Abstract
Tim44 is a protein of the mitochondrial inner membrane and serves as an adaptor protein for mtHsp70 that drives the import of preproteins in an ATP-dependent manner. In this study we have modified the interaction of Tim44 with mtHsp70 and characterized the consequences for protein translocation. By deletion of an 18-residue segment of Tim44 with limited similarity to J-proteins, the binding of Tim44 to mtHsp70 was weakened. We found that in the yeast Saccharomyces cerevisiae the deletion of this segment is lethal. To investigate the role of the 18-residue segment, we expressed Tim44Δ18 in addition to the endogenous wild-type Tim44. Tim44Δ18 is correctly targeted to mitochondria and assembles in the inner membrane import site. The coexpression of Tim44Δ18 together with wild-type Tim44, however, does not stimulate protein import, but reduces its efficiency. In particular, the promotion of unfolding of preproteins during translocation is inhibited. mtHsp70 is still able to bind to Tim44Δ18 in an ATP-regulated manner, but the efficiency of interaction is reduced. These results suggest that the J-related segment of Tim44 is needed for productive interaction with mtHsp70. The efficient cooperation of mtHsp70 with Tim44 facilitates the translocation of loosely folded preproteins and plays a crucial role in the import of preproteins which contain a tightly folded domain.
Keywords: mitochondria, inner membrane, protein translocation, Tim44, Hsp70
The biogenesis of mitochondria requires the import of nuclear-encoded preproteins from the cytosol. The translocation of preproteins across the mitochondrial membranes is mediated by Tom proteins in the mitochondrial outer membrane and by Tim proteins in the inner membrane (Pon and Schatz, 1991; Ryan and Jensen, 1995; Schatz, 1996; Neupert, 1997; Pfanner et al., 1997; Pfanner and Meijer, 1997). Tim441 was the first Tim protein identified (Maarse et al., 1992; Scherer et al., 1992). The gene encoding Tim44 is essential. Tim44 is synthesized as a precursor protein of 49 kD and processed inside mitochondria to a mature protein of 44 kD (Maarse et al., 1992). It behaves like a peripheral inner membrane protein and is exposed to the matrix (Blom et al., 1993; Rassow et al., 1994). Tim44 was found as a component of complexes together with the integral membrane proteins Tim23 and Tim17 (Berthold et al., 1995; Blom et al., 1995; Bömer et al., 1997) which are putative components of a protein translocation channel (Bauer et al., 1996; Dekker et al., 1997; Jensen and Kinnally, 1997; Lohret et al., 1997; Ryan et al., 1998; Rassow et al., 1999).
Recent efforts to determine the function of Tim44 have concentrated on the finding that Tim44 binds to mitochondrial Hsp70 (mtHsp70) in a 1:1 complex (Kronidou et al., 1994; Rassow et al., 1994; Schneider et al., 1994). Cells combining mutations in the genes encoding these proteins show synthetic growth defects (Rassow et al., 1994). mtHsp70 is essential for driving preproteins across the membranes into the matrix (Kang et al., 1990) and seems to constitute the motor unit of mitochondrial protein import (Pfanner and Meijer, 1995; Rassow and Pfanner, 1995; Schatz, 1996). Nucleotides and cochaperones are involved in the complex formation between both proteins. mtHsp70 initially binds to Tim44 in the ATP-bound state, but the ATP is rapidly hydrolyzed (von Ahsen et al., 1995; Horst et al., 1996; Schneider et al., 1996; Ungermann et al., 1996). The interaction with nucleotides is modulated by Mge1, the mitochondrial homologue of the prokaryotic heat shock protein GrpE. Mge1 is associated with the Tim44 complex in substoichiometric amounts, acts as a nucleotide release factor, and is required for the efficient function of the import motor (Nakai et al., 1994; Voos et al., 1994; Laloraya et al., 1995; Westermann et al., 1995; Deloche and Georgopoulos, 1996; Schneider et al., 1996; Dekker and Pfanner, 1997; Deloche et al., 1997a; Miao et al., 1997). Mdj1p and Mdj2p, the mitochondrial homologues of prokaryotic DnaJ, are not associated with the Tim44 complex and there is no indication of an involvement in preprotein translocation (Rowley et al., 1994; Westermann et al., 1996; Deloche et al., 1997b; Westermann and Neupert, 1997).
The binding site of Tim44 for mtHsp70 is not known. Therefore, it is interesting to note that the sequence of Tim44 contains a short stretch of 18 amino acids (residues 185–202) which shows some similarity to a part of the J-domain which is characteristic of DnaJ and related modulators of Hsp70 proteins (Silver and Way, 1993; Cyr et al., 1994; Rassow et al., 1994; Hartl, 1996; Bukau and Horwich, 1998; Cheetham and Caplan, 1998; Kelley, 1998). This observation is reminiscent of several systems in which J-domains are the structures by which Hsp70s are bound to partner proteins and customized for specific functions (Rassow et al., 1995). It is tempting to speculate that the 18 residues of the J-similarity region of Tim44 may contribute to the interaction with mtHsp70. In this case Tim44 could function in analogy to Sec63p, the membrane protein of the ER membrane which binds BiP and thereby plays an important role in the transport of proteins into this organelle (Scidmore et al., 1993; Schlenstedt et al., 1995; Brodsky, 1996; Rapoport et al., 1996; Corsi and Schekman, 1997). However, the similarity of Tim44 to J-domains is very limited, raising the question of whether the 18-residue segment in the sequence of Tim44 is of functional relevance for the protein.
Similarly unclear is the role of Tim44 in the mechanism of mtHsp70-driven protein import. A complete inactivation of Tim44 retains the mitochondrial protein import channels intact but causes defects in the translocation of the mature parts of matrix-targeted preproteins (Bömer et al., 1998). Therefore, it is possible that the only function of Tim44 is the cooperation with mtHsp70. Since up to now no method was available to specifically modulate the interaction between Tim44 and mtHsp70, suggestions on the involvement of Tim44 in the mechanism of protein transport were often based on circumstantial evidence or referred to studies using temperature-sensitive mutants of the SSC1 gene encoding mtHsp70. In these mutants, the interactions between Tim44 and mtHsp70 are impaired, but additional defects and thus indirect effects could not be ruled out. In fact, there is evidence that the mitochondrial Tim machinery contains two binding sites for mtHsp70, one site directly at Tim44 and a second binding site at the Tim23/Tim17 complex (Rassow et al., 1995; Voos et al., 1996; Bömer et al., 1997). Therefore, it was unclear whether the defects in mitochondrial protein import which are observed with the ssc1-mutant strains are due to the impaired interaction of mtHsp70 with Tim44 or due to other functions of mtHsp70.
In this study, we asked whether residues 185–202 of Tim44 are required for the function of Tim44. We found that Tim44 lacking this segment (Tim44Δ18) is unable to substitute for authentic Tim44 in a yeast strain lacking the wild-type TIM44 gene although Tim44Δ18 is correctly imported into mitochondria and assembled into the inner membrane. Our results indicate that the 18-residue segment of Tim44 is required for the efficient interaction of Tim44 with mtHsp70. Moreover, we find that this interaction is necessary for the efficient action of mtHsp70 on translocating folded preproteins and thus for the full activity of the mitochondrial protein import motor.
Materials and Methods
Construction of Strains and Plasmids
For expression of Tim44 in vivo, a 2.7-kb HindIII fragment containing the TIM44 gene (Maarse et al., 1992) was cloned into YEplac33 and YEplac181 (Gietz and Sugino, 1988). tim44 Δ18 was constructed using the Promega Altered Sites System. The 2.7-kb HindIII TIM44 fragment was cloned into pSELECT-1 (= pALTER-1) and mutagenized with the mutagenic oligonucleotide 5′CAAAGGAGACTTAAACGTGCAGGAACAGCAGTGG3′. The 54-bp deletion was verified by DNA sequence analysis. The mutagenized HindIII fragment was subsequently cloned into YEplac181 (as a multi-copy vector) or YCplac111 (as a single-copy vector), respectively. The following Saccharomyces cerevisiae strains were used: PK82 (MATα his4-713 lys2 ura3-52 Δtrp1 leu2-3,112; Gambill et al., 1993), PK81 (MATα ade2-101 lys2 ura3-52 leu2-3,112 Δtrp1 ssc1-2(LEU2); Gambill et al., 1993), MB3 (MATα ade2-101 his3-Δ200 leu2-Δ1 lys2-801 ura3::LYS2; Maarse et al., 1992), and MB20 (MATa ade2- trp1- ura3- leu2- TIM44::LYS2 + YEplac33::TIM44(URA3); this study). For plasmid shuffling, double transformed cells were grown in rich liquid broth (YPD) and then plated on solid medium containing 5-fluoro-orotic acid according to Boeke et al. (1987).
Import of Preproteins into Isolated Mitochondria, Cross-linking by EGS, and Blue Native Electrophoresis
The cDNA constructs encoding mitochondrial preproteins were cloned into pGEM4 (Promega), the transcription was performed using SP6-RNA-polymerase (Stratagene), and the precursor proteins were subsequently synthesized in rabbit reticulocyte lysate in the presence of [35S]methionine and [35S]cysteine (Amersham). For import experiments, mitochondria were isolated from strains of S. cerevisiae following a standard protocol (Daum et al., 1982). The wild-type strain PK82 or the strain MB3 containing different expression plasmids was used, as indicated in the figure legends. The import assays were carried out following standard procedures as published previously (Söllner et al., 1991; Alconada et al., 1995). In brief, the mitochondria (20 μg protein/100-μl assay) were incubated with reticulocyte lysate containing the preproteins in import buffer (3% [wt/vol] BSA, 250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 10 mM MOPS-KOH, pH 7.2) at 25°C for up to 30 min. Treatment with proteinase K (50 μg/ml) was performed at 0°C for 10 min. The protease was stopped by addition of 1 mM PMSF. The mitochondria were reisolated by centrifugation at 16,000 g at 2°C for 10 min.
