Abstract
Meiotic recombination requires pairing of homologous chromosomes, the mechanisms of which remain largely unknown. When pairing occurs during meiotic prophase in fission yeast, the nucleus oscillates between the cell poles driven by astral microtubules. During these oscillations, the telomeres are clustered at the spindle pole body (SPB), located at the leading edge of the moving nucleus and the rest of each chromosome dangles behind. Here, we show that the oscillatory nuclear movement of meiotic prophase is dependent on cytoplasmic dynein. We have cloned the gene encoding a cytoplasmic dynein heavy chain of fission yeast. Most of the cells disrupted for the gene show no gross defect during mitosis and complete meiosis to form four viable spores, but they lack the nuclear movements of meiotic prophase. Thus, the dynein heavy chain is required for these oscillatory movements. Consistent with its essential role in such nuclear movement, dynein heavy chain tagged with green fluorescent protein (GFP) is localized at astral microtubules and the SPB during the movements. In dynein-disrupted cells, meiotic recombination is significantly reduced, indicating that the dynein function is also required for efficient meiotic recombination. In accordance with the reduced recombination, which leads to reduced crossing over, chromosome missegregation is increased in the mutant. Moreover, both the formation of a single cluster of centromeres and the colocalization of homologous regions on a pair of homologous chromosomes are significantly inhibited in the mutant. These results strongly suggest that the dynein-driven nuclear movements of meiotic prophase are necessary for efficient pairing of homologous chromosomes in fission yeast, which in turn promotes efficient meiotic recombination.
Keywords: meiosis, homologous chromosome pairing, dynein, fission yeast, telomere
Meiotic recombination contributes to the diversity in genetic information of eukaryotic cells. This event requires a physical association of homologous regions on each pair of homologous chromosomes. It occurs during meiotic prophase. In most organisms, the pairing takes place along the entire length of homologous chromosomes, and is followed by the formation of the synaptonemal complex (SC)1, a tripartite structure that appears to link the chromosomes. To accomplish successful chromosome pairing, cells possess a mechanism(s) that allows an efficient encounter of the homologous regions followed by their association without chromosome entangling. How cells accomplish proper pairing of homologous chromosomes remains an unsolved problem.
Cytological observations in many organisms have demonstrated that during meiotic prophase chromosomes form a characteristic bouquet-like arrangement by clustering their telomeres (Chikashige et al., 1994; Scherthan et al., 1996; Bass et al., 1997; Trelles-Sticken et al., 1999; reviewed in Fussell, 1987; John, 1990; Loidl, 1990; Therman and Susman, 1992; Dernburg et al., 1995). It is frequently observed that the centrosome is located near the cluster of telomeres (Trelles-Sticken et al., 1999; reviewed in Loidl, 1990; Therman and Susman, 1992; Dernburg et al., 1995). In addition to the clustering of telomeres, a specific pattern of nuclear movement has been observed during meiotic prophase in some organisms. For example, the nucleus rotates and oscillates during the bouquet stage in rodent spermatocytes (Yao and Ellingson, 1969; Parvinen and Söderström, 1976; Salonen et al., 1982). Some plant cells may have similar nuclear movements, since their nuclei have been observed at positions away from the center of the cell during this stage (Hiraoka, 1941, 1952a,b). Since these nuclear dynamics are observed around the time when homologous chromosome pairing occurs, it has been inferred that they play some role in the pairing. However, this has not been directly demonstrated.
Studies in the fission yeast, Schizosaccharomyces pombe, have presented one of the most striking examples of the bouquet structure and nuclear movement. In these cells, the whole nucleus, which is extensively elongated, migrates back and forth between the two poles of the cell during meiotic prophase (Chikashige et al., 1994). This movement continues for some hours through the entire meiotic period preceding chromosome segregation. This motion is driven by astral microtubules radiating from the spindle pole body (SPB; a centrosome-equivalent in fungi); these microtubules interact with the cell cortex (Svoboda et al., 1995; Ding et al., 1998). During movements, all the telomeres remain clustered near the SPB, which is located at the leading edge of the moving nucleus (Chikashige et al., 1994). It has been speculated that nuclear movement, in conjunction with telomere clustering, facilitates the spatial alignment of homologous chromosomes required for their proper pairing during meiotic prophase (Chikashige et al., 1994, 1997; Kohli, 1994; Ding et al., 1998).
Recent studies have provided evidence showing an important role of telomere clustering in the pairing of homologous chromosomes. Meiotic recombination is significantly reduced in mutants which fail to form a telomere cluster due to aberrant telomere structure (Cooper et al., 1998; Nimmo et al., 1998). However, these mutants still show nuclear movement (Nimmo et al., 1998; A. Yamamoto and Y. Hiraoka, unpublished observation), suggesting that telomere clustering and the nuclear movement are distinct biological events. Specific inhibition of the nuclear movement without affecting telomere clustering would provide direct evidence for the role of this process in homologous chromosome pairing. To achieve specific inhibition, we have initiated a search for microtubule motor proteins that are involved in the microtubule-mediated nuclear movement. Cytoplasmic dynein is a likely candidate for such a motor, since it is required for microtubule-mediated nuclear movement in other fungi (Eshel et al., 1993; Li et al., 1993; Plamann et al., 1994; Xiang et al., 1994, 1995; Inoue et al., 1998). The dynein holoenzyme is a complex of several molecules with an ATP-dependent motility towards the minus end of microtubules. The largest subunit, cytoplasmic dynein heavy chain (DHC), contains the motor domain with ATPase activity (reviewed in Holzbaur and Vallee, 1994).
In this study, we have identified a cytoplasmic DHC as a motor that is necessary for the nuclear movement of meiotic prophase in fission yeast. Cells disrupted for the gene encoding a cytoplasmic DHC lacked the meiotic oscillatory nuclear movement, but most of them completed meiosis successfully to form four viable spores. In the cells lacking the nuclear movement, meiotic recombination was significantly reduced. Moreover, the colocalization of homologous regions on a pair of homologous chromosomes were significantly inhibited in the meiotic stage preceding chromosome segregation, strongly suggesting that the mutant cells failed to achieve efficient pairing of homologous chromosomes. Our study provides the first evidence that the nuclear movement of meiotic prophase, together with telomere clustering, facilitates the homologous chromosome pairing that is required for meiotic recombination.
Materials and Methods
Strains, Media, and Basic Technique
The strains used in this study are listed in Table I and were created in this study. Media were prepared as previously described (Moreno et al., 1991), except that YE medium containing adenine sulfate at the concentration of 75 μg/ml (YEA medium) was used for routine growth of cells. fur1 mutation was tested by the growth on the YEA medium containing 5-fluorouracil (Sigma Chemical Co.) at the concentration of 100 μg/ml. Genetic manipulations were carried out as described by Moreno et al. (1991).
Table I.
Strain List
| Strain | Genotype | |||
|---|---|---|---|---|
| AY122-1A | h− | ade6-M210 leu1 | ||
| AY122-1C | h+ | his2 ura4 | ||
| AY122-11C | h− | leu1 ura4 | ||
| AY126-1B | h+ | his2 leu1 lys3 ura1 | ||
| AY141-3D | h+ | ade6-M210 leu1 dhc1-d3 | ||
| AY1491-1A | h+ | ade6-M216 his2 ura4 dhc1-d4 | ||
| AY1491-1B | h− | ade6-M210 lys1 ura4 dhc1-d4 | ||
| AY1491-1C | h− | ade6-M216 ura4 | ||
| AY1491-1D | h+ | ade6-M210 his2 lys1 ura4 | ||
| AY153-19B | h+ | ade6-469 his2 | ||
| AY154-12A | h− | ade6-M375 ura4 | ||
| AY155-3A | h− | ade6-M26 leu1 ura4 dhc1-d3 | ||
| AY156-1B | h− | ade6-M375 leu1 ura4 dhc1-d3 | ||
| AY157-3C | h+ | ade6-469 leu1 his2 dhc1-d3 | ||
| AY160-30A | h90 | leu1 lys1 ura4 dhc1-d4 | ||
| AY161-2C | h− | ade6-M26 ura4 | ||
| AY177-14A | h+ | ade8 leu1 | ||
| AY179-8A | h− | leu1 trp1 | ||
| AY180-1B | h− | ade6-M210 fur1 leu1 | ||
| AY180-1C | h+ | his2 leu1 dhc1-d3 | ||
| AY180-2C | h+ | his2 leu1 | ||
| AY180-2D | h− | ade6-M210 fur1 leu1 dhc1-d3 | ||
| AY181-8C | h+ | ade8 leu1 dhc1-d3 | ||
| AY182-1A | h− | leu1 trp1 dhc1-d3 | ||
| CRL152 | h90 | leu1 lys1 ura4 | ||
| CRL1521 | h90 | leu1 lys1 ura4 dhc1-d2 | ||
| CRL1522 | h90 | leu1 lys1 ura4 dhc1-d3 | ||
| CRL1526 | h90 | leu1 lys1 ura4 dhc1::GFP-LEU2 | ||
| DHC102 | h90 | ade6-M216 leu1 ura4 dhc1-d1 | ||
| DHC105 | h+ | his2 ura4 dhc1-d2 | ||
| DHC106 | h− | leu1 ura4 dhc1-d2 | ||
| DHC110 | h − | his2 leu1 lys3 ura1 dhc1-d3 | ||
| GT1521 | h90 | leu1 ura4 lys1+::pnmt1-GFP-atb2+ dhc1-d2 | ||
| GT160-30A | h90 | leu1 ura4 lys1+::pnmt1-GFP-atb2+ dhc1-d4 | ||
| YY105 | h90 | leu1 ura4 lys1+::pnmt1-GFP-atb2+ | ||
| AY162 | h+/h− | ade6-M210/ade6-M216 his2/his2+ leu1+/leu1 | ||
| AY163 | h+/h− | his2/his2+ leu1+/leu1 ura4/ura4 dhc1-d2/dhc1-d2 |
Cloning and Sequencing of Dynein Heavy Chain Gene
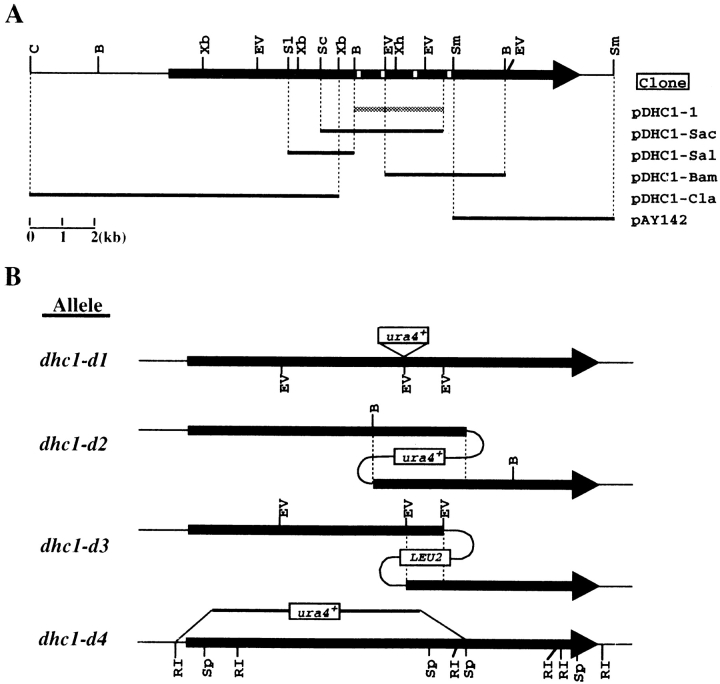
A portion of the DHC was isolated by PCR using degenerated primers based on regions of highly conserved amino acid sequence that lie in the neighborhood of the P-loop ATP-binding site lying nearest to the NH2 terminus of the DHC (Vaisberg et al., 1993). A 320-bp genomic DNA fragment encoding a partial open reading frame (ORF) was obtained which had ∼52% identity to the DHC sequence of Dictyostelium (Koonce et al., 1992). A genomic DNA library (Barbet et al., 1992) was screened using the PCR-amplified DNA fragment as a probe, and a 2.7-kb genomic clone (pDHC1-1) was obtained (Fig. 1 A). This cloned fragment was mapped between the mei2+ and leu2+ loci on the right arm of chromosome I from two different cosmid libraries (Hoheisel et al., 1993, Mizukami et al., 1993).
Figure 1.
Cloning and disruption of dhc gene. (A) Restriction map and cloned genomic DNA fragments of dhc1+. Arrow, position of the ORF for Dhc1p; white boxes on the arrow, sites corresponding to four P-loops; shaded bar, the original 2.7-kb genomic DNA clone isolated from genomic DNA libraries; solid bars, clones obtained by recovering plasmids from the genome that had been integrated at the dhc1 locus. Names of clones are shown on the right. B, BamHI; C, ClaI; EV, EcoRV; Sc, SacI; Sl, SalI; Sm, SmaI; Xb, XbaI; Xh, XhoI. (B) A diagram of the disruption schemes used for the dhc1+ gene. Solid arrows, ORF; dotted lines, overlapping regions; open boxes, integrated markers; thin lines, plasmid DNA. Allele names of each disruption are shown on the left. Sp, SphI; RI, EcoRI.
