Abstract
The cell nucleus is organized as discrete domains, often associated with specific events involved in chromosome organization, replication, and gene expression. We have examined the spatial and functional relationship between the sites of heat shock gene transcription and the speckles enriched in splicing factors in primary human fibroblasts by combining immunofluorescence and fluorescence in situ hybridization (FISH). The hsp90α and hsp70 genes are inducibly regulated by exposure to stress from a low basal level to a high rate of transcription; additionally the hsp90α gene contains 10 introns whereas the hsp70 gene is intronless. At 37°C, only 30% of hsp90α transcription sites are associated with speckles whereas little association is detected with the hsp70 gene, whose constitutive expression is undetectable relative to the hsp90α gene. Upon exposure of cells to heat shock, the heavy metal cadmium, or the amino acid analogue azetidine, transcription at the hsp90α and hsp70 gene loci is strongly induced, and both hsp transcription sites become associated with speckles in >90% of the cells. These results reveal a clear disconnection between the presence of intervening sequences at specific gene loci and the association with splicing factor–rich regions and suggest that subnuclear structures containing splicing factors are associated with sites of transcription.
Keywords: heat shock genes, nuclear organization, splicing, transcription
Understanding the spatial relationship between nuclear architecture and genomic function represents a major objective of cell biology. Discrete subnuclear sites for transcription and splicing have been described; however, the structure/function relationships of these organized structures remain unresolved. For example, several studies using labeled precursors for RNA synthesis have shown that the sites of RNA polymerase II (Pol II)1 transcription are widely distributed in the nucleus (for reviews see Fakan, 1994; Moen et al., 1995; Clemson and Lawrence, 1996; Huang and Spector, 1996a; Misteli and Spector, 1998). Similarly, active Pol II has been found randomly distributed in the nucleus (Jiménez-Garcia and Spector, 1993; Zeng et al., 1997), thus suggesting that transcription occurs at multiple sites throughout the nucleus. However, other studies using different reagents have shown that a subpopulation of the active Pol II is concentrated in specific nuclear regions (Thibodeau and Vincent, 1991; Bregman et al., 1995; Blencowe et al., 1996; Mortillaro et al., 1996; Patturajan et al., 1998). A recent study by Wei et al. (1998) has shown that individual Pol II transcription sites, detected by BrUTP incorporation, are spatially segregated in the nucleus into ∼16 higher order domains, providing support for the compartmentalization of RNA Pol II transcription within a discrete number of nuclear factories. Consistent with the organization of the transcription sites into domains, total poly(A) RNAs detected by fluorescence in situ hybridization (FISH) appear to concentrate in 20–40 nuclear foci (Carter et al., 1991, 1993).
The small nuclear ribonucleoproteins (snRNPs) and various non-snRNP splicing factors such as SC35, U2AF, U2B″, or SF2/ASF detected by immunofluorescence distribute in the nucleus in 20–40 foci of varying size termed “speckles,” in addition to a diffuse nucleoplasmic staining of lesser intensity (for reviews see Moen et al., 1995; Clemson and Lawrence, 1996; Huang and Spector, 1996a; Lamond and Earnshaw, 1998; Misteli and Spector, 1998). Three classes of nuclear structures enriched in splicing factors have been described by electron microscopy (for reviews see Fakan, 1994; Puvion and Puvion-Dutilleul, 1996): (a) the interchromatin granules (IGs), which correspond to the speckles described by immunofluorescence and represent compact structures containing high concentrations of both snRNPs and non-snRNPs splicing factors; (b) the perichromatin fibrils (PFs), which are less concentrated in splicing factors and may correspond to the diffuse nucleoplasmic staining; and (c) the coiled bodies (CBs), which are compact structures highly enriched in snRNPs but devoid of the essential splicing factor SC35 (for review see Matera, 1998).
To what extent do nuclear domains enriched in RNA Pol II or splicing factors reflect gene expression, sites of storage, and assembly of macromolecular structures, or recycling centers? In contrast to available methodologies which have demonstrated the presence of organizing sites of transcription, it has been difficult to establish the relationship between structure and function for RNA splicing, specifically to discriminate between actively engaged and inert splicing complexes. Based on in vitro and in vivo evidence that intron excision is spatially and temporally associated with transcription (Huang and Spector, 1991; Raap et al., 1991; Xing et al., 1993; Wuarin and Schibler, 1994; Zhang et al., 1994), an indirect measure of the role of these different nuclear structures in transcription and splicing has thus been to analyze the relative distribution of splicing factors and active sites of transcription. Electron microscopic studies have shown that sites of [3H]uridine incorporation occur preferentially in the vicinity of PFs (for review see Fakan, 1994; Puvion and Puvion-Dutilleul, 1996). Similarly, sites of transcription detected by BrUTP incorporation reveal little colocalization with the nuclear speckles (Wansink et al., 1993; Pombo and Cook, 1996; Fay et al., 1997), and active RNA Pol II is distributed randomly relative to the speckles enriched in splicing factors (Zeng et al., 1997). Finally, several viral and endogenous transcription sites detected by FISH display little or no association relative to the speckles (Zhang et al., 1994; Lampel et al., 1997; Smith et al., 1999). Altogether, these observations suggest that transcription by RNA Pol II and splicing would occur in domains scattered throughout the nucleus but distinct from SC35/snRNP-rich speckles.
In contrast, others have shown that the 20–40 foci enriched in poly(A) RNA colocalize with speckles (Carter et al., 1991, 1993; Visa et al., 1993). In addition, several specific transcription sites detected by FISH have been shown to be spatially associated with these regions (Huang and Spector, 1991, 1996a,b; Wang et al., 1991; Lawrence et al., 1993; Xing et al., 1993), and a study of five endogenous genes has shown that inactive genes are randomly distributed relative to the speckles enriched in splicing factors, whereas active genes were closely associated with these regions (Xing et al., 1995). Similarly, the cytomegalovirus immediate early gene clusters (Dirks et al., 1997) as well as the stably transfected homeobox gene pem (Misteli et al., 1998) are found associated with nuclear speckles only when they are actively transcribed, thus providing support for a role of the speckles in transcription and splicing.
Three models for the role of speckles in transcription and splicing have been proposed (Clemson and Lawrence, 1996): (a) speckles represent sites of storage and/or assembly-disassembly of splicing factors; (b) speckles are often associated with pre-mRNA metabolism of specific genes; and (c) speckles represent distinct functional entities with some representing storage sites and corresponding to the IGs, while others associated with active genes would represent sites of transcription and splicing and more likely correspond to large accumulations of PFs. In agreement with the latter model, Huang and Spector (1996a) have proposed that upon activation of RNA Pol II transcription, splicing factors would be recruited from their sites of storage and/or reassembly, the IGs, to the sites of transcription where nascent transcripts would be spliced. When the transcription rate is very high, a substantial amount of splicing factors would be recruited to the sites of transcription, resulting in a granular IG-like appearance which would be functionally distinct from IGs. This assumption can explain the colocalization observed between several specific transcripts and the speckles. Consistent with this proposal, the splicing factor SF2/ASF fused to the green fluorescent protein is recruited to new sites of viral transcription upon transcriptional activation in living cells, thus demonstrating that one function of the speckles is to supply splicing factors to neighboring active genes (Misteli et al., 1997). The recruitment of splicing factors to the sites of transcription can be intron-dependent as demonstrated by Huang and Spector (1996b) using a set of intron-containing and intronless constructs. However, a caveat of these studies is that the genes which were studied were either integrated viral genomes or transfected constructs.
To address key issues concerning the involvement of the nuclear speckles in transcription and splicing activities, we used a combination of FISH and immunofluorescence to examine whether the speckles are associated with sites of transcription of a class of coregulated cellular heat shock genes including members which are intron-containing (hsp90α) (Hickey et al., 1986) and intronless (hsp70) (Wu et al., 1985), and whose expression can be inducibly regulated (for reviews see Morimoto, 1993; Morimoto et al., 1996). This system allows the examination of both the dynamics of transcription of well studied cellular genes and the role of introns in association with the speckles. Our results reveal that both hsp90α and hsp70 transcription sites associate with the speckles upon stress-induced transcriptional activation, independent of the presence of introns.
Materials and Methods
Cell Culture and Stress Induction
Human normal primary fibroblasts were obtained from a skin biopsy performed on a healthy female donor. They were grown in RPMI medium supplemented by 10% fetal calf serum and 100 μg/ml ampicillin. For in situ analysis, cells were grown directly on two-chamber glass slides (Labtek). Heat treatment was performed by immersing the slides or the flasks in a waterbath set up at 42 or 45°C. Cadmium was used at a final concentration of 75 μM for 4 h and azetidine was used at a final concentration of 10 mM for 2 h.
