Abstract
The mitotic checkpoint blocks cell cycle progression before anaphase in case of mistakes in the alignment of chromosomes on the mitotic spindle. In budding yeast, the Mad1, 2, 3, and Bub1, 2, 3 proteins mediate this arrest. Vertebrate homologues of Mad1, 2, 3, and Bub1, 3 bind to unattached kinetochores and prevent progression through mitosis by inhibiting Cdc20/APC-mediated proteolysis of anaphase inhibitors, like Pds1 and B-type cyclins. We investigated the role of Bub2 in budding yeast mitotic checkpoint. The following observations indicate that Bub2 and Mad1, 2 probably activate the checkpoint via different pathways: (a) unlike the other Mad and Bub proteins, Bub2 localizes at the spindle pole body (SPB) throughout the cell cycle; (b) the effect of concomitant lack of Mad1 or Mad2 and Bub2 is additive, since nocodazole-treated mad1 bub2 and mad2 bub2 double mutants rereplicate DNA more rapidly and efficiently than either single mutant; (c) cell cycle progression of bub2 cells in the presence of nocodazole requires the Cdc26 APC subunit, which, conversely, is not required for mad2 cells in the same conditions. Altogether, our data suggest that activation of the mitotic checkpoint blocks progression through mitosis by independent and partially redundant mechanisms.
Keywords: budding yeast, Bub2, mitotic checkpoint, anaphase, anaphase-promoting complex
In eukaryotic cells, the correct transmission of genetic information relies on surveillance mechanisms, called checkpoints, that block or delay cell cycle progression in response to errors concerning the integrity, replication, and segregation of the genome (Hartwell and Weinert, 1989). Checkpoint loss results in genome instability and has been implicated in the evolution of normal cells into cancer cells.
Once a cell's chromosomes have been duplicated, their proper segregation to the daughter cells in anaphase requires the prior execution of a number of processes: a bipolar mitotic spindle must be assembled and the chromosomes must attach via their kinetochores to microtubules that associate with opposite poles of this spindle. Subsequently, each pair of sister chromatids is aligned on the metaphase plate by means of the tension exerted on sister kinetochores. Chromosome alignment during metaphase depends not only on pulling forces exerted by microtubules on kinetochores, but also on an opposing force exerted by tethers that hold sister chromatids together. This is the means by which cells determine which DNA molecules are sisters, and it is therefore essential that anaphase is not initiated before all pairs of sister chromatids have aligned on the mitotic spindle. The mitotic checkpoint prevents the onset of anaphase and the entry into the next cell cycle until alignment of chromosomes on the spindle has been properly accomplished. In the budding yeast Saccharomyces cerevisiae, several genes implicated in the mitotic checkpoint have been identified by genetic screens. These include BUB1-3 (budding uninhibited by benzimidazole; Hoyt et al., 1991)1 and MAD1-3 (mitotic arrest deficient; Li and Murray, 1991). The products of these genes are required to delay cell cycle progression in response to defects in spindle assembly and, except for Bub2, in kinetochore and centromere structure (Hoyt et al., 1991; Li and Murray, 1991; Wang and Burke, 1995; Pangilinan and Spencer, 1996). Strikingly, homologues of Mad1, Mad2, Bub1, and Bub3 have recently been discovered in fission yeast (He et al., 1997; Bernard et al., 1998) and in higher eukaryotes (Chen et al., 1996, 1998; Li and Benezra, 1996; Taylor and McKeon, 1997; Taylor et al., 1998) and found to be involved in the mitotic checkpoint also in these organisms. The vertebrate counterparts appear to be localized at unattached kinetochores during prophase and prometaphase; as soon as kinetochores bind to microtubules, centromeric staining of these proteins rapidly vanishes (Chen et al., 1996, 1998; Li and Benezra, 1996; Taylor and McKeon, 1997; Taylor et al., 1998; Gorbsky et al., 1998). SpBub1, which is so far the only yeast mitotic checkpoint protein whose localization has been characterized, is also recruited to centromeric regions during the early stages of mitosis; however, a fraction of SpBub1 remains at kinetochores also during metaphase and anaphase (Bernard et al., 1998). Altogether, these observations account for previous evidence that implicated kinetochores in transmitting an inhibitory signal in the presence of monooriented chromosomes (Campbell and Gorbsky, 1995; Nicklas et al., 1995; Rieder et al., 1995). Moreover, the fact that a mutation affecting the yeast kinetochore protein Ndc10 causes defects in the mitotic checkpoint (Tavormina and Burke, 1998) further strengthens the notion of a critical role of kinetochores in this checkpoint.
Mad and Bub proteins are also likely to be involved in detecting endogenous errors during mitosis; in fact, budding yeast mad mutants, as well as fission yeast bub1 mutants, show an increased frequency of spontaneous chromosome loss (Li and Murray, 1991; Bernard et al., 1998), and microinjection of anti-Mad2 antibodies into mammalian cells results in a premature anaphase onset (Gorbsky et al., 1998). Furthermore, overexpression of a dominant negative version of the murine Bub1 causes precocious exit from mitosis (Taylor and McKeon, 1997). Bub1 is a protein kinase that can bind and phosphorylate Bub3 (Roberts et al., 1994), whereas Bub3 is required in human cells for the correct localization of Bub1 at kinetochores (Taylor et al., 1998). Mad1 forms a complex with Mad2 (Chen et al., 1998) and becomes phosphorylated when the mitotic checkpoint is activated; its phosphorylation depends on Mad2, Bub1, and Bub3, but not on Bub2 and Mad3 (Hardwick et al., 1995). Mps1, a protein kinase required for spindle pole body duplication and mitotic checkpoint function (Weiss and Winey, 1996), appears to phosphorylate Mad1 directly (Hardwick et al., 1996). A combination of genetics and biochemistry based on Mad1 phosphorylation has partially defined a signal transduction cascade that is activated in response to spindle damage and results in cell cycle arrest (Hardwick et al., 1995; Rudner and Murray, 1996; Elledge, 1996). This analysis places Bub1, Bub3, and Mps1 upstream of Mad1 and Mad2, while Bub2 and Mad3 might act downstream. Altogether, the above data suggest that Mad1, 2, and 3 and Bub1 and Bub3 might form a complex at unattached kinetochores and work in concert to inhibit cell cycle progression, while the role of Bub2 has not yet been defined.
Mad and Bub proteins are thought to be signal transducers of the mitotic checkpoint pathway. Recently, a great deal of evidence points at the anaphase-promoting complex (APC) as the major target of the mitotic checkpoint. The APC is required for ubiquitination and subsequent degradation via the proteasome of two sets of proteins: anaphase inhibitors, like Pds1 in S. cerevisiae (Cohen-Fix et al., 1996) and Cut2 in S. pombe (Funabiki et al., 1996, 1997), and several other substrates ubiquitinated at the end of mitosis, like Ase1 (Juang et al., 1997), Cdc5 (Charles et al., 1998; Shirayama et al., 1998), and B-type cyclins (Irniger et al., 1995; King et al., 1995; Sudakin et al., 1995). While degradation of Pds1/Cut2 at the metaphase/ anaphase transition is required for sister chromatid separation (Yamamoto et al., 1996; Ciosk et al., 1998), degradation of the budding yeast B-type cyclins Clb1-4 contributes to the inactivation of the Clb1-4/Cdc28 kinases, which is necessary for exit from mitosis and the subsequent round of budding and replication (Surana et al., 1993; Schwob et al., 1994; Dahmann et al., 1995; Piatti et al., 1996). The APC substrate specificity towards the two sets of proteins is conferred by cofactors containing WD-40 motifs, such as Cdc20 and Hct1/Cdh1, which bind to the APC and activate its ubiquitin ligase activity (Townsley and Ruderman, 1998). Cdc20 (called Slp1 in fission yeast, Fizzy in Drosophila, and p55Cdc in vertebrates) is responsible for ubiquitination of Pds1 and entry into anaphase (Philp and Glover, 1997; Fang et al., 1998a; Kallio et al., 1998; Lim et al., 1998; Visintin et al., 1998; Shirayama et al., 1998), whereas Hct1/Cdh1 (homologous to the Drosophila Fizzy-related protein and Ste9 in S. pombe; Sigrist and Lehner, 1997; Kitamura et al., 1998; Kominami et al., 1998) promotes ubiquitination of B-type cyclins, Ase1 and Cdc5 (Schwab et al., 1997; Visintin et al., 1997; Charles et al., 1998). CDC20 is an essential gene; temperature-sensitive cdc20 mutations cause cells to arrest before anaphase, due to a failure in degrading Pds1 (Yamamoto et al., 1996; Shirayama et al., 1998). However, when the PDS1 gene is deleted, cdc20 mutants arrest in telophase with high levels of Clb2/Cdc28 kinase, suggesting that Cdc20 might also play a role in promoting ubiquitination of mitotic cyclins or other APC substrates at the end of mitosis (Lim et al., 1998; Shirayama et al., 1998). Accordingly, Drosophila Fizzy is involved in the degradation of cyclins A and B (Dawson et al., 1995; Sigrist et al., 1995; Lorca et al., 1998). Unlike CDC20, HCT1 is totally dispensable for cell viability, despite the inability of hct1 mutants to trigger degradation of cyclins B in G1 (Schwab et al., 1997; Visintin et al., 1997; Zachariae et al., 1998). On the other hand, viability of hct1 cells depends on the presence of Sic1, a specific inhibitor of Clb1-4/Cdc28 kinases (Mendenhall, 1993; Schwob et al., 1994; Schwab et al., 1997; Visintin et al., 1997). How are Cdc20/APC and Hct1/APC complexes regulated during the cell cycle? Cdc20 is an unstable protein, which accumulates early in mitosis and gets degraded in G1 and S phases with a process at least partially dependent on the APC (Fang et al., 1998a; Prinz et al., 1998; Shirayama et al., 1998). Conversely, Hct1 is present at roughly constant levels throughout the cell cycle (Fang et al., 1998b; Prinz et al., 1998), but its association with the APC is negatively regulated by the phosphorylation state of the protein (Zachariae et al., 1998). Both Cln- and Clb-dependent CDKs promote Hct1 phosphorylation during S, G2, and M phases, thereby preventing its binding to the APC. Dephosphorylation of Hct1 at the end of mitosis is triggered both by Sic1-dependent inactivation of Clb1-6/ Cdc28 kinases and directly by the activity of the Cdc14 phosphatase, which, by dephosphorylating and activating Sic1 at the same time, engages a feedback loop (Zachariae et al., 1998; Visintin et al., 1998). These results strongly support the notion that inactivation of Clb/Cdc28 kinases is a key event in exiting the cell cycle, not only because of its self-sustaining nature, but also because it leads to Hct1 dephosphorylation and activation, and, therefore, to the Hct1/APC-dependent degradation of other substrates.