The complete coding sequence was obtained by recovering plasmids that had been integrated at the dhc1 locus. Plasmids containing different portions of the DHC coding region were integrated at the dhc1 locus of strain CRL152. The genomic DNA of the integrant was isolated, digested with a restriction enzyme, and ligated. The ligated DNA was transformed into Escherichia coli strain STBL2 for subsequent analysis (GIBCO BRL). Cloned genomic DNA fragments on the recovered plasmids are schematically shown in Fig. 1 A.
Integration plasmids and restriction enzymes used for obtaining dynein clones are as follows: Plasmids of pAY119, pAY123, pAY120, pAY131, and pAY130 were used for the integration to obtain pDHC1-Sac, pDHC1-Sal, pDHC1-Bam, pDHC1-Cla, and pAY142, respectively. pAY-119 was constructed by deleting a 1.2-kb ClaI fragment containing ars1 from pDHC1-1. pAY123 was constructed by inserting an Ecl136II– BamHI fragment of pDHC1-Sac between the SmaI and BamHI sites of pRS405 (Stratagene). pAY120 was constructed by moving an EcoRV fragment of pDHC1-1 into the SmaI site of pRS405. pAY131 was constructed by inserting an XbaI fragment of pDHC1-Sal, of which ends were filled in by Klenow fragments, into the SmaI site of pRS306 (Sikorski and Hieter, 1989). pAY130 was constructed by inserting a BamHI–SmaI fragment of pDHC1-Bam between the Ecl136II and BamHI sites of pRS-306. Restriction enzymes used for obtaining pDHC1-Sac, pDHC1-Sal, pDHC1-Bam, pDHC1-Cla, and pAY142 were SacI, SalI, BamHI, ClaI, and SmaI, respectively. DNA sequences of the cloned fragments were determined by the method recommended by Applied Biosystems Inc., using synthetic DNA primers complementary to the sequence.
Disruption of Dynein Heavy Chain Gene
The dhc1-d1 disruption allele was generated as follows: a 1.8 kb DNA fragment bearing the ura4+ gene was inserted at an EcoRV site located between the BamHI and XhoI sites in the DHC coding region of pDHC1-1. A linear DNA fragment of the heavy chain coding region between the BamHI and EcoRV (located between XhoI and SmaI) sites bearing the ura4+ gene was integrated at the dhc1+ locus of a diploid strain (h+/h− ade6-210/ade6-216 leu1/leu1 ura4/ura4) by one-step gene replacement (Moreno et al., 1991). The diploid integrants were sporulated and one of the ura+ progenies of integrants was further crossed with an h90 homothallic strain to obtain a dhc1-d1 homothallic strain (DHC102). Homologous integration was confirmed by Southern blot and PCR analyses.
To construct strains bearing the dhc1-d2 and the dhc1-d3 mutant alleles, pAY119 and pAY120 were integrated at the dhc1 locus in a haploid strain. Strains bearing dhc1-d2 (CRL1521) and dhc1-d3 (CRL1522) were used for most of the phenotypic analyses. The dhc1-d4 allele was created as follows: a BamHI–EcoRI DNA fragment of pDHC1-Cla was inserted between EcoRI and BamHI sites of an integration vector, which was constructed by deleting the ClaI fragment containing ars1 from pUR19 (Barbet et al., 1992). Then, a BamHI–SphI DNA fragment of pDHC1–Bam was inserted between BamHI and SphI sites. The resulting plasmid, pAY144, was linearized by digesting with BamHI and then integrated at dhc1 gene in a diploid strain (h+/h− ade6-M210/ade6-M216 his2/his2+ lys1/ lys1+ ura4/ura4) by one-step gene replacement. The diploid integrants were sporulated and one of the ura+ progenies of integrants was further crossed with an h90 homothallic strain to create a dhc1-d4 homothallic strain (AY160-30A). Homologous integration was confirmed by Southern blot analysis.
Synchronization of Mitotic Cell Cycle
Cells were grown in YEA medium at 30°C at the cell density of 4 to 6 × 106 cells/ml. They were arrested in S phase by incubating for 3 h in the presence of 15 mM hydroxyurea at 30°C. After washing twice with YEA medium, arrested cells were resuspended in fresh YEA medium at the cell density of 2 × 106 cells/ml and further incubated at 30°C to synchronously resume mitotic cell cycle. A portion of the culture was taken at intervals and cells were fixed by adding 1/10 vol of 37% formaldehyde. Progression of mitosis was monitored by analyzing chromosomal DNA of fixed cells stained with 4',6-diamidino-2-phenylindole (DAPI; Sigma Chemical Co.).
Analysis of Microtubule Morphology and Dynamics of Mitotic Spindles in Living Cells
To examine microtubule morphology, cells were transformed with pDQ105, a multicopy plasmid which carries a green fluorescent protein (GFP)–α-tubulin fusion gene (GFP–atb2+) placed under the nmt1 promoter (Ding et al., 1998). The transformed cells were grown or sporulated in the medium containing 5 μM thiamine for the repression of the GFP-tubulin expression at low levels. The GFP-labeled microtubules were examined under a microscope as described by Ding et al. (1998). To examine dynamics of the mitotic spindle, cells were used which express the GFP– α-tubulin from the fusion gene integrated at the lys1+ locus. To integrate the fusion gene, PstI–SacI fragment of pDQ105, which contains the GFP– α-tubulin gene placed under the nmt1 promoter, was moved into pYC36, an integration plasmid containing a partial lys1+ gene. The resultant plasmid was introduced into strains CRL152, CRL1521, and AY160-30A to produce strains YY105, GT1521, and GT160-30A, respectively. Integration was confirmed by Southern analysis. Cells of these strains were grown in YEA medium containing 5 μM thiamine at the cell density of 2 to 4 × 106 cells/ml and examined for spindle dynamics at 30°C.
Live Cell Analysis of Chromosome Dynamics
Live cell analysis of chromosome dynamics was carried out using a computer-operated microscope system, previously described by Chikashige et al. (1994 and 1997).
Synchronization of the Meiotic Process
Wild-type (AY162) and dhc1 (AY163) diploid strains were grown in liquid YEA medium at 33°C to 5 × 107 cells/ml. We found that sporulation efficiency was high when meiosis was induced in cells at late log or early stationary phase. The cells were then washed twice and resuspended in EMM medium lacking a source of nitrogen (EMM − N). Meiosis was synchronously induced by incubating the culture at 26°C with shaking. Progression of meiosis was monitored as described in Synchronization of Mitotic Cell Cycle.
Chromosome Missegregation Assay
Chromosome III missegregation was examined using intragenic complementation of two alleles of the ade6 gene on chromosome III, ade6-M210 and ade6-M216, previously described in Molnar et al. (1995). Strains AY1491-1A and AY1491-1B, or AY1491-1C and AY1491-1D, were mated and sporulated on ME plates at 26°C for 3 d. Sporulated cells were suspended in 400 μl of water containing 2 μl of Glusulase (Sigma Chemical Co.) and incubated for 16 to 18 h at 30°C to liberate spores and kill vegetative cells. Liberated spores were washed twice with water and plated on YEA and EMM plates supplemented with appropriate amino acids for selecting ade+ progenies. More than 100 ade+ progenies were tested for disomic chromosome III by streaking on YE plates after growing for several generations on YEA plates. Cells disomic for chromosome III give rise to red colonies on YE plates, since they frequently generate offspring monosomic for chromosome III (Niwa and Yanagida, 1985). Frequency of spores disomic for chromosome III was determined by the frequency of ade+ progenies and the ratio of the disomic progenies.
Construction of a Strain Bearing GFP–DHC Fusion Gene
The coding sequence of GFP-S65T (Heim et al., 1995) cloned in pRSET-B vector (Invitrogen Corp.; a gift from Dr. Tsien, University of California, San Diego, CA) was amplified by PCR using primers, CGCGGATCCATGAGTAAAGGAGAAGAACTT and GAAGGCCTATTTGTATAGTTCATCCATGCC. The PCR product was digested with BamHI and StuI and then inserted between the BamHI and SmaI sites of pREP1 vector (Maundrell, 1993) to form pEG3. A BamHI–SacI fragment of pEG3 containing GFP-S65T with a nmt1-polyadenylation sequence was cloned into the corresponding polylinker sites of pRS405 to form pAY147. A DNA fragment encoding a COOH-terminal portion of DHC was amplified by PCR using pAY142 as a template and synthetic oligonucleotides, TGGATGCCCTAAGAGATTT and CGGGATCCAATGCACATACTAAAATAAATATC. The PCR product was digested with NsiI and BamHI and inserted between the PstI and BamHI sites of pAY147 to form pAY150, which encodes the COOH-terminal portion of the DHC protein fused with GFP. pAY150 was integrated at the dhc1 locus of CRL152 to make strain CRL1526, replacing the wild-type dhc1 gene with the GFP-fused dhc1 gene.
Indirect Immunofluorescence Microscopy
Cells induced into meiosis on solid ME medium were fixed in methanol for 8 min at −30°C. The fixed cells were washed once with 0.1 M potassium phosphate buffer, pH 6.5, and resuspended in the same buffer containing 1 M sorbitol (spheroplast buffer). They were spheroplasted by adding 100 μg/ml of Zymolyase 100T (Seikagaku Kogyo) and 0.1% 2-mercaptoethanol. The spheroplasted cells were washed twice with spheroplast buffer and permeabilized with 1% Triton X-100 in spheroplast buffer. After permeabilization, the cells were washed twice with the phosphate buffer and resuspended in solution F (10 mM potassium phosphate buffer, pH 7.0, 150 mM NaCl, and 1% BSA). Antibody reactions with permeabilized cells were carried out in a microtube at 23°C as described in Hagan and Hyams (1988) using solution F for antibody incubations. α-Tubulin and GFP were stained using TAT1 mouse monoclonal anti–α-tubulin antibody (Woods et al., 1989) and rabbit polyclonal anti-GFP antibody (1:2,000 dilution, CLONTECH Laboratories, Inc.), respectively. FITC-conjugated goat polyclonal anti–mouse antibody (1:100 dilution, Jackson ImmunoResearch Laboratories, Inc.) and rhodamine-conjugated goat polyclonal anti–rabbit antibody (1:100 dilution, Cappel Laboratories/Organon Teknika Corp.) were used for secondary antibodies. DNA was stained with 100 ng/ml DAPI.
Analysis of Recombination Frequency
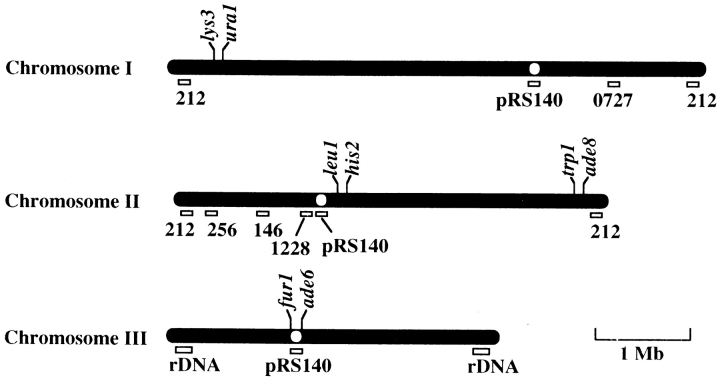
To examine the frequency of meiotic recombination, two strains bearing appropriate genetic markers were grown on the solid YEA medium at 33°C, mated, and then sporulated on the solid ME or EMM − N medium at 26°C (zygotic meiosis). Alternatively, after mating, diploid cells were selected on the solid EMM medium supplemented with appropriate amino acids. The diploid cells were grown on the solid YEA medium at 33°C and then sporulated on the solid ME medium at 26°C (azygotic meiosis). The frequency of intergenic recombination was examined by tetrad analysis. The frequency of intragenic recombination at the ade6 locus was analyzed, previously described by Ponticelli and Smith (1989). An approximate map of genetic markers used in this study is shown in Fig. 9.
Figure 9.
A chromosome map of genetic markers and hybridization sites for DNA probes. Bars and open circles indicate three chromosomes and centromeres, respectively. The positions of genetic markers and hybridization sites for DNA probes are approximately indicated according to the maps presented by Kohli et al. (1977), Gygax and Thuriaux (1984), Hoheishel et al. (1993), and Mizukami et al. (1993). Open rectangles indicate hybridization sites for DNA probes. 146, 212, 256, 1228, and 0727 represent cosmids, cos146, cos212, cos256, cos1228, and ICRFc60D0727, respectively, used for DNA probes. rDNA indicates chromosomal regions encoding ribosomal RNA genes. A thin line at the bottom right indicates an approximate chromosome length corresponding to 1 Mb of genomic DNA.