Probes and Antibodies
The pH 2.3 genomic probe covering the entire coding sequence (2.3 kb) of the human hsp70 gene was used to detect hsp nuclear transcripts (Wu et al., 1985). The hsp70 gene was detected using the cosmid clone 12HI which contains a portion of the coding sequence of hsp70 (kindly provided by Dr. R.D. Campbell, University of Cambridge, Cambridge, UK). cDNA probes specific for hsp90α (pHS 801) and hsp90β (pHS 811) genes were obtained from Dr. E. Hickey (University of Nevada, Reno, NV). pHS 801 and 811 probes contain, respectively, 1.3 and 0.9 kb of the coding region (Hickey et al., 1986). All probes were labeled by random priming with biotin-14-dATP (GIBCO BRL).
The mouse monoclonal antibody specific for the non-snRNP splicing factor SC35 (Sigma) was used at a dilution of 1:250 for immunofluorescence (Fu and Maniatis, 1990). The mouse monoclonal Y12 directed against the Sm protein of snRNPs was obtained from Dr. J.A. Steitz (Yale University, New Haven, CT) and used at 1:250 (Lerner et al., 1981). The mouse monoclonal antibody against the U2B″ splicing component (Cappel) was used at 1:50 (Mattaj et al., 1986). The mouse monoclonal POL3/3 antibody against RNA Pol II was obtained from Dr. E.K. Bautz (University of Heidelberg, Heidelberg, Germany) and used at 1:200 (Krämer et al., 1980). The mouse monoclonal CC-3 antibody against RNA Pol II was obtained from Dr. M. Vincent (University of Laval, Quebec, Canada) and used at 1:500 (Thibodeau and Vincent, 1991). The mouse monoclonal MARA3 antibody against RNA Pol II was obtained from Dr. B.M. Sefton (Salk Institute, La Jolla, CA) and used at 1:200 (Patturajan et al., 1998).
Combined Immunofluorescence and FISH
Probe Preparation.
100 ng of the cDNA probe or 100 ng of the cosmid probe was precipitated with 30 μg of salmon sperm DNA. 3 μg of human Cot I competitor DNA (GIBCO) was also added to the cosmid probes. The pellets were resuspended in 50% formamide/10% dextran sulfate/2× SSC, and denatured for 5 min at 75°C. Cosmid probes were incubated 1 h at 37°C to allow suppression of repeated sequences (Lichter et al., 1988).
Sample Preparation for Detection of RNA and Proteins.
Immunofluorescence combined to FISH was performed as described previously (Jolly et al., 1997a). FISH was performed first to enhance the efficiency of hybridization. Briefly, cells were fixed in 4% formaldehyde/PBS. A first permeabilization step was performed by three successive incubations of 5 min each in 0.5% saponin/0.5% Triton X-100/PBS. After an equilibration step in 20% glycerol/PBS for 20 min, cells were freeze-thawed three times successively by briefly dipping in liquid nitrogen as a second permeabilization step. Cells were subsequently dehydrated through sequential incubations in 70%, 90%, and 100% ethanol baths for 5 min each. The denatured probe was applied to the dry slide and hybridization was allowed to run overnight. After hybridization, probes were detected using avidin-FITC (Sigma). After postdetection washes in 4× SSC/0.1% Tween 20, a 45-min blocking step in 10% FCS/0.3% Triton X-100/PBS was performed, followed by incubation for 90 min at 37°C with the anti-SC35 or the Y12 antibody. Antibody staining was revealed using an anti–mouse-TRITC antibody (Sigma) and nuclei were counterstained with 250 ng/ml DAPI (4′, 6-diamidino-2-phenylindole/2 HCl) (Sigma) diluted in an antifading solution consisting in 90% glycerol, 20 mM Tris-HCl, pH 8.0, 2.33% DABCO (1–4 diazabicyclo [2,2,2] octane) (Sigma).
Sample Preparation for Detection of DNA and Proteins.
Immunofluorescence was performed first in this case. After detection with the secondary antibody, cells were rinsed three times with PBS, and subsequently denatured by a 3-min incubation in 70% formamide/2× SSC at 75°C, followed by a 1-min incubation in 50% formamide/2× SSC at the same temperature. The denatured cosmid probe was then applied to the slide. After hybridization, cells were washed three times for 5 min in 50% formamide/2× SSC at 45°C, and three times for 5 min in 0.5× SSC at 60°C. Detection was then performed as described above for nuclear transcript detection.
In Vivo Incorporation of BrUTP
BrUTP incorporation was performed according to a protocol derived from Wansink et al. (1993). Briefly, cells were rinsed once with PBS for 3 min, and once with a glycerol buffer (20 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 25% glycerol, 0.5 mM PMSF, 0.5 mM EGTA) for 3 min. Cells were then permeabilized by incubation for 3 min in the same buffer complemented with 0.05% Triton X-100 and 10 U/ml RNAsin, and subsequently incubated with the transcription cocktail (100 mM KCl, 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 0.5 mM EGTA, 25% glycerol, 1 mM PMSF, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 0.2 mM BrUTP, 160 μM S-adenosyl-methionine, 25 U/ml RNAsin) for 15 min at room temperature. At the end of the reaction, cells were rinsed once in PBS containing 0.05% Triton X-100 and 5 U/ml RNAsin, once in PBS added with 0.5 mM PMSF and 5 U/ml RNAsin, and subsequently fixed in 4% formaldehyde in PBS. Incorporation sites were revealed using a mouse anti-BrdU antibody (Sigma) and anti–mouse FITC (Sigma).
Fluorescence Microscopy and Image Analysis
Images were acquired using a confocal laser scanning microscope (Zeiss LSM 410) using a 63×, 1.25 NA oil immersion objective. Confocal images were analyzed for the relative distribution of the speckles and hsp transcription sites using software developed at the University of Grenoble (Monier et al., 1996). Deconvolution was used to revert the distortion of fluorescent signals due to the point spread function of the microscope which allowed our ability to define the limits of the speckles. Transcription sites were defined as associated with a speckle when no pixels were separating the two fluorescent signals. Percentages were determined based on the analysis of 100 nuclei which corresponds to 200 sites of gene transcription.
RT-PCR Reaction
The RT-PCR reaction was performed as described in Wang et al. (1999). The reaction was internally controlled by including known amounts of internal control transcripts corresponding to the same genes carrying small deletions to distinguish from wild-type, thus allowing us to precisely quantify the levels of transcripts (see Wang et al., 1999 for preparation of internal control transcripts).
RNA Extraction.
Total RNAs were extracted using the procedure described by Gough (1988). Briefly, cells at 80% confluency were scraped and spun down. The cell pellet was resuspended in 200 μl of buffer A (10 mM Hepes, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, and 2.5 mM DTT), spun again, and the supernatant was added to 200 μl of ice-cold buffer B (7 M urea, 0.35 M NaCl, 10 mM EDTA, 10 mM Tris-HCl, pH 7.5, 1% SDS). 400 μl of phenol/chloroform (1:1) was then added, and RNAs were precipitated as described.
Primers.
Specific sense and antisense primers for hsp70, hsp90α, and hsp90β transcripts (GIBCO BRL) were designed using the MacVector program as follows (the numbers indicate positions on the wild-type transcripts): hsp70 sense: 5′-TTCCGTTTCCAGCCCCCAATC-3′ (nucleotides [nt] 435–455); hsp70 antisense: 5′-CGTTGAGCCCCGCGATCACA-3′ (nt 993–974); hsp90α sense: 5′-AAAAGTTGAAAAGGTGGTTG-3′ (nt 1803–1822); hsp90α antisense: 5′-TATCCACAGCATCACTTAGTA-3′ (nt 2426–2405); hsp90β sense: 5′-AGAAGGTTGAGAAAGGTGACAA-3′ (nt 1803–1822); hsp90β antisense: 5′-AAGAAGTTAGAGAGGGAATAAA-3′ (nt 2444–2425). The expected sizes of PCR products are 641, 625, and 558 bp for wild-type hsp90β, hsp90α, and hsp70 transcripts, respectively, and 531, 499, and 438 bp for in vitro transcribed hsp90β, hsp90α, and hsp70 transcripts, respectively.
Reverse Transcription.
The reaction was performed in a total volume of 20 μl. 2 μg of total RNAs was incubated for 1 h at 37°C with 3.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl, pH 8.3, 10 mM DTT, 0.5 mM dNTP, 20 pmol of each antisense primer, 30 pg of each internal control transcript, 400 U RNAsin, and 400 U of Moloney murine leukemia virus reverse transcriptase (Pharmacia).
Polymerase Chain Reaction.
PCR reactions were performed in a final volume of 50 μl. To the 20 μl of the reverse transcription reaction were added (to final concentrations): 3.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl, pH 8.3, 20 pmol of each sense primer, 0.5 mM dNTP, 1 μCi [α-32P]dATP, 0.01 μg/μl DNase-free RNAse A, and 50 U Taq polymerase (Pharmacia). The reactions were performed in a Pelletier effect thermal cycler (MJ Research, PTC100) for 35 cycles (each cycle: 1 min at 92°C, 1 min at 56°C, 1 min at 72°C) with an initial denaturation of 1 min at 94°C and a final extension at 72°C for 10 min.
