Abstract
The Saccharomyces cerevisiae CHS7 gene encodes an integral membrane protein located in the ER which is directly involved in chitin synthesis through the regulation of chitin synthase III (CSIII) activity. In the absence of CHS7 product, Chs3p, but not other secreted proteins, is retained in the ER, leading to a severe defect in CSIII activity and consequently, to a reduced rate of chitin synthesis. In addition, chs7 null mutants show the yeast phenotypes associated with a lack of chitin: reduced mating efficiency and lack of the chitosan ascospore layer, clear indications of Chs7p function throughout the S. cerevisiae biological cycle.
CHS3 overexpression does not lead to increased levels of CSIII because the Chs3p excess is retained in the ER. However, joint overexpression of CHS3 and CHS7 increases the export of Chs3p from the ER and this is accompanied by a concomitant increase in CSIII activity, indicating that the amount of Chs7p is a limiting factor for CSIII activity. Accordingly, CHS7 transcription is increased when elevated amounts of chitin synthesis are detected.
These results show that Chs7p forms part of a new mechanism specifically involved in Chs3p export from the ER and consequently, in the regulation of CSIII activity.
Keywords: chitin synthesis, chitin synthase III, endoplasmic reticulum export, morphogenesis, cell wall
Chitin is a structural polymer present in most fungal cell walls. Although its amount varies from 1–30% of the cell wall, it has been shown to be indispensable for the maintenance of the fungal cell (reviewed in Cabib et al., 1996). In Saccharomyces cerevisiae, chitin accounts for only 3% of the cell wall mass. However, some evidence indicates that S. cerevisiae cannot survive without chitin (Bowers et al., 1974; Shaw et al., 1991). S. cerevisiae contains three different chitin synthases (CSs)1 responsible for the synthesis of this polymer: CSI acts as a repair enzyme at the time of cytokinesis (Cabib et al., 1989, 1992); CSII makes the chitin disk of the primary septum that separates mother and daughter cells (Sburlati and Cabib, 1986; Shaw et al., 1991); and CSIII makes 90–95% of the cellular chitin, including the chitin synthesized during mating and sporulation (Roncero et al., 1988b; Valdivieso et al., 1991; Pammer et al., 1992). Each CS contains at least one catalytic subunit encoded respectively by CHS1, CHS2, and CHS3 gene products (reviewed in Bulawa, 1993; Cabib et al., 1996). These genes show significant similarity, but the differences among them should account for the different kinetics and regulatory requirements of each CS (Choi and Cabib, 1994).
The fact that each CS has a different cellular function suggests that the activity of each enzyme must be spatially and temporally regulated. In the case of CSI and CSII, it has been proposed that these would be regulated at the transcriptional (Pammer et al., 1992) and posttranslational (Choi et al., 1994a; Uchida et al., 1996) levels, the latter mechanism achieved through proteolytic processing (reviewed in Cabib et al., 1996). By contrast, CSIII is not proteolytically regulated and transcriptional regulation of CHS3, although it does exist (Pammer et al., 1992), does not control CSIII activity (Choi et al., 1994a; Chuang and Schekman, 1996; Cos et al., 1998). In addition to CHS3, three other genes, CHS4, CHS5, and CHS6, have been shown to be required for CSIII activity (reviewed in Bulawa, 1993). Chs4p has a dual function: it is a direct activator of CSIII activity (Trilla et al., 1997) and at the same time is responsible for anchoring, through Bni4p, of Chs3p to the septin ring (DeMarini et al., 1997). Chs5p is involved in the polarized transport of Chs3p in specialized vesicles (Santos and Snyder, 1997). However, its function is not restricted to Chs3p transport, since it is also involved in the transport of other proteins during mating (Santos et al., 1997). Chs6p is required for Chs3p transport in a later step than Chs5p (Ziman et al., 1998). It also has been shown that Chs3p is subject to endosomal internalization (Ziman et al., 1996), although the biological meaning of this process is not known. It therefore seems clear that the function of CSIII mainly depends on proper Chs3p transport and localization.
In addition to temporally and spatially regulated chitin synthesis during vegetative growth, there are several circumstances in the life cycle of S. cerevisiae where chitin synthesis is altered. Chitin synthesis is significantly increased during mating and Calcofluor treatment and in both instances this increase depends on CSIII activity (Roncero et al., 1988a,b). During sporulation, CSIII is required to make chitin the precursor of chitosan, a component of the outermost layer of the ascospore wall (Pammer et al., 1992). However, it is not yet known if chitin/chitosan synthesis is increased during this process. Despite all these findings, there is still no clear picture of how CHS genes can regulate chitin synthesis in vivo, and to date only Chs4p can be envisaged as a direct regulator of CSIII activity.
This paper addresses CHS7, a new S. cerevisiae gene required for CSIII activity. CHS7 is involved in the regulation of CSIII activity by controlling Chs3p export from the ER. The significance of this specific control mechanism is discussed within the framework of the hierarchy of proteins involved in the control of CSIII activity.
Materials and Methods
Strains, Growth Conditions, and Genetic Methods
Yeast strains used in this study are listed in Table I. S. cerevisiae strains were grown in YEPD (1% Bacto yeast extract, 2% peptone, 2% glucose) or SD medium (2% glucose, 0.7% Difco yeast nitrogen base without amino acids, 0.07% amino acids solid mix). Growth supplements were added to SD medium when required. Escherichia coli was grown in LB medium (1% bactotriptone, 0.5% bacto yeast extract, 1% NaCl) supplemented with 100 μg/ml ampicillin. Standard methods were used for DNA manipulations (Sambrook et al., 1989) and yeast genetics (Rose et al., 1990).
Table I.
S. cerevisiae Strains Used in this Study
| Strain | Relevant genotype | Source | ||
|---|---|---|---|---|
| X2180-1A | MATa, SUC2, mal, mel, gal2, CUP1 | American Type Culture Collection | ||
| JAY1 | X2180-1A ura3 | This study | ||
| W303-1A | MATa can1-100 ade2-1 his3-11,15 leu2-3,12 trp1-1 ura3-1 | Lab Collection | ||
| JAY6 | W303-1A chs7::HIS3 | This study | ||
| YPA24 | MATα ade2 ura3 leu1 cyh2 can1 | Lab Collection | ||
| MATa ade2-1 ura3-1 leu2-3 his3-11 can1-100 | ||||
| 15D | MATa leu2 ade1 his1 trp1 | Richardson et al., 1989 | ||
| 15Daub | 15D Δbar1 ura3 Δns | Richardson et al., 1989 | ||
| HVY374 | 15D ura3 Δns ADE1 | Dr. Valdivieso* | ||
| HVY376 | 15DMAT α ura3 Δns TRP1 | Dr. Valdivieso* | ||
| JAY21 | HVY374 chs3::LEU2 | This study | ||
| JAY22 | HVY376 chs3::LEU2 | This study | ||
| JAY25 | HVY374 chs7::HIS2 | This study | ||
| JAY26 | HVY376 chs7::HIS2 | This study | ||
| Y604 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3 Δ200 | Dr. Santos‡ (Santos and Snyder, 1997) | ||
| Y1306 | Y604 CHS3::3xHA | Dr. Santos‡ (Santos and Snyder, 1997) | ||
| JAY27 | Y1306 chs7::HIS3 | This study | ||
| JAY28 | Y1306 chs6::URA3 | This study | ||
| JAY29 | Y1306 chs4::URA3 | This study | ||
| JAY30 | Y1306 chs5::ADE2 | This study | ||
| JAY31 | Y1306 chs4::URA3 chs7::HIS3 | This study | ||
| JAY32 | Y1306 chs6::URA3chs7::HIS3 | This study | ||
| JAY33 | Y1306 chs5::ADE2 chs7::HIS3 | This study |
Departmento de Microbiología y Genética/Instituto de Microbiología Bioquímica, Consejo Superior de Investigaciones Cientificas/Universidad de Salamanca.
Yale University, New Haven, CT.
Calcofluor resistance was tested by a plate assay (Valdivieso et al., 1991) in SD medium buffered with 50 mM potassium hydrogen phthalate, pH 6.1, and supplemented with different Calcofluor concentrations (50– 1,000 μg/ml; Bayer Industrial Corporation). Calcofluor staining was observed after growing cells in the presence of 75 μg/ml Calcofluor for 2 h at 30°C.