Mad1, 2, and 3 have been found to interact with Cdc20 in yeast cells (Hwang et al., 1998); furthermore, Mad2 has been shown to associate to Slp1/p55Cdc (Fang et al., 1998b; Kallio et al., 1998; Kim et al., 1998; Wassmann and Benezra, 1998) and the APC (Li et al. 1997; Wassmann and Benezra, 1998) when the checkpoint is active. Overexpression of CDC20 or mutated versions of the protein that are no longer able to bind Mad1, 2, and 3 cause an inability to halt the cell cycle in response to microtubule depolymerization (Lim and Surana, 1996; Hwang et al., 1998; Schott and Hoyt, 1998), suggesting that Mad proteins block cell cycle progression by inhibiting Cdc20. Due to the similar subcellular localization of Mad1, 2, 3, Bub1, and Bub3, current models envision these five proteins to monitor kinetochore attachment to microtubules and to prevent, through inhibition of Cdc20, the APC-dependent ubiquitination of Pds1 and cyclins, thereby blocking the onset of anaphase and exit from mitosis.
Very little is known about the role of Bub2 in the mitotic checkpoint and how it interacts with the other Mad and Bub proteins. Both Bub2 and its fission yeast counterpart, Cdc16, are required to maintain high levels of histone H1 kinase activity in response to spindle defects (Hoyt et al., 1991; Fankhauser et al., 1993). In addition, Cdc16 has been implicated in coordinating mitosis with septation and cytokinesis (Fankhauser et al., 1993). Some evidence suggests that Bub2 might play a different role from the other Mad and Bub proteins in activating the mitotic checkpoint. As mentioned above, unlike the other Mad and Bub proteins, Bub2 is not required for the anaphase delay generated by centromere DNA mutations or impaired kinetochore function (Wang and Burke, 1995; Pangilinan and Spencer, 1996). Thus, it has been proposed that Bub2 might be part of a separate checkpoint that responds to abnormal spindle structure rather than to the lack of kinetochore-microtubule attachment (Wang and Burke, 1995).
To gain new insights into the role of Bub2 in the mitotic checkpoint, we have investigated the subcellular localization of Bub2 in budding yeast and its relationships with the other checkpoint proteins. We find that Bub2 is constitutively localized at spindle pole bodies throughout the cell cycle and in the presence of microtubule depolymerizing drugs. Furthermore, we show that BUB2 belongs to a different epistasis group from MAD1 and MAD2, since bub2 mad2 and bub2 mad1 double mutants are much more defective in the mitotic checkpoint than either single mutant or mad1 mad2 cells. Finally, we find that cell cycle progression of bub2 mutant cells in the presence of nocodazole depends on Cdc26, a dispensable APC subunit neither required in wild-type cells for anaphase and exit from mitosis during an unperturbed cell cycle at 25°C (Zachariae et al., 1996), nor in mad2 cells in the presence of microtubule depolymerizing drugs. Together these data suggest that Bub2 activates the checkpoint via a distinct pathway from that involving the other Mad and Bub proteins characterized so far.
Materials and Methods
Strains, Media, and Reagents
All yeast strains were derivatives of or were backcrossed at least three times to W303 (ade2-1, trp1-1, leu2-3,112, his3-11,15, ura3, and ssd1). Strains used for this work are listed in Table I. Cells were grown in YEP medium (1% yeast extract, 2% bactopeptone, and 50 mg/liter adenine) supplemented with 2% glucose (YEPD). α factor was used at 2 μg/ml, unless otherwise stated, nocodazole at 15 μg/ml, benomyl at 12.5 μg/ml and hydroxyurea at 150 mM. All the experiments were performed at 25°C, with the exception of those involving the ts mutants mps1-1 and ndc10-1, which were performed at 37°C.
Table I.
Yeast Strains Used in This Study
| Strain | Relevant genotype | |
|---|---|---|
| ySP460 | MATa, ndc10::NDC10myc6::TRP1 | |
| ySP464 | MATa, mad2::URA3, leu2::LEU2::tetR-GFP, ura3::URA3::224XtetO | |
| ySP520 | MATa, cdc26::K.l.URA3, leu2::LEU2::tetR-GFP, ura3::URA3::224XtetO | |
| ySP601 | MATa, leu2::LEU2::tetR-GFP, ura3::URA3::224XtetO | |
| ySP710 | MATa, bub2::BUB2myc9::TRP1 | |
| ySP862 | MATa, bub2::URA3, leu2::LEU2::tetR-GFP, ura3::URA3::224XtetO | |
| ySP874 | MATa, mad1::LEU2, bub2::BUB2myc9::TRP1 | |
| ySP875 | MATa, mad2::URA3, bub2::BUB2myc9::TRP1 | |
| ySP876 | MATa, mad3, bub2::BUB2myc9::TRP1 | |
| ySP877 | MATa, bub1-1, bub2::BUB2myc9::TRP1 | |
| ySP879 | MATa, ndc10-1, bub2::BUB2myc9::TRP1 | |
| ySP880 | MATa, mps1-1, bub2::BUB2myc9::TRP1 | |
| ySP906 | MATa, bub3::LEU2, bub2::BUB2myc9::TRP1 | |
| ySP921 | MATa, mad2::URA3, bub2::URA3, leu2::LEU2::tetR-GFP, ura3::URA3::224XtetO | |
| ySP978 | MATa, cdc26::K.l.URA3, bub2::URA3, leu2::LEU2::tetR-GFP, ura3::URA3::224XtetO | |
| ySP984 | MATa, mad1::LEU2, leu2::LEU2::tetR-GFP, ura3::URA3::224XtetO | |
| ySP987 | MATa, mad2::URA3, mad1::LEU2, leu2::LEU2::tetR-GFP, ura3::URA3::224XtetO | |
| ySP1016 | MATa, mad2::TRP1, bub2::HIS3, leu2::LEU2::tetR-GFP, ura3::URA3::224XtetO, PDS1myc18::LEU2 | |
| ySP1072 | MATa, bub2::HIS3, leu2::LEU2::tetR-GFP, ura3::URA3::224XtetO, PDS1myc18::LEU2 | |
| ySP1094 | MATa, mad2::URA3, cdc26::K.l.URA3, leu2::LEU2::tetR-GFP, ura3::URA3::224XtetO | |
| ySP1104 | MATa, mad1::LEU2, bub2::HIS3, leu2::LEU2::tetR-GFP, ura3::URA3::224XtetO | |
| ySP1125 | MATa, leu2::LEU2::tetR-GFP, ura3::URA3::224XtetO, PDS1myc18::LEU2 | |
| ySP1145 | MATa, mad2::TRP1, leu2::LEU2::tetR-GFP, ura3::URA3::224XtetO, PDS1myc18::LEU2 |
Plasmid Constructions and Genetic Manipulations
Standard genetic techniques were used to manipulate yeast strains (Sherman, 1991) and standard protocols were used for genetic manipulations (Maniatis et al., 1992). For tagging BUB2 at the COOH terminus, a NotI cassette containing nine tandem repeats of the myc epitope (myc9; Piatti et al., 1996) was inserted before the termination codon of the HindIII-EcoRI COOH-terminal fragment of BUB2 subcloned in Yiplac204. The resulting plasmid (pSP49) was cut with BclI for integration at the BUB2 locus of W303, thus generating a full-length tagged version of BUB2 flanked by a truncated untagged version of the gene (strain ySP710). For tagging NDC10 at the COOH terminus, a NotI cassette containing six copies of the myc epitope (myc6; Piatti et al., 1996) was inserted before the termination codon of the NdeI/XbaI COOH-terminal fragment of NDC10 subcloned in pRS304. The resulting plasmid (pSP18) was cut with BclI for integration at the NDC10 locus of W303, thus generating a full-length tagged version of NDC10 flanked by a truncated untagged version of the gene (strain ySP460).
Western Blot Analysis
For Western blot analysis, protein extracts were prepared by TCA precipitation (see Fig. 3) as previously described (Piatti et al., 1996) or as described in Surana et al., 1993 (see Fig. 6 B). Proteins were transferred to Protran membranes (Schleicher and Schuell). myc-tagged Bub2 and Pds1 were detected with 9E10 mAb, whereas polyclonal antibodies (Amon et al., 1992) were used to detect Clb2. Anti-actin antibodies were purchased from Sigma Chemical Co. Secondary antibodies were purchased from Amersham and proteins were detected by an enhanced chemiluminescence system according to the manufacturer.
Figure 3.
