Abstract
The 17S U2 small nuclear ribonucleoprotein particle (snRNP) represents the active form of U2 snRNP that binds to the pre-mRNA during spliceosome assembly. This particle forms by sequential interactions of splicing factors SF3b and SF3a with the 12S U2 snRNP. We have purified SF3b and the 15S U2 snRNP, an intermediate in the assembly pathway, from HeLa cell nuclear extracts and show that SF3b consists of four subunits of 49, 130, 145, and 155 kD. Biochemical analysis indicates that both SF3b and the 12S U2 snRNP are required for the incorporation of SF3a into the 17S U2 snRNP. Nuclease protection studies demonstrate interactions of SF3b with the 5′ half of U2 small nuclear RNA, whereas SF3a associates with the 3′ portion of the U2 snRNP and possibly also interacts with SF3b. Electron microscopy of the 15S U2 snRNP shows that it consists of two domains in which the characteristic features of isolated SF3b and the 12S U2 snRNP are conserved. Comparison to the two-domain structure of the 17S U2 snRNP corroborates the biochemical results in that binding of SF3a contributes to an increase in size of the 12S U2 domain and possibly induces a structural change in the SF3b domain.
Keywords: electron microscopy, pre-mRNA splicing, spliceosome, splicing factor, U2 small nuclear ribonucleoprotein particle
Introns are removed from nuclear pre-mRNAs in large complexes, termed spliceosomes, that are assembled by a network of interactions between the pre-mRNA, small nuclear ribonucleoprotein particles (snRNPs),1 and non-snRNP proteins (for review see Moore et al., 1993; Madhani and Guthrie, 1994). The RNA moieties of the snRNPs base pair with conserved sequences at the splice sites and the intron branch site and also with one another, thus generating the catalytic core of the spliceosome. snRNP and non-snRNP proteins are essential for splicing, although they are probably not directly involved in catalysis (for review see Krämer, 1996).
snRNPs were initially isolated from mammalian cells under high salt conditions as complexes of 10–12S, containing a set of eight common (Sm or core) proteins and a limited number of proteins that are characteristic for each snRNP (for review see Will and Lührmann, 1997). Milder purification procedures have later led to the detection of larger snRNP complexes containing varying numbers of additional polypeptides.
The U2 snRNP has been isolated in two forms of 12 and 17S. The 12S U2 snRNP contains the Sm proteins and two characteristic proteins, A′ and B′′. The Sm proteins are bound to a structural motif, the Sm binding site, which consists of a single-stranded region of the sequence RA(U)nGR (n ≥ 3) flanked by double-stranded stems (see Fig. 4 B; Branlant et al., 1982; Mattaj and De Robertis, 1985). The A′ and B′′ proteins are bound to the 3′ terminal stem-loop IV of U2 small nuclear RNA (snRNA) (Scherly et al., 1990; Bentley and Keene, 1991; Boelens et al., 1991; Price et al., 1998). The 17S U2 snRNP, which was isolated at salt concentrations <200 mM, contains nine additional proteins of 35, 53, 60, 66, 92, 110, 120, 150, and 160 kD (Behrens et al., 1993b).
Figure 4.
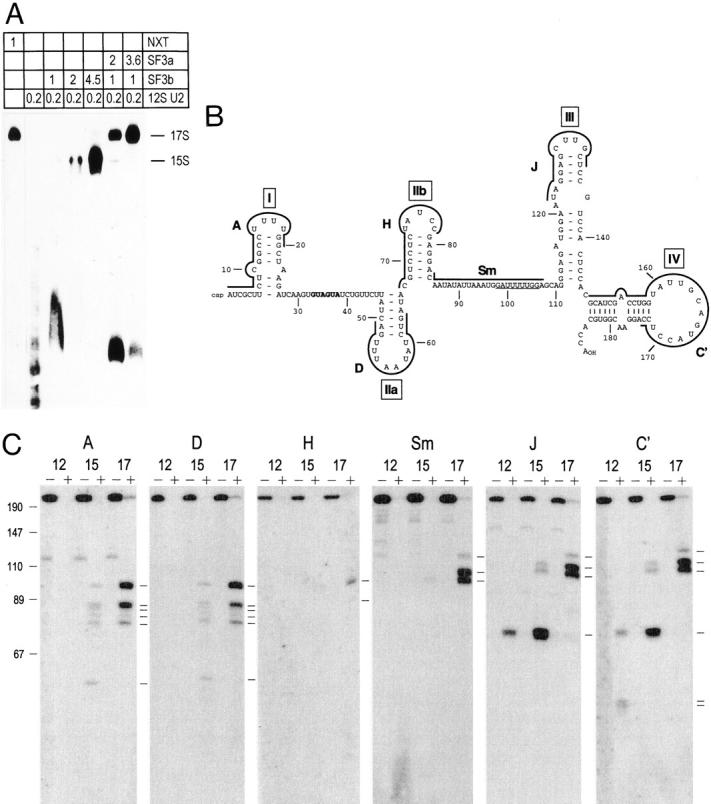
Analysis of micrococcal nuclease-resistant regions in U2 snRNA in the 12S, 15S, and 17S U2 snRNPs. (A) In vitro reconstitution of the 15S and 17S U2 snRNPs. The indicated amounts (in microliters) of nuclear extract (NE), SF3a, SF3b, and the 12S U2 snRNP were incubated with 5′ endlabeled oligonucleotide U2A and separated in a 4% polyacrylamide gel. The position of the 15S and 17S U2 snRNPs is indicated on the right. (B) Secondary structure of mammalian U2 snRNA (according to Ares and Igel, 1990; Hartshorne and Agabian, 1990). The Sm-binding site is underlined, the branch site interaction sequence is shown in bold, and the stem-loops are indicated by roman numerals. Sequences complementary to oligonucleotides are shown by thick lines. (C) Analysis of protected fragments of U2 snRNA in the 12S, 15S, and 17S particles by Northern blotting. The 12S U2 snRNP (Mono Q) and reconstituted 15S and 17S U2 snRNPs were incubated in the absence (−) or presence (+) of micrococcal nuclease for 10 min at 30°C. RNA was isolated, separated in a 14% polyacrylamide/8.3 M urea gel, and blotted onto a nylon membrane, followed by detection with oligonucleotides complementary to different portions of U2 snRNA as indicated above each panel. The migration of DNA length markers is indicated on the left of the figure. Please note that the DNA fragments migrate faster than the RNA fragments. Protected U2 snRNA fragments are indicated by lines on the right side of each panel.
Electron microscopy revealed two tightly attached domains in the 12S U2 snRNP (Kastner et al., 1990). A main body (or core domain) of ∼8 nm in diameter comprises the Sm proteins. The A′ and B′′ proteins are present in an additional domain 4 nm in length and 6 nm in width which is directly attached to the core domain. The 17S U2 snRNP consists of two distinct globular domains of ∼10–12 nm which differ in their appearance and are connected by a short filamentous structure that is sensitive to RNase (Behrens et al., 1993b). The fine structure of the 12S U2 snRNP was not evident in either of these domains and it appeared that both domains of the 17S U2 snRNP contained one or more of the additional 17S U2 snRNP-specific proteins.
In vitro, the 17S U2 snRNP is reconstituted by incubation of the 12S U2 snRNP with splicing factors (SF) 3a and 3b (Brosi et al., 1993a) that were initially isolated as non-snRNP proteins (Brosi et al., 1993b). The 12S U2 snRNP and SF3b associate to form a particle of 15S; addition of SF3a to the 15S U2 snRNP results in the assembly of the 17S particle. Biochemical and immunological analyses have confirmed that the three subunits of SF3a (SF3a60, SF3a66, and SF3a120) represent the 17S U2 snRNP-specific proteins of 60, 66, and 110 kD (Brosi et al., 1993a). In a separate study, a U2 snRNP-specific patient serum precipitated seven spliceosome-associated proteins (SAPs) from a fraction enriched in U2 snRNP (Staknis and Reed, 1994). Three of these (SAP61, SAP62, and SAP114) correspond to the SF3a subunits, whereas the other proteins (SAP49, SAP130, SAP145, and SAP155) are similar in size to the 17S U2-specific polypeptides of 53, 120, 150, and 160 kD. Together these studies suggested that SAP49, SAP130, SAP145, and SAP155 and possibly the two remaining 17S U2 snRNP-specific proteins of 35 and 92 kD are constituents of SF3b.
U2 snRNP functions in splicing by converting the early complex E into presplicing complex A (for review see Krämer, 1996; Reed, 1996). Complex E is formed by binding of U1 snRNP to the 5′ splice site and interaction of splicing factors U2AF and SF1 with the polypyrimidine tract upstream of the 3′ splice site and the branch site, respectively. The formation of complex A occurs by subsequent binding of U2 snRNP to the pre-mRNA, which involves the formation of a short helix between a single-stranded region in U2 snRNA (the branch site interaction sequence; shown in bold in Fig. 4 B) and the intron branch site. This reaction requires the 12S U2 snRNP plus SF3a and SF3b (Brosi et al., 1993b) or the isolated 17S U2 snRNP (Behrens et al., 1993a); the 12S U2 snRNP alone is inactive in complex assembly. Thus, SF3a and SF3b are essential for prespliceosome assembly in combination with the 12S U2 snRNP. It has furthermore been shown that SF3a and three of the presumptive SF3b polypeptides cross-link to the pre-mRNA in the vicinity of the branch site during A complex assembly (Gozani et al., 1996, 1998). These interactions are thought to stabilize the U2 snRNA/branch site helix and may also aid to incorporate the U2 snRNP into the spliceosome.