Construction of Plasmids and Strains
Gene replacement was performed basically as described by Rothstein (1983). The Δchs7-1 disruption was constructed by replacing the 1.2-kb NdeI–NdeI fragment containing the CHS7 coding region (from nucleotide +24–297, downstream from the stop codon) with the HIS3 gene, thus creating pTM10. The NdeI downstream site is still 724 bp ahead of the next ATG, therefore this deletion should not affect the YHR143 open reading frame (ORF). Strains W303-1A and Y1306 were transformed with a linear 3.5-kb EcoRV–EcoRV fragment from pTM10, containing the chs7::HIS3 gene, giving rise to strains JAY6 and JAY27. The Δchs7-2 disruption was constructed by replacement of the 1.2-kb NdeI–NdeI fragment with the HIS2 gene, affording pTM100. By digestion of pTM100 with SacI, a 4.1-kb fragment containing the chs7::HIS2 gene was used to transform strains HVY374 and HVY376, creating strains JAY25 and JAY26. The Δchs6-1 disruption was constructed using a pGEM-CHS6 plasmid, in which the 1.6-kb BalI–EcoRI internal fragment of CHS6 was replaced with the URA3 gene, thus creating pTM12. By digestion of pTM12 with SalI–SphI, a 3.0-kb fragment containing the chs6::URA3 gene was used to transform strain Y1306, obtaining JAY28. Correct replacement of the CHS7 and CHS6 loci was determined by PCR analysis and tests of Calcofluor resistance. To construct a fusion gene encoding the full-length Chs7p fused to the green fluorescent protein (GFP; Martin-Chalfie et al., 1994), a NotI site was created by directed mutagenesis at the end of the CHS7 coding region to give pTM14. A 0.7-kb NotI–NotI fragment from pTM13 containing the GFP-encoding sequence (Fernandez-Abalos et al., 1998) was ligated into NotI-digested pTM14, thus creating pTM15 (pRS316::CHS7-GFP). CHS7::3XHA construction (pTM16) was achieved by inserting a 115-bp NotI–NotI DNA fragment containing three copies of the hemagglutinin epitope (HA) at the same NotI site of pTM14. The functionality of the hybrid proteins was determined by complementation of the phenotypes associated with Δchs7. For intracellular localization of Sec63p, 15Daub cells were transformed with plasmid pRS315::SEC63-MYC, kindly provided by R. Schekman (University of California, Berkeley, CA; Lyman and Schekman, 1997).
Multicopy plasmids pRS423::CHS3 or pRS423::CHS3-3XHA were obtained by inserting the EcoRI–SalI fragment of pHV7 that contains either CHS3 or CHS3-3XHA genes into pRS423 (Cos et al., 1998). pRS425:: CHS4 was done by cloning the CHS4 gene as a BamHI fragment into pRS425 (Trilla et al., 1997). Similarly, for construction of pRS426::CHS7, CHS7 gene was cloned as a EcoRV–StuI fragment (pTM11; this work) into pRS426. Plasmids pRS423, pRS425, and pRS426 have been previously described (Christianson et al., 1992).
Chitin and Chitin Synthase Activity Determinations
Chitin measurements were performed as described (Bulawa et al., 1986) using chitinase from Serratia marcescens (Sigma Chemical Co.). GlcNAc release was determined colorimetrically by the method of Reissig et al. (1955). Total amounts of chitin are expressed as nanomoles of GlcNAc liberated per 100 mg of cells. For CS activity assays, total cell membranes were prepared from 250 ml of exponentially growing cells (2 × 107 cells/ml) as described by Valdivieso et al. (1991). CS activity was measured essentially as described in Choi and Cabib (1994): CSI activity was assayed in 50 mM Tris-HCl, pH 6.5, and 5 mM magnesium acetate; CSII activity was assayed in 50 mM Tris-HCl, pH 8.0, and 5 mM cobalt acetate; and CSIII was assayed in 50 mM Tris-HCl, pH 8.0, with 5 mM cobalt acetate and nickel acetate. For the proteolytic activation step, 2 μl of trypsin (1–3 mg/ml) was added to the reaction medium and proteolysis activation was stopped after 15 min of incubation by adding 2 μl of soybean trypsin inhibitor solution. 1.1 mM UDP[14C]GlcNAc (Nycomed Amersham; 400 cpm/nmol) was used as substrate for the reaction. Newly synthesized chitin was determined by measuring the radioactivity incorporated into the insoluble material after the addition of 10% trichloroacetic acid and filtration through glass fiber filters (Whatman Inc.; Choi and Cabib, 1994). Specific activity is expressed as nanomoles of GlcNAc incorporated per hour per milligram of protein.
Direct and Indirect Immunofluorescence
Localization of Chs7p-GFP was observed in exponentially growing cells containing pTM15. After mounting, images were captured with a Zeiss laser-confocal microscope (LSM 510) with a 63× objective and processed with Adobe Photoshop software. Localization of Chs3p-3XHA by indirect immunofluorescence was carried out as described in Bähler and Pringle (1998) with slight modifications. Spheroplasts were obtained by treatment with 5 μg/ml Zymolyase 100T (Seikagaku) for 30–45 min at 30°C in PEMS (100 mM Pipes, 1 mM EGTA, 1 mM MgSO4, 1.2 M sorbitol, pH 6.9) buffer. Mouse HA.11 anti-HA mAb (Berkeley Antibody Co.) was used at a 1:100 dilution for 16 h at 4°C. Cy3-conjugated anti-mouse secondary antibody (Sigma Chemical Co.) was used at a 1:300 dilution for 45 min at 25°C. After mounting, images were captured with a Zeiss laser-confocal microscope (LSM 510) with a 63× objective. For these experiments, an epitope-tagged version of Chs3p in cells whose chromosomal locus had been replaced with CHS3-3XHA was used (Strain Y1306; Santos and Snyder, 1997). All strains used for immunofluorescence were derived from Y1306. Sec63p-Myc was immunolocalized with a similar protocol, but using supernatants of 9E10 hybridome at 1:100 dilution as primary antibodies (Evan et al., 1985).
Northern Blot Analysis
For Northern blot analysis, total RNA was prepared as described by Sambrook et al. (1989) from cells grown under different conditions. 12.5 μg of total RNA was loaded per lane and after electrophoresis, transferred to Hybond membranes (Nycomed Amersham). Hybridization was carried out as described (Sambrook et al., 1989) using a 1.2-kb NdeI–NdeI fragment from CHS7 as a probe. For quantitative analysis, Northern blots were exposed to PhosphorImager screens (BASS 1500; FujiFilm). α-Factor treatment for Northern blot experiments was carried out in exponentially growing cells using YEPD medium supplemented with 5 μM α-factor (Choi et al., 1994a).
Subcellular Fractionation Experiments
Cell lysates were prepared (as described in Santos and Snyder, 1997) from a JAY25 strain transformed with pRS315::SEC63-MYC and pRS316:: CHS7-3XHA plasmids. In brief, exponentially growing cells in SD medium (1.5 g wet wt), were resuspended in 5 ml of 17% sucrose (wt/vol) in 50 mM Tris-HCl, pH 7.5, and 1 mM EDTA containing the protease inhibitor cocktail (1 mM phenylmethanesulfonil fluoride and 1 mg/ml each of leupeptin, pepstatin, and aprotinin), and broken by vortexing with glass beads. Lysated cells were centrifuged at 1,500 g for 10 min. The cleared supernatant was layered on top of 33 ml of a linear sucrose gradient (10– 65% wt/vol) in 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, and centrifuged in a SW28 rotor at 25,000 rpm for 20 h at 4°C. 1.25 ml fractions were collected from the bottom using a peristaltic pump. Equal volumes of each fraction were examined for their content of Pma1p, Kre2p, Sec63p-MYC, and Chs7p-HA, determined by SDS-PAGE and immunoblot with their respective antibodies. Rabbit polyclonal antibodies (1:30,000 dilution; kindly provided by Dr. R. Serrano, Universidad de Valencia, Valencia, Spain) were used to detect Pma1p, a typical plasmatic membrane marker. Kre2p, a medial-Golgi compartment protein (Lussier et al., 1995), was detected using polyclonal antibodies (1:1,000 dilution) kindly supplied by H. Bussey (McGill University, Montreal, Qúebec, Canada). Sec63-MYC, an ER marker, was detected using monoclonal anti-MYC antibodies (supernatants of 9E10 hybridome, 1:5,000 dilution). Mouse HA.11 anti-HA mAb (Berkeley Antibody Co.; 1:2,000 dilution) was used to detect Chs7p-HA. In all cases, Western blots were developed using ECL (Nycomed Amersham).
Western Blot Analysis of Chs3p
Chs3p-3XHA expression levels were determined in total cellular extracts by Western blot analysis as described in Cos et al. (1998). Blotted proteins were incubated with mouse HA.11 anti-HA antibody (Berkeley Antibody Co.; 1:2,000 dilution) and developed using ECL (Nycomed Amersham).
Endoglycosidase H (EndoH) treatment was carried out on Chs3p-3XHA after immunoprecipitation under denaturing conditions as described by Cos et al. (1998). The immunoprecipitated complex was subjected to Western blot analysis.
Other Methods
Protein was measured by the method of Lowry et al. (1951). Yeast strains were mated quantitatively as described in Santos et al. (1997). The frequency of diploid formation was estimated as the number of diploids formed out of the total number of cells. Exoglucanase activity was assayed as described by San Segundo et al. (1993); the method is based on the release of reducing sugar groups from laminarin, which were quantified by the method of Somogyi (1952) and Nelson (1957). The α-factor secretion assay was developed as described in Michaelis and Herskowitz (1988).
