Abstract
A collection of yeast strains surviving with mutant 5S RNA has been constructed. The mutant strains presented alterations of the nucleolar structure, with less granular component, and a delocalization of the 25S rRNA throughout the nucleoplasm. The 5S RNA mutations affected helix I and resulted in decreased amounts of stable 5S RNA and of the ribosomal 60S subunits. The shortage of 60S subunits was due to a specific defect in the processing of the 27SB precursor RNA that gives rise to the mature 25S and 5.8S rRNA. The processing rate of the 27SB pre-rRNA was specifically delayed, whereas the 27SA and 20S pre-rRNA were processed at a normal rate. The defect was partially corrected by increasing the amount of mutant 5S RNA. We propose that the 5S RNA is recruited by the pre-60S particle and that its recruitment is necessary for the efficient processing of the 27SB RNA precursor. Such a mechanism could ensure that all newly formed mature 60S subunits contain stoichiometric amounts of the three rRNA components.
Keywords: 5S ribosomal RNA, pre-rRNA, ribosomes, yeasts, cell nucleolus
Eukaryotic ribosome biogenesis requires the coordinate assembly of 70–80 ribosomal proteins with four ribosomal RNAs to yield the two mature ribosomal subunits. It takes place in the nucleolus where the large 35S rRNA precursor encoding the 18S, 5.8S, and 25S rRNAs (the sizes are from Saccharomyces cerevisiae) is synthesized by RNA polymerase I (for review see Woolford and Warner, 1991). The fourth RNA, 5S RNA, is encoded by independent genes transcribed by RNA polymerase III. In most eukaryotic organisms the 5S RNA genes are located outside the nucleolus (Hadjiolov, 1985), with the exception of S. cerevisiae in which the 5S RNA genes are located in the nucleolus between the large ribosomal transcription units and transcribed in the opposite orientation (Warner, 1989). The synthesis of the 5S ribosomal RNA by a distinct form of RNA polymerase raises the question of its coordinated production with the other rRNA components. As yet, there is no indication that coregulation exists at the level of transcription, even in S. cerevisiae with its special form of ribosomal gene organization.
5S ribosomal RNA, before its incorporation into the ribosome, can bind to different proteins and be translocated in different cellular compartments. In eukaryotes, newly synthesized 5S RNA interacts transiently with the protein La before its binding to the ribosomal protein rpL5 (Steitz et al., 1988; Guddat et al., 1990; Yoo and Wolin, 1994). In Xenopus oocytes, because of the extraordinary demand for ribosome production, 5S RNA is produced in large quantities and is complexed, in addition to rpL5, with other proteins including the transcription factor TFIIIA (Picard and Wegnez, 1979; Picard et al., 1980). The 5S/ RNP particles are exported to the cytoplasm where they are stored. During vitellogenesis, rpL5 replaces TFIIIA in association with 5S RNA, which migrates back to the nucleus where it is directed to the nucleolus for incorporation into ribosomes (Rudt and Pieler, 1996). The transit of the 5S RNA through the cytoplasm is likely due to the necessity to store large amounts of 5S RNA during the early stages of the Xenopus development. In somatic cells, the nuclear rpL5–5S particle is probably directed immediately to the nucleolus and does not transit through the cytoplasm (Allison et al., 1995; Michael and Dreyfuss, 1996). Similarly, in S. cerevisiae, where 5S RNA is synthesized in the nucleolus, the RNA is probably directly incorporated into ribosomes and does not transit through the cytoplasm, as suggested by the fact that the rpL5–5S complex is found mostly in the nucleus upon the disruption of the large ribosomal subunit assembly in rpL16 mutants (Deshmukh et al., 1993).
Ribosome assembly occurs simultaneously with the maturation of the 35S precursor rRNA that includes methylation, pseudouridylation, and cleavage reactions. The cleavage steps of the rRNA have been particularly well studied in S. cerevisiae and serve as landmarks to describe the whole pathway, but the step at which the 5S–rpL5 particle is incorporated to the ribosome is not well known (Venema and Tollervey, 1995). In HeLa cells, 5S RNA can be found in ribosomal precursor particles that have not yet undergone the processing step that makes 5.8S rRNA (Warner and Soeiro, 1967). Similarly, indirect methods using yeast-labeled cells have suggested that 5S RNA is present in pre-60S particles, precursors of the large subunits, and may already be present in the less matured pre-90S particles that contain 35S pre-rRNA (Trapman et al., 1976). These two studies suggest that 5S RNA incorporation is an early event of the ribosome assembly/maturation pathway. On the other hand, the fact that a 5S–rpL5 complex could be easily dissociated from the large 60S ribosomal subunit without disrupting it, suggested that the 5S– rpL5 complex was added at a late step on the surface of the subunit (Blobel, 1971; Nazar et al., 1979). In vitro ribosome reconstitution experiments using proteins and mature rRNA purified from Escherichia coli did not clarify the situation as the 5S RNA could be incorporated at any step, but was, nevertheless, necessary at a late step to form an active ribosomal particle (Dohme and Nierhaus, 1976). The location of 5S RNA in mature ribosomal particles has been the subject of intensive studies in E. coli (Bogdanov et al., 1995). The 5S RNA is located in the central protuberance of the large subunit, in close proximity to the small ribosomal subunit. Cross-linking experiments suggest a direct interaction between residues in the D-loop of the 5S RNA and at least two regions of 23S rRNA that are of major functional importance: the GTPase-associated region, involved in EF-G binding, and the peptidyltransferase center (Dontsova et al., 1994; Sergiev et al., 1998).
In the present study, by using mutant 5S RNA we show that 5S RNA is incorporated at a late step of preribosomal RNA maturation and that the recruitment of the 5S RNA is necessary for the efficient processing of the 27SB pre-rRNA. The participation of 5S RNA in the pre-rRNA processing pathway would ensure that stoichiometric amounts of the three rRNAs of the large subunit are produced. In addition, we found that alterations of the 60S subunit formation are accompanied by modifications of the nuclear localization of these subunits and of the nucleolar granular compartment.
Materials and Methods
Strains, Plasmids, and Media
Yeast genetic techniques and media were as described by Ausubel et al. (1987). The wild-type strain YRW1 was MATa can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 ade2-1 tfc2::LEU2 pJA230 (URA3, TFC2, and CEN3) (Archambault et al., 1992). The YSC14 strain was derived from YRW1; it survived without the TFIIIA factor because of the presence of a multicopy plasmid (pRSC3) containing a 5S RNA gene under the control of the RNA polymerase III RPR1 promoter as described in Camier et al. (1995). The RPR1-5S gene was mutagenized as described below to create a collection of pOK plasmids. Mutant cells carrying these new plasmids were obtained by transforming YRW1 and selecting the cells that survived without pJA230. The mutant strains contained the following plasmids: YOK69 contained pOK1 plasmid; YOK71 (pOK3); YOK72 (pOK4); YOK73 (pOK5); YOK 74 (pOK6); YOK76 (pOK7); and YOK77 (pOK8).
Mutagenesis of the RPR1-5S Gene
The four nucleotides (GGCA) at the 5′ end of the mature 5S RNA in YSC14 strain were mutagenized according to the two-step PCR method described by Higuchi et al. (1988). The sequence at the junction between the RPR1 promoter and the 5S DNA was GATTGGCA GGTT; the italic sequence corresponds to the 5′ end of the wild-type 5S RNA and the underlined sequence corresponds to the sequence that was mutagenized. Two overlapping PCR fragments, one corresponding to the 5′ part of the RPR1-5S (from the 5′ end of the gene to downstream of the mutagenesis site), and the other to the 3′ part of the RPR1-5S (from upstream of the mutagenesis site to the 3′ end of the gene) were synthesized. These two PCR products were obtained using mutagenic primers and 5′ and 3′ primers containing a BamHI restriction site. Mutagenic PCR products were used as templates for a third PCR reaction to synthesize a complete RPR1-5S gene with mutations on both strands. The mutagenic primers for the synthesis of the 5′ fragments were: d(TGCGATT(T/C)(T/C)CT GGTTGCGGCCATAT) for the preparation of plasmids pOK1, pOK3, pOK5, and pOK6, or d(TGCGATTGGTTGCGGCCATAT) for the deletion of the GGCA sequence (plasmid pOK7). For the synthesis of the 3′ PCR fragments, the primers were d(GCAACC AG(A/G)(A/G)AATCGCAGCTCCC) for pOK1, 4, 5, and 6, or d(GCAACCAATCGCAGCTCCC) for the preparation of pOK7. The third PCR combined 1 pmol of each 5′ and 3′ PCR products and the 5′ and 3′ primers containing the BamHI restriction site. The resulting PCR fragments were digested with BamHI. The pOK plasmids derived from the pRSC3 plasmid by exchange of the BamHI fragment containing the wild-type RPR1-5S gene with the BamHI fragment containing a mutated RPR1-5S gene. The nature of the mutations was determined by sequencing. The pOK3 and pOK8 plasmids contained two repeats of the BamHI fragment from pOK4 and pOK1, respectively.
RNA Analysis
RNAs were extracted as described by Schmitt et al. (1990) with some modifications. Cells corresponding to 10 ml of culture at an OD600nm 0.4–0.6 were resuspended in 250 μl of 50 mM sodium acetate, pH 5.3, 10 mM EDTA, plus 25 μl of 10% SDS, mixed with 150 μl of acid-washed glass beads and 250 μl of phenol, vortexed vigorously, and incubated for 5 min at 65°C with intermittent vortexing. RNAs were precipitated with 0.3 M sodium acetate, pH 5.3, and 3 vol of ethanol at −20°C overnight. Small RNAs were separated on a denaturing 8% polyacrylamide gel and visualized by ethidium bromide staining.