Functions for U2 snRNP-associated proteins at later stages of the splicing reaction have not been reported. However, the U2 snRNA plays a vital role in the formation of the active site of the spliceosome. Sequences upstream and downstream of the branch site interaction sequence engage in base pairing interactions with U6 snRNA that are essential for intron removal (for review see Madhani and Guthrie, 1994; Yu et al., 1999). Base pairing between U2 and U6 snRNA necessitates the destabilization of stem-loops I and IIa in U2 snRNA as well as sequences in U6 that are base-paired to U4 snRNA, which is most likely achieved by the action of RNA-dependent ATPases (for review see Staley and Guthrie, 1998).
The purpose of the present study was to determine the subunit composition of SF3b and the roles of SF3a and SF3b in the formation of the 17S U2 snRNP. Protein–protein interactions between individual SF3a subunits and SAP49 and SAP145 have been described and are conserved in Saccharomyces cerevisiae (Krämer, 1996; Fromont-Racine et al., 1997; Igel et al., 1998). However, it is not known whether SF3a and SF3b interact with one another in the absence of other components of the 17S U2 snRNP. In addition, several studies have addressed the interaction of proteins with the U2 snRNA. First, Temsamani et al. (1991) showed that sequences in the 5′ half of U2 snRNA and stem-loop IV are required for the formation of a complex of low mobility that most likely corresponds to the 17S U2 snRNP. Second, biochemical and electron microscopic studies by Behrens et al. (1993b) suggested that most of the 17S U2 snRNP-specific proteins associated with the 5′ portion of the U2 particle. Third, studies in S. cerevisiae indicated genetic interactions between stem-loops IIa and IIb and homologues of the SF3a polypeptides and between stem-loop IIa and Cus1p, the homologue of SAP145 (Ruby et al., 1993; Wells and Ares, 1994; Wells et al., 1996; Yan and Ares, 1996).
We have used a biochemical approach to analyze interactions that occur during the assembly of the 17S U2 snRNP and to position SF3a and SF3b within this particle. These studies were complemented by a comparison of the morphology of the 15S U2 snRNP and SF3b by electron microscopy. Based on these results we propose a model of the principle architecture of the U2 snRNP particle.
Materials and Methods
RNAs and Oligonucleotides
The template for the pre-mRNA substrate (T3-RNA1) has been described (Arning et al., 1996). Human antisense U2 snRNA was transcribed with T7 polymerase from a pGem-U2 construct digested with EcoRI (Lamond et al., 1989). 2′-O-allyl oligoribonucleotides complementary to U2 snRNA were generous gifts from Angus Lamond (University of Dundee, Dundee, UK) and have been described by Barabino et al. (1992). The Sm and C′ oligodeoxynucleotides are complementary to nucleotides 84–107 and 148–174 of U2 snRNA, respectively.
Preparation of Splicing Factors and snRNPs
Splicing factors and snRNPs were fractionated from HeLa cell nuclear extracts by DEAE-Sepharose, heparin-Sepharose, and Mono Q chromatography as described (Krämer et al., 1987; Krämer and Utans, 1991). SF3a and SF3b were further purified by spermine-agarose and Mono S chromatography (Brosi et al., 1993b). In addition, SF3b (Mono S), U1, and U2 snRNPs (Mono Q) were sedimented in glycerol gradients. Fractions were adjusted to 10% glycerol and layered onto 4-ml gradients of 15–40% glycerol in 100 mM KCl, 20 mM Hepes-KOH (pH 7.9), 1 mM MgCl2, 0.5 mM PMSF, 0.5 mM DTT, and 0.2 mM EDTA. Sedimentation was performed in a TST60.4 rotor (Kontron) for 8 h at 55,000 rpm. Fractions of 200 μl were collected from the top.
In Vitro Assembly of U2 snRNP Complexes and Prespliceosomes
U2 snRNPs complexes were assembled in the presence of SF3a and SF3b (Mono S fractions) and the 12S U2 snRNP (Mono Q) as described (Brosi et al., 1993a). The formation of presplicing complexes was performed in the presence of HeLa cell nuclear extract or partially purified splicing factors (Krämer, 1988): SF1, ammonium sulfate–concentrated DS100 (Krämer, 1992); SF3a, Mono S (Brosi et al., 1993b); U2AF, HS1000 (Krämer and Utans, 1991), U1 snRNP purified by glycerol gradient centrifugation (see above), and the fractions indicated in the figure legends.
Immunoprecipitation
IgG from monoclonal anti-SF3a66 antibody (mAb66; Brosi et al., 1993b) was bound to protein A–Sepharose (PAS) in NET-2 buffer (50 mM Tris-HCl, pH 7.9, 150 mM NaCl, 0.5 mM DTT, 0.5% NP-40) for 1 h at 4°C. The material was washed with NET-2 to remove unbound IgG. PAS-coupled antibodies were incubated with combinations of SF3a, SF3b, and U2 snRNP for 90 min at 4°C. Unbound material was removed by washing with NET-2 and the samples were divided into three aliquots. Bound components were eluted with SDS sample solution from two aliquots each and separated by SDS-PAGE. Proteins were either stained with silver or blotted onto nitrocellulose membranes followed by detection of the U2 snRNP-specific protein B′′ with mAb4G3 (a gift from W. van Venrooij, University of Nijmegen, Nijmegen, The Netherlands) and Sm proteins with mAbY12 (a gift from J. Steitz, Yale University, New Haven, CT) as described (Brosi et al., 1993b). The third set of aliquots was treated with proteinase K (0.35 mg/ml proteinase K, 2% sarcosyl, 0.1 M Tris-HCl, pH 7.5, 20 mM EDTA) for 30 min at 60°C to release bound U2 snRNA, followed by phenol-chloroform extraction and ethanol precipitation. RNA was separated in a 14% polyacrylamide/8.3 M urea gel followed by Northern blotting and detection with radiolabeled antisense U2 snRNA as described below.
Protection from Micrococcal Nuclease Digestion and Analysis by Northern Blotting
U2 snRNP fractions isolated from HeLa cell nuclear extracts or reconstituted from individual purified components were digested with micrococcal nuclease for 10 min at 30°C (Krämer et al., 1987). RNA was isolated by proteinase K digestion, separated in 14% polyacrylamide/8.3 M urea gels, and blotted onto GeneScreen membranes (Schleicher and Schüll) as described (Utans et al., 1992). RNA was cross-linked to the membranes by UV irradiation in a Stratalinker (Stratagene) and the membranes were prehybridized in 5× SSC, 5× Denhardt's solution, 1% SDS, 2.5% dextran sulfate, 50 mM sodium phosphate (pH 6.5), and 1 mg/ml salmon sperm DNA. The membranes were hybridized overnight in the same solution with uniformly labeled antisense U2 snRNA or 5′ endlabeled oligonucleotides at 25–40°C (depending on the oligonucleotides) followed by several washes in 2× SSC/1% SDS at 25°C.
Electron Microscopy
Glycerol gradient fractions containing purified U2 snRNPs or SF3b were negatively stained with 2.5% uranyl formate by the double carbon film method as described (Kastner and Lührmann, 1989; Kastner, 1998). The preparations were examined under a Zeiss EM109 electron microscope with an acceleration voltage of 80 kV and electron micrographs were taken with a magnification of 85,000.
Results
Different Forms of the U2 snRNP Are Separated by Mono Q Chromatography
The majority of the U2 snRNP in HeLa cell nuclear extracts sediments with ∼17S in glycerol gradients (at 100 mM KCl) and most SF3a is found in the same gradient fractions (Brosi et al., 1993a). In contrast, after chromatography of nuclear extracts on DEAE-Sepharose, heparin-Sepharose, and Mono Q, SF3a and SF3b activity was detected in snRNP-free fractions eluting at ∼0.2–0.28 M KCl (Krämer et al., 1987). In Western blots with antibodies directed against SF3a or SF3b subunits, the bulk of these proteins is found in the same fractions and only small amounts elute at higher salt concentrations (data not shown). U2 snRNP elutes from Mono Q in a highly reproducible heterogeneous pattern at salt concentrations >0.3 M (Fig. 1 A; Krämer et al., 1987). To test whether this heterogeneity resulted from a separation of the different forms of U2 snRNP, snRNPs of the Mono Q fractions were separated by nondenaturing PAGE (Fig. 1 B). To visualize U2 snRNPs by autoradiography Mono Q fractions were incubated with a 5′ endlabeled oligoribonucleotide complementary to the 5′ end of U2 snRNA (U2A; see Fig. 4 B) before gel electrophoresis. U2 snRNP eluting at ∼0.3 M KCl (fractions 34–38) migrated similar to U2 snRNP in nuclear extracts and a 17S U2 snRNP control obtained after glycerol gradient sedimentation. U2 snRNP eluting at 0.4 M KCl (fractions 48–50) corresponded in mobility to a particle formed in vitro with the 12S U2 snRNP and a SF3b-containing fraction (see Fig. 4 A; Brosi et al., 1993a), thus representing the 15S U2 snRNP. Most U2 snRNP eluted at high salt concentration (∼0.48 M KCl; fractions 54–60) and its migration corresponded to that of the 12S U2 particle. Thus, a large portion of SF3a and SF3b dissociates from the 17S U2 snRNP during the fractionation procedure, which is consistent with the observation that the 17S U2 snRNP is disrupted at salt concentrations >200 mM (Behrens et al., 1993b; Brosi et al., 1993a).