Results
Isolation of CHS7
A new screening for S. cerevisiae mutants resistant to Calcofluor was carried out to identify new genes involved in chitin synthesis. Ethyl methanesulfonate-mutagenized JAY1 cells (∼20% of survival) were plated in YEPD media supplemented with 1 mg/ml Calcofluor exactly as described in Roncero et al. (1988b). After 4 d of incubation, 53 bona fide independent clones which were able to grow were selected (mutation frequency 1.1 × 10−5). These mutants were back-crossed with previously known Calcofluor resistant and chitin-deficient mutants (chs3, chs4, and chs5). Diploid analysis indicated that most mutants belonged to chs3 (83%), chs4 (5.7%), or chs5 (5.7%) complementation groups. Only three of them, cwr6, cwr12, and crw19 (Calcofluor white resistant), defined new complementation groups. Further genetic testing indicated that mutants cwr12 and cwr19 belonged to the same group. A preliminary characterization of these mutants indicated that all showed defects in chitin synthesis.
The genes affected in cwr12 and cwr6 mutants were cloned by complementation of the Calcofluor resistant phenotype after transformation with a centromeric library (Rose et al., 1987). Complementation was confirmed to be dependent on the presence of plasmid and complementing plasmids were isolated in E. coli for further work. Originally isolated plasmids were subjected to endonuclease restriction mapping and the results obtained indicated that the plasmid complementing cwr12 contained the previously described CHS6/CSD3 gene (Bulawa, 1992). Therefore, its study was discontinued. The DNA fragment that complemented the cwr6 mutant contains several ORFs (Fig. 1 A). Subcloning experiments indicated that the minimum fragment able to complement was the StuI–EcoRV fragment included in pTM11. With these results, we were able to obtain a preliminary identification of YHR0142w as the gene that complements cwr6 mutation. Due to the defect in CS observed in cwr6 mutant, we named this gene CHS7 following accepted nomenclature (Cabib, 1994).
Figure 1.

Identification and characteristics of Chs7p. (A) Subcloning strategy used in the identification of YHR142 as complementing gene for the cwr6 mutants. Complementation was determined by analyzing the reversal of Calcofluor resistance in cwr6 cells. Useful restriction sites are indicated: E, EcoRI; C, ClaI; V, EcoRV; X, XbaI; St, StuI; B, BamHI; P, PstI; K, KpnI; and S, SalI. (B) Amino acid sequence of Chs7p. Membrane-spanning domains are underlined. (C) Kyte-Doolittle hydrophobicity plot of Chs7p.
CHS7 encodes a protein of 316 amino acids that contains six or seven putative transmembrane domains (Fig. 1, B and C). It has no significant homology with any known protein, although the search programs showed limited homologies with other transmembrane regions.
Characterization of chs7 Mutants
We made several chs7 null mutants by replacing the CHS7 ORF by different auxotrophic markers (Materials and Methods). Table II shows some of the characteristic phenotypes associated with this mutation. chs7 null mutants had only 13.6% of the wild-type chitin, a defect that was associated with a comparable decrease in CSIII activity. Due to this defect in chitin synthesis, chs7 cells were resistant to 1 mg/ml Calcofluor and they did not show enlarged septa after Calcofluor treatment. In addition, Δchs7/Δchs7 diploids showed compact ascospores, a defect associated with the absence of the chitosan layer (Pammer et al., 1992). chs7 mutations also reduced their mating efficiency to a level similar to that of chs3 mutants (Table II). It should be noted that the original cwr6 mutants showed similar, but less pronounced phenotypes. As expected, wild-type CHS7 (YHR0142w) in a centromeric plasmid complemented the phenotypes observed in the null (Table II, last column) or point (not shown) chs7 mutants. Once we had constructed null mutants, we confirmed the allelism between YHR142w and CHS7 genetically. Diploid strain yhr142::HIS3/cwr6 was resistant to Calcofluor and analysis of 18 tetrads in this cross showed a Calcofluor resistance segregation of 4:0. Therefore, CHS7 and YHR142w must be the same gene.
Table II.
Characteristic Phenotypes of cwr6 (chs7) Mutants
| Phenotype | Strains* | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT | Δchs3 | cwr6 (chs7) | Δchs7 | Δchs7 (CHS7) | ||||||
| Calcofluor resistance‡ | − | + | + | + | − | |||||
| Septa enlargement§ | + | − | − | − | + | |||||
| Chitin ¶ | 100 | 6.5 | 21.6 | 13.6 | 87.6 | |||||
| CSIII ¶ | 100 | 15.8 | 28.2 | 14.6 | 103.4 | |||||
| Mating efficiency ¶ | 100 | 69.3 | ND | 65.3 | ND | |||||
| Compact ascospores‖ | No | Yes | ND | Yes | ND | |||||
All strains used, except the original cwr6 mutant, are isogenic derivatives of HVY374 obtained by gene replacement (Table I).
Growth in buffered minimal media supplemented with 1 mg/ml Calcofluor.
Presence (+) or absence (−) of enlarged septa after 2 h of growth in YEPD media supplemented with 75 μg/ml Calcofluor.
Numeric values represent the percentage of the values obtained for each strain compared to that of wild-type. Chitin, CSIII, and mating efficiency were determined as described in Materials and Methods.
Determined by observation under phase-contrast microscope.
Analysis of CHS7 Expression
To analyze CHS7 expression levels, we determined CHS7 mRNA levels in cells harvested after growth under different conditions. CHS7 was detected in all conditions (Fig. 2) as a single band of ∼1.1 kbp, in clear agreement with sequence data. Therefore, CHS7 is expressed constitutively. However, as shown in Fig. 2 A, α-factor treatment (mimicking the mating process) increased CHS7 expression 3.5 times after only 15 min; shortly after, vegetative levels were resumed. Calcofluor treatment also increased CHS7 expression twofold (Fig. 2 A, right). Despite its constitutive expression during vegetative growth, CHS7 mRNA levels were strongly increased during sporulation, reaching a maximum (∼24-fold more) after 10 h of sporulation induction. Thereafter, CHS7 mRNA levels slowly decreased. The typical expression pattern observed resembled that of a middle sporulation gene (Krisak et al., 1994; Hepworth et al., 1998). These results allowed us to conclude that the expression of CHS7 is transcriptionally regulated. In addition, CHS7 transcription increased under all growth conditions in which chitin synthesis was increased.
Figure 2.

Transcription levels of CHS7. Northern blots were done with RNAs obtained from cells grown under different conditions and loaded in equivalent amounts. CHS7 mRNA was detected using the NdeI–NdeI fragment as a probe (Materials and Methods). In all cases, autoradiographic images and quantitative determinations by PhosphorImager (Materials and Methods) are shown. Relative amounts of CHS7 mRNA are referred to actin levels (not shown) and normalized with respect to the value obtained at the respective control points (left lane of each figure). (A) RNA was obtained from X2180-1A cells incubated for the indicated times on 5 μM α-factor (left) or from cells grown for 2 h in the absence (−) or presence (+) of 50 μg/ml Calcofluor. (B) RNA was obtained from YPA24 diploid strain incubated in sporulation medium for the indicated times.
Chs7p Is a Membrane Protein Localized in the ER
Chs7p was tagged at the COOH terminus either with the 3XHA epitope or GFP (Materials and Methods) to determine the subcellular localization of Chs7p. Chimeric proteins Chs7p-3XHA or Chs7p-GFP complemented the Calcofluor resistance phenotype of chs7 null mutant (JAY6 strain). In addition, the chimeric proteins restored wild-type levels of chitin synthesis in this strain, as determined by chitin staining with Calcofluor (Cos et al., 1998). It can be concluded that Chs7p-3XHA and Chs7p-GFP are fully functional. Western blot analysis indicated that Chs7p-3XHA runs as a 35-kD protein that is localized in the particulate fraction (data not shown). Molecular size and subcellular localization are in clear agreement with sequence predictions. Logarithmically growing cells expressing Chs7p-GFP from its own promoter showed the fluorescent staining depicted in Fig. 3 A. Chs7p-GFP was uniformly localized in the nuclear periphery and in discrete patches associated with the cytoplasmic membrane. This pattern coincides with that of Sec63p-Myc (Fig. 3 B), a typical ER marker (Sadler et al., 1989). The vacuolar staining observed seems to be an artifact because it does not appear in all the cells and it does not show the typical green color of GFP (data not shown). A similar distribution in the ER was observed by indirect immunofluorescence in cells expressing Chs7p-3XHA (data not shown). No polarized distribution of Chs7p was observed in either case. To confirm the immunofluorescence results a subcellular fractionation experiment was carried out. Total cellular extract were loaded into the top of a linear sucrose density gradient and after centrifugation, Pma1p, Sec63p-MYC, Kre2p, and Chs7p-3XHA distribution was analyzed by Western blot as described in Materials and Methods. Fig. 3 C shows that membrane protein Pma1p is localized at the bottom of the gradient, mainly between fraction 1 to 4. Sec63p is localized from fractions 5 to 17, with a significant peak between the 11 to 15 fractions. Chs7p distribution is quite similar with that of Sec63, also showing maximum accumulation between 11 to 15 fractions. Golgi compartment distribution, marked by Kre2p (Lussier et al., 1995), is displaced to lighter fractions. Therefore, it can be concluded that Chs7p is an ER membrane protein.