Quantification of Small RNA.
RNAs ranging from 1 to 5 μg were analyzed on 8% denaturing polyacrylamide gels and visualized by ethidium bromide staining. The gel picture was registered with an enhanced analysis system (E.A.S.Y.; Herolab) and the amount of each species was measured by densitometry using the NIH Image program. The curves for each RNA species were used to measure the average ratio between the amount of two RNA species (5S RNA and tRNA and 5.8S rRNA and 5S RNA). These experiments were reproduced at least three times for each strain.
Analysis of High Molecular Weight rRNAs.
High molecular weight rRNAs were analyzed on 1.2% agarose gels after denaturation with glyoxal and dimethylsulfoxide as described in Ausubel et al. (1987). After a run of 14 h at 55 V in 10 mM NaHPO4, pH 7.0, RNAs were transferred to a positively charged nylon membrane (Boehringer Mannheim) and cross-linked with UV. rRNAs were visualized by staining the membrane with 0.04% methylene blue in 0.5 M sodium acetate and 3H-labeled rRNAs were detected by spraying the membrane with EN3HANCE® (DuPont) and autoradiographed at −70°C.
Analysis of the 5′ and 3′ Ends of 5S RNA.
The 5′ end of 5S RNAs was identified by primer extension using the MMLV reverse transcriptase and the internal 5S RNA oligonucleotide GTCAGGCTCTTACCAGCTTAAC as described in Jacquet et al. (1989). The identification of the 3′ end of the mutant 5S RNAs was carried out by sequencing using either chemical modifications or ribonuclease digestions as described below. The mutant 5S RNAs were isolated from denaturing gels and 3′ end was labeled with [32P]pCp using T4 RNA ligase. The two labeled bands were separated on an 8% sequencing polyacrylamide gel, isolated, and analyzed separately. The chemical sequence was carried out using standard reactions as described in England et al. (1980) using diethylpyrocarbonate for the modification of A, dimethylsulfate for G, aqueous hydrazine for U, and 3 M NaCl in anhydrous hydrazine for C. After the modifications of the bases, the RNAs were digested with aniline and separated on a sequencing gel. The ribonuclease sequencing of the 3′ end–labeled 5S RNAs was performed as described in Donis-Keller et al. (1977) with modifications: 5S RNAs were partially digested with RNase T1 (that cleaves next to G residues) or RNase A (that cleaves after C and U). The products resulting from the two digestions were analyzed on a 6% denaturing polyacrylamide gel. The results obtained with the two methods were identical.
Analysis of rRNA Extremities Resulting from Processing in ITS2.
The 5′ end of the 25S rRNA was analyzed by primer extension and the 3′ end of the 7S pre-rRNA precursor was analyzed by an RNase protection assay. Primer extension analysis was done according to the method described by Hong et al. (1997). The 5′ end 32 P-labeled oligonucleotide for the analysis of the 5′ end of the 25S rRNA (cleavage at site C1) was TACTAAGGCAATCCCGGTTGG (Hong et al., 1997). The RNase protection assay was carried out using a 35S-labeled transcript complementary to the ITS2 sequence, prepared with T7 RNA polymerase. The T7 template corresponds to a DNA fragment encoding the major part of the ITS2 sequence (nucleotides 1–215), amplified by PCR, using oligonucleotides CCTTCTCAAACATTCTGTTTGGTAG and GCCTAGACGCTCTCTTCTTA as primers and cloned under a T7 RNA polymerase promoter. Total yeast RNA was hybridized with T7 RNA transcripts and the resulting RNA duplexes were digested with a mixture of RNase A and RNase T1 and analyzed on a denaturing gel (Ausubel et al., 1987).
Ribosome Analysis
Yeast polyribosomes were fractionated by sucrose gradients (Baim et al., 1985). Cells were grown in 100 ml of yeast extract/peptone/glucose (YPD)1 medium at an OD600nm 0.6–0.8. 50 μg/ml of cycloheximide was added to the culture, which was immediately transferred to ice. After 10 min, cells were collected by centrifugation and washed with lysis buffer containing 10 mM Tris-Cl, pH 7.4, 100 mM NaCl, 30 mM MgCl2, 50 μg/ml of cycloheximide, and 200 μg/ml of heparin. They were resuspended in two volumes of lysis buffer and one volume of acid-washed glass beads. The cells were disrupted by vortexing the suspension eight times for 15 s with a 30 s period of cooling on ice after each vortexing step. The samples were spun at 5,000 rpm for 5 min and the supernatants were centrifuged at 10,000 rpm for 10 min. A portion of the supernatant corresponding to 10 OD units at 260 nm was layered over an 11-ml linear sucrose gradient (7–47% wt/vol) containing 50 mM Tris-acetate, pH 7.6, 50 mM NH4Cl, 12 mM MgCl2, and 1 mM DTT. The sucrose gradients were centrifuged at 39,000 rpm for 2.5 h in an SW41 rotor (Beckman) and analyzed at 254 nm using a density gradient fractionator (model 640; Isco).
Ribosomal subunit quantification was done according to the method described by Moritz et al. (1991). Cycloheximide, heparin, and MgCl2 were omitted from all buffers, and ribosomal subunits were separated on high salt (0.5 M KCl) sucrose velocity gradients.
Pulse-Chase Experiments
For pulse-chase labeling of pre-rRNA, 10 ml of cells growing in a synthetic medium without methionine at an OD600nm 0.3–0.4 were labeled with 70 μCi/ml of [methyl-3H]methionine for 1 min at 30°C. Unlabeled methionine was added to a final concentration of 5 mM and incubation was continued for various periods of time as indicated in the figures. Samples (2 ml) were centrifuged at room temperature. Cell pellets were immediately frozen in a dry ice/ethanol bath and stored at −20°C until RNA extraction and analysis as described above. The time of harvesting and freezing of the cells (1 min) was included in the chase times.
Overexpression of RPL5 and LHP1 Genes
Overproduction of the rpL5 protein was obtained by transforming the wild-type and mutant strains with the multicopy plasmid JW2601 carrying the RPL5 gene under the control of its own promoter. JW2601 was a gift of Dr. J.L. Woolford (Carnegie-Mellon University, Pittsburgh, PA), and it was derived from YEp352 (URA3, 2 μm).
The open reading frame encoding the Lhp1 protein was a gift of Dr. S.G. Clarkson (Département de Génétique et de Microbiologie, Genève, Switzerland) (LinMarq and Clarkson, 1998). The LHP1 gene was cloned into the multicopy vector pFL44L (URA3, 2 μm) under the control of its own promoter (pFL44-LHP1) (Arrebola, R., personal communication). As a control, the wild-type and mutant strains were transformed with pFL44L (URA3, 2 μm) without insert.
Strains were grown under selective conditions in a synthetic medium without uracil (−Ura) at an OD600nm 0.2, diluted 10–103-fold and spotted on −Ura plates. The plates were incubated at 30°C or 37°C for various periods of time as indicated in the figure legend. Photographs were scanned and arranged with Adobe Photoshop to have a homogenous black background.
Electron Microscopy
The preparation of cells for electron microscopy and freeze electron substitution was as described by Leger-Silvestre et al. (1997). For all observations, the grids were contrasted with saturated aqueous uranyl acetate alone or combined with lead citrate (freeze substitution grids) and imaged in a JEOL-1200EX electron microscope operating at 80 kV.
The grids for immunoelectron microscopy were pretreated for 15 min with PBS buffer, pH 7.6, containing 2% BSA and incubated for 2 h at room temperature with anti-Nop1 mAbs (1:10) provided by Dr. J. Aris (University of Florida, Gainesville, FL). The grids were washed for 30 min with PBS buffer containing 1% BSA and transferred for 1 h to colloidal gold-conjugate goat anti–mouse diluted 1:80 in the same buffer. After incubation, the grids were washed for 20 min in PBS buffer and 10 min in water, and air-dried. Controls were performed using gold-labeled antiserum alone. No labeling was detected on these grids.
For in situ detection of ribosomal RNAs, a 35S pre-rRNA probe was synthesized by nick-translation in the presence of digoxigenin-11dUTP (Boehringer Mannheim) from a plasmid containing an rDNA unit. The probes for the 25S and 18S rRNA were synthesized by random priming using a DIG-High Prime kit (Boehringer Mannheim) from isolated restriction DNA fragments: the 3,533-bp KpnI-HindIII fragment for the 25S rRNA probe, or the 1,954-bp EcoRI-EcoRI fragment for the 18S rRNA probe. The probes were diluted (final, 2 ng/ml) in a hybridization medium containing 50% formamide and 10% dextran sulfate in 1× SSC. 20 μl of probe was added on the grids and denatured for 5 min at 75°C. Hybridization was carried out overnight in a wet chamber at 37°C. The grids were washed three times for 5 min in 50% formamide/2× SSC at 45°C, three times for 5 min in 0.1× SSC at 60°C, and a few seconds in 4× SSC at room temperature. Nonspecific immunological sites were blocked for 30 min with 5% BSA in 4× SSC. The probe was detected with an antidigoxigenin antibody gold-conjugate diluted 1:20 in 4× SSC. The grids were washed three times for 5 min each with 4× SSC and air-dried. For the specific detection of the ribosomal DNA, grids were pretreated before hybridization with DNase-free RNase at 100 μg/ml in 2× SSC for 1 h at 37°C. Grids were washed three times for 5 min in 2× SSC at room temperature and dehydrated through an ethanol series of 70, 90, and 100%.