Gel.
PCR products were analyzed on a 4% acrylamide, 42% (wt/vol) urea denaturing gel.
Quantification.
The levels of wild-type hsp70, hsp90α, and hsp90β transcripts were quantified using the PhosphorImager analyzer system (Molecular Dynamics) and normalized by the amount of the corresponding internal control transcripts.
Transcriptional Run-on Assay
Run-on transcription reactions were performed with isolated cell nuclei in the presence of 50 μCi of [α-32P]UTP (Amersham) as described previously (Banerji et al., 1984). After precipitation, radioactive RNA was hybridized to DNA probes for the human hsp70 gene (pH 2.3), human hsp90α (pHS 801), pBR322 as a control for nonspecific hybridization, and the rat gapdh gene (Fort et al., 1985) as a normalization control for transcription. The intensities of radioactive signals were quantitated using the PhosphorImager analyzer system (Molecular Dynamics).
Results
Relative Distribution of Splicing Factors and hsp Genes in Unstressed Cells
We investigated the relative distribution of SC35 splicing factor and sites of hsp70 or hsp90α genes in normal human fibroblasts. Our rationale for selection of the hsp90α and hsp70 genes was based on three criteria: (a) both genes are transcribed at a low basal rate in cells at normal growth temperatures; (b) the transcription rates of both genes are induced to high levels upon exposure to heat shock and other stresses; and (c) the hsp90α gene contains 10 introns whereas the hsp70 gene is intronless. The relative distribution of hsp70 or hsp90α transcription sites and SC35 splicing factor was analyzed by using a procedure combining immunofluorescence for the detection of splicing factors and FISH for the detection of hsp nuclear transcripts (Jolly et al., 1997a).
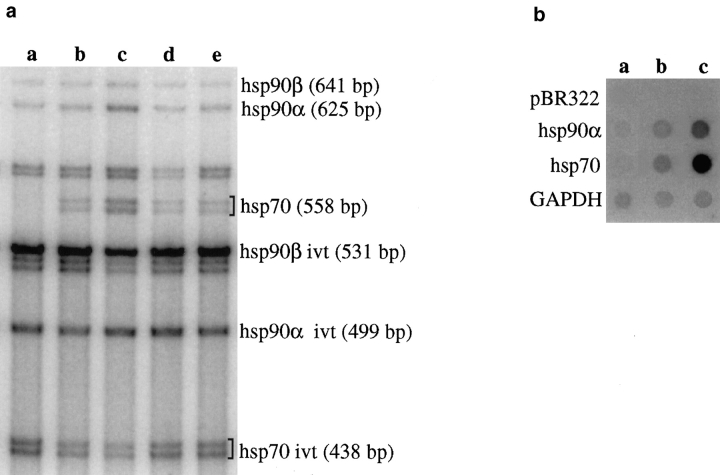
We have demonstrated previously that hsp70 and hsp90α gene expression is induced by heat shock and other stresses (Watowich and Morimoto, 1988; Abravaya et al., 1991; Shi et al., 1998; for review see Morimoto et al., 1996). At 37°C, hsp90α transcripts are constitutively detected whereas hsp70 mRNAs were undetectable (Fig. 1 a, lane a). This corresponded, by transcriptional run-on analysis, to a very low basal rate of hsp90α gene transcription at 37°C while hsp70 gene transcription was repressed (Fig. 1 b, lane a). Within the nucleus of diploid fibroblasts, hsp transcripts detected by FISH appear as two foci (Fig. 2). Because hsp90α transcription rate is low, the foci detected by FISH may correspond partially to nascent transcripts which are retained at the site of transcription, as has been shown for hsp70 transcripts (Jolly et al., 1998). Codetection of the transcripts and the corresponding gene by FISH showed a complete overlap of the two hybridization signals at the level of light microscopy (data not shown). For the hsp70 gene whose constitutive expression is too low (Fig. 1), we chose to detect the gene itself by FISH.
Figure 1.
(a) RT-PCR analysis of total mRNA extracts from normal human fibroblasts submitted to different conditions: 37°C (lane a), 42°C for 1 h (lane b), 45°C for 1 h (lane c), azetidine 10 mM for 2 h (lane d), and cadmium 75 μM for 4 h (lane e). The relative levels of hsp70, hsp90α, and hsp90β transcripts were obtained by normalizing the amount of each wild-type RNA by the amount of the corresponding internal control transcript. At 37°C, hsp90α transcripts are detectable, whereas hsp70 mRNAs are undetectable. Under heat shock at 42°C or 45°C, azetidine or cadmium treatment, an increase in the total amount of both transcripts is observed. PCR products of the expected size were obtained for each transcript, thus attesting that splicing occurs efficiently in all stress conditions. The bands appearing between hsp70 and hsp90α bands correspond to non–stress-responsive members of the hsp gene family. (b) Run-on analysis of the transcription rate of hsp70 and hsp90α genes in cells submitted to different conditions: 37°C (lane a), 42°C for 1 h (lane b), and 45°C for 1 h (lane c). The transcription rate of both genes is low at 37°C, but shows a marked increase after heat shock at 42°C or 45°C.
Figure 2.
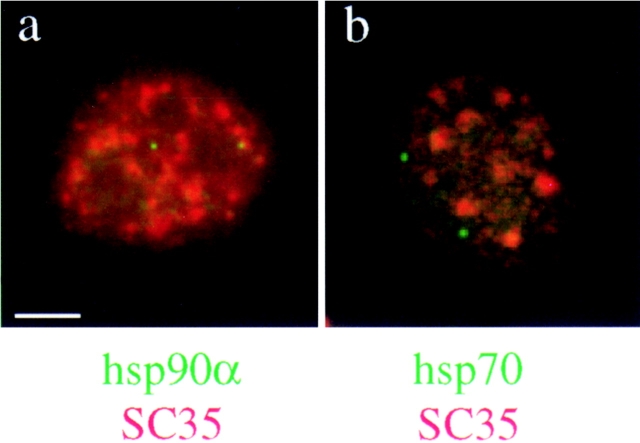
Codetection of SC35 splicing factor (red) and hsp90α transcripts (a) or hsp70 gene (b) (green) by combined immunofluorescence and FISH in human normal fibroblasts at 37°C. hsp90α transcription sites are found associated with SC35 speckles in 30% of the cells (a). In this example a nucleus is shown in which only one of the two hsp90α transcription sites associates with a speckle. In contrast to hsp90α, the hsp70 gene is found associated with the speckles in only 10% of the cells (b). Bar, 5 μm.
In control cells, SC35 speckles were associated with 30% of the hsp90α transcription sites, averaging over 200 transcription sites (Fig. 2 a and Table I). In contrast, only 10% of the signals corresponding to the hsp70 gene were associated with SC35 speckles (Fig. 2 b). No differences in the size or intensity of associated versus nonassociated transcription sites were observed. Nuclear speckles were found by quantitative digital imaging analyses to occupy 5–17% of the nuclear volume (Huang and Spector, 1991; Carter et al., 1993) whereas hsp transcripts occupy <1% of the nuclear volume. The low percentage of association between SC35 speckles and hsp70 transcription sites consequently reflects random distribution, whereas the 30% association with hsp90α transcription sites is significant and likely reflects the higher basal transcription rate of the hsp90α gene.
Table I.
Summary of the Percentages of Association Observed in Each Condition between Single hsp Transcription Sites and Speckles (Averaging on 200 Hybridization Signals)
| Conditions | hsp70 gene | hsp90α gene | ||
|---|---|---|---|---|
| 37°C | 10% | 30% | ||
| 42°C, 1 h | 92% | 92% | ||
| 45°C, 1 h | 93% | 94% | ||
| Azetidine 10 mM, 2 h | 90% | 94% | ||
| Cadmium 75 μM, 4 h | 93% | 95% |
Relative Distribution of SC35 Speckles and hsp Genes in Heat-shocked Cells
To address whether the distribution of SC35 speckles and hsp genes is a reflection of introns or of the transcription rate, we exposed the cells to a heat shock at 42°C or 45°C, conditions which result in a dramatic elevation of heat shock gene transcription (Watowich and Morimoto, 1988; Abravaya et al., 1991; Shi et al., 1998; for review see Morimoto et al., 1996). The analysis of hsp90α and hsp70 gene transcription rates, by nuclear run-on analysis, revealed that both genes were induced strongly following a 42°C or 45°C heat shock (5- and 12-fold induction for the hsp90α gene, and 14- and 35-fold induction for the hsp70 gene at 42°C and 45°C, respectively) (Fig. 1 b). Quantification of the mRNA levels by RT-PCR revealed a 11.4-fold (42°C) and 29.7-fold (45°C) induction of hsp70 mRNA levels (Fig. 1 a, lanes b and c) and a 1.5-fold (42°C) and 2.5-fold (45°C) (Fig. 1 a, lanes b and c) induction of hsp90α mRNA. As expected, the fold-induction of hsp90α transcripts determined by measuring mRNA levels was lower than for hsp70 due to the higher basal levels of hsp90α transcripts in control cells.