Figure 3.
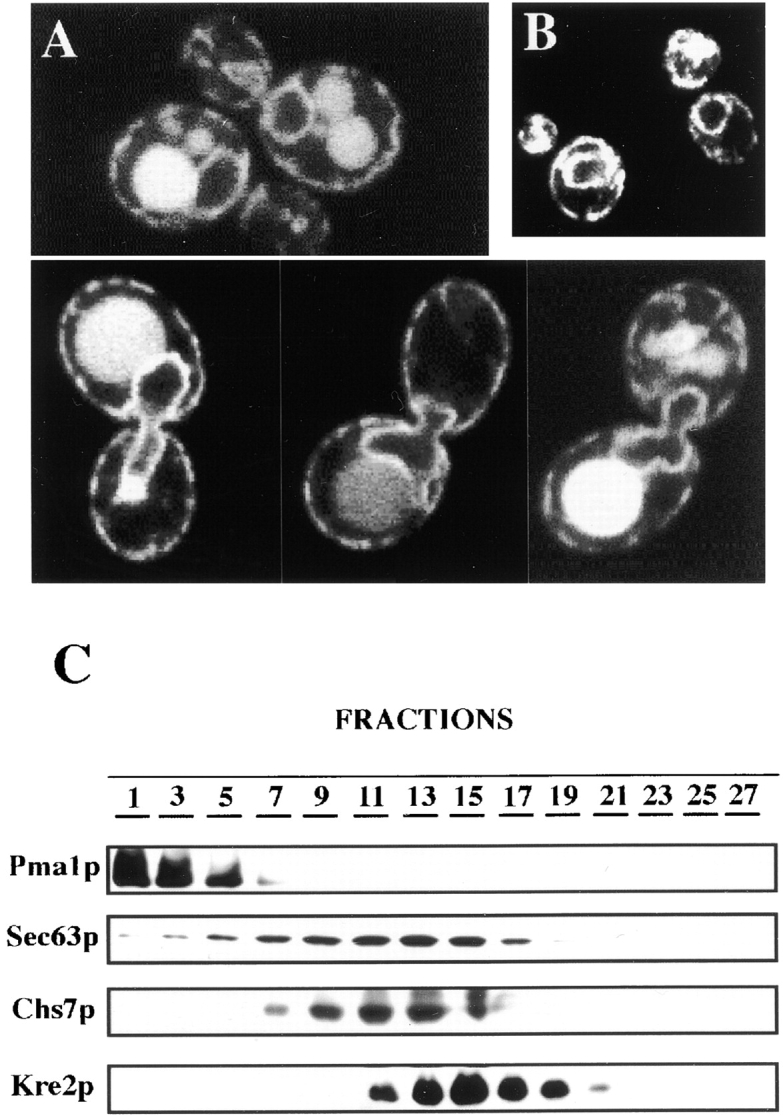
Localization of Chs7p. (A) JAY25 cells transformed with pTM15 (pRS316::CHS7-GFP) were visualized directly in a Zeiss laser-confocal microscope under appropriate excitation conditions. Note the continuous staining along the nuclear periphery and the patches associated with cytoplasmic membrane. Cells shown on the lower part of the figure are in the process of nuclear division. (B) Indirect immunofluorescence localization of Sec63p-Myc in wild-type (15Daub) cells transformed with pRS315::SEC63-MYC. (C) Western blot experiments showing subcellular distribution of several proteins after equilibrium centrifugation in a sucrose gradient. Fractions were collected from the bottom (lower fractions, higher densities). Equal volumes of each fraction were loaded per lane (Materials and Methods).
Chs7p Is Required for Chs3p Export from the ER
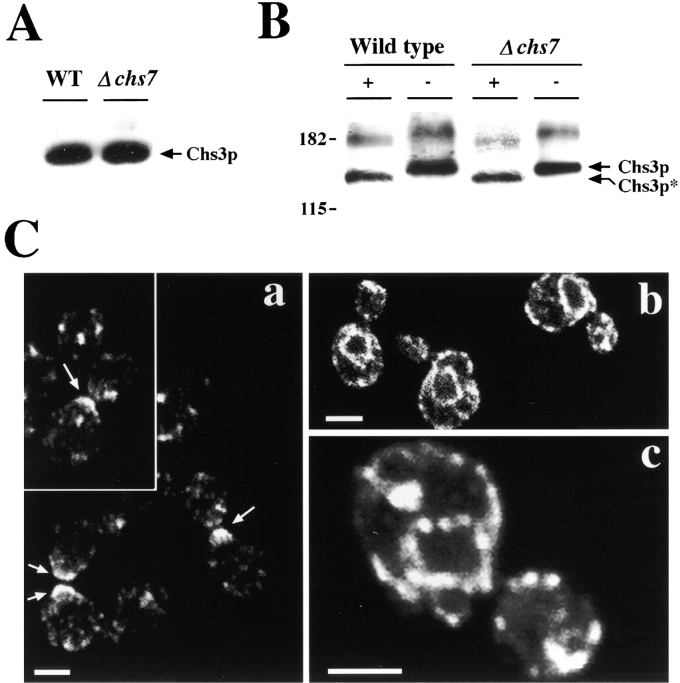
Functional CSIII activity depends on the appropriate synthesis and transport of Chs3p, its catalytic subunit. Chs7p does not seem involved in the synthesis of Chs3p, since Chs3p levels in Δchs7 mutants are similar to those observed in wild-type (Fig. 4 A). Therefore, Chs7p could be involved in the transport of Chs3p, a process that is mediated by Chs4p, Chs5p, and Chs6p. Following a strategy similar to that used in the characterization of these genes, we localized an HA-tagged version of Chs3p (Santos and Snyder, 1997) in wild-type and chs7-null mutants. As previously reported (Chuang and Schekman, 1996; Santos and Snyder, 1997), Chs3p-3XHA localized in the base of the emerging bud in wild-type strains (Fig. 4 C, a). However, in Δchs7 mutants all the Chs3p-3XHA remained in the ER, showing a perinuclear localization with partial association with the plasma membrane (Fig. 4 C, b, compare to Fig. 3 A). No polarized distribution of Chs3p-3XHA was observed in Δchs7 mutants. Higher magnification (Fig. 4 C, c) indicated that the perinuclear staining was punctuated, suggesting possible Chs3p aggregation. In addition, it is important to notice that although retained in the ER, Chs3p-3XHA is correctly core-glycosylated in chs7 mutants (Fig. 4 B). Therefore, it can be concluded that Chs7p is required for Chs3p export from the ER and Chs7p must be the initial step in the hierarchy of proteins required for CSIII activity. To confirm this hypothesis, we analyzed the localization of Chs3p-3XHA in Δchs4 Δchs7, Δchs5 Δchs7, or Δchs6 Δchs7 double mutants (Fig. 5, B, D, and F). In all cases, Chs3p was retained in the ER and did not show the localization described for Δchs4, Δchs5, or Δchs6 single mutants. As previously reported (DeMarini et al., 1997), the distribution of Chs3p in Δchs4 mutant was polarized (Fig. 5 A). However, it was not as densely packed at the base of the bud as in the wild-type (Fig. 4 C, a). In Δchs5 and Δchs6 mutants, Chs3p was localized in internal vesicles (Fig. 5, C and E) without polarized distribution (Santos and Snyder, 1997; Ziman et al., 1998). From these results, we conclude that the chs7 mutation is epistatic to the chs mutations and that Chs7p is necessary for Chs3p export from the ER.
Figure 4.
Characterization of Chs3p in wild-type and Δchs7 mutants. (A) Expression levels of Chs3p-3XHA in wild-type and Δchs7 strains. (B) Mobility of Chs3p-3XHA from wild-type and Δchs7 before (−) and after (+) EndoH treatment. Chs3p* indicates mobility of the protein after N-glycosylation removal. Protein extracts (A and B) were prepared from HVY374 (wild-type) and JAY25 (Δchs7) strains transformed with plasmid pHV7-CHS3::3XHA and detected by Western blot (Materials and Methods). (C) Chs3p-3XHA detected by indirect immunofluorescence with HA.11 antibody. Early logarithmically growing cells were processed as described in Materials and Methods. Strains: a, Y1306 (CHS3::3XHA); b and c, JAY27 (CHS3::3XHA, chs7::HIS3). (a) Polarized Chs3p distribution in wild-type cells. (b and c) Typical ER distribution of Chs3p in Δchs7 mutant. Note the irregular distribution (dotted staining) observed after higher magnification in c. Bars, 2 μm.
Figure 5.