Results
5S Mutations Result in Less Stable 5S RNA and Decreased Growth Rate
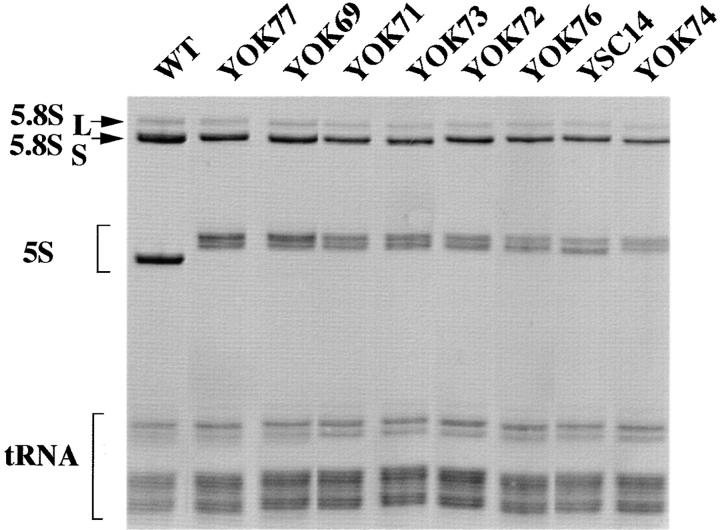
A collection of yeast strains surviving with mutated 5S RNA has been constructed using a genetic system described previously (Camier et al., 1995). In these strains, transcription of the endogenous 5S RNA genes was abolished by inactivating the gene coding for the transcription factor TFIIIA. The cells survive with 5S RNA expressed from a chimeric gene that does not require TFIIIA. The RPR1 promoter fused to the 5S RNA gene is transcribed by RNA polymerase III to give an RPR1-5S RNA precursor that is processed into 5S RNA molecules with extended 5′ and 3′ ends. The 5′ extremity contains four additional 5′ nucleotides GGCA (Table I, strain YSC14) and the 3′ extremity is heterogeneous, with 1–3 additional residues (data not shown). In wild-type 5S RNA, the 5′ and 3′ ends of the molecules are paired in a stem called helix I; the additional nucleotides could be part of an extended helix I and protected from degradation during RNA processing (Moore, 1996). To remove the extra nucleotides, mutations, which could potentially decrease interactions between the 5′ and 3′ ends of the 5S RNA, were introduced in the sequence upstream of the 5S RNA gene (Table I, strains YOK69, YOK73, YOK72, and YOK74). A deletion mutant was also constructed that removed the last four nucleotides of the RPR1 sequence at the junction with the 5S RNA gene (Table I, strain YOK76). All the mutations gave rise to functional 5S RNA, since the cells could grow in the absence of TFIIIA, at 30°C (Table I). Small RNAs were prepared from all mutants and analyzed on a denaturing polyacrylamide gel (Fig. 1). No mutation restored the normal size of the 5S RNA. The 5S RNA of all mutants migrated as two RNA bands of 125- and 127-nt long. The analysis of their 5′ and 3′ extremities by primer extension and RNA sequencing, respectively, showed that all the mutated 5S RNA still contained the 5′ and 3′ nucleotide extensions (data not shown).
Table I.
Characteristics of Yeast Strains Surviving with 5S rRNA Mutants
| Strain | 5′ sequence* | Number of RPR1-5S repeats | Doubling time at 30°C | Growth at 37°C | tRNA/5S ratio‡ | 5.8S/5S ratio‡ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| h | ||||||||||||
| WT | − | − | 1.5 | ++++ | 2.1 ± 0.4 | 1.2 ± 0.1 | ||||||
| YSC14§ | GGCA | 1 | 3.8 | − | 5.8 ± 0.5 | 1.4 ± 0.1 | ||||||
| YOK69 | CCCT | 1 | 2.3 | ++ | 3.4 ± 0.4 | 1.1 ± 0.1 | ||||||
| YOK73 | TCCT | 1 | 3.4 | − | 4.7 ± 0.6 | 1.1 ± 0.1 | ||||||
| YOK72 | CTCT | 1 | 3.4 | − | 4.8 ± 0.5 | 1.1 ± 0.2 | ||||||
| YOK76‖ | GATT | 1 | 3.7 | − | 6.1 ± 0.2 | 1.3 ± 0.1 | ||||||
| YOK74 | TTCT | 1 | 5.3 | − | 7.2 ± 0.5 | 1.4 ± 0.1 | ||||||
| YOK77 | CCCT | 2 | 2 | +++ | 3.1 ± 0.2 | 1.1 ± 0.1 | ||||||
| YOK71 | CTCT | 2 | 3.3 | + | 4.1 ± 0.4 | 1.1 ± 0.1 |
Sequence immediately upstream of the 5S rRNA gene in the RPR1-5S construct.
5.8S rRNA, 5S rRNA, and tRNA were separated by electrophoresis in polyacrylamide denaturing gels and their amounts were determined by scanning the gels after ethidium bromide staining (see Materials and Methods). The results are from at least three independent experiments.
The original 5S rRNA mutant derived from the RPR1-5S construct with nonmutagenized 5′ sequence.
Deletion mutant.
Figure 1.
Analysis of the small stable RNA in 5S RNA mutants. Wild-type and mutant strains were grown in YPD medium to an OD600nm of 0.5 at 30°C. Total RNA was extracted, 5 μg were analyzed on an 8% polyacrylamide/8 M urea gel, and visualized by ethidium bromide staining. 5.8S rRNA, 5S RNA (mutant and wild-type), and tRNA are indicated by arrows or brackets. The picture corresponds to a negative exposure.
The cells surviving with mutant 5S RNA displayed very different growth rates both at 30°C and 37°C and presented defects in cell and colony morphology (Table I and Fig. 2). The cells had a hyperpolarized growth and did not separate well after division, which resulted in filaments. These defects were particularly pronounced in slow growing mutants (Fig. 2, strains YSC14 and YOK74). The fact that the growth rate at 30°C and 37°C, and that the cell morphology could be improved by varying the mutations in the RPR1-5S gene indicated that these defects were due to the mutated 5S RNA rather than to the absence of TFIIIA.
Figure 2.
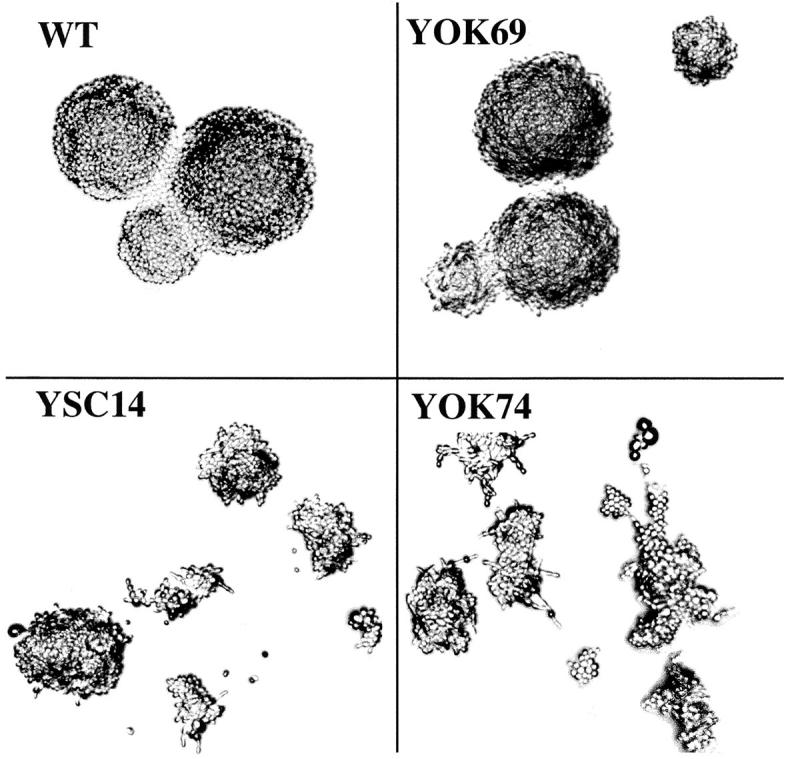
Morphology defects in 5S RNA mutants. Single cells of wild-type (WT), YOK69, YSC14, and YOK74 strains were isolated on YPD medium and incubated at 30°C for 1 d. Developing clones were observed using a Zeiss microscope with a 10× objective lens and Nomarski DIC optics.
