Abstract
We have examined the process of spindle pole body (SPB) duplication in Saccharomyces cerevisiae by electron microscopy and found several stages. These include the assembly, probably from the satellite, of a large plaque-like structure, the duplication plaque, on the cytoplasmic face of the half-bridge and its insertion into the nuclear envelope. We analyzed the role of the main SPB components in the formation of these structures by identifying them from an SPB core fraction by mass spectrometry. Temperature-sensitive mutants for two of the components, Spc29p and Nud1p, were prepared to partly define their function. The composition of two of the intermediates in SPB duplication, the satellite and the duplication plaque, was examined by immunoelectron microscopy. Both contain cytoplasmic SPB components showing that duplication has already been partly achieved by the end of the preceding cell cycle when the satellite is formed. We show that by overexpression of SPB components the structure of the satellite can be changed and SPB duplication inhibited by disrupting the attachment of the plaque-like intermediate to the half-bridge. We present a model for SPB duplication where binding of SPB components to either end of the bridge structure ensures two separate SPBs.
Keywords: spindle pole body, duplication, Spc42p, Spc29p, Nud1p
The controlled duplication of the microtubule organizing centers which form the spindle pole in eukaryotic cells is an important step in ensuring that the mitotic spindle is bipolar. This step has been studied extensively by EM (Gall, 1961; Robbins et al., 1968; Rattner and Phillips, 1973) and, despite much structural diversity, some common features are apparent in the initial events. Duplication takes place in the immediate vicinity of the existing or parent spindle pole and at a particular position in relation to that pole, suggesting that a template was laid down by the existing pole for the assembly of the new or daughter pole. Thus, provided assembly was confined to the template and restricted to one round per cell cycle, only one new pole would be produced.
In Saccharomyces cerevisiae the structure which forms the spindle pole is called the spindle pole body (SPB).1 The SPB is embedded in the nuclear envelope which remains intact during mitosis (Byers, 1981a). Duplication of the SPB is associated with the half-bridge, a specialization of the nuclear membrane attached to one side of the SPB. In early G1 an apparently spherical structure called the satellite assembles on the distal end of the cytoplasmic side of the half-bridge, ∼150 nm from the existing SPB (Byers, 1981a). Although there is no direct evidence, it seems likely that the new SPB assembles from this satellite and is then inserted into the nuclear envelope to give side-by-side SPBs connected by the bridge (Byers, 1981a). In Schizosaccharomyces pombe the duplication process is quite similar, in that the new SPB also assembles on the end of a bridge structure attached to the existing SPB (Ding et al., 1997). In higher eukaryotes the initial steps in centrosome duplication take place during S phase with the assembly of a procentriole from the parent centriole. Here there is no structural evidence for a parental template but it seems likely that one exists because of the fairly precise arrangement between the procentriole and the parent centriole. Procentriole assembly usually commences in an orthogonal arrangement ∼70 nm from the proximal end of the parent centriole (Gall, 1961; Robbins et al., 1968; Rattner and Phillips, 1973). This precise arrangement between the procentriole and the parent centriole is particularly apparent during basal body formation where multiple procentrioles assemble at a set distance around the parent centriole or basal body (Gall, 1961; Sorokin, 1968; Anderson and Brenner, 1971).
We have little understanding in molecular terms about the duplication of spindle poles. The best studied system for spindle poles is the SPB of S. cerevisiae. Here there is increasing evidence that this is a good model system for the spindle pole of higher eukaryotes because some SPB components have homologues in the centrosome. Thus, Cdc31p (Byers, 1981b; Spang et al., 1993) is homologous to the mammalian centrosomal component centrin (Salisbury, 1995). Two other proteins, Spc98p (Rout and Kilmartin, 1990; Geissler et al., 1996) and Spc97p (Knop et al., 1997), which are present in a complex with the γ-tubulin homologue Tub4p (Knop and Schiebel, 1997), also have homologues in the vertebrate centrosomal γ-tubulin complex (Martin et al., 1998; Murphy et al., 1998; Tassin et al., 1998). In S. cerevisiae most mutants which show defects in SPB duplication either show these at very early or very late stages. Thus, cdc31 (Byers, 1981b), kar1 (Rose and Fink, 1987), mps1 (Winey et al., 1991), dsk2Δ rad23Δ (Biggins et al., 1996), and pcs1 (McDonald and Byers, 1997) fail to form daughter SPBs, while most but not all of the daughter SPBs are assembled in mps2 (Winey et al., 1991), ndc1 (Winey et al., 1993), and cdc37-1 (Schutz et al., 1997). A mutant which may arrest at a more intermediate stage is spc42-10 (Donaldson and Kilmartin, 1996). Here cells have a single SPB with disordered material associated with the cytoplasmic face of the half-bridge, suggesting an attempt at SPB duplication. Spc42p probably forms a crystalline layer in the cytoplasmic part of the central plaque of the SPB (Bullitt et al., 1997), and it has been proposed that SPB assembly would commence with a lateral expansion of this crystalline layer followed by a vertical expansion to produce the various layers in the SPB (Bullitt et al., 1997).