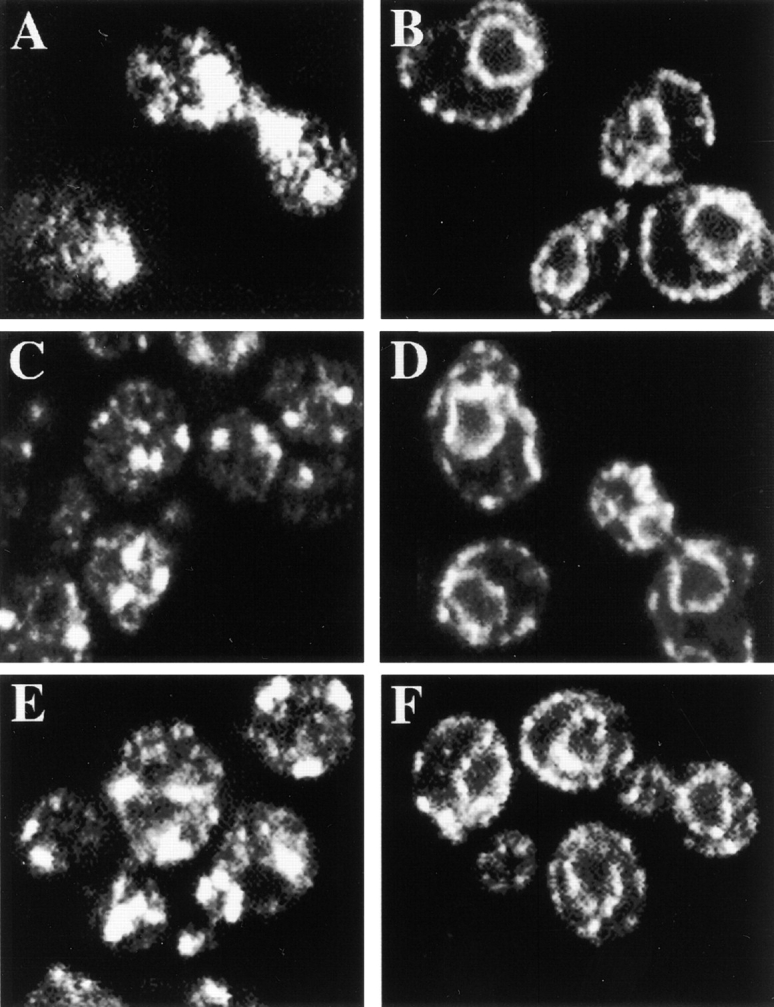
Immunolocalization of Chs3p-3XHA in chs mutants. All cells are Y1306 (CHS3::3XHA) with different chs mutations. (A) JAY29, chs4::URA3. (B) JAY31, chs4::URA3 chs7::HIS3. (C) JAY30, chs5::ADE2. (D) JAY33, chs5::ADE2 chs7::HIS3. (E) JAY28, chs6::URA3. (F) JAY32, chs6::URA3 chs7::HIS3. (A, C, and E) Polarized distribution of Chs3p at the base of the emerging bud (Fig. 4 A) is lost in chs mutants. Each chs mutant shows the typical Chs3p localization as described. (B, D, and F) In Δchs7 mutants, Chs3p-3XHA shows ER localization.
Chs7p Is Not Required for the Transport of other Proteins
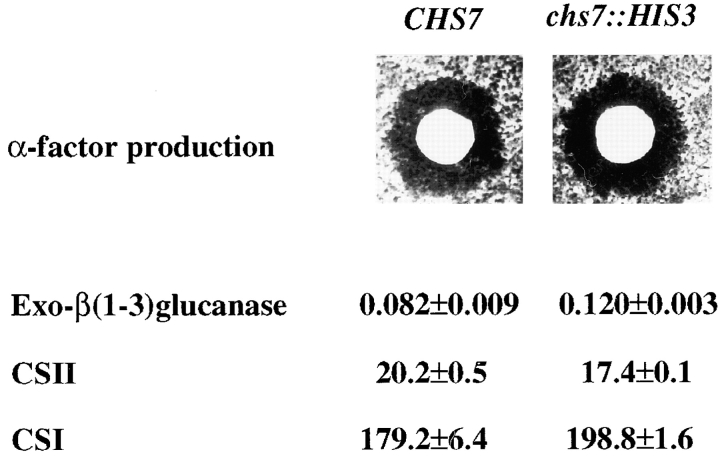
Chs7p could play a general role in secretion or protein sorting rather than being specific for chitin synthesis. To test this hypothesis, we analyzed the behavior of several secreted or sorted proteins in Δchs7 mutants. α-factor and exo-β(1-3)glucanase are two extracellular S. cerevisiae proteins that follow the normal secretion route. Fig. 6 shows that the expression levels of these two proteins are unaffected in Δchs7 mutants. Similarly, CSI and CSII, the other two CS activities present in S. cerevisiae, showed wild-type levels in Δchs7 mutants. Apparently, Chs7p is not required for secretion or sorting of other cellular proteins.
Figure 6.
Secretion phenotypes of Δchs7 mutants. (A) Production of α-factor measured by the size of the growth inhibition halo. (B) Activities of several enzymatic activities in wild-type and chs7 null mutants. Activities were determined in HVY374 (WT) and JAY25 (chs7::HIS2) isogenic strains and expressed as described in Materials and Methods.
In addition to the regular secretion mechanism, S. cerevisiae contains several other mechanisms involved in the specific assembly and transport of certain proteins. Sorting of vacuolar ATPase (v-ATPase) or several amino acid permeases to cytoplasmic membrane has been shown to depend on specific mechanisms of export from the ER (reviewed in Kaiser et al., 1997), raising the possibility of the involvement of Chs7p is such processes. However, chs7 null mutants grew in glycerol and did not show synthetic lethality with other amino acid auxotrophies, clear indications that v-ATPase and amino acid permeases function properly in Δchs7 mutants.
Chs7p Is a Limiting Factor in the Constitution of Functional CSIII Activity
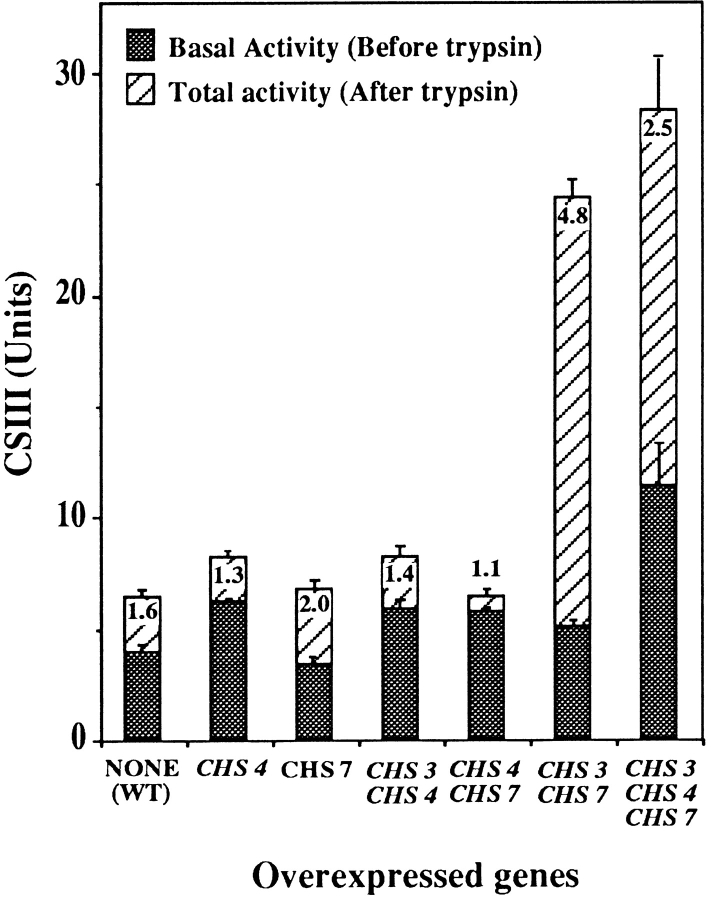
So far we have shown that Chs7p is required for Chs3p export. However, from Fig. 2 it is also apparent that CHS7 expression increases when more chitin is synthesized. These observations suggest that the level of expression of CHS7 could be involved in the control of CSIII activity. If so, this would explain why overexpression of CHS3 does not increase CSIII levels. To confirm this possibility, we measured CSIII activity in strains overexpressing CHS genes in different combinations. Overexpression of CHS3 (Cos et al., 1998) or CHS7 (Fig. 7) alone had no significant effect on CSIII. As previously reported (Trilla et al., 1997), overexpression of CHS4 increased CSIII activity and reduced its trypsin dependence. Joint overexpression of CHS4 and CHS3 or CHS4 and CHS7 did not alter the effect of CHS4 on CSIII (Fig. 7). Cooverexpression of CHS3 and CHS7 increased total CSIII almost four times. However, basal activity was only increased 1.3 times and, as a consequence, the activation factor by trypsin treatment was almost five times, in comparison with 1.6–2 times, for the controls. Joint overexpression of CHS4, CHS3, and CHS7 did not further increase total CSIII activity. However, basal activity was increased more than twofold. Therefore, activation by trypsin treatment was reduced to 2.5 in this case. Taken together, these results indicate that the amounts of Chs7p and Chs3p act as limiting factors to total CSIII activity. In addition, Chs4p levels are also limiting in the reconstitution of an in vitro fully active CSIII. Overexpression of CHS3, CHS4, and CHS7 genes also induced a modest, but significant, increase in chitin synthesis in vivo because a strain overexpressing these three genes contained ∼68 ± 11% more chitin than the corresponding wild-type. This increase in chitin synthesis was not observed if only CHS3 and CHS7 were overexpressed (not shown).