Fluorescence In Situ Hybridization Analysis of Centromere and Telomere Positions
Fluorescence in situ hybridization (FISH) analysis was performed as described (Chikashige et al., 1994) with the following modifications. Cells grown on the YEA agar medium were transferred on the ME agar medium to induce meiosis. After incubation at 23°C for 12 to 13 h, they were fixed in 3% paraformaldehyde and 0.2% glutaraldehyde in PEM buffer (100 mM Pipes-KOH, pH 6.9, 1 mM EGTA, 1 mM MgCl2) for 30 min at 23°C. After fixation, the cells were permeabilized and treated with RNase A as described (Funabiki et al., 1993). The RNase A-treated cells were then processed for FISH. For simultaneous staining of the SPB and telomeres, the cells were immunostained for the SPB before being processed for FISH. The cells were reacted with rabbit polyclonal anti–γ-tubulin antibody (1:10,000 dilution; a gift from Dr. Masuda, The Institute of Physical and Chemical Research, Wako, Saitama, Japan) and then with Cy5-conjugated anti-rabbit antibody (1:100 dilution, Nycomed Amersham). After antibody reactions, the cells were postfixed with 3% paraformaldehyde in PEM buffer for 10 min at room temperature and then washed twice with PEM buffer.
For FISH, the cells were suspended in 2× SSC and incubated for 10 min at 23°C. Next, they were resuspended in 2× SSC containing 10% formamide and incubated for another 10 min at 23°C. The concentration of formamide was increased to 20% and then to 40%, step-wise with 10 min incubations at each step. After incubating cells in a 40% formamide solution, they were suspended in hybridization buffer (50% formamide, 5× Denhardt's solution, and 5% dextran sulfate in 2× SSC) containing fluorescent-labeled probes. Fluorescent-labeled probes were prepared as follows: Ribosomal RNA genes, which hybridize to both ends of chromosome III (Uzawa and Yanagida, 1992), were used for telomere probes. Alternatively, cosmid cos212 was used; this hybridizes to both ends of chromosome I and II (Funabiki et al., 1993). Plasmid pRS140, which contains centromeric repeats common to all three chromosomes, was used to hybridize to the centromeres (Chikashige et al., 1989). Cosmids, cos256, cos146, cos1228 (from Dr. Yanagida, Kyoto University, Kyoto, Japan) and ICRFc60D0727 (from Resource Center of the German Human Genome Project at the Max-Planck-Institut for Molecular Genetics) were used to hybridize to regions on the chromosome arms (Hoheisel et al., 1993, Mizukami et al., 1993). Approximate position of the chromosome regions to which the probes hybridize are shown in Fig. 9. The DNA used for telomere and chromosome-arm probes was fragmented with RsaI, TaqI, AluI, Sau3AI, and HaeIII, and then labeled with Cy3- or Cy5-dUTP (Nycomed Amersham Inc.) using an oligonucleotide tailing kit containing a terminal deoxynucleotidyl transferase (Boehringer Mannheim Co.). Similarly, the DNA used for centromere probes was digested with RsaI, TaqI, and AluI and labeled with Cy3-dUTP. Concentrations of the fluorescent-labeled DNA probes in the hybridization buffer were ∼15 μg/ml for telomere probes and 30 μg/ml for centromere and chromosome-arm probes. Before the hybridization step the cell suspension containing fluorescent probes was preheated to 70°C for 5 min to denature the DNA. Hybridization was carried out at 37°C for 16 h. After hybridization, the formamide concentration in the hybridization buffer was decreased to 40, 25, 15, and finally 7.5% by step-wise dilution with 2× SSC. For each dilution step, the cell suspension was incubated for 10 min at 23°C. After the concentration was decreased to 7.5%, the cells were resuspended in fresh 2× SSC and incubated for another 10 min at 23°C. Lastly, the cells were washed twice with 2× SSC and then stained for DNA with DAPI.
Results
Cloning and Sequence Analysis of the dhc Gene
Since the meiotic oscillatory nuclear movement is driven by the dynamic activity of microtubules (Svoboda et al., 1995; Ding et al., 1998), microtubule-dependent motor enzymes seemed to be involved. As a first step in examining this possibility, we cloned the dhc gene (Fig. 1 A; see Materials and Methods). A total of 17.3 kb of genomic DNA from the locus containing a candidate gene was examined. Sequence analysis of the cloned fragments revealed that they contained a continuous ORF of 12.6 kb. The ORF encodes a predicted protein of 4,196 amino acids with a deduced molecular weight of 484 kD (Fig. 2 A; GenBank/ EMBL/DDBJ accession number AB006784). A comparison of the amino acid sequence with the protein sequences in the SwissProt database using the BLASTP algorithm (Altschul et al., 1990) revealed a high similarity to the DHC sequences of other organisms. The cytoplasmic dhc gene of Aspergillus nidulans was most similar to the fission yeast sequence (27.4% identity). Dot-plot analysis showed that they are highly similar in their entire length (Fig. 2 B). These results indicate that the cloned ORF encodes the cytoplasmic DHC of fission yeast. Thus, the gene was named dhc1+. Southern blot and PCR analyses suggested that it is the only dhc gene in this organism (data not shown).
Figure 2.
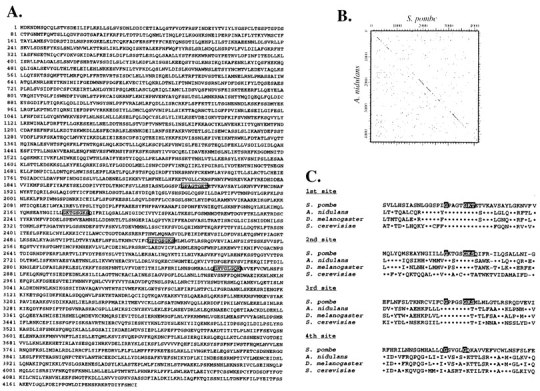
Deduced amino acid sequence of the dhc1+ gene and its comparison to other dhc genes. (A) Deduced amino acid sequence of Dhc1p. The four sites corresponding to the P-loops of other DHCs are shown by boxes. (B) A dot-plot matrix comparing the predicted DHC of fission yeast with that of A. nidulans with a stringency of 7 in a window of 23. Numbers indicate amino acid numbers. (C) Sequence comparison of the four regions containing putative ATP-binding sites from several different organisms. The ATP-binding motifs are shown by shadowed letters (GXXXXGKS/T). Dots and hyphens indicate identical amino acids and spaces, respectively. GenBank/EMBL/DDBJ accession numbers of the heavy chain sequences of A. nidulans, Drosophila melanogaster, and S. cerevisiae are U03904, L23195, and L15626, respectively.
The central motor domain of most DHC molecules contains four P-loops, which are putative ATP-binding sites comprising the consensus sequence, GXXXXKS/T (Walker et al., 1982; Saraste et al., 1990). Similarly, the fission yeast DHC contained the consensus sequences at the first three sites corresponding to other DHCs (Fig. 2 A, boxed sequences, and C). The fourth site lacked the typical consensus sequence, although sequence around this site was similar to those of other DHCs (Fig. 2 C). The existence of ATP-binding motifs suggests that fission yeast dynein generates motor activity by ATP hydrolysis, like other dynein molecules.
Cytoplasmic DHC Is Not Required for Proper Positioning and Orientation of the Mitotic Spindle
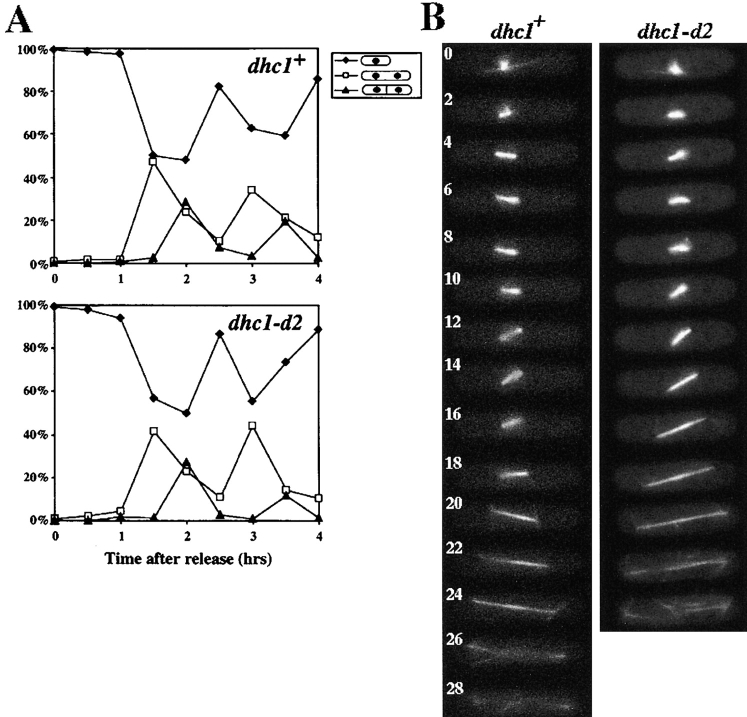
To examine the role of the DHC molecule in fission yeast, we disrupted the coding sequence by integrating either the ura4+ gene of fission yeast or the LEU2 gene of budding yeast at the dhc1+ locus by homologous recombination (Fig. 1 B; see Materials and Methods). Cells containing the integration that truncates the putative motor domain (dhc1-d1, dhc1-d2, and dhc1-d3) or a deletion of the NH2-terminal two-thirds of the heavy chain gene (dhc1-d4) grew in rich and minimal media at temperatures from 20°C to 36°C and their doubling times were similar to those of wild-type cells. Indirect immunofluorescence and living cell analysis using GFP-tagged α-tubulin (Ding et al., 1998) revealed that nuclear and microtubule morphology of all the mutants were similar to those of wild-type cells. In addition, the number of cytoplasmic microtubules extending along the cell axis in G2 cells (Hagan and Hyams, 1988) were not significantly different, 2.9 ± 0.7 (mean ± SD) for wild-type and 2.7 ± 0.7 for the dhc1-d2 mutant (n = 40). Cytokinesis took place in the middle of the cell, just as in wild-type cells. In addition, when cells were synchronized in S phase by a DNA synthesis inhibitor, hydroxyurea, the mutant cells underwent nuclear division and cytokinesis with a similar timing to that of wild-type cells (Fig. 3 A). These results indicated that the DHC is not essential for the mitotic growth and the loss of its function does not cause gross defects in mitosis.
Figure 3.
Mitotic cell-cycle progression and spindle behavior of wild-type and the dynein mutant. (A) Cell-cycle progression of wild-type and the mutant cells synchronized by hydroxyurea. Wild-type (upper graph) and the dynein mutant (lower graph) cells grown in YEA medium were arrested in S phase with hydroxyurea (Materials and Methods), and synchronously released from the arrest by removing the drug from the medium. After release from arrest, a portion of the cells were fixed at intervals and analyzed for cell types. More than 100 cells were examined at each time point. Percentage of cells containing one nucleus (closed diamonds) and those containing two nuclei, with (closed triangles) or without (open squares) a septum are shown. (B) Time-lapse series of wild-type (left) and dhc1-d2 (right) cells expressing GFP-tagged α-tubulin on a multicopy plasmid. Wild-type (strain CRL152) and mutant (strain CRL1521) cells bearing a GFP-tubulin plasmid, pDQ105 (Ding et al., 1998), were grown in EMM medium supplemented with appropriate amino acids for growth and 5 μM thiamine for the low-level expression of GFP-tubulin, and examined for spindle dynamics at 30°C. Representative series from wild-type and mutant cells are shown. Numbers indicate time in minutes.
It has been reported that dynein plays an important role in maintaining proper positioning, orientation, and kinetics of a mitotic spindle through astral microtubules in budding yeast (Eshel et al., 1993; Li et al., 1993; Yeh et al., 1995; Carminati and Stearns, 1997; Cottingham and Hoyt, 1997; DeZwaan et al., 1997; Shaw et al., 1997). To seek a possible role of dynein in dynamics of the mitotic spindle, we carried out detailed analysis of spindle behavior in wild-type and the dynein-disrupted cells using GFP-tagged α-tubulin (Ding et al., 1998). In wild-type cells, the mitotic spindle was formed at the center of the cell and elongated to ∼1 μm within 2 min (Fig. 3 B, left, 0 and 2 min). During this early stage of the spindle formation, cytoplasmic microtubules extending along the cell axis disappeared. The spindle continuously elongated to ∼3 μm and its length remained constant until further elongation (anaphase B) begins (Fig. 3 B, left, 2–16 min; Table II), as previously reported by Nabeshima et al., (1998). The spindle was always located at the middle of the cell and often changed its orientation (see spindle orientation in Fig. 3 B, left, 2, 10, and 14 min). In addition, short astral microtubules radiating from the spindle pole(s) were often observed (Fig. 3 B, left, 6 and 14 min). The time length of this stage was between 8 and 26 min, varying among cells. After this stage, the spindle elongated along the cell axis at the constant rate to reach its maximum length (anaphase B; Fig. 3 B, left, 18–26 min; Table II), and eventually disappeared (Fig. 3 B, left, 28 min). Astral microtubules radiating from the spindle poles were observed during elongation (Fig. 3 B, left, 20 and 24 min). When the dhc1-d2 cells were examined, no significant differences were observed in positioning, orientation, or kinetics of the mitotic spindles or in the behavior of astral-microtubules (Fig. 3 B, right; Table II). Therefore, we conclude that DHC is not required for maintaining proper positioning, orientation, or kinetics of mitotic spindles in fission yeast.
Table II.