In this paper we examine the role of core SPB components in SPB duplication. First we look at the mechanism of SPB duplication by EM and show that the satellite is replaced by a plaque-like structure attached to the cytoplasmic side of the half-bridge at intermediate stages. We show by immunoEM that cytoplasmic SPB components, which we have identified in highly enriched SPB cores, are present in the satellite and the plaque-like intermediate, suggesting a precursor–product relationship. Finally, we show that by overexpression of SPB components we can either alter the structure of the satellite or inhibit SPB duplication by disrupting the attachment of the plaque-like intermediate to the half-bridge.
Materials and Methods
Preparation of Heparin-extracted SPB Cores
Percoll SPBs (Wigge et al., 1998) were pelleted twice to remove Percoll (Rout and Kilmartin, 1990), then diluted ∼50-fold into bis-tris (bt) buffer (Rout and Kilmartin, 1990) containing 1 mg/ml heparin, 0.1 mM DTT, and left overnight in ice. Extracted SPBs were separated on gradients of 1.75, 2.0, 2.25, and 2.5 M sucrose-bt in a Beckman SW40 rotor at 40,000 rpm for 4 h. SPB cores (no band was visible) were taken from the 2.0 M layer and the 2–2.25 M interface, then diluted with 1:1 bt-DMSO (Rout and Kilmartin, 1990), pelleted (330,000 g, 15 min), and treated with either alkaline phosphatase (Wigge et al., 1998) or λ phosphatase (New England Biolabs; 90 U/μl, 3 h, 37°C) before SDS gel electrophoresis. For thin section EM the very thin pellet of SPB cores was located with a hand magnifier and encapsulated in 2% agarose after fixation to prevent breakup during processing.
Mass Spectrometry
SDS gel bands were digested with trypsin (Wigge et al., 1998) and the tryptic peptide masses determined by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry in a PerSeptive Biosystems Voyager-DE STR mass spectrometer using matrix peaks and trypsin peptides as internal standards. The NCBI nonredundant database of >300,000 proteins was searched using MS-fit (http://prospector.ucsf.edu) set at 50 parts per million, 0–300 kD and with all modifications and extra cleavages excluded. This decreased the number of peptides matched but increased the specificity of the match. All the proteins identified in the SPB core gel were the top match with MOWSE (Pappin et al., 1993) scores (P factor 0.4) of between 2 × 105 and 3 × 109 apart from Spc29p which had a score of 1 × 104. A second search allowing methionine oxidation, protein NH2-terminal acetylation, and two missed tryptic cleavages was then carried out to match further peptides. For the identifications in this paper the first number in brackets is the number of tryptic peptides identified followed by the percentage sequence covered. These were: Spc110p dimer (18, 21%), Pom152p (13, 10%), Spc110p (42, 33%), Nud1p (12, 21%), Cnm67p (23, 33%), Spc42p (22, 36%), Spc29p (9, 42%), Sec53p (14, 38%), and Cmd1p (9, 61%).
Yeast Strains
All yeast strains were prepared in Nasmyth's (IMP, Vienna) K699 background or the isogenic diploid K842, all vectors used were the pRS series (Sikorski and Hieter, 1989). Most strains were made using the direct PCR method (Baudin et al., 1993) in K842 using as markers either the Schizosaccharomyces pombe HIS5 or the Kluyveromyces lactis URA3 genes followed by sporulation at 23°C. Standard yeast genetic methods were used (Guthrie and Fink, 1991). All strains were checked by colony PCR. All base pair numbers start from the A of the presumed initiator methionine. All tags were placed either at the NH2 terminus (written as for example green fluorescent protein GFP-Spc110p) or at the COOH terminus (written as Spc42p-GFP). All tagged strains had the wild-type gene removed and grew at the same rate as untagged strains.