Figure 7.
CSIII activity in strains overexpressing CHS genes. CSIII activity was determined in cell extracts with (total activity) or without (basal activity) trypsin activation (Materials and Methods). Numbers indicate the ratio between total and basal activity. The results are the average of three independent experiments. Standard deviation bars are shown. On the abscissa, the names of overexpressed CHS genes are indicated in each case. Overexpression was achieved by transforming wild-type (15Daub) cells with multicopy plasmids: pRS423::CHS3, pRS425:: CHS4, or pRS426::CHS7 (Materials and Methods).
These results clearly suggest that the amount of Chs7p is a limiting factor in vivo for CSIII activity and therefore, could be the reason for the absence of an increase in CSIII activity after CHS3 overexpression. In fact, immunostaining of cells overexpressing CHS3-3XHA alone revealed a severe retention of Chs3p in the ER even in the presence of normal amounts of Chs7p (Fig. 8 A). Cooverexpression of CHS3 with CHS7 dramatically reduced Chs3p accumulation in the ER (Fig. 8 B) and only residual staining was observed at ER level. In this case, Chs3p was mainly localized in discrete patches in the cytoplasm, which in some cells showed a polarized distribution (Fig. 8 B, arrows). This distribution resembled an intermediate stage between Δchs6, Δchs5, and Δchs4 mutants (Fig. 5, A, C, and E). In sum, Chs3p excess is not retained in the ER in the presence of high levels of Chs7p and hence, higher levels of functional CSIII activity are achieved.
Figure 8.
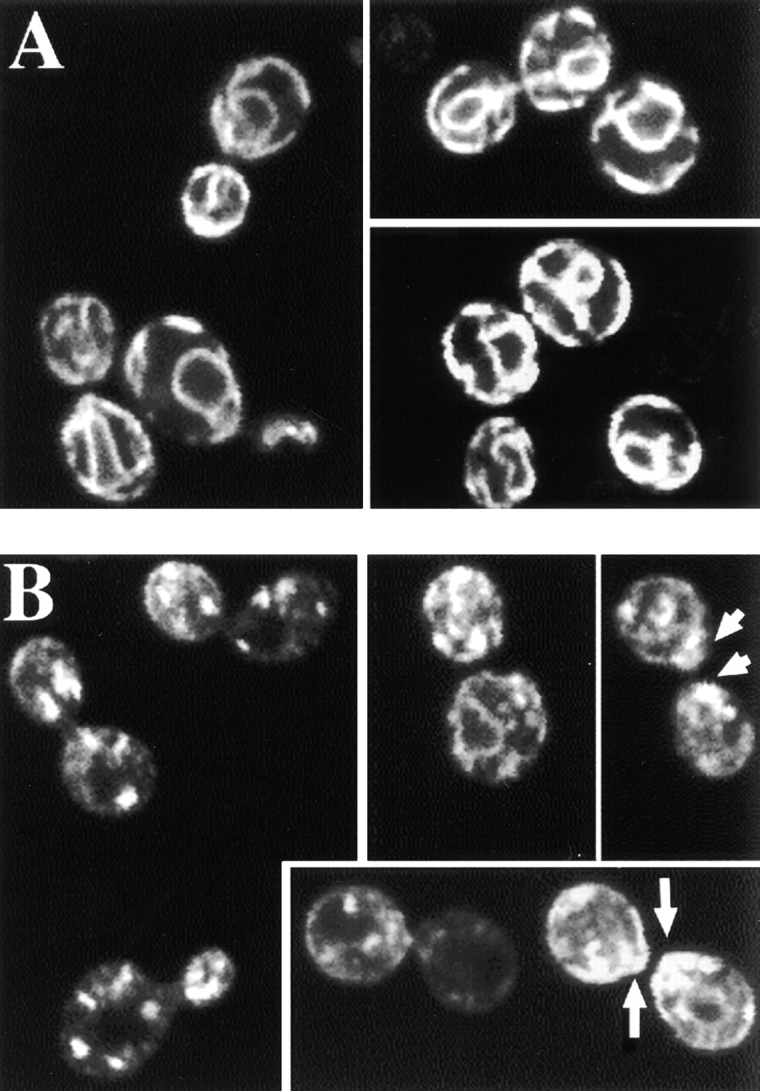
Immunolocalization of overexpressed Chs3p-3XHA. (A) Characteristic ER staining of strain Y604 transformed with pRS423::CHS3-3XHA after immunodetection of Chs3p-3XHA. (B) Chs3p-3XHA localization in Y604 cotransformed with pRS423::CHS3-3XHA and pRS426::CHS7. Note the dotted staining in most cells, some showing polarized staining (arrows). Some cells also show residual perinuclear staining.
Discussion
Chitin is a minor, but essential polymer in the S. cerevisiae cell wall. Despite its low abundance, three different CS activities have been described in this yeast (Cabib et al., 1996). From a quantitative point of view, CSIII activity is the most important, since it is involved in the synthesis of 90–95% of cell wall chitin during vegetative growth (Roncero et al., 1988b; Valdivieso et al., 1991; Bulawa, 1992). In addition, it is required for the synthesis of new chitin that takes place during the mating and sporulation processes (Roncero et al., 1988b; Pammer et al., 1992).
Recently, it has been shown that the transcriptional regulation of CHS3, the gene that encodes the catalytic subunit of CSIII, is not the control mechanism for CSIII activity (Choi et al., 1994a; Chuang and Schekman, 1996; Cos et al., 1998). Therefore, the identification of genes involved in the control of CSIII and chitin synthesis gains interest. Three proteins, Chs4p, Chs5p, and Chs6p, have been identified, so far, as factors involved in the control of CSIII activity. This report deals with the isolation of a new gene, CHS7, also involved in the control of CSIII activity. CHS7 was isolated by complementation of a mutant resistant to Calcofluor, showing that this strategy still has potential, although most (83%) of the mutants isolated belong to the chs3 complementation group.
Δchs7 mutants have reduced levels of CSIII activity and chitin in their cell walls (Table II). This defect is comparable to that observed in chs3 null mutants (Cos et al., 1998) and stronger than that detected in chs4 (Trilla et al., 1997), chs5 (Santos et al., 1997), or chs6 mutants (Bulawa, 1993), underscoring the relevance of this gene in the control of CSIII activity. CHS7 encodes a small protein with six or seven putative transmembrane domains (Kyte and Doolittle, 1982). Repeated database searches using different algorithms did not reveal any protein with significant homology to this protein. Some Chs7p regions with similarity to other integral membrane proteins probably reflect the hydrophobicity of the membrane-spanning domains rather than protein conservation. Chs7p colocalized with Chs3p in a crude particulate cell fraction and we therefore expected to find colocalization of Chs3p and Chs7p in the plasma membrane. However, Chs7p was localized exclusively in the ER (Fig. 3), showing the typical localization of other ER proteins such as Sec63p (Sadler et al., 1989; Fig. 3 B). This localization does not seem to be an artifact of the chimeric Chs7p-GFP because similar results were obtained with Chs7p-3XHA either in immunofluorescence (data not shown) or in subcellular fractionation (Fig. 3 C) experiments.
A functional explanation to account for this localization, unique among Chs proteins, became apparent when we observed that Chs3p accumulates at the ER in the absence of Chs7p (Fig. 4 C, b). However, the Chs3p protein accumulated in the chs7 mutant is correctly core-glycosylated (Fig. 4 B), a clear indication that Chs3p is translocated and correctly localized in the ER compartment in this mutant. Therefore, Chs7p should be required for Chs3p export from the ER. Moreover, the chs7 mutation is epistatic over other chs mutations, suggesting that Chs7p is the initial element in the hierarchy of proteins involved in CSIII activity. These results explain the stronger defect in CSIII activity observed in Δchs7, as compared with Δchs4, Δchs5, or Δchs6 mutants. It should be noted that Chs4p, Chs5p, or Chs6p is not required for Chs3p export from the ER (Fig. 5). At this point, all the data points to the specific involvement of Chs7p in the export of Chs3p from the ER. However, the concomitant participation of other CHS products in this export cannot be ruled out. Despite this, preliminary evidence indicates that Chs5p and Chs4p are efficiently exported in Δchs7 mutants (data not shown) and therefore, their participation is rather unlikely.
