Abstract
We recently developed an assay in which nuclear export of the shuttling transcription factor NFAT (nuclear factor of activated T cells) can be reconstituted in permeabilized cells with the GTPase Ran and the nuclear export receptor CRM1. We have now used this assay to identify another export factor. After preincubation of permeabilized cells with a Ran mutant that cannot hydrolyze GTP (RanQ69L), cytosol supports NFAT export, but CRM1 and Ran alone do not. The RanQ69L preincubation leads to accumulation of CRM1 at the cytoplasmic periphery of the nuclear pore complex (NPC) in association with the p62 complex and Can/Nup214. RanGTP-dependent association of CRM1 with these nucleoporins was reconstituted in vitro. By biochemical fractionation and reconstitution, we showed that RanBP1 restores nuclear export after the RanQ69L preincubation. It also stimulates nuclear export in cells that have not been preincubated with RanQ69L. RanBP1 as well as Ran-binding domains of the cytoplasmic nucleoporin RanBP2 promote the release of CRM1 from the NPC. Taken together, our results indicate that RanGTP is important for the targeting of export complexes to the cytoplasmic side of the NPC and that RanBP1 and probably RanBP2 are involved in the dissociation of nuclear export complexes from the NPC in a terminal step of transport.
Keywords: nuclear transport, CRM1, Ran, RanBP1
Molecular transport between the cytoplasm and the nucleus is mediated by nuclear pore complexes (NPCs;1 for reviews see Doye and Hurt, 1997; Nigg, 1997), large supramolecular structures that span the nuclear envelope (NE). Molecules of up to 20-40 kD can passively diffuse through the NPC, whereas larger molecules, including most proteins and RNAs, are transported through the NPC by signal- and energy-dependent mechanisms. In most cases, signal-mediated nuclear import and export appear to involve nucleocytoplasmic shuttling receptors of the importin β family, which bind to their cargoes either directly or via adaptor molecules (for reviews see Görlich and Mattaj, 1996; Nigg, 1997; Mattaj and Englmeier, 1998). After cargo molecules interact with their cognate receptors in the originating compartment, the transport complexes appear to undergo stepwise translocation through the NPC to the destination compartment, whereupon the cargo dissociates from the receptor and the latter is recycled. Several different classes of transport signals have been described for nuclear import (nuclear localization signals, or NLSs) and export (nuclear export signals, or NESs). A common type of NLS (the “classical” NLS) comprises a short amino acid sequence enriched in basic amino acids, and interacts with the import receptor importin β via importin α, which acts as an adaptor. A common type of NES is a short peptide segment enriched in leucine residues (for review see Gerace, 1995), which interacts directly with the export receptor CRM1 (Fornerod et al., 1997a; Fukuda et al., 1997; Stade et al., 1997).
The NPC (for reviews see Rout and Wente, 1994; Doye and Hurt, 1997) has an estimated mass of ∼125 MD in vertebrate cells. It consists of nucleoplasmic and cytoplasmic rings flanking eight central spokes, which surround a central gated channel that is the site of signal-mediated transport. Emanating outward from the nucleoplasmic and cytoplasmic rings are ∼50–100-nm-long fibrils, which may represent initial binding sites for transport complexes at the NPC. The NPC is thought to be composed of ∼50–100 different proteins (nucleoporins). A substantial number of different nucleoporins have been molecularly characterized in vertebrates and yeast. Of particular interest are a group of NPC proteins containing multiple copies of FG (phenylalanine, glycine) repeats dispersed throughout a portion of their sequence. Some members of this group have been found to bind directly to nuclear transport factors in vitro, including importin β-like receptors (Iovine et al., 1995; Radu et al., 1995; Rexach and Blobel, 1995; Hu et al., 1996). Moreover, some FG repeat proteins have been implicated in nuclear import/export in functional studies (see Nigg, 1997). This indicates that FG repeat nucleoporins constitute at least some of the binding sites for nuclear transport complexes at various stages of movement through the NPC. In vertebrate cells, some FG repeat nucleoporins are localized to the nucleoplasmic fibrils (Nup153 [Cordes et al., 1993; Sukegawa and Blobel, 1993], and Nup98 [Radu et al., 1995]), some are found in the cytoplasmic fibrils (RanBP2 [Yokoyama et al., 1995; Wilken et al., 1995; Wu et al., 1995] and Can/Nup214 [Panté et al., 1994]) and one (the p62 complex, consisting of four different FG repeat proteins; Finlay et al., 1991; Hu et al., 1996; Hu and Gerace, 1998) is localized near the central gated channel on both sides of the NPC (Guan et al., 1995). However, detailed roles of individual FG repeat proteins in specific transport pathways are not understood.
The small GTPase Ran, which shuttles between the nucleus and the cytoplasm, plays a key role in determining the vectoriality of nuclear transport (for reviews see Melchior and Gerace, 1998; Moore, 1998). Many importin β-related nuclear transport receptors are RanGTP binding proteins (Wozniak et al., 1998), but the binding of RanGTP has distinct effects on import and export receptors in vitro: RanGTP dissociates cargo from nuclear import receptors (Rexach and Blobel, 1995; Izaurralde et al., 1997), whereas it promotes the association of cargo with nuclear export receptors (Fornerod et al., 1997a; Kutay et al., 1997, 1998; Arts et al., 1998). Since the GTPase-activating protein for Ran (RanGAP; Coutavas et al., 1993; Bischoff et al., 1995) is sequestered in the cytoplasm and its guanine nucleotide exchange factor (RCC1) is restricted to the nucleus (Ohtsubo et al., 1989), most nuclear Ran is probably in the GTP-bound form, whereas most cytoplasmic Ran is likely to be in the GDP-bound form. Thus, the nucleocytoplasmic compartmentalization of Ran effectors in cells and the resulting asymmetric distribution of RanGTP appear to be important for the loading/unloading of nuclear transport receptors in the nucleus. It remains possible that Ran has additional functions for specifying the vectoriality of nuclear transport, although these are not yet characterized. Nuclear import of the import receptors importin β and transportin (by themselves) does not strictly depend on Ran (Kose et al., 1997; Nakielny and Dreyfuss, 1998), and transportin-mediated import of cargo also does not require Ran under certain in vitro conditions (Englmeier at al., 1999; Ribbeck et al., 1999). On the other hand, since RanGTP and cargo molecules bind cooperatively in vitro to several export receptors, including CRM1 (Fornerod et al., 1997a), exportin-t (Arts et al., 1998; Kutay et al., 1998), and CAS (Kutay et al., 1997), a trimeric cargo/RanGTP/receptor complex is likely to be the form that is actually translocated through the NPC to the cytoplasm. Whether RanGTP has a role in the translocation itself is unknown.
A major fraction of RanGAP is bound to the cytoplasmically oriented nucleoporin RanBP2 due to its modification by the ubiquitin-related modifier SUMO-1 (Matunis et al., 1996; Mahajan et al., 1997), and the remainder of RanGAP is soluble in the cytoplasm. RanBP2 has four RanGTP-binding domains (RBDs), which are similar in sequence to the RBD of the cytosolic protein RanBP1, a RanGTP-binding protein of 28 kD that stimulates the activity of RanGAP (Coutavas et al., 1993; Bischoff et al., 1995b). A number of studies in yeast (Schlenstedt et al., 1995) and in higher eukaryotes (Chi et al., 1996; Pu and Dasso, 1997), support the notion that RanBP1 has a role in some aspect of nuclear trafficking, but its precise mechanism of action is not known. RanBP1 can release RanGTP from certain importin β-type receptors, and it has been proposed that one function of this protein is to promote the termination of nuclear export pathways in the cytosol by disassembling export complexes and triggering RanGTP hydrolysis (Bischoff and Görlich, 1997).
We recently developed an in vitro assay for nuclear protein export (Kehlenbach et al., 1998), which involves HeLa cells that are stably transfected with the shuttling transcription factor NFAT (nuclear factor of activated T cells) fused to the green fluorescent protein (GFP). NFAT contains a leucine-rich NES similar to that found in Rev, PKI, and a growing number of other proteins. After preincubation of permeabilized cells with ATP to deplete endogenous export factors, we found that the addition of the GTPase Ran and the nuclear export receptor CRM1 is sufficient to reconstitute the nuclear export of GFP-NFAT. We have now developed a new preincubation condition to make other export factors rate limiting. This involves treatment of permeabilized cells with RanQ69L, a Ran mutant that is deficient in GTP hydrolysis (Klebe et al., 1995). Under these conditions, we identified RanBP1 and the RanBP1-like domains of RanBP2 as potent stimulators of nuclear export in vitro. RanBP1 also stimulates nuclear export in cells that have not been preincubated with RanQ69L. Our data indicate that RanBP1 and probably RanBP2 are involved in releasing transport complexes from the cytoplasmic side of the NPC in a terminal step of export. In addition, our work suggests that RanGTP is important for targeting export complexes to the cytoplasmic side of the NPC, delineating a new potential role for Ran in promoting the vectoriality of nuclear export.
Materials and Methods
Nuclear Export Assay
HeLa cells, stably transfected with GFP-NFAT (Kehlenbach et al., 1998), were grown in Dulbecco's modified Eagle medium (GIBCO BRL) with 10% fetal bovine serum on plastic dishes. Treatment of intact cells with trichostatin A (Wako BioProducts) to induce expression of GFP-NFAT, and with ionomycin (Calbiochem Corp.) to induce import of GFP-NFAT into the nucleus was done as described (Kehlenbach et al., 1998). As indicated, permeabilized cells were subjected to a preincubation step in the presence of ATP, either including or excluding RanQ69L and cytochrome c-NES (cc-NES) at final concentrations of 20–25 and 100 μg/ml, respectively. After preincubation, cells were washed twice with transport buffer (20 mM Hepes-KOH, pH 7.3, 110 mM KOAc, 2 mM Mg[OAc]2, 2 mM DTT, and 1 μg/ml each of leupeptin, pepstatin, and aprotinin). Nuclear export reactions and quantitation of nuclear export by flow cytometry were performed as described (Kehlenbach et al., 1998).