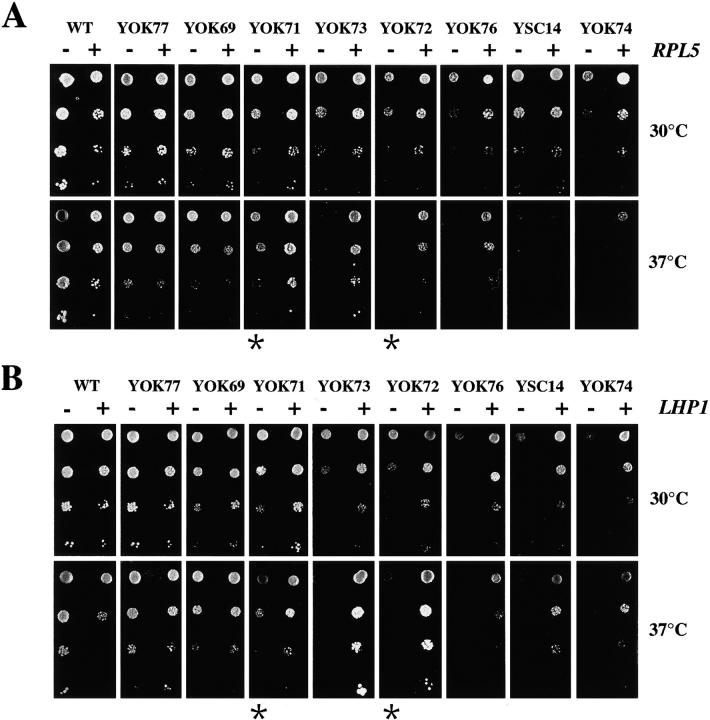
Duplication of the RPR1-5S insert in two different mutants resulted in a growth rate improvement both at 30°C and 37°C (Table I and Fig. 3, compare YOK69 and YOK77, or YOK72 and YOK71, indicated by asterisks in Fig. 3). This result suggested that the different growth rates reflected in part the amount of stable 5S RNA in the cell. The ratio between the amounts of tRNA and 5S RNA was determined by densitometry and found to be ∼2 in the wild-type cells and between 3 and >7 in the mutants (Table I). The decrease in the amount of 5S RNA correlated well with the decrease in the cell growth rate. Apparently, mutated 5S RNA accumulated at different levels depending on the sequence located at the 5′ end of the molecule, and the different levels of 5S RNA determined the growth rate of the mutant cells.
Figure 3.
Overexpression of RPL5 or LHP1 improves the growth of 5S RNA mutants. Dilutions of cultures from wild-type and mutant strains transformed with the RLP5 (A) or the LHP1 gene (B) were spotted on −Ura plates and incubated at 30°C or 37°C. (A) WT, YOK77, YOK69, and YOK71 strains were incubated at 30°C for 2 d or at 37°C for 3 d. YOK73, YOK72, YOK76, YSC14, and YOK74 strains were incubated at 30°C or 37°C for 6 d. (B) All strains were incubated for 2 d at 30°C. At 37°C, WT, YOK77, and YOK69 strains were incubated for 3 d, YOK71 strain for 4 d, and YOK 73, YOK72, YOK76, YSC14, and YOK74 strains for 6 d. Asterisks (*) indicate two strains containing the same mutated 5S rDNA but present at a different copy number.
Overproduction of Two 5S RNA Binding Proteins, rpL5 and Lhp1p, Partially Rescues 5S RNA Defects
Woolford and co-workers have shown that free 5S RNA was not stable in vivo and that its stabilization required the binding of ribosomal protein rpL5 (Deshmukh et al., 1993). One of the major binding sites of rpL5 to 5S RNA is helix I; the presence of extra nucleotides adjacent to that helix could perturb the binding of rpL5 and, therefore, result in decreased amount of stable 5S RNA. We found that in the slow growing mutants (YOK74, YOK76, YOK72, YOK73, and YOK71), the overexpression of the gene encoding rpL5 improved the growth rate at 30°C and 37°C (Fig. 3). The amount of 5S RNA, analyzed in the most altered mutant (YOK74), was increased upon RPL5 overexpression, which is likely the cause of the improved growth rate (Table II). Similar experiments were conducted with the LHP1 gene encoding the yeast homologue of the La protein, known to interact with the 3′ end of RNA polymerase III transcripts (Stefano, 1984). La overexpression also led to an improved growth rate and resulted in an increased amount of stable 5S RNA (Fig. 3, Table II). In this case, the nature of the mutant 5S RNA was modified, and the 5S RNA now migrates as a single band, corresponding to the slow migrating band of the doublet on denaturing polyacrylamide gels (data not shown). The 5′ end of 5S RNA, as determined by primer extension, was unchanged, suggesting that the increase in size was due to the protection of the extra 3′ nucleotides by bound Lhp1p. The increase in the amount of 5S RNA by overxpressing the LHP1 gene could reflect the existence of an in vivo pool of 5S RNA-Lhp1p. Alternatively, the slow migrating RNA band generated by LHP1 overexpression could have a higher affinity for rpL5 than the fastest migrating band and, therefore, be better stabilized.
Table II.
Overexpression of RPL5 or LHP1 Increases Both the Amount of 5S rRNA and the Growth Rate
| Strain | Doubling time at 30°C | Growth at 37°C | tRNA/5S ratio | 5.8S/5S ratio | ||||
|---|---|---|---|---|---|---|---|---|
| h | ||||||||
| YOK74 + vector | 5.5 | − | 7.2 ± 0.5 | 1.4 ± 0.1 | ||||
| YOK74 + RPL5-2μ | 4 | + | 5.8 ± 0.5 | 1.4 ± 0.1 | ||||
| YOK74 + LHP1-2μ | 2.9 | ++ | 4.6 ± 0.4 | 1.3 ± 0.1 |
Mutations in 5S RNA Result in Decreased Synthesis of the Ribosomal 60S Subunit
When analyzing the small RNA species present in 5S mutants, we noticed that whereas the ratio between tRNA and 5S RNA increased severalfold in slow growing cells, the ratio between 5.8S rRNA and 5S RNA remained practically constant, similar to the wild-type value (Table I). This result indicated that the amount of 5.8S rRNA decreased concomitantly with the amount of 5S RNA. Since 5S RNA and 5.8S rRNA are part of the same large 60S ribosomal subunit, we analyzed the content in ribosomal subunits in the mutant strains. Polyribosomes and free ribosomal particles were separated on sucrose velocity gradients from wild-type cells and two different 5S mutants (Fig. 4).
Figure 4.
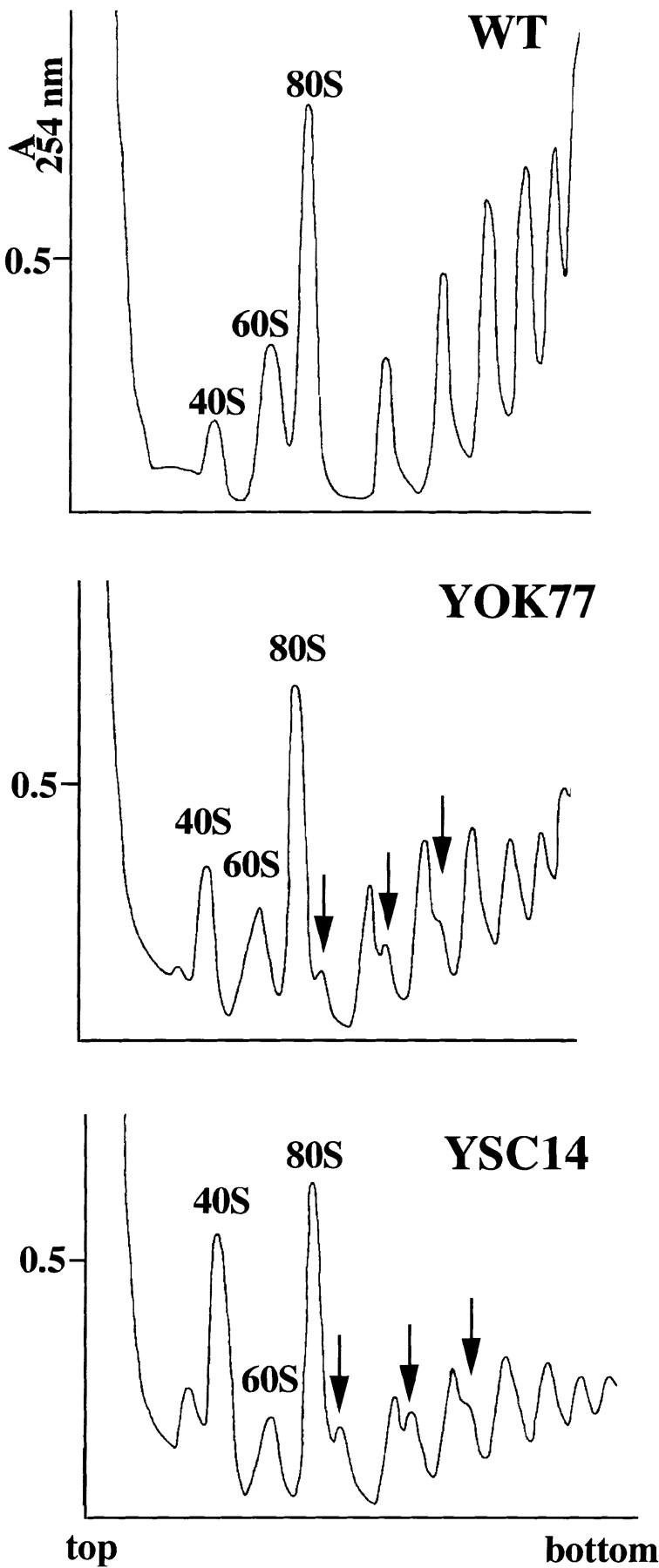
Mutations in 5S RNA lead to a shortage in 60S ribosomal subunits and to the accumulation of halfmer polysomes. WT, YOK77, and YSC14 strains were grown in YPD medium to an OD600nm 0.6–0.8 at 30°C. Cell extracts were resolved on 7–47% sucrose gradients. The gradients were analyzed by continuous monitoring at A254nm. The peaks of free 40S and 60S ribosomal subunits and 80S ribosomes (free couples and monosomes) are indicated. Halfmer polysomes are indicated by arrows.