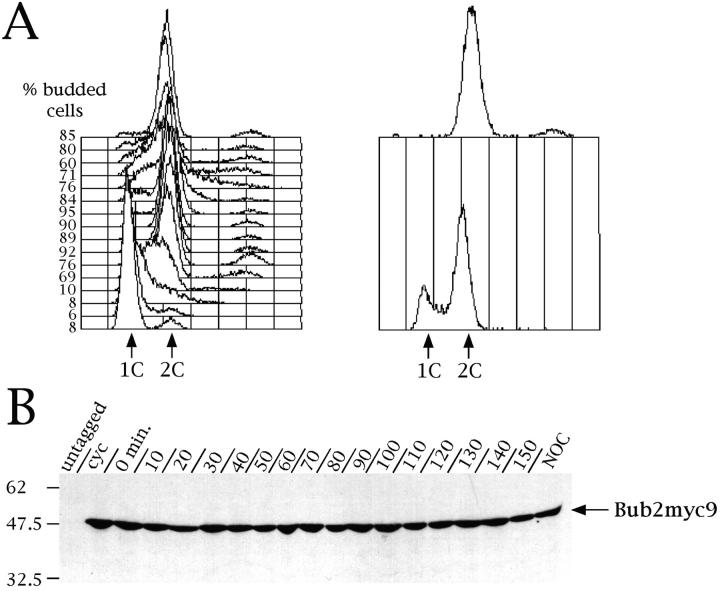
Bub2 protein levels do not vary during the cell cycle. A cycling culture of ySP710 strain (Bub2myc9) was arrested with α factor (time = 0) and then released from the G1 block. An aliquot of the synchronized culture was also incubated for 3 h with nocodazole. At different time points cell samples were collected to monitor cell cycle progression by flow cytometry analysis of the DNA content and to follow the kinetics of budding (A), or to study Bub2 protein levels by Western analysis using anti-myc (9E10) antibodies (B). During the course of this experiment localization of Bub2myc9 in chromosome spreads was also investigated and did not show any qualitative difference with staining shown in Fig. 2. cyc, cycling cells; NOC, nocodazole.
Figure 6.
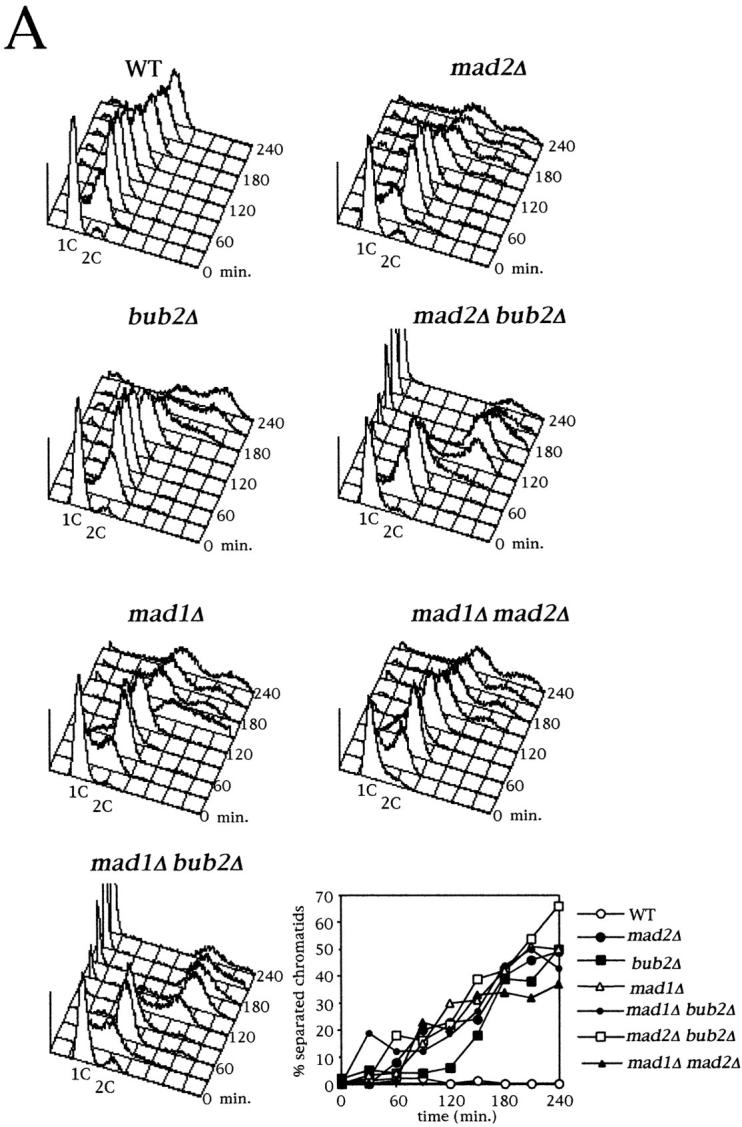
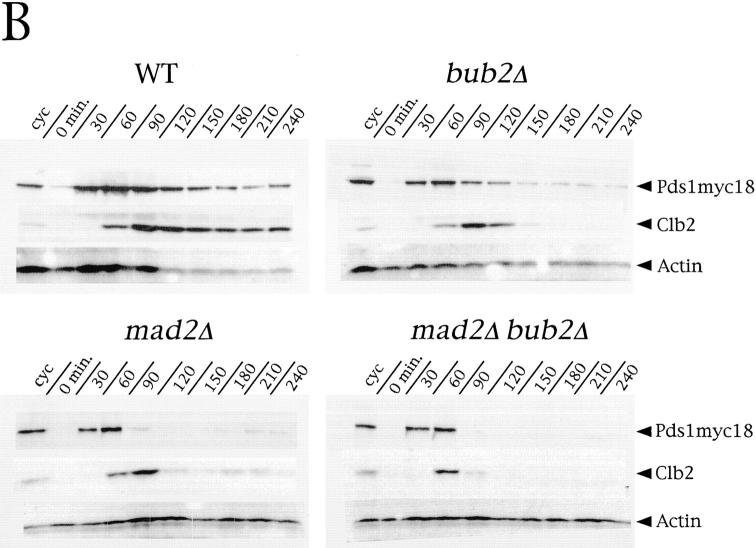
Bub2 prevents exit from mitosis and rereplication in nocodazole via a distinct pathway from Mad1/2. (A) Logarithmically growing cultures of strains ySP601 (WT), ySP464 (mad2Δ), ySP862 (bub2Δ), ySP984 (mad1Δ), ySP1104 (mad1Δ bub2Δ), ySP921 (mad2Δ bub2Δ), and ySP984 (mad1Δ mad2Δ) were synchronized in G1 with α factor (time = 0) and then released into medium containing or lacking (not shown) nocodazole. At the indicated time points DNA contents and loss of cohesion between sister chromatids were analyzed. (B) Cell cultures of strains ySP1125 (WT), ySP1145 (mad2Δ), ySP1016 (bub2Δ), and ySP1072 (mad2Δ bub2Δ), all expressing a Pds1 version tagged with 18 copies of the myc epitope (Pds1myc18; Shirayama et al., 1998), were synchronized in G1 with α factor (time = 0) and released in the presence of nocodazole. To prevent reaccumulation of Pds1 and Clb2, after 60′ α factor was added back to the cultures at a concentration of 10 μg/ml. Cell samples were collected at the indicated times for Western blot analysis of Pds1myc18 and Clb2. Actin was used as loading control. cyc, cycling culture.
Other Techniques
Flow cytometric DNA quantitation was determined according to Epstein and Cross (1992) on a Becton Dickinson FACScan®. In situ immunofluorescence was performed according to Nasmyth et al. (1990). Visualization of Tet operators using GFP and chromosome spreading were performed as described in Michaelis et al., 1997. Immunostaining of Bub2myc9 and Ndc10myc6 were detected by incubation with the 9E10 mAb followed by indirect immunofluorescence using CY3-conjugated goat anti–mouse Ab (1:500; Amersham); signals were amplified using a CY3-conjugated donkey anti–goat Ab (1:500; Amersham). Spindle pole bodies were visualized with polyclonal anti-Spc72 (Knop and Schiebel, 1998) followed by incubation with FITC-conjugated anti–rabbit Ab (1:50, Jackson ImmunoResearch Labs.).
Results
Deletion of BUB2 or MAD2 Does Not Cause Detectable Advancement of Mitosis during a Normal Cell Cycle
Since the mitotic checkpoint is thought to play an important role while chromosomes try to reach a bipolar attachment to the spindle, and inactivation of proteins involved in the mitotic checkpoint increases the frequency of chromosome loss (Li and Murray, 1991; Bernard et al., 1998), we asked whether the checkpoint proteins Mad2 and Bub2 had any role in the timing of sister chromatid separation during a normal cell cycle. For this purpose, we constructed wild-type, bub2Δ, and mad2Δ strains carrying the tetR-GFP/tetO constructs which allow to monitor sisters separation at the centromeric regions of chromosome V (Michaelis et al., 1997). Cell cultures of the above strains were synchronized in G1 by α factor treatment and released into fresh medium either lacking or containing the microtubule depolymerizing drug nocodazole, which activates the mitotic checkpoint. As previously reported (Hoyt et al., 1991; Li and Murray, 1991; Straight et al., 1996), in the presence of nocodazole both bub2Δ and mad2Δ cells lost the cohesion between sister chromatids, rebudded, and rereplicated, accumulating with DNA contents higher than 2C, whereas wild-type cells arrested in G2 as dumbbells with duplicated but unseparated chromosomes (Fig. 1 B). Interestingly, we reproducibly found that in these conditions bub2Δ mutants separated sister chromatids and started rereplicating later than mad2Δ, suggesting that, in the absence of Bub2, entry into anaphase can be still somewhat delayed by other mechanisms. Without nocodazole, all three strains underwent anaphase and proceeded through the cell cycle with similar kinetics (Fig. 1 A), thus indicating that either Mad2 and Bub2 have no role in the timing of sister chromatid separation during a normal cell cycle or bipolar chromosome attachment to spindle fibers in budding yeast might be a process too fast and efficient in order to allow detection of subtle differences in the timing of anaphase entry in the absence of surveillance mechanisms.
Figure 1.