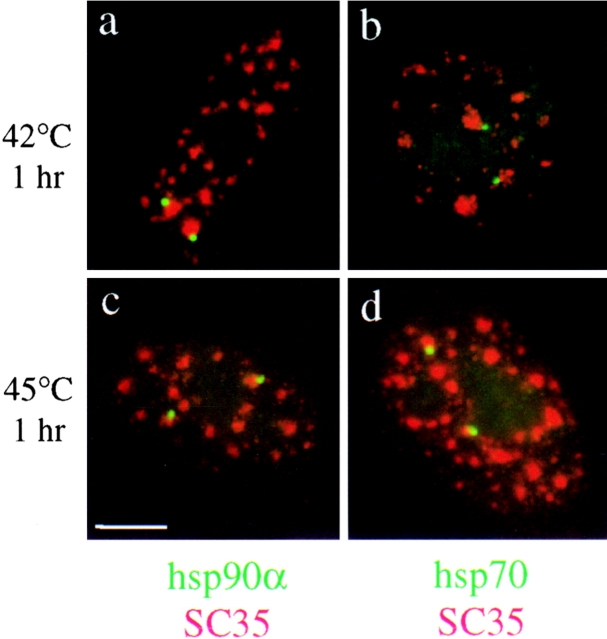
hsp70 or hsp90α transcripts were detected together with the SC35 splicing factors in heat-shocked cells. As shown in Fig. 3, 92% of hsp90α transcription sites were observed to be adjacent to SC35 speckles in the 42°C treated cells (Fig. 3 a) and 94% in the 45°C treated cells (Fig. 3 c). Likewise, 92% and 93% of the chromosomal sites of hsp70 transcription were associated with a SC35 speckle in cells exposed to 42°C (Fig. 3 b) and to 45°C (Fig. 3 d), respectively. Identical results were obtained in cells exposed at 42°C or 45°C for only 10 min (data not shown), attesting that the association of splicing factors with transcribing genes is a very rapid process, directly correlated to the transcriptional activity of the gene and not due to major rearrangements of the nuclear architecture as a consequence of heat shock.
Figure 3.
Codetection of SC35 (red) and hsp90α or hsp70 transcripts (green) in human fibroblasts submitted to a 1-h heat shock at 42°C (a and b) or 45°C (c and d). hsp90α transcripts are associated with SC35 speckles in 92% of the cells at 42°C (a) and 94% of the cells at 45°C (c). hsp70 transcripts are associated with speckles in 92% of the cells at 42°C (b) and in 93% of the cells at 45°C (d). Bar, 5 μm.
The high degree of spatial coincidence between SC35 speckles and sites of hsp gene transcription following activation of the heat shock response reveals that a key feature of recruitment of SC35 splicing factors relates to the dynamics of transcription. A similar spatial association of the heat-activated hsp90β gene with the speckles has been reported previously (Lampel et al., 1997). Our results demonstrate, however, that the splicing factors do not distinguish between intron-containing and intronless genes. Activated hsp genes were not found to be preferentially associated with larger speckles as has been observed for fibronectin transcripts (Xing et al., 1993, 1995), perhaps reflecting a gene specificity in the pattern of association with the speckles.
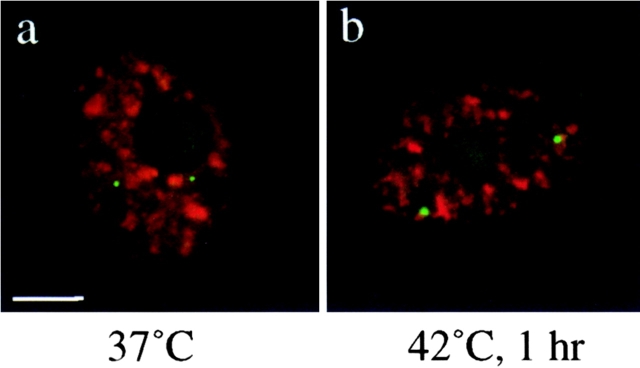
To what extent do our observations reflect features of SC35 which are not general to splicing complexes? To ensure that our results were not limited to the SC35 splicing factor or due to the fact that the anti-SC35 antibody only recognizes a phosphoepitope of the protein, we performed the same experiments on control and heat-shocked cells with the Y12 antibody to detect snRNPs (Lerner et al., 1981) (Fig. 4) or with an anti-U2B′′antibody (Mattaj et al., 1986) (data not shown). The results obtained for both antibodies revealed the association of both hsp70 (Fig. 4) and hsp90α transcription sites (data not shown) with the speckles only in cells exposed to 42°C.
Figure 4.
Codetection of hsp70 transcription sites (green) and snRNPs (red) in human fibroblasts. At 37°C, hsp70 genes are associated with snRNPs speckles in 32% of cells (a). After a 1-h heat shock at 42°C, hsp70 transcription sites are found associated with snRNPs speckles in 93% of the cells (b).
Other Stresses Also Induce a Tight Association of Active hsp Genes with SC35 Speckles
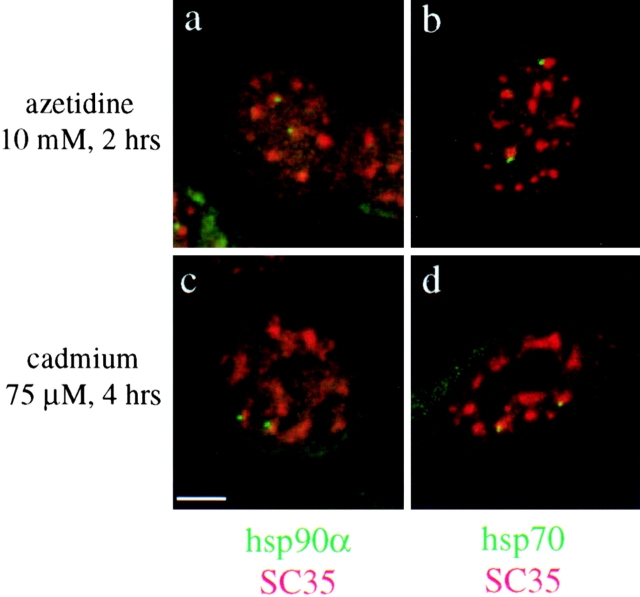
To exclude that the redistribution of splicing factors following heat shock was due solely to the effects of elevated temperatures on nuclear organization, heat shock gene transcription was activated by exposure to azetidine or cadmium (Mosser et al., 1988; Williams and Morimoto, 1990). Both treatments resulted in an increase in hsp70 and hsp90α mRNA levels comparable to those induced by a 42°C heat shock (i.e., a 10.9- and 12.1-fold increase in hsp70 mRNA levels, and a 1.4- and 1.5-fold increase in hsp90α mRNA levels in azetidine- and cadmium-treated cells, respectively) (Fig. 1 a, lanes d and e). This was corroborated by transcriptional run-on assay showing that both genes were actively induced in cadmium- and azetidine-treated cells (data not shown). As shown in Fig. 5, 94% and 95% of the hsp90α transcripts were associated with SC35 speckles in azetidine (Fig. 5 a) and cadmium-treated cells (Fig. 5 c), respectively. Similarly, 90% of the hsp70 transcription sites were associated with SC35 speckles in azetidine-treated cells (Fig. 5 b) and 93% in cadmium-treated cells (Fig. 5 d). These observations confirm and extend the results of the heat-induced association of hsp70 and hsp90α transcription sites with the speckles, and demonstrate that the dynamic relocalization of splicing factors is not caused by the thermal effects of heat shock but is primarily a reflection of the elevated rates of transcription of both gene loci.
Figure 5.
Codetection of SC35 (red) and hsp90α or hsp70 transcripts (green) in human fibroblasts treated with 10 mM azetidine for 2 h (a and b) or with cadmium 75 μM for 4 h (c and d). hsp90α transcription sites are associated with SC35 speckles in 94% of the azetidine-treated cells (a) and in 95% of the cadmium-treated cells (c), whereas hsp70 transcription sites are associated with the speckles in 90% of the azetidine-treated cells (b) and in 93% of the cadmium-treated cells (d). Bar, 5 μm.
Effect of Stress on the Transcription and Splicing Activities of the Cells
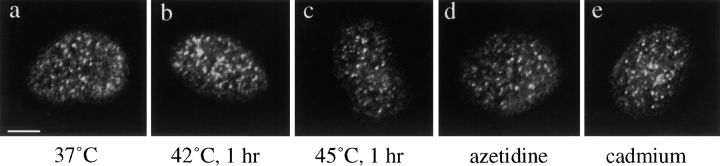
Human primary fibroblasts are relatively resistant to heat shock and display a very low percentage of cell death following a 1-h heat shock at 45°C (Jolly et al., 1997b). In addition, general features of nuclear morphology visualized by light microscopy do not appear to be altered by heat exposure (Jolly et al., 1997a). To address whether the different inducers of heat shock gene expression employed here caused a transcriptional arrest, we monitored general transcriptional activity by visualizing the sites of BrUTP incorporation into nascent transcripts (Wansink et al., 1993). As shown in Fig. 6, there was no detectable change in the transcriptional pattern in cells treated with either heat shock, cadmium, or azetidine when compared to control cells.