To date, there is no clear relationship between the levels of expression of different CHS genes and chitin levels, although regulation of chitin synthesis has been observed during the yeast life cycle (Roncero et al., 1988a,b; Pammer et al., 1992). It should especially be stressed that overexpression of CHS3 several times (∼30) did not increase CSIII activity or chitin levels (Cos et al., 1998) and that Chs3p levels remain stable during vegetative growth (Choi et al., 1994a; Chuang and Schekman, 1996). Under these circumstances, only Chs4p can be envisaged as a controller of CSIII activity (Bulawa, 1993; Trilla et al., 1997). Might Chs7p also be involved in the control of CSIII? Fig. 2 shows some results in favor of this hypothesis, since CHS7 is the only CHS gene whose transcription is increased under all conditions in which chitin synthesis is seen to be increased: mating, Calcofluor treatment, and sporulation. The increase in CHS7 expression during sporulation is especially relevant and its pattern of induction resembles that of a middle-late sporulation gene (Hepworth et al., 1998). The timing of expression places CHS7 among the sporulation genes involved in spore wall formation (Krisak et al., 1994; Hepworth et al., 1998). Further and more direct confirmation of this hypothesis was achieved after joint overexpression of CHS3 and CHS7 genes. CSIII activity rose dramatically after the overexpression (Fig. 7), reaching values several-fold higher than those of the control strain. This increase depends on joint overexpression, suggesting the requirement of a balanced expression of both genes.
Biochemically, we have shown that Chs7p acts as a limiting factor when Chs3p levels are increased, but the reason at an intracellular level for this limitation remains unknown. As shown in Fig. 8 A, the cells cannot process an excess of Chs3p and therefore, overexpression of CHS3 leads to Chs3p retention in the ER. However, if CHS7 is overexpressed in strains with high levels of Chs3p, this protein is released from the ER (Fig. 8 B). In addition, the localization of Chs3p in this strain resembles that observed in Δchs6 (Ziman et al., 1998) or Δchs5 (Santos and Snyder, 1997) mutants (bright internal spots) with partial polarization (Fig. 8 B, arrows), as in chs4 mutants (Fig. 5 B; DeMarini et al., 1997). These results indicate that although Chs3p is efficiently exported from the ER, its transport to its physiological site of action is impaired, probably due to some other limiting factors. In this condition, the high amount of active CSIII detected suggests that the limiting factor, if it exists, should appear at the time of or after CHS6 participation, because mutations in genes that function early on the pathway show severe defects in CSIII activity (Bulawa, 1993; Santos et al., 1997).
Chitin synthesis increases in S. cerevisiae during α-factor treatment and this process is associated with the increase in CSIII activity (Roncero et al., 1988a). Recently, it has been shown that during α-factor treatment Chs3p levels increase significantly (Cos et al., 1998). However, no Chs3p is localized at the ER (Santos and Snyder, 1997). We have shown that the effect of increased Chs3p levels on chitin synthesis depends mainly on correct Chs3p export from the ER. Therefore, the increase in CHS7 transcription observed during α-factor treatment (Fig. 2 A) guarantees the export of excess Chs3p and the increase in chitin synthesis required during this process.
Overexpression of CHS3 and CHS7 leads to a significant increase in CSIII activity. However, the activity obtained under these conditions was highly zymogenic. When we increased Chs4p levels in the same experiment, the CSIII dependence on trypsin decreased significantly. Therefore, it seems that Chs4p is also a limiting factor for this activity. We cannot be sure if Chs4p function is mediated by proteolytic processing of CSIII, but it does seem clear that in vitro trypsin treatment of CSIII resembles the activation carried out in vivo by Chs4p. Moreover, the strain carrying the triple overexpression also showed a significant increase in cellular chitin levels (68% higher than controls) indicating that these genes not only participate in the control of CSIII, but also in the in vivo control of chitin synthesis. We cannot expect a perfect correlation between CSIII and chitin levels, since there could be as yet undescribed cellular determinants participating in this process. In addition, problems of interference could arise in the replication of three independent plasmids that could mask quantitative interpretation of these data. By contrast, strains overexpressing only CHS3/CHS7, although they showed high CSIII activity, did not show any increase in chitin levels. This result is the first experimental evidence that Chs4p is an in vivo regulator of chitin synthesis because until now, the Chs4p function has only been determined in CSIII measurements in vitro (Bulawa, 1993; Choi et al., 1994b; Trilla et al., 1997).
With this evidence in mind, it can be concluded that the extent of CSIII activity in vivo depends on a delicate balance between the levels of Chs3p, Chs4p, and Chs7p proteins, a balance that ensures the level of chitin synthesis required at each moment of the S. cerevisiae life cycle, besides the specific roles of CSI and CSII activities.
At this point, we are still unable to pinpoint the exact role of Chs7p in the export of Chs3p. However, we can compare this system with the assembly of v-ATPase or the transport of amino acid permeases, the two previously reported cases in S. cerevisiae that involve specific proteins in their exit from the ER (reviewed in Kaiser et al., 1997). v-ATPase mature complex is assembled in the ER with the help of several proteins, such as Vma12p or Vma22p, which do not leave the ER and therefore do not participate in the active v-ATPase complex (Graham et al., 1998). Likewise, Shr3p is required for the release of several amino acid permeases from the ER (Ljungdahl et al., 1992). If this protein is not present, the amino acid permeases cannot be loaded into COPII vesicles (Kuehn et al., 1996, 1998). We can exclude the participation of CHS7 in any of these processes because chs7 mutants did not show phenotypes associated with the lack of v-ATPase or the amino acid permeases. The role of Chs7p in the export of Chs3p could be similar to that reported in any of these cases. In the absence of Chs7p, the CSIII complex, including Chs3p, is either not assembled or not loaded into secretory vesicles. There is indirect evidence suggesting that Chs7p could have a similar role to Shr3p. First of all, Shr3p and Chs7p, although lacking significant homology, have a very similar secondary structure with several transmembrane domains. This type of secondary structure is typical of resident ER proteins (outfitters) rather than proteins loaded into cargo vesicles (escorts or guides; Herrmann et al., 1999). Chs7p also lacks the protein sequences involved in COPI mediated retrograde transport. In addition, we have been unable to detect Chs7p outside of the ER compartment. However, we cannot exclude that Chs7p is rapidly recycled between the ER and Golgi compartments. The answer to this question will have to await the description of the nature of the secretory vesicles involved in the transport of Chs3p.
The results reported here indicate that CHS7 is part of a specific mechanism for CS export and its presence in vertebrates is therefore unlikely. This opens a new possibility in the design of an antifungal agent that selectively inhibits chitin synthesis.
Acknowledgments
We thank M.H. Valdivieso, B. Santos, and R. Schekman for plasmids and strains, A. Pandiella and members of the A. Durán laboratory for critical comments on the manuscript, and N. Skinner for language revision. Special thanks are due to D. Baggot (R. Schekman Laboratory) for informing us that Chs3p is retained in the ER when overexpressed and to H. Bussey for providing the anti-Kre2p antibody.
This work has been supported by a Ministerio de Educacion y Ciencia predoctoral fellowship to J.A. Trilla and Comision Interministerial de Ciencia y Tecnologia grants BIO95-0500 and BIO98-0814 to C. Roncero.
Abbreviations used in this paper
- CS
chitin synthase
- cwr
Calcofluor white resistant
- GFP
green fluorescent protein
- HA
hemagglutinin epitope
- ORF
open reading frame
- v-ATPase
vacuolar ATPase
References
- Bähler J, Pringle JR. Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev. 1998;12:1356–1370. doi: 10.1101/gad.12.9.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers B, Levin G, Cabib E. Effect of polyoxin D on chitin synthesis and septum formation in Saccharomyces cerevisiae. . J Bacteriol. 1974;119:564–575. doi: 10.1128/jb.119.2.564-575.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa CE. CSD2, CSD3, and CSD4, genes required for chitin synthesis in Saccharomyces cerevisiae: the CSD2 gene product is related to chitin synthases and to developmentally regulated proteins in Rhizobium species and Xenopus laevis. . Mol Cell Biol. 1992;12:1764–1776. doi: 10.1128/mcb.12.4.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa CE. Genetics and molecular biology of chitin synthesis in fungi. Annu Rev Microbiol. 1993;47:505–534. doi: 10.1146/annurev.mi.47.100193.002445. [DOI] [PubMed] [Google Scholar]
- Bulawa CE, Slater M, Cabib E, Au-Young J, Sburlati A, Adair WL, Jr, Robbins PW. The S. cerevisiaestructural gene for chitin synthase is not required for chitin synthesis in vivo. Cell. 1986;46:213–225. doi: 10.1016/0092-8674(86)90738-5. [DOI] [PubMed] [Google Scholar]
- Cabib, E. 1994. Nomenclature of genes related to chitin synthesis. Yeast Newsl. XLIII:58.