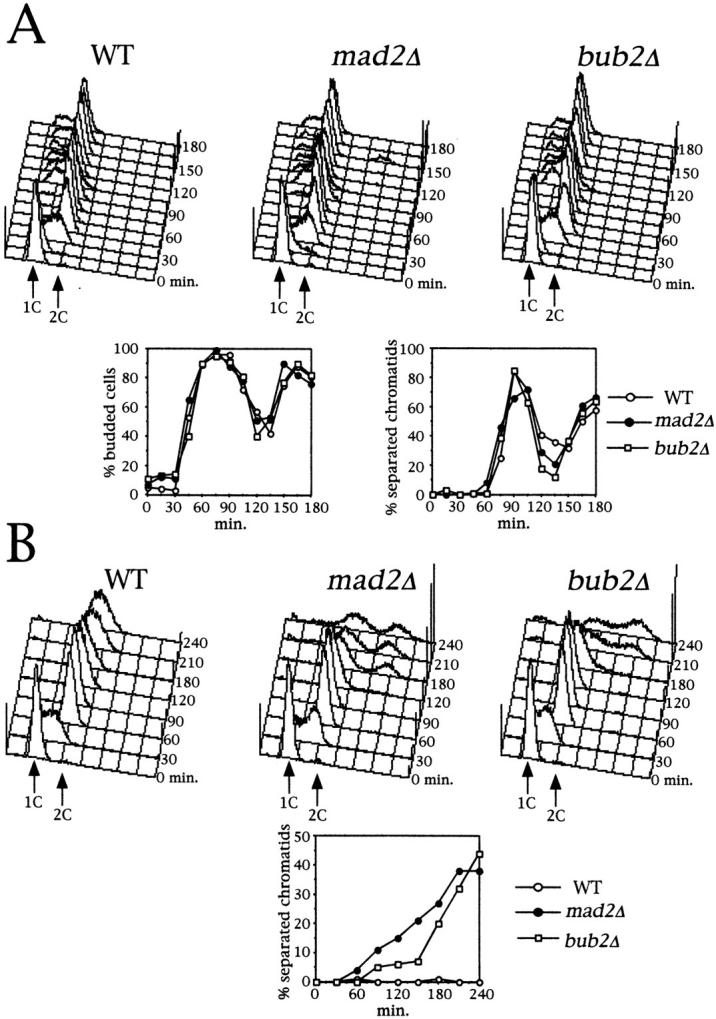
Lack of Mad2 or Bub2 does not advance anaphase during an unperturbed cell cycle. Log phase cultures of ySP601 (WT), ySP464 (mad2Δ), and ySP862 (bub2Δ) strains were arrested with α factor (time = 0) and then released into fresh medium either lacking (A) or containing (B) nocodazole. At the indicated time points cell samples were withdrawn for flow cytometry analysis of the DNA contents and to score budding and separation of sister chromatids. The latter was determined by scoring the signals from tetR-GFP in ethanol-fixed cells. Reproducible results were obtained in three independent experiments.
Bub2p Is Localized at the Spindle Pole Body
Since Mad1, Mad2, Bub1, and Bub3 have been found associated to unattached kinetochores during prophase and prometaphase in higher eukaryotic cells (Chen et al., 1996, 1998; Li and Benezra, 1996; Taylor and McKeon, 1997; Taylor et al., 1998), while the intracellular localization of Bub2 had not been established and little was known about its relationships with other mitotic checkpoint proteins, we analyzed the subcellular localization of Bub2. We tagged Bub2 at the COOH terminus with nine copies of the myc epitope, thus generating the Bub2myc9 variant, which was fully functional, as judged by the checkpoint proficiency of BUB2myc9 cells in nocodazole (see Fig. 3 A and not shown). Since Bub2myc9 staining in whole cells was fairly homogeneous and it was difficult to distinguish specific staining from the background observed with the same antibody on cells lacking tagged Bub2, we studied the protein localization by indirect immunofluorescence on chromosome spreads (Klein et al., 1992; Tanaka et al., 1997). By this technique, squashed nuclei stick to the slide and nucleoplasmic proteins are washed away, allowing detection of proteins bound to subnuclear insoluble structures, such as chromatin and nuclear cytoskeleton. We found that in all nuclei, Bub2myc9 staining was concentrated in one or two dots, very reminiscent of spindle pole bodies (SPBs). We, therefore, repeated the experiment by double staining the spreads with both anti-myc and anti-Spc72 antibodies, the latter of which recognize a constitutive SPB component (Knop and Schiebel, 1998). As shown in Fig. 2 A, the Bub2myc9 and Spc72 stainings completely overlapped, indicating that Bub2 resides at SPBs. As a control, we performed a double staining of the kinetochore protein Ndc10 and Spc72. To this purpose, we used a strain where the only functional copy of NDC10 was expressing a protein tagged with six copies of the myc epitope at the COOH terminus (Ndc10myc6). As it was previously shown for yeast centromeres (Funabiki et al., 1993; Guacci et al., 1997; Jin et al., 1998b), we found that Ndc10myc6 formed clusters near the SPBs, both in cycling and in nocodazole-arrested cells (Fig. 2 B). However, not only Ndc10 clusters had a different shape compared with that of SPBs and Bub2, but not all Ndc10myc6 clusters colocalized with Spc72, while Bub2myc9 always did. Although we cannot fully prove that the Ndc10myc6 clusters correspond to kinetochores, these data strongly suggest that Bub2 does not localize at kinetochores.
Figure 2.
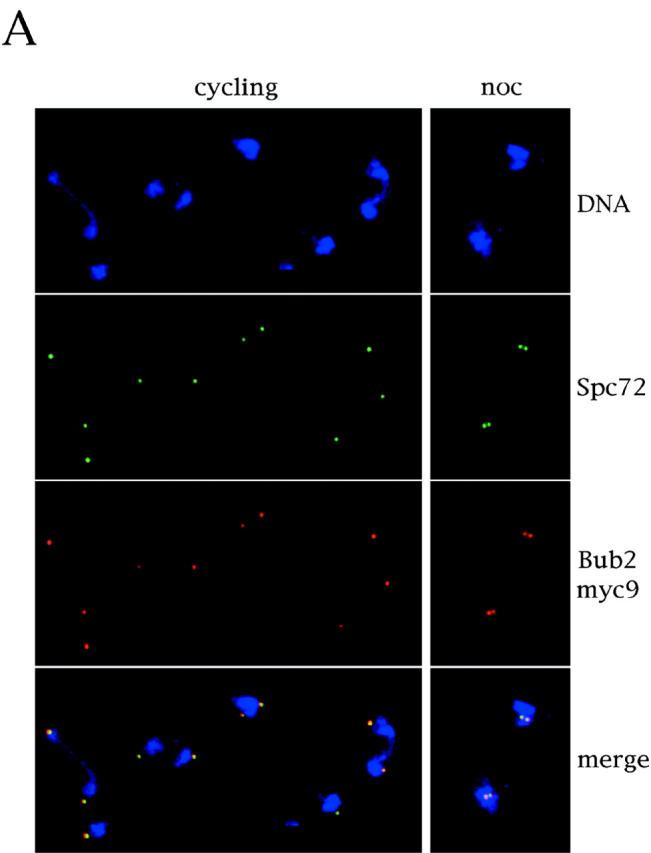
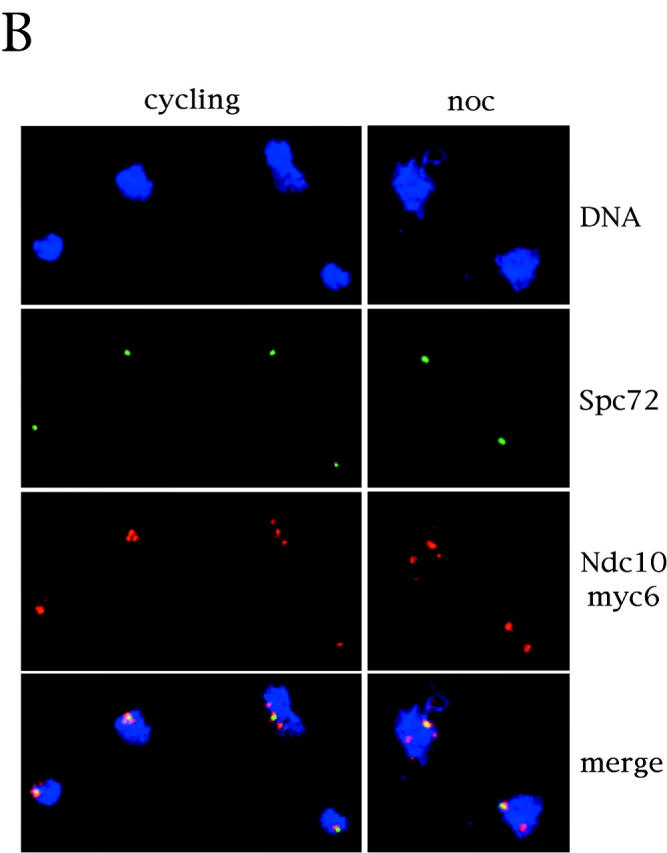
Bub2 is localized at SPBs. (A) Cycling ySP710 (Bub2myc9) cells and nocodazole arrested cells (noc) of the same strain, obtained from the experiment in Fig. 3, were processed for chromosome spreading. Chromosome spreads were decorated with anti-myc antibodies (9E10), followed by detection with CY3-conjugated anti–mouse antibodies, and with anti-Spc72 polyclonal antibodies, followed by detection with FITC-conjugated anti–rabbit antibodies. (B) Cycling ySP460 (Ndc10myc6) cells were treated with nocodazole for three hours. Both cycling and nocodazole arrested cells were processed as in A.