Antibodies, SDS-PAGE, and Immunoblotting
The anti–CRM1 peptide antibody was described in Kehlenbach et al. (1998). For immunoprecipitation, immunofluorescence, and immunogold-EM, the antibody was affinity purified on a column consisting of the COOH-terminal peptide against which it was raised, coupled to CNBr-Sepharose 4B (Pharmacia LKB Biotechnology Inc.). The anti–RanBP1 antibody was raised in rabbits and affinity purified using recombinant RanBP1 coupled to CNBr-Sepharose 4B. Anti–Can/Nup214 antibody was kindly provided by Dr. Gerard Grosveld. The monoclonal antibodies RL1 and mAb414 were from Affinity BioReagents, Inc. and BAbCO, respectively. The monoclonal anti-importin β-antibody (ascites fluid; Affinity BioReagents, Inc.) used for immunoprecipitations was diluted 1:1 with 5 mg/ml BSA in transport buffer. The polyclonal anti-importin β-antibody used for immunoblots was raised against recombinant glutathione- S-transferase (GST)–tagged protein and purified using recombinant 6× His-tagged protein.
For immunoblotting, proteins were separated by SDS-PAGE and electrophoretically transferred onto nitrocellulose using standard methods. Blots were blocked with 5% milk powder in TBST (10 mM Tris-HCl, pH 8, 150 mM NaCl, 0.05% Tween 20) overnight. HRP-coupled goat anti– mouse or donkey anti–rabbit IgG (Pierce Chemical Co.; 1:10,000 in 10 mM Tris-HCl, pH 8, 150 mM NaCl, 0.05% Tween 20) were used as secondary antibodies. The enhanced chemiluminescence system (Pierce Chemical Co.) was used for visualization of proteins.
Nuclear Transport Factors
Recombinant wild-type Ran and RanQ69L were prepared as described (Melchior et al., 1995). For preparation of GST-Ran, the cDNA was cloned into a pGEX-2T (Pharmacia LKB Biotechnology Inc.)–derived expression vector. The protein was expressed in Escherichia coli and purified using glutathione-Sepharose beads (Pharmacia LKB Biotechnology Inc.). To load Ran with nucleotides, soluble protein (1 mg/ml) was treated with 10 mM EDTA, 2 mM DTT, and 2 mM GDP or GMP-PNP. Nucleotides were added from a 20-mM stock in 20 mM Hepes-KOH, pH 7.2, and 20 mM MgCl2. After 30 min at 30°C, MgCl2 was added to a final concentration of 20 mM and the sample was diluted with an equal volume of transport buffer to 0.5 mg Ran/ml. For loading of GST-Ran, the protein was first bound to glutathione-Sepharose beads (Pharmacia LKB Biotechnology Inc.) at 3 mg/ml of packed beads. After incubation with 2 mM GMP-PNP, 10 mM EDTA, and 2 mM DTT for 30 min at 30°C, 20 mM MgCl2 was added and the beads were washed with transport buffer.
Recombinant RanBP1 (purified by ammonium sulfate precipitation, ion exchange chromatography, and gel filtration) was kindly provided by Dr. R. Mahajan (The Scripps Research Institute). GST-tagged p62, p58, and p54 were expressed and purified as described in Hu et al. (1996). A 6× His-tagged COOH-terminal fragment of Can/Nup214 (amino acids 1861–2090) was cloned into pTrcHisA (Invitrogen Corp.). 6× His-Can/ Nup214 and the 6× His-tagged RBD1 of RanBP2 (amino acids 1155–1321; in pET28; Novagen, Inc.) were purified using Ni-NTA Superflow-beads (QIAGEN Inc.), according to the manufacturer's instructions. Native CRM1 was partially purified by ammonium sulfate precipitation and ion-exchange chromatography as described (Kehlenbach et al., 1998), dialyzed against transport buffer and frozen in liquid N2 at ∼160 μg CRM1/ ml (as compared by SDS-PAGE to a BSA standard). Aliquots were stored at −80°C. When samples were analyzed by SDS-PAGE followed by Coomassie staining, CRM1 appeared as the most abundant protein with an estimated purity of ∼60–70%.
Preparation of Cytochrome c-NES
To prepare cc-NES, 5 mg cytochrome c (Sigma Chemical Co.) in 1.4 ml PBS was activated with 7 mM sulfo-SMCC (Pierce Chemical Co.) for 1 h at room temperature. Free crosslinker was removed by chromatography over a PD10 column (Pharmacia LKB Biotechnology Inc.) equilibrated with PBS. 2 mg of NES-peptide (the Rev-sequence CLPPLERLTL, synthesized at the Scripps Core Facility) was added and the reaction was incubated overnight at 4°C. Free peptide was then removed by chromatography over a PD10 column equilibrated with transport buffer, and the protein (∼2 mg/ml) was stored at 4°C.
Fractionation of Cytosol
Cytosol was prepared as described (Kehlenbach et al., 1998) and dialyzed into triethanolamine buffer (20 mM triethanolamine, pH 7.4, 2 mM MgCl2). To isolate the cytosolic factor that restores transport after preincubation of cells in the presence of RanQ69L, cytosol (10 mg of protein) was loaded on a FPLC Mono Q column (HR 5/5; Pharmacia LKB Biotechnology Inc.) and 1-ml fractions were eluted with an 18-ml linear gradient of 0 to 500 mM NaCl in triethanolamine buffer at a flow rate of 0.4 ml/ min. Maximal nuclear export activity was recovered around 280 mM NaCl. 900 μl of the fraction containing peak activity (as determined by the nuclear export assay) was concentrated to 300 μl by vacuum dialysis and applied to a gel filtration column (Superdex 200 HR 10/30; Pharmacia LKB Biotechnology Inc.) equilibrated in transport buffer. The sample was chromatographed at 0.4 ml/min and 0.5-ml fractions were collected. The nuclear export activity eluted with an apparent molecular mass of ∼30 kD.
Immunofluorescence Microscopy and Immunogold-EM
For immunofluorescence staining, digitonin-permeabilized cells were allowed to bind to poly lysine–coated coverslips for 15 min at 4°C, fixed for 6 min with 4% formaldehyde in PBS at 20°C, and then treated with 0.2% Triton X-100 in PBS for 5 min at 20°C. After washing, cells were incubated with purified anti–CRM1 antibody at 1–2 μg/ml for 1 h at 20°C in 2% BSA in PBS. Texas red donkey anti–rabbit IgG (Pierce) was used as secondary antibody. Confocal microscopy was performed using an MRC1024 (Bio-Rad Laboratories) attached to an Axiovert S100TV (Carl Zeiss, Inc.). Microscopic parameters were identical for pairs of pictures. Digital data were processed identically, using Adobe Photoshop (Adobe Systems Inc.).
To carry out immunogold-localization, cells were permeabilized by one cycle of freeze-thawing and incubated in transport buffer with anti–CRM1 antibody (2 μg/ml) for 2–3 h. Samples were washed and incubated with anti–rabbit IgG coupled to 5 nm gold (Sigma Chemical Co.) as secondary antibody. After washing with transport buffer, cells were fixed with 2% glutaraldehyde for 15 min, postfixed with 1% osmium tetroxide, and stained en block with 1% uranyl acetate. Cells were then dehydrated in serial ethanol concentrations and embedded in Eponate 812 (Ted Pella, Inc.). Thin sections were analyzed using a Phillips EM 208, without further staining. All steps were performed at room temperature.
Immunoprecipitation
For immunoprecipitation, 4 × 106 digitonin-permeabilized cells were solubilized for 20 min on ice in 1 ml NP40 buffer (1% NP40, 50 mM Tris-HCl, pH8, 5 mM EDTA, 5 mM EGTA, 15 mM MgCl2, 60 mM β-glycerophosphate, 100 μM KF, 100 μM NaVO4, 2 mM DTT, 1 μg/ml each of leupeptin, pepstatin, aprotinin, and 300 mM NaCl). After centrifugation at 100,000 g for 30 min at 4°C, purified anti–CRM1 antibody (10 μg/ml) or anti–importin β antibody (ascites fluid, 1:100) was added. Samples were incubated at 4°C for 2 h and immunoreactive proteins were collected with protein A–Sepharose (Pharmacia LKB Biotechnology Inc.). The beads were washed four times with NP40 buffer and precipitated proteins were analyzed by SDS-PAGE, followed by immunoblotting or silver staining.
In Vitro Binding and Release Experiments
To analyze the binding of NE-proteins to immobilized CRM1, 30 μg of purified anti–CRM1 antibody was first bound to 60 μl protein A–Sepharose beads. 2 × 107 digitonin-permeabilized HeLa cells were solubilized in NP40 buffer containing 300 mM NaCl and centrifuged at 100,000 g for 30 min. To load the protein A-anti–CRM1 beads with CRM1, beads were incubated with the permeabilized cell supernatant for 90 min at 20°C, and then washed four times with NP40 buffer. Only bands corresponding to CRM1 and IgG could be detected when a sample of these beads was analyzed by SDS-PAGE, followed by blotting and Ponceau staining. Nuclear envelopes were prepared as described (Gerace et al., 1978) and stored at −80°C. 200 OD260 U of NE (derived from ∼6 × 108 nuclei) were solubilized on ice for 20 min in 1 ml NP40 buffer containing 450 mM NaCl. After centrifugation at 100,000 g for 30 min, the supernatant was diluted with NP40 buffer without NaCl to 150 mM NaCl. 60 μl of this extract was added to 6 μl protein A-CRM1 beads in the absence or presence of 25 μg/ml RanGDP or RanGMP-PNP and 100 μg/ml cc-NES. NP40 buffer containing 150 mM NaCl, and BSA (final concentration of 10 mg/ml) was added to give a final volume of 250 μl. After incubation at 20°C for 1 h, beads were washed four times with NP40 buffer containing 150 mM NaCl and bound proteins were analyzed by SDS-PAGE, followed by immunoblotting.
For binding experiments using purified CRM1 and GST-p62, GST-p58, GST-p54, or 6×His-Can/Nup214, the recombinant proteins were bound to glutathione-Sepharose beads or Ni-NTA-superflow beads (QIAGEN Inc.) at 50–100 μg/ml. The beads were then treated with 30 mg/ml BSA in transport buffer for 20 min to block nonspecific binding sites. 5 μl CRM1 (∼160 μg/ml) was added to 6 μl of beads in the absence or presence of 25 μg/ml RanGDP or RanGMP-PNP and 1 mg/ml NES-peptide (CLPPLERLTL; from a 2-mg/ml stock in transport buffer) in a total volume of 100 μl containing 10 mg/ml BSA. After 90 min at 20°C, the beads were washed with transport buffer three times. All buffers involving Ni-beads contained 20 mM imidazole. The amount of bound CRM1 was analyzed by SDS-PAGE and immunoblotting.