For cross-linking experiments, the hybrid protein Su9-DHFR was accumulated in import sites by import into ATP-depleted mitochondria. Mitochondria (in the presence of 20 μM oligomycin to inhibit the F0F1-ATPase) and reticulocyte lysate were depleted of ATP by incubation with apyrase (10 U/ml; Sigma) for 10 min at 0°C. The import was performed in the presence of 20 μM oligomycin. The mitochondria were washed by centrifugation through a sucrose cushion (500 mM sucrose, 1 mM EDTA, 10 mM MOPS, pH 7.2) and resuspended in 250 mM sucrose, 1 mM EDTA, 10 mM MOPS-KOH, pH 7.2 (SEM buffer). The mitochondria (100 μg protein) were incubated at 0°C for 20 min in 1 ml SEM containing 0.1 mg/ml ethylene glycolbis succinimidylsuccinate (EGS). The reaction was stopped by addition of 100 mM Tris-HCl, pH 7.2, and a second incubation of 20 min at 0°C. Proteins were precipitated by addition of 10% trichloroacetic acid in the presence of 0.0125% deoxycholate. For immunoprecipitation, the precipitates were lysed in 20 μl 1% SDS, 60 mM Tris-HCl, pH 6.8. The samples were diluted 40-fold by addition of 1% (wt/vol) Triton X-100, 0.3 M NaCl, 10 mM Tris-HCl, pH 7.5, and the antibodies bound to protein A–Sepharose (Pharmacia) were added.
Blue native electrophoresis was essentially performed following the protocols of Schägger and von Jagow (1991). The samples were prepared as described by Dekker et al. (1997).
Fractionation of Mitochondria
The procedures of protease treatment, swelling of mitochondria, and carbonate extraction were published previously (Blom et al., 1993; Rassow et al., 1994). The tendency of mitochondrial proteins to form aggregates was tested by lysis of the organelles and subsequent centrifugation. Mitochondria from the wild-type and from the strain expressing Tim44Δ18 in addition to the authentic Tim44 (40 μg protein) were lysed in 0.1% Triton X-100, 250 mM sucrose, 5 mM EDTA, 80 mM KCl, 10 mM MOPS, pH 7.5. The samples were divided into three parts, one part was incubated for 1 h at 0°C, the second part at 25°C, the third part was immediately mixed with trichloroacetic acid. After 1 h, the samples which had been left without trichloroacetic acid were subjected to a centrifugation of 10 min at 16,000 g and 2°C. Proteins were precipitated from the supernatants by addition of 10% trichloroacetic acid. For sonication the mitochondria (40 μg protein) were suspended in 500 μl 1 mM EDTA, 1 mM EGTA, 30 mM Tris, pH 7.5, 1 mM PMSF, 0.5 mM o-phenantroline and sonicated by a Branson Sonifier 10 times for 10 s (30% duty cycle, output 3) with intervals of 6 min for cooling of the samples. The membranes were pelleted by centrifugation at 100,000 g for 30 min. Proteins were precipitated from the supernatants by addition of 10% trichloroacetic acid.
Coimmunoprecipitations
For coimmunoprecipitations, mitochondria (25 μg/sample) were lysed in 200 μl 250 mM sucrose, 80 mM KCl, 20 mM MOPS-KOH, pH 7.2, 0.1% (vol/vol) Triton X-100, 5 mM EDTA, 0.5 mM PMSF. In the experiment shown in Fig. 6 F, the mitochondria were lysed in 150 mM NaCl, 0.1% Triton X-100, 0.5 mM PMSF, 10 mM Tris-HCl, pH 7.4. After a spin of 16,000 g for 10 min, the lysates were incubated with antibodies directed against mtHsp70. The antibodies were covalently coupled to protein A–Sepharose as described previously (Voos et al., 1994).
Figure 6.
Deletion of the 18-residue segment of Tim44 reduces complex formation with mtHsp70. (A) Coimmunoprecipitation of Tim44 and Tim44Δ18 with antibodies directed against mtHsp70. Mitochondria were lysed in the presence of detergent and protein complexes were precipitated during an incubation of 1 h by antibodies raised against mtHsp70. The precipitates were analyzed by SDS-PAGE, Western blotting, and immunostaining, using polyclonal antisera raised against Ssc1p (Hsp70) or Tim44. Sample 1 is total protein of wild-type mitochondria (10 μg protein). Sample 2 is Tim44-mtHsp70 complex precipitated from wild-type mitochondria (100 μg protein). (B) Sample 1 is total protein (10 μg protein); sample 2 is complex precipitated from mitochondria (100 μg protein) of the strain coexpressing Tim44Δ18 together with authentic Tim44. (C) Samples 1 and 2 are total protein (10 μg) of mitochondria from the same strain as in B. To compare the stability of Tim44Δ18 and Tim44 in the lysis buffer, the proteins of sample 1 were precipitated by trichloroacetic acid immediately after lysis of the mitochondria; the proteins of sample 2 were precipitated after the lysed mitochondria had been incubated for 1 h at the same temperature as the coimmunoprecipitations. Samples 1 and 3 are similar to the samples of B, except that the lysis buffer contained 80 mM KCl instead of 10 mM KCl. The different stability of the complexes formed between mtHsp70 and Tim44 versus mtHsp70 and Tim44Δ18 is indicated by the ratio of the bands corresponding to both forms of Tim44 in the immunoprecipitation (lane 3) in comparison to the mitochondria (lanes 1 and 2). (D) Ratio of the amounts of Tim44Δ18 and Tim44 as determined by quantifications of coimmunoprecipitations by antibodies against mtHsp70 in comparison to the ratio of both proteins in total mitochondria (as shown in B and C). Columns 1 and 2 are coimmunoprecipitation in the presence of 10 mM KCl; columns 3 and 4 are coimmunoprecipitation in the presence of 80 mM KCl. (E) Release of Tim44 and Tim44Δ18 from mtHsp70 in the presence of ATP. The complex of Tim44 and Tim44Δ18 with mtHsp70 was precipitated by protein A–Sepharose-bound antibodies against mtHsp70 (−ATP). Parallel samples were incubated in the presence of 2 mM ATP and 5 mM MgCl2 (+ATP). The protein A–Sepharose was separated from the supernatant by centrifugation and the bound complex was eluted at pH 2.5. The proteins were precipitated by trichloroacetic acid and analyzed by SDS-PAGE and immunoblotting. The relative amounts of coprecipitated Tim44 and Tim44Δ18 were quantified; the combined amounts of bound and free protein were set to 100% (total). Pellet is the coimmunoprecipitated protein (released from the protein A–Sepharose at pH 2.5); Sup. is protein in the supernatant of the coimmunoprecipitation. (F) Interaction of Tim44 and Tim44Δ18 with wild-type mtHsp70 and with mutant mtHsp70 encoded by the allele ssc1-2. Tim44Δ18 was expressed from a single-copy vector in the strain ssc1-2; mitochondria were isolated (containing both the authentic Tim44 and Tim44Δ18), lysed, and analyzed by coimmunoprecipitation using the antibodies raised against mtHsp70. Mitochondria from a strain containing wild-type mtHsp70 were analyzed in parallel (WT). Shown are the relative amounts of Tim44 and Tim44Δ18, respectively. The relative amounts of coprecipitated protein obtained in the strain expressing wild-type mtHsp70 were set to 100%. To determine the affinity of both forms of mtHsp70 to a preprotein, Su9-DHFR was imported into mitochondria of the strain ssc1-2 and the corresponding wild-type strain. The imported Su9-DHFR was coimmunoprecipitated by the antibodies raised against mtHsp70. After quantification, the amounts of coprecipitated Su9-DHFR were calculated relative to the amounts of imported Su9-DHFR. The ratio found for binding of Su9-DHFR to Ssc1-2p was set to 100%.
Assessment of the Mitochondrial Membrane Potential
The membrane potential (Δψ) of isolated yeast mitochondria was determined by recording the fluorescence decrease of the voltage-sensitive dye 3,3′-dipropylthiacarbocyanine iodide [DiSC3(5); Molecular Probes; Sims et al., 1974]. The assays were performed using a Perkin-Elmer 640-40 fluorescence spectrometer at 25°C (excitation at 622 nm, slit width 5 nm). The mitochondria (100 μg protein) were incubated in 1 ml of 0.6 M sorbitol, 0.1% (wt/vol) BSA, 80 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, pH 7.4. The final concentration of the fluorescent dye DiSC3(5) was 3.6 μM. The membrane potential was dissipated by the addition of 3 μM (final concentration) KCN. The difference between the fluorescences before and after the addition of KCN represents a rough assessment to the membrane potential.
Results
A Segment of Tim44 Essential for Viability of S. cerevisiae
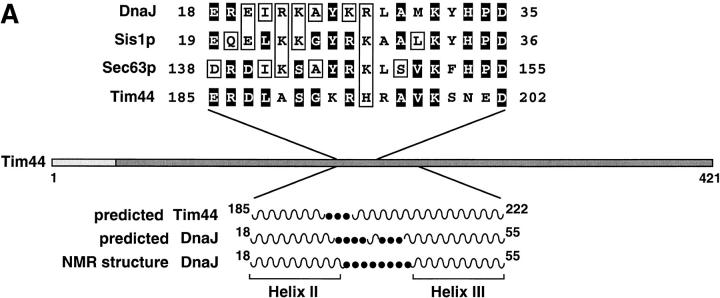
The region of sequence similarity between Tim44 and characteristic J-domains is shown in Fig. 1 A. The similarity extends from residue 185 to residue 202 and is depicted for the two yeast proteins Sec63p (Sadler et al., 1989; Feldheim et al., 1992) and Sis1p (Luke et al., 1991; Zhong and Arndt, 1993), and for the Escherichia coli protein DnaJ (Ohki et al., 1986; Liberek et al., 1991). The structure of the J-domain of DnaJ has been determined by NMR spectroscopy (Szyperski et al., 1994; Hill et al., 1995). The J-similarity segment of Tim44 corresponds to α helix II of DnaJ (residues 18–30) and the first half of the following turn region. In DnaJ, the turn is followed by helix III comprising residues 41–55. The secondary structures which are predicted for the corresponding part of Tim44 similarly show two hydrophilic α helices which are connected by a putative turn element. However, it should be emphasized that the similarity of Tim44 to J-domains is still very limited. Tim44 obviously does not belong to the family of homologous J-proteins.