Dynamic Parameters of the Mitotic Spindle
| dhc1 allele | Short spindle length* | Maximum spindle length‡ | Anaphase-B rate§ | Examined spindles‖ | ||||
|---|---|---|---|---|---|---|---|---|
| μm | μm | μm/min | ||||||
| dhc1+ ¶ | 2.9 ± 0.2 | 12.9 ± 1.0 | 1.1 ± 0.1 | 11 | ||||
| dhc1-d2** | 2.9 ± 0.4 | 12.8 ± 0.9 | 1.1 ± 0.2 | 8 | ||||
| dhc1-d4 †† | 2.8 ± 0.3 | 12.9 ± 1.5 | 1.1 ± 0.1 | 7 |
Cells were grown in YEA medium containing 5 μM thiamine and examined for the spindle dynamics at 30°C, as described in Materials and Methods. Average and standard deviation are shown.
Maximum length of short spindles preceding anaphase B.
Maximum spindle length after completion of anaphase B.
The elongation rate of the spindle during anaphase B.
Number of spindles examined.
Strain YY105.
Strain GT1521.
Strain GT160-30A.
Cytoplasmic DHC Is Required for Meiotic Prophase Nuclear Movement
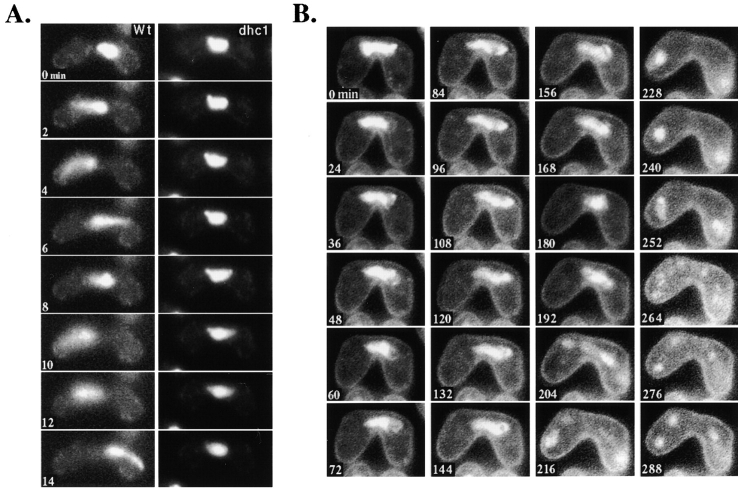
We then examined the phenotype of the dynein-disrupted cells during meiotic progression. Haploid cells of opposite mating types are induced to conjugate upon depletion of nitrogen from the growth medium. In conjugating cells, the two haploid nuclei migrate towards each other and fuse at the site of their SPBs (karyogamy). Cells bearing the dhc1-d2 mutation in both mating types underwent conjugation and nuclear fusion to form a zygotic cell containing a single chromosomal DNA mass and one SPB (Figs. 4 and 10 C). However, the fraction of conjugated cells with unfused nuclei was significantly larger in the mutant than in the wild-type: of 160 conjugated cells in the stage before meiotic chromosome segregation, unfused nuclei were observed in 21.9% of the mutant cells, whereas only 2.5% of wild-type cells had this phenotype. When unfused nuclei in 10 conjugated cells of the mutant were followed using a portion of DNA-polymerase-α tagged with GFP as a nuclear marker (D.-Q. Ding and Y. Hiraoka manuscript in preparation), all of the nuclei eventually fused to form a single nucleus. These results strongly suggest that nuclear fusion was delayed to some extent in cells lacking a cytoplasmic DHC, but not abolished.
Figure 4.
Chromosome dynamics in the dynein mutant cells during meiosis. Chromosomal DNA of conjugated cells was monitored by staining with Hoechst 33342 (Materials and Methods). (A) Chromosome behavior of wild-type (strain CRL152) and the dhc1-d2 (strain CRL1521) mutant in single-nuclear zygotes. The left and right columns are time-lapse series of a single wild-type or of the dynein mutant cell, respectively. The numbers indicate time in minutes. (B) Sequential observation of chromosomal DNA in the dhc1-d2 (strain CRL1521) cell during meiosis. The dynamics of chromosomal DNA in a single cell was followed by time-lapse observation immediately after nuclear fusion. The numbers indicate time in minutes.
Figure 10.
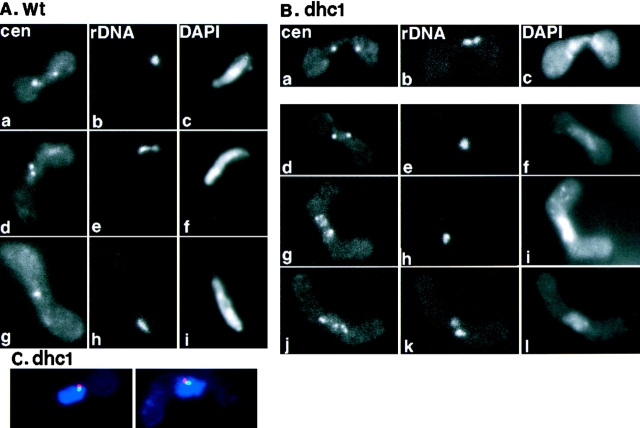
FISH analysis of centromere and telomere position in wild-type and the dynein mutant cells. (A) Centromere and telomere position in wild-type cells. Wild-type (strain CRL152) cells were induced into meiosis and processed for FISH using DNA probes against centromeric repetitive DNA sequences (a, d, and g) and a ribosomal DNA sequence, which is located near the telomeres of chromosome III (b, e, and h). Chromosomal DNA was stained with DAPI (c, f, and i). Single-nuclear zygotes are shown. (B) Centromere and telomere position in the dynein mutant cells. The dhc1-d2 mutant (strain CRL1521) cells were treated as in A. A two-nuclear zygote in karyogamy (a–c) and single-nuclear zygotes (d–l) are shown. a, d, g, and j, centromere staining; b, e, h, and k, ribosomal DNA staining; c, f, i, and l, DNA staining. (C) Simultaneous staining of the SPB and telomeres in the dynein mutant cells. The single-nuclear zygotes of the dynein mutant were processed for indirect immunofluorescence staining of the SPB using anti–γ-tubulin antibody, followed by FISH staining using DNA probes against telomere-associated sequences on both ends of chromosome I and II. Chromosomal DNA was stained with DAPI. Staining is shown in merged images: red, γ-tubulin; green, telomere; and blue, DNA staining.
The most striking consequence of the dynein disruption was observed after nuclear fusion. In wild-type cells, the fused nucleus immediately started oscillatory movement between the cell poles (Fig. 4 A, left), as previously reported (Chikashige et al., 1994). In contrast, the fused nucleus of the dynein mutant cells did not show oscillatory movement and remained in the middle of the cell (Fig. 4 A, right, and B). These observations indicated that the DHC is required for the oscillatory nuclear movement.
Despite the lack of oscillatory nuclear movement, many mutant cells underwent two rounds of meiotic chromosome segregation, as observed by the appearance of two and then four nuclei (Fig. 4 B). In wild-type cells, the nucleus performs some hours of oscillatory movement through meiotic prophase and then initiates two rounds of chromosome segregation (Chikashige et al., 1994). Live observations of the nuclear behavior in the dynein-disrupted mutant showed that the nucleus remained in the middle of the cell for ∼3 h after nuclear fusion and then initiated the first chromosome segregation. Unlike wild-type cells, chromosomes sometimes moved separately and failed to form two distinct chromosome masses during the first chromosome segregation (Fig. 4 B, 204 min; compare to Figure 2 C in Chikashige et al., 1994). Despite this aberrant behavior, two rounds of chromosome segregation were accomplished within 2 h, as in wild-type cells (Fig. 4 B).
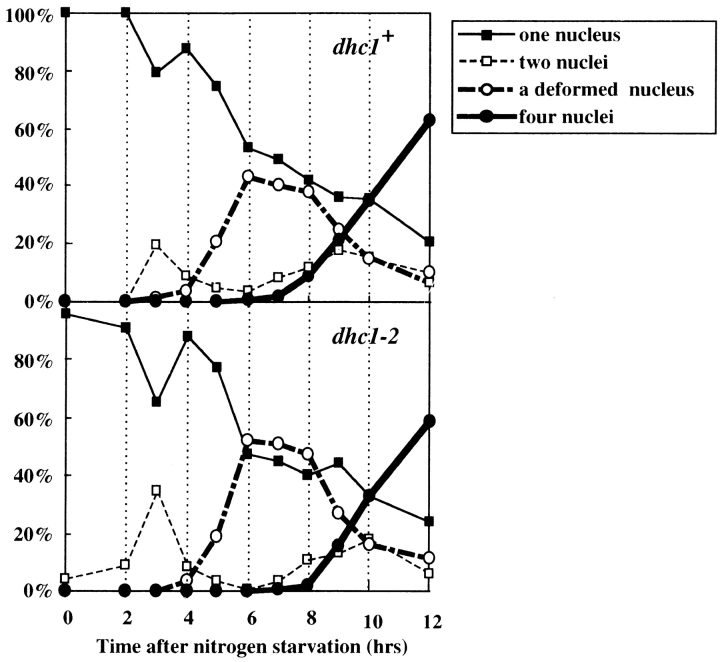
Analysis of azygotic diploid cells, which synchronously entered meiosis without undergoing nuclear fusion, suggest that both wild-type and mutant cells accomplished meiotic processes with a similar timing. As shown in Fig. 5, both wild-type and mutant diploid cells showed similar profiles after nitrogen starvation. Specifically, cells containing a deformed nucleus, which is characteristic of the meiotic stage preceding chromosome segregation, increased at ∼5 h and decreased at ∼8 h. Live observations confirmed that the deformed nucleus of the mutant lacked the oscillatory movement, whereas that of wild-type moved back and forth between cell poles (data not shown). These results suggested that the mutant cells accomplished the meiotic process preceding meiotic chromosome segregation with a timing similar to that of wild-type, despite the lack of nuclear movement.
Figure 5.
Time course changes in a number of different cell types of azygotic diploid cells after nitrogen starvation. Diploid wild-type (strain AY162) and dhc1-d2 (strain AY163) cells were synchronously induced into meiosis by depleting nitrogen from the growth medium and examined at the times indicated. More than 100 cells were examined at each time point. Upper and lower graphs indicate time course of changes in cell types of wild-type and mutant cells, respectively. Closed squares, open squares, open circles, and closed circles indicate percentages of cells containing one round nucleus, two nuclei, deformed nucleus, and four nuclei, respectively.
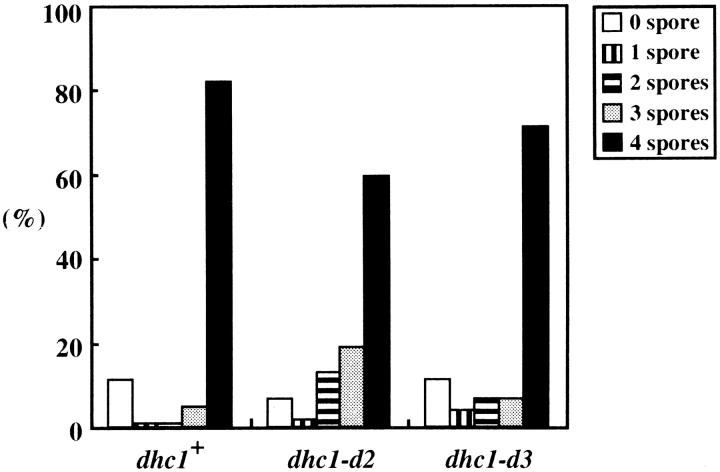
After accomplishing two rounds of chromosome segregation, many mutant cells formed four spores (Fig. 6), most of which were viable like those of wild-type cells (Table III), indicating that chromosomes were properly segregated in those cells. The slight increase in the number of the mutant cells that failed to form four spores, however, suggested increased chromosome missegregation in the mutant (Fig. 6). To know if chromosome missegregation is increased in the mutant, we examined the frequency of spores disomic for chromosome III. The frequencies of the disomic spores were 8.0 × 10−4 for wild-type and 1.1 × 10−2 for the mutant. The ∼14-fold increase of the disomic spores indicated that chromosome missegregation increased during meiosis I in the mutant. Thus, the dynein function is required for faithful chromosome segregation.
Figure 6.
Spore formation efficiency of wild-type and dhc1 mutant cells. Wild-type (strain CRL152) and dhc1 mutant (strains CRL1521 and CRL1522) cells were sporulated on the ME solid medium and scored for spore numbers in zygotic cells. More than 100 zygotes were examined for each strain. Bars indicate the population of cells that formed different numbers of spores.
Table III.