We then investigated further whether Bub2 protein levels and localization varied during the cell cycle and after nocodazole treatment. To this purpose, the BUB2myc9 strains were released from an α factor G1 arrest, and the Bub2myc9 protein levels and localization were analyzed at different time points. An aliquot of the synchronized culture was also incubated for 3 h in nocodazole. Bub2myc9 protein levels remained constant throughout the cell cycle and when the mitotic checkpoint was activated (Fig. 3 B). Furthermore, the protein is constitutively localized at SPBs: G1 cells showed a single dot of Bub2myc9 staining, whereas two bright dots started appearing at the time of bud emergence and entry into S phase, which coincides with the time of SPB duplication (data not shown). About 30–40% of G1 nuclei displayed a fainter staining of Bub2 at SPBs than during the other cell cycle phases, regardless of growth conditions. About 55% of nuclei from nocodazole arrested cells showed two bright dots of Bub2myc9 staining side by side (Fig. 2 A). This is, in these conditions, the typical spatial arrangement observed by electron microscopy for SPBs, which are duplicated but do not migrate apart in the absence of microtubules (Goetsch, L., and B. Byers, personal communication). Therefore, the nuclei showing single SPB signals by immunofluorescence (∼40%) under the same conditions likely had duplicated SPBs, which could not be resolved by this technique. Finally, we found that the localization of Bub2myc9 at SPBs was unaffected in the mitotic checkpoint mutants mad1Δ, mad2Δ, mad3, bub1-1, bub3Δ, mps1-1, and ndc10-1 (data not shown). Therefore, Bub2 is constitutively present at SPBs during the cell cycle and its localization does not require any of the analyzed mitotic checkpoint proteins.
Bub2 Checkpoint Function Is Also Required after SPBs Separation
The finding that Bub2 is localized at SPBs, and SPBs are mostly unseparated in nocodazole, prompted us to investigate whether Bub2 could be required to delay cell cycle progression only before SPB separation. For this purpose, bub2Δ, mad2Δ, and wild-type cells were synchronized by hydroxyurea (HU) treatment, which blocks DNA synthesis but allows duplication and separation of SPBs, thus arresting cells in S phase with short bipolar spindles. Cell cultures were then released from the HU block into nocodazole containing medium, in order to disassemble the previously formed spindles, and cell cycle progression of the three strains was analyzed. Formation and disassembly of the spindle in HU and in nocodazole, respectively, was confirmed by immunostaining of tubulin (data not shown). As shown in Fig. 4, under these conditions wild-type cells arrested in G2, with 2C DNA contents and unseparated sister chromatids, while mad2Δ and bub2Δ cells attempted to undergo anaphase and entered a new cell cycle, as indicated by their ability to separate sister chromatids and rereplicate DNA. Therefore, both Mad2 and Bub2 checkpoint functions are required also after SPBs separation and bipolar spindle formation.
Figure 4.

Bub2 is required for checkpoint activation also after separation of SPBs. Log phase cultures of ySP601 (WT), ySP464 (mad2Δ), and ySP862 (bub2Δ) strains were arrested in early S phase by HU treatment (time = 0) and then released into nocodazole-containing medium. Distribution of DNA contents obtained by flow cytometry analysis at the indicated times and kinetics of sister chromatids separation are shown. cyc, cycling cells.
Bub2 and Mad1/Mad2 Activate the Mitotic Checkpoint via Separate Pathways
Since we found Bub2 concentrated at SPBs, whereas Mad1 and Mad2 are localized, at least in higher eukaryotes, at unattached kinetochores, we asked whether Bub2 and Mad1, 2 might activate the mitotic checkpoint via different pathways.
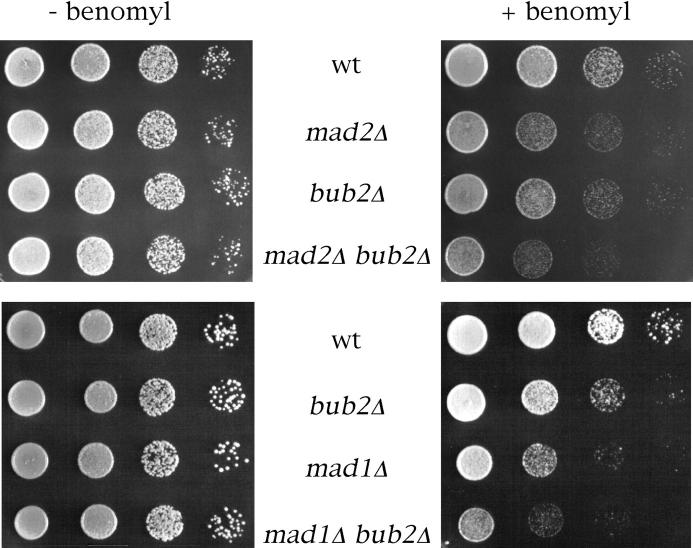
To address this question, we first constructed mad1Δ bub2Δ and mad2Δ bub2Δ double mutants, and compared their sensitivity to the microtubule depolymerizing drug benomyl to that of the single mutants. In fact, hypersensitivity to benomyl of mad and bub mutants correlates with their mitotic checkpoint defect (Hoyt et al., 1991; Li and Murray, 1991), and inactivation of two proteins that lead to checkpoint arrest by different routes should result in a more severe checkpoint defect, and therefore in an increased benomyl sensitivity, compared with inactivation of either one. As shown in Fig. 5, mad1Δ bub2Δ and mad2Δ bub2Δ cells were much more sensitive to benomyl than mad1Δ, mad2Δ, and bub2Δ cells, suggesting that BUB2 belongs to an epistasis group different from that of MAD1 and MAD2.
Figure 5.
BUB2 belongs to a different epistasis group from MAD1 and MAD2. Serial dilutions of YEPD-saturated cultures of strains ySP601 (wt), ySP464 (mad2Δ), ySP862 (bub2Δ), ySP984 (mad1Δ), ySP921 (mad2Δ bub2Δ), and ySP1104 (mad1Δ bub2Δ) were spotted on YEPD plates either containing or lacking benomyl (12.5 μg/ml).
We then verified whether the increased benomyl sensitivity of mad2Δ bub2Δ and mad1Δ bub2Δ double mutants correlated with an enhanced checkpoint defect. Cultures of mad1Δ, mad2Δ, bub2Δ, mad1Δ bub2Δ, mad2Δ bub2Δ, mad1Δ mad2Δ and wild-type strains were arrested with α factor and released in either the presence or the absence of nocodazole. Progression through the cell cycle in the absence of nocodazole was very similar in all strains (data not shown); however, in the presence of the drug mad1Δ bub2Δ and mad2Δ bub2Δ cells started rereplicating much faster and more efficiently than the single mutants and the mad1Δ mad2Δ double mutant (Fig. 6 A). Since the timing and efficiency of rereplication likely reflect the kinetics by which cyclin B–dependent kinases are inactivated, these data suggest that mad1Δ bub2Δ and mad2Δ bub2Δ double mutants advance inactivation of Clb-kinases with respect to the other mutants. Conversely, deletion of BUB2 in mad1Δ or mad2Δ cells did not accelerate their timing of sister chromatids separation (Fig. 6 A), indicating that Bub2 might play only a minor role in controlling degradation of the anaphase inhibitor Pds1. We then measured directly by Western blot analysis the levels of Pds1 and Clb2 during a similar experiment, where mad2Δ, bub2Δ, mad2Δ bub2Δ and wild-type strains expressed a myc-tagged Pds1 protein (Shirayama et al., 1998). As shown in Fig. 6 B, Clb2 was degraded in both mad2Δ and bub2Δ cells in nocodazole, suggesting that rereplication in these mutants is likely to result from their inability to prevent cyclin proteolysis. In addition, Clb2 disappeared more rapidly in the mad2Δ bub2Δ double mutant than in either single mutant, thus confirming that acceleration in the kinetics of rereplication in the double mutant correlates with an advanced inactivation of Clb/Cdk1 kinases. Although bub2Δ cells did also degrade Pds1, they did it very slowly, whereas Pds1 disappeared very rapidly and with nearly identical kinetics in both mad2Δ and mad2Δ bub2Δ cells. Thus, Pds1 degradation and the onset of anaphase appear to be mainly controlled by a Mad2-dependent pathway that does not seem to require Bub2. Altogether these data, while confirming that Mad1 and Mad2 are involved in the same pathway, indicate that Bub2 and Mad2 prevent B-type cyclin proteolysis by different routes, since the concomitant lack of Mad2 (or Mad1) and Bub2 has an additive effect on Clb2 degradation, exit from mitosis and entry into a new cell cycle. Conversely, Bub2 seems to play only a minor role, if any, in controlling the onset of anaphase.
The Cdc26 APC Subunit Is Required for Cell Cycle Progression in Nocodazole of bub2Δ but Not mad2Δ Cells
Since mitotic checkpoint arrest involves APC inactivation (He et al., 1997; Li et al., 1997; Hwang et al., 1998; Kim et al., 1998; Schott and Hoyt, 1998), it was possible that different APC subunits might be alternatively involved in the Mad2- and the Bub2-dependent checkpoint response. To address this question, we asked whether various APC subunits were required for cell cycle progression of mad2Δ or bub2Δ mutants in the presence of microtubule depolymerizing drugs. Unfortunately, most apc mutations are temperature sensitive and cause a cell cycle arrest in metaphase already during an otherwise unperturbed cell cycle (Irniger et al., 1995; Zachariae et al., 1996; Hwang and Murray, 1997). Furthermore, bub2Δ mutants show a poor checkpoint defect at 37°C (data not shown). However, we were able to test whether CDC26, which is totally dispensable in a normal cell cycle at 25°C (Zachariae et al., 1996), might become indispensable for cell cycle progression of either mad2Δ or bub2Δ cells in the presence of nocodazole. The role of Cdc26 in the mitotic checkpoint was also of particular interest as we found that cdc26Δ mutants are hypersensitive to benomyl at the permissive temperature (Amato, S., and S. Piatti, unpublished results). Cell cycle progression of mad2Δ cdc26Δ and bub2Δ cdc26Δ double mutants was compared with that of each single mutant after release from α factor in the presence of nocodazole at 25°C. As shown in Fig. 7, bub2Δ cdc26Δ cells failed to proceed in the cell cycle and arrested mostly in G2 with duplicated and unseparated chromosomes. Conversely, deletion of CDC26 did neither affect the time of anaphase onset nor cell cycle progression of mad2Δ cells. These results indicate that under these conditions Cdc26 becomes strictly required for both the onset of anaphase and exit from mitosis in the absence of Bub2 but not of Mad2, again consistently with the involvement of these two proteins in different mitotic checkpoint pathways.
Figure 7.