Mutant ribosomal profiles showed marked alterations that were more accentuated in the slow growth mutant, YSC14, than in a better growing mutant, YOK77. First, the total amount of polyribosomes was significantly reduced in both mutants, suggesting a less efficient protein synthesis rate compared with wild-type cells. Second, the ratio between the amount of free 60S and free 40S ribosomal subunits was reversed in the mutants because of the accumulation of 40S subunits and a reduction in 60S particles. The imbalance between the 60S and 40S subunits was also evidenced by the presence of additional peaks sedimenting more quickly than the 80S monoribosomes or the polyribosomes. These peaks (Fig. 4, arrows) are likely to represent halfmers polyribosomes that contain mRNA associated with an integral number of ribosomes plus a stalled 48S preinitiation complex. The presence of halfmers can be an indication of a decreased amount of 60S subunits. To confirm the deficiency in 60S subunits, ribosomal profiles were run under conditions that dissociate the ribosomes. The ratio between the 60S and 40S subunits, which is ∼2 for wild-type cells, decreased to 1 in YSC14 cells (data not shown). Therefore, the decreased amount of 5S RNA observed in the mutant cells is accompanied by a decrease in the total amount of 60S subunits. This decrease very likely alters the protein synthesis rate that results in decreased growth rate.
The Level of 5S RNA Affects Pre-rRNA Processing
The shortage in mature 60S subunits could reflect the misincorporation of the mutant 5S RNA in the pre-60S particles, which in turn would lead to the accumulation of partially assembled 60S subunits. This possibility was particularly interesting since in all the mutant profiles analyzed, we reproducibly observed an additional peak sedimenting as a 35S particle, absent in the wild-type cells and whose intensity was particularly high in the YSC14 strain. However, the analysis of the RNA content of the peak showed that it did not contain any ribosomal RNA, but instead an RNA species of 2600 nt that corresponded to the 20S single-strand viral RNA (data not shown) (Wejksnora and Haber, 1978; Matsumoto et al., 1990; Rodriguez-Cousino et al., 1991). We did not identify any other peak that could represent the partially assembled 60S subunit. The analysis of the 5S RNA content in the 60S subunits showed that the different mutant 5S RNA species were all incorporated in the subunits, which shows that the decrease in 60S was not due to the selective incorporation of only some of these species.
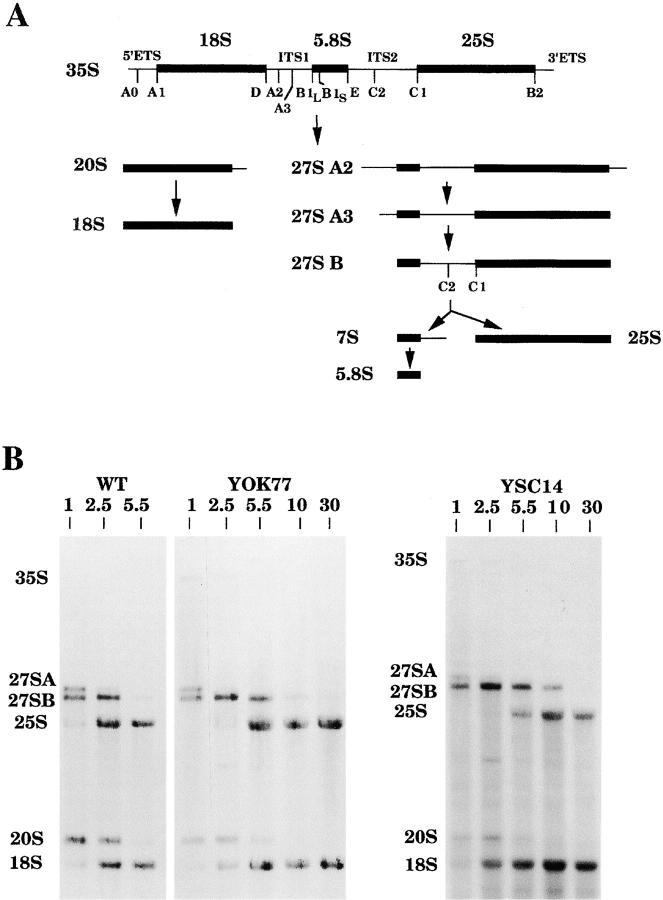
We explored the possibility that 5S RNA could play a role in the biogenesis of the 60S subunit and analyzed the preribosomal RNA processing steps in the mutants (Fig. 5). The fate of the ribosomal precursors was followed in the WT strain and the YOK77 and YSC14 mutants by pulse-chase experiments after labeling with methyl- [3H]methionine. The methylation patterns were quantified and the efficiencies of maturation of the different species were measured (Table III). The efficiency of maturation of the 20S pre-RNA into 18S mature rRNA was similar in the mutants and the WT, ∼70% after a 2.5-min chase and 90% after a 5.5-min chase (Table III, ε18). In sharp contrast, the accumulation of the mature 25S rRNA took much longer in the mutant cells compared with the WT, with a more pronounced defect in the YSC14 strain: after a 2.5-min chase no mature 25S rRNA was detected in YSC14 cells, only 15% in YOK77 cells, whereas the maturation was 70% complete in the WT cells (Table III, ε25). The delay in the formation of the mature 25S rRNA was due to the delay of one specific step corresponding to the maturation of the 27SB precursor. The processing of the 27SA pre-RNA was not detectably altered since after a 2.5-min chase, most of the 27SA pre-RNA had been converted into 27SB pre-RNA in the three cell types (Table III, εB). No 32S pre-RNA or 35S pre-RNA was detected in the experiments, which suggests that the maturation of these species was not significantly altered in the mutants.
Figure 5.
Alteration of the 27SB pre-rRNA processing in 5S RNA mutants. (A) The S. cerevisiae pre-rRNA processing pathway. The sequences corresponding to the mature 18S, 5.8S, and 25S rRNAs are represented as black bars, embedded in the 5′ and 3′ external transcribed spacers and in the ITS1 and ITS2. Processing of the primary transcript starts at site A0 in the 5′ external transcribed spacers. The resulting 33S pre-rRNA is processed at site A1 and A2 to give rise, successively, to the 32S pre-rRNA (not represented) and to the 20S and 27SA2 precursors. Cleavage at site A2 separates the pre-rRNA for the small and the large ribosomal subunits. The 20S precursor is cleaved at site D to yield mature 18S rRNA. Cleavage of 27SA2 pre-rRNA at site A3 is rapidly followed by digestion to site B1S generating the 27SBS precursor. The mature 25S rRNA and the 7S pre-rRNA are released from 27SBS after cleavages at sites C1 and C2. The 7S pre-rRNA digestion generates the mature 3′ end of the 5.8S rRNA. For simplicity only the major processing pathway from 27SA2 pre-rRNA to the 5.8SS and the 25S rRNA is shown, an alternative pathway generates the minor 5.8SL rRNA, which is 7–9 nt longer at its 5′ end. (B) The processing of the 27SB pre-rRNA is slowed down in 5S mutants. WT, YOK77, and YSC14 strains were pulse-labeled for 1 min with methyl- [3H]methionine and chased for 1, 2.5, 5.5, 10, or 30 min with an excess of unlabeled methionine. Total RNA was extracted, denatured with glyoxal, run on a 1.2% agarose gel, transferred to a nylon membrane, and visualized by fluorography. The specific activities of the 3H-labeled RNA for WT, YOK77, and YSC14 were ∼7,000, 2,000,and 850 cpm/μg, respectively. 10,000 cpm were loaded on the first gel (WT and YOK77) and 8,000 cpm on the second gel (YSC14). Autoradiography was 10 times longer for the second gel than for the first one. The position of the pre-rRNAs and mature rRNA are indicated.
Table III.
Maturation Efficiencies (ε) of 27SA RNA into 27SB RNA (εB), 27S (A+ B) RNA into 25S RNA (ε25), and 20S RNA into 18S RNA (ε18)
| Time | 1 min | 2.5 min | 5.5 min | 10 min | 30 min | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain ε | εB | ε25 | ε18 | εB | ε25 | ε18 | εB | ε25 | ε18 | εB | ε25 | ε18 | εB | ε25 | ε18 | |||||||||||||||
| WT | 70 | 25 | 35 | 95 | 70 | 75 | 95 | 95 | 95 | — | 100 | 95 | — | 100 | 100 | |||||||||||||||
| YOK77 | 60 | 15 | 35 | 100 | 15 | 60 | 100 | 70 | 90 | 100 | 90 | 100 | — | 100 | 100 | |||||||||||||||
| YSC14 | 75 | 0 | 60 | 100 | 0 | 70 | 100 | 50 | 90 | 100 | 75 | 100 | — | 100 | 100 | |||||||||||||||
This table is a quantification of pulse-chase experiments realized as in Fig. 5, from two sets of experiments. The time points refer to the chase times. The maturation efficiency was calculated as the percentage of matured species relative to the amount of unmatured and matured product. εB, amount of 27SB RNA/amount of (27SA + 27SB) RNA × 100; ε25, amount of 25S RNA/amount of (27S + 25S) RNA × 100; ε18, amount of 18S RNA/amount of (20S + 18S) RNA × 100.