The construct for Spc42p-GFP was prepared by PCR amplification of GFP (S65T) and insertion into the ClaI site of SPC42 at the 3′ end. This was designed to insert a short polypeptide linker (GA)5 between the COOH terminus of Spc42p and the NH2 terminus of GFP. The construct was integrated into the TRP1 locus of the covered knockout strain AY4 (Donaldson and Kilmartin, 1996) as either a single or about three copies (IAY18) and the wild-type gene removed by plasmid shuffle.
Strains containing a deletion of CNM67 marked with HIS5 and using the K. lactis URA3 marker to tag either NUD1 or SPC72 with GFP at the 3′ end (Wigge et al., 1998) were prepared, together with the same strains rescued with CNM67 on a LEU2 CEN vector. A strain containing a deletion of CNM67 and SPC42-GFP was prepared by mating with IAY18.
Temperature-sensitive (ts) mutants in NUD1 and SPC29 were prepared (Muhlrad et al., 1992) first by cloning the wild-type genes by gap repair to give bases −1501 to 4054 for NUD1 and bases −666 to 1382 for SPC29 and preparing the covered knockout strains from the heterozygous knockout diploid strains (Wigge et al., 1998). The two genes were amplified under error prone conditions using primers at −5 to 16 and 2726 to 2705 for NUD1 and −666 to −645 and 1382 to 1371 for SPC29 (the 3.1-kb open reading frame at the 5′ end of SPC29 terminates at −269). These were transformed into the covered knockout strains together with the gapped plasmids, cut at PflMI (165) and BsiWI (2572) for NUD1 and BsmI (−495) and ClaI (684) for SPC29. Plasmids were recovered from ts strains, and integrated at the TRPI locus as single copies to give nud1-44 and spc29-20 after removal of the wild-type gene. Both alleles were recessive, grew at normal rates on plates at 23°C, did not grow above 30°C, and were fully rescued by the wild-type genes on CEN plasmids at 37°C. Both alleles were sequenced between the PCR primers and the changes found for nud1-44 were Q96L, E127V, I418V, S419P, V504G, S534C, N600I, N613D, and I633F. For spc29-20 they were A-70G, T-15C (these two changes are in the 5′ noncoding region), E166D, and I220T. Two additional alleles were also examined nud1-52, I353F and Y696N, and spc29-10, A-389Δ, T-143Δ, A-121G, and L165Q. The changes in the promoter region in the two spc29 alleles were not responsible for the ts phenotype since the phenotype remained when this region was replaced by the wild-type promoter.
The constructs for GFP-Spc110p and GFP-Spc29p were prepared by PCR amplification of GFP between BspLU11 I sites and ligation of this into NcoI sites at the initiator methionine of SPC110 in pPY133 (Kilmartin and Goh, 1996) and SPC29 (the NcoI site was made by PCR). The initiator methionines were replaced with a short polypeptide linker (GAGA) between it and the COOH terminus of GFP. These constructs replaced the wild-type gene after integration at the TRP1 locus as multiple copies for GFP-SPC110 (JKY1143) and as either single (for immunoEM) or multiple copies for GFP-SPC29.
Versions of the earlier ts alleles of SPC42 (Donaldson and Kilmartin, 1996) and SPC110 (Kilmartin and Goh, 1996) integrated at the SPC42 or SPC110 loci without markers were prepared either by the pop-in/pop-out procedure to give spc42-10i, or by replacement of the disrupted SPC110 with the PCR-amplified ts allele (Tyers et al., 1993) to give spc110-1i and spc110-2i.
A tetraploid version of K699 homozygous at the MAT a locus was prepared by transformation of K842 with GAL-HO to make two diploid strains homozygous at either the MAT a or MATα locus. These strains were mated and GAL-HO used again to prepare the tetraploid strain homozygous for MAT a. The ploidy of this strain was confirmed by flow cytometry.
Strains containing GFP-labeled Spc42p, Spc110p, and Spc29p in mps2-1 (Winey et al., 1991) were prepared by backcrossing mps2-1 with K699 once, then crossing in IAY18, JKY1143, and a strain containing multiple integrated copies of GFP-SPC29. These particular strains would not be completely in K699 background.