To analyze the release of CRM1 from RanGMP-PNP by RanBP1, ∼1 μg of purified CRM1 was bound to 6 μl GST-RanGMP-PNP beads in the presence of 1 mg/ml NES peptide and 10 mg/ml BSA in a total volume of 100 μl transport buffer. After extensive washing with transport buffer, the beads were incubated at 0° or 30°C with 5 mg/ml BSA and 1 mg/ml NES-peptide in the absence or presence of 120 μg/ml RanBP1 for 20 min. These high concentrations of RanBP1 were used because the matrix contains a high concentration of RanGTP, to which RanBP1 and CRM1 can bind. Released proteins were precipitated with 10% TCA for 5 min on ice and pelleted by centrifugation at 14,000 g for 15 min at 4°C. Samples were analyzed by SDS-PAGE, followed by immunoblotting.
To analyze the release of CRM1 from permeabilized cells after the RanQ69L preincubation, they were subjected to a second incubation for 15 min in the absence or presence of RanBP1 or RBD1 of RanBP2. The cells were collected by centrifugation and equivalent amounts of pellet and supernatant were analyzed by SDS-PAGE, followed by immunoblotting.
Results
An Assay for an Additional Nuclear Export Factor(s)
We previously found that the NES receptor CRM1 and the small GTPase Ran reconstitute efficient nuclear export of GFP-NFAT in digitonin-permeabilized HeLa cells that have been preincubated with ATP at 30°C in order to deplete shuttling export factors (Kehlenbach et al., 1998). To identify additional cytosolic export factors besides CRM1 and Ran, we had to find conditions to make them rate limiting in our assay. We therefore compared the ability of CRM1 and cytosol to promote NFAT export in HeLa cells after various preincubation steps that might more completely deplete export factors from the permeabilized cells.
After our standard preincubation with ATP alone, we found that either CRM1 alone or cytosol alone stimulated nuclear export of GFP-NFAT to equivalent levels in the presence of an excess of wild-type Ran (Fig. 1 a), consistent with our previous work (Kehlenbach et al., 1998). We then examined the effects of preincubation with RanQ69L, a mutant form of Ran that is insensitive to RanGAP and therefore is predominantly in the GTP-bound form (Klebe et al., 1995). RanQ69L has been shown to promote nuclear export of substrates carrying a leucine-rich NES (Richards et al., 1997; Kehlenbach et al., 1998) and to strongly inhibit NLS-mediated nuclear protein import (Dickmanns et al., 1996; Palacios et al., 1996). After preincubation of permeabilized cells with ATP and RanQ69L, the ability of CRM1 plus wild-type Ran to promote nuclear export was clearly reduced (Fig. 1 b). In contrast, cytosol still retained its strong stimulatory effect on nuclear export. We next examined the effect of including an excess of NES substrate in the preincubation, which would be expected to more fully mobilize the endogenous export factors. For the NES substrate, we coupled the Rev NES to cytochrome c, a 12-kD protein that is expected to enter the nucleus by passive diffusion. Consistent with this possibility, the Rev-NES conjugate (cc-NES), but not a conjugate containing an export-defective NES, was able to compete for nuclear export of GFP-NFAT in vitro (data not shown). After preincubation of permeabilized cells with ATP, RanQ69L, and cc-NES, CRM1 and Ran were no longer sufficient to support any nuclear export of GFP-NFAT (Fig. 1 c). Under these conditions, the GFP-NFAT remained in the nucleoplasm (see Fig. 2 b). Again, the addition of cytosol after this preincubation resulted in a strong stimulation of nuclear export. However, preincubation of cells with ATP and cc-NES alone did not affect the ability of CRM1 and Ran to support nuclear export (data not shown).
Figure 1.
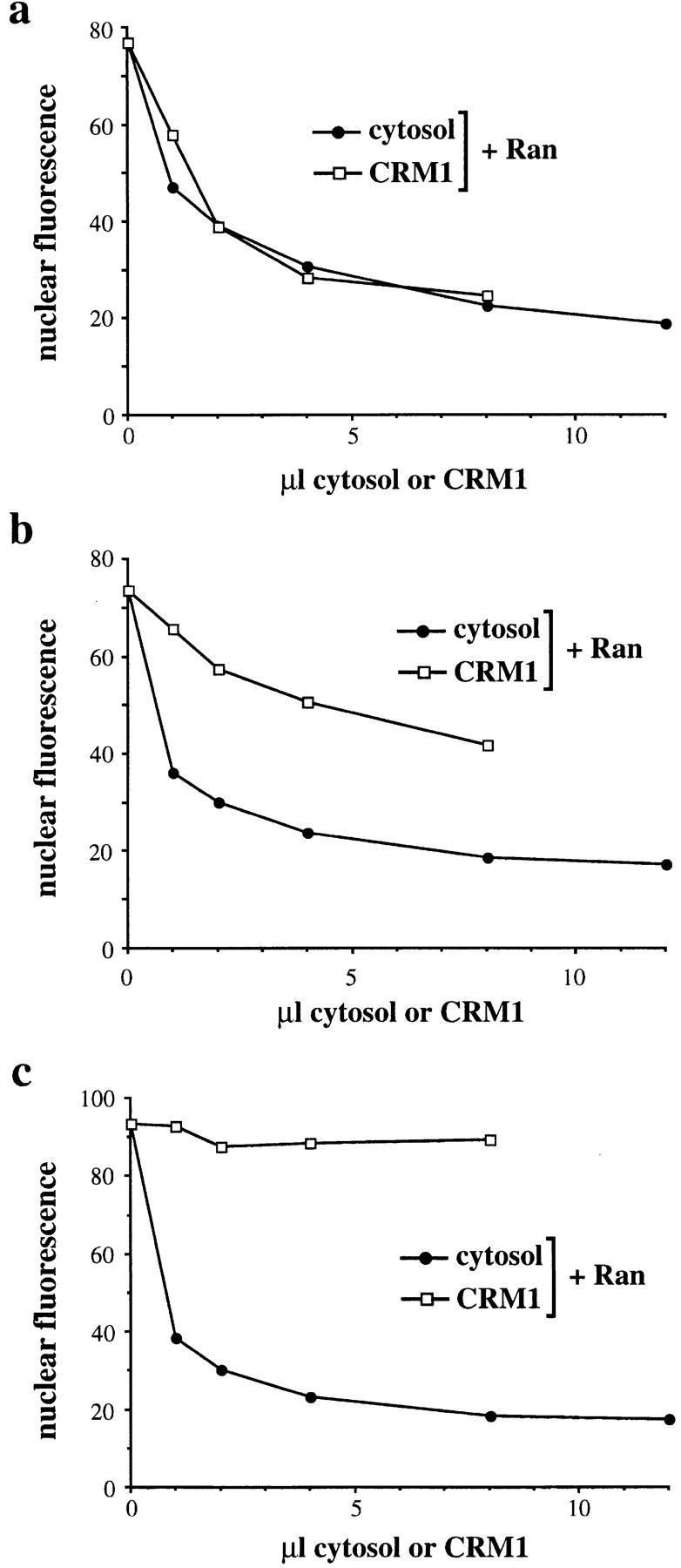
Preincubation of cells with RanQ69L and cc-NES reveals a new cytosolic export factor. Permeabilized cells were preincubated with ATP (a), ATP and RanQ69L (b), or ATP, RanQ69L, and cc-NES (c). Subsequent export reactions were performed with increasing amounts of cytosol (•) or partially purified CRM1 (□). All reactions contained 25 μg/ml Ran. Addition of 100 μg/ml Ran gave essentially the same result (data not shown). Note that in c the initial level of nuclear fluorescence is slightly higher than in a and b, presumably because the added cc-NES prevented export of GFP-NFAT during the preincubation by competing with the GFP-NFAT.
Figure 2.
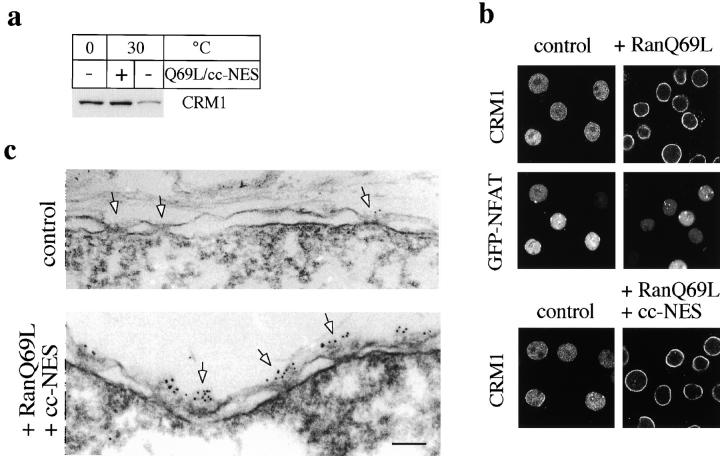
RanQ69L targets CRM1 to the cytoplasmic periphery of the NPC. (a) Permeabilized HeLa cells expressing GFP-NFAT were incubated on ice or at 30°C in either the absence or presence of a combination of RanQ69L and cc-NES. The amount of CRM1 associated with the cells after the incubations was analyzed by immunoblotting. (b and c) Permeabilized cells were kept on ice (control) or incubated at 30°C with RanQ69L (B, top two panels) or RanQ69L and cc-NES (b, bottom, and c as indicated). Cells were analyzed by fluorescence microscopy to detect CRM1 and GFP-NFAT (b) or immunogold EM (c) using purified anti–CRM1 antibodies. Note that some cells in b do not express GFP-NFAT (see Kehlenbach et al., 1998). Arrows in c point to NPCs. Bar, 100 nm.
These results clearly demonstrate that cytosol contains an activity distinct from CRM1 and Ran that is required for nuclear export of GFP-NFAT after preincubation of permeabilized cells with RanQ69L. The export substrate cc-NES potentiates this requirement for an additional factor when added with RanQ69L in the preincubation. The cytosolic activity that stimulates export under these conditions could be a nucleocytoplasmic shuttling factor that is lost from the nuclei during the preincubation. Alternatively, RanQ69L and cc-NES could impose a block on nuclear export that is efficiently relieved by a cytosolic factor. In an attempt to distinguish between these possibilities, we decided to characterize the localization of GFP-NFAT and of CRM1, the NES receptor for NFAT, after incubation of permeabilized cells with RanQ69L.