Tetrad Analysis of Genetic Linkage in Dynein Mutants
| Genetic markers* | Genotype | Number of tetrads | Genetic distance ¶ | Spore viability** | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T‡ | NPD§ | PD‖ | Total | |||||||||||
| A. Zygotic meiosis | cM | % | ||||||||||||
| lys3-ura1 | dhc1+ †† | 42 | 2 | 66 | 110 | 24.5 | 91.1 | |||||||
| dhc1-d3 §§ | 12 | 0 | 99 | 111 | 5.4 | 96.0 | ||||||||
| leu1-his2 | dhc1+ ‖‖ | 38 | 0 | 69 | 107 | 17.8 | 85.5 | |||||||
| dhc1-d2 ¶¶ | 8 | 0 | 109 | 117 | 3.4 | 96.5 | ||||||||
| trp1-ade8 | dhc1+ ** | 39 | 4 | 85 | 128 | 24.6 | 94.9 | |||||||
| dhc1-d3 170 | 10 | 0 | 101 | 111 | 4.5 | 93.8 | ||||||||
| fur1-ade6 | dhc1+ 173 | 38 | 1 | 105 | 144 | 15.3 | 96.7 | |||||||
| dhc1-d3 ‖‖ ‖ | 6 | 1 | 119 | 126 | 4.8 | 94.5 | ||||||||
| B. Azygotic meiosis | ||||||||||||||
| lys3-ura1 | dhc1+ †† | 33 | 0 | 97 | 130 | 12.7 | 92.2 | |||||||
| dhc1-d3 §§ | 20 | 1 | 109 | 130 | 10.0 | 91.0 | ||||||||
| leu1-his2 | dhc1+ ‖‖ | 13 | 2 | 114 | 129 | 9.7 | 91.6 | |||||||
| dhc1-d2 ¶¶ | 7 | 0 | 134 | 117 | 2.5 | 95.0 | ||||||||
| trp1-ade8 | dhc1+ ** | 23 | 1 | 87 | 111 | 13.1 | 91.9 | |||||||
| dhc1-d3 170 | 18 | 1 | 97 | 116 | 10.3 | 93.8 | ||||||||
| fur1-ade6 | dhc1+ §§ | 20 | 1 | 80 | 101 | 12.9 | 89.6 | |||||||
| dhc1-d3 ‖‖ ‖ | 16 | 0 | 106 | 126 | 6.3 | 94.5 | ||||||||
Spores formed in zygotic (A) or azygotic (B) diploid cells (see Materials and Methods) were analyzed for a linkage of two genetic markers.
Genetic markers: markers examined for genetic distance.
Tetratype.
Non-parental ditype.
Parental ditype.
Genetic distance = 50(T+6NPD)/Total.
Spore viability = number of viable spores in the dissected tetrads/total number of spores in the tetrads.
AY126-1B × AY122-1A.
AY141-3D × DHC110.
AY122-1C × AY122-11C.
DHC105 × DHC106.
AY179-8A × AY177-14A.
AY182-1A × AY181-8C.
AY180-1B × AY180-2C.
∥∥
∥ AY180-2D × AY180-1C.
Meiotic phenotypes similar to those described were observed in cells bearing each of the different mutations in the dynein gene (Fig. 1 B). In addition, the dynein mutations were recessive for the phenotypes described, since nuclear movement was normal in conjugates of mutant and wild-type cells. In summary, the DHC is required for the oscillatory nuclear movement of meiotic prophase and faithful chromosome segregation during meiosis. Moreover, the successful completion of meiotic processes in the absence of oscillatory nuclear movement in many mutant cells indicates that the movement itself is not essential for the progression of meiosis.
Microtubule Morphology in the Dynein Mutant during Meiosis
Since the oscillatory nuclear movement is dependent upon astral microtubules radiating from the SPB (Ding et al., 1998), it is possible that the lack of the movement in the dynein mutants is due to a loss of astral microtubules. We examined microtubule morphology in living meiotic dhc1-d2 cells using GFP-tagged α-tubulin (Ding et al., 1998). In a zygote containing a single nucleus, astral microtubules radiated from the SPB (Fig. 7, c–f), although the oscillatory nuclear movement was not observed. Therefore, the lack of the oscillatory nuclear movement in the mutant is not caused by a loss of astral microtubules. The microtubule morphology of the mutant cells was not significantly different from that observed in wild-type cells during the remainder of meiosis (Hagan and Yanagida, 1995; Svoboda et al., 1995; Ding et al., 1998). During karyogamy, astral microtubules radiated from the SPB as each of two nuclei converged to bring the two SPBs together (Fig. 7, a and b) and the meiotic spindles were normal during meiotic chromosome segregation (Fig. 7, g–j).
Figure 7.
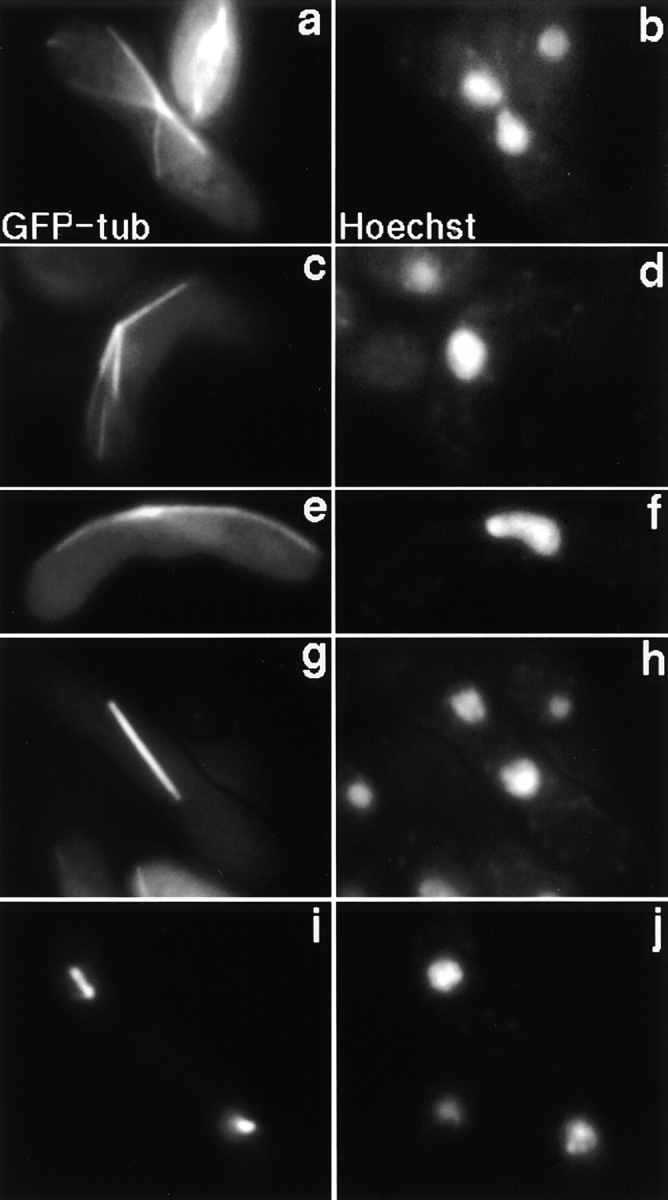
Microtubule morphology in dhc1-d2 mutant during karyogamy and meiosis. dhc1-d2 (strain CRL1521) cells expressing GFP-tagged α-tubulin were induced into meiosis in the presence of 5 μM thiamine to repress the expression of the tubulin-GFP to low levels, as described by Ding et al. (1998). Chromosomal DNA was stained with a DNA specific dye, Hoechst 33342. A two-nuclear zygote in karyogamy (a and b), a single-nuclear zygote before meiotic division (c–f), and two-nuclear zygotes in first (g and h) and second (i and j) meiotic divisions are shown. a, c, e, g, and i, α-tubulin GFP; and b, d, f, h, and j, DNA signal of Hoechst 33342.
Cytoplasmic DHC Is Localized at the SPB and Astral Microtubules
To further understand the functions of the DHC, we examined its intracellular localization by tagging the COOH terminus of Dhc1p with GFP. The dhc1+ gene was replaced with the tagged gene by chromosomal integration of the DNA encoding GFP, resulting in the expression of tagged Dhc1p under the endogenous promoter on the chromosome. The GFP-tagged Dhc1p (dynein-GFP) was functional, since cells expressing solely the tagged protein showed no aberrant phenotypes in meiosis, particularly in the oscillatory nuclear movement and intragenic recombination at the ade6 locus (data not shown).
In mitotically growing cells, the dynein-GFP was not detected (data not shown). Meiotic cells, on the other hand, showed specific intracellular localization. During nuclear fusion through meiotic prophase, the dynein-GFP was localized at the SPB (Fig. 8 A, b, d, and f, large arrows, and B, large arrowheads), and astral microtubules (Fig. 8 A, b, d, and f, and C, e). The localization of the dynein-GFP to the SPB was demonstrated by the position of the GFP dot at the protruding edge of the nucleus (Fig. 8 A, a and b, and B, large arrowheads) and its colocalization with the focal point of astral microtubules (Fig. 8 C). This localization was apparent until anaphase of the first meiotic division (Fig. 8 A, g and h), but was not detected through the rest of meiosis (Fig. 8 A, i and j). The localization to astral microtubules was demonstrated by colocalization of the GFP line with microtubules (Fig. 8 C, e, f, and h) and further confirmed by the disappearance of the GFP lines by treating the cells with a microtubule-depolymerizing drug, thiabendazole (data not shown). Interestingly, during the oscillatory nuclear movement, a dynein-GFP dot was frequently observed on an astral microtubule(s) at a point where the microtubule(s) contacts the cell cortex in front of the moving nucleus, and the nucleus moved towards it (Fig. 8 A, d and f, small arrows, and B, small arrowheads). The dot appeared during the approach of the SPB to the cortex (Fig. 8 B, lower, 15 s) and disappeared after the SPB reached it and reversed the direction of its movement (Fig. 8 B, lower, 90 s). This observation suggests the presence of a novel structure on the cell cortex that promotes a transient accumulation of the dynein-GFP and is involved in the nuclear movement (see Discussion).
Figure 8.
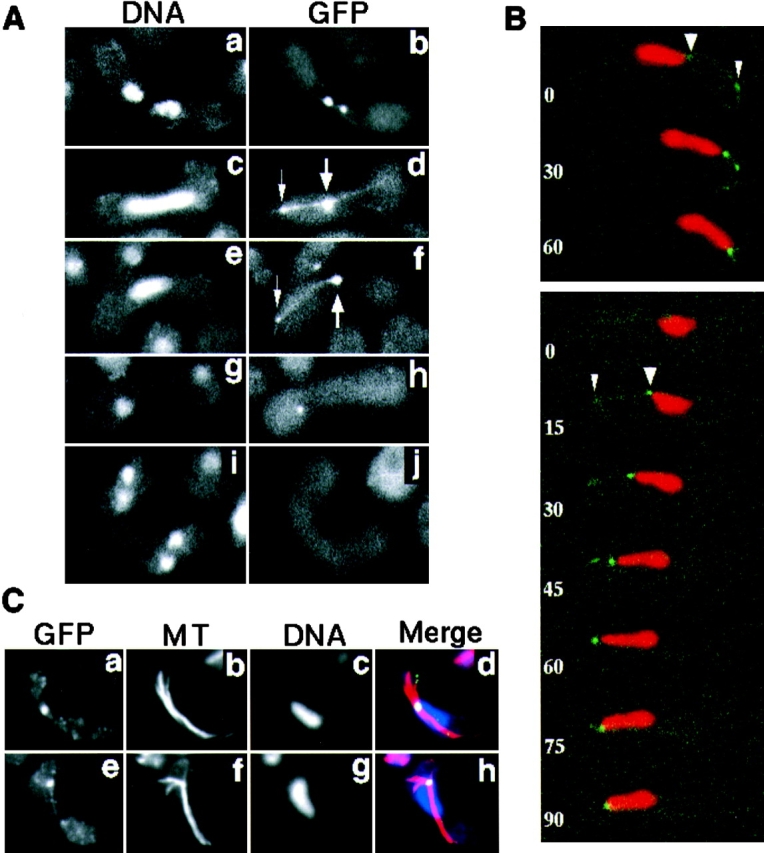
Localization of GFP-tagged dynein during karyogamy and meiosis. (A) Localization of dynein-GFP in living cells. Homothallic cells bearing the dynein-GFP fusion gene (strain CRL1526) were induced into meiosis and observed for localization of dynein-GFP. Chromosomal DNA was stained with Hoechst 33342. Cells undergoing karyogamy (a and b), the oscillatory nuclear movement (c–f), first meiotic division (g and h), and second meiotic division (i and j) are shown. a, c, e, g, and i, DNA staining; b, d, f, h, and j, GFP signal. The large arrows indicate the GFP dot at the leading edge of the moving nucleus where the SPB is expected. The small arrows indicate a GFP dot at a point where a GFP line contacts the cell cortex. (B) Time-lapse series of meiotic cells expressing dynein-GFP. Two zygotic cells undergoing the oscillatory nuclear movement are shown. Red and green indicate DNA staining and GFP signal, respectively. Numbers indicate time in seconds. Large and small arrowheads indicate the GFP dots at the leading edge of the moving nucleus and at the point where a GFP line contacts the cell cortex, respectively. (C) Immunofluorescence staining of dynein-GFP and microtubules in fixed cells. Cells induced to meiosis, as in A, were fixed and stained using antibodies against GFP (a and e) and α-tubulin (b and f). They were also stained with DAPI (c and g). Merged images are shown in d and h. Yellow/green, GFP; red, tubulin; and blue, DNA staining.