Figure 6.
Transcription sites detected by BrUTP incorporation in human fibroblasts submitted to different conditions: 37°C (a), 42°C for 1 h (b), 45°C for 1 h (c), azetidine 10 mM for 2 h (d), and cadmium 75 μM for 4 h (e). The patterns of transcription are similar in unstressed (a) and in stressed cells (b–e), attesting that the cells are not dramatically affected by stress. Bar, 5 μm.
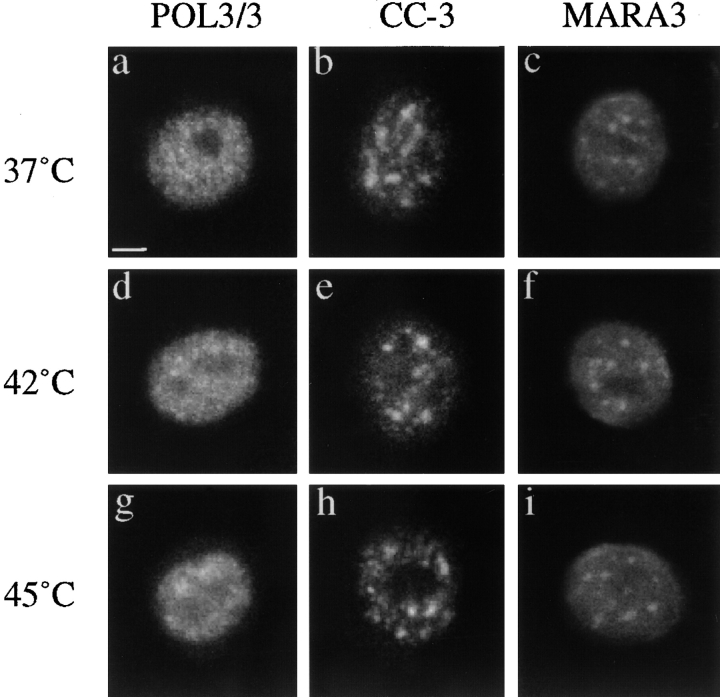
We next investigated the distribution of RNA Pol II for its presence within the speckles and to determine whether the various stress conditions influenced its distribution. Since substantial variations in Pol II distribution depending on the cell type and the specific antibody used have been reported (Thibodeau and Vincent, 1991; Jiménez-Garcia and Spector, 1993; Bregman et al., 1995; Blencowe et al., 1996; Mortillaro et al., 1996; Zeng et al., 1997; Patturajan et al., 1998), we used three characterized antibodies recognizing different epitopes on the RNA Pol II. As previously reported for other cell types, the POL3/3 antibody, which recognizes an epitope outside of the COOH-terminal domain (CTD) of Pol II and is independent of the phosphorylation state of the enzyme, shows a diffuse nucleoplasmic staining at 37°C (Fig. 7 a) (Krämer et al., 1980; Kontermann et al., 1995). In contrast, the CC-3 antibody, which recognizes a phosphoepitope in the CTD, stains a subpopulation of Pol II concentrated in speckles (Fig. 7 b) (Thibodeau and Vincent, 1991). The MARA3 antibody, which recognizes a phosphoepitope in the CTD different from CC-3, stains both a diffuse population and a subpopulation of the RNA Pol II concentrated in the speckles (Fig. 7 c) (Patturajan et al., 1998). As previously shown by others, these different patterns correspond to different subpopulations of RNA Pol II. In our human primary fibroblasts, at least two distinct hyperphosphorylated forms of RNA Pol II appear to concentrate in the speckles; however, whether these subpopulations represent active forms of the enzyme is still unknown. None of these patterns were altered by a 42°C (Fig. 7, d–f) or 45°C heat shock (Fig. 7, g–i), or by cadmium and azetidine treatments (data not shown). These observations showed that in human fibroblasts at least a subpopulation of RNA Pol II was localized to the speckles and that the overall distribution of the enzyme was not affected dramatically by stress.
Figure 7.
Immunodetection of RNA Pol II using three antibodies in normal fibroblasts submitted to different conditions: 37°C (a–c), 42°C for 1 h (d–f), and 45°C for 1 h (g–i). At 37°C, POL3/3 antibody recognizes a diffuse population of RNA Pol II (a) while CC-3 recognizes primarily a subpopulation of the enzyme present in the speckles (b). MARA3 antibody recognizes both a diffuse population and a subpopulation concentrated in speckles (c). The distribution of RNA Pol II revealed by the three antibodies is not affected by heat shock at 42°C (d–f) or 45°C (g–i).
As RNA splicing is affected by heat shock in many organisms (for review see Jolly and Morimoto, 1999), we chose to examine whether the differential distribution of hsp genes relative to SC35 speckles during stress was a consequence of a potential stress-induced arrest in RNA splicing. In human cells, heat shock results in a redistribution of snRNPs from the speckles to a diffuse nucleoplasmic pattern (Spector et al., 1991). It has also been shown that extracts from HeLa cells heat-shocked at 43°C or higher temperatures are unable to form a functional spliceosome; however, the putative factor(s) inactivated by heat remains unidentified (Shukla et al., 1990). To examine whether the association of hsp transcription sites with speckles was due to a heat-induced retention of unprocessed hsp transcripts at the sites of transcription, we analyzed the transcripts of the two intron-containing hsp90α and hsp90β genes from cells exposed to heat shock at 42°C or 45°C, cadmium, or azetidine by RT-PCR using primers surrounding an intron. As shown in Fig. 1 a, the hsp90α and hsp90β transcripts detected under all conditions corresponded only to the expected processed species with no detection of the predicted precursor species.
Discussion
This study is the first to investigate, in mammalian cells, the relative distribution of splicing factors and endogenous intronless or intron-containing genes with relation to their inducible transcriptional activity. At 37°C the inactive hsp70 gene was associated randomly relative to the distribution of nuclear SC35 speckles, whereas the hsp90α gene was weakly associated with splicing speckles, consistent with a low but detectable basal transcription. When the cells were exposed to various stressors which resulted in the inducible transcription of the hsp genes, both hsp70 and hsp90α genes became associated with the speckles. The association of splicing factors with the new sites of transcription is a rapid process, occuring immediately upon gene activation. In addition, at least two subpopulations of hyperphosphorylated RNA Pol II were found to concentrate in the speckles, and this distribution was unaffected by stress. Altogether our data demonstrate that the association of specific genes with splicing factors is a reflection principally of the transcription rate of the endogenous cellular gene and does not depend upon the presence of introns in the primary transcript. These observations complement the recent findings by Smith et al. (1999) that some intron-containing pre-mRNAs are poorly associated with increased concentrations of SC35, both demonstrating a disconnection between the presence of introns and the spatial association with splicing factors.
Is the Association with Splicing Speckles a Measure of Active Gene Transcription?
An important conclusion of our results is that the association of splicing factors with the sites of transcription of endogenous genes is best indicated by the level of transcriptional activity. Under control conditions where heat shock genes are either repressed or transcribed at low basal levels, they are randomly distributed in regards to splicing factors or weakly associated with them, whereas upon gene activation, splicing factors accumulate rapidly at the sites of abundant nascent transcripts. The significance of the 30% association of hsp90α transcription sites with the speckles at 37°C is uncertain. Since we did not observe a significant difference in the fluorescent intensity of associated versus nonassociated transcription sites, this may reflect variation in the relative rate of transcription at the chromosomal loci. Alternatively, this could reflect variation in the local concentration of splicing factors and the limitations of light microscopy. Finally, high concentrations of splicing factors may not be required at the sites of transcription, which may at least in part correspond to sites of accumulation of full-length mature nascent transcripts rather than growing RNA molecules (Jolly et al., 1998).
Our results are in good agreement with several previous studies, showing a redistribution of factors involved in transcription, pre-mRNA processing, and RNA packaging in response to changes in gene activity. Indeed, splicing factors have been shown to relocalize in response to RNA Pol II inhibition (Carmo-Fonseca et al., 1992; Zeng et al., 1997), to viral infection (Martin et al., 1987; Jiménez-Garcia and Spector, 1993; Spector et al., 1993; Pombo et al., 1994; Puvion-Dutilleul et al., 1994; Bridge et al., 1995), or to inhibition of pre-mRNA splicing (O'Keefe et al., 1994). Moreover, the localization of splicing factors to Balbiani ring genes occurs in a transcription-dependent manner in the nuclei of Chironomus tentans (Baurén et al., 1996), and splicing factors are associated with the loops of lampbrush chromosomes in amphibian germinal vesicles (Wu et al., 1991). In human cells, several in situ studies have shown that the nuclear speckles are associated predominantly with transcriptionally active cellular and viral genes (Lawrence et al., 1993; Xing et al., 1995; Dirks et al., 1997; Misteli et al., 1997, 1998).