Figure 1.
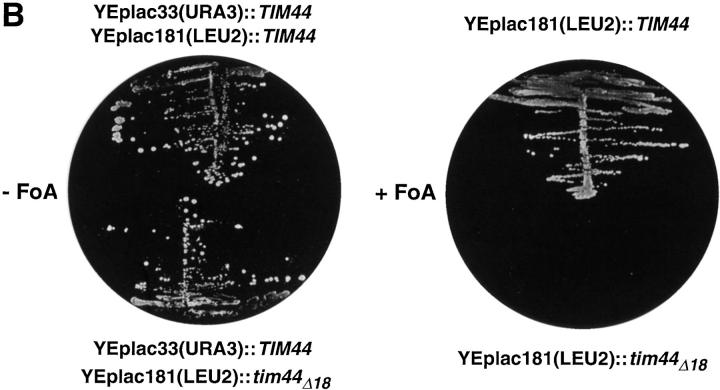
Deletion of residues 185–202 in Tim44 is lethal for S. cerevisiae. (A) The J-similar segment of Tim44. Shown is a comparison of the amino acid sequence of S. cerevisiae Tim44 residues 185–202 with sequences in E. coli DnaJ, S. cerevisiae Sis1p, and Sec63p. Dark boxes are identical residues; white boxes are similar amino acids. Secondary structure predictions were performed according to Garnier and Robson using the program Protean. Indicated is the number of the residues in the corresponding amino acid sequences of the compared proteins. Serpentine lines are the predicted α helix; dark dots are predicted unstructured amino acids (turn or coil). Helix II and helix III of DnaJ were defined according to the NMR structure. (B) The segment of residues 185–202 in Tim44 is essential for viability of yeast. Strain MB20 contains a deletion of chromosomal TIM44 sequences and harbors the multicopy plasmid YEplac33 (with URA3 as a selective marker) encoding authentic Tim44. (Left plate) MB20 was transformed with the LEU2 marked 2-μm plasmid YEplac181 carrying the wild-type TIM44 gene (upper half of the plate) or tim44 Δ18 (lower part of the plate). (Right plate) Double transformants were grown in nonselective rich broth (YPD) and then plated on solid medium containing glucose as a carbon source and 5-fluoro-orotic acid (FOA) for selection of cells which have lost the URA3 marked expression plasmids. The cells retaining YEplac181 containing the authentic TIM44 coding sequence grew normally (upper half of the plate). Cells retaining only the Tim44Δ18-encoding construct were not viable (lower half of the plate).
To determine the relevance of the J-similar segment for the function of Tim44 in vivo, we constructed a plasmid encoding a Tim44 with a deletion of residues 185–202 (Tim44Δ18). We then tested whether Tim44Δ18 can substitute for authentic Tim44 in a genetic assay (Fig. 1 B). In a strain of S. cerevisiae expressing both forms of Tim44 from different plasmids, we used the URA3/FOA technique to deplete the authentic Tim44. It turned out that the cells lacking the authentic Tim44 were not viable. Tim44Δ18 could not substitute for wild-type Tim44, and neither on glycerol nor on glucose was any growth observed. Thus, this result demonstrates that the 18-residue segment of Tim44 is essential for viability of yeast.
Tim44 Lacking Residues 185–202 Is Imported into Mitochondria and Reaches Its Functional Location
The lethality of the 18-residue deletion could be caused by two different reasons. Either the 18-residue segment is crucial for the biogenesis of Tim44, or this segment is required for the function of Tim44 within the mitochondria. To address the first possibility, we expressed Tim44Δ18 in reticulocyte lysate and tested whether the protein could be imported into isolated mitochondria. In parallel we imported the authentic Tim44 (Fig. 2 A). Both preproteins were processed to a mature form (Fig. 2 A, lanes 1 and 5) and translocated into a location protected against externally added proteinase K (lanes 3 and 7). The import was dependent on the mitochondrial membrane potential, confirming the specificity of the reaction. The result demonstrates that the 18-residue segment of Tim44 is not required for efficient transport into mitochondria in vitro.
Figure 2.
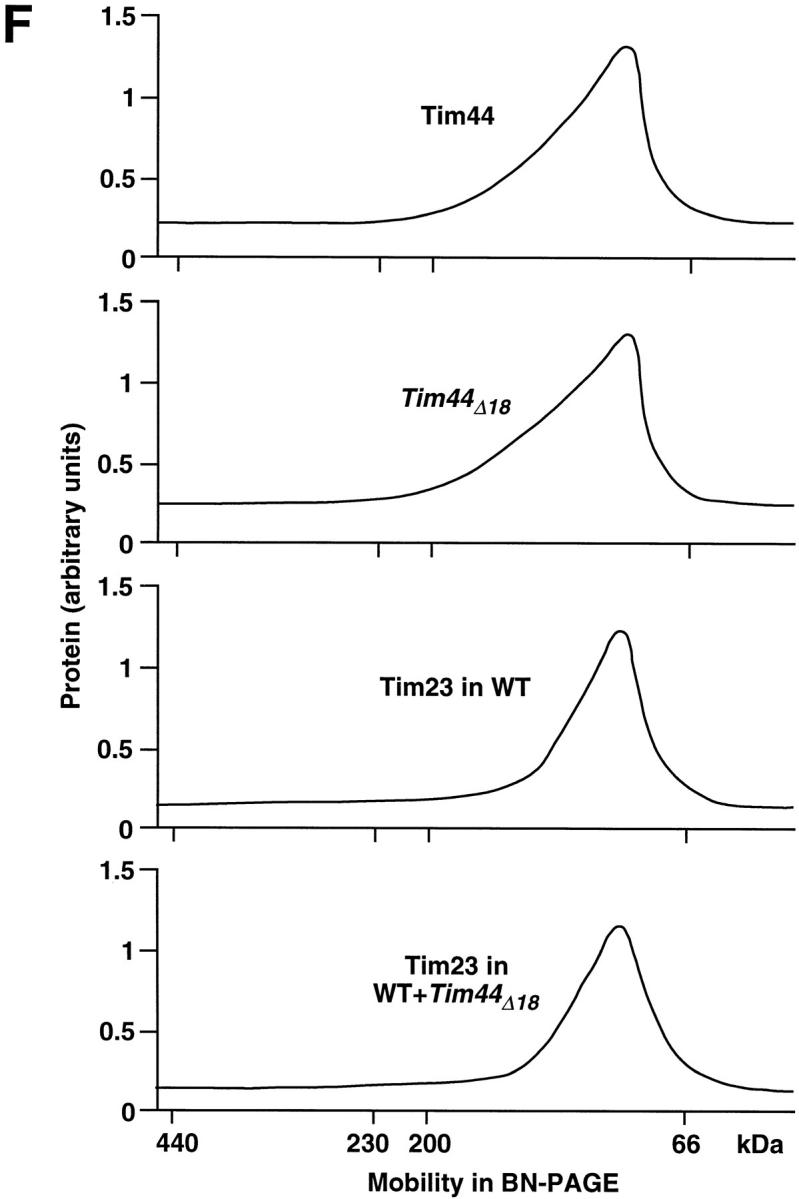

Tim44Δ18 is efficiently imported into mitochondria. (A) Import of Tim44Δ18 in vitro and in vivo. Lanes 1–8 show the import of 35S-labeled proteins into isolated mitochondria. Wild-type Tim44 and Tim44Δ18 (containing a deletion of residues 185–202) were synthesized in reticulocyte lysate and incubated with mitochondria isolated from a S. cerevisiae wild-type strain. Samples 2, 4, 6, and 8 contained valinomycin to dissipate the membrane potentia (Δψ). The mitochondria were reisolated and treated with proteinase K (Prot. K) as indicated. The imported proteins were analyzed by SDS-PAGE and fluorography. p, precursor protein; m, mature protein. Lanes 9 and 10 show the import of Tim44 and Tim44Δ18 in vivo. The multi-copy plasmid YEplac181 encoding Tim44Δ18 was transformed into the yeast wild-type strain MB3. Mitochondria were isolated to determine the expression and import of Tim44Δ18 and of the authentic Tim44. The mitochondrial proteins were separated by SDS-PAGE and analyzed by Western blotting. Tim44 and mtHsp70 were visualized by specific antibodies. WT, wild-type mitochondria; WT + Tim44Δ18, mitochondria from the strain MB3 expressing Tim44Δ18 in addition to the authentic Tim44. The same mitochondria as in lane 10 were analyzed in the experiments shown in B–E. (B) Fractionation of mitochondria. Mitochondria isolated from the yeast strain expressing both authentic Tim44 and Tim44Δ18 were swollen to open the intermembrane space (lanes 4–6) or sonified to open the matrix (lanes 3 and 6) and tested for the accessibility of Tim44 by trypsin. Both forms of Tim44 became accessible for the protease only after sonication. (C) Solubility of Tim44Δ18 after lysis of the mitochondria. Mitochondria containing authentic Tim44 together with Tim44Δ18 were lysed in the presence of 0.1% Triton X-100 and 80 mM KCl, and after a spin of 5 min at 16,000 g the supernatants were incubated for 1 h at 0 or 25°C, respectively. The samples were subsequently subjected to a centrifugation of 1 h at 100,000 g av. Supernatants and pellets were separated and analyzed by SDS-PAGE and Western blotting. The blots were decorated with antibodies directed against Tim44 and mtHsp70. (D) Carbonate extraction. Mitochondria containing Tim44 and Tim44Δ18 were suspended in 100 mM sodium carbonate (Na2CO3) and incubated at 0°C for 20 min. Subsequently, the membranes were pelleted by centrifugation and the proteins of the supernatant were precipitated by trichloroacetic acid. Control, 50% of the total material applied in the carbonate extraction. AAC, ADP/ATP carrier. (E) Sonication of mitochondria containing authentic Tim44 and Tim44Δ18. The mitochondria were sonified at different concentrations of KCl as described in Materials and Methods. The membranes were pelleted by centrifugation and the distribution of proteins between membranes and supernatant was determined by SDS-PAGE, Western blotting, and immunostaining. Shown is the amount of proteins retained in the pelleted membranes in percentages of the total amount of protein applied to the samples. (F) Two-dimensional electrophoresis of mitochondrial protein complexes. Mitochondria isolated from a strain expressing Tim44Δ18 in addition to authentic Tim44 were lysed in the presence of digitonin and subjected to blue native electrophoresis (first dimension). After separation, a lane was cut out and analyzed by SDS-PAGE to determine the distribution of individual proteins (second dimension). The gel was blotted and analyzed by decoration with antibodies raised against Tim44. The pattern of labeled protein was quantified by densitometry. The upper panel is authentic Tim44; the second panel is Tim44Δ18. Complex formation of Tim23 was analyzed essentially following the same procedure. The third panel is mitochondria isolated from the wild-type strain; the lower panel is mitochondria isolated from the strain expressing Tim44Δ18 in addition to authentic Tim44. The blots were decorated using an antiserum raised against a peptide corresponding to an amino-terminal segment of Tim23.