RanQ69L Targets CRM1 to the Cytoplasmic Periphery of the NPC
An export substrate containing a leucine-rich NES has been shown to bind cooperatively with RanGTP to CRM1 in vitro (Fornerod et al., 1997a), forming a putative nuclear export complex. Preincubation of permeabilized cells with an excess of RanQ69L and cc-NES substrate should efficiently mobilize CRM1 into export complexes and perhaps trigger the transport of these complexes out of the nucleus. As we observed previously (Kehlenbach et al., 1998), incubation of permeabilized cells at 30°C with ATP alone (Fig. 2 a, − Q69L/cc-NES) led to a depletion of CRM1 from the cells, as compared with the 0°C control incubation (Fig. 2 a, 0°C). In contrast, incubation with RanQ69L (Fig. 2 a, + Q69L/cc-NES), but not with wild-type Ran (data not shown), largely prevented the loss of CRM1 from the permeabilized cells. The high level of CRM1 retained by preincubation with RanQ69L could result either from inhibition of export of CRM1 out of the nucleus or from facilitated recycling/reimport back into the nucleus. Since incubation of the same number of cells in a 30-fold larger volume at the same concentration of RanQ69L did not change the level of CRM1 compared with the standard RanQ69L incubation as detected by immunoblotting (data not shown), RanQ69L appears to act by inhibiting export of CRM1, rather than by promoting its reimport. This notion is further validated by the data shown below. Thus, although RanQ69L promotes efficient nuclear export of NES-containing substrates in vivo (Richards et al., 1997), and also a moderate level of nuclear export in vitro (Kehlenbach et al., 1998), the release of the nuclear export receptor CRM1 from the permeabilized cells is impaired by RanQ69L.
We next used immunocytochemical approaches to examine the localization of CRM1 in the nucleus with and without preincubation of the cells with RanQ69L. In previous work, CRM1 was found to be localized throughout the nucleoplasm as well as at the cytoplasmic and nucleoplasmic sides of the NPC in cultured cells (Fornerod et al., 1997b). Consistent with this, by indirect immunofluorescence microscopy, we detected CRM1 predominantly in the nucleoplasm of digitonin-permeabilized cells (Fig. 2 b, control). Incubation of cells with RanQ69L, but not with wild-type Ran (data not shown), led to a strong increase in staining at the NE together with a decrease in intranuclear staining (Fig. 2 b, top). The localization of the export substrate GFP-NFAT did not substantially change upon incubation with RanQ69L (Fig. 2 b, middle). Very similar results were obtained when the cells were incubated with a combination of RanQ69L and cc-NES (Fig. 2 b, bottom). When the Triton permeabilization was omitted after the fixation step before antibody labeling, a similar level of CRM1 was detected at the NE, even though the NE remained intact, as demonstrated by the inaccessibility of nuclear lamins to anti-lamin antibodies (data not shown). This suggests that CRM1 resides at the cytoplasmic side of the NPC after the RanQ69L cc-NES incubation.
To further examine the localization of CRM1 under these conditions, we performed immunogold labeling of digitonin-permeabilized cells that had been incubated either at 0°C in buffer, or at 30°C with RanQ69L and cc-NES (Fig. 2 c). The nuclei were permeabilized by freeze-thawing before immunolabeling. Under these conditions, the nucleoplasmic side of the NPC was freely accessible to gold probes, as demonstrated by strong labeling of the nucleoplasmic side of the NPC with antibodies to Tpr (data not shown), which is found on the nuclear side of the NPC (Cordes et al., 1997). In the control cells, some anti-CRM1 gold labeling was seen throughout the nucleoplasm (not shown) and gold particles were occasionally found at the cytoplasmic side of the NPC (e.g., Fig. 2 c, control, rightmost arrow). Strikingly, however, after cells were incubated with RanQ69L, the cytoplasmic periphery of every NPC was strongly decorated with gold, while essentially no labeling of the nucleoplasmic side of the NPC was evident. Labeling was specific, because it was completely abolished by preincubating the anti-CRM1 antibody with the peptide against which it had been raised. These results indicate that CRM1 crosses the NE and accumulates at the cytoplasmic periphery of the NPC when RanQ69L is present. Hence, RanQ69L appears to inhibit export by inducing the stable trapping of CRM1 at the cytoplasmic periphery of the NPC. Since this accumulation is observed only in the presence of RanQ69L but not wild-type Ran, the release of CRM1 from this binding site is likely to require GTP hydrolysis on Ran and/or a cytosolic release factor that is absent from the permeabilized cells.
Can/Nup214 Is the Major Binding Site for CRM1 at the Cytoplasmic Site of the NPC
We next carried out biochemical analyses to identify the NPC proteins with which CRM1 associates in the presence of RanQ69L. Previous work showed that immunoprecipitation of an epitope-tagged version of the nucleoporin Can/Nup214 from an extract of cultured HeLa cells resulted in the coprecipitation of some CRM1 (Fornerod et al., 1997). Can/Nup214 is one of the FG-repeat–containing nucleoporins that is detected by the monoclonal antibodies RL1 (Snow et al., 1987) and mAb414 (Davis and Blobel, 1986). In addition to Can/Nup214, RL1 detects the nucleoporins RanBP2, Nup153, Pom121, Nup98, p62, p58, and p54 (Snow et al., 1987).
As shown in Fig. 3 a, on silver-stained gels, we detected two proteins with apparent molecular weights of ∼70 and ∼220 kD (marked by asterisks) that specifically coprecipitated with CRM1 from a cell lysate after preincubation with RanQ69L at 30°C. These bands were not seen after preincubation at 30°C without RanQ69L (when the level of precipitated CRM1 was reduced due to the loss of CRM1 from the permeabilized cells under this condition; compare with Fig. 2 a). Addition of the peptide that had been used to prepare the anti–CRM1-antibody abolished the precipitation of CRM1 and also led to the loss of the coprecipitated proteins, indicating that the latter are in a complex with CRM1.
Figure 3.
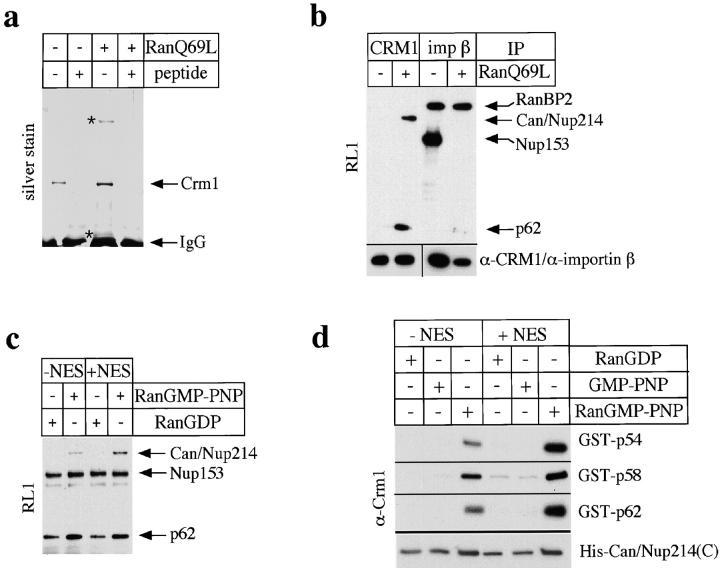
RanQ69L promotes binding of CRM1 to Can/ Nup214 and p62. (a) Permeabilized HeLa cells were incubated at 30°C in the absence (−) or presence (+) of RanQ69L. Proteins were immunoprecipitated from an NP40 lysate (containing 300 mM NaCl) with the anti– CRM1 antibody in the absence (−) or presence (+) of the peptide that had been used to raise the antibody. Precipitated proteins were analyzed by SDS-PAGE followed by silver staining. Two coprecipitating proteins are marked by asterisks. (b) Permeabilized HeLa cells were incubated in the absence (−) of RanQ69L and cc-NES at 0°C or the presence (+) of RanQ69L and cc-NES at 30°C. Proteins were immunoprecipitated from an NP40 lysate (containing 300 mM NaCl) using antibodies against CRM1 or importin β (imp β). Precipitated proteins were detected on immunoblots using the RL1 antibody (top) or anti–CRM1 (bottom left) and anti–importin β antibodies (bottom right). Essentially the same result as shown in this figure was obtained when the NP40 buffer contained either 150 or 450 mM NaCl (data not shown). (c) Solubilized rat liver NE proteins were incubated in the absence (−) or presence (+) of RanGDP, RanGMP-PNP, and cc-NES, with CRM1 that had been immobilized on anti–CRM1–Protein A beads. Bound proteins were detected on immunoblots with the RL1 antibody. No binding of Can/Nup214 was detected with GMP-PNP alone (i.e., when Ran was omitted in the loading step; data not shown). (d) GST-p54, -p58, -p62, and a 6× His-tagged COOH-terminal fragment of Can/ Nup214 were incubated in the absence (−) or presence (+) of RanGDP, GMP-PNP (Ran was omitted in the loading step), RanGMP-PNP, and NES peptide. Bound CRM1 was detected on immunoblots using the anti–CRM1 antibody.
In a similar experiment we used immunoblotting with the RL1 monoclonal antibody to analyze the NPC proteins that coprecipitated with CRM1 (Fig. 3 b). As a control, we examined the proteins that coprecipitated with the import receptor importin β. No RL1-reactive proteins were detectable in the anti–CRM1 immunoprecipitate from cells that had been incubated in buffer at 0°C. In contrast, after incubation with ATP, RanQ69L, and cc-NES at 30°C, two RL1-positive proteins with the same apparent molecular weights as the coprecipitated proteins detected on silver-stained gels (∼70 and ∼220 kD; compare with Fig. 3 a) were coprecipitated with CRM1. The latter was identified as Can/Nup214, using a specific antibody against this protein (data not shown). The 70-kD protein is p62, as judged by its mobility on SDS-PAGE and by the specificity of RL1. The monoclonal antibody mAb414 detected the same two proteins as RL1 in these immunoprecipitates (data not shown). The p58, p54, and p45 subunits of the p62 complex were not detected in these immunoprecipitation experiments by either RL1 or mAb414 with our immunoblotting conditions, probably because larger amounts of these proteins are required for detection compared with p62 (see Davis and Blobel, 1986; Snow et al., 1987). However, since these proteins remain tightly associated with p62 under similar extraction conditions (Guan et al., 1995), they most likely are present in the immunoprecipitates with p62. The level of immunoprecipitated CRM1 itself in this experiment was the same with or without the RanQ69L preincubation (Fig. 3 b, bottom left; note the difference from Fig. 3 a, where both, the + and − RanQ69L samples were incubated at 30°C), consistent with the finding that CRM1 is retained in the permeabilized cells during the RanQ69L treatment.