Figure 1.
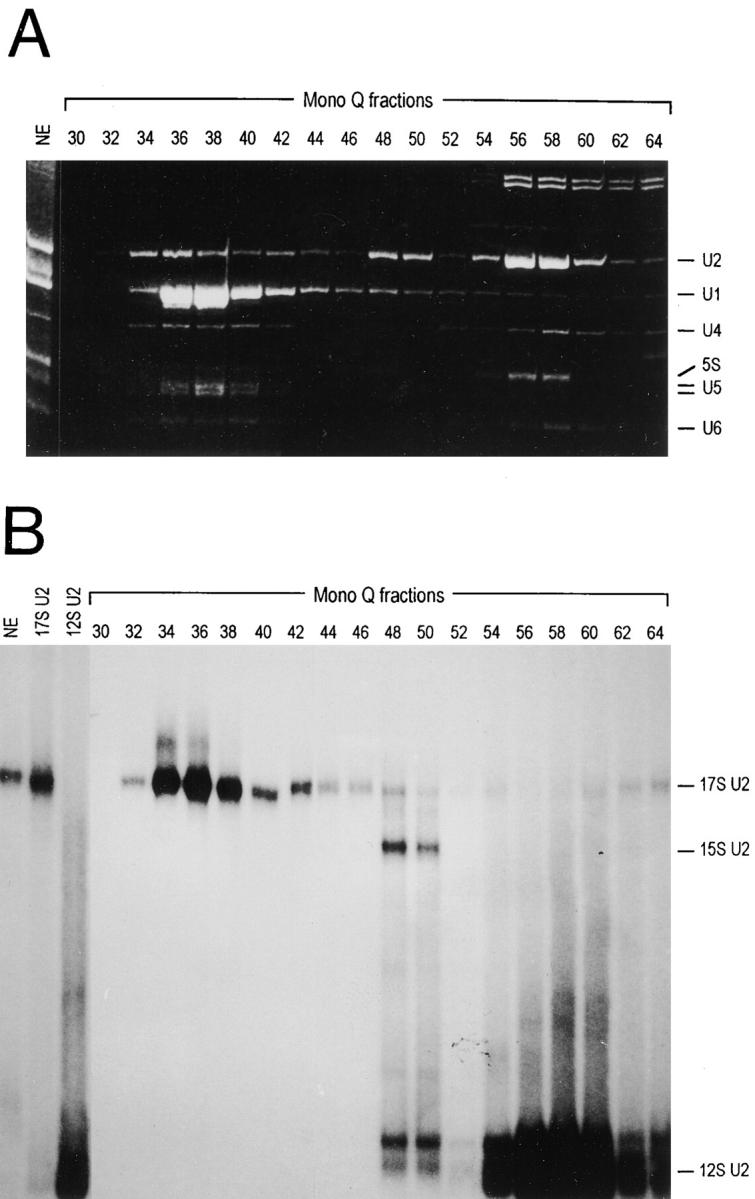
Separation of different forms of U2 snRNP by Mono Q chromatography. (A) RNA from 20-μl aliquots of HeLa cell nuclear extract and Mono Q fractions was isolated by proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation, separated in a 10% polyacrylamide/8.3 M urea gel, and stained with ethidium bromide. snRNAs and 5S RNA are indicated on the right. (B) Aliquots (5 μl) of HeLa cell nuclear extract (NE), glycerol gradient–purified 17S and 12S U2 snRNPs, and Mono Q fractions were incubated with 5′ endlabeled oligonucleotide U2A (see Fig. 4 B) and separated in a 4% polyacrylamide gel (Brosi et al., 1993a). The different forms of U2 snRNP are indicated on the right.
Identification of Polypeptides Associated with SF3b
As a first step in the identification of SF3b-associated polypeptides, Mono Q fractions containing the 15S U2 snRNP (which by definition should contain the Sm proteins, A′, B′′, and SF3b; see Introduction) were sedimented in a glycerol gradient. Individual gradient fractions were analyzed for their RNA and protein composition. The majority of U2 snRNA is detected in fractions 10–13 (Fig. 2 A) and the same fractions are highly enriched in polypeptides of ∼130–150 kD (Fig. 2 B, arrowheads). In addition, a polypeptide of ∼50 kD cofractionates with U2 snRNA. We next tested whether the glycerol gradient fractions containing U2 snRNA were active in the assembly of presplicing complex A, i.e., could substitute for both, the 12S U2 snRNP and SF3b. Reactions were performed in the presence of SF1, SF3a, U2AF, and U1 snRNP which are required for presplicing complex formation in addition to SF3b and U2 snRNP (Krämer and Utans, 1991; Brosi et al., 1993b). Complex A was efficiently assembled in the presence of partially purified U2 snRNP and SF3b or the Mono Q 15S U2 snRNP fraction used as the input for glycerol gradient sedimentation, but complex A was not formed in the absence of U2 snRNP and SF3b (Fig. 2 C). Addition of glycerol gradient fractions 10–14 to the reconstitution reaction promoted the assembly of the complex. Complex formation was not observed when only SF3b was omitted from a spliceosome assembly reaction (Brosi et al., 1993b); thus, we conclude that the 15S U2 snRNP fractions contain functional U2 snRNP and SF3b.
Figure 2.
Analysis of glycerol gradient–purified 15S U2 snRNP. The 15S U2 snRNP (Mono Q) was sedimented through a 15–40% glycerol gradient. (A) RNA from individual gradient fractions was separated in a 10% polyacrylamide/8.3 M urea gel and stained with ethidium bromide. snRNAs are indicated on the left. (B) Proteins were separated in a 10% SDS polyacrylamide gel and stained with silver. The SF3b subunits are indicated by arrows. (C) The 12S U2 snRNP (Mono Q) and SF3b (Mono S), the 15S U2 snRNP (Mono Q; IP), and glycerol gradient fractions of the 15S U2 snRNP were analyzed for presplicing complex formation in the presence of SF1, SF3a, U2AF, and U1 snRNP (see Materials and Methods). A control reaction with nuclear extract (NE) is shown in the second lane. The migration of pre-mRNA and presplicing complex A is indicated on the left. (D) Glycerol gradient–purified SF3b and 15S U2 snRNP were separated in a 10% SDS polyacrylamide gel. Arrows indicate the SF3b subunits. The U2-specific proteins A′ and B′′ are indicated by lines; size markers are shown on the left.
We also sedimented partially purified SF3b (Mono S fraction; Brosi et al., 1993b) in glycerol gradients. This step yielded fractions with extremely low activity in A complex formation (data not shown). Loss of activity could have been caused by dilution or inactivation of SF3b during centrifugation or by loss of another component(s) required for spliceosome assembly (see Discussion). As shown in Fig. 2 D, however, the protein content of glycerol gradient–purified SF3b with residual activity in complex formation and the purified 15S U2 snRNP was similar. Four proteins of ∼50, 130, 140, and 155 kD are present in both fractions. A polypeptide of 120 kD present in the 15S U2 snRNP is barely visible in SF3b. This protein is consistently coimmunoprecipitated with the 15S U2 snRNP, but is absent from the 17S U2 particle and does not represent SF3a120 (Nesic, D., and A. Krämer, unpublished observation). At present, it is unknown whether this protein represents a contamination or a 15S U2 snRNP-associated protein. A candidate for a specifically bound protein is Tat-SF1, the human orthologue of Cus2p which has been found associated with the U2 snRNP and interacts with SF3a (Yan et al., 1998). Two proteins of 29.5 and 28.5 kD present in the 15S U2 snRNP represent the U2 snRNP-specific A′ and B′′ proteins. These proteins are absent from SF3b fractions as confirmed by Western blotting with specific antibodies (see Fig. 3 B, lane 2; data not shown). From these results we conclude that SF3b comprises four subunits of 50, 130, 140, and 155 kD, which is further corroborated by immunoprecipitation experiments (see below). It is highly likely that the SF3b subunits correspond to the polypeptides of 53, 120, 150, and 160 kD found associated with the 17S U2 snRNP (Behrens et al., 1993b) and to SAP49, 130, 145, and 155 detected by immunoprecipitation with anti-U2 snRNP antibodies (Staknis and Reed, 1994). In accordance with the sizes determined for the U2 snRNP-associated SAPs (Bennett et al., 1992; Staknis and Reed, 1994), the SF3b subunits will be referred to as SF3b49, 130, 145, and 155.
Figure 3.