To test whether Tim44Δ18 is similarly imported in vivo, Tim44Δ18 was expressed in a S. cerevisiae wild-type strain from a multi-copy vector, and mitochondria were isolated to determine the amount of imported protein (Fig. 2 A, lanes 9 and 10). We found that the mitochondria had imported both the authentic Tim44 and Tim44Δ18. The amount of authentic Tim44 was not significantly reduced as compared with the original wild-type strain; Tim44Δ18 was overexpressed about threefold. The location of the imported Tim44 proteins was confirmed by a fractionation experiment. Both proteins were resistant against externally added trypsin in intact mitochondria (Fig. 2 B, lane 2) and after opening of the intermembrane space by swelling (Fig. 2 B, lane 5). Only after disruption of the inner membrane by sonication did the proteins become accessible for the protease (Fig. 2 B, lanes 3 and 6), in agreement with results which were obtained previously for the wild-type Tim44 (Blom et al., 1993; Rassow et al., 1994). After lysis of the mitochondria by detergent, both proteins were similarly soluble (Fig. 2 C). Even after prolonged incubation at 0 or 25°C, no formation of aggregates was observed (Fig. 2 C, lanes 2 and 4). We then tested whether the deletion of the 18-residue segment may affect the association of Tim44 with the inner membrane. The extraction of Tim44 and Tim44Δ18 was monitored by sodium carbonate and again both proteins showed the same behavior (Fig. 2 D). In contrast to the ADP/ATP carrier (AAC) which was resistant against this treatment, both Tim44 proteins were extracted. The association of Tim44 with the inner membrane was also determined in the presence of different salt concentrations (Fig. 2 E). Mitochondria were sonicated in the presence of up to 500 mM KCl and the membranes were subsequently pelleted by centrifugation. Tim44 and Tim44Δ18 were found stably associated with the membranes; only minor amounts of both proteins were released at higher ionic strength. As a control, we followed the distribution of Mge1p which was soluble in all samples. The AAC was completely resistant against extraction.
Eventually, we investigated the involvement of both forms of Tim44 in the formation of high molecular weight complexes using the method of blue native electrophoresis (BN-PAGE; Schägger and von Jagow, 1991; Dekker et al., 1997). As shown in Fig. 2 F, Tim44 and Tim44Δ18 both showed the same distribution, suggesting that both proteins participate in the same interactions with the components of the Tim machinery. In a previous study we found that Tim44 is mainly associated with Tim23 (Bömer et al., 1997). Therefore, we also determined the complex formation of Tim23 and found that it showed the same running behavior in the BN-PAGE, irrespective if only the authentic Tim44 was present or if Tim44Δ18 was overexpressed in addition.
We conclude from these experiments that Tim44Δ18 is efficiently imported into mitochondria and acquires the correct topology at the inner side of the inner membrane. The results of the BN-PAGE indicate that Tim44Δ18 adopts the native folding state and engages in the same interactions with the Tim machinery as the authentic Tim44. These conclusions are corroborated by the interactions of Tim44Δ18 with different forms of mtHsp70 (see below, Fig. 6 F).
Tim44Δ 18 Interacts with Mitochondrial Preproteins
If Tim44Δ18 is recruited by the Tim machinery, is it also present at import sites during translocation of preproteins? We addressed this question by chemical cross-linking (Fig. 3). As a substrate we synthesized the hybrid protein Su9-DHFR, containing the presequence of subunit 9 of the mitochondrial ATP synthase fused to the complete DHFR. The import of this protein is dependent on the membrane potential Δψ (Fig. 3 A, lanes 1–4) and ATP (Fig. 3 A, lanes 5–8). After depletion of ATP, Su9-DHFR is accumulated in import sites as a membrane-spanning translocation intermediate. Following this protocol, Su9-DHFR was accumulated in mitochondria of both the Tim44Δ18 overproducing strain and the wild-type (Fig. 3 B, lanes 1 and 2). The translocation intermediates were cross-linked to the proteins in the vicinity by addition of the reagent EGS. In wild-type mitochondria, two products were formed, corresponding to the precursor form and the processing intermediate of Su9-DHFR (Fig. 3 B, lane 4, bands labeled Tim44* and Tim44**; Blom et al., 1993). In mitochondria, which in addition contained Tim44Δ18, a third cross-linking product was formed which could be precipitated by antibodies against Tim44 (Fig. 3 B, lane 7, band labeled Tim44***). No reaction product was precipitated by the preimmune serum. According to its size in the SDS-PAGE, the additional product corresponds to cross-linking of Tim44Δ18 to the processing intermediate of Su9-DHFR. The cross-linking of a protein in transit across the mitochondrial membranes confirms that Tim44Δ18 is present at protein import sites.
Figure 3.
Tim44Δ18 can be chemically cross-linked to a precursor in transit through the mitochondrial membranes. (A) Import of the preprotein Su9-DHFR. The preprotein contains the first 70 amino acid residues of subunit 9 of the mitochondrial ATP synthase fused to the complete DHFR. Radiolabeled Su9-DHFR was incubated with isolated wild-type mitochondria at 25°C for the times indicated. In samples 1 and 2 the membrane potential (Δψ) was dissipated by addition of valinomycin. Samples 1–4 were supplemented with 2 mM ATP, the reticulocyte lysate of samples 5–8 was pretreated with apyrase to deplete the ATP. After incubation at 25°C, the samples were divided into halves, and one-half was treated with 50 μg/ml proteinase K (Prot. K). The samples were analyzed by SDS-PAGE and digital autoradiography. The precursor form of Su9-DHFR (p) is processed in two steps, first between residues 35 and 36 to the intermediate form (i) and subsequently between residues 66 and 67 to the mature form (m). (B) Cross-linking of a membrane spanning intermediate of Su9-DHFR to Tim44Δ18. Radiolabeled Su9-DHFR was accumulated as a translocation intermediate by import into isolated apyrase-pretreated mitochondria from the wild-type (lanes 1 and 3–5) and from the strain coexpressing Tim44Δ18 (lanes 2 and 6–8). For cross-linking, the samples were incubated with EGS as described in Materials and Methods (lanes 4, 5, 7, and 8). After cross-linking, mitochondria were lysed and reaction products were immunoprecipitated with antiserum against Tim44 (lanes 3, 4, 6, and 7) or preimmune serum (lanes 5 and 8). The immunoprecipitates were analyzed by SDS-PAGE and fluorography. The cross-link products of lanes 4 and 7 are separately shown in magnification. Tim44*, Tim44**, and Tim44*** are reaction products of radiolabeled Su9-DHFR (precursor form and processing intermediate) precipitated by the serum against Tim44. Tim44*** corresponds to cross-linking of processed Su9-DHFR to Tim44Δ18.
Coexpression of Tim44Δ 18 Causes Membrane Potential–independent Defects in Protein Import
To determine the possible role of the 18-residue segment in Tim44, additional preproteins were imported and tested for defects in distinct steps of translocation across the mitochondrial membranes. In a first series of experiments, the β subunit of the mitochondrial ATP synthase (F1β) was synthesized in reticulocyte lysate in the presence of [35S]methionine/[35S]cysteine and imported into mitochondria which were isolated from the Tim44Δ18-overexpressing strain and the corresponding wild-type (Fig. 4 A). It is known from previous studies that the import of F1β is very sensitive against defects in the import machinery and requires the mtHsp70-dependent unfolding machinery of the mitochondria (Kang et al., 1990; Rassow and Pfanner, 1991; Gambill et al., 1993). We now observed only a slight reduction in processing of F1β, suggesting that the deletion of the 18-residue segment of Tim44 does not cause major changes in the insertion of the presequence into the Tim machinery of the inner membrane (Fig. 4 A, top). However, a protease-protection assay revealed a delay in the translocation of the mature part of the preprotein (Fig. 4 A, bottom), indicating that the deletion affected the completion of translocation. After an import time of 20 min the efficiency of translocation was reduced by 75–80% (Fig. 4 B, column 2 vs. column 3). Some reduction was also observed after overexpression of the authentic Tim44, but the effect was much less pronounced (Fig. 4 B, column 3, and see below, Fig. 5 C).
Figure 4.