Frequencies of Meiotic Recombination Are Significantly Reduced in the Dynein Mutant
If the oscillatory nuclear movement of meiotic prophase facilitates homologous chromosome pairing, as postulated previously (Chikashige et al., 1994, 1997; Kohli, 1994; Ding et al., 1998), the lack of this movement should affect meiotic recombination. To test this idea, we examined meiotic recombination in the dynein mutant cells. Tetrad analysis investigating the genetic linkage at four regions on three chromosomes (Fig. 9) revealed that the frequencies of intergenic recombination at the regions were significantly reduced by up to fivefold in the mutant (Table III). Thus, the intergenic recombination was reduced in the dynein mutant. Interestingly, the recombination frequencies in meioses initiated from the haploid state through karyogamy (zygotic meiosis) and from the diploid state without undergoing karyogamy (azygotic meiosis) were significantly different. In the dynein-disrupted cells, recombination occurs more frequently in azygotic meiosis than in zygotic meiosis at almost every locus. In contrast, in wild-type cells recombination for azygotic meiosis is less frequent. These results indicated that processes of intergenic recombination in azygotic and zygotic meiosis are distinct.
We then examined the frequency of intragenic recombination. It has been known that gene conversion occurs at high frequency at the ade6 locus on chromosome III. That is, a strain carrying a mutant allele of the ade6 gene, ade6-M26 or ade6-M375, spontaneously gives rise to ade+ recombinants in high frequency when crossed with a strain carrying another ade6 allele, ade6-469 (Gutz, 1971; Ponticelli et al., 1988; Schuchert and Kohli, 1988; Szankasi et al., 1988; Mizuno et al., 1997). As shown in Table IV, the frequency of intragenic recombination of both ade6-M26 and ade6-M375 was reduced by about nine- and fivefold, respectively, through zygotic meiosis in the dynein mutant. Thus, intragenic recombination was also reduced in the dynein mutant and this reduction was not allele-specific. Taken together, these results demonstrated that the dynein function is required for efficient meiotic recombination. In addition, the reduced frequency of the distinct types of recombination suggest that the reduced meiotic recombination results from inefficient pairing of homologous chromosomes, rather than defects in recombination machinery in the dynein mutant.
Table IV.
Effect of the dhc1 Mutations on Intragenic Recombination at the ade6 Locus
| Cross* | dhc1 allele‡ | ade6 allele§ | ade+ frequency‖ | |||
|---|---|---|---|---|---|---|
| × 10−4 | ||||||
| A | dhc1+ | M26 | 77.6 (± 4.0) | |||
| B | dhc1-d3 | M26 | 8.9 (± 1.7) | |||
| C | dhc1+ | M375 | 5.4 (± 2.7) | |||
| D | dhc1-d3 | M375 | 1.2 (± 0.4) |
Strains bearing the ade6 mutations, together with or without the dhc1-d3 mutation, were mated and sporulated on ME plates. Spores were plated on both YEA and EMM plates and analyzed for recombination frequency at the ade6 locus.
Cross: A, AY161-2C × AY153-19B; B, AY155-3A × AY157-3C; C, AY154-12A × AY153-19B; D, AY156-1B × AY157-3C.
dhc1 allele, dhc1 alleles of the two strains in the crosses.
ade6 allele, ade6 mutations analyzed for gene conversion frequency when crossed with ade6-469 mutation.
ade+ Frequency, frequency is expressed as the ratio of ade+ recombinants by viable spores. Shown is the mean of three independent experiments. Numbers in parentheses indicate the standard deviation.
Homologous Regions on a Pair of homologous Chromosomes Fail to Colocalize Efficiently in the Dynein Mutant Cells
To characterize chromosome pairing in the dynein mutant, we examined the nuclear position of chromosomes by FISH using probes that hybridize to several different chromosomal regions (Fig. 9). First, we examined the positions of centromeres and telomeres. It has been shown that centromeres and telomeres have distinct positions during the progression of meiosis (Chikashige et al., 1994, 1997). Centromeres locate away from the SPB forming one cluster in each nucleus during nuclear fusion and one or two clusters in most of the elongated zygotic nuclei (Fig. 10 A, a, d, and g). In contrast, telomeres locate near the SPB forming a single cluster (Fig. 10 A, b, e, and h). It has been inferred that one or two clusters of centromeres in a nucleus reflect the paired or unpaired state of homologous centromeres, respectively (Chikashige et al., 1994). Thus, inefficient pairing of homologous chromosomes should result in a reduced population of cells containing a single centromere cluster.
Cells were induced into meiosis and the positions of centromeres and telomeres were determined in zygotic nuclei by FISH analysis. When centromeres were examined the population of cells containing a single centromere cluster was significantly reduced in the dhc1 mutant. The fraction of single-nuclear cells containing a single centromere cluster was about threefold smaller in mutant than in wild-type cells (Table V). On the other hand, the fraction of those containing dispersed centromeres was significantly larger in the mutant (Table V; Fig. 10 B, g and j). Dispersed centromeres were not observed during karyogamy in the mutant, as in wild-type. Each of the two nuclei in the fusion process contained a single centromere signal (Fig. 10 B, a). These results suggest that centromeres fail to form a single cluster and disperse in many dhc1-d2 cells. The reduced population of cells containing a single centromere cluster strongly suggest that the dynein mutant cells failed to achieve efficient pairing of homologous centromeres.
Table V.
Centromere and Telomere Distribution in Single-nuclear Zygotes of Wild-type and the Dynein Mutant
| Cell types* (%) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
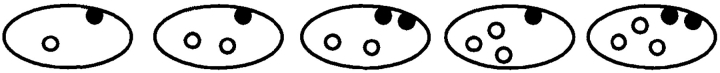
|
||||||||||||||||
| Experiment | Genotype | telo 1 cen 1 | 1 2 | 2 2 | 1 >2 | 2 >2 | Others | Examined cells‡ | ||||||||
| 1 | dhc1+ | 40.0 | 46.4 | 0.0 | 13.6 | 0 | 0 | 110 | ||||||||
| dhc1-d2 | 16.4 | 35.5 | 7.3 | 33.6 | 6.4 | 0.9 | 110 | |||||||||
| 2 | dhc1+ | 58.0 | 34.0 | 2.0 | 6.0 | 0 | 0 | 50 | ||||||||
| dhc1-d2 | 10.2 | 34.7 | 12.5 | 36.7 | 6.1 | 0 | 49 | |||||||||
Wild-type (strain CRL152) and the dynein mutant (strain CRL1521) cells, which were induced for meiosis on ME agar medium, were fixed and processed for FISH analysis (see Materials and Methods). Centromeres and telomeres were visualized using probes against centromere repetitive and ribosomal DNA sequences, respectively. Single-nuclear zygotes were examined. Results obtained from two independent experiments are shown.
Cell types. Cells were classified into six different types according to their numbers of centromere (cen, open circles) and telomere (telo, closed circles) signals in the nucleus.
Number of cells examined.
When telomeres were examined the fraction of cells containing two telomere signals was larger in the mutant than in wild-type (Table V; Fig. 10 B, k). This mutant phenotype may result from delayed SPB fusion in the mutant, which is likely to be a cause of the delayed nuclear fusion during karyogamy. Otherwise, telomere behavior in dhc1 cells was not significantly different from that of wild-type cells; they form a single cluster near the SPB during karyogamy (Fig. 10 B, b and c) and during meiotic prophase (Fig. 10 C) in most cells.
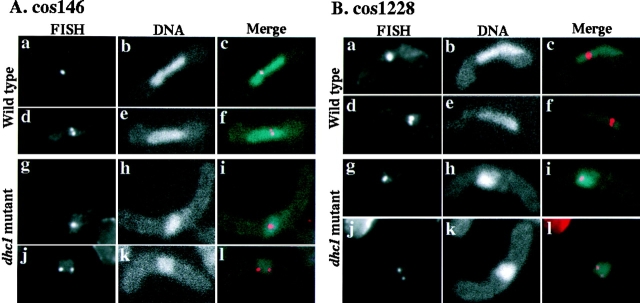
We then examined four different chromosomal regions on the arms of chromosome I and II in the mutant (Fig. 9). Greater than 90% of the single-nuclear cells showed one or two nuclear FISH signals in both wild-type and the mutant (Fig. 11). However, in the mutant cells the population of cells containing a single FISH signal, which probably indicates the paired state of the regions, was significantly smaller than in wild-type cells (Table VI). These results strongly suggested that the mutant cells failed to achieve efficient pairing of homologous regions on a pair of homologous chromosomes. Taken together, these results support the idea that homologous chromosomes failed to pair efficiently in the dynein mutant, due to the lack of the oscillatory nuclear movement.
Figure 11.
FISH analysis of nuclear position of chromosome-arm regions in wild-type and the dynein mutant cells. (A) Position of chromosomal regions in single-nuclear zygotes, to which cos146 DNA probe hybridized. a–f, wild-type (strain CRL152) cells; g–l, dhc1-d2 (strain CRL1521) cells; a, d, g, and j, FISH signals; b, e, h, and k, DNA staining; c, f, i, and l, merged images of FISH signals and DNA staining. (B) Position of chromosomal regions in single-nuclear zygotes, to which cos1228 DNA probe hybridized. a–f, wild-type (strain CRL152) cells; g–l, dhc1-d2 (strain CRL1521) cells; a, d, g, and j, FISH signals; b, e, h, and k, DNA staining; c, f, i, and l, merged images of FISH signals and DNA staining.
Table VI.
Colocalization of Homologous Regions in Single-nuclear Zygotes
| Probe* | Genotype | Cells containing a single FISH signal‡ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Exp. 1 | Exp. 2 | |||||||||
| % | % | |||||||||
| ICRFc60D | dhc1+ | 85.0 | (80) | 74.0 | (50) | |||||
| 0727 | dhc1-2 | 37.5 | (80) | 20.0 | (50) | |||||
| cos256 | dhc1+ | 57.5 | (80) | 52.0 | (50) | |||||
| dhc1-2 | 28.0 | (82) | 30.0 | (40) | ||||||
| cos146 | dhc1+ | 43.8 | (80) | 48.0 | (50) | |||||
| dhc1-2 | 15.0 | (80) | 27.5 | (40) | ||||||
| cos1228 | dhc1+ | 47.0 | (83) | 62.0 | (50) | |||||
| dhc1-2 | 23.8 | (80) | 30.0 | (40) | ||||||
Wild-type (strain CRL152) and the dynein mutant (strain CRL1521) cells were fixed and processed for FISH analysis, as in Table V. Homologous regions on a pair of homologous chromosomes were visualized using cosmid DNA probes (see Fig. 9) and single-nuclear zygotes were examined. Results of two independent experiments are shown. Figures in parentheses indicate number of cells examined.
Cosmid DNA probes used for FISH analysis.
Percentage of cells containing a single FISH signal. Two FISH signals were scored as a single FISH signal if the distance between the signal centers was < 0.5 μm.
Discussion
Functions of Cytoplasmic Dynein in Meiotic Prophase Nuclear Movement of Fission Yeast
We have cloned a fission yeast gene encoding a cytoplasmic DHC and demonstrate that it plays an essential role in the oscillatory nuclear movement of meiotic prophase. In addition, it is probably important for nuclear fusion during karyogamy, since nuclear fusion appeared to be delayed in the dynein mutant cells. Consistent with its role in meiosis-specific nuclear movement, dynein was detected at the SPB and astral microtubules from karyogamy through anaphase of the first meiotic division. It remains to be determined if localization was only detected during meiosis due to a meiosis-specific enhancement of dynein's expression or to its meiosis-specific accumulation at the SPB and microtubules.
Involvement of cytoplasmic dynein in microtubule-mediated nuclear migration has been reported in other fungi. In Neurospora crassa, A. nidulans, and Nectria haematococca, dynein is required for the migration of nuclei in the germ tube (Plamann et al., 1994; Xiang et al., 1994, 1995; Inoue et al., 1998). The budding yeast, Saccharomyces cerevisiae, requires dynein function for nuclear positioning and migration at the bud neck in mitosis (Eshel et al., 1993; Li et al., 1993; Yeh et al., 1995; Carminati and Stearns, 1997; Cottingham and Hoyt, 1997; DeZwaan et al., 1997; Shaw et al., 1997). Specifically, the nucleus penetrates into the bud neck at the onset of anaphase, oscillates several times at the neck, and then divides into mother and daughter cells (Yeh et al., 1995; DeZwaan et al., 1997). In the absence of dynein function, the anaphase nucleus often fails to penetrate into the neck and there are no oscillations (Yeh et al., 1995; DeZwaan et al., 1997). In those cells, the nucleus divides within the mother cell and eventually segregates into mother and bud cells. Thus, cytoplasmic dynein generates forces required for the nuclear penetration and oscillation at the bud neck during anaphase in budding yeast.