As a control, we examined whether RL1 antigens coimmunoprecipitated with the import receptor importin β under these conditions. In contrast to the results obtained with CRM1, a distinct set of nucleoporins coprecipitated with importin β in this experiment. RanBP2 and Nup153 were the only detectable nucleoporins that appeared in anti–importin β immunoprecipitates from extracts of cells that had been incubated at 0°C (Fig. 3 b). When cells were preincubated with ATP, RanQ69L, and cc-NES at 30°C, Nup153 no longer coprecipitated with importin β, although the amount of coimmunoprecipitated RanBP2 did not change significantly. The amount of importin β in the immunoprecipitate (Fig. 3 b, bottom right) was clearly reduced after the RanQ69L incubation, indicating that importin β, unlike CRM1, is depleted from the nucleus under these conditions. Thus, the export receptor CRM1 and the import receptor importin β behave differently during preincubation with RanQ69L: RanQ69L promotes the association of CRM1 with a subset of cytoplasmic nucleoporins, but does not promote its release from nuclei, whereas RanQ69L induces the dissociation of importin β from a nucleoplasmic nucleoporin (Nup153) and stimulates release of importin β from the permeabilized cells.
To investigate the binding of CRM1 to proteins of the NPC under defined in vitro conditions, we immobilized CRM1 on anti–CRM1 antibodies coupled to Protein A beads. The beads were then incubated with an NP40 lysate derived from purified rat liver NEs in the presence of RanGDP or RanGMP-PNP, a nonhydrolyzable GTP analogue. In some reactions, we added cc-NES, which enhances the binding of RanGMP-PNP to CRM1 (data not shown). Proteins bound to the CRM1 beads were detected on immunoblots using the RL1 antibody. In the absence of added cc-NES, the nucleoporins p62 and Nup153 and a faint band migrating below Nup153 bound to the beads in the presence of RanGDP (Fig. 3 c). Very similar levels of these proteins bound when no exogenous Ran was added (data not shown). When the incubation contained Ran that had been loaded with GMP-PNP, Can/Nup214 and an increased level of p62 bound to the beads. Neither the level of Nup153 nor of the minor band below Nup153 changed upon addition of RanGMP-PNP. Although addition of cc-NES to the incubations had no effect when the samples contained RanGDP, cc-NES increased the amount of Can/Nup214 and p62 bound to the beads in the presence of RanGMP-PNP. As a control for the specificity of binding, we used Protein A beads that had been coupled to rabbit IgG instead of anti–CRM1 antibody, but were otherwise treated identically. Only a very faint band corresponding to Nup153 could be detected (data not shown), indicating that the binding of nucleoporins to the beads was specific for CRM1. The differential binding of Nup153 to CRM1 in Fig. 3, b and c, probably results from the different experimental approaches: in the immunoprecipitation experiment (Fig. 3 b), we examined CRM1 that is associated with proteins of intact NPCs at the end point of a transport reaction in a permeabilized cell assay. This reaction leads to accumulation of CRM1 on the cytoplasmic side of the NPC, whereas Nup153 is localized to the nucleoplasmic side. In contrast, in the in vitro binding experiment (Fig. 3 c), solubilized proteins of the NPC are allowed to bind to immobilized CRM1.
We next investigated the binding of partially purified CRM1 to recombinant proteins of the p62 complex (p54, p58, and p62) and to a COOH-terminal fragment of Can/ Nup214 that has previously been shown to bind to CRM1 in vivo (Fornerod et al., 1996). In this experiment, the nucleoporins (instead of CRM1) were immobilized on affinity matrices for the binding reactions. As shown in Fig. 3 d, RanGMP-PNP, but not RanGDP or GMP-PNP alone, strongly promoted binding of CRM1 to all three proteins of the p62 complex. The binding was further increased if an NES peptide was included in the reaction. We detected substantial binding of CRM1 to the COOH-terminal fragment of Can/Nup214 in the presence of RanGDP or free GMP-PNP (i.e., in the absence of Ran), and only a modest increase in binding when RanGMP-PNP was added. No binding was detected when we used Ni beads that had not been coupled with the Can/Nup214 fragment (data not shown).
In the two different binding reconstitution experiments (Fig. 3, c and d), RanGMP-PNP stimulated the nucleoporin/CRM1 interactions to different levels, depending on the source of nucleoporins and the nature of the affinity matrix. It strongly enhanced the interaction between Can/Nup214 and CRM1 when solubilized NEs were used as a source of nucleoporins (Fig. 3 c), but only modestly enhanced the interaction when a recombinant fragment of Can/Nup214 was used (Fig. 3 d). Conversely, RanGMP-PNP only modestly enhanced the association of p62 with CRM1 when p62 came from solubilized NEs, but strongly enhanced the association when recombinant p62, p58, and p54 were used (Fig. 3 d). Aside from the different nature of the affinity matrices in the two cases (see Fig. 3, legend), these differences may be due to the presence of components in the solubilized NEs that differentially affect the binding of Can/Nup214 versus p62 to CRM1. In addition, the short recombinant fragment from the COOH terminus of Can/Nup214 might be folded in a way that results in a different binding activity of the fragment compared with the full-length protein from NEs.
Despite these quantitative differences, the results of these in vitro binding reconstitution experiments are in agreement with the results of the immunoprecipitation experiments involving extracts of permeabilized cells (Fig. 3, a and b), which showed that RanQ69L induces the stable binding of CRM1 to Can/Nup214 and p62 in permeabilized cells. The stimulation of binding of CRM1 to these nucleoporins by RanQ69L/RanGMP-PNP suggests that these interactions reflect the association of an export complex separately with Can/Nup214 and p62. It remains possible that a ternary complex of CRM1, p62, and Can/ Nup214 can be formed. However, in light of our EM data, we think it unlikely that the end point of the transport reaction in the presence of RanQ69L, which is analyzed in the immunoprecipitation experiments (Fig. 3, a and b), involves such a complex. This is because much of the CRM1 was found at the periphery of the NPC where Can/Nup214 is localized (Panté et al., 1994), which is distinct from the central location where p62 is found (Guan et al., 1995).
Significantly, the above results imply that RanGTP not only promotes the formation of an export complex, but also targets it to specific sites in the NPC during transport. Since Can/Nup214 is located in a more peripheral cytoplasmic location in the NPC than the p62 complex, it is likely that the association of the export complex with Can/ Nup214 represents a late step in nuclear export. Release from this site is mediated by a cytosolic factor, at least in cells that have been preincubated with RanQ69L, thereby allowing efficient nuclear export in a subsequent reaction (see Fig. 1).
RanBP1 Promotes Nuclear Export of GFP-NFAT
To identify the cytosolic factor that restores export of GFP-NFAT after preincubation of permeabilized cells with RanQ69L and cc-NES, we fractionated cytosol by column chromatography and tested individual fractions for their ability to promote export in the presence of exogenous Ran and partially purified CRM1. With analysis by ion exchange chromatography using a Mono Q column, we obtained a major peak of export activity in fraction 20, which eluted from the column at ∼280 mM NaCl (Fig. 4 a). This peak fraction was further analyzed by gel filtration on an S200 column. A small peak of export activity was observed in fraction 19, at the position of a globular protein of ∼30 kD (Fig. 4 b). A much larger peak eluting at the same position was obtained when total cytosol instead of a Mono Q fraction was used for gel filtration (data not shown). The apparent size of the active protein eluting from the gel filtration column prompted us to probe fractions of both purification steps for the presence of RanBP1, a previously characterized cytosolic RanGTP-binding protein of 28 kD (Coutavas et al., 1993; Bischoff et al., 1995b). As shown in Fig. 4, a and b (insets), the peak of RanBP1 detectable by immunoblotting coincides with the peak of export activity on both the Mono Q and the gel filtration columns. When the S200 samples were analyzed by silver staining, a protein with the same mobility as RanBP1 was the only species that peaked in fraction 19 (data not shown). This protein accounted for ∼10% of the total protein in that fraction.
Figure 4.
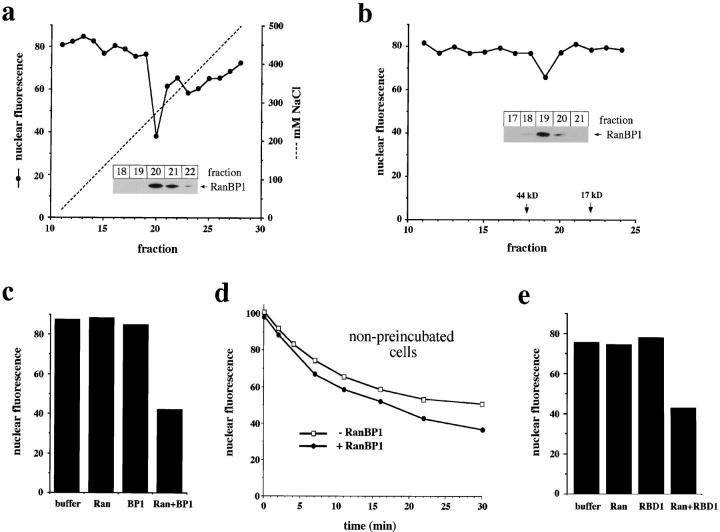
RanBP1 and RanBP1-related domains promote nuclear export in vitro. (a) Activity profile of Mono Q fractions. HeLa cytosol was chromatographed on a Mono Q ion exchange column. 20 μl of individual fractions was dialyzed against transport buffer and added to nuclear export reactions. (b) Activity profile of Superdex 200 fractions. Nuclear export reactions contained 25 μl of Superdex 200 fractions. (a and b) All reactions contained 25 μg/ml Ran. (Insets) Immunoblots of indicated fractions to detect RanBP1. RanBP1 was not detected in any other fraction. (c and e) Stimulation of nuclear export by RanBP1 (c) and RBD1 (amino acids 1155–1321) of RanBP2 (e). Reactions contained 25 μg/ml Ran, 25 μg/ml RanBP1, and 7.5 μg/ml RBD1 as indicated. (d) Time course of nuclear export in the absence (□) or presence (•) of 15 μg/ml RanBP1 using cells that have not been preincubated with RanQ69L. (a–c, and e) RanQ69L cc-NES–preincubated cells were used in all reactions.