- Cabib E, Sburlati A, Bowers B, Silverman SJ. Chitin synthase 1, an auxiliary enzyme for chitin synthesis in Saccharomyces cerevisiae. . J Cell Biol. 1989;108:1665–1672. doi: 10.1083/jcb.108.5.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E, Silverman SJ, Shaw JA. Chitinase and chitin synthase 1: counterbalancing activities in cell separation of Saccharomyces cerevisiae. . J Gen Microbiol. 1992;138:97–102. doi: 10.1099/00221287-138-1-97. [DOI] [PubMed] [Google Scholar]
- Cabib, E., J.A. Shaw, P.C. Mol, B. Bowers, and W.J. Choi. 1996. Chitin biosynthesis and morphogenetic processes. In The Mycota. Biochemistry and Molecular Biology, Vol. III. R. Brambl, G.A. Marzluf, editors. Springer-Verlag, Berlin. 243–267.
- Choi W, Cabib E. The use of divalent cations and pH for the determination of specific yeast chitin synthases. Anal Biochem. 1994;219:368–372. doi: 10.1006/abio.1994.1278. [DOI] [PubMed] [Google Scholar]
- Choi W, Santos B, Duran A, Cabib E. Are yeast chitin synthases regulated at the transcriptional or the posttranslational level? . Mol Cell Biol. 1994a;14:7685–7694. doi: 10.1128/mcb.14.12.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W, Sburlati A, Cabib E. Chitin synthase 3 from yeast has zymogenic properties that depend on both the CAL1 and CAL3genes. Proc Natl Acad Sci USA. 1994b;91:4727–4730. doi: 10.1073/pnas.91.11.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Chuang JS, Schekman RW. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cos T, Ford RA, Trilla JA, Duran A, Cabib E, Roncero C. Molecular analysis of Chs3p participation in chitin synthase III activity. Eur J Biochem. 1998;256:419–426. doi: 10.1046/j.1432-1327.1998.2560419.x. [DOI] [PubMed] [Google Scholar]
- DeMarini DJ, Adams AEM, Fares H, De Virgilio C, Valle G, Chuang JS, Pringle JR. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiaecell wall. J Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogen product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Abalos JM, Fox H, Pitt C, Wells B, Doonan JH. Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus, Aspergillus nidulans. . Mol Microbiol. 1998;27:121–130. doi: 10.1046/j.1365-2958.1998.00664.x. [DOI] [PubMed] [Google Scholar]
- Graham LA, Hill KJ, Stevens TH. Assembly of the yeast vacuolar H+-ATPase occurs in the endoplasmic reticulum and requires a Vma12p/Vma22p assembly complex. J Cell Biol. 1998;142:39–49. doi: 10.1083/jcb.142.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SH, Friesen H, Segall J. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. . Mol Cell Biol. 1998;18:5750–5761. doi: 10.1128/mcb.18.10.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JM, Malkus P, Schekman R. Out of the ER: outfitters, escorts and guides. Trends Cell Biol. 1999;9:5–7. doi: 10.1016/s0962-8924(98)01414-7. [DOI] [PubMed] [Google Scholar]
- Kaiser, C.A., R.E. Gimeno, and D.A. Shaywitz. 1997. Proteins secretion, membrane biogenesis, and endocytosis. In The Molecular and Cellular Biology of the Yeast Saccharomyces, Vol. III. J. Pringle, J.R. Broach, E.W. Jones, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 91–228.
- Krisak L, Strich R, Winters RS, Hall JP, Mallory MJ, Kreitzer D, Tuan RS, Winter E. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. . Genes Dev. 1994;8:2151–2161. doi: 10.1101/gad.8.18.2151. [DOI] [PubMed] [Google Scholar]
- Kuehn MJ, Schekman RW, Ljungdahl PO. Amino acid permeases require COPII components and the ER resident Shr3p for packaging into transport vesicles in vitro. J Cell Biol. 1996;135:585–595. doi: 10.1083/jcb.135.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn MJ, Herrmann JM, Schekman R. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature. 1998;391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathy of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Ljungdahl PO, Gimeno CJ, Styles CA, Fink GR. SHR3:a novel component of the secretory pathway specifically required for localization of amino acid permeases in yeast. Cell. 1992;71:463–478. doi: 10.1016/0092-8674(92)90515-e. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough J, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Lussier M, Sdicu A, Ketela T, Bussey H. Localization and targeting of the Saccharomyces cerevisiae Kre2p/Mnt1 α1,2-mannosyltransferase to a medial-Golgi compartment. J Cell Biol. 1995;131:913–927. doi: 10.1083/jcb.131.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman SK, Schekman R. Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell. 1997;88:85–96. doi: 10.1016/s0092-8674(00)81861-9. [DOI] [PubMed] [Google Scholar]
- Martin-Chalfie YT, Euskirchen G, Ward WW, Prasher CD. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Michaelis S, Herskowitz I. The a-factor pheromone of Saccharomyces cerevisiaeis essential for mating. Mol Cell Biol. 1988;8:1309–1318. doi: 10.1128/mcb.8.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MJ. Colorimetric analysis of sugars. Methods Enzymol. 1957;3:85–86. [Google Scholar]
- Pammer M, Briza P, Ellinger A, Schuster T, Stucka R, Brteintenbach M. DIT101 (CSD2, CAL1), a cell cycle-regulated yeast gene required for synthesis of chitin in cell walls and chitosan in spore walls. Yeast. 1992;9:1089–1099. doi: 10.1002/yea.320081211. [DOI] [PubMed] [Google Scholar]
- Reissig JL, Strominger JL, Leloir LF. A modified colorimetric method for the estimation of N-acetyl-aminosugars. J Biol Chem. 1955;217:959–966. [PubMed] [Google Scholar]
- Richardson HE, Wittenberg C, Cross FR, Reed SI. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- Roncero C, Valdivieso MH, Ribas JC, Duran A. Effect of Calcofluor white on chitin synthases from Saccharomyces cerevisiae. . J Bacteriol. 1988a;170:1945–1949. doi: 10.1128/jb.170.4.1945-1949.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncero C, Valdivieso MH, Ribas JC, Duran A. Isolation and characterization of Saccharomyces cerevisiaemutants resistant to Calcofluor white. J Bacteriol. 1988b;170:1950–1954. doi: 10.1128/jb.170.4.1950-1954.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Novick P, Thomas J, Bostein D, Fink G. A Saccharomyces cerevisiaegenomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., F. Wisnton, and P. Hieter. 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sadler I, Chiang A, Kurihara T, Rothblatt J, Way J, Silver P. A yeast gene important for protein assembly into the endoplasmic reticulum and the nucleus has homology to DnaJ and Escherichia coliheat shock protein. J Cell Biol. 1989;109:2665–2675. doi: 10.1083/jcb.109.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Manniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- San Segundo, P., J. Correa, C.R. Vazquez de Aldana, and F. del Rey. SSG1, a gene encoding a sporulation-specific 1,3-B-glucanase in Saccharomyces cerevisiae. . J Bacteriol. 1993;175:3823–3837. doi: 10.1128/jb.175.12.3823-3837.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Duran A, Valdivieso MH. CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae. . Mol Cell Biol. 1997;17:2485–2496. doi: 10.1128/mcb.17.5.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sburlati A, Cabib E. Chitin synthase 2, a presumptive participant in septum formation in Saccharomyces cerevisiae. . J Biol Chem. 1986;261:15147–15152. [PubMed] [Google Scholar]
- Shaw JA, Mol PC, Bowers B, Silverman SJ, Valdivieso MH, Duran A, Cabib E. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiaecell cycle. J Cell Biol. 1991;114:111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi M. Notes on sugar determination. J Biol Chem. 1952;195:19–23. [PubMed] [Google Scholar]
- Trilla JA, Cos T, Duran A, Roncero C. Characterisation of CHS4 (CAL2), a gene of Saccharomyces cerevisiae involved in chitin biosynthesis and allelic to SKT5 and CSD4. . Yeast. 1997;13:795–807. doi: 10.1002/(SICI)1097-0061(199707)13:9<795::AID-YEA139>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Uchida Y, Shimmi O, Sudoh M, Arisawa M, Yamada-Okabe H. Characterization of Chitin synthase 2 of Saccharomyces cerevisiaeII: both full size and processed enzymes are active for chitin synthesis. J Biochem. 1996;119:659–666. doi: 10.1093/oxfordjournals.jbchem.a021293. [DOI] [PubMed] [Google Scholar]
- Valdivieso MH, Mol PC, Shaw JA, Cabib E, Duran A. CAL1, a gene required for activity of chitin synthase 3 in Saccharomyces cerevisiae. . J Cell Biol. 1991;114:101–109. doi: 10.1083/jcb.114.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M, Chuang JS, Schekman RW. Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiaeendocytic pathway. Mol Biol Cell. 1996;7:1909–1919. doi: 10.1091/mbc.7.12.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M, Chuang JS, Tsung M, Hamamoto S, Schekman R. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. . Mol Biol Cell. 1998;9:1565–1576. doi: 10.1091/mbc.9.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]