Immunoprecipitation analysis of interactions between SF3a, SF3b, and the 12S U2 snRNP. Combinations of fractions containing SF3a (Mono S), SF3b (Mono S), and the 12S U2 snRNP (Mono Q) were incubated for 15 min at 30°C (Brosi et al., 1993a) followed by precipitation with mAb66 bound to PAS. (A) Aliquots of untreated components or material bound to PAS were separated in a 10% SDS polyacrylamide gel and stained with silver. Lanes 1–3, input fractions of SF3a, SF3b, and the 12S U2 snRNP (representing one-third of the material used in the immunoprecipitations). Lane 4, control reaction of reconstituted 17S U2 snRNP bound to PAS in the absence of mAb66. Lanes 5–12, immunoprecipitation in the presence of the fractions indicated. Lane 13, the 12S U2 snRNP was treated with micrococcal nuclease (MN) before incubation with SF3a and SF3b. Lane 14, SF3b and the 12S U2 snRNP were incubated for 15 min at 30°C, and treated with micrococcal nuclease followed by addition of SF3a and immunoprecipitation. Lane 15, SF3a, SF3b, and the 12S U2 snRNP were incubated for 15 min at 30°C, and treated with micrococcal nuclease followed by immunoprecipitation with mAb66-PAS. Lane 16, control immunoprecipitation of endogenous 17S U2 snRNP. Lane 17, glycerol gradient–purified 15S U2 snRNP. The SF3a and SF3b subunits are indicated on the right of the figure by arrowheads and arrows, respectively. The protein indicated by H represents the heavy chain of mAb66 IgG and two proteins indicated by closed circles are contaminants present in the mAb66 preparation. Protein size standards are shown on the left. (B and C) Aliquots of untreated components or material bound to PAS were separated in a 13% SDS polyacrylamide gel, blotted to nitrocellulose and incubated with mAb4G3 (anti-B′′, panel B) or mAbY12 (anti-Sm, panel C; only the staining of the Sm proteins B and B′ is shown). The migration of U2B′′, Sm proteins B and B′, and the mAb66 light chain (L) is indicated. (D) RNA from HeLa cell nuclear extract (NE), the 12S U2 snRNP, the 12S U2 snRNP treated with micrococcal nuclease (MN-12S), and from the material bound to PAS was separated in a 14% polyacrylamide/8.3 M urea gel and blotted to a nylon membrane. U2 snRNA was detected with radiolabeled antisense U2 snRNA.
Interactions Required for the Assembly of the 17S U2 snRNP
By nondenaturing PAGE we have shown previously that incorporation of SF3a into the U2 snRNP requires the presence of SF3b (Brosi et al., 1993a). In these experiments weak contacts between the 12S U2 snRNP and SF3a could have been disrupted during gel electrophoresis. Moreover, interactions between SF3a and SF3b could not be tested. We have addressed these issues in an immunoprecipitation experiment. A monoclonal antibody directed against SF3a66 (mAb66; Brosi et al., 1993b) was coupled to PAS and incubated with different combinations of partially purified SF3a, SF3b, and the 12S U2 snRNP (Fig. 3 A, lanes 1–3). Immunoprecipitates were analyzed for the presence of SF3a and SF3b subunits by SDS PAGE (Fig. 3 A). Other U2 snRNP-associated proteins were detected by Western blotting with antibodies directed against the U2 snRNP-specific protein B′′ (Fig. 3 B) and the Sm proteins (Fig. 3 C). The presence of U2 snRNA was analyzed by Northern blotting with radioactively labeled antisense U2 snRNA (Fig. 3 D). mAb66 specifically precipitated the proteins of 60, 66, and 120 kD from the SF3a fraction (lane 6), whereas no polypeptides were precipitated from control reactions containing SF3b and the 12S U2 snRNP (lanes 7 and 8). When SF3a, SF3b, and the 12S U2 snRNP were combined, the SF3a subunits, the proteins found in purified SF3b, B, B′, and B′′ and U2 snRNA were precipitated by mAb66, consistent with the formation of the 17S U2 snRNP (lane 12). In contrast, from reactions in which SF3a was incubated with either SF3b or the 12S U2 snRNP, only the SF3a subunits were precipitated (lanes 9 and 10). These results confirm that both the 12S U2 snRNP and SF3b are essential for the incorporation of SF3a into the particle (Brosi et al., 1993a). Furthermore, SF3a and SF3b do not detectably associate with one another in the absence of the 12S U2 snRNP. Thus, the SF3a interaction site is generated upon binding of SF3b to the 12S U2 snRNP.
A requirement of U2 snRNA for the formation of the 17S U2 snRNP was tested by digestion of U2 snRNA with micrococcal nuclease at different stages of the assembly reaction. When the 12S U2 snRNP (Fig. 3, lane 13) or a combination of SF3b and the 12S U2 snRNP (lane 14) was digested with micrococcal nuclease before the addition of the remaining components, neither the SF3b subunits nor the 12S U2 snRNP proteins were precipitated by mAb66. However, when the digestion was performed after preincubation of SF3a, SF3b, and the 12S U2 snRNP (lane 15), i.e., after the 17S U2 snRNP had been formed, SF3a, SF3b, and 12S U2 snRNP-associated proteins were found in the mAb66 precipitates and no intact U2 snRNA was detected. These results indicate that the U2 snRNA is required for the formation of the 15S and 17S U2 snRNP particles, but once the 17S U2 snRNP is formed, SF3a, SF3b, and the 12S U2 snRNP proteins remain associated with one another in the absence of intact U2 snRNA.
Nuclease Protection Analysis Positions SF3b in the 5′ Half and SF3a in the 3′ Half of the U2 snRNP
In the Northern blot shown in Fig. 3 D the U2 snRNA was not completely digested, but distinct RNA fragments were protected from micrococcal nuclease digestion (not shown). We exploited this observation to analyze the portions in U2 snRNA that are inaccessible to micrococcal nuclease in the 12S, 15S, and 17S U2 snRNPs due to the binding of proteins. We first determined concentrations of SF3a, SF3b, and the 12S U2 snRNP that allow for the efficient assembly of the 15S and 17S U2 snRNPs without cross-contamination. Fig. 4 A shows that the 12S U2 snRNP was converted quantitatively into the 15S U2 snRNP at the highest concentration of SF3b. Efficient assembly of the 17S U2 snRNP was achieved at the highest concentration of SF3a. No 15S U2 snRNP was detected and only a minor contaminating complex that migrated slightly slower in the native gel than the 12S U2 snRNP was seen. This complex has not been analyzed further.
For the protection experiment the 12S U2 snRNP (Mono Q) and 15S and 17S U2 snRNPs assembled under the conditions established above were subjected to micrococcal nuclease digestion. RNA was isolated from untreated and micrococcal nuclease-digested samples and processed for Northern blotting with radioactively labeled oligonucleotides complementary to different regions of U2 snRNA (Fig. 4 C; see Fig. 4 B for oligonucleotides). Protected RNA fragments in the 12S U2 snRNP were only observed with oligonucleotides that are complementary to the 3′ half of U2 snRNA. The Sm oligonucleotide detected a smear of RNA at the bottom of the gel, possibly resulting from protection of the Sm-binding site by the Sm proteins (Mattaj and De Robertis, 1985). A smaller doublet and a larger fragment hybridized to oligonucleotide C′ (stem-loop IV); the larger fragment was also detected with oligonucleotide J (stem-loop III). The protection of stem-loop IV of U2 snRNA in the 12S U2 snRNP is consistent with the binding of the U2 snRNP-specific proteins A′ and B′′ to this region (Scherly et al., 1990; Bentley and Keene, 1991; Boelens et al., 1991). A protection of stem-loop III was not necessarily expected. However, Hamm et al. (1989) found that deletion of stem-loop III of U2 snRNA resulted in a destabilization of the binding of the A′ and B′′ proteins in Xenopus oocytes, suggesting that stem-loop III influences the binding of these proteins. In addition, Reveillaud et al. (1984) observed a comparable protection even at higher micrococcal nuclease concentrations than those used here.
In the 15S U2 snRNP several protected fragments were detected with oligonucleotides A (stem-loop I) and D (stem-loop IIa). The largest of these fragments also hybridized very weakly with oligonucleotide H (stem-loop IIb). (The weak hybridization of oligonucleotide H to this fragment as compared with full-length U2 snRNA could be explained if micrococcal nuclease cleaved within the region complementary to the oligonucleotide, thereby reducing the efficiency of hybridization to the remaining sequences.) Thus, binding of SF3b leads to a protection of the U2 snRNA in a region from stem-loop I to stem-loop IIa, which possibly extends into stem-loop IIb, indicating that the SF3b-binding site is located in the 5′ half of U2 snRNA. In addition, the protection in the 3′ half of U2 snRNA changed. The short doublet of fragments that hybridized to oligonucleotide C′ in the 12S U2 snRNP was no longer apparent and the fragment detected with C′ and J appeared more intense. Furthermore, oligonucleotides J and C′ hybridized to a larger doublet of fragments in the 15S U2 snRNP. A faint hybridization was also detected with oligonucleotide Sm, suggesting that the protection extended from stem-loop IV to the Sm-binding site. These changes could be caused either by direct contacts of SF3b with this region or a conformational change in the 3′ half of the U2 snRNP upon binding of SF3b that results in increased protection from micrococcal nuclease digestion by the U2A′ and B′′ and/or the Sm proteins.
Binding of SF3a to the 15S U2 snRNP resulted in qualitative changes in the protection pattern of the 3′ half of U2 snRNA as apparent after digestion of the 17S U2 particle. Oligonucleotides Sm, J, and C′ efficiently hybridized to larger RNA fragments and the smaller fragment observed in the 15S U2 snRNP disappeared. Changes in the protection of the 5′ half of U2 snRNA (oligonucleotides A and D) were mainly quantitative in nature, with the exception of the smallest fragment present in the micrococcal nuclease digest of the 15S U2 snRNP but absent from the digest of the 17S U2 snRNP. Moreover, oligonucleotide H hybridized more efficiently to the two largest fragments; thus, the protection in the 5′ half of U2 snRNA extended into the region encompassing nucleotides 70–78 of U2 snRNA. These results suggest that the major site for interaction of SF3a is the 3′ portion of the U2 snRNP. The block of the micrococcal nuclease-sensitive site between the sequences complementary to oligonucleotides Sm and J may position SF3a relatively close to the Sm proteins. The binding of SF3b is affected as well. Because incorporation of SF3a into the U2 snRNP requires both SF3b and the 12S U2 snRNP (see above) it is possible that binding of SF3b to the 5′ half of U2 snRNA is stabilized by interaction with SF3a. Major micrococcal nuclease-sensitive sites were detected only in the region between oligonucleotides H and Sm, i.e., between stem-loop IIb and the Sm-binding site. Thus together, SF3a, SF3b, A′, B′′, and the Sm proteins appear to cover the U2 snRNA in the 17S U2 snRNP almost entirely.