We next tested the ability of recombinant RanBP1 to stimulate nuclear export of GFP-NFAT after the RanQ69L cc-NES preincubation. We found that a combination of wild-type Ran and RanBP1 strongly stimulated nuclear export (Fig. 4 c). Neither Ran nor RanBP1 alone had a significant effect. No further stimulation of export was obtained by adding purified CRM1 to Ran and RanBP1 in the reconstitution (data not shown). In a titration experiment (data not shown), we found that 50% of the maximal stimulation was obtained with ∼2 μg/ml RanBP1. RanBP1 was maximally active at a concentration of ∼15 μg/ml, whereas a higher concentration resulted in inhibition of nuclear export. These results demonstrate that RanBP1 stimulates nuclear export in cells that have been preincubated with RanQ69L and cc-NES, indicating that it relieves the block imposed by these reagents. This raises the possibility that RanBP1 or related proteins have a role in nuclear export under normal conditions.
As discussed above (see Fig. 1), Ran and CRM1 support efficient nuclear export in permeabilized cells that have been preincubated with ATP alone. Nevertheless, RanBP1 does have a modest stimulatory effect on nuclear export in nonpreincubated cells, as demonstrated by the time course experiment shown in Fig. 4 d (note that RanQ69L-preincubated cells were used in all other panels of Fig. 4). Here, incubation of the permeabilized cells for 30 min reduced the nuclear fluorescence from 100 to 37 U in the presence of RanBP1, but only to 51 U in its absence. This RanBP1-mediated stimulation of nuclear export by ∼30% was obtained in four independent experiments.
The observation that RanBP1 is not absolutely required for nuclear export under these conditions raised the possibility that another factor might provide RanBP1-like functions when the cells have not been preincubated with RanQ69L. RanBP2 is a giant nucleoporin that has four RBDs very similar to that of RanBP1 (Yokoyama et al., 1995). RanBP2 is localized in close proximity to Can/ Nup214 on the cytoplasmic side of the NPC (Wilken et al., 1995; Wu et al., 1995; Yokoyama et al., 1995) and is well situated to interact with an export complex bound to Can/ Nup214. In RanQ69L preincubated cells, the RBDs of RanBP2 are likely to be stably occupied by RanQ69L, which cannot be readily released because the RanQ69L mutant is insensitive to RanGAP. This situation would necessitate the addition of exogenous RanBP1 to promote export, even though the RBDs of RanBP2 would be available under normal conditions.
To test whether the RBDs of RanBP2 can function like RanBP1 in our assay, we expressed them as 6× His-tagged fusion proteins and tested their ability to stimulate nuclear export of GFP-NFAT after preincubation of permeabilized cells with RanQ69L and cc-NES. As shown in Fig. 4 e, the combination of RBD1 of RanBP2 and Ran stimulates nuclear export to a similar extent as RanBP1 and Ran (compare Fig. 4 c). RBDs 2–4 of RanBP2 were equally effective (data not shown). These results show that either RanBP1 or the RBDs of RanBP2 can release the block of nuclear export imposed by preincubating cells with RanQ69L and cc-NES.
RanBP1 and RBDs of RanBP2 Release CRM1 from Can/Nup214 at the Cytoplasmic Periphery of the NPC
RanBP1 has been shown to promote the dissociation of RanGTP from the importin β-related proteins transportin and CAS, as well as from importin β itself (in the presence of importin α; Bischoff and Görlich, 1997). To test whether RanBP1 has a similar effect on the RanGTP-CRM1 complex, we bound purified CRM1 to GST-RanGMP-PNP and examined whether RanBP1 was able to release the CRM1 in a subsequent incubation. As shown in Fig. 5 a, RanBP1 released the majority of CRM1 into the supernatant (compare the levels of released and bound CRM1), whereas very little CRM1 was released in the absence of RanBP1.
Figure 5.
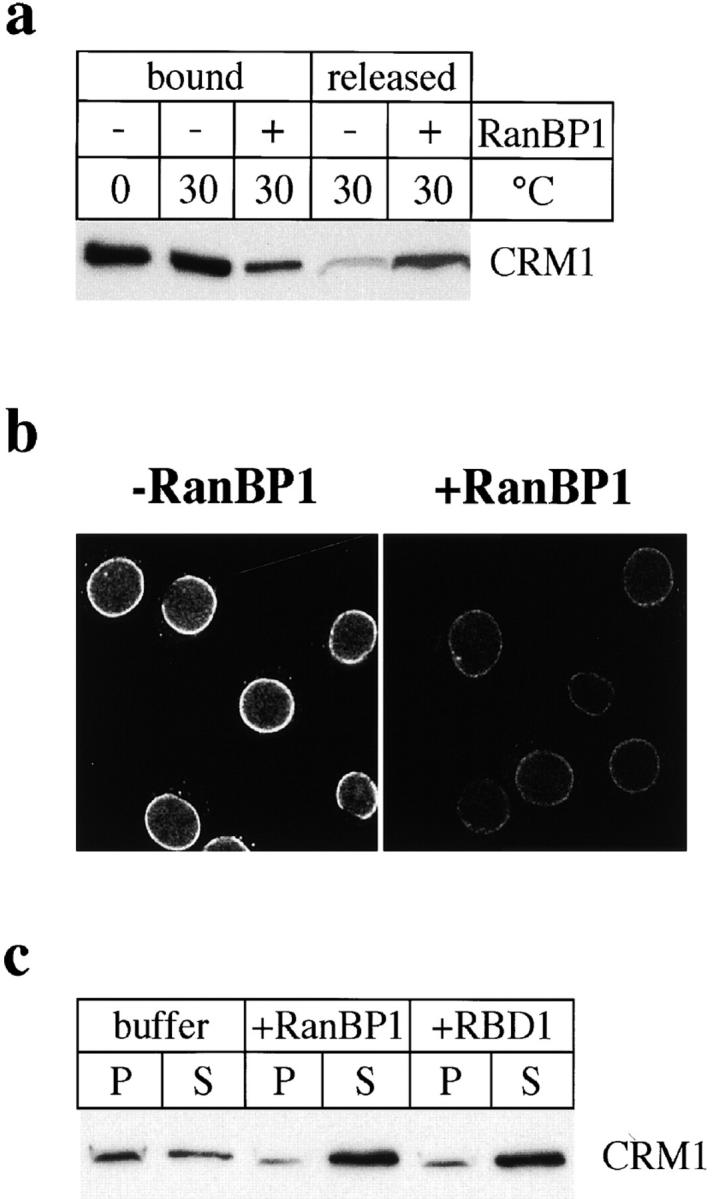
RanBP1 and RBDs of RBP2 release CRM1 from the NE. (a) Release of CRM1 from GST-Ran by RanBP1. Purified CRM1 was bound to GST-RanGMP-PNP. Beads were either kept on ice (0°C) or incubated at 30°C in the absence (−) or presence (+) of 120 μg/ml RanBP1. Equivalent amounts of proteins bound to the beads or released into the supernatant were analyzed by immunoblotting to detect CRM1. (b) Detection of CRM1 by immunofluorescence of RanQ69L cc-NES–preincubated cells after a second incubation in the absence (− RanBP1) or presence (+ RanBP1) of 20 μg/ml RanBP1. (c) RanQ69L cc-NES–preincubated cells were subjected to a second incubation with buffer, 40 μg/ml RanBP1, or 10 μg/ml RBD1 of RanBP2, as indicated. Equivalent amounts of fractions containing pelleted cells/nuclei (P) or proteins released into the supernatant (S) were analyzed by immunoblotting to detect CRM1.
We next tested whether RanBP1 might have a similar effect on CRM1 that is trapped in a complex with Can/ Nup214 at the cytoplasmic side of the NPC as a consequence of incubation of cells with RanQ69L and cc-NES. Since the binding of CRM1 to Can/Nup214 is strongly promoted by RanGTP, the dissociation of RanGTP from CRM1 would be expected to promote the release of the latter from the NPC. Indeed, after a first incubation of permeabilized cells with RanQ69L, a second incubation with RanBP1 strongly reduced the amount of CRM1 at the NE as seen by immunofluorescence microscopy (Fig. 5 b). Without RanBP1, cells exhibited a strong nuclear rim staining for CRM1, very similar to the one obtained without a second incubation (compare with Fig. 2 b). The release of CRM1 from NE of RanQ69L-preincubated cells was analyzed also by immunoblotting (Fig. 5 c). Whereas a major proportion of CRM1 remained associated with the cells (pellet, P) in the absence of RanBP1, the addition of RanBP1 resulted in the recovery of most CRM1 in the released fraction (supernatant, S). A similar effect was obtained with RBD1 of RanBP2 (Fig. 5 c).
Taken together, these results suggest that RanBP1, and probably the RBDs of RanBP2, are involved in the disassembly of the export complex at a terminal nucleoporin site (Can/Nup214). RanBP1 and RanBP2 apparently act by releasing RanGTP from CRM1, thereby promoting release of the export complex from the NPC and dissociation of the cargo-receptor complex. If the RBDs of RanBP2 are occupied by RanQ69L, exogenously added RanBP1 (or cytosolic RanBP1 in intact cells) or soluble RBDs of RanBP2 are required to fulfill this function.
Discussion
Targeting of CRM1 to the Cytoplasmic Periphery of the NPC by RanGTP during Nuclear Export
In this study, we used biochemical and functional approaches to search for new components involved in CRM1-mediated nuclear export. Our studies were based on the initial observation that incubation of permeabilized cells with RanQ69L and cc-NES results in strong inhibition of CRM1-mediated nuclear export. During this treatment, the nuclear export receptor CRM1 becomes highly concentrated at the cytoplasmic side of the NPC in association with Can/Nup214 and the p62 complex, concomitant with CRM1 depletion from the nuclear interior. We found that the cytosolic protein RanBP1 and the RBDs of the nucleoporin RanBP2 can relieve the transport inhibition imposed by RanQ69L by releasing CRM1 from the cytoplasmic nucleoporins. As discussed below, this indicates that RanBP1 and probably RanBP2 play a key role in a terminal step of nuclear export involving export complex dissociation from the NPC.