The dynein-dependent nuclear migration in budding yeast during mitosis and in fission yeast during meiosis share several features. First, both types of nuclear movement are driven by astral microtubules interacting with the cell cortex (Palmer et al., 1992; Sullivan and Huffaker, 1992; Svoboda et al., 1995; Carminati and Stearns, 1997; Shaw et al., 1997; Ding et al., 1998). Second, both are driven by dynein localized at the SPB and/or astral microtubules (Yeh et al., 1995; Shaw et al., 1997). Finally, both are oscillatory, though the amplitude of the oscillation is greater in fission yeast (Yeh et al., 1995; DeZwaan et al., 1997). These similarities strongly suggest that these nuclear movements in two evolutionary divergent organisms are driven by similar mechanisms.
Despite many similarities, dynein is not required for mitotic spindle dynamics in fission yeast, as demonstrated by the lack of significant defects in spindle behavior of the mutant. The difference may be due to the different geometry of their cell division. In budding yeast, divided nuclei are segregated into the mother and daughter cells through a narrow path at the bud neck. However, the fission yeast cell is divided by a septum formed at the middle of the cell after the divided nuclei have been segregated to opposite ends of the cell. Thus, the positioning of the nucleus in mitosis may be more important to budding yeast. Alternatively, another DHC, which has not been detected by our analysis, may play a role in mitosis of fission yeast.
During the oscillatory nuclear movements, GFP-tagged dynein is localized to the SPB and astral microtubules. It is also concentrated at the point where astral microtubules contact the cell cortex that faces an approaching nucleus. From analogy with other organisms, astral microtubules in fission yeast are probably oriented with their plus ends distal and minus ends proximal to the SPB (Bergen et al., 1980; Euteneuer and McIntosh, 1981; McIntosh and Euteneuer, 1984; Toriyama et al., 1988; Yamamoto et al., 1990). Given the localization of dynein and the polarity of the microtubules, we suggest two models for dynein function in the nuclear movement. In the first model, dynein immobilized on the cell cortex generates a pulling force by walking along astral microtubules toward their minus ends. The accumulation of dynein at the point where microtubules contact with the cortex facing an approaching nucleus is consistent with this model. A similar model was proposed for the function of dynein in budding yeast (Carminati and Stearns, 1997; Cottingham and Hoyt, 1997; DeZwaan et al., 1997). However, a cortical accumulation of dynein was not observed in this organism. In the second model, dynein motors at the SPB generate a pushing force on the SPB by walking in the minus end direction on microtubules that extend rearward. The localization of dynein at the SPB is consistent with this model. A contribution of pushing forces to create nuclear movement was previously proposed, since the movement was associated with the elongation of microtubules extending rearward (Ding et al., 1998). Supporting this model, it has been reported that a pushing by astral microtubules drives preanaphase nuclear migration in budding yeast. However, in this case the pushing force is not generated by dynein (Shaw et al., 1997). In either model, dynein may also play a role in dynamic changes of microtubule length, which are required for the oscillatory movement. Preliminary results showed that the rate of microtubule shortening, which occurs mostly in front of the moving nucleus in wild-type cells, was significantly altered in the dynein mutant at the meiotic stage preceding chromosome segregation (A. Yamamoto, unpublished results). Future analysis will be required to clarify the functions of DHC in the oscillatory nuclear movement.
Biological Significance of the Oscillatory Nuclear Movement during Meiotic Prophase
Most dynein mutant cells completed meiosis and formed four viable spores, despite the lack of oscillatory nuclear movement, and the duration of the meiotic stage preceding chromosome segregation is not obviously different in those cells. These results indicate that the movement is not essential for the progression of meiosis, and that the duration of the stage preceding chromosome segregation is determined independently of nuclear movement. However, the dynein mutant showed significant reduction in meiotic recombination suggesting that nuclear movement is required for efficient meiotic recombination. Our FISH observations of chromosome positioning strongly suggest that the reduced recombination results from inefficient pairing of homologous chromosomes. That is, the formation of a single cluster of centromeres and the colocalization of homologous regions in single-nuclear zygotes, which likely reflects the paired state of homologous chromosomes, were significantly inhibited in the dynein mutant. Preliminary observations of a centromere-linked locus in living cells using lac repressor/operator recognition (Robinett et al., 1996, Straight et al., 1996; Nabeshima et al., 1998) also support this idea. During oscillatory nuclear movement, the centromere-proximal loci on a pair of homologous chromosomes frequently contact each other in wild-type cells, but rarely in the dynein mutant (A. Yamamoto, K. Nabeshima, M. Yanagida, and Y. Hiraoka, unpublished observation). In addition to this idea, the frequency of both intergenic and intragenic recombination was reduced significantly in the mutant. Given the cytoplasmic role of dynein in nuclear movement, it is likely that the lack of the nuclear movement causes inefficient pairing of homologous chromosomes in the dynein mutant, which brings significant reduction in meiotic recombination.
In the mutant, intergenic recombination occurred more frequently in azygotic meiosis than in zygotic meiosis (Table III). The higher recombination frequencies in azygotic meiosis could be due to the positioning of homologous chromosomes in proximity during mitosis of diploid cells (Scherthan et al., 1994), which may facilitate pairing of homologous chromosomes in subsequent meiosis. However, despite this positioning the recombination frequencies of wild-type cells were lower in azygotic meiosis than in zygotic. Perhaps, a process(es) during karyogamy, which is absent in azygotic meiosis, is required for efficient meiotic recombination in fission yeast.
In addition to reduced frequencies of recombination, the dynein mutant showed increased missegregation of chromosomes, probably resulting in the slight increase of cells that failed to form four spores in the mutant. It is most likely that the increased missegregation of homologous chromosomes is caused by reduced crossing over on pairs of homologous chromosomes as a consequence of reduced recombination. The aberrant chromosome behavior that sometimes was observed during the first meiotic chromosome segregation of the dynein mutant (Fig. 3 B, 204 min) may also result from reduced crossing over. Future analysis is required to clarify the relation between crossing over and reductional segregation of homologous chromosomes in the dynein mutant.
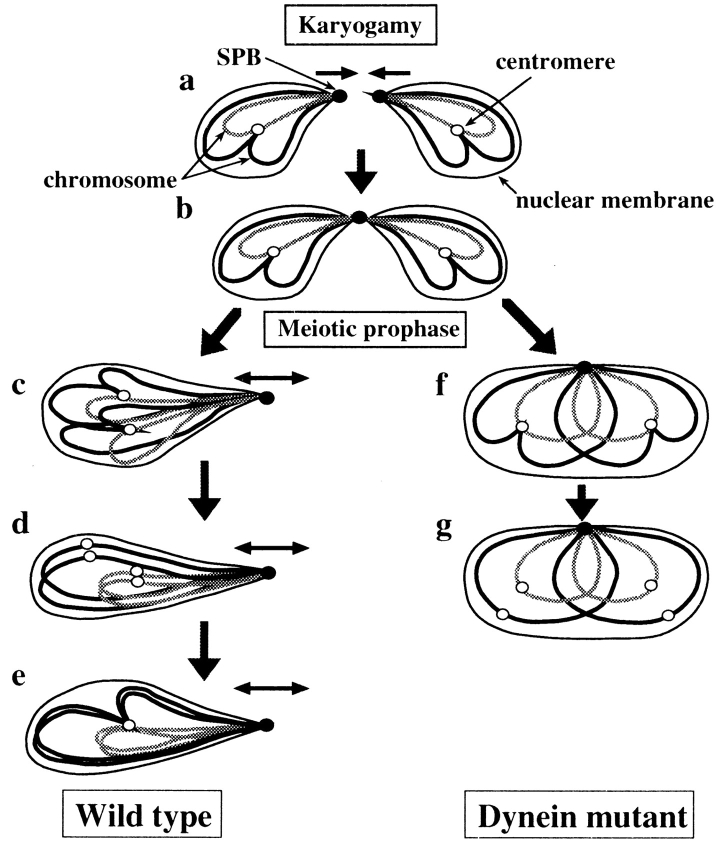
When the fission yeast nucleus oscillates between the cell poles, its chromosomes form a bouquet structure, i.e., the telomeres remain clustered near the SPB at the leading edge of the moving nucleus (Chikashige et al., 1994, 1997). The reduction of the recombination frequency in several mutants of fission yeast in which telomeres failed to form a cluster suggests that formation of the telomere cluster is required for proper pairing of homologous chromosomes (Shimanuki et al., 1997; Cooper et al., 1998; Nimmo et al., 1998). Although the dynein mutants showed a similar phenotype, the defect was not caused by failure to form a telomere cluster, since telomeres were clustered near the SPB in most dhc1 cells (Fig. 10 C). Conversely, mutants defective in telomere clustering still showed nuclear movement (Nimmo et al., 1998; A. Yamamoto and Y. Hiraoka, unpublished observation) indicating that the lack of the nuclear movement is not a cause of the reduced recombination in these mutant cells. These results strongly suggest that telomere clustering and microtubule-mediated nuclear movement are distinct biological events, both of which are required for proper pairing of homologous chromosomes.
Requirement of both telomere clustering and nuclear movement for efficient chromosome pairing is consistent with a previously published model (Chikashige et al., 1994, 1997; Kohli, 1994; Ding et al., 1998). In this model, the nuclear dynamics produce chromosome movement by pulling the clustered telomeres and this chromosome movement aligns homologous chromosomes side by side to promote their efficient pairing (Fig. 12, a–e). Subsequently, pairing of homologous chromosomes leads to the formation of a single centromere cluster. The nuclear dynamics also cause an elongation of the nucleus and we speculate that the constricted intranuclear space contributes to the chromosome pairing by promoting an efficient encounter of homologous chromosomes. In this scenario, when the nuclear movement is abolished, as in the dynein mutants, homologous chromosomes fail to achieve efficient pairing because they lack the necessary spatial alignment and inefficient pairing results in the reduced meiotic recombination and frequent failure to form a single centromere cluster (Fig. 12, f and g). The scattering of centromeres observed in dhc1 cells (Fig. 12 g) probably corresponds to the chromosomal state, which transiently appears in wild-type cells before the formation of a single centromere cluster (Fig. 12 d), given that a small population of wild-type cells contains dispersed centromeres (Table V). Similarly, when the telomere clustering is abolished homologous chromosomes fail to pair efficiently and meiotic recombination is reduced, as observed in the mutants defective in telomere clustering (Shimanuki et al., 1997; Cooper et al., 1998; Nimmo et al., 1998).
Figure 12.
Model for a role of the oscillatory nuclear movement in homologous chromosome pairing. Behaviors of two pairs of homologous chromosomes (solid and shaded lines), the SPB (closed circles) and centromeres (open circles), before meiotic chromosome segregation are shown. During karyogamy, chromosomes are positioned opposite to each other, with telomeres at the leading edges of the approaching nuclei (a and b). The subsequent SPB-led nuclear movement changes the telomere-centromere orientation of a homologous set of chromosomes from an antiparallel to a parallel relationship, thus placing homologous regions of homologous chromosomes in proximity (c). The centromeres fall apart transiently before homologous centromeres pair (d), and then, pairing of centromeres results in the formation of a single centromere cluster, which may also facilitate pairing of other homologous regions (e). In the dynein mutant, homologous chromosomes fail to align properly due to the lack of the nuclear movement (f) and centromeres disperse as a consequence of inefficient pairing of centromeres (g). The small arrows indicate direction of chromosome movement.
The bouquet configuration of homologous chromosomes in meiotic prophase observed in fission yeast has been described for a wide variety of other organisms, including human, mouse, and maize (Scherthan et al., 1996; Bass et al., 1997; reviewed in Fussell, 1987; John, 1990; Loidl, 1990; Therman and Susman, 1992; Dernburg et al., 1995). It also seems likely that some types of nuclear movement occur during meiotic prophase in organisms other than S. pombe (Hiraoka, 1941, 1952a,b; Yao and Ellingson, 1969; Parvinen and Söderström, 1976; Salonen et al., 1982). In addition, it was recently found that chromosomes form the bouquet configuration during meiotic prophase and a microtubule-dependent motor protein is required for proper meiotic recombination and the SC formation in the budding yeast, S. cerevisiae (Bascom-Slack and Dawson, 1997; Trelles-Sticken et al., 1999). These observations imply the presence of a similar mechanism to create the spatial alignment of homologous chromosomes in other organisms. Although other organisms seem likely to employ a similar mechanism, the tightly clustered telomeres and the striking nuclear movement, which are characteristic of fission yeast, have not been observed among them. Unlike other organisms, fission yeast does not form the SC during meiotic prophase (Olson et al., 1978; Hirata and Tanaka, 1982; Bähler et al., 1993). This structure is thought to promote alignment and intimate association of homologous chromosomes through their entire length. Supporting the different mode of the chromosome association in fission yeast, FISH analysis of homologous regions in azygotic diploid cells suggested that fission yeast lacks the SC-like stage when all chromosomal regions are engaged in pairing (Scherthan et al., 1994). Due to the lack of the SC, fission yeast may depend more heavily than other organisms on the mechanism consisting of the bouquet arrangement and the nuclear movement for aligning homologous chromosomes. To understand the mechanisms of homologous chromosome pairing, it may be important to examine nuclear dynamics during meiotic prophase from this perspective.