Translocation of a preprotein across the mitochondrial membranes is impaired in the presence of Tim44Δ18. (A) Import of the β subunit of the mitochondrial ATP synthase (F1β). Mitochondria were isolated from the strain overexpressing Tim44Δ18 in addition to the authentic Tim44 (WT + Tim44Δ18) and the corresponding wild-type strain MB3 containing only the authentic Tim44 (WT). 35S-labeled F1β was synthesized in reticulocyte lysate and incubated with isolated mitochondria at 25°C for the times indicated. In samples 4 and 8 the membrane potential (Δψ) was dissipated by addition of valinomycin. After incubation at 25°C, the samples were cooled to 0°C and divided into halves. One-half was treated with 50 μg/ml proteinase K for 20 min (+ Prot. K), the other half was left without protease (− Prot. K). After incubation with PMSF, the samples were analyzed by SDS-PAGE and digital autoradiography using a PhosphorImaging system. (B) Import of F1β into mitochondria isolated from the wild-type (column 1), in mitochondria isolated from the strain expressing Tim44Δ18 in addition to authentic Tim44 (column 2), and in mitochondria from the strain overexpressing the authentic Tim44 (column 3). F1β was imported for 20 min at 25°C following the protocol described in A. The amounts of processed and protease-protected F1β were quantified using the PhosphorImager. The value obtained for import into wild-type mitochondria was set to 100% (control). (C) Assessment of the mitochondrial membrane potential. The membrane potential Δψ was determined at 25°C, using the fluorescence dye 3,3′-dipropylthiadi-carbocyanine iodide, DiSC3(5). The Δψ is indicated by the difference in fluorescence before and after addition of potassium cyanide (KCN).
Figure 5.
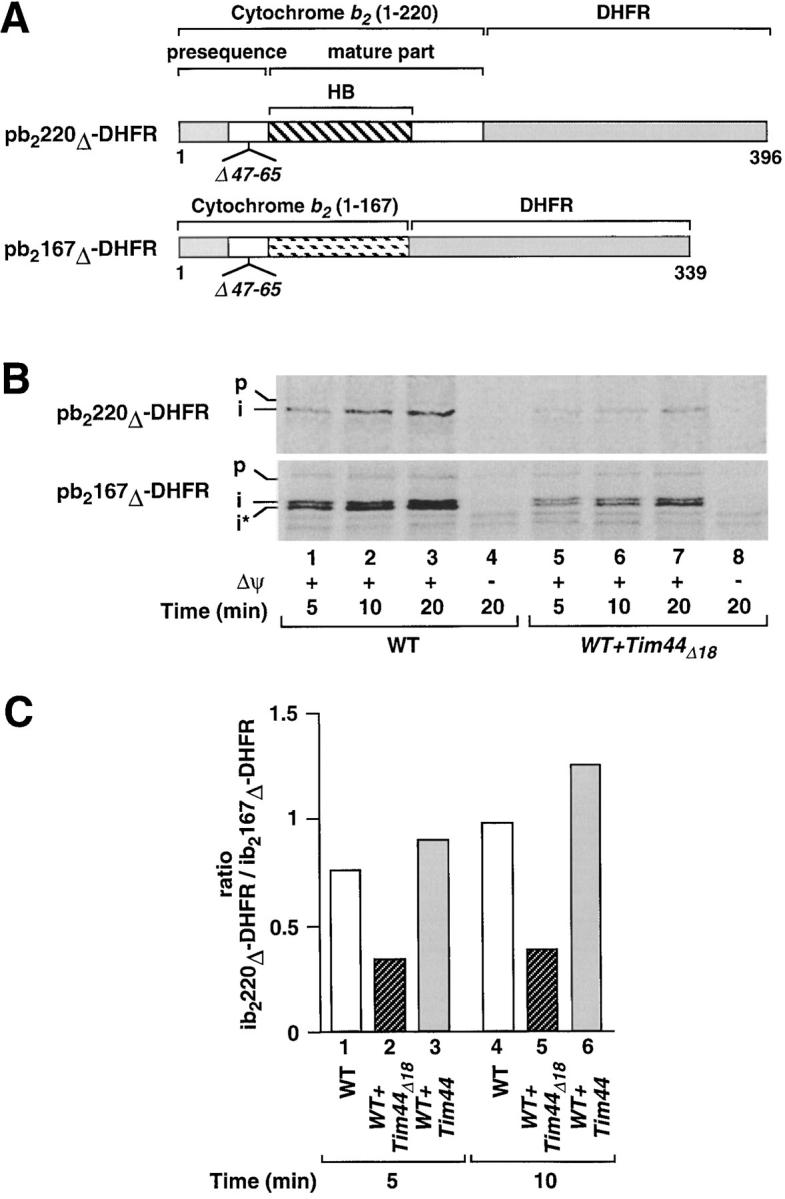
The reduction in the efficiency of protein translocation caused by the deletion of the 18-residue segment of Tim44 mainly affects the import of tightly folded domains. (A) The hybrid protein pb2220Δ-DHFR contains the first 220 amino acid residues of cytochrome b 2, including the complete heme-binding domain (HB; position 81–179), fused to DHFR. In pb2167Δ-DHFR the first 167 amino acid residues of cytochrome b 2 (lacking the carboxy-terminal part of the heme-binding domain) are fused to DHFR. In both constructs a part of the sorting signal (residues 47–65) is deleted within the presequence. (B) Import of pb2220Δ-DHFR and pb2167Δ-DHFR. The radiolabeled preproteins were synthesized in reticulocyte lysate and incubated with mitochondria isolated from the Tim44Δ18-overproducing strain (WT + Tim44Δ18; the same strain as applied in the experiments shown in Figs. 2–4) or the corresponding wild-type strain MB3 (WT). Lanes 4 and 8 contained valinomycin to dissipate the membrane potential. Subsequently, all samples were treated with proteinase K. The imported proteins were analyzed by SDS-PAGE and fluorography. (C) Ratio of the import efficiencies of pb2220Δ-DHFR and pb2167Δ-DHFR after overexpression of Tim44Δ18 and after overexpression of authentic Tim44. Both versions of Tim44 were expressed in the strain MB3 using the vector YEplac181(LEU2). The preproteins pb2220Δ-DHFR and pb2167Δ-DHFR were imported as described in the legend to Fig. 4 B and quantified for calculation of the import efficiencies (imported protein in percentage of total preprotein added). Shown are the ratios of the efficiencies of both proteins after 5 or 10 min of import at 25°C. Standard deviations of the means (five independent determinations) were below 9% of the reported values. In the presence of Tim44Δ18 the import efficiency of pb2220Δ-DHFR is about two times more reduced as compared with the import of pb2167Δ-DHFR, demonstrating that the deletion of the 18-residue segment mainly affects the import of the preprotein containing the tightly folded heme-binding domain.
Modifications of mitochondrial inner membrane proteins can easily lead to a reduction of the mitochondrial membrane potential and thereby indirectly cause reduced efficiencies of protein transport. To address this possibility, we compared the membrane potential of the mitochondria which had been used in the previous experiments. As a sensitive assay we determined the membrane potential-dependent uptake of the dye DiSC3(5) (Sims et al., 1974). The uptake is reversible and can be quantified by following the change in the fluorescence of the dye. With mitochondria from the Tim44Δ18 mutant strain no reduction in the membrane potential was observed (Fig. 4 C). An indirect effect of the mutation on mitochondrial protein import mediated by a weakened membrane potential can thus be excluded.
Coexpression of Tim44Δ 18 Leads to Reduced Import Efficiencies of Folded Protein Domains
We then asked if the effect of the deletion of the 18-residue segment on protein import is dependent on the folding state of the preprotein. Previous studies have shown that the heme-binding domain of cytochrome b 2 is tightly folded and requires an intact mtHsp70 system to drive the unfolding of this domain (Glick et al., 1993; Voos et al., 1993, 1996). Following this principle, we now imported two different preproteins, b2(167)Δ-DHFR and b2(220)Δ-DHFR (Fig. 5 A). Both constructs contain amino-terminal parts of cytochrome b 2 of different length fused to DHFR. The intermembrane space sorting signal is deleted to allow passage into the mitochondrial matrix. The hybrid protein b2(167)Δ-DHFR only contains an incomplete and therefore loosely folded heme-binding domain. A step of active unfolding of this preprotein is not required to allow the import reaction. The situation is different with the longer construct b2(220)Δ-DHFR. This protein contains the complete heme-binding domain (residues 81–179) and requires an unfolding reaction to allow the import. Thereby, the heme-binding domain causes restrictions if imported by a weakened translocation machinery (Glick et al., 1993; Voos et al., 1993, 1996).
We compared the import kinetics of b2(167)Δ-DHFR and b2(220)Δ-DHFR in wild-type mitochondria and in mitochondria containing Tim44Δ18 in addition to the intact Tim44 (Fig. 5 B, lanes 1–8). With both preproteins, the import was clearly reduced in the Tim44Δ18 mitochondria (Fig. 5 B, lanes 5–7). To assess the effect of the heme-binding domain on the import efficiency we calculated the relative amounts of the imported proteins (Fig. 5 C). We found that after 10 min of import, both constructs were imported into wild-type mitochondria with the same efficiency (Fig. 5 C, column 4). With mitochondria from a Tim44-overproducing strain, the relative import efficiency of b2(220)Δ-DHFR was slightly improved (Fig. 5 C, column 6). However, with mitochondria from the strain overproducing Tim44Δ18, the relative import efficiency of b2(220)Δ-DHFR was reduced to ∼35%, as compared with the import efficiency of b2(167)Δ-DHFR (Fig. 5 C, column 5). Similar ratios were found at other time points of the import reaction (Fig. 5 C, columns 1–3). The 18-residue segment of Tim44 appears to be required to permit the efficient import of partially folded preproteins which require the full activity of the import machinery. The effect on loosely folded preproteins is less pronounced but also in this case the translocation is clearly facilitated if the activity of mtHsp70 is exclusively mediated by intact Tim44.