Previous studies have indicated that CRM1 binds cooperatively to RanGTP and export cargo in the nucleus (Fornerod et al., 1997b) to form a putative export complex that is transported through the NPC. Several criteria indicate that the accumulation of CRM1 at the cytoplasmic side of the NPC during treatment of permeabilized cells with RanQ69L and cc-NES reflects terminal stages of nuclear export, as opposed to initial stages of reimport. First, the in vitro binding of CRM1 to the cytoplasmic nucleporins Can/Nup214 and to the p62 complex is promoted by components known to be involved in the formation of an export complex (RanGTP, or analogues thereof, and export substrate). Similarly, the accumulation of CRM1 at the cytoplasmic nucleoporins in permeabilized cells is dependent on RanQ69L, which is effectively locked in the GTP-bound form, and the block of export by RanQ69L is potentiated by the export substrate cc-NES. Furthermore, CRM1 is released from these cytoplasmic sites by RanBP1 and RBDs, a reaction that is compatible with an export intermediate but not with an import intermediate. RanBP1 has previously been proposed to have a role in disassembly of nuclear export complexes in the cytoplasm (Bischoff and Görlich, 1997; also see below).
Whereas Can/Nup214 is localized in the fibrils at the cytoplasmic periphery of the NPC, the p62 complex is situated on both the cytoplasmic and nucleoplasmic sides of the NPC near the central gated channel (Guan et al., 1995). Our data are consistent with the possibility that Can/Nup214 is the terminal binding site for the CRM1 export complex at the NPC, to which it is transferred after binding to the cytoplasmically oriented p62 complex. The apparent flexibility of the cytoplasmic fibrils (Panté and Aebi, 1996), might allow them to interact with the NPC region containing the p62 complex, enabling direct transfer of the export complex to Can/Nup214. The fact that CRM1 is associated with the p62 complex as well as with Can/Nup214 in permeabilized cells preincubated with RanQ69L may be due to the saturation of all available CRM1 binding sites at Can/Nup214, resulting in the accumulation of CRM1 at a more proximal site (the p62 complex) in the export pathway.
The movement of the CRM1 export complex through the NPC first must involve its association with nucleoplasmic nucleoporins. CRM1 from yeast (Crm1p) has been shown to interact by a two-hybrid assay with a number of nucleoporins, including mammalian Nup98 (Neville et al., 1997), which is localized to the nucleoplasmic side of the NPC (Radu et al., 1995). Nup153, a protein localized to the nucleoplasmic fibrils (Sukegawa and Blobel, 1993), may be another one of the nucleoplasmic binding sites for CRM1 in mammals, since we found that it associates with CRM1 in vitro (albeit not in a RanGTP-stimulated fashion). An additional site probably is the population of p62 complexes found on the nucleoplasmic side of the gated channel.
It is striking that CRM1 was not detected on the nucleoplasmic side of the NPC after preincubation with RanQ69L. This result may be explained, at least in part, by the possibility that the affinity of the CRM1 export complex for cytoplasmic nucleoporins is higher than that for nucleoplasmic nucleoporins. In this respect, the p62 complex found on the cytoplasmic side of the NPC may not be biochemically identical to the nucleoplasmic p62 complex. It also is possible that RanGTP in export complexes acts at the central gated channel of the NPC to promote vectorial movement to the cytoplasm. Whatever the mechanism, the finding that RanGTP (RanQ69L) strongly promotes the binding/accumulation of the CRM1 export complex at the cytoplasmic side of the NPC indicates that RanGTP is important for the vectorial transfer of the export complex through the NPC. Thus, these results point to a second important function for RanGTP in nuclear export, in addition to its role in promoting the association of export cargo with CRM1 inside the nucleus.
The behavior of the import receptor importin β was very different from CRM1 in our studies, arguing that the nucleoporin associations seen for CRM1 are specific and functionally relevant. Importin β appears to be more highly associated with the NPC than CRM1 in cultured cells at steady state, since substantial association of importin β with RanBP2 and Nup153 was seen in immunoprecipitates of permeabilized cell extracts, whereas no significant association of CRM1 with Can/Nup214 or the p62 complex was detected unless cells were pretreated with RanQ69L. High levels of importin β remained associated with RanBP2 after treatment with RanQ69L, but the association with Nup153 was no longer seen. These findings are consistent with previous work showing that adding GMP-PNP to Xenopus egg lysates results in loss of coprecipitation of importin β with Nup153 (Shah et al., 1998), and that RanGTP strongly promotes the binding of importin β to RanBP2 at the cytoplasmic side of the NPC in vitro (Delphin et al., 1997). Incubation of cells with RanQ69L appears to promote the export (recycling) of importin β, since substantial amounts of the latter are released from the cells under these conditions. Thus, RanBP2 may be a terminal NPC binding site for the export of importin β, much like Can/Nup214 appears to be a terminal site for CRM1 export. This suggests that the recycling (export) of at least one import receptor uses nucleoporin binding sites that are at least partially distinct from those used by an export complex. Teleologically, it makes sense that these two pathways would be distinct, so that protein export would not compete with recycling of import factors.
Role of RanBP1 and RanBP2 in a Terminal Step of Nuclear Export
We have identified RanBP1 as the major cytosolic activity that relieves the block of nuclear export imposed by RanQ69L and cc-NES, and showed that it also stimulates export in cells that have not been preincubated with RanQ69L. RanBP1 has previously been suggested to have a role in nuclear import (Chi et al., 1996; Pu and Dasso, 1997) and nuclear assembly (Nicolas et al., 1997; Pu and Dasso, 1997) in higher eukaryotes. The yeast homologue of RanBP1, Yrb1p, also has been suggested to play a role in nuclear protein import and RNA export (Schlenstedt et al., 1995). However, the molecular basis for these previously observed functional effects, and whether they are direct or indirect, is not entirely clear.
Recent biochemical studies have provided new insight on a potential role for RanBP1 in nuclear export. In addition to its role in activating RanGAP (Coutavas et al., 1993; Bischoff et al., 1995b), RanBP1 has been found to reduce the inhibitory effect of certain importin β–related transport receptors (including CAS, transportin, and importin β itself) on RanGAP (Lounsbury and Macara, 1997; Deane et al., 1997; Görlich et al., 1997). This is a consequence of the ability of RanBP1 to release RanGTP from these transport receptors (Bischoff and Görlich, 1997). This release reaction could allow the cytosolic RanBP1 both to promote RanGTP hydrolysis and to stimulate the dissociation of cargo from export complexes, thereby irreversibly dissociating the export complex. In this study, we have found that RanBP1 releases RanGTP from the export receptor CRM1 in a manner similar to other importin β–like receptors that have been examined. This not only is likely to promote cargo release from CRM1, but probably also is responsible for the release of CRM1 from the NPC, since RanGTP strongly enhances the association of CRM1 with cytoplasmic nucleoporins. We also observed some release of CRM1 from immobilized GST-RanGTP using very high concentrations of RanGAP in vitro (our unpublished observations). Thus, we cannot rule out that RanGAP also contributes to the release of CRM1 from the NPC. However, exogenously added RanGAP, even at high concentrations, gave no stimulation of nuclear export in nonpreincubated cells (our unpublished observations). Similarly, in a recent study, Englmeier et al. (1999) have described a small stimulatory effect of RanBP1, but not of RanGAP, on nuclear export in vitro. The isolation of a native complex consisting of a nucleoporin, CRM1, export cargo, and RanGTP will be required for further studies on the function of RanBP1 and RanGAP.
RanBP1 stimulates nuclear export to a modest level (∼30%) in our in vitro assay when the permeabilized cells have not been preincubated with RanQ69L and cc-NES. Under these conditions, RanBP2 most likely fulfills similar functions as RanBP1, since RBDs of RanBP2 are active both in releasing CRM1 from NEs and in stimulating nuclear export after preincubation with RanQ69L. It is interesting to note that there is no RanBP2 homologue in yeast. Yeast cells therefore rely on the activity of RanBP1-like proteins for nuclear import and export (Schlenstedt et al., 1995). In fact, Yrb1p appears to be the only essential RanBP1-like protein in yeast.
Richards et al. (1997) detected efficient export of an NES-containing reporter protein after injecting RanG19V into the nuclei of cultured cells. RanG19V, like RanQ69L, is insensitive to RanGAP and is therefore predominantly in the GTP-bound form. Unlike our in vitro studies involving incubation of permeabilized cells (depleted of most of the endogenous Ran) with RanQ69L, the microinjection studies were done in cells containing a high concentration of wild-type Ran. Thus, the RBDs of endogenous RanBP1 and RanBP2 may not have been completely blocked by RanG19V in this study, and this could explain how efficient export could occur in this situation, in contrast to the results we have obtained in our studies.
We were unable to detect any accumulation of the export cargo GFP-NFAT at the NE under conditions of RanQ69L preincubation. This could be due to the vast excess of cc-NES and/or other endogenous export substrates in the reaction, which would compete with NFAT for binding to CRM1. However, even when export complexes are arrested at the cytoplasmic side of the NPC by RanQ69L, cargo may slowly dissociate from CRM1 even though CRM1 remains bound to the NPC. This could explain results we obtained in our previous study (Kehlenbach et al., 1998), in which we found a low but significant level of nuclear export of GFP-NFAT in the presence of RanQ69L, allowing a single round of nuclear export but no recycling of the export receptor.
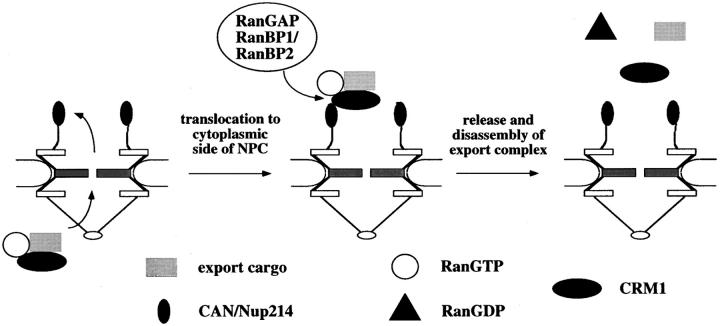
Our data suggest a working model for CRM1-mediated nuclear export that is shown in Fig. 6. We propose that an export complex of cargo, CRM1, and RanGTP that forms in the nucleus first binds to the nucleoplasmic fibrils of the NPC. It subsequently is translocated through the central gated channel via the p62 complex on the nucleoplasmic and cytoplasmic sides of the NPC, and then associates with the peripheral nucleoporin Can/Nup214 at a terminal NPC site. The binding of RanBP2 and/or cytosolic RanBP1 to the RanGTP in the export complex at this site then dissociates RanGTP from CRM1, leading to the release of the cargo from CRM1 and, concomitantly, the dissociation of CRM1 from Can/Nup214. GTP hydrolysis on Ran by cytosolic or RanBP2-associated RanGAP would make export essentially irreversible. Our data suggest a new function for RanGTP in promoting the vectoriality of nuclear export: in addition to stimulating the binding of cargo to CRM1 in the nucleus, RanGTP in an export complex promotes targeting to the cytoplasmic side of the NPC. It will be interesting to examine whether RanGDP, which might be part of a nuclear import complex (see Melchior and Gerace, 1998), could have an analogous role in targeting the import complex to the nucleoplasmic side of the NPC.