Similar pulse-chase experiments performed in the presence of [3H]uracil instead of [3H]methionine also showed a specific delay in the processing of the 27SB precursor, which was again more pronounced in the YSC14 strain as compared with the YOK77 strain (data not shown). Therefore, the delay observed was not due to a delay in the methylation reaction. The processing of the 27SB precursor consists in the removal of a part of the internal transcribed spacer 2 (ITS2) between sites C1 and C2, through a still unknown mechanism (Venema and Tollervey, 1995). We analyzed the C1 and C2 cleavage sites by primer extension and RNase protection experiments, respectively. No difference was found between the 5S mutants and the wild-type cells (data not shown). Therefore, the 5S RNA mutations did not alter the cleavage sites in the precursor but had a strong effect on the processing reaction rate. The decreased processing rate of the RNA of the large subunit is likely at the origin of the decreased growth rate. There is indeed a good correlation between the processing rate and the growth rate, as shown for example in the YSC14 mutant, which grows four times more slowly than the WT, and processes the 27SB pre-RNA four times more slowly (half complete maturation of the 25S rRNA is observed after a 6-min chase compared with 1.5-min in the WT).
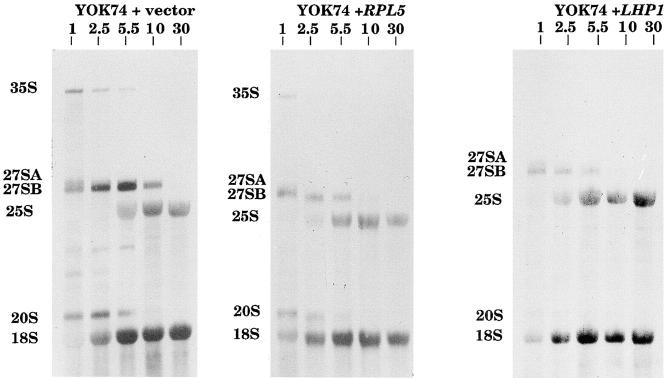
As described above, one main consequence of the 5S mutations was to decrease the amount of stable 5S RNA in the cells. If the low processing rate in the mutants originated at least in part from the lower amount of stable 5S RNA, increasing the amount of 5S RNA by overexpression of the RPL5 or LHP1 genes should speed up the processing reaction. We analyzed the ribosomal processing in the strain YOK74 (the most affected mutant) transformed with either gene (Fig. 6). When either rpL5 or Lhp1p was overproduced, the processing rate of the 27SB precursor was improved, since it was 67–90% complete after a 5.5-min chase compared with only half complete in the absence of overproduced protein. We conclude that the amount of stable 5S RNA in the cell influenced the rate of processing of the 27SB precursor, which suggests that the 5S RNA is recruited at this step of the processing pathway and that its recruitment is necessary for the processing.
Figure 6.
Overexpression of RPL5 or LHP1 in YOK74 improves the processing of the 27SB precursor. YOK74 mutant cells transformed with a vector sequence, the RPL5 gene, or the LHP1 gene in multicopy were pulse-labeled for 1 min with [methyl-3H]methionine and chased for 1, 2.5, 5.5, 10, and 30 min with an excess of unlabeled methionine. Total RNA was extracted, denatured with glyoxal, run on a 1.2% agarose gel, transferred to a nylon membrane, and visualized by fluorography. The specific activities of the 3H-labeled RNA for YOK74 + vector, + RPL5, or + LHP1 were ∼1,500, 12,000, or 12,200 cpm/ μg, respectively. Equal amounts of radioactivity were loaded on each gel corresponding to 22,000 cpm for YOK74 + vector, 140,000 cpm for YOK74, or 120,000 cpm for YOK74 + LHP1. Autoradiography was for 12 d for YOK74 + vector, 9 d for YOK74 + RPL5, or 7 d for YOK74 + LHP1. The positions of the pre-rRNAs and mature rRNAs are indicated.
Nucleolar Alterations and Abnormal Nuclear Localization of the 60S Subunit RNA
In S. cerevisiae, 5S RNA genes are present in the nucleolus. The absence of endogenous chromosomal 5S transcription and the defects in 60S subunit formation prompted us to analyze the structure of the nucleolus in these mutants. The small size of yeast and its tough cell wall considerably hindered precise structural analysis of its nucleolus by fluorescence and electron microscopy. Recently, by combining cryofixation and cryosubstitution, it has been possible to identify distinct substructures similar to the components of nucleoli of higher eukaryotes: the fibrillar centers, the dense fibrillar component, and the granular component (Leger-Silvestre et al., 1997). These techniques coupled with conventional in situ techniques at the electron microscopy level were applied to the analysis of some of the 5S mutants described here.
In all mutants, a nucleolus could be morphologically identified. Its structure was similar for all the mutants but differed from a wild-type nucleolus since it appeared less dense, mostly fibrillar with less granular component (Fig. 7 A). These characteristics were more pronounced in slow growing mutants. To appreciate the degree of disorganization of the nucleolus in the 5S mutants, we analyzed the localization of a nucleolar marker, the Nop1 protein, and the ribosomal RNA. The immunolocalization of Nop1p was normal and corresponded to the dense region of the nucleus referred to as the nucleolus (Fig. 7 B). Therefore, the nucleolus of the mutants was not strongly disorganized. The ribosomal RNA was detected by in situ hybridization with digoxigenin-labeled probes complementary either to the 35S rRNA precursor (Fig. 8 A) or to the mature 25S rRNA (Fig. 8 B). The results were identical for both probes but differed depending on the strain analyzed. For the wild-type strain, the nuclear labeling was, as expected, concentrated in the nucleolus. No significant labeling appeared in the nucleoplasm except for a few gold particles in the nuclear pores, which likely corresponded to preribosomal particles exported to the cytoplasm. For one of the most altered strains, YSC14, the precursor or the 25S rRNA was totally dispersed throughout the nucleus. For YOK69, one of the fast growing mutants, the situation was intermediate, with the nucleolus being mostly labeled plus regions at the periphery of the nucleoplasm. An accumulation of gold particles near the nuclear envelope was observed with both probes for all the 5S mutants analyzed. When similar experiments were conducted with a probe hybridizing to the 18S rRNA of the small ribosomal subunit, its localization was exclusively nucleolar for both wild-type and 5S mutant cells (data not shown). The ribosomal RNA mislocalized in the nucleus of 5S mutants corresponded, therefore, to the RNA of the large ribosomal subunit. The nuclear localization of the small subunit RNA remained unaffected.
Figure 7.
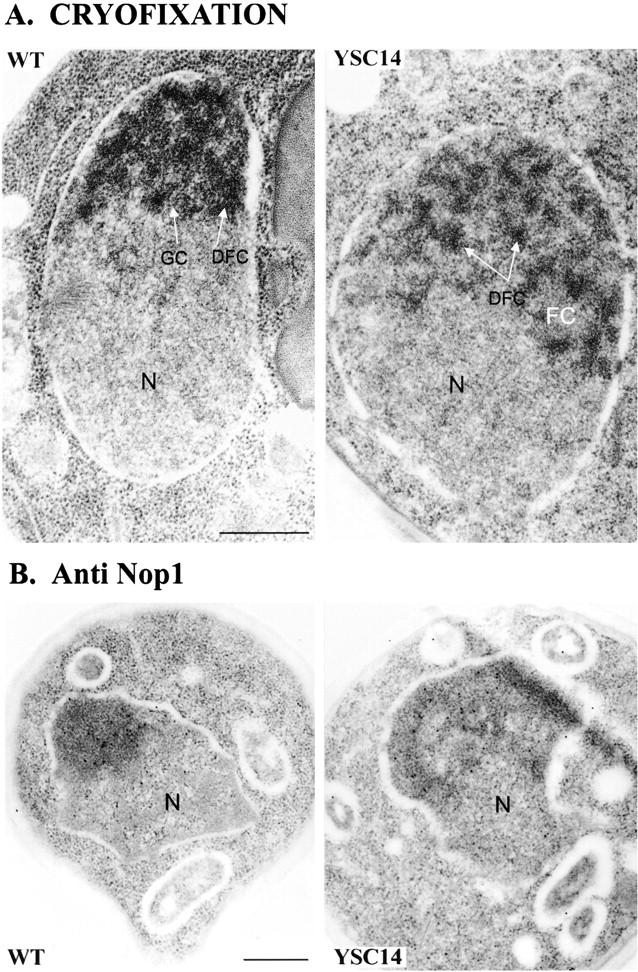
The nucleolus of 5S RNA mutants is partially disorganized. (A) Electron micrograph after freeze electron substitution of the wild-type and YSC14 strains grown at 30°C. Substructures similar to the components of the nucleolus of higher eukaryotes are visualized: the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC). The fibrillar centers are not visible on the WT section. N, nucleoplasm. (B) Immunogold localization of the nucleolar protein Nop1 in the wild-type and YSC14 strains. Nop1 was detected using anti-Nop1 mAbs revealed with a secondary gold-conjugated antibody. Gold particles colocalized with the nucleolus in the two strains. N, nucleoplasm. Bars, 500 nm.
Figure 8.