Other observations, however, indicate that certain highly spliced endogenous pre-mRNAs are poorly associated with increased concentrations of SC35 (Smith et al., 1999). Similarly, some viral transcripts display little or no association with speckles (Zhang et al., 1994; Lampel et al., 1997). Altogether, these findings suggest that transcription is not sufficient for the association of specific genes with nuclear speckles and reveal a more complex view of the nuclear compartimentation of transcription and splicing activities. The most likely hypothesis to integrate our observations in the context of other results is the existence of a gene specificity and/or cell type specificity in the distribution of splicing factors regions (Smith et al., 1999). Likewise since some active genes predominantly associate with larger speckles (Xing et al., 1993, 1995), we can imagine that other active genes preferentially associate with very low amounts of splicing factors which can be missed due to the limitations of microscopic resolution. The concentration of splicing factors associated with specific transcription sites may be gene-specific and may depend on the combined effect of several factors such as the size and complexity of the gene, its transcription rate, and its position in the nucleus and/or its chromosomal environment which can impose structural constraints, thus limiting the access of splicing factors to these regions (Moen et al., 1995; Xing et al., 1995; Smith et al., 1999). Our work now adds to this by demonstrating that the presence or absence of introns is not a determining factor for the association of active genes with splicing factor–rich regions.
Role of Nuclear Speckles in Transcription and Splicing
The functional significance of splicing factors organized into subnuclear structures has been uncertain, although much of the data in the literature are consistent with a role of splicing factor complexes associated with the transcription and processing of intron-containing genes. The association of splicing factors with intron-dependent sites of transcription was demonstrated in experiments transiently or stably expressing intronless and intron-containing genes in HeLa cells (Huang and Spector, 1996b). The recruitment of splicing factors to sites of transcription based on a phosphorylation cycle which is tightly correlated with splicing activity also supports this conclusion (Misteli et al., 1998). Consequently, how do we incorporate the observations of the present study, that splicing factors can also be associated with sites of transcription independent of introns?
In our analysis, we examined the sites of transcription of endogenous cellular genes, in contrast to genes reintroduced by transfection or expressed following viral infection (Huang and Spector, 1996b; Misteli et al., 1998). The introduction of exogenous nucleic acids, by transfection, could potentially influence aspects of nuclear organization (Zhang et al., 1994) with consequences on endogenous transcriptional activities and a redistribution of splicing factors (Huang and Spector, 1996b). Likewise, the genomic site of integration in stably transfected constructs or integrated viral genomes may also influence their expression (Al-Shawi et al., 1990).
The association of the intronless hsp70 gene with speckles could be the result of the physical proximity on the chromosome of a distinct, highly transcribed, intron-containing gene, although this seems unlikely given that our data show a very high percentage of association between hsp70 transcription sites and the speckles. In addition, this association is strictly observed in stressed cells, suggesting that the putative neighboring gene would need to be stress-responsive. The only known hsp gene located in the vicinity of the hsp70 gene, which is located in the 6p21.3 region (Harrison et al., 1987), is the hsp90β gene which maps to the 6p12 locus (Durkin et al., 1993); the physical distance between these two genes allows a clear discrimination between the two transcription sites by light microscopy (Jolly et al., 1997c). Thus, the association observed between the intronless hsp70 transcription sites and the speckles is significant.
A more likely explanation for the different interpretations is that the distribution of transcription sites relative to nuclear speckles varies in a gene-specific manner. Indeed, differential distributions of viral or endogenous transcripts relative to nuclear speckles have been reported already, and they may reflect differences in the organization of nuclear RNAs derived from endogenous or from integrated viral genomes (Lampel et al., 1997). For example, some transcriptionally active genes containing introns display little or no association with SC35 speckles (Zhang et al., 1994; Lampel et al., 1997; Smith et al., 1999), whereas the collagen gene which has numerous introns is associated with speckles independent of its transcriptional activity (Xing et al., 1995). In that respect, we cannot rule out the possibility of a gene-specific organization of transcription sites with respect to nuclear regions enriched in splicing factors.
Of potential interest is the observation that active hsp transcription sites are found adjacent to the speckles rather than colocalizing with them, suggesting that transcription and splicing activities occur preferentially at the edges of the speckles where PFs seem to be localized as described by electron microscopy (for review see Fakan, 1994). A similar observation has been reported for several other genes (for review see Schul et al., 1998a). This may represent a mechanism to accelerate the release of nascent transcripts from the sites of transcription, which would indeed be affected if transcription and splicing were to take place in the core of the speckle where the concentration in RNA Pol II and processing factors may be elevated. Alternatively, this situation may simply reflect gene-specific differences in the association with speckles. Whatever the hypothesis, our data show that the distribution of a specific active gene as adjacent or overlapping with speckles does not rely on the presence of intronic sequences in the gene.
Why are splicing factors present at sites in the nucleus where they are apparently not required? A first hypothesis to explain the recruitment of splicing factors to an intronless gene is that complexes containing splicing factors could have other functions at the site of transcription in addition to intron excision. This suggestion is supported by a recent work in which we have shown that full-length nascent hsp70 transcripts are retained at the site of transcription for a period of <15 min after their completion (Jolly et al., 1998). Perhaps splicing factors could be involved in a primary step of the transcription/splicing process to scan nascent transcripts for the presence of introns. An alternative explanation is that active transcription sites would in fact associate with a subset of active RNA Pol II to which splicing factors are bound. This is supported by observations that splicing factors and other mRNA processing enzymes interact transiently with the hyperphosphorylated CTD of RNA Pol II (Mortillaro et al., 1996; Vincent et al., 1996; Yuryev et al., 1996; Cho et al., 1997; Du and Warren, 1997; McCracken et al., 1997a,b; Schul et al., 1998b). Even in mitosis when transcription is arrested, the association between splicing factors and RNA Pol II persists (Kim et al., 1997). At the cell biological level, at least a subset of RNA Pol II colocalizes with splicing factors within the speckles (this paper and Thibodeau and Vincent, 1991; Bregman et al., 1995; Blencowe et al., 1996; Mortillaro et al., 1996; Patturajan et al., 1998), as well as factors involved in the 5′ capping (Cho et al., 1997; McCracken et al., 1997a) and in the 3′ mRNA processing (Krause et al., 1994; Schul et al., 1998b). The corecruitment of splicing factors and RNA Pol II, together with the movement of genes towards speckles, could be beneficial for the cell in several ways. First, it would accelerate the splicing reaction. Second, by ensuring a sufficient amount of splicing factors at the site of transcription, this system would decrease the risk of producing unspliced transcripts which may generate aberrant and nonfunctional proteins. Third, this system would provide a feedback control mechanism to arrest transcription if splicing is interrupted (Yuryev et al., 1996). Fourth, it would minimize the information quantity required to displace the different factors to the sites of transcription, since the polymerase and splicing factors would all be displaced together as a single complex. This would suggest the existence of signals both in the transcription and processing apparatus and at the level of the chromosome and/or within the primary transcript, which determine the recognition between active transcription sites and proteins involved in RNA biogenesis. Such targeting and/or retention signals, if localized to the nascent transcripts, are not contained within the introns as demonstrated by our results. Identifying these signaling pathways will provide clues to understand the mechanisms of production and trafficking of RNAs within the nucleus.
While a growing number of observations support the idea of a compartimentation of transcription and processing activities in the nucleus, we may still have a simplified view of a more complex biological situation generated by the combination of multiple factors varying in a gene-specific and/or cell type–specific manner, therefore proving to be invalid for certain transcripts. Of particular importance may be the chromosomal context of the gene, which could dramatically influence the supply of splicing factors to the gene because of physical constraints (Smith et al., 1999). We are convinced that further understanding of the complex organization of transcription and splicing activities within the cell nucleus will come from a large scale analysis of specific endogenous genes, in particular genes with distinctive transcriptional and splicing characteristics such as heat shock genes or genes expressed in a tissue-specific manner.
Acknowledgments
We are grateful to Sui Huang, Tom Misteli, Philip T. Moen, and David L. Spector for helpful suggestions; to Li Tai for excellent technical assistance on RT-PCR; to San Ming Wang for providing the RT-PCR method; to Pernette Verschure for help in the BrUTP experiments; to Drs. E.K. Bautz, E. Hickey, B. Sefton, J.A. Steitz, and M. Vincent for providing us with the probes and antibodies used in this study; and to Robert A. Lamb and Robert Holmgren for providing access to the light microscopy facilities.
