1 at the NE nor accumulation of poly(A)+ RNA in the nucleus, indicating that the mRNP export defect induced by the GLEBS-like motif is not an overexpression artifact, but rather a result of impaired RAE1 function. How could RAE1 overexpression neutralize the mRNA export defect induced by the GLEBS-like motif? If the GLEBS-like motif indeed interacts only with RAE1 that is bound to mRNP, as proposed in possibility one (above), then RAE1 overexpression is expected to have little or no effect on the formation of anchoring-defective complexes. However, if the GLEBS-like motif only associates with a free RAE1 molecule, as outlined in possibility two, then RAE1 overexpression could simply titrate GLEBS-like motifs and restore the pool of free RAE1 to a level sufficient for proper mRNP export. Thus, the data from our co-overexpression studies are apparently more consistent with possibility b than a.
Additional data presented in this report are consistent with the idea that RAE1 and the RAE1–NUP98 interaction directly serve in the nuclear export pathway for mRNA. First, NUP98 fragments with point mutations in the GLEBS-like motif that abolish its association with RAE1 failed to inhibit poly(A)+ RNA export from the nucleus. Thus, there is a correlation between nuclear accumulation of poly(A)+ RNA and binding of the GLEBS-like motif to RAE1. Second, there is no evidence to suggest that the observed poly(A)+ RNA accumulation phenotype results from impaired NLS-mediated protein import, loss, clustering, or herniation of nuclear pores, or release of NUP98 from the NE. In particular, the absence of any gross defects in NPC structure and distribution is of importance with respect to earlier work in the yeast system. In gle2 mutant yeast, as well as gle2 or nup116 null alleles (Wente and Blobel, 1993; Murphy et al., 1996), a block in poly(A)+ RNA export is always accompanied by severe clustering and herniation of the nuclear pores. Because of these NPC perturbations, it has been difficult to ascertain whether Gle2p and Nup116p are mediators of RNA transport. Nup116(ΔGLEBS) mutants (Bailer et al., 1998), which are defective in the docking of Gle2p to Nup116p at the NPC, display spatio-structural NPC defects very similar to gle2 or nup116p mutants. Because of these defects, it is difficult to study the export function of the Gle2p– Nup116p interaction in the nup116(ΔGLEBS) mutants. An interesting difference between S. cerevisiae Gle2p and S. pombe Rae1p is that a lack of Rae1p function results in poly(A)+ RNA accumulation in the absence of any detectable NPC defects (Brown et al., 1995; Whalen et al., 1997). It is conceivable that Gle2p and Rae1p functions have diverged, and that Gle2p may perform separate functions in both mRNA transport and NPC structure. Insight into how Gle2p and Rae1p function in the pathway for nuclear export of mRNA is required to address such issues. In S. cerevisiae, it will be important to establish whether the Nup116p–Gle2p interaction at the NPC serves directly in the RNA export pathway. Perhaps GLEBS-overexpression studies similar to those presented in this paper can yield such information. To gain insight into how Rae1p functions in fission yeast, a crucial step will be to identify whether S. pombe contains an S. cerevisiae Nup116p homologue that has a GLEBS-like binding site, and, if so, whether this site directly functions in mRNA export.
Could RAE1 serve as the exportin for mRNA or as a factor that provides the NES signal to mRNA (Ohno et al., 1998; Ullman et al., 1997)? Unlike Rae1p in S. pombe (Brown et al., 1995), Gle2p in S. cerevisiae (Murphy et al., 1996) is not a strictly essential protein indicating that mRNA export is impaired rather than completely blocked in the absence of functional Gle2p. Thus, Gle2p does not seem to operate as the sole exportin or NES-containing factor for mRNA export. Instead, Gle2p and its homologues in fission yeast (Rae1p) and mammals (RAE1) more likely participate in mRNA export as mRNP-interacting proteins necessary for efficient anchoring of the transport complex to the NPC.