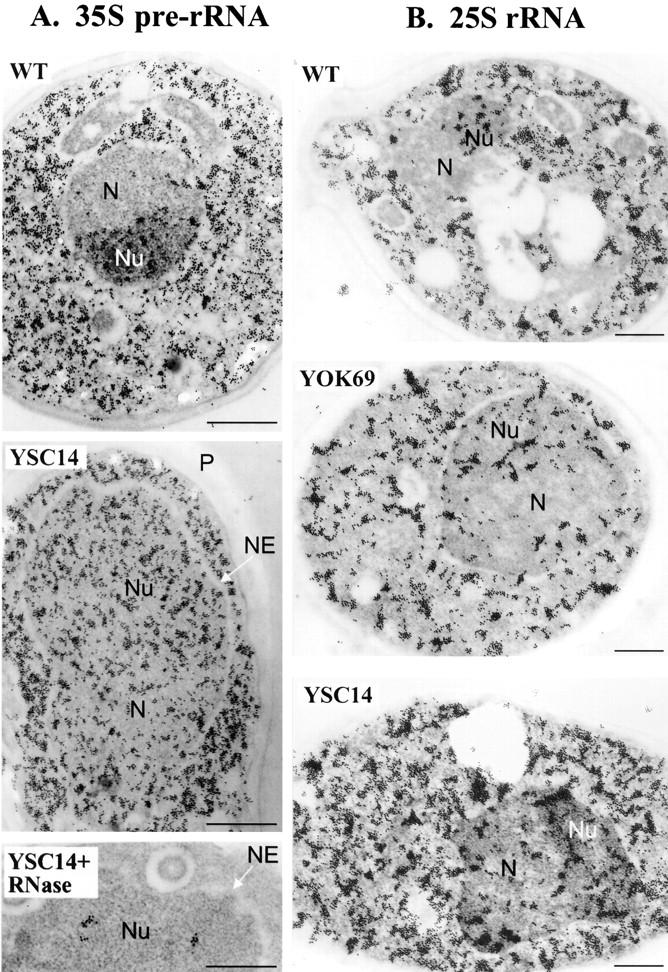
Nuclear delocalization of the 25S rRNA in 5S RNA mutants. The localization of the 35S pre-rRNA and the 25S mature rRNA was analyzed by in situ hybridization using digoxigenin-11dUTP-labeled probes specific for the 35S pre-rRNA (A) or the 25S rRNA (B) in the wild-type (WT), YOK69, and YSC14 strains. The probes were detected with antidigoxigenin antibodies gold conjugate. In A, as a control, the grids were pretreated with DNase-free RNase (YSC14 + RNase) before hybridization. N, nucleoplasm; Nu, nucleolus; NE, nuclear envelope; and P, cell wall. Bars, 500 nm.
Discussion
The novelty of this paper consists in the observation that 5S RNA plays a role in the processing of the large ribosomal subunit RNA. We show that there is a direct correlation between the rate of processing of the 27SB RNA precursor and the amount of 5S RNA present in the cells. We propose that 5S RNA binds to the preparticle containing the 27SB pre-rRNA and that this binding is necessary for the processing to proceed at a normal rate and give rise to the mature rRNA of the large subunit. This mechanism could participate in a quality control process ensuring that all newly formed mature 60S ribosomal subunits contain stoichiometric amounts of the three rRNA components.
We have studied a collection of yeast strains surviving with mutant 5S RNA. The 5S mutations that correspond to nucleotide extensions at the 5′ and 3′ ends of 5S RNA result in decreased accumulation of 5S RNA in the cells, which is possibly due to the lower affinity of the ribosomal protein rpL5 for the mutant 5S RNA. In all the 5S mutants studied here, the decrease in the amount of 5S RNA was paralleled by a decrease in the amount of 60S subunits. No partially assembled 60S subunits devoid of 5S RNA were detected, suggesting that either they were degraded or their formation was impaired. The analysis of the preribosomal processing in the mutants favors the second hypothesis. We found that the rate of processing of 27SB pre-rRNA leading to mature 25S rRNA and 7S pre-rRNA (a precursor to the 5.8S rRNA) was considerably decreased in the 5S mutants. The slower rate was, at least in part, a consequence of the decrease in the amount of 5S RNA since it could be improved by the overproduction of rpL5 or Lhp1p that increased the amount of stable 5S RNA in the cells. We propose that 5S RNA is recruited by the preribosomal particles containing the 27SB precursor and that its binding allows processing to proceed at a normal rate. In the presence of limiting amounts of 5S RNA, 27SB pre-rRNA does not appear to accumulate, as judged by Northern analysis (data not shown) showing that it is eventually degraded.
Our observation that 5S RNA plays a role in the formation of the mature 60S subunits is in agreement with previous observations made by other groups. Nazar and collaborators found that expressing mutant 5S RNA in otherwise wild-type cells (i.e., also containing wild-type 5S RNA) led to a decrease in the production of 60S subunits suggesting that the 5S RNA was necessary for the formation or the stability of the subunit (Van Ryk et al., 1992). Additional evidence came from the work of Woolford and collaborators who found that in the absence of rpL5 synthesis, no mature 60S subunits were produced, and of Lee and collaborators who observed a decreased production of 60S subunits in the presence of mutant rpL5 (Deshmukh et al., 1993; Yeh and Lee, 1995a,b; Yeh et al., 1996). Although these experiments altered rpL5 levels, they probably reflected the role of 5S RNA as well since rpL5 and 5S RNA are associated in an RNP that binds to the preribosomes. In yeast cells depleted of rpL5 no specific defect of the large ribosomal RNA processing was detected, but the whole processing pathway was slowed down. The delay of all the processing steps probably does not mean that rpL5 plays a role in all these steps. It may rather be due to a feedback mechanism, as proposed for two other yeast mutants (Nip7 and Spb4) altered in the processing of 27SB pre-rRNA and that displayed a delay in the processing of 35S and 32S pre-rRNAs (Zanchin et al., 1997; de la Cruz et al., 1998b).
The maturation step affected by 5S RNA corresponds to two cleavage steps in the ITS2, at site C1 (the 5′ end of mature 25S rRNA) and site C2 (located within ITS2) that release the mature 25S rRNA and the 7S pre-rRNA, which is further processed to yield the mature 5.8S rRNA. No mutant has been isolated that presented a defect in only one of the cleavage steps, which suggests that the two cleavages are connected. The maturation of 27SB pre-rRNA is one of the slowest steps of the pathway, reflecting probably important structural rearrangements and the need to recruit several components (Gautier et al., 1997), among which is probably the 5S–rpL5 complex. 5S RNA probably does not have a direct role in the cleavage reactions but could well be involved in the correct assembly of a maturation-competent preparticle. Other yeast mutants slowed down in the processing of 27SB pre-rRNA are assembly mutants altered in the ribosomal proteins rpL16 or rpL32 (Moritz et al., 1991; Vilardell and Warner, 1997), mutants in putative RNA helicases (Ripmaster et al., 1992; de la Cruz et al., 1998a), and mutants in a methylase (Hong et al., 1997).
In parallel with the ribosomal processing defects, the 5S RNA mutants presented alterations of the nucleolar structure. Their nucleolus appeared less dense and with a less granular component than a wild-type nucleolus. The modification of the granular compartment is expected since this compartment corresponds to a concentration of preribosomal particles in the late stages of maturation (Shaw and Jordan, 1995), and the 5S mutants are altered in one of these stages. More unexpected was the delocalization of the large ribosomal subunit RNA throughout the nucleoplasm, which shows that the transport of the preparticles from the nucleolus to the cytoplasm is altered in these mutants. There is not a general defect in the nuclear transport since the localization of the small ribosomal subunit RNA was normal. Interestingly, in yeast mutants altered in a nucleoporin and, therefore, in the transport across the nuclear pores, a similar delocalization of 35S ribosomal RNA throughout the nucleus has been observed (Gas, N., unpublished observations). We propose that mutant preparticles not properly processed are not exported to the cytoplasm, which perturbs the whole trafficking of the presubunits in the nucleus. Very few data are available on the mechanism of transport of the preribosomal particles across the nucleoplasm and the nuclear envelope. There are some indications that transport and nucleolar organization could be linked, since mutants affected in proteins of the mRNA export pathway exhibit alterations of the nucleolus and defects in the preribosomal RNA processing (Kadowaki et al., 1994).
Acknowledgments
This work was supported in part by Association pour la Recherche sur le Cancer and the Region Midi-Pyrénées (to N. Gas).
Abbreviations used in this paper
- ITS2
internal transcribed spacer 2
- YPD
yeast extract/peptone/glucose.
Footnotes
We are very thankful to André Sentenac (CEA/Saclay, France) for his critical review of the manuscript and his constant support during this work. We also thank Pierre Thuriaux (CEA/Saclay, France) for his comments and interest in the study. We are very grateful to Françoise Wyers from François Lacroute's lab (Centre de Génétique Moléculaire, Centre de la Recherche Scientifique, Gif-sur-Yvette, France) for her help in the set up of the analysis of the ribosomal profiles. We also thank John Woolford, Jacqueline Segall (University of Toronto, Canada), Stuart Clarkson, Rosalia Arrebola (CEA/Saclay, France), and John Aris for their gifts of plasmids, yeast strains, and antibodies.
References
- Allison LA, North MT, Neville LA. Differential binding of oocyte-type and somatic-type 5S rRNA to TFIIIA and ribosomal protein L5 in Xenopus oocytes: specialization for storage versus mobilization. Dev Biol. 1995;168:284–295. doi: 10.1006/dbio.1995.1080. [DOI] [PubMed] [Google Scholar]
- Archambault J, Milne CA, Schappert KT, Baum B, Friesen JD, Segall J. The deduced sequence of the transcription factor TFIIIA from Saccharomyces cerevisiae reveals extensive divergence from XenopusTFIIIA. J Biol Chem. 1992;267:3282–3288. [PubMed] [Google Scholar]
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1987. Current Protocols in Molecular Biology. Vols. 1 and 2. G.P.A. Wiley-Interscience, New York. 1.0.1–16.21.9.