Deletion of the 18-Residue Segment Causes Reduced Complex Formation of Tim44 with mtHsp70
The experiments shown in Figs. 2 and 3 have indicated that the interactions of Tim44Δ18 with the Tim machinery are not disturbed by the deletion of the 18-residue segment. We now ask if Tim44Δ18 shows an altered interaction with mtHsp70. We lysed mitochondria from the wild-type in the presence of detergent and performed coimmunoprecipitations using specific antibodies raised against mtHsp70. The precipitates were analyzed by immunoblotting and demonstrated the association of mtHsp70 with Tim44 (Fig. 6 A). We then lysed mitochondria from the strain expressing both forms of Tim44. The mitochondria contained about three- to fourfold more Tim44Δ18 than Tim44 (Fig. 6 B, lane 1, and Fig. 6 C, lane 1). By coimmunoprecipitates from these lysates we compared the association of mtHsp70 with Tim44Δ18 and the authentic Tim44. The ratio of Tim44Δ18 to Tim44 in the precipitates was close to 1:1, demonstrating that complex formation of Tim44Δ18 to mtHsp70 was reduced about three- to fourfold by the deletion of the 18-residue segment (Fig. 6 B, lane 2, and Fig. 6 C, lane 3). Both forms of Tim44 were stable upon prolonged incubation after lysis, confirming that the reduced amount of Tim44Δ18 in the immunoprecipitates was due to a reduced complex formation with mtHsp70 (Fig. 6 C, lanes 1 and 2).
The reduced affinity of mtHsp70 to Tim44Δ18 as compared with the authentic Tim44 was confirmed by systematic quantifications (Fig. 6 D). Complex formation of mtHsp70 with Tim44Δ18 was reduced by ∼70%. In the presence of ATP, Tim44Δ18 and Tim44 were both efficiently released from mtHsp70, confirming the specificity of the precipitations (Fig. 6 E). ATP at a concentration of 2.5 nM was sufficient to cause the dissociation of 50% of the complexes with Tim44Δ18 as well as with Tim44 (not shown). We conclude that the 18-residue segment of Tim44 is not the only structure which is involved in complex formation with mtHsp70. But the segment appears to be required to allow binding of sufficient efficiency and, as suggested by the results of the import experiments, for optimal cooperation of both proteins in mitochondrial protein import.
The allele ssc1-2 encodes a mutant form of mtHsp70 which shows a reduced affinity for Tim44 but an enhanced affinity for substrate proteins (Kang et al., 1990; Gambill et al., 1993; Voos et al., 1993, 1996; Schneider et al., 1994; von Ahsen et al., 1995). We expressed Tim44Δ18 in a ssc1-2 strain and determined the interactions of the mutant mtHsp70 with both Tim44 proteins and a substrate protein by coimmunoprecipitations (Fig. 6 F). While the efficiency of binding of the mutant mtHsp70 to the substrate protein Su9-DHFR was more than fourfold higher than that of wild-type mtHsp70 (Fig. 6 F, columns 5 and 6), the association of the mutant mtHsp70 with both Tim44 and Tim44Δ18 was blocked (Fig. 6 F, columns 2 and 4). This result implies that not only the authentic Tim44 but also the truncated Tim44Δ18 is recognized by mtHsp70 as a partner protein of special properties, and not as a substrate protein.
Discussion
In this study we have characterized the role of complex formation of mtHsp70 with Tim44 in mitochondrial protein import. A system of reduced binding between both proteins was created by the deletion of an 18-residue segment in Tim44 (residues 185–202) which shows a limited similarity to one of the two α helices of J-domains. The intracellular localization of Tim44Δ18 is not altered by the deletion. In all fractionation experiments Tim44Δ18 showed the same behavior as the authentic Tim44, and by chemical cross-linking we found that Tim44Δ18 is localized at the inner membrane protein import sites.
The correct topology of Tim44Δ18 within the mitochondria allowed us to test whether the 18-residue segment of Tim44 is required to recruit mtHsp70 to the Tim machinery of the inner membrane. We found that binding of mtHsp70 to Tim44Δ18 was reduced by ∼70% as compared with the authentic Tim44. The 18-residue segment of Tim44 is obviously not the only site for binding to mtHsp70. In contrast to the integral membrane protein Sec63p which is exposed to the ER lumen only by short segments of its sequence (Feldheim et al., 1992), Tim44 is a peripheral protein and in larger parts exposed to the matrix (Blom et al., 1993; Rassow et al., 1994). This topology may allow the formation of multiple binding sites.
While this manuscript was in preparation, a publication appeared by Greene et al. (1998) showing that the binding site of DnaJ for DnaK is the helix II of the J-domain, which corresponds exactly to the segment of similarity to Tim44. Since Tim44 seems not to belong to the family of J-proteins (Wada and Kanwar, 1998) we assume that the J-related segment of Tim44 does not represent a J-homology in the strict sense but rather a J-analogous development to facilitate the interaction with mtHsp70. According to Greene et al. (1998), the helix II of DnaJ interacts with the ATPase domain of DnaK. Following the analogy between Tim44 and DnaJ, Tim44 should similarly bind to the ATPase domain of mtHsp70. However, other J-proteins were found to interact with the carboxy-terminal domain of Hsp70s (Freeman et al., 1995; Demand et al., 1998) or to require both domains for binding (Ungewickell et al., 1997). Therefore, it may be speculated that the interaction between mtHsp70 and Tim44 is mediated by multiple attachment sites, as was shown recently by the x-ray structure for the complex of DnaK with GrpE (Harrison et al., 1997). We cannot completely rule out allosteric effects of the deletion of the 18-residue segment. However, the only difference to the wild-type protein we observed was restricted to the interaction with mtHsp70. The very sensitive assays of chemical cross-linking (Fig. 3 B) and blue native electrophoresis (Fig. 2 F) demonstrate that the oligomeric state of Tim44, and the direct interactions with preproteins and other components of the Tim machinery were retained. The comparison to the DnaJ-DnaK complex as analyzed by Greene et al. (1998) suggests that the 185–202 segment of Tim44 provides the major binding site for mtHsp70.
Several data indicate that Tim44 binds to Tim23 and provides a dynamic link between the Tim proteins which form the protein import channel and the soluble mtHsp70 system of the matrix (Bömer et al., 1997; Dekker et al., 1997). With Tim44Δ18 the function of this link is specifically impaired in the interactions of Tim44Δ18 with mtHsp70. Our import experiments demonstrate that the presence of Tim44Δ18 causes a significant reduction in the import efficiencies of different preproteins, including proteins which are regarded as loosely folded. The import of all of these preproteins is strictly dependent on mtHsp70 as demonstrated by previous studies using temperature-sensitive strains of SSC1 (encoding mtHsp70) (Gambill et al., 1993; Voos et al., 1993). The strongest inhibition of import was observed with preproteins which contain a tightly folded domain. Such domains cause restrictions in the translocation across the mitochondrial membranes which are due to the requirement of unfolding within the import channel (Gambill et al., 1993; Glick et al., 1993; Voos et al., 1993, 1996; Matouschek et al., 1997). To overcome these restrictions, the mtHsp70 system of the matrix has to exert a force on the translocating protein which is sufficient to pull the protein across the membranes.
Studies to elucidate the mechanism by which this force is generated made use of the ssc1-2 mutant of mtHsp70 (Kang et al., 1990; Schneider et al., 1994; von Ahsen et al., 1995; Voos et al., 1996). The mtHsp70 of this mutant binds efficiently to translocating preproteins but is impaired in binding to Tim44. This defect correlates with an inhibition in the import of tightly folded protein domains. However, conclusions could only be drawn with reservation. The Tim machinery seems to contain at least two binding sites for mtHsp70, one at Tim44 and a second site at the Tim23/ Tim17 import channel, and both interactions are inhibited by the ssc1-2 mutation (Bömer et al., 1997). The principle which governs the mechanism of mtHsp70-dependent protein import is still unknown. A Brownian ratchet mechanism (Simon et al., 1992; Ungermann et al., 1994; Gaume et al., 1998) and a mechanism of mtHsp70/Tim44-mediated pulling (Glick, 1995; Pfanner and Meijer, 1995) have been suggested. In this context it is remarkable that the effect of Tim44Δ18 on the import of different preproteins and on the viability of yeast resembles the effects of ssc1-2. This similarity in the phenotype thus corroborates and specifies the concept that the cooperation of Tim44 with mtHsp70 is of particular importance in the import of tightly folded protein domains. In a previous study on a complete inactivation of functional Tim44 in isolated mitochondria we showed that Tim44 acts at the inner side of the inner membrane (Bömer et al., 1998). The results obtained with the Tim44Δ18 construct suggest that in this location the functions of Tim44 in protein import may be confined to specific interactions with mtHsp70.
In summary, the results of this study demonstrate that the J-related segment of Tim44 (residues 185–202) is required for the essential functions of Tim44 in mitochondria. This segment is not the only element involved in the interaction of Tim44 with mtHsp70, but it is required for productive cooperation of both proteins and the optimal efficiency of mitochondrial protein import. mtHsp70 is an essential motor protein in the translocation of all proteins which are imported into the mitochondrial matrix, irrespective of whether or not they contain tightly folded domains (Schatz, 1996; Neupert, 1997; Pfanner et al., 1997). In contrast, the requirement for an interaction of mtHsp70 with Tim44 seems to be less strict and appears to play an important role primarily in situations which require the full activity of the import motor, for example in overcoming stronger restrictions in the translocation of preproteins. The import of loosely folded preproteins is facilitated by Tim44, but the effect is much more pronounced in the case of tightly folded domains.
Acknowledgments
We thank Drs. Oliver von Ahsen, Michael Kübrich, and Martin Moczko for technical advice, Nicole Zufall for help in cloning procedures, Dr. Bernard Guiard for the b2-DHFR plasmids, Dr. Elizabeth Craig for the ssc1-2 strain, and Drs. Bernd Bukau, Ulrich Hartl, and Gabriel Schlenstedt for helpful discussions.
This work was supported by the Deutsche Forschungsgemeinschaft, the Sonderforschungs-bereich 388, and the Müller-Fahnenberg-Stiftung.