Evidence for Specialization of WD-repeats in RAE1
RAE1 is a member of the superfamily of WD-repeat proteins. Given the high conservation of the WD-repeats, it is likely that they all fold into a propeller structure (Garcia-Higuera et al., 1996a). Members of this family do not have an immediately obvious common function. Rather, the common thread between WD-repeat proteins seems to be that they make up part of large macromolecule assemblies. The capacity to assemble multiple proteins may be an essential part of their function, including that of RAE1 (Neer et al., 1994a). The signal-transducing WD-repeat protein Gβ binds to Gγ to form a heterodimer that in turn interacts with Gα or a variety of different effector proteins (Clapham and Neer, 1997). The various interactions with Gβ are established through unique as well as overlapping contact sites involving specific blades of the Gβ propeller (Li et al., 1998). Similarly, the blades of the RAE1 propeller might serve as contact sites for multiple distinct RAE1 partners. This idea has some support from the observation that the interaction of RAE1 and the GLEBS-like motif of NUP98 is highly sensitive to a point mutation in blade 4, but not those in blades 2 and 3. Additionally, studies in S. pombe have demonstrated that a conserved 10-residue basic motif at the carboxy terminus of Rae1p is necessary for Rae1p function. Here, we found that this motif is not essential for the RAE1–NUP98 interaction, which is in keeping with the idea that RAE1 may be involved in an additional protein–protein interaction. A future goal to provide further insight into the complex mechanism of mRNA export from the nucleus is to determine whether RAE1 preferentially interacts with specific kinds of mRNA molecules and to define how RAE1 interacts with mRNP particles. Preliminary in vivo cross-linking studies suggest that RAE1 indeed interacts with at least one other protein besides NUP98.
Acknowledgments
These studies were supported by Cancer Center CORE grant CA-21765 and the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children's Research Hospital. J. van Deursen, L.H. Kasper, and C.E.J. Pritchard were also supported by the National Institutes of Health grant RO1 CA77262-01. M. Fornerod was supported by a European Molecular Biology Organization long-term fellowship.
Abbreviations used in this paper
- AMD
actinomycin D
- DSS
disuccinimidyl suberate
- GST
glutathione-S-transferase
- HA
hemagglutinin
- NE
nuclear envelope
- NES
nuclear export signal
- NLS
nuclear localization signal
- NPC
nuclear pore complex
Footnotes
We are very grateful to Iain Mattaj for his constructive criticisms and generous support throughout this study. We thank Haruhiko Siomi, Susanne Bailer, Richard Bram, Mike Matunis, Eva Neer, and Xiaosheng Wu for helpful discussions, and Paul Brindle and Mutsuhito Ohno for critical reading of the manuscript. Scott Kuersten, Brian Burke, Gerard Grosveld, Hermann Bujard, Derrick Person, Arthur Nienhuis, and Albert Reynolds kindly provided materials. This work would not have been possible without the confocal microscope of Jim Ihle.