Figure 6.
Model for the termination of nuclear export by RanBP1 or RanBP2. The high intranuclear concentration of RanGTP allows the formation of the export complex, which consists of export cargo, CRM1, and RanGTP. The complex then is translocated through the gated channel of the NPC and binds to Can/Nup214. RanBP1 or RanBP2 releases CRM1 from RanGTP, which leads to the dissociation of the complex. RanGTP is converted to RanGDP by soluble cytoplasmic and/or RanBP2-associated RanGAP. See text for details.
Acknowledgments
We thank G. Grosveld (St. Jude Children's Research Hospital, Memphis, TN) for the kind gift of Can/Nup214-cDNA and of anti–Can/Nup214 antibody and T. Nishimoto (Kyushu University, Kyushu, Japan) for expression vectors for RBDs of RanBP2. We further thank C. Delphin for the preparation of nuclear envelopes, R. Mahajan for the preparation of RanBP1, J. Bednenko for the purification of GST-p54, -p58, and-p62, and S. Lyman for the cloning of GST-Ran, the purification of anti–importin β antibodies, and for critically reading the manuscript (all four from The Scripps Research Institute).
This work was supported by a grant from Novartis Pharmaceuticals to L. Gerace and by fellowships from the Human Frontier Science Program to R.H. Kehlenbach (LT-400/96) and the Deutsche Forschungsgemeinschaft to A. Dickmanns (Di 676/1-1). This is TSRI manuscript No. 12173-CB.
Abbreviations used in this paper
- cc
cytochrome c
- GFP
green fluorescent protein
- GST
glutathione-S-transferase
- NE
nuclear envelope
- NES
nuclear export sequence
- NFAT
nuclear factor of activated T cells
- NLS
nuclear localization sequence
- NPC
nuclear pore complex
- RBD
Ran-binding domain
Footnotes
Dr. Dickmanns' present address is Max Planck Institut für Biochemie, Am Klopferspitz 18a, D-82152 Martinsried, Germany.
References
- Arts GJ, Fornerod M, Mattaj IW. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Görlich D. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Kempf T, Hermes I, Ponstingl H. Human RanGTPase-activating protein RanGAP1 is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc Natl Acad Sci USA. 1995a;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Smirnova E, Dong W, Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO (Eur Mol Biol Organ) J. 1995b;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam EJ, Visser GD, Adam SA. RanBP1 stabilizes the interaction of Ran with p97 nuclear protein import. J Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Koehler A, Stuurman N, Van Driel R, Franke WW. Intranuclear filaments containing a nuclear pore complex protein. J Cell Biol. 1993;123:1333–1344. doi: 10.1083/jcb.123.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Rackwitz HR, Franke WW. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutavas E, Ren M, Oppenheim JD, D'Eustachio P, Rush MG. Characterization of proteins that interact with the cell-cycle regulatory protein Ran/TC4. Nature. 1993;366:585–587. doi: 10.1038/366585a0. [DOI] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- Deane R, Schäfer W, Zimmermann HP, Mueller L, Görlich D, Prehn S, Ponstingl H, Bischoff FR. Ran-binding protein 5 (RanBP5) is related to the nuclear transport factor importin-beta but interacts differently with RanBP1. Mol Cell Biol. 1997;17:5087–5096. doi: 10.1128/mcb.17.9.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delphin C, Guan T, Melchior F, Gerace L. RanGTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex. Mol Biol Cell. 1997;8:2379–2390. doi: 10.1091/mbc.8.12.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickmanns A, Bischoff FR, Marshallsay C, Lührmann R, Ponstingl H, Fanning E. The thermolability of nuclear protein import in tsBN2 cells is suppressed by microinjected Ran-GTP or Ran-GDP, but not by RanQ69L or RanT24N. J Cell Sci. 1996;109:1449–1457. doi: 10.1242/jcs.109.6.1449. [DOI] [PubMed] [Google Scholar]
- Doye V, Hurt E. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- Englmeier L, Olivo J-C, Mattaj I. Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr Biol. 1999;9:30–41. doi: 10.1016/s0960-9822(99)80044-x. [DOI] [PubMed] [Google Scholar]
- Finlay DR, Meier E, Bradley P, Horecka J, Forbes DJ. A complex of nuclear pore proteins required for pore function. J Cell Biol. 1991;114:169–183. doi: 10.1083/jcb.114.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Boer J, van Baal S, Morreau H, Grosveld G. Interaction of cellular proteins with the leukemia specific fusion proteins DEK-CAN and SET-CAN and their normal counterpart, the nucleoporin CAN. Oncogene. 1996;13:1801–1808. [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997a;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO (Eur Mol Biol Organ) J. 1997b;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Gerace L. Nuclear export signals and the fast track to the cytoplasm. Cell. 1995;82:341–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- Gerace L, Blum A, Blobel G. Immunocytochemical localization of the major polypeptides of the nuclear pore complex-lamina fraction. Interphase and mitotic distribution. J Cell Biol. 1978;79:546–566. doi: 10.1083/jcb.79.2.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Görlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan T, Muller S, Klier G, Panté N, Blevitt JM, Haner M, Paschal B, Aebi U, Gerace L. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol Biol Cell. 1995;6:1591–1603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, Gerace L. cDNA cloning and analysis of the expression of nucleoporin p45. Gene. 1998;221:245–253. doi: 10.1016/s0378-1119(98)00467-3. [DOI] [PubMed] [Google Scholar]
- Hu T, Guan T, Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, Watkins JL, Wente SR. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J Cell Biol. 1995;131:1699–1713. doi: 10.1083/jcb.131.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO (Eur Mol Biol Organ) J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehlenbach RH, Dickmanns A, Gerace L. Nucleocytoplasmic shuttling factors including Ran and CRM1 mediate nuclear export of NFAT in vitro. J Cell Biol. 1998;141:863–874. doi: 10.1083/jcb.141.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe C, Bischoff FR, Ponstingl H, Wittinghofer A. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry. 1995;34:639–647. doi: 10.1021/bi00002a031. [DOI] [PubMed] [Google Scholar]
- Kose S, Imamoto N, Tachibana T, Shimamoto T, Yoneda Y. Ran-unassisted nuclear migration of a 97-kD component of nuclear pore-targeting complex. J Cell Biol. 1997;139:841–849. doi: 10.1083/jcb.139.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Görlich D. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- Lounsbury KM, Macara IG. Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin beta and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin beta. J Biol Chem. 1997;272:551–555. doi: 10.1074/jbc.272.1.551. [DOI] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase–activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Gerace L. Two-way trafficking with Ran. Trends Cell Biol. 1998;8:175–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- Melchior F, Sweet DJ, Gerace L. Analysis of Ran/TC4 function in nuclear protein import. Methods Enzymol. 1995;257:279–291. doi: 10.1016/s0076-6879(95)57032-2. [DOI] [PubMed] [Google Scholar]
- Moore MS. Ran and nuclear transport. J Biol Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Import and export of the nuclear protein import receptor transportin by a mechanism independent of GTP hydrolysis. Curr Biol. 1998;8:89–95. doi: 10.1016/s0960-9822(98)70039-9. [DOI] [PubMed] [Google Scholar]
- Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- Nicolas F, Zhang C, Hughes M, Goldberg M, Watton S, Clarke P. Xenopus Ran-binding protein 1: molecular interactions and effects on nuclear assembly in Xenopusegg extracts. J Cell Sci. 1997;110:3019–3030. doi: 10.1242/jcs.110.24.3019. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios I, Weis K, Klebe C, Mattaj IW, Dingwall C. RAN/TC4 mutants identify a common requirement for snRNP and protein import into the nucleus. J Cell Biol. 1996;133:485–494. doi: 10.1083/jcb.133.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panté N, Aebi U. Sequential binding of import ligands to distinct nucleopore regions during their nuclear import. Science. 1996;273:1729–1732. doi: 10.1126/science.273.5282.1729. [DOI] [PubMed] [Google Scholar]
- Panté N, Bastos R, McMorrow I, Burke B, Aebi U. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J Cell Biol. 1994;126:603–617. doi: 10.1083/jcb.126.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu RT, Dasso M. The balance of RanBP1 and RCC1 is critical for nuclear assembly and nuclear transport. Mol Biol Cell. 1997;8:1955–1970. doi: 10.1091/mbc.8.10.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Kutay U, Paraskeva E, Görlich D. The translocation of transportin-cargo complexes through nuclear pores is independent of both Ran and energy. Curr Biol. 1999;9:47–50. doi: 10.1016/s0960-9822(99)80046-3. [DOI] [PubMed] [Google Scholar]
- Richards SA, Carey KL, Macara IG. Requirement of guanosine triphosphate-bound Ran for signal-mediated nuclear protein export. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- Rout MP, Wente SR. Pores for thought: nuclear pore complex proteins. Trends Cell Biol. 1994;4:357–365. doi: 10.1016/0962-8924(94)90085-x. [DOI] [PubMed] [Google Scholar]
- Schlenstedt G, Wong DH, Koepp DM, Silver PA. Mutants in a yeast Ran binding protein are defective in nuclear transport. EMBO (Eur Mol Biol Organ) J. 1995;14:5367–5378. doi: 10.1002/j.1460-2075.1995.tb00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Tugendreich S, Forbes D. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J Cell Biol. 1998;141:31–49. doi: 10.1083/jcb.141.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow CM, Senior A, Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol. 1987;104:1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- Wilken N, Senecal JL, Scheer U, Dabauvalle MC. Localization of the Ran-GTP binding protein RanBP2 at the cytoplasmic side of the nuclear pore complex. Eur J Cell Biol. 1995;68:211–219. [PubMed] [Google Scholar]
- Wozniak RW, Rout MP, Aitchison JD. Karyopherins and kissing cousins. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- Yokoyama N, Hayashi N, Seki T, Panté N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]