Abbreviations used in this paper
- EGS
ethylene glycolbis succinimidylsuccinate
- mt
mitochondrial
- Su9-DHFR
fusion protein between presequence of F0-ATPase subunit 9 and dihydrofolate reductase
- Tim44
44-kD protein of the mitochondrial inner membrane import machinery
References
- Alconada A, Gärtner F, Hönlinger A, Kübrich M, Pfanner N. Mitochondrial receptor complex from Neurospora crassa and Saccharomyces cerevisiae. . Methods Enzymol. 1995;260:263–287. doi: 10.1016/0076-6879(95)60144-9. [DOI] [PubMed] [Google Scholar]
- Bauer MF, Sirrenberg C, Neupert W, Brunner M. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell. 1996;87:33–41. doi: 10.1016/s0092-8674(00)81320-3. [DOI] [PubMed] [Google Scholar]
- Berthold J, Bauer MF, Schneider H-C, Klaus C, Dietmeier K, Neupert W, Brunner M. The MIM complex mediates preprotein translocation across the mitochondrial inner membrane and couples it to the mt-Hsp70/ ATP driving system. Cell. 1995;81:1085–1093. doi: 10.1016/s0092-8674(05)80013-3. [DOI] [PubMed] [Google Scholar]
- Blom J, Kübrich M, Rassow J, Voos W, Dekker PJT, Maarse AC, Meijer M, Pfanner N. The essential yeast protein MIM44 (encoded by MPI1) is required at an early step of preprotein translocation across the mitochondrial inner membrane. Mol Cell Biol. 1993;13:7364–7371. doi: 10.1128/mcb.13.12.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom J, Dekker PJT, Meijer M. Functional and physical interactions of components of the yeast mitochondrial inner-membrane import machinery (MIM) Eur J Biochem. 1995;232:309–314. doi: 10.1111/j.1432-1033.1995.tb20813.x. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Bömer U, Meijer M, Maarse AC, Dekker PJT, Pfanner N, Rassow J. Multiple interactions of components mediating preprotein translocation across the inner mitochondrial membrane. EMBO (Eur Mol Biol Organ) J. 1997;16:2205–2216. doi: 10.1093/emboj/16.9.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bömer U, Maarse AC, Martin F, Geissler A, Merlin A, Schönfisch B, Meijer M, Pfanner N, Rassow J. Separation of structural and dynamic functions of the mitochondrial translocase: Tim44 is crucial for the inner membrane import sites in translocation of tightly folded domains, but not of loosely folded preproteins. EMBO (Eur Mol Biol Organ) J. 1998;17:4226–4237. doi: 10.1093/emboj/17.15.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL. Post-translational protein translocation: not all hsc70s are created equal. Trends Biochem Sci. 1996;21:122–126. [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi AK, Schekman R. The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. . J Cell Biol. 1997;137:1483–1493. doi: 10.1083/jcb.137.7.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr D, Langer T, Douglas M. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- Daum G, Gasser SM, Schatz G. Import of proteins into mitochondria: energy-dependent, two-step processing of the intermembrane space enzyme cytochrome b2by isolated yeast mitochondria. J Biol Chem. 1982;257:13075–13080. [PubMed] [Google Scholar]
- Dekker PJT, Pfanner N. Role of mitochondrial GrpE and phosphate in the ATPase cycle of matrix Hsp70. J Mol Biol. 1997;270:321–327. doi: 10.1006/jmbi.1997.1131. [DOI] [PubMed] [Google Scholar]
- Dekker PJT, Martin F, Maarse AC, Bömer U, Müller H, Guiard B, Meijer M, Rassow J, Pfanner N. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO (Eur Mol Biol Organ) J. 1997;16:5408–5419. doi: 10.1093/emboj/16.17.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloche O, Georgopoulos C. Purification and biochemical properties of Saccharomyces cerevisiae's Mge1p, the mitochondrial cochaperone of Ssc1p. J Biol Chem. 1996;271:23960–23966. doi: 10.1074/jbc.271.39.23960. [DOI] [PubMed] [Google Scholar]
- Deloche O, Kelley WL, Georgopoulos C. Structure-function analysis of the Ssc1p, Mdj1p, and Mge1p Saccharomyces cerevisiae mitochondrial proteins in Escherichia coli. . J Bact. 1997a;179:6066–6075. doi: 10.1128/jb.179.19.6066-6075.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloche O, Liberek K, Zylicz M, Georgopoulos C. Purification and biochemical properties of Saccharomyces cerevisiaeMdj1p, the mitochondrial DnaJ homologue. J Biol Chem. 1997b;272:28539–28544. doi: 10.1074/jbc.272.45.28539. [DOI] [PubMed] [Google Scholar]
- Demand J, Lüders J, Höhfeld J. The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol Cell Biol. 1998;18:2023–2028. doi: 10.1128/mcb.18.4.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldheim D, Rothblatt J, Scheckman R. Topology and functional domains of Sec63p, an ER membrane protein required for secretory protein translocation. Mol Cell Biol. 1992;12:3288–3296. doi: 10.1128/mcb.12.7.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Myers MP, Schumacher R, Morimoto RI. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO (Eur Mol Biol Organ) J. 1995;14:2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambill D, Voos W, Kang PJ, Miao B, Langer T, Craig EA, Pfanner N. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J Cell Biol. 1993;123:109–117. doi: 10.1083/jcb.123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaume B, Klaus C, Ungermann C, Guiard B, Neupert W, Brunner M. Unfolding of preproteins upon import into mitochondria. EMBO (Eur Mol Biol Organ) J. 1998;17:6497–6507. doi: 10.1093/emboj/17.22.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitromutagenized yeast lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Glick BS. Can Hsp70 proteins act as force-generating motors? . Cell. 1995;80:11–14. doi: 10.1016/0092-8674(95)90444-1. [DOI] [PubMed] [Google Scholar]
- Glick BS, Wachter C, Reid GA, Schatz G. Import of cytochrome b2to the mitochondrial intermembrane space: the tightly folded heme-binding domain makes import dependent upon matrix ATP. Protein Sci. 1993;2:1901–1917. doi: 10.1002/pro.5560021112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene MK, Maskos K, Landry SJ. Role of the J-domain in the cooperation of hsp40 with hsp70. Proc Natl Acad Sci USA. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CJ, Hayer-Hartl M, Liberto MD, Hartl F-U, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- Hartl F-U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hill RB, Flanagan JM, Prestegard JH. 1H and 15N magnetic resonance assignments, secondary structure, and tertiary fold of Escherichia coliDnaJ (1-78) Biochemistry. 1995;34:5587–5596. doi: 10.1021/bi00016a033. [DOI] [PubMed] [Google Scholar]
- Horst M, Oppliger W, Feifel B, Schatz G, Glick BS. The mitochondrial protein import motor: dissociation of mitochondrial hsp70 from its membrane anchor requires ATP binding rather than ATP hydrolysis. Protein Sci. 1996;5:759–767. doi: 10.1002/pro.5560050421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RE, Kinnally KW. The mitochondrial import pathway: are precursors imported through membrane channels? . J Bioenerg Biomembr. 1997;29:3–10. doi: 10.1023/a:1022470303365. [DOI] [PubMed] [Google Scholar]
- Kang PJ, Ostermann J, Shilling J, Neupert W, Craig EA, Pfanner N. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990;348:137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- Kelley WL. The J-domain family and the recruitment of chaperone power. Trends Biochem Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- Kronidou NG, Oppliger W, Bolliger L, Hannavy K, Glick BS, Schatz G, Horst M. Dynamic interaction between Isp45 and mitochondrial hsp70 in the protein import system of the yeast mitochondrial inner membrane. Proc Natl Acad Sci USA. 1994;91:12818–12822. doi: 10.1073/pnas.91.26.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloraya S, Dekker PJT, Voos W, Craig EA, Pfanner N. Mitochondrial GrpE modulates the function of matrix Hsp70 in translocation and maturation of preproteins. Mol Cell Biol. 1995;15:7098–7105. doi: 10.1128/mcb.15.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C. Escherichia coliDnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohret TA, Jensen RE, Kinnally KW. Tim23, a protein import component of the mitochondrial inner membrane, is required for normal activity of the multiple conductance channel, MCC. J Cell Biol. 1997;137:377–386. doi: 10.1083/jcb.137.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke MM, Sutton A, Arndt KT. Characterization of SIS1, a Saccharomyces cerevisiaehomologue of bacterial dnaJ proteins. J Cell Biol. 1991;114:623–638. doi: 10.1083/jcb.114.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maarse AC, Blom J, Grivell LA, Meijer M. MPI1, an essential gene encoding a mitochondrial membrane protein, is possibly involved in protein import into yeast mitochondria. EMBO (Eur Mol Biol Organ) J. 1992;11:3619–3628. doi: 10.1002/j.1460-2075.1992.tb05446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouschek A, Azem A, Ratliff K, Glick BS, Schmid K, Schatz G. Active unfolding of precursor proteins during mitochondrial protein import. EMBO (Eur Mol Biol Organ) J. 1997;16:6727–6736. doi: 10.1093/emboj/16.22.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao B, Davis JE, Craig EA. Mge1 functions as a nucleotide release factor for Ssc1, a mitochondrial Hsp70 of Saccharomyces cerevisiae. . J Mol Biol. 1997;265:541–552. doi: 10.1006/jmbi.1996.0762. [DOI] [PubMed] [Google Scholar]
- Nakai M, Kato Y, Ikeda E, Toh-e A, Endo T. YGE1p, a eukaryotic GrpE homolog, is localized in the mitochondrial matrix and interacts with mitochondrial Hsp70. Biochem Biophys Res Commun. 1994;200:435–442. doi: 10.1006/bbrc.1994.1468. [DOI] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Ohki M, Tamura F, Nishimura S, Uchida H. Nucleotide sequence of the Escherichia colidnaJ gene and purification of the gene product. J Biol Chem. 1986;261:1778–1781. [PubMed] [Google Scholar]
- Pfanner N, Meijer M. Protein sorting: pulling in the proteins. Curr Biol. 1995;5:132–135. doi: 10.1016/s0960-9822(95)00033-9. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Meijer M. Mitochondrial biogenesis: the Tom and Tim machine. Curr Biol. 1997;7:100–103. doi: 10.1016/s0960-9822(06)00048-0. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Craig EA, Hönlinger A. Mitochondrial preprotein translocase. Annu Rev Cell Dev Biol. 1997;13:25–51. doi: 10.1146/annurev.cellbio.13.1.25. [DOI] [PubMed] [Google Scholar]
- Pon, L., and G. Schatz. 1991. Biogenesis of yeast mitochondria. In The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae. J.R. Broach, J. Pringle, and E. Jones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 333–406.