SF3b Represents a Distinct Globular Domain of the 15S and 17S U2 snRNPs
The morphology of the 12S and 17S U2 snRNPs has been studied previously by electron microscopy. The 12S U2 snRNP consists of two tightly attached domains (Fig. 5 B; Kastner et al., 1990). A core domain of ∼8 nm in diameter contains the Sm proteins and a slightly smaller head domain comprises the A′ and B′′ proteins. The 17S U2 snRNP displays a bipartite structure in which two globular domains of 10–12 nm each are connected by a thin RNase-sensitive filament (Fig. 5 A; Behrens et al., 1993b). Based on the morphology and additional biochemical data, it was suggested that one of these domains comprises a large part of the 17S U2-specific proteins. The second domain should contain the 12S U2 domain; however, characteristic features of the 12S U2 snRNP were not apparent in either globular structure of the 17S U2 particle. Based on the results presented above, one might expect that one of the domains is built from the SF3b subunits whereas the second domain could represent SF3a in association with the 12S domain of the U2 snRNP.
Figure 5.
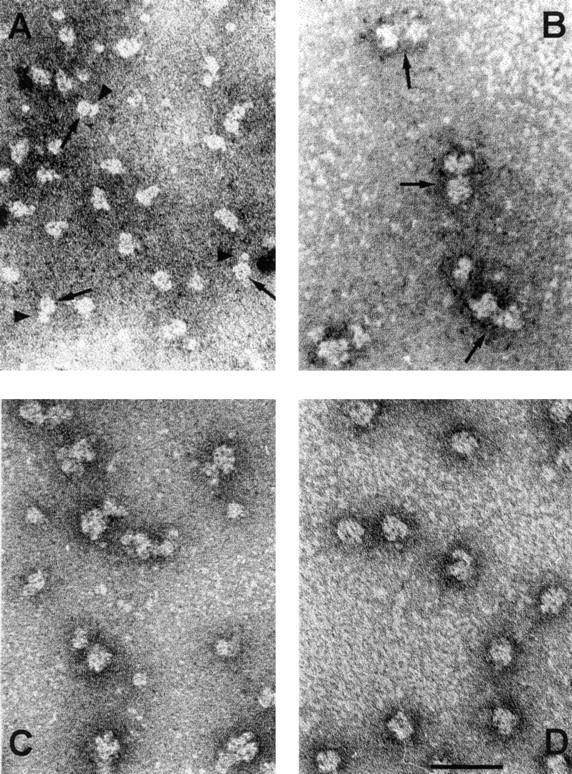
General view of the morphology of the 12S, 15S, and 17S U2 snRNPs and SF3b. Preparations of U2 snRNP particles and SF3b were negatively contrasted with uranyl formate and viewed by electron microscopy. (A) 17S U2 snRNP. Arrowheads indicate the filamentous connection between the two globular domains of the 17S U2 snRNP. (B) 12S U2 snRNP. Large arrows point to the core domain of the 12S U2 snRNP and small arrows point to the additional domain containing the A′ and B′′ proteins. (C) 15S U2 snRNP. (D) SF3b. Images of the 12S and 17S U2 snRNPs were taken from previous studies (Kastner et al., 1990; Behrens et al., 1993b).
To test this assumption, glycerol gradient–purified SF3b and 15S U2 snRNP were adsorbed onto carbon film, negatively contrasted with uranyl formate and subjected to electron microscopy. Preparations of SF3b displayed globular structures of uniform size with a diameter of 10–12 nm (Fig. 5 D). Close inspection of the shape of the particles and the accumulation of stain revealed structural details such as indentations and dot-like or elongated high intensity staining. Upon examination of ∼500 detailed views seven representative forms of SF3b that account for ∼80% of the images were distinguished. The images in Fig. 6 A (forms I and II) showed a typical dot-like high intensity stain, which was never located in the center but found in the periphery of the particle. The images are oriented such that the dot is visible in the upper third of the structure. The dot was relatively small (<2 nm) with intensive contrast (forms I and II) or larger (up to 5 nm) with an extended appearance that followed the contour of the particle (form III). A thin filamentous structure protruding from the left side was apparent in images of form III. Structures with a line of stain (oriented horizontally in the images shown) that bisects the particles were observed in forms IV–VI. The line either separated the SF3b particle into roughly two halves (form IV) or an upper third and a lower two-thirds (forms V and VI). Forms V and VI appeared as approximate mirror images with asymmetric bisecting lines that ended in an indentation on the left or right side of the particles, respectively. The last class of particles (from VII) was characterized by a single wedge-shaped indentation oriented upwards in the images shown. In summary, the SF3b protein complex has a globular appearance with a highly structured surface. The dot-like accumulation of stain most likely represents a deep hole or tunnel. The bisecting lines seen in other images could represent side views of such structure. On the other hand, the bisecting line could result from accumulation of stain in a surface furrow at the interface of two proteins.
Figure 6.
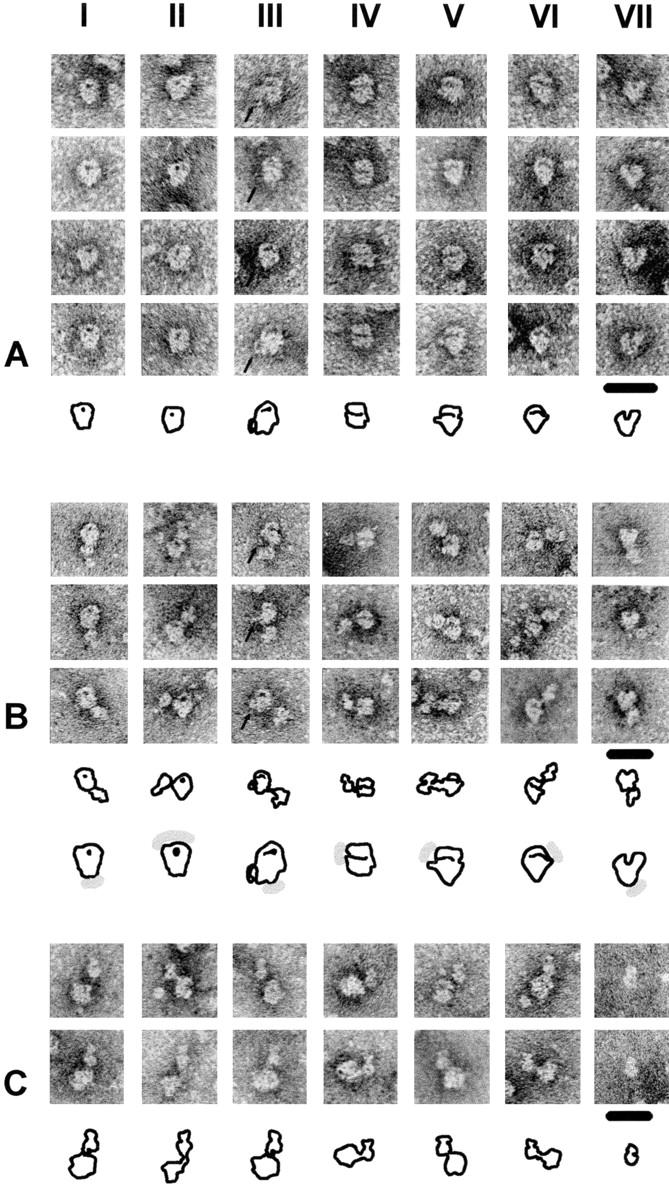
Characteristic images of SF3b and the 15S U2 snRNP. (A) Each panel (I–VII) shows four examples of the seven principal forms of SF3b with interpretative sketches. The arrows in images of form III indicate a peripheral filamentous structure that can only be observed in this type of projection. (B) Panels I–VII show three examples each of the 15S U2 snRNP with characteristic views of SF3b. The electron micrographs are oriented such that they are comparable to those of SF3b shown in A (except for form II). The arrows in images of form III indicate the filamentous structure that is typical for this type of projection. Interpretative sketches of the 15S U2 snRNPs are shown below the images. These sketches are compared with those of SF3b (A); the shaded area indicates the expected position of the 12S U2 snRNP domain. (C) Images of the 15S U2 snRNP. Panels I–VI show images of the 15S U2 snRNP in which the characteristic morphology of the 12S U2 snRNP is visible. For comparison, panel VII shows two images of the 12S U2 snRNP. The roughly round core domain of the 12S U2 snRNP with the central dot (visible only in some images) is oriented close to the SF3b domain in the 15S U2 snRNP and the domain containing A′ and B′′ is oriented upwards. Interpretative sketches are shown below the images. Bar, 20 nm.