Abbreviations used in this paper
- CTD
COOH-terminal domain
- FISH
fluorescence in situ hybridization
- IG
interchromatin granule
- PF
perichromatin fibril
- Pol II
polymerase II
- snRNP
small nuclear ribonucleoprotein
Footnotes
C. Jolly was supported by the Association pour la Recherche sur le Cancer and by the Daniel F. and Ada L. Rice Foundation. R.I. Morimoto was supported by the National Institutes of Health grant GM38109. M. Robert-Nicoud and C. Vourc'h were supported by the Université Joseph Fourier and the Ministère de l'Enseignement Supérieur et de la Recherche (ACC-SV No. 5).
References
- Abravaya K, Phillips B, Morimoto RI. Attenuation of the heat shock response in HeLa cells is mediated by the release of bound heat shock transcription factor and is modulated by changes in growth and in heat shock temperatures. Genes Dev. 1991;5:2117–2127. doi: 10.1101/gad.5.11.2117. [DOI] [PubMed] [Google Scholar]
- Al-Shawi R, Kinnaird J, Burke J, Bishop JO. Expression of a foreign gene in a line of transgenic mice is modulated by a chromosomal position effect. Mol Cell Biol. 1990;10:1192–1198. doi: 10.1128/mcb.10.3.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji SS, Laing K, Morimoto RI. Erythroid lineage-specific expression and inducibility of the major heat shock protein Hsp70 during avian embryogenesis. Genes Dev. 1984;1:946–953. doi: 10.1101/gad.1.9.946. [DOI] [PubMed] [Google Scholar]
- Baurén G, Jiang W-Q, Bernholm K, Gu F, Wieslander L. Demonstration of a dynamic, transcription-dependent organization of pre-mRNA splicing factors in polytene nuclei. J Cell Biol. 1996;133:929–941. doi: 10.1083/jcb.133.5.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Mortillaro MJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R. A hyperphosphorylated form of the largest subunit of RNA polymerase II is associated with splicing complexes. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman DB, Du L, van der Zee S, Warren SL. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge E, Xia D-X, Carmo-Fonseca M, Cardinali B, Lamond AI, Pettersson U. Dynamic organization of splicing factors in adenovirus-infected cells. J Virol. 1995;69:281–290. doi: 10.1128/jvi.69.1.281-290.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter KC, Taneja KL, Lawrence JB. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J Cell Biol. 1991;115:1191–1202. doi: 10.1083/jcb.115.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter KC, Bowman D, Carrington W, Fogarty K, McNeil JA, Fay FS, Lawrence JB. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993;259:1330–1335. doi: 10.1126/science.8446902. [DOI] [PubMed] [Google Scholar]
- Cho E-J, Tagaki T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Lawrence JB. Multifunctional compartments in the cell nucleus: insights from DNA and RNA localization. J Cell Biochem. 1996;62:181–190. doi: 10.1002/(SICI)1097-4644(199608)62:2%3C181::AID-JCB6%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Dirks RW, de Pauw ESD, Raap AK. Splicing factors associate with nuclear HCMV-IE transcripts after transcriptional activation of the gene, but dissociate upon transcription inhibition: evidence for a dynamic organization of splicing factors. J Cell Sci. 1997;110:515–522. doi: 10.1242/jcs.110.4.515. [DOI] [PubMed] [Google Scholar]
- Du L, Warren SL. A functional interaction between the carboxy-terminal domain of RNA polymerase II and pre-mRNA splicing. J Cell Biol. 1997;136:5–18. doi: 10.1083/jcb.136.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin AS, Maglott DR, Vamvakopoulos NC, Zoghbi HY, Nierman WC. Assignment of an intron-containing human heat-shock protein gene (hsp90β, HSPCB) to chromosome 6 near TCTE1 (6p21) and two intronless pseudogenes to chromosomes 4 and 15 by polymerase chain reaction amplification from a panel of hybrid cell lines. Genomics. 1993;18:452–454. doi: 10.1006/geno.1993.1498. [DOI] [PubMed] [Google Scholar]
- Fakan S. Perichromatin fibrils are in situforms of nascent transcripts. Trends Cell Biol. 1994;4:86–90. doi: 10.1016/0962-8924(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Fay FS, Taneja KL, Shenoy S, Lifshitz L, Singer RH. Quantitative digital analysis of diffuse and concentrated nuclear distributions of nascent transcripts, SC35 and poly(A) Exp Cell Res. 1997;231:27–37. doi: 10.1006/excr.1996.3460. [DOI] [PubMed] [Google Scholar]
- Fort P, Marty L, Piechaczyk M, El S, Sabrouty, Dani C, Jeanteur P, Blanchard JM. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate dehydrogenase multigenic family. Nucleic Acids Res. 1985;13:1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Gough NM. Rapid and quantitative preparation of cytoplasmic RNA from small number of cells. Anal Biochem. 1988;15:93–95. doi: 10.1016/0003-2697(88)90164-9. [DOI] [PubMed] [Google Scholar]
- Harrison GS, Drabkin HA, Kao FT, Hartz J, Hart IM, Chu EHY, Wu BJ, Morimoto RI. Chromosomal location of human genes encoding major heat-shock protein HSP70. Somat Cell Mol Genet. 1987;13:119–130. doi: 10.1007/BF01534692. [DOI] [PubMed] [Google Scholar]
- Hickey E, Brandon SE, Sadis S, Smale G, Weber LA. Molecular cloning of sequences encoding the human heat shock proteins and their expression during hyperthermia. Gene. 1986;43:147–154. doi: 10.1016/0378-1119(86)90018-1. [DOI] [PubMed] [Google Scholar]
- Huang S, Spector DL. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in spicing factors. Genes Dev. 1991;5:2288–2302. doi: 10.1101/gad.5.12a.2288. [DOI] [PubMed] [Google Scholar]
- Huang S, Spector DL. Dynamic organization of pre-mRNA splicing factors. J Cell Biochem. 1996a;62:191–197. doi: 10.1002/(sici)1097-4644(199608)62:2<191::aid-jcb7>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Huang S, Spector DL. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J Cell Biol. 1996b;133:719–732. doi: 10.1083/jcb.133.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Garcia LF, Spector DL. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell. 1993;73:47–59. doi: 10.1016/0092-8674(93)90159-n. [DOI] [PubMed] [Google Scholar]
- Jolly, C., and R.I. Morimoto. 1999. Stress and the cell nucleus: dynamics of gene expression and structural reorganization. Gene Expr. In press. [PMC free article] [PubMed]
- Jolly C, Mongelard F, Robert-Nicoud M, Vourc'h C. Optimization of nuclear transcripts detection by FISH and combination with fluorescence immunocytochemical detection of transcription factors. J Histochem Cytochem. 1997a;45:1585–1592. doi: 10.1177/002215549704501201. [DOI] [PubMed] [Google Scholar]
- Jolly C, Morimoto RI, Robert-Nicoud M, Vourc'h C. Nuclear foci enriched in HSF1 transcription factor: relationship with heat shock gene transcription sites. J Cell Sci. 1997b;110:2935–2941. doi: 10.1242/jcs.110.23.2935. [DOI] [PubMed] [Google Scholar]
- Jolly C, Michelland S, Rocchi M, Robert-Nicoud M, Vourc'h C. Analysis of the transcriptional activity of amplified genes in tumor cells by fluorescence in situhybridization. Hum Genet. 1997c;101:81–87. doi: 10.1007/s004390050591. [DOI] [PubMed] [Google Scholar]
- Jolly C, Robert-Nicoud M, Vourc'h C. Contribution of the growing RNA molecules to the nuclear transcripts foci observed by FISH. Exp Cell Res. 1998;238:299–304. doi: 10.1006/excr.1997.3838. [DOI] [PubMed] [Google Scholar]
- Kim E, Du L, Bregman DB, Warren SL. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontermann RE, Liu Z, Schulze RA, Sommer KA, Queitsch I, Dubel S, Kipriyanov SM, Breitling F, Bautz EK. Characterization of the epitope recognized by a monoclonal antibody directed against the largest subunit of DrosophilaRNA polymerase II. Biol Chem Hoppe-Seyler. 1995;376:473–481. doi: 10.1515/bchm3.1995.376.8.473. [DOI] [PubMed] [Google Scholar]
- Krämer A, Haars R, Kabisch R, Will H, Bautz FA, Bautz EK. Monoclonal antibody directed against RNA polymerase II of Drosophila melanogaster. . Mol Gen Genet. 1980;180:193–199. doi: 10.1007/BF00267369. [DOI] [PubMed] [Google Scholar]
- Krause S, Fakan S, Weis K, Wahle E. Immunodetection of poly(A) binding protein II in the cell nucleus. Exp Cell Res. 1994;214:75–82. doi: 10.1006/excr.1994.1235. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lampel S, Bridger JM, Zirbel RM, Mathieu UR, Lichter P. Nuclear RNA accumulations contain released transcripts and exhibit specific distributions with respect to Sm antigen foci. DNA Cell Biol. 1997;16:1133–1142. doi: 10.1089/dna.1997.16.1133. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Carter KC, Xing X. Probing functional organization within the nucleus: is genome structure integrated with RNA metabolism? . Cold Spring Harbor Symp Quant Biol. 1993;58:807–818. doi: 10.1101/sqb.1993.058.01.088. [DOI] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CAJ, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter P, Cremer T, Borden J, Manuelidis L, Ward DC. Delineation of individual chromosomes in metaphase and interphase cells by in situsuppression hybridization using recombinant DNA libraries. Hum Genet. 1988;80:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- Martin TE, Barghusen SC, Leser GP, Spear PG. Redistribution of nuclear ribonucleoprotein antigens during herpes simplex virus infection. J Cell Biol. 1987;105:2069–2082. doi: 10.1083/jcb.105.5.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG. Of coiled bodies, gems, and salmon. J Cell Biochem. 1998;70:181–192. [PubMed] [Google Scholar]
- Mattaj IW, Habets WJ, van Venrooij WJ. Monospecific antibodies reveal details of U2 snRNP structure and interaction between U1 and U2 snRNPs. EMBO (Eur Mol Biol Org) J. 1986;5:997–1002. doi: 10.1002/j.1460-2075.1986.tb04314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997a;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997b;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Misteli T, Spector DL. The cellular organization of gene expression. Curr Opin Cell Biol. 1998;10:322–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- Misteli T, Cáceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Misteli T, Cáceres JF, Clement JQ, Krainer AR, Wilkinson MF, Spector DL. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J Cell Biol. 1998;143:297–307. doi: 10.1083/jcb.143.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen PT, Jr, Smith KP, Lawrence JB. Compartmentalization of specific pre-mRNA metabolism: an emerging view. Hum Mol Genet. 1995;4:1779–1789. doi: 10.1093/hmg/4.suppl_1.1779. [DOI] [PubMed] [Google Scholar]
- Monier K, Usson Y, Mongelard F, Szepetowski P, Robert-Nicoud M, Vourc'h C. Metaphase and interphase mapping by FISH: improvement of chromosome banding and signal resolution in interphase nuclei by means of iterative deconvolution. Cytogenet Cell Genet. 1996;72:200–204. doi: 10.1159/000134189. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Cells in stress: transcriptional regulation of heat shock genes. Science. 1993;259:1409–1410. doi: 10.1126/science.8451637. [DOI] [PubMed] [Google Scholar]
- Morimoto, R.I., P.E. Kroeger, and J.J. Cotto. 1996. The transcriptional regulation of heat shock genes: a plethora of heat shock factors and regulatory conditions. In Stress-inducible Cellular Responses. U. Feige, R.I. Morimoto, I. Yahara, and B.S. Polla, editors. Birkhaüser Verlag, Basel, Switzerland. 139–163.