- Rapoport TA, Rolls MM, Jungnickel B. Approaching the mechanism of protein transport across the ER membrane. Curr Opin Cell Biol. 1996;8:499–504. doi: 10.1016/s0955-0674(96)80027-5. [DOI] [PubMed] [Google Scholar]
- Rassow J, Pfanner N. Mitochondrial preproteins en route from the outer membrane to the inner membrane are exposed to the intermembrane space. FEBS Lett. 1991;293:85–88. doi: 10.1016/0014-5793(91)81157-4. [DOI] [PubMed] [Google Scholar]
- Rassow, J., and N. Pfanner. 1995. The motor of mitochondrial protein import: a mitochondrial analog of the Sec63p-Kar2p system. In 30 Years of Mitochondrial Research. E. Qagliariello and F. Palmieri, editors. Elsevier Science Publishers, Amsterdam. 113–118.
- Rassow J, Maarse AC, Krainer E, Kübrich M, Müller H, Meijer M, Craig EA, Pfanner N. Mitochondrial protein import: biochemical and genetic evidence for interaction of matrix hsp70 and the inner membrane protein MIM44. J Cell Biol. 1994;127:1547–1556. doi: 10.1083/jcb.127.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow J, Voos W, Pfanner N. Partner proteins determine multiple functions of Hsp70. Trends Cell Biol. 1995;5:207–212. doi: 10.1016/s0962-8924(00)89001-7. [DOI] [PubMed] [Google Scholar]
- Rassow J, Dekker PJT, van der Wilpe S, Meijer M, Soll J. The preprotein translocase of the mitochondrial inner membrane: function and evolution. J Mol Biol. 1999;286:105–120. doi: 10.1006/jmbi.1998.2455. [DOI] [PubMed] [Google Scholar]
- Rowley N, Prip-Buus C, Westermann B, Brown C, Schwarz E, Barrell B, Neupert W. Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell. 1994;77:249–259. doi: 10.1016/0092-8674(94)90317-4. [DOI] [PubMed] [Google Scholar]
- Ryan KR, Jensen RE. Protein translocation across mitochondrial membranes: what a long, strange trip it is. Cell. 1995;83:517–519. doi: 10.1016/0092-8674(95)90089-6. [DOI] [PubMed] [Google Scholar]
- Ryan KR, Leung RS, Jensen RE. Characterization of the mitochondrial inner membrane translocase complex: the Tim23p hydrophobic domain interacts with Tim17p but not with other Tim23p molecules. Mol Cell Biol. 1998;18:178–187. doi: 10.1128/mcb.18.1.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler IA, Chiang A, Kurihara T, Rothblatt J, Way J, Silver P. A yeast gene essential for protein assembly in the endoplasmic reticulum and the nucleus has homology to DnaJ, an E. coliheat shock protein. J Cell Biol. 1989;109:2665–2675. doi: 10.1083/jcb.109.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271:31763–31766. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- Scherer PE, Manning-Krieg UC, Jenö P, Schatz G, Horst M. Identification of a 45-kD protein at the protein import site of the yeast mitochondrial inner membrane. Proc Natl Acad Sci USA. 1992;89:11930–11934. doi: 10.1073/pnas.89.24.11930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Harris S, Risse B, Lill R, Silver P. A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum with BiP/ Kar2p via a conserved domain that specifies interaction with Hsp70s. J Cell Biol. 1995;129:979–988. doi: 10.1083/jcb.129.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H-C, Berthold J, Bauer MF, Dietmeier K, Guiard B, Brunner M, Neupert W. Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature. 1994;371:768–774. doi: 10.1038/371768a0. [DOI] [PubMed] [Google Scholar]
- Schneider H-C, Westermann B, Neupert W, Brunner M. The nucleotide exchange factor MGE exerts a key function in the ATP-dependent cycle of mtHsp70-Tim44 interaction driving mitochondrial protein import. EMBO (Eur Mol Biol Organ) J. 1996;15:5796–5803. [PMC free article] [PubMed] [Google Scholar]
- Scidmore MA, Okamura HH, Rose MD. Genetic interactions between KAR2 and SEC63, encoding eukaryotic homologues of DnaK and DnaJ in the endoplasmic reticulum. Mol Biol Cell. 1993;4:1145–1159. doi: 10.1091/mbc.4.11.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver PA, Way JC. Eucaryotic DnaJ homologs and the specificity of Hsp70 activity. Cell. 1993;74:5–6. doi: 10.1016/0092-8674(93)90287-z. [DOI] [PubMed] [Google Scholar]
- Simon SM, Peskin CS, Oster GF. What drives the translocation of proteins? . Proc Natl Acad Sci USA. 1992;89:3770–3774. doi: 10.1073/pnas.89.9.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims PJ, Waggoner AS, Wang C-H, Hoffmann JF. Studies on the mechanism by which cyanine dyes measure membrane potential in red blood cells and phosphatidylcholine vesicles. Biochemistry. 1974;13:3315–3330. doi: 10.1021/bi00713a022. [DOI] [PubMed] [Google Scholar]
- Söllner T, Rassow J, Pfanner N. Analysis of mitochondrial protein import using translocation intermediates and specific antibodies. Methods Cell Biol. 1991;34:345–357. doi: 10.1016/s0091-679x(08)61689-1. [DOI] [PubMed] [Google Scholar]
- Szyperski T, Pellecchia M, Wall D, Georgopoulos C, Wüthrich K. NMR structure determination of the Escherichia coliDnaJ molecular chaperone: secondary structure and backbone fold of the N-terminal region (residues 2-108) containing the highly conserved J domain. Proc Natl Acad Sci USA. 1994;91:11343–11347. doi: 10.1073/pnas.91.24.11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C, Neupert W, Cyr DM. The role of Hsp70 in conferring unidirectionality on protein translocation into mitochondria. Science. 1994;266:1250–1253. doi: 10.1126/science.7973708. [DOI] [PubMed] [Google Scholar]
- Ungermann C, Guiard B, Neupert W, Cyr DM. The ΔΨ- and Hsp70/MIM44-dependent reaction cycle driving early steps of protein import into mitochondria. EMBO (Eur Mol Biol Organ) J. 1996;15:735–744. [PMC free article] [PubMed] [Google Scholar]
- Ungewickell E, Ungewickell H, Holstein SEH. Functional interaction of the auxilin J domain with the nucleotide- and substrate-binding modules of Hsc70. J Biol Chem. 1997;272:19594–19600. doi: 10.1074/jbc.272.31.19594. [DOI] [PubMed] [Google Scholar]
- von Ahsen O, Voos W, Henninger H, Pfanner N. The mitochondrial protein import machinery. Role of ATP in dissociation of the Hsp70-Mim44 complex. J Biol Chem. 1995;270:29848–29853. doi: 10.1074/jbc.270.50.29848. [DOI] [PubMed] [Google Scholar]
- Voos W, Gambill D, Guiard B, Pfanner N, Craig EA. Presequence and mature part of preproteins strongly influence the dependence of mitochondrial protein import in heat shock protein 70 in the matrix. J Cell Biol. 1993;123:119–126. doi: 10.1083/jcb.123.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W, Gambill D, Laloraya S, Ang D, Craig EA, Pfanner N. Mitochondrial GrpE is present in a complex with Hsp70 and preproteins in transit across membranes. Mol Cell Biol. 1994;14:6627–6634. doi: 10.1128/mcb.14.10.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W, von Ahsen O, Müller H, Guiard B, Rassow J, Pfanner N. Differential requirement for the mitochondrial Hsp70-Tim44 complex in unfolding and translocation of preproteins. EMBO (Eur Mol Biol Organ) J. 1996;15:2668–2677. [PMC free article] [PubMed] [Google Scholar]
- Wada J, Kanwar YS. Characterization of mammalian translocase of inner mitochondrial membrane (Tim44) isolated from diabetic newborn mouse kidney. Proc Natl Acad Sci USA. 1998;95:144–149. doi: 10.1073/pnas.95.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B, Neupert W. Mdj2p, a novel DnaJ homolog in the mitochondrial inner membrane of the yeast Saccharomyces cerevisiae. . J Mol Biol. 1997;272:477–483. doi: 10.1006/jmbi.1997.1267. [DOI] [PubMed] [Google Scholar]
- Westermann B, Prip-Buus C, Neupert W, Schwarz E. The role of the GrpE homologue, Mge1p, in mediating protein import and protein folding in mitochondria. EMBO (Eur Mol Biol Organ) J. 1995;14:3452–3460. doi: 10.1002/j.1460-2075.1995.tb07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B, Gaume B, Herrmann JM, Neupert W, Schwarz E. Role of the mitochondrial DnaJ homolog Mdj1p as a chaperone for mitochondrially synthesized and imported proteins. Mol Cell Biol. 1996;16:7063–7071. doi: 10.1128/mcb.16.12.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong T, Arndt KT. The yeast SIS1 protein, a DnaJ homolog, is required for the initiation of translation. Cell. 1993;73:1175–1186. doi: 10.1016/0092-8674(93)90646-8. [DOI] [PubMed] [Google Scholar]