References
- Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiaerequired for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Arts GJ, Fornerod M, Mattaj IW. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- Audigier Y. Assays for studying functional properties of in vitro translated Gs alpha subunit. Methods Enzymol. 1994;237:239–254. doi: 10.1016/s0076-6879(94)37066-4. [DOI] [PubMed] [Google Scholar]
- Bailer SM, Siniossoglou S, Podtelejnikov A, Hellwig A, Mann M, Hurt E. Nup116p and nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor gle2p. EMBO (Eur Mol Biol Organ) J. 1998;17:1107–1119. doi: 10.1093/emboj/17.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R, Lin A, Enarson M, Burke B. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J Cell Biol. 1996;134:1141–1156. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathi A, Ghosh A, Whalen WA, Yoon JH, Pu R, Dasso M, Dhar R. The human RAE1 gene is a functional homologue of Schizosaccharomyces pomberae1 gene involved in nuclear export of Poly(A)+ RNA. Gene. 1997;198:251–258. doi: 10.1016/s0378-1119(97)00322-3. [DOI] [PubMed] [Google Scholar]
- Brown JA, Bharathi A, Ghosh A, Whalen W, Fitzgerald E, Dhar R. A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+RNA export and in the cytoskeleton. J Biol Chem. 1995;270:7411–7419. doi: 10.1074/jbc.270.13.7411. [DOI] [PubMed] [Google Scholar]
- Clapham DE, Neer EJ. G protein beta gamma subunits. Annu Rev Pharmacol Toxicol. 1997;37:167–203. doi: 10.1146/annurev.pharmtox.37.1.167. [DOI] [PubMed] [Google Scholar]
- Cole CN, Hammell CM. Nucleocytoplasmic transport: driving and directing transport. Curr Biol. 1998;8:R368–R372. doi: 10.1016/s0960-9822(98)70239-8. [DOI] [PubMed] [Google Scholar]
- Doye V, Hurt E. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Piñol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997a;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO (Eur Mol Biol Organ) J. 1997b;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Fenoglio J, Li Y, Lewis C, Panchenko MP, Reiner O, Smith TF, Neer EJ. Folding of proteins with WD-repeats: comparison of six members of the WD-repeat superfamily to the G protein beta subunit. Biochemistry. 1996a;35:13985–13994. doi: 10.1021/bi9612879. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Gaitatzes C, Smith TF, Neer EJ. Folding a WD repeat propeller. Role of highly conserved aspartic acid residues in the G protein beta subunit and Sec13. J Biol Chem. 1998;273:9041–9049. doi: 10.1074/jbc.273.15.9041. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Thomas TC, Yi F, Neer EJ. Intersubunit surfaces in G protein alpha beta gamma heterotrimers. Analysis by cross-linking and mutagenesis of beta gamma. J Biol Chem. 1996b;271:528–535. doi: 10.1074/jbc.271.1.528. [DOI] [PubMed] [Google Scholar]
- Görlich D. Nuclear protein import. Curr Opin Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- Huang S, Deerinck TJ, Ellisman MH, Spector DL. In vivo analysis of the stability and transport of nuclear poly(A)+RNA. J Cell Biol. 1994;126:877–899. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Jarmolowski A, Beisel C, Mattaj IW, Dreyfuss G, Fischer U. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 1997a;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO (Eur Mol Biol Organ) J. 1997b;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. A cap-binding protein complex mediating U snRNA export. Nature. 1995a;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Mattaj IW. RNA export. Cell. 1995;81:153–159. doi: 10.1016/0092-8674(95)90323-2. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, McGuigan C, Mattaj IW. Nuclear localization of a cap-binding protein complex. Cold Spring Harbor Symp Quant Biol. 1995b;60:669–675. doi: 10.1101/sqb.1995.060.01.072. [DOI] [PubMed] [Google Scholar]
- Jarmolowski A, Boelens WC, Izaurralde E, Mattaj IW. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper LH, Brindle PK, Schnabel CA, Pritchard CEJ, Cleary ML, van Deursen JMA. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19:764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer D, Blobel G. mRNA binding protein mrnp 41 localizes to both nucleus and cytoplasm. Proc Natl Acad Sci USA. 1997;94:9119–9124. doi: 10.1073/pnas.94.17.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Görlich D. Identification of a tRNA specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- Li Y, Sternweis PM, Charnecki S, Smith TF, Gilman AG, Neer EJ, Kozasa T. Sites for Galpha binding on the G protein beta subunit overlap with sites for regulation of phospholipase Cbeta and adenylyl cyclase. J Biol Chem. 1998;273:16265–16272. doi: 10.1074/jbc.273.26.16265. [DOI] [PubMed] [Google Scholar]