- Baim SB, Pietras DF, Eustice DC, Sherman F. A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiaeiso-1-cytochrome c. Mol Cell Biol. 1985;5:1839–1846. doi: 10.1128/mcb.5.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. Isolation of a 5S RNA–protein complex from mammalian ribosomes. Proc Natl Acad Sci USA. 1971;68:1881–1885. doi: 10.1073/pnas.68.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov AA, Dontsova OA, Dokudovskaya SS, Lavrik IN. Structure and function of 5S rRNA in the ribosome. Biochem Cell Biol. 1995;73:869–876. doi: 10.1139/o95-094. [DOI] [PubMed] [Google Scholar]
- Camier S, Dechampesme AM, Sentenac A. The only essential function of TFIIIA in yeast is the transcription of 5S rRNA genes. Proc Natl Acad Sci USA. 1995;92:9338–9342. doi: 10.1073/pnas.92.20.9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Rojo M, Tollervey D, Linder P. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. . RNA. 1998a;4:1268–1281. doi: 10.1017/s1355838298981158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1998b;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh M, Tsay Y-F, Paulovich AG, Woolford JLJ. Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol Cell Biol. 1993;13:2835–2845. doi: 10.1128/mcb.13.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohme F, Nierhaus KH. Role of 5S RNA in assembly and function of the 50S subunit from Escherichia coli. . Proc Natl Acad Sci USA. 1976;73:2221–2225. doi: 10.1073/pnas.73.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H, Maxam AM, Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977;4:2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontsova O, Tishkov V, Dokudovskaya S, Bogdanov A, Doring T, Rinke-Appel J, Thamm S, Greuer B, Brimacombe R. Stem-loop IV of 5S rRNA lies close to the peptidyltransferase center. Proc Natl Acad Sci USA. 1994;91:4125–4129. doi: 10.1073/pnas.91.10.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England TE, Bruce AG, Uhlenbeck OC. Specific labeling of 3′ termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65:65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Gautier T, Berges T, Tollervey D, Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol Cell Biol. 1997;17:7088–7098. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guddat U, Bakken AH, Pieler T. Protein-mediated nuclear export of RNA: 5S rRNA containing small RNPs in Xenopusoocytes. Cell. 1990;60:619–628. doi: 10.1016/0092-8674(90)90665-2. [DOI] [PubMed] [Google Scholar]
- Hadjiolov, A.A. 1985. The Nucleolus and Ribosome Biogenesis. Springer-Verlag New York Inc., New York. 268 pp.
- Higuchi R, Krummel B, Saiki RK. A general method of in vitropreparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B, Brockenbrough JS, Wu P, Aris JP. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol Cell Biol. 1997;17:378–388. doi: 10.1128/mcb.17.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet MA, Ehrlich R, Reiss C. In vivo gene expression directed by synthetic promoter constructions restricted to the -10 and -35 consensus hexamers of E. coli. . Nucleic Acids Res. 1989;17:2933–2945. doi: 10.1093/nar/17.8.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff AM. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants [published erratum appears in J. Cell Biol.1994. 126: 1627] J Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger-Silvestre I, Noaillac-Depeyre J, Faubladier M, Gas N. Structural and functional analysis of the nucleolus of the fission yeast Schizosaccharomyces pombe. . Eur J Cell Biol. 1997;72:13–23. [PubMed] [Google Scholar]
- LinMarq N, Clarkson SG. Efficient synthesis, termination, and release of RNA polymerase III transcripts in Xenopusextracts depleted of La protein. EMBO (Eur Mol Biol Organ) J. 1998;17:2033–2041. doi: 10.1093/emboj/17.7.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Fishel R, Wickner RB. Circular single-stranded RNA replicon in Saccharomyces cerevisiae. . Proc Natl Acad Sci USA. 1990;87:7628–7632. doi: 10.1073/pnas.87.19.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Dreyfuss G. Distinct domains in ribosomal protein L5 mediate 5 S rRNA binding and nucleolar localization. J Biol Chem. 1996;271:11571–11574. doi: 10.1074/jbc.271.19.11571. [DOI] [PubMed] [Google Scholar]
- Moore, P.B. 1996. The structure and function of 5S ribosomal RNA. In Ribosomal RNA: Structure, Evolution, Processing and Function in Protein Biosynthesis. R.A. Zimmermann and A.E. Dahlberg, editors. CRC Press, Boca Raton, FL. 199–236.
- Moritz M, Pulaski BA, Woolford JL., Jr Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol Cell Biol. 1991;11:5681–5692. doi: 10.1128/mcb.11.11.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazar RN, Yaguchi M, Willick GE, Rollin CF, Roy C. The 5S RNA binding protein from yeast (Saccharomyces cerevisiae)ribosomes. Evolution of the eukaryotic 5S RNA binding protein. Eur J Biochem. 1979;102:573–582. doi: 10.1111/j.1432-1033.1979.tb04274.x. [DOI] [PubMed] [Google Scholar]
- Picard B, Wegnez M. Isolation of a 7S particle from Xenopus laevisoocytes: a 5S RNA-protein complex. Proc Natl Acad Sci USA. 1979;76:241–245. doi: 10.1073/pnas.76.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard B, le Maire M, Wegnez M, Denis H. Biochemical research on oogenesis. Composition of the 42-S storage particles of Xenopus laevisoocytes. Eur J Biochem. 1980;109:359–368. doi: 10.1111/j.1432-1033.1980.tb04802.x. [DOI] [PubMed] [Google Scholar]
- Ripmaster TL, Vaughn GP, Woolford JL., Jr A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiaeribosome assembly. Proc Natl Acad Sci USA. 1992;89:11131–11135. doi: 10.1073/pnas.89.23.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Cousino N, Esteban LM, Esteban R. Molecular cloning and characterization of W double-stranded RNA, a linear molecule present in Saccharomyces cerevisiae. Identification of its single-stranded RNA form as 20 S RNA. J Biol Chem. 1991;266:12772–12778. [PubMed] [Google Scholar]
- Rudt F, Pieler T. Cytoplasmic retention and nuclear import of 5S ribosomal RNA containing RNPs. EMBO (Eur Mol Biol Organ) J. 1996;15:1383–1391. [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. . Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergiev P, Dokudovskaya S, Romanova E, Topin A, Bogdanov A, Brimacombe R, Dontsova O. The environment of 5S rRNA in the ribosome: cross-links to the GTPase-associated area of 23S rRNA. Nucleic Acids Res. 1998;26:2519–2525. doi: 10.1093/nar/26.11.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Jordan EG. The nucleolus. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Stefano JE. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell. 1984;36:145–154. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- Steitz J-A, Berg C, Hendrick JP, La Branche-Chabot H, Metspalu A, Rinke J, Yario T. A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J Cell Biol. 1988;106:545–556. doi: 10.1083/jcb.106.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapman J, Planta RJ, Raue HA. Maturation of ribosomes in yeast. II. Position of the low molecular weight rRNA species in the maturation process. Biochim Biophys Acta. 1976;442:275–284. doi: 10.1016/0005-2787(76)90302-6. [DOI] [PubMed] [Google Scholar]
- Van Ryk DI, Lee Y, Nazar RN. Unbalanced ribosome assembly in Saccharomyces cerevisiaeexpressing mutant 5 S rRNAs. J Biol Chem. 1992;267:16177–16181. [PubMed] [Google Scholar]
- Venema J, Tollervey D. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. . Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- Vilardell J, Warner JR. Ribosomal protein L32 of Saccharomyces cerevisiaeinfluences both the splicing of its own transcript and the processing of rRNA. Mol Cell Biol. 1997;17:1959–1965. doi: 10.1128/mcb.17.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. Synthesis of ribosomes in Saccharomyces cerevisiae. . Microbiol Rev. 1989;53:256–271. doi: 10.1128/mr.53.2.256-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, Soeiro R. Nascent ribosomes from HeLa cells. Proc Natl Acad Sci USA. 1967;58:1984–1990. doi: 10.1073/pnas.58.5.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wejksnora PJ, Haber JE. Ribonucleoprotein particle appearing during sporulation in yeast. J Bacteriol. 1978;134:246–260. doi: 10.1128/jb.134.1.246-260.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford, J.L., and J.R. Warner. 1991. The ribosome and its synthesis. In The Molecular and Cellular Biology of the yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics. J.R. Broach, J.R. Pringle, and E.W. Jones, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 1:587–626.
- Yeh LC, Lee JC. Contributions of multiple basic amino acids in the C-terminal region of yeast ribosomal protein L1 to 5 S rRNA binding and 60 S ribosome stability. J Mol Biol. 1995a;246:295–307. doi: 10.1006/jmbi.1994.0085. [DOI] [PubMed] [Google Scholar]
- Yeh LC, Lee JC. Involvement of multiple basic amino acids in yeast ribosomal protein L1 in 5S rRNA recognition. Nucleic Acids Symp Ser. 1995b;33:63–65. [PubMed] [Google Scholar]
- Yeh LC, Deshmukh M, Woolford JL, Lee JC. Involvement of lysine 270 and lysine 271 of yeast 5S rRNA binding protein in RNA binding and ribosome assembly. Biochim Biophys Acta. 1996;1308:133–141. doi: 10.1016/0167-4781(96)00085-1. [DOI] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: a yeast homolog of the La autoantigen is dispensable for growth. Mol Cell Biol. 1994;14:5412–5424. doi: 10.1128/mcb.14.8.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanchin NIT, Roberts P, De Silva A, Sherman F, Goldfarb DS. Saccharomyces cerevisiaeNip7p is required for efficient 60S ribosome subunit biogenesis. Mol Cell Biol. 1997;17:5001–5015. doi: 10.1128/mcb.17.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]