Purified 15S U2 snRNP displayed a bipartite structure in the electron microscope (Fig. 5 C). A globular domain of ∼10–12 nm in diameter was connected by a thin filament to a second domain which was up to 12 nm long and 8 nm wide. The distance between the two domains measured up to 4 nm. The larger domain showed dot-like and elongated areas of high intensity staining, very similar to those observed with isolated SF3b (Fig. 6, A and B). In addition, the contours of different images of this domain were comparable to those of SF3b, suggesting that the large domain represents the SF3b component of the 15S U2 snRNP. The smaller domain revealed many similarities to the structure of the 12S U2 snRNP determined by Kastner et al. (1990) (Fig. 6 C, panel VII). It consisted of a main body of ∼8 nm in diameter and a closely attached additional structure of 4 nm length and 6 nm width (Fig. 6 C, panels I–VI; the smaller domain of the 15S U2 snRNP is oriented upwards). In addition, a central dot, which is a characteristic feature of the snRNP core domain (Kastner et al., 1990), was often visible in the main body.
A very close similarity was observed between the structures of the isolated 12S U2 snRNP and SF3b and the two domains in the 15S U2 snRNP (Fig. 6). Therefore, it is highly likely that the association between SF3b and the 12S U2 snRNP does not result in dramatic structural changes within either of the individual components. Considering the distance between the 12S U2 and SF3b domains visible in most images of the 15S U2 snRNP, direct communication between the individual domains is probably limited. In the 15S U2 particle a thin filament always connects SF3b to the 12S U2 snRNP core domain (Fig. 6 C). In the images shown, the connecting filament protrudes from the left (forms I−IV) or right (forms V and VI) bottom part of the core domain. The smaller structure containing the A′ and B′′ proteins is more distantly located and found opposite of the SF3b domain in most projections. Thus, direct contacts between the SF3b and A′/B′′ proteins are unlikely.
The connection to the 12S U2 domain with respect to SF3b can be described as follows. In the SF3b images that display dot-like staining the 12S U2 domain was either found close to (Fig. 6 B, form I) or distant from (form II) the stained dot. A close location was usually apparent when the stained dot was large. The 12S U2 domain was located at a distance from the extended structure of high intensity stain in projections of form III. Similar to the corresponding images of isolated SF3b (Fig. 6 A, form III), the filamentous protrusion was visible on the left side of the particle. This protrusion was located in the lower part of the particle but was clearly separated from the filament that connects the 12S U2 to the SF3b domain. In forms IV–VI, which are characterized by a bisecting line, the 12S U2 domain was usually found close to the end of the line (Fig. 6 B). Finally, in form VII the 12S U2 domain was located opposite to the wedge-like indentation.
The distance between the SF3b and 12S U2 snRNP domains varies considerably between different forms. The connecting filament is clearly visible in those 15S U2 snRNP images that display a maximal distance between the two domains. In the example shown at higher magnification in Fig. 7 A the connecting filament (with a width of ∼2 nm) exits the core snRNP domain and appears to branch out before connecting to the SF3b domain. Compared with the 15S U2 snRNP the two domains of the 17S U2 particle are closer to one another and the connecting filament appears shorter and thicker (Fig. 7 D). The typical features of the 15S U2 snRNP cannot be recognized in the 17S U2 domains, suggesting structural changes in both domains upon conversion of the 15S into the 17S U2 snRNP.
Figure 7.
Interpretation of the structure of the 15S and 17S U2 snRNP. (A and D) Electron micrographs of representative 15S and 17S U2 snRNPs with a well visible connection between the two domains. (B and E) Interpretative sketches of the 15S and 17S U2 snRNP images. (C and F) Models of the architecture of the 15S and 17S U2 snRNPs indicating the localization of the SF3b protein complex, the 12S U2 snRNP core, and A′/B′′ domains, and SF3a in the 17S U2 snRNP particle. In addition, a projection of the structure of the U2 snRNA in the RNP is shown. Sm-site, binding site of the core proteins; bp-site, region complementary to the intron branch site; cap, tri-methylated cap structure of U2 snRNA. Bar, 20 nm.
In summary, the positioning of SF3a and SF3b inferred from the nuclease protection experiment agrees well with the morphology of the different particles visualized in the electron microscope. In the 12S U2 snRNP only the 3′ portion of U2 snRNA is protected, consistent with the binding of the Sm and A′/B′′ proteins to the Sm-binding site and stem-loop IV, respectively. Protection of the 5′ half upon binding of SF3b corresponds to the appearance of a novel structural domain in the 15S U2 snRNP that is very similar in size and shape to isolated SF3b (Fig. 7, A–C). Binding of SF3a induces an extended protection in the 3′ half of U2 snRNA, which is reflected in a different appearance of the 12S domain in the 17S U2 snRNP. In addition, the shape of the SF3b portion of the U2 snRNP is changed in the 17S U2 snRNP, which correlates with a more efficient protection from micrococcal nuclease digestion in the 5′ half of the U2 snRNA (Fig. 7, D–F).
Discussion
Comparison of polypeptides that are associated with the purified 17S U2 snRNP and SF3a or precipitated from spliceosomes with anti-U2 snRNP antibodies has previously suggested that SF3b consists of at least four subunits of 49, 130, 145, and 155 kD (Behrens et al., 1993b; Brosi et al., 1993a; Staknis and Reed, 1994). Here we have demonstrated that SF3b purified from HeLa cell nuclear extracts comprises four subunits of the expected sizes. Polypeptides of identical apparent molecular mass are highly enriched in glycerol gradient fractions of the 15S U2 snRNP and these fractions substitute for the 12S U2 snRNP and SF3b activity in presplicing complex formation. Moreover, only the presumptive SF3b subunits are precipitated from purified or reconstituted 17S U2 snRNP with an antibody directed against SF3a66 in addition to the SF3a subunits, 12S U2 snRNP-associated proteins, and U2 snRNA.
Together, the SF3a and SF3b subunits account for seven of the 17S U2 snRNP-specific proteins (Behrens et al., 1993b). Similarly, anti-U2 snRNP antibodies precipitated seven polypeptides in addition to the 12S U2 snRNP proteins from a fraction enriched in U2 snRNP (Staknis and Reed, 1994). This raises questions about the 35 and 92-kD polypeptides found associated with the purified 17S U2 snRNP by Behrens et al. (1993b). It is possible that these proteins are even less tightly bound to the U2 snRNP than SF3a and SF3b (Behrens et al., 1993b; Brosi et al., 1993a) and dissociate from the remainder of the particle during purification or immunoprecipitation procedures. The 35- and 92-kD proteins have not been detected in mAb66 immunoprecipitates of the 17S U2 snRNP, suggesting that they are not required for SF3a, SF3b, and the 12S U2 snRNP to remain stably associated with one another. However, this does not exclude the possibility that these proteins function in the assembly of the 17S U2 snRNP or the prespliceosome. Loss of the 35- and 92-kD proteins from the 15S U2 snRNP or SF3b fractions during glycerol-gradient sedimentation could, for example, explain the low activity of these fractions in presplicing complex formation.
Brosi et al. (1993a) previously established that the 17S U2 snRNP is formed in vitro by stepwise interactions of SF3b and SF3a with the U2 snRNP. The results presented here strongly suggest that the assembly pathway is initiated by contacts of SF3b with the 5′ half of U2 snRNA. First, in the 12S U2 snRNP only the 3′ terminal stem-loops and the Sm-binding site are protected from micrococcal nuclease digestion. In contrast, the region extending from stem-loop I to stem-loop IIa (and to a lesser extent stem-loop IIb) of U2 snRNA, which is fully accessible to micrococcal nuclease digestion in the 12S U2 snRNP, is protected in the reconstituted 15S U2 particle. Second, treatment of the 12S U2 snRNP with micrococcal nuclease before incubation with SF3a and SF3b prevents the assembly of the 17S U2 snRNP, indicating that at least part of the U2 snRNA is required for stable contacts. Moreover, visualization by electron microscopy demonstrated that the 15S U2 particle contains two structurally distinct domains. The smaller domain exhibits the typical features of the single domain of the 12S U2 snRNP, whereas the characteristic structure of SF3b is conserved in the larger domain. The domains are connected by a thin filament, which most likely represents U2 snRNA, in analogy to the 17S U2 snRNP where a corresponding connection is RNase-sensitive (Behrens et al., 1993b).
The interpretation of these data is in agreement with and extends results of previous studies. (Although the following results were obtained with the 17S U2 snRNP, we believe that they reflect the situation of the 15S U2 particle, i.e., the binding of SF3b, because the differences in micrococcal nuclease protection in the 5′ half of U2 snRNA in the 15S and 17S U2 snRNP were quantitative rather than qualitative in nature [see Fig. 4 C].) Temsamani et al. (1991) reconstituted the U2 snRNP in a cytoplasmic S100 extract from synthetic U2 snRNA. Upon incubation in nuclear extract the particles (which exhibited a high mobility in native polyacrylamide gels) were converted to low mobility complexes that almost certainly correspond to the 17S U2 snRNP. Oligonucleotide-directed RNase H cleavage of the 5′ end of U2 snRNA and the branch site interaction sequence before incubation in the nuclear extract abolished 17S U2 snRNP formation. Behrens et al. (1993b) reported that the 5′ end of the U2 snRNA was cleaved more efficiently by RNase H in the presence of a complementary oligonucleotide in the 17S than in the 12S U2 snRNP. In addition, nucleotides C8 and C10 were more accessible to chemical modification in the 17S than in the 12S U2 snRNP. Together these data suggested a structural change in the 5′ portion of U2 snRNA upon binding of the 17S U2-specific proteins that results in a (partial) melting of stem I and thus an increase in the potential for intermolecular base pairing. Furthermore, strengthening of stem-loop I by mutation inhibited splicing in a mammalian extract (Wu and Manley, 1992), which could reflect an interference with the binding of SF3b. U2 snRNA sequences on both sides of stem I engage in base pairing interactions with U6 snRNA (for review see Staley and Guthrie, 1998). Therefore, it is intriguing to speculate that one or more SF3b subunits are directly involved in presenting this region of U2 to U6 snRNA for the formation of helix Ia/Ib which is an essential element of the catalytic center of the spliceosome both in mammals and yeast.