- Mehlin H, Daneholt B, Skoglund U. Structural interaction between the nuclear pore complex and a specific translocating RNP particle. J Cell Biol. 1995;129:1205–1216. doi: 10.1083/jcb.129.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Michael WM, Eder PS, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO (Eur Mol Biol Organ) J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Spector DL. The cellular organization of gene expression. Curr Opin Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- Murphy R, Watkins JL, Wente SR. GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombeexport factor RAE1, is required for nuclear pore complex structure and function. Mol Biol Cell. 1996;7:1921–1937. doi: 10.1091/mbc.7.12.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Nuclear export of proteins and RNAs. Curr Opin Cell Biol. 1997;9:420–429. doi: 10.1016/s0955-0674(97)80016-6. [DOI] [PubMed] [Google Scholar]
- Nakielny S, Fischer U, Michael WM, Dreyfuss G. RNA transport. Annu Rev Neurosci. 1997;20:269–301. doi: 10.1146/annurev.neuro.20.1.269. [DOI] [PubMed] [Google Scholar]
- Neer EJ, Denker BM, Thomas TC, Schmidt CJ. Analysis of G-protein alpha and beta gamma subunits by in vitro translation. Methods Enzymol. 1994a;237:226–239. doi: 10.1016/s0076-6879(94)37065-6. [DOI] [PubMed] [Google Scholar]
- Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994b;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- Neer EJ, Smith TF. G protein heterodimers: new structures propel new questions. Cell. 1996;84:175–178. doi: 10.1016/s0092-8674(00)80969-1. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Persons DA, Allay JA, Allay ER, Smeyne RJ, Ashmun RA, Sorrentino BP, Nienhuis AW. Retroviral-mediated transfer of the green fluorescent protein gene into murine hematopoietic cells facilitates scoring and selection of transduced progenitors in vitro and identification of genetically modified cells in vivo. Blood. 1997;90:1777–1786. [PubMed] [Google Scholar]
- Richards SA, Carey KL, Macara IG. Requirement of guanosine triphosphate-bound ran for signal-mediated nuclear protein export. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- Saavedra CA, Hammell CM, Heath CV, Cole CN. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes Dev. 1997;11:2845–2856. doi: 10.1101/gad.11.21.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+RNA and nuclear pores. EMBO (Eur Mol Biol Organ) J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. Dbp5p/ Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO (Eur Mol Biol Organ) J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Stutz F, Kantor J, Zhang D, McCarthy T, Neville M, Rosbash M. The yeast nucleoporin rip1p contributes to multiple export pathways with no essential role for its FG-repeat region. Genes Dev. 1997;11:2857–2868. doi: 10.1101/gad.11.21.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- Tseng SS, Weaver PL, Liu Y, Hitomi M, Tartakoff AM, Chang TH. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+RNA export. EMBO (Eur Mol Biol Organ) J. 1998;17:2651–2662. doi: 10.1093/emboj/17.9.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman KS, Powers MA, Forbes DJ. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- van Deursen J, Boer J, Kasper L, Grosveld G. G2 arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the proto-oncogene CAN/Nup214. EMBO (Eur Mol Biol Organ) J. 1996;15:5574–5583. [PMC free article] [PubMed] [Google Scholar]
- Watkins JL, Murphy R, Emtage JL, Wente SR. The human homologue of Saccharomyces cerevisiae Gle1p is required for poly(A)+RNA export. Proc Natl Acad Sci USA. 1998;95:6779–6784. doi: 10.1073/pnas.95.12.6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K. Importins and exportins: how to get in and out of the nucleus. Trends Biochem Sci. 1998;23:185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- Wente SR, Blobel G. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen WA, Bharathi A, Danielewicz D, Dhar R. Advancement through mitosis requires rae1 gene function in fission yeast. Yeast. 1997;13:1167–1179. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1167::AID-YEA154>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Yi F, Denker BM, Neer EJ. Structural and functional studies of cross-linked Go protein subunits. J Biol Chem. 1991;266:3900–3906. [PubMed] [Google Scholar]