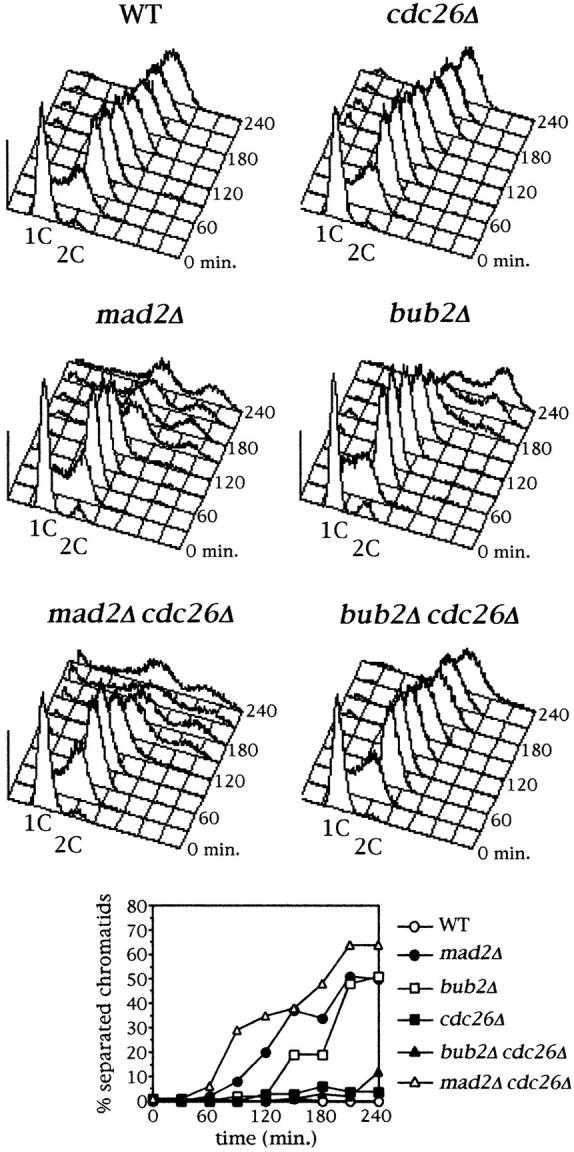
Cdc26 is required for cell cycle progression in nocodazole of bubΔ but not mad2Δ cells. Cyclin cultures of strains ySP601 (WT), ySP464 (mad2Δ), ySP862 (bub2Δ), ySP520 (cdc26Δ), ySP978 (bub2Δ cdc26Δ) and ySP1094 (mad2Δ cdc26Δ) were arrested in G1 by α factor (time = 0) and then released from the block in the presence of nocodazole. Kinetics of replication analyzed by flow cytometry and of sister chromatids separation during the synchronous cell cycles are shown.
Discussion
Are Mitotic Checkpoint Proteins Involved in Controlling the Timing of Mitosis during a Normal Cell Cycle?
The mitotic checkpoint is likely to play a crucial role also during an unperturbed cell cycle, delaying the onset of anaphase until all chromosomes are properly aligned on the bipolar spindle and thereby preventing the occurrence of aneuploidies. Mistakes made during mitosis may be responsible for the abnormal karyotype of many human tumour cells and have an important role in oncogenesis. Loss of function of the p53 tumour suppressor gene impairs the mitotic checkpoint and the normal regulation of centrosome duplication (Fukasawa et al., 1996). In addition, a number of human tumour cell lines have been recently found to be defective in the activation of the mitotic checkpoint in response to microtubule depolymerizing drugs (Li and Benezra, 1996; Cahill et al., 1998). In addition, the chromosomal instability observed in two colorectal cancers is associated to mutational inactivation of BUB1 (Cahill et al., 1998). Finally, expression of dominant negative mutant versions of either murine Bub1 or human Mad1 accelerates exit from mitosis and causes accumulation of multinucleate cells, respectively, even in the absence of microtubule depolymerizing drugs, whereas microinjection of anti-Mad2 antibodies into mammalian cells induces the onset of anaphase before chromosomes have congressed to the metaphase plate (Gorbsky et al., 1998), suggesting that these proteins play an important role in regulating the normal timing of mitosis (Taylor and McKeon, 1997; Jin et al., 1998). We tested directly whether loss of function of MAD2, BUB2, or both, might accelerate progression through mitosis of yeast cells. We found that the kinetics of anaphase and exit from mitosis were unaffected in mad2Δ, bub2Δ (Fig. 1) and mad2Δ bub2Δ cells (data not shown) compared with wild-type. Since mad mutants were shown to increase the frequency of chromosome loss (Li and Murray, 1991), our data suggest that either Bub and Mad proteins are not required for the correct timing of mitosis during an unperturbed cell cycle, or bipolar attachment of yeast chromosomes to spindle fibers is so fast and efficient that does not enable to detect subtle differences in the absence of surveillance mechanisms. In agreement with the latter hypothesis is the finding that in the absence of fission yeast Bub1 cells do not prematurely enter mitosis despite showing massive chromosome missegregation (Bernard et al., 1998).
Two Independent Pathways Involved in the Mitotic Checkpoint
The mechanism by which Mad1, 2, 3 block cell cycle progression in response to spindle depolymerization or monopolar attachment has been recently uncovered. Mad2 has been shown by several groups to bind and inactivate Cdc20/APC, thus causing stabilization of Pds1 and cyclins B with subsequent block of anaphase and mitotic exit (Fang et al., 1998b; Hwang et al., 1998; Kallio et al., 1998; Kim et al., 1998; Wassman and Benezra, 1998). This physical interaction might occur directly at the kinetochore; consistently, the mammalian homologue of budding yeast APC1, Tsg24, was constitutively localized at centromeres (Jörgensen et al., 1998) and p55CDC, homologous to Cdc20, was also found at kinetochores during mitosis (Kallio et al., 1998). However, APC components, p55CDC and HsMad1 have also been localized at centrosomes (Tugendreich et al., 1995; Jin et al., 1998a; Kallio et al., 1998). Our finding that BUB2 and MAD1, 2 belong to different epistasis groups, as indicated by the accelerated degradation of Clb2, exit from mitosis and rereplication of mad1 bub2 and mad2 bub2 compared with that of the corresponding single mutants, suggests that Bub2 might block cell cycle progression in case of monopolar attachment of chromosomes or spindle damage by a pathway distinct from the one dependent on Mad1, 2. This conclusion is further strengthened by the observation that cell cycle progression of bub2Δ, but not mad2Δ, cells in nocodazole depends on the unessential APC subunit Cdc26. Although Cdc26 seems to be constitutively part of the APC, it is required for anaphase and ubiquitination of B-type cyclins only at 37°C; furthermore, it is induced after heat shock (Zachariae et al., 1996). In S. pombe, overexpression of the Cdc26 homologue Hcn1 suppresses a cut9 mutation which causes defects in APC assembly (Yamada et al., 1997). Thus, Cdc26/Hcn1 is a dispensable subunit that might be only required to modulate APC activity under stress conditions or when other APC subunits are defective (Townsley and Ruderman, 1998). The fact that cell cycle progression of mad2, but not bub2, mutants tolerates loss of Cdc26 function suggests that either Bub2 acts by finally inhibiting a special form of APC which requires Cdc26, or that the function of a Cdc26-less APC is somewhat compromised already at 25°C and the lack of Bub2 is not sufficient to circumvent the defect.
The conclusion that Mad1, 2 and Bub2 both contribute to the activation of the mitotic checkpoint via distinct routes can only be drawn for what concerns mitotic exit, which is a consequence of the inactivation of cyclin B–dependent kinases. Conversely, loss of cohesion between sister chromatids, which instead depends on Pds1 degradation, is mostly driven by the inactivation of the Mad1, 2 pathway, as the kinetics of sister chromatids separation of mad1Δ bub2Δ and mad2Δ bub2Δ double mutants is very similar to those displayed by mad1Δ or mad2Δ cells. Obviously, bub2Δ cells are able to degrade Pds1 and attempt to undergo anaphase to a certain extent in the absence of a functional spindle, but this might be a consequence, rather than a primary effect, of the inactivation of mitotic CDKs. Indeed, we found that degradation of Pds1 is very slow and inefficient in bub2Δ cells in the presence of nocodazole, suggesting that Bub2 plays only a minor role in controlling the onset of anaphase. The loss of sister chromatids cohesion we observe in bub2Δ cells treated with nocodazole might just reflect a pathological situation rather than shedding light on the actual role of Bub2. We therefore propose that Bub2's major, if not sole, role in the activation of the checkpoint is to prevent, by inhibiting B-type cyclins proteolysis, inactivation of mitotic CDKs and, therefore, cytokinesis and entry into a new cell cycle. The finding that the fission yeast homologue of Bub2, Cdc16, in addition to having a role in the mitotic checkpoint, is implicated in regulating septation and cytokinesis during a normal cell cycle (Fankhauser et al., 1993; Furge et al., 1998) supports this hypothesis. We currently do not know the molecular mechanism by which the Bub2-dependent pathway contributes to maintaining high levels of these CDKs in nocodazole. In fact, Bub2 might inhibit directly the APC-dependent degradation of cyclins B, by inhibiting Hct1 or Cdc5 or even by interfering with the integrity of the APC subunit composition; alternatively, it could prevent Sic1 accumulation or Cdc28 inhibitory phosphorylations on Thr18 and Tyr19, which have been previously shown to play a role in the mitotic checkpoint (Minshull et al., 1996; Wang and Burke, 1997). Whatever the mechanism is, the absence of Bub2 in cells treated with nocodazole might cause a drop in Clb1-4/Cdc28 kinase activity, which would in turn lead to dephosphorylation and unscheduled activation of Hct1; Hct1/APC, possibly specifically involving the Cdc26 subunit, would then promote degradation of both mitotic cyclins and Pds1, despite Cdc20 is kept inactive by the Mad1, 2 pathway. Several lines of evidence strongly support this model: overexpression of Hct1 in nocodazole arrested cells is able to promote degradation of Pds1, albeit inefficiently (Visintin et al., 1997), whereas deletion of BUB2 is sufficient to drive a slow degradation of Pds1 and continuous cell cycle progression in cdc20 mutants (Shirayama et al., 1998; Tavormina and Burke, 1998).