The protection of the region encompassing stem-loop IIa in the 15S (and 17S) U2 snRNP is in good agreement with the result that chemical modification of nucleotides C40, G42, and C45 is less efficient in the 17S than the 12S U2 snRNP (Behrens et al., 1993b). Moreover, deletion of nucleotides 46–49 prevents the assembly of the 17S U2 snRNP (Temsamani et al., 1991). In addition, a mutation in Cus1p, the S. cerevisiae homologue of SF3b145 (Gozani et al., 1996; Wells et al., 1996), suppresses mutations in yeast U2 snRNA that correspond to nucleotides 39–44, 52, and 61 of human U2 snRNA (Wells et al., 1996; Yan and Ares, 1996). Based on these results and the evolutionary conservation of SF3b145 and Cus1p (Gozani et al., 1996; Wells et al., 1996) we assume that the protection of stem-loop IIa reflects the binding of SF3b145. Preliminary evidence indicates that SF3b145, SF3b49, and one of the other large SF3b subunits cross-link to U2 snRNA (Gröning, K., and A. Krämer, unpublished observation). Moreover, Cus1p is apparently associated with an RNA-binding activity (see Igel et al., 1998). Thus, it is highly likely that SF3b145 binds directly to stem-loop IIa of U2 snRNA.
Whether or not the branch site interaction sequence of U2 snRNA is involved in interactions with the SF3b subunits is unclear. We have not detected any micrococcal nuclease-sensitive sites between stem-loops I and IIa in the 15S or 17S U2 snRNPs. However, this region is equally well accessible to oligonucleotide-targeted RNase H digestion and chemical modification in the 12S and 17S U2 snRNPs (Behrens et al., 1993b). This discrepancy could be explained by a different availability of the branch site interaction region to oligonucleotide-targeted RNase H or micrococcal nuclease digestion or to relatively mild conditions used for nuclease treatment in this report. The observation that U2 snRNAs carrying mutations within the branch site interaction region fail to assemble into the 17S U2 snRNP (Temsamani et al., 1991) argues in favor of a contribution of these sequences to the assembly of the active U2 snRNP. Clearly, this issue needs further clarification.
During the formation of the 15S U2 snRNP we also observed changes in the protection pattern in the 12S domain, in that the protection of stem-loops III and IV is more efficient and a weak protection of the Sm-binding site is apparent. Equivalent changes in the structure of the subdomains of the 15S U2 snRNP compared with isolated SF3b or the 12S U2 snRNP are not visible in the electron microscope. In fact, the domain containing the A′ and B′′ proteins appears to be located at a distance from the SF3b domain; therefore, we believe that direct contacts between these proteins are limited. Although we cannot rule out interactions between SF3b and the Sm proteins that may not be resolved in the electron microscope, we favor the notion that binding of SF3b to the 5′ half of U2 snRNA results in a subtle conformational change in the 12S domain.
SF3a does not stably interact with either SF3b or the 12S U2 snRNP alone (Brosi et al., 1993a; data presented here), indicating that both components contribute to the binding site for SF3a. Consistent with this, in the 17S U2 snRNP we observed an extended protection from micrococcal nuclease digestion that encompasses the Sm binding site and stem-loops III and IV of U2 snRNA as well as a more efficient protection of the 5′ half. In addition, we observed differences in the morphology of the 17S U2 snRNP in the electron microscope. Like the 15S U2 snRNP, the 17S particle is composed of two domains, both of which approximately correspond in size to the SF3b domain of the 15S U2 snRNP. However, structural details of neither the SF3b nor 12S domain are apparent in the two globular structures of the 17S U2 particle. Together, the results from micrococcal nuclease protection and electron microscopic analysis suggest that the main binding site for SF3a is the 12S domain of the U2 snRNP, thus contributing to the increase in size. The chemical modification pattern in the 3′ half of U2 snRNA was identical in the 12S and 17S U2 snRNP (Behrens et al., 1993b), which was interpreted to mean that most 17S U2 snRNP-specific proteins associate with the 5′ portion of the U2 snRNP. On the other hand, it is possible that binding of SF3a does not occur by tight protein–RNA interactions or induce substantial structural changes in the RNA, but may rely mainly on protein–protein interactions. Good candidates for interacting proteins are the A′ and/or B′′ proteins, because deletion of their binding site (nucleotides 154–167) prevents the assembly of the 17S U2 snRNP (Temsamani et al., 1991). Thus, the integrity of the 3′ end of U2 snRNA and/or the presence of the A′ and B′′ proteins may be necessary for binding of SF3a. An interaction of SF3a with these proteins (or this region of U2 snRNA) may also explain why U2 snRNAs carrying mutations in loop IV were inactive in mammalian splicing (Wu and Manley, 1992). Contacts with the Sm proteins are also possible, given that no micrococcal nuclease sensitive sites were detected between stem-loop IV and the Sm-binding site. In fact, the major micrococcal nuclease-sensitive site between sequences complementary to oligonucleotides Sm and J in the 15S U2 snRNP is completely protected in the 17S U2 particle, which locates SF3a close to the Sm proteins.
The increased efficiency of micrococcal nuclease protection in the 5′ half of U2 snRNA observed upon binding of SF3a can be interpreted by stabilization of the interaction of SF3b with the U2 snRNA. This could either be the result of a structural change induced by binding of SF3a to the 3′ portion of U2 snRNP or by direct contacts between the SF3a and SF3b subunits. The idea of direct contacts between these proteins is supported by the result that the SF3a and SF3b subunits as well as the B′′, B, and B′ proteins are coprecipitated after micrococcal nuclease digestion of the reconstituted 17S U2 snRNP. Evidence for interactions between SF3a and SF3b or U2 snRNA comes from studies in S. cerevisiae. First, mutations in homologues of the SF3a subunits (Prp9p, Prp11p, and Prp21p) are synthetic lethal with a number of mutations in stem-loops IIa and IIb of U2 snRNA and the region between the branch site interaction sequence and stem-loop IIa (Ruby et al., 1993; Wells and Ares, 1994; Yan and Ares, 1996). The same pattern of phenotype was observed with all three mutant proteins, which suggested that they interacted with the U2 snRNP as a functional unit. A candidate for interaction is Cus1p, the homologue of SF3b145. Mutations in Cus1p suppressed mutations in U2 snRNA downstream of the branch site interaction sequence and in stem-loop IIa (Wells et al., 1996; Yan and Ares, 1996). The synthetic lethality between mutations in the SF3a subunits and stem IIb (Wells and Ares, 1994) may also explain the weak but apparent protection within this region. Second, extra copies of wild-type Cus1p partially suppressed the growth defect of prp11-1, but not prp9-1 or prp21-1, suggesting interactions between SF3b145 and SF3a66 (Wells et al., 1996). Third, in a yeast two-hybrid screen the gene product of Yml049c, which represents the yeast orthologue of SF3b130 (Caspary et al., 1999; Krämer, A., unpublished data), was found as a partner of Prp9p (SF3a60; Fromont-Racine et al., 1997).
Given these possible interactions, it may be surprising that the 17S U2 snRNP consists of two distinct globular domains in the electron microscope (Fig. 7 D; Behrens et al., 1993b). On the one hand, this discrepancy could be explained if interactions between SF3a and SF3b were indirect and merely resulted in a structural change of the SF3b domain, which may be reflected in a loss of the typical features of this domain upon formation of the 17S U2 snRNP. On the other hand, interactions between SF3a and SF3b could be relatively weak and thus not be visible in the electron microscope. Another possibility is that protein–protein contacts involve structures that are beyond the detection limit of the electron microscope.
In summary, the combined data from biochemical and electron microscopic analyses can be incorporated into a model in which SF3b initiates the assembly of the active U2 snRNP by interactions with the 5′ half of U2 snRNA, which positions this splicing factor close to U2 snRNA sequences that are essential for the catalysis of splicing. SF3a then associates with the 3′ portion of the U2 snRNP and, most likely, directly interacts with one or more of the SF3b subunits.
Acknowledgments
We thank Angus Lamond for generously supplying oligoribonucleotides and the human U2 construct, Walther van Venrooij for mAb4G3, Joan Steitz for the Y12 hybridoma cell line, and Dobrila Nesic and Jeffrey Patton for comments on the manuscript. B. Kastner gratefully acknowledges the excellent technical assistance of Beate Rückert.
This work was supported by grants from the Schweizerischer Nationalfonds and the Canton of Geneva to A. Krämer, by the Boehringer Ingelheim Fonds to K. Gröning, and by a grant from the Deutsche Forschungsgemeinschaft (Ka 805/2) to B. Kastner.
Abbreviations used in this paper
- PAS
protein A–Sepharose
- SAP
spliceosome-associated protein
- SF
splicing factor
- snRNA
small nuclear RNA
- snRNP
small nuclear ribonucleoprotein particle
Footnotes
Patric Grüter's present address is 5 rue Cherbuliez, CH-1207 Genève, Switzerland. Karsten Gröning's present address is Nikolaus-Otto-Str. 8, D-59557 Lippstadt, Germany.