Acknowledgments
We thank Dr. R. Tsien for the plasmid bearing S65T GFP, Dr. K. Gull for TAT1 antibody, Dr. H. Masuda for anti–γ-tubulin antibody, Drs. O. Niwa, K. Okazaki, and K. Ohta for strains, and Dr. M. Yanagida for the cosmid DNA. We thank Dr. D.-Q. Ding for many helpful suggestions for live cell analysis and construction of the GFP-tubulin integration plasmid and strain YY105, and Dr. Y. Chikashige for the modified FISH method and many helpful suggestions. We thank Y. Tomita and R. Kurokawa for DNA sequencing, and C. Tsutsumi for assisting tetrad analysis and constructing a strain containing a portion of DNA polymerase α tagged with GFP. We thank Drs. D.-Q. Ding, A. Nabetani, T. Haraguchi, H. Masuda, and especially H. Renauld for critically reading the manuscript and many helpful comments to improve the manuscript.
This work was supported by grants from the Japanese Ministry of Culture, Science, and Education and the Science and Technology Agency of Japan to Y. Hiraoka, and by National Institutes of Health grant GM36663 to J.R. McIntosh. R.R. West was supported by the American Cancer Society (PF-4035) and J.R. McIntosh is a Research Professor of the American Cancer Society.
Abbreviations used in this paper
- DAPI
4',6-diamidino-2-phenylindole
- DHC
dynein heavy chain
- GFP
green fluorescent protein
- FISH
fluorescence in situ hybridization
- ORF
open reading frame
- SC
synaptonemal complex
- SPB
spindle pole body
References
- Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bähler J, Wyler T, Loidl J, Kohli J. Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J Cell Biol. 1993;121:241–256. doi: 10.1083/jcb.121.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet N, Muriel WJ, Carr AM. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. . Gene. 1992;114:59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- Bascom-Slack CA, Dawson DS. The yeast motor protein, Kar3p, is essential for meiosis I. J Cell Biol. 1997;139:459–467. doi: 10.1083/jcb.139.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass HK, Marshall WF, Sedat JW, Agard DA, Cande WZ. Telomeres cluster de novobefore the initiation of synapsis: a three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J Cell Biol. 1997;137:5–18. doi: 10.1083/jcb.137.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen L, Kuriyama R, Borisy GG. Polarity of microtubules nucleated by centrosomes and chromosomes of chinese hamster ovary cells in vitro. J Cell Biol. 1980;84:151–159. doi: 10.1083/jcb.84.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. Microtubule orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M. Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotIrestriction sites. Cell. 1989;57:739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Ding D-Q, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264:270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Ding D-Q, Imai Y, Yamamoto M, Haraguchi T, Hiraoka Y. Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. . EMBO (Eur Mol Biol Organ) J. 1997;16:193–202. doi: 10.1093/emboj/16.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Watanabe Y, Nurse P. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature. 1998;392:828–831. doi: 10.1038/33947. [DOI] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiaeis accomplished by antagonistically acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg, A.F., J.W. Sedat, W.Z. Cande, and H.W. Bass. 1995. Cytology of telomeres. In Telomeres. E.H. Blackburn and C.W. Greider, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 295–338.
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin-related KIP3 of Saccharomyces cerevisiaeis required for a distinct step in nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D-Q, Chikashige Y, Haraguchi T, Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci. 1998;111:701–712. doi: 10.1242/jcs.111.6.701. [DOI] [PubMed] [Google Scholar]
- Eshel D, Urrestarazu LA, Vissers S, Jauniaux J-C, van Vliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneuer U, McIntosh JR. Polarity of some motility-related microtubules. Proc Natl Acad Sci USA. 1981;78:372–376. doi: 10.1073/pnas.78.1.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centrosome and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussell, C.P. 1987. The Rabl orientation: a prelude to synapsis. In Meiosis. P.B. Moens, editor. Academic Press, Orlando, FL. 275–299.
- Gutz H. Site specific induction of gene conversion in Schizosaccharomyces pombe. . Genetics. 1971;69:317–337. doi: 10.1093/genetics/69.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygax A, Thuriaux P. A revised chromosome map of the fission yeast Schizosaccharomyces pombe. Curr. . Genetics. 1984;8:85–92. doi: 10.1007/BF00420223. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene sad1+associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS. The use of cell cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. . J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Hiraoka T. Studies of mitosis and meiosis in comparison. IV. A contribution to the study of the origin of the “bouquet” and its formation. Cytologia. 1941;11:483–492. [Google Scholar]
- Hiraoka T. Observational and experimental studies of meiosis with special reference to the bouquet stage. XI. Locomotory movement of the nucleolus in the bouquet stage. Cytologia. 1952a;17:201–209. [Google Scholar]
- Hiraoka T. Observational and experimental studies of meiosis with special reference to the bouquet stage. XIV. Some considerations on a probable mechanism of the bouquet formation. Cytologia. 1952b;17:292–299. [Google Scholar]
- Hirata A, Tanaka K. Nuclear behavior during conjugation and meiosis in the fission yeast Schizosaccharomyces pombe. . J Gen Appl Microbiol. 1982;28:263–274. [Google Scholar]
- Hoheisel JD, Maier E, Mott R, McCarthy L, Grigoriev AV, Schalkwyk LC, Nizetic D, Francis F, Lehrach H. High resolution cosmid and P1 maps spanning the 14 Mb genome of the fission yeast S. pombe. . Cell. 1993;73:109–120. doi: 10.1016/0092-8674(93)90164-l. [DOI] [PubMed] [Google Scholar]
- Holzbaur ELF, Vallee RB. Dyneins: molecular structure and cellular function. Annu Rev Cell Biol. 1994;10:339–372. doi: 10.1146/annurev.cb.10.110194.002011. [DOI] [PubMed] [Google Scholar]
- Inoue S, Turgeon BG, Yorder OC, Aist JR. Role of fungal dynein in hyphal growth, microtubule organization, spindle pole body motility and nuclear migration. J Cell Sci. 1998;111:1555–1566. doi: 10.1242/jcs.111.11.1555. [DOI] [PubMed] [Google Scholar]
- John, B. 1990. Meiosis. Cambridge University Press, Cambridge, UK.
- Kohli J. Telomeres lead chromosome movement. Curr Biol. 1994;4:724–727. doi: 10.1016/s0960-9822(00)00160-3. [DOI] [PubMed] [Google Scholar]
- Kohli J, Hottinger H, Munz P, Strauss A, Thuriaux P. Genetic mapping in Schizosaccharomyces pombeby mitotic and meiotic analysis and induced haplodization. Genetics. 1977;87:471–489. doi: 10.1093/genetics/87.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonce MP, Grissom PM, McIntosh JR. Dynein from Dictyostelium: primary structure comparisons between a cytoplasmic motor enzyme and flagellar dynein. J Cell Biol. 1992;119:1597–1604. doi: 10.1083/jcb.119.6.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J. The initiation of meiotic chromosome pairing: the cytological view. Genome. 1990;33:759–778. doi: 10.1139/g90-115. [DOI] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- McIntosh JR, Euteneuer U. Tubulin hooks as probes for microtubule polarity: an analysis of the method and an evaluation of data on microtubule polarity in the mitotic spindle. J Cell Biol. 1984;98:525–533. doi: 10.1083/jcb.98.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami T, Chang WI, Garkavtsev I, Kaplan N, Lombardi D, Matsumoto T, Niwa O, Kounosu A, Yanagida M, Marr TG, et al. A 13 kb resolution cosmid map of the 14 Mb fission yeast genome by nonrandom sequence-tagged site mapping. Cell. 1993;73:121–132. doi: 10.1016/0092-8674(93)90165-m. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Emura Y, Baur M, Kohli J, Ohta K, Shibata T. The meiotic recombination hot spot created by the single-base substitution ade6-M26results in remodeling of chromatin structure in fission yeast. Genes Dev. 1997;11:876–886. doi: 10.1101/gad.11.7.876. [DOI] [PubMed] [Google Scholar]
- Molnar M, Bähler J, Sipiczki M, Kohli J. The rec8 gene of Schizosaccharomyces pombeis involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics. 1995;141:61–73. doi: 10.1093/genetics/141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. . Methods Enzymol. 1991;194:793–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Nakagawa T, Straight AF, Murray A, Chikashige Y, Yamashita Y, Hiraoka Y, Yanagida Y. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: dis1 is implicated in force balance in metaphase bipolar spindle. Mol Biol Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo ER, Pidoux AL, Perry PE, Allshire RC. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. . Nature. 1998;392:825–828. doi: 10.1038/33941. [DOI] [PubMed] [Google Scholar]
- Niwa O, Yanagida M. Triploid meiosis and aneuploidy in Schizosaccharomyces pombe: an unstable aneuploid disomic for chromosome III. Curr Genet. 1985;9:463–470. [Google Scholar]
- Olson LW, Eden U, Egel-Mitani M, Egel R. Asynaptic meiosis in fission yeast? . Hereditas. 1978;89:189–199. [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvinen M, Söderström K-O. Chromosome rotation and formation of synapsis. Nature. 1976;260:534–535. doi: 10.1038/260534a0. [DOI] [PubMed] [Google Scholar]
- Plamann M, Minke PF, Tinsley JH, Bruno KS. Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli AS, Smith GR. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. . Genetics. 1989;123:45–54. doi: 10.1093/genetics/123.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli AS, Sena EP, Smith GR. Genetic and physical analysis of the M26 recombination hotspot of Schizosaccharomyces pombe. . Genetics. 1988;119:491–497. doi: 10.1093/genetics/119.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. In vivolocalization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen K, Paranko J, Parvinen M. A colcemid-sensitive mechanism involved in regulation of chromosome movements during meiotic pairing. Chromosoma. 1982;85:611–618. doi: 10.1007/BF00330775. [DOI] [PubMed] [Google Scholar]
- Saraste M, Sibbald PR, Wittinghofer A. The P-loop: a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Bähler J, Kohli J. Dynamics of chromosome organization and pairing during meiotic prophase in fission yeast. J Cell Biol. 1994;127:273–285. doi: 10.1083/jcb.127.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan HS, Weich S, Schwegler H, Heyting C, Härle M, Cremer T. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol. 1996;134:1109–1125. doi: 10.1083/jcb.134.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchert P, Kohli J. The ade6-M26 mutation of Schizosaccharomyces pombeincreases the frequency of crossing over. Genetics. 1988;119:507–515. doi: 10.1093/genetics/119.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanisms for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimanuki M, Miki F, Ding D-Q, Chikashige Y, Hiraoka Y, Horio T, Niwa O. A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol Gen Genet. 1997;254:238–249. doi: 10.1007/s004380050412. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. . J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda A, Bähler J, Kohli J. Microtubule-driven nuclear movements and linear elements as meiosis-specific characteristics of the fission yeasts Schizosaccharomyces versatilis and Schizosaccharomyces pombe. . Chromosoma. 1995;104:203–214. doi: 10.1007/BF00352185. [DOI] [PubMed] [Google Scholar]
- Szankasi P, Heyer W-D, Schuchert P, Kohli J. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe: wild-type and mutant alleles including the recombination hot spot allele ade6-M26. . J Mol Biol. 1988;204:917–925. doi: 10.1016/0022-2836(88)90051-4. [DOI] [PubMed] [Google Scholar]
- Therman, E., and M. Susman. 1992. Human chromosomes. Springer-Verlag, New York.
- Toriyama M, Ohta K, Endo S, Sakai H. 51-kd protein, a component of microtubule-organizing granules in the mitotic apparatus involved in aster formation in vitro. Cell Motil Cytoskel. 1988;9:117–128. doi: 10.1002/cm.970090204. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken E, Loidl J, Scherthan H. Bouquet formation in budding yeast: initiation of recombination is not required for meiotic telomere clustering. J Cell Sci. 1999;112:651–658. doi: 10.1242/jcs.112.5.651. [DOI] [PubMed] [Google Scholar]
- Uzawa S, Yanagida M. Visualization of centromeric and nucleolar DNA in fission yeast by fluorescence in situhybridization. J Cell Sci. 1992;101:267–275. doi: 10.1242/jcs.101.2.267. [DOI] [PubMed] [Google Scholar]
- Vaisberg EA, Koonce MP, McIntosh JR. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO (Eur Mol Biol Organ) J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K. Definition of individual components within the cytoskeleton of Trypanosoma bruceiby a library of monoclonal antibodies. J Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Xiang X, Beckwith SM, Morris NR. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. . Proc Natl Acad Sci USA. 1994;91:2100–2104. doi: 10.1073/pnas.91.6.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Roghi C, Morris NR. Characterization and localization of the cytoplasmic dynein heavy chain in Aspergillus nidulans. . Proc Natl Acad Sci USA. 1995;92:9890–9894. doi: 10.1073/pnas.92.21.9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Nagai K, Yamasaki M, Matsuhashi M. Solubilization of aster-forming proteins from yeast: possible constituents of spindle pole body and reconstitution of asters in vitro. . Cell Struct Funct. 1990;15:221–228. doi: 10.1247/csf.15.221. [DOI] [PubMed] [Google Scholar]
- Yao KTS, Ellingson DJ. Observations on nuclear rotation and oscillation in chinese hamster germinal cells in vitro. Exp Cell Res. 1969;55:39–42. doi: 10.1016/0014-4827(69)90451-0. [DOI] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]