- Mortillaro MJ, Blencowe BJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser D, Theodorakis NG, Morimoto RI. Coordinate changes in heat shock element binding activity and hsp70 gene transcription rates in human cells. Mol Cell Biol. 1988;8:4736–4744. doi: 10.1128/mcb.8.11.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe RT, Mayeda A, Sadowski CL, Krainer AR, Spector DL. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J Cell Biol. 1994;124:249–260. doi: 10.1083/jcb.124.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- Pombo A, Cook PR. The localization of sites containing nascent RNA and splicing factors. Exp Cell Res. 1996;229:201–203. doi: 10.1006/excr.1996.0360. [DOI] [PubMed] [Google Scholar]
- Pombo A, Ferreira J, Bridge E, Carmo-Fonseca M. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO (Eur Mol Biol Org) J. 1994;13:5075–5085. doi: 10.1002/j.1460-2075.1994.tb06837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puvion E, Puvion-Dutilleul F. Ultrastructure of the nucleus in relation to transcription and splicing: roles of perichromatin fibrils and interchromatin granules. Exp Cell Res. 1996;229:217–225. doi: 10.1006/excr.1996.0363. [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul F, Bachellerie J-P, Visa N, Puvion E. Rearrangements of intranuclear structures involved in RNA processing in response to adenovirus infection. J Cell Sci. 1994;107:1457–1468. doi: 10.1242/jcs.107.6.1457. [DOI] [PubMed] [Google Scholar]
- Raap AK, van de Rijke FM, Dirks RW, Sol CJ, Boom R, van der Ploeg M. Bicolor fluorescence in situhybridization to intron and exon mRNA sequences. Exp Cell Res. 1991;197:319–322. doi: 10.1016/0014-4827(91)90439-2. [DOI] [PubMed] [Google Scholar]
- Schul W, de Jong L, van Driel R. Nuclear neighbours: the spatial and functional organization of genes and nuclear domains. J Cell Biochem. 1998a;70:159–171. doi: 10.1002/(sici)1097-4644(19980801)70:2<159::aid-jcb2>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Schul W, van Driel R, de Jong L. A subset of poly(A) polymerase is concentrated at sites of RNA synthesis and is associated with domains enriched in splicing factors and poly(A) RNA. Exp Cell Res. 1998b;238:1–12. doi: 10.1006/excr.1997.3808. [DOI] [PubMed] [Google Scholar]
- Shi Y, Mosser DD, Morimoto RI. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla RR, Dominski Z, Zwierzynski T, Kole R. Inactivation of splicing factors in HeLa cells subjected to heat shock. J Biol Chem. 1990;265:20377–20383. [PubMed] [Google Scholar]
- Smith KP, Moen PT, Jr, Wydner KL, Coleman JR, Lawrence JB. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene-specific. J Cell Biol. 1999;144:617–629. doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Fu X-D, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO (Eur Mol Biol Org) J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, O'Keefe RT, Jiménez-Garcia LF. Dynamics of transcription and pre-mRNA splicing within the mammalian cell nucleus. Cold Spring Harbor Symp Quant Biol. 1993;58:799–805. doi: 10.1101/sqb.1993.058.01.087. [DOI] [PubMed] [Google Scholar]
- Thibodeau A, Vincent M. Monoclonal antibody CC-3 recognizes phosphoproteins in interphase and mitotic cells. Exp Cell Res. 1991;195:145–153. doi: 10.1016/0014-4827(91)90510-2. [DOI] [PubMed] [Google Scholar]
- Vincent M, Lauriault P, Dubois M-F, Lavoie S, Bensaude O, Chabot B. The nuclear matrix protein p255 is a highly phosphorylated form of RNA polymerase II largest subunit which associates with spliceosomes. Nucleic Acids Res. 1996;24:4649–4652. doi: 10.1093/nar/24.23.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N, Puvion-Dutilleul F, Harper F, Bachellerie JP, Puvion E. Intranuclear distribution of poly(A) RNA determined by electron microscope in situhybridization. Exp Cell Res. 1993;208:19–34. doi: 10.1006/excr.1993.1218. [DOI] [PubMed] [Google Scholar]
- Wang J, Cao L-G, Wang Y-L, Pederson T. Localization of pre-messenger RNA at discrete nuclear sites. Proc Natl Acad Sci USA. 1991;88:7391–7395. doi: 10.1073/pnas.88.16.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S.M., J.D. Khandekar, K.L. Kaul, D.J. Winchester, and R.I. Morimoto. 1999. A method for the quantitative analysis of human heat shock gene expression using a multiplex RT-PCR assay. Cell Stress Chaperones. In press. [DOI] [PMC free article] [PubMed]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watowich SS, Morimoto RI. Complex regulation of heat shock- and glucose-responsive genes in human cells. Mol Cell Biol. 1988;8:393–405. doi: 10.1128/mcb.8.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Samarabandu J, Devdhar RS, Siegel AJ, Acharya R, Berezney R. Segregation of transcription and replication sites into higher order domains. Science. 1998;281:1502–1505. doi: 10.1126/science.281.5382.1502. [DOI] [PubMed] [Google Scholar]
- Williams GT, Morimoto RI. Maximal stress-induced transcription of the human HSP70 promoter involves interactions with the basal promoter independent of rotational alignment. Mol Cell Biol. 1990;10:3125–3146. doi: 10.1128/mcb.10.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Hunt C, Morimoto RI. Structure and expression of the human gene encoding major heat shock protein hsp70. Mol Cell Biol. 1985;5:330–341. doi: 10.1128/mcb.5.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZA, Murphy C, Callan HG, Gall JG. Small nuclear ribonucleoproteins and heterogeneous nuclear ribonucleoproteins in the amphibian germinal vesicle: loops, spheres, and snurposomes. J Cell Biol. 1991;113:465–483. doi: 10.1083/jcb.113.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol Cell Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Dobner PR, Lawrence JB. Higher level organization of individual gene transcription and RNA splicing. Science. 1993;259:1326–1329. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Moen PT, Jr, McNeil JA, Lawrence JB. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, Gebara M, Corden JL. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng CQ, Kim E, Warren SL, Berget SM. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO (Eur Mol Biol Org) J. 1997;16:1401–1412. doi: 10.1093/emboj/16.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Taneja KL, Singer RH, Green MR. Localization of pre-mRNA splicing in mammalian nuclei. Nature. 1994;372:809–812. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]