Relationships between the Function of Mitotic Checkpoint Proteins and Their Subcellular Localization
Many mitotic checkpoint proteins, like Mad1-3, Bub1, and Bub3, have been localized at kinetochores not attached to spindle fibers, such as in prophase and prometaphase of a normal mitosis or in the presence of microtubule disrupting agents (Chen et al., 1996, 1997; Li and Benezra, 1996; Taylor and McKeon, 1997; Taylor et al., 1998). Their localization is fully consistent with previous lines of evidence implicating the kinetochore in transmitting an inhibitory signal to the cell cycle machinery in case of monopolar attachment (Rieder et al., 1994, 1995). Other observations suggested that the mitotic checkpoint also monitors the lack of tension at kinetochores (Li and Nicklas, 1995; Nicklas et al., 1995). Phosphorylation of the kinetochore-associated 3F3/2 epitope correlates with the activation of the mitotic checkpoint (Campbell and Gorbsky, 1995) and is activated by tension (Nicklas et al., 1995). Conversely, localization of Mad2 to kinetochores depends only on microtubule attachment but not on tension (Waters et al., 1998), suggesting that different proteins might be involved in sensing and transducing different signals. Localization of Bub2 at SPBs does not correlate with the activation of the mitotic checkpoint, but is fairly constitutive throughout the cell cycle, raising the questions of how Bub2 function is regulated, whether it plays any role during a normal cell cycle, and which signals it monitors. Whatever the signals are, we have shown that Bub2, as well as Mad2, is required to arrest cell cycle progression also after SPB separation. One possibility is that signals coming from single unattached kinetochores are integrated at SPBs, thus activating the Bub2 dependent checkpoint control. It is worth noting that, unlike in other organisms (Chen et al., 1998), human Mad1 is localized at kinetochores during interphase but at centrosomes during mitosis (Jin et al., 1998a), suggesting that microtubule organizing centers are likely to play a central role in the mitotic checkpoint. Another alternative is that the signal detected by Bub2 might be different from that sensed by the other Mad and Bub proteins; for instance, it has been proposed that Bub2 might be involved in detecting alterations in the spindle structure, since it is not required to arrest the cell cycle in response to impaired kinetochore function (Wang and Burke, 1995). Unfortunately, this hypothesis cannot be tested directly because spindle defects would necessarily also compromise the kinetochore-microtubule attachment. Finally, since we propose that Bub2 only plays a minor role, if any, in regulating the degradation of Pds1 and separation of sister chromatids, we speculate that Bub2 might be involved in the activation of the mitotic checkpoint only later than the other Mad and Bub proteins, by preventing cytokinesis and exit from mitosis until anaphase has taken place. An intriguing hypothesis is that SPBs can somehow monitor when anaphase has been completed, and the inhibitory signal generated by Bub2 is rapidly extinguished when sister kinetochores have reached the poles.
Which proteins does Bub2 interact with at SPBs in order to prevent cytokinesis? Recent work in fission yeast has started to elucidate the mechanism by which the Bub2 homologue Cdc16 might function. In fact, Cdc16 has been found to physically interact with Byr4, which was previously implicated in the control of septation (Song et al., 1996; Furge et al., 1998). The Cdc16/Byr4 complex displays GAP activity towards Spg1 (Furge et al., 1998), a GTPase homologous to S. cerevisiae Tem1 (Shirayama et al., 1994; Schmidt et al., 1997). Remarkably, Spg1 is constitutively bound to SPBs and in the GTP-bound form seems to recruit at SPBs the Cdc7 protein kinase, which is involved in promoting cytokinesis (Fankhauser and Simanis, 1994; Sohrmann et al., 1998). Mutations affecting budding yeast TEM1 cause cells to arrest in telophase and have been recently found to impair the APC-dependent ubiquitination of mitotic cyclins (Shirayama et al., 1994; Jaspersen et al., 1998), thus providing a link between SPBs, APC function, and cytokinesis. The phenotype of tem1 mutants is very similar to that of several mutants defective in late mitotic events, like cdc15, dbf2, and cdc14. Their characterization has uncovered a complex network of genetic interactions which led to the hypothesis that all the corresponding genes might be involved in the same process. In addition, Cdc14, which directly dephosphorylates both Sic1 and Hct1, has been postulated to be the downstream effector of the whole cascade (Visintin et al., 1998).
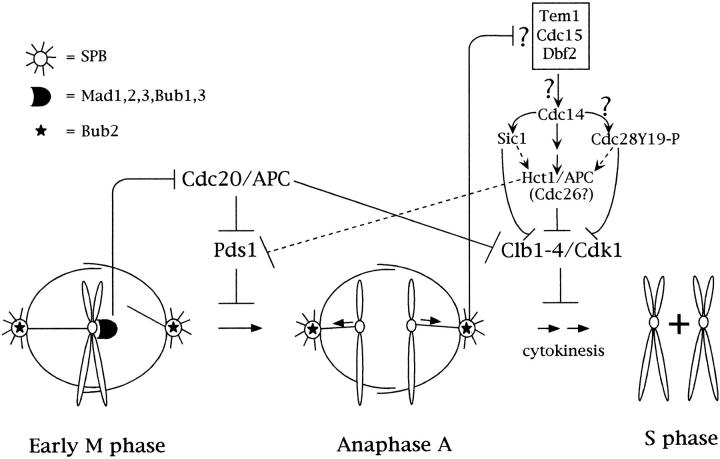
In conclusion, our current model of how the mitotic checkpoint could work is depicted in Fig. 8. During prophase, while chromosomes start to attach to spindle fibers, Mad1, 2, 3, Bub1, and Bub3 present at unattached kinetochores prevent activation of Cdc20 and, therefore, degradation of Pds1 and inactivation of mitotic CDKs. At the same time Bub2, localized at SPBs, also prevents inhibition of Clb1-4/Cdc28 kinases and cytokinesis by an independent mechanism, which might involve inactivation of late mitotic proteins, like Tem1, Cdc15, Dbf2, and Cdc14. Exit from mitosis and cytokinesis are therefore blocked until anaphase has taken place, a process which might switch Bub2 function off. In the absence of Bub2 the ectopic activation of Hct1 triggered by Cdc14 would bring about Pds1 degradation and sister chromatid separation even if Cdc20 is kept inactive.
Figure 8.
A model for the mitotic checkpoint. Early in mitosis, Mad and Bub proteins localized at unattached kinetochores prevent degradation of Pds1 and the separation of sister chromatids by inhibiting Cdc20. Once all chromosomes are attached to spindle fibers in a bipolar way the inhibitory signal is no longer sent by Mad1, 2, 3, Bub1, 3, and anaphase takes place. Meanwhile, Bub2, which resides at SPBs, blocks exit from mitosis and cytokinesis by acting on effectors which so far have not been identified but might include the late mitotic proteins Tem1, Cdc15, Dbf2, and Cdc14. Bub2-mediated inactivation of its targets would inhibit the APC-dependent degradation of B-type cyclins, as well as the accumulation of Sic1 or the inhibitory phosphorylations on Cdc28. The lack of Clb1-4 degradation combined with the lack of inhibition of the corresponding kinases would prevent cytokinesis. After anaphase has taken place, presumably the Bub2-dependent inhibitory signal is shut off. In cells treated with nocodazole inactivation of Bub2 would lead, one way or the other, to a drop in Clb1-4/Cdc28 kinases, which might cause activation of Hct1 and, therefore, B-type cyclins proteolysis, unscheduled degradation of Pds1, and the onset of anaphase. Dashed lines indicate secondary pathways.
Acknowledgments
We are grateful to Rafal Ciosk and Kim Nasmyth for providing the tetR-GFP system and other strains; Michael Knop and Elmar Schiebel for antibodies against Spc72; Loretta Goetsch and Breck Byers for communicating unpublished results; Etienne Schwob for anti-Clb2 antiserum; Pier Carlo Marchisio and Simona Tognin for help with photomicroscopy; Léon Dirick, Marco Muzi Falconi, and Maria Pia Longhese for critical reading of the manuscript.
This work was supported by grants from Associazione Italiana Ricerca sul Cancro to S. Piatti, Cofinanziamento 1997 by Ministero Università e Ricerca Scientifica e Tecnologica, Università di Milano to G. Lucchini, Training and Mobility of Researcher contract ERBFMRXCT98-0212 to S. Piatti and CNR Target Project on Biotechnology grant CT.97.01180.PF49(F).
Abbreviations used in this paper
- APC
anaphase-promoting complex
- BUB
budding uninhibited by benzimidazole
- cyc
cycling
- exp
exponential phase
- HU
hydroxyurea
- MAD
mitotic arrest-deficient
- SPB
spindle pole body
Footnotes
The first two authors contributed equally to this work.