References
- Ares M, Jr, Igel AH. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 1990;4:2132–2145. doi: 10.1101/gad.4.12a.2132. [DOI] [PubMed] [Google Scholar]
- Arning S, Grüter P, Bilbe G, Krämer A. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA. 1996;2:794–810. [PMC free article] [PubMed] [Google Scholar]
- Barabino SM, Sproat BS, Lamond AI. Antisense probes targeted to an internal domain in U2 snRNP specifically inhibit the second step of pre-mRNA splicing. Nucleic Acids Res. 1992;20:4457–4464. doi: 10.1093/nar/20.17.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens SE, Galisson F, Legrain P, Lührmann R. Evidence that the 60-kD protein of 17S U2 small nuclear ribonucleoprotein is immunologically and functionally related to the yeast PRP9 splicing factor and is required for the efficient formation of prespliceosomes. Proc Natl Acad Sci USA. 1993a;90:8229–8233. doi: 10.1073/pnas.90.17.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens SE, Tyc K, Kastner B, Reichelt J, Lührmann R. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol Cell Biol. 1993b;13:307–319. doi: 10.1128/mcb.13.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M, Michaud S, Kingston J, Reed R. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 1992;6:1986–2000. doi: 10.1101/gad.6.10.1986. [DOI] [PubMed] [Google Scholar]
- Bentley RC, Keene JD. Recognition of U1 and U2 small nuclear RNAs can be altered by a 5-amino-acid segment in the U2 small nuclear ribonucleoprotein particle (snRNP) B′′ protein and through interactions with U2 snRNP-A′ protein. Mol Cell Biol. 1991;11:1829–1839. doi: 10.1128/mcb.11.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelens W, Scherly D, Beijer RP, Jansen EJ, Dathan NA, Mattaj IW, van Venrooij WJ. A weak interaction between the U2A′ protein and U2 snRNA helps to stabilize their complex with the U2B′′ protein. Nucleic Acids Res. 1991;19:455–460. doi: 10.1093/nar/19.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant C, Krol A, Ebel J-P, Lazar E, Haendler B, Jacob M. U2 RNA shares a structural domain with U1, U4, and U5 snRNAs. EMBO (Eur Mol Biol Org) J. 1982;1:1259–1265. doi: 10.1002/j.1460-2075.1982.tb00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosi R, Gröning K, Behrens S-E, Lührmann R, Krämer A. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science. 1993a;262:102–105. doi: 10.1126/science.8211112. [DOI] [PubMed] [Google Scholar]
- Brosi R, Hauri HP, Krämer A. Separation of splicing factor SF3 into two components and purification of SF3a activity. J Biol Chem. 1993b;268:17640–17646. [PubMed] [Google Scholar]
- Caspary, F., A. Shevchenko, M. Wilm, and B. Séraphin. 1999. Partial purification of the yeast U2 snRNP reveals novel yeast pre-mRNA splicing factor required for pre-spliceosome assembly. EMBO (Eur. Mol. Biol. Org.) J. In press. [DOI] [PMC free article] [PubMed]
- Fromont-Racine M, Rain J-C, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- Gozani O, Potashkin J, Reed R. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol. 1998;18:4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J, Dathan NA, Mattaj IW. Functional analysis of mutant XenopusU2 snRNAs. Cell. 1989;59:159–169. doi: 10.1016/0092-8674(89)90878-7. [DOI] [PubMed] [Google Scholar]
- Hartshorne T, Agabian N. A new U2 RNA secondary structure provided by phylogenetic analysis of trypanosomatid U2 RNAs. Genes Dev. 1990;4:2121–2131. doi: 10.1101/gad.4.12a.2121. [DOI] [PubMed] [Google Scholar]
- Igel H, Wells S, Perriman R, Ares M., Jr Conservation of structure and subunit interactions in yeast homologues of splicing factor 3b (SF3b) subunits. RNA. 1998;4:1–10. [PMC free article] [PubMed] [Google Scholar]
- Kastner, B. 1998. Purification and electron microscopy of spliceosomal snRNPs. In Springer Lab Manual: RNP Particles, Splicing and Autoimmune Diseases. J. Schenkel, editor. Springer Verlag, Heidelberg. 95–140.
- Kastner B, Lührmann R. Electron microscopy of U1 small nuclear ribonucleoprotein particles: shape of the particle and position of the 5′ RNA terminus. EMBO (Eur Mol Biol Org) J. 1989;8:227–286. doi: 10.1002/j.1460-2075.1989.tb03374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B, Bach M, Lührmann R. Electron microscopy of small nuclear ribonucleoprotein (snRNP) particles U2 and U5: evidence for a common structure-determining principle in the major U snRNP family. Proc Natl Acad Sci USA. 1990;87:1710–1714. doi: 10.1073/pnas.87.5.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A. Pre-splicing complex formation requires two proteins and U2 snRNP. Genes Dev. 1988;2:1155–1167. doi: 10.1101/gad.2.9.1155. [DOI] [PubMed] [Google Scholar]
- Krämer A. Purification of splicing factor SF1, a heat-stable protein that functions in the assembly of a pre-splicing complex. Mol Cell Biol. 1992;12:4545–4552. doi: 10.1128/mcb.12.10.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A. The structure and function of proteins involved in nuclear pre–mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Krämer A, Utans U. Three protein factors (SF1, SF3 and U2AF) function in pre-splicing complex formation in addition to snRNPs. EMBO (Eur Mol Biol Org) J. 1991;10:1503–1509. doi: 10.1002/j.1460-2075.1991.tb07670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A, Frick M, Keller W. Separation of multiple components of HeLa cell nuclear extracts required for pre-messenger RNA splicing. J Biol Chem. 1987;262:17630–17640. [PubMed] [Google Scholar]
- Lamond AI, Sproat B, Ryder U, Hamm J. Probing the structure and function of U2 snRNP with antisense oligonucleotides made of 2′-OMe RNA. Cell. 1989;58:383–390. doi: 10.1016/0092-8674(89)90852-0. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- Mattaj IW, De Robertis EM. Nuclear segregation of U2 snRNA requires binding of specific snRNP proteins. Cell. 1985;40:111–118. doi: 10.1016/0092-8674(85)90314-9. [DOI] [PubMed] [Google Scholar]
- Moore, M.J., C.C. Query, and P.A. Sharp. 1993. Splicing of precursors to mRNA by the spliceosome. In The RNA World. R. Gesteland and J. Atkins, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 303–357.
- Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B′′-U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- Reed R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- Reveillaud I, Lelay-Taha MN, Sri-Widada J, Brunel C, Jeanteur P. Mg2+ induces a sharp and reversible transition in U1 and U2 small nuclear ribonucleoprotein configurations. Mol Cell Biol. 1984;4:1890–1899. doi: 10.1128/mcb.4.9.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby SW, Chang T-H, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- Scherly D, Boelens W, Dathan NA, van Venrooij WJ, Mattaj IW. Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B′′ and their cognate RNAs. Nature. 1990;345:502–506. doi: 10.1038/345502a0. [DOI] [PubMed] [Google Scholar]
- Staknis D, Reed R. Direct interactions between pre-mRNA and six U2 small nuclear ribonucleoproteins during spliceosome assembly. Mol Cell Biol. 1994;14:2994–3005. doi: 10.1128/mcb.14.5.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Temsamani J, Rhoadhouse M, Pederson T. The U2 small nuclear ribonucleoprotein particle associates with nuclear factors in a pre-mRNA independent reaction. J Biol Chem. 1991;266:20356–20362. [PubMed] [Google Scholar]
- Utans U, Behrens S-E, Lührmann R, Kole R, Krämer A. A splicing factor that is inactivated during in vivo heat shock is functionally equivalent to the [U4/U6.U5] triple snRNP-specific proteins. Genes Dev. 1992;6:631–641. doi: 10.1101/gad.6.4.631. [DOI] [PubMed] [Google Scholar]
- Wells SE, Ares M., Jr Interactions between highly conserved U2 small nuclear RNA structures and Prp5p, Prp9p, Prp11p, and Prp21p proteins are required to ensure integrity of the U2 small nuclear ribonucleoprotein in Saccharomyces cerevisiae. . Mol Cell Biol. 1994;14:6337–6349. doi: 10.1128/mcb.14.9.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Neville M, Haynes M, Wang J, Igel H, Ares M., Jr Cus1, a suppressor of cold-sensitive U2 snRNA mutations, is a novel yeast splicing factor homologous to human SAP145. Genes Dev. 1996;10:220–232. doi: 10.1101/gad.10.2.220. [DOI] [PubMed] [Google Scholar]
- Will, C., and R. Lührmann. 1997. SnRNP structure and function. In Eukaryotic mRNA Processing. A. Krainer, editor. IRL Press, Oxford. 130–173.
- Wu J, Manley JL. Multiple functional domains of human U2 small nuclear RNA: strengthening conserved stem I can block splicing. Mol Cell Biol. 1992;12:5464–5473. doi: 10.1128/mcb.12.12.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Ares M., Jr Invariant U2 RNA sequences bordering the branchpoint recognition region are essential for interaction with yeast SF3a and SF3b subunits. Mol Cell Biol. 1996;16:818–828. doi: 10.1128/mcb.16.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Perriman R, Igel H, Howe KJ, Neville M, Ares M., Jr CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol Cell Biol. 1998;18:5000–5009. doi: 10.1128/mcb.18.9.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.-T., E.C. Scharl, C.M. Smith, and J.A. Steitz. 1999. The growing world of small nuclear ribonucleoproteins. In The RNA World. 2nd edition. R. Gesteland, T. Cech, and J. Atkins, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 487–524.