References
- Amon A, Surana U, Muroff I, Nasmyth K. Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae. . Nature. 1992;355:368–371. doi: 10.1038/355368a0. [DOI] [PubMed] [Google Scholar]
- Bernard P, Hardwick K, Javerzat JP. Fission yeast Bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy in mitosis. J Cell Biol. 1998;143:1775–1787. doi: 10.1083/jcb.143.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- Campbell MS, Gorbsky GJ. Microinjection of mitotic cells with the 3F3/2 anti-phosphoepitope antibody delays the onset of anaphase. J Cell Biol. 1995;129:1195–1204. doi: 10.1083/jcb.129.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. . Curr Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- Chen R, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint XMAD2 with unattached kinetochores. Science. 1996;274:242–245. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Chen R, Shevchenko A, Mann M, Murray AW. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase/anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiaeis controlled by the APC-dependent degradation of the anaphase inhibitor Pds1. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Diffley JFX, Nasmyth K. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- Dawson IA, Roth S, Artavanis-Tsakonas S. The Drosophila cell cycle gene fizzy is required for normal degradation of cyclins A and B during mitosis and has homology to the CDC20 gene of Saccharomyces cerevisiae. . J Cell Biol. 1995;129:725–737. doi: 10.1083/jcb.129.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Epstein CB, Cross FR. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998a;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998b;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Marks J, Reymond A, Simanis V. The S. pombe cdc16 gene is required both for maintenance of p34cdc2 kinase activity and regulation of septum formation: a link between mitosis and cytokinesis? . EMBO (Eur Mol Biol Organ) J. 1993;12:2697–2704. doi: 10.1002/j.1460-2075.1993.tb05931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande GF, Woude Abnormal centrosome amplification in the absence of p53. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Hamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis is required for sister chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Nagao K, Tanaka H, Yasuda H, Hunt T, Yanagida M. Fission yeast Cut2 required for anaphase has two destruction boxes. EMBO (Eur Mol Biol Organ) J. 1997;16:5977–5987. doi: 10.1093/emboj/16.19.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furge KA, Wong K, Armstrong J, Balasubramanian M, Albright CF. Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr Biol. 1998;8:947–954. doi: 10.1016/s0960-9822(98)70394-x. [DOI] [PubMed] [Google Scholar]
- Gorbsky GJ, Chen R, Murray AW. Microinjection of antibody to Mad2 protein into mammalian cells in mitosis induces premature anaphase. J Cell Biol. 1998;141:1193–1205. doi: 10.1083/jcb.141.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Centromere position in budding yeast: evidence for anaphase A. Mol Biol Cell. 1997;8:957–972. doi: 10.1091/mbc.8.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Murray AW. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- He X, Patterson TE, Sazer S. The Schizosaccharomyces pombespindle checkpoint protein mad2 blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl Acad Sci USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiaegenes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Murray AW. A novel yeast screen for mitotic arrest mutants identifies DOC1, a new gene involved in cyclin proteolysis. Mol Biol Cell. 1997;8:1877–1887. doi: 10.1091/mbc.8.10.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–277. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Tinker-Kulberg RL, Morgan DO. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. . Mol Biol Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D, Spencer F, Jeang K. Human T cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell. 1998a;93:81–91. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- Jin Q, Trelles-Sticken E, Scherthan H, Loidl J. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J Cell Biol. 1998b;141:21–29. doi: 10.1083/jcb.141.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörgensen PM, Brundell E, Starborg M, Höög C. A subunit of the anaphase-promoting complex is a centromere-associated protein in mammalian cells. Mol Cell Biol. 1998;18:468–476. doi: 10.1128/mcb.18.1.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang YL, Huang J, Peters JM, McLaughlin ME, Tai CY, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- Kallio M, Weinstein J, Daum JR, Burke DJ, Gorbsky GJ. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. J Cell Biol. 1998;141:1393–1406. doi: 10.1083/jcb.141.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Maekawa H, Shimoda C. Fission yeast Ste9, a homolog of Hct1/Cdh1 and Fizzy-related, is a novel negative regulator of cell cycle progression during G1-phase. Mol Biol Cell. 1998;9:1065–1080. doi: 10.1091/mbc.9.5.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Laroche T, Cardenas ME, Hofmann JF, Schweizer D, Gasser SM. Localization of Rap1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO (Eur Mol Biol Organ) J. 1998;17:3952–3967. doi: 10.1093/emboj/17.14.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami K, Seth-Smith H, Toda T. Apc10 and Ste9/Srw1, two regulators of the APC-cyclosome, as well as the CDK inhibitor Rum1 are required for G1 cell-cycle arrest in fission yeast. EMBO (Eur Mol Biol Organ) J. 1998;17:5388–5399. doi: 10.1093/emboj/17.18.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. . Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Li Y, Gorbea C, Mahaffey D, Reichsteiner M, Benezra R. MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc Natl Acad Sci USA. 1997;94:12431–12436. doi: 10.1073/pnas.94.23.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HH, Surana U. Cdc20, a b-transducin homologue, links RAD9-mediated G2/M checkpoint control to mitosis in Saccharomyces cerevisiae. . Mol Gen Genet. 1996;253:138–148. doi: 10.1007/s004380050306. [DOI] [PubMed] [Google Scholar]
- Lim HH, Goh P, Surana U. Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr Biol. 1998;8:231–234. doi: 10.1016/s0960-9822(98)70088-0. [DOI] [PubMed] [Google Scholar]
- Lorca T, Castro A, Martinez AM, Vigneron S, Morin N, Sigrist S, Lehner C, Dorée M, Labbé JC. Fizzy is required for activation of the APC/cyclosome in Xenopusegg extracts. EMBO (Eur Mol Biol Organ) J. 1998;17:3565–3575. doi: 10.1093/emboj/17.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., E.F. Fritsch, and J. Sambrook. 1992. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Mendenhall MD. An inhibitor of p34CDC28 protein kinase activity from Saccharomyces cerevisiae. . Science. 1993;259:216–219. doi: 10.1126/science.8421781. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Minshull J, Straight A, Rudner AD, Dernburg AF, Belmont A, Murray AW. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr Biol. 1996;6:1609–1620. doi: 10.1016/s0960-9822(02)70784-7. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Adolf G, Lydall D, Seddon A. The identification of a second cell cycle control in the HOpromoter in yeast: cell cycle regulation of SWI5 nuclear entry. Cell. 1990;62:631–647. doi: 10.1016/0092-8674(90)90110-z. [DOI] [PubMed] [Google Scholar]
- Nicklas RB, Ward S, Gorbsky GJ. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J Cell Biol. 1995;130:929–939. doi: 10.1083/jcb.130.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangilinan F, Spencer F. Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol Biol Cell. 1996;7:1195–1208. doi: 10.1091/mbc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philp AV, Glover DM. Mutations affecting chromatid separation in Drosophila: the fizzy metaphase arrest persists in pimples fizzy and fizzy three rowsdouble mutants. Exp Cell Res. 1997;230:103–110. doi: 10.1006/excr.1996.3396. [DOI] [PubMed] [Google Scholar]
- Piatti S, Böhm T, Cocker JH, Diffley JFX, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- Prinz S, Hwang ES, Visintin R, Amon A. The regulation of Cdc20 proteolysis reveals a role for the APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr Biol. 1998;8:750–760. doi: 10.1016/s0960-9822(98)70298-2. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Schultz A, Cole R, Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J Cell Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts BT, Farr KA, Hoyt MA. The Saccharomyces cerevisiae checkpoint gene BUB1encodes a novel protein kinase. Mol Cell Biol. 1994;14:8282–8291. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner AD, Murray AW. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- Schmidt S, Sohrmann M, Hofmann K, Woollard A, Simanis V. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. . Genes Dev. 1997;11:1519–1534. doi: 10.1101/gad.11.12.1519. [DOI] [PubMed] [Google Scholar]
- Schott EJ, Hoyt MA. Dominant alleles of Saccharomyces cerevisiae CDC20reveal its role in promoting anaphase. Genetics. 1998;148:599–610. doi: 10.1093/genetics/148.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Schwob E, Böhm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. . Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Matsui Y, Toh A -E. The yeast TEM1gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol Cell Biol. 1994;14:7476–7482. doi: 10.1128/mcb.14.11.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5 and the WD-repeat protein Cdc20/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist SJ, Lehner CF. Drosophila fizzy-relateddown-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell. 1997;90:671–681. doi: 10.1016/s0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- Sigrist S, Jacobs J, Stratmann R, Lehner CF. Exit from mitosis is regulated by fizzy and the sequential destruction of cyclins A, B and B3. EMBO (Eur Mol Biol Organ) J. 1995;14:4827–4838. doi: 10.1002/j.1460-2075.1995.tb00164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann M, Schmidt S, Hagan I, Simanis V. Asymmetric segregation on spindle poles of the Schizosaccharomyces pombeseptum-inducing protein kinase Cdc7p. Genes Dev. 1998;12:84–94. doi: 10.1101/gad.12.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Mach KE, Chen C, Reynolds T, Albright CF. A novel suppressor of ras1 in fission yeast, byr4, is a dosage-dependent inhibitor of cytokinesis. J Cell Biol. 1996;133:1307–1319. doi: 10.1083/jcb.133.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Herschko J, Luca FC, Ruderman JV, Herschko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–198. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO (Eur Mol Biol Organ) J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Tavormina PA, Burke DJ. Cell cycle arrest in cdc20 mutants of Saccharomyces cerevisiaeis independent of Ndc10p and kinetochore function but requires a subset of spindle checkpoint genes. Genetics. 1998;148:1701–1713. doi: 10.1093/genetics/148.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley FM, Ruderman JV. Proteolytic ratchets that control progression through mitosis. Trends in Cell Biol. 1998;8:238–244. doi: 10.1016/s0962-8924(98)01268-9. [DOI] [PubMed] [Google Scholar]
- Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Wang Y, Burke DJ. Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. . Mol Cell Biol. 1995;15:6838–6844. doi: 10.1128/mcb.15.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Burke DJ. Cdc55p, the B-type regulatory subunit of protein phosphatase 2A, has multiple functions in mitosis and is required for the kinetochore/spindle checkpoint in Saccharomyces cerevisiae. . Mol Cell Biol. 1997;17:620–626. doi: 10.1128/mcb.17.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassman K, Benezra R. Mad2 transiently associates with an APC/ p55Cdc complex during mitosis. Proc Natl Acad Sci USA. 1998;95:11193–11198. doi: 10.1073/pnas.95.19.11193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Chen R, Murray AW, Salmon ED. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Kumada K, Yanagida M. Distinct subunit functions and cell cycle regulated phosphorylation of 20S APC/cyclosome required for anaphase in fission yeast. J Cell Sci. 1997;110:1793–1804. doi: 10.1242/jcs.110.15.1793. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Shin TH, Galova M, Obermaier B, Nasmyth K. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. . Science. 1996;274:1201–1204. doi: 10.1126/science.274.5290.1201. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]