A version of Spc42p with all 34 serines changed to alanine (S:A Spc42p) was prepared by a combination of site-directed mutagenesis, directed mutagenesis by PCR, and reconstruction of the gene by overlapping 80-mer oligos. This sequence is available from the authors. The S:A Spc42p gene was integrated as a single copy at the TRP1 locus of AY4 and the wild-type gene removed by plasmid shuffle. The integrated gene was amplified by PCR and sequenced to confirm all serines were replaced by alanine. The copy number in this transformant was further increased by integration of multiple copies of the S:A Spc42p gene at the URA3 locus. A version of this gene under the control of the GAL promoter and with a hemagglutinin tag at the NH2 terminus was prepared and integrated as a single copy at the URA3 locus in K699.
We removed a potential bipartite nuclear localization sequence (NLS) in Spc110p situated between residues 24–59 by replacing K24, 26, 54, and 55, and R27, 56, 58, and 59 with alanine using overlapping oligos in a construct designed to overexpress SPC110 (Kilmartin and Goh, 1996) under the control of the GAL promoter and with a myc tag at the NH2 terminus. This construct was integrated at the URA3 locus of K699 and IAY18 as a single copy. As a control to test the function of ΔNLS-Spc110p we inserted the SV-40 T-antigen NLS (MARPKKKRKVA) between the myc tag and the NH2 terminus and used the wild-type promoter. Strains containing only this compensated version of Spc110p grew normally.
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were performed essentially as described by Clontech. Two-hybrid constructs were made by PCR with a (GA)5 polypeptide linker at the NH2-terminal end, cloned into plasmids pGAD424 and pGBT9 (full-length Spc42p in the higher expression vectors pACT2 and pAS-1 was toxic in yeast), transformed into Y190 and Y187, respectively, and lacZ assays performed after mating. The fragments used were: Cnm67p-Cterm (amino acids 442–581), Spc42p-coil-Cterm (49–363), Spc42p-Nterm-coil (1–141), and Spc110p-Cterm (781–944). Constructs containing only the coiled-coil domains of Spc42p (49–141) or Spc110p (161–800) were not detected by Western blotting, and constructs containing the NH2 terminus of Cnm67p (1–180) in pGBT9 showed nonspecific activation of the reporter gene.
ImmunoEM
The immunoEM protocol described in this paper was developed specifically to detect transient SPB duplication intermediates which might be rarely present even in synchronized cells. We used a preembedding staining method because of its greater sensitivity; however, a major problem with SPB antigens is that even short periods of formaldehyde fixation (1 min in the case of Spc42p) can abolish their reactivity with both polyclonal and monoclonal antibodies. In the case of Spc42p we tried a number of different epitope tags, including single and multiple myc and hemagglutinin tags at the NH2 and COOH termini, but found GFP to be far superior in retaining reactivity after formaldehyde fixation. We found that during processing for immunoEM the cells fractured open; this was very advantageous because we could avoid detergent permeabilization and thus preserve the nuclear membrane which was necessary for the proper identification of the half-bridge, satellite, and duplication plaque.
Cells were harvested by centrifugation, washed once with H2O, then fixed in 3.7% formaldehyde solution (BDH Chemicals), 0.1 M potassium phosphate, pH 6.5, for 20 min at 22°C. After three washes with 0.1 M potassium phosphate, pH 6.5, and one with 0.1 M phosphate citrate buffer, pH 5.8 (PC), the cells were incubated with 10% vol/vol glusulase (DuPont NEN) and 0.1 mg/ml zymolyase 20T (ICN Biomedicals) in PC at 30°C for 1 h. The samples were then washed once with PC, incubated with 50 mM glycine in PBS at 4°C for 5 min, washed twice with 0.5 ml PBS-BSA, and incubated with either affinity-purified rabbit anti-GFP antibody (a gift from K. Sawin, ICRF, London, United Kingdom) diluted 1:150 in PBS-BSA, 1% solution P (90 mg PMSF, 2 mg pepstatin in 5 ml absolute ethanol), 30–100 μg/ml of purified 9E10 or 12CA5, or with affinity-purified rabbit anti-Tub4p (Wigge et al., 1998) diluted 1:3,000 for 1 h at 22°C. After washing three times with PBS-BSA, the samples were incubated with a 1:50 dilution of Nanogold-labeled goat anti–rabbit Fab′ fragment or 1:20 dilution of goat anti–mouse Fab′ fragment (Nanoprobes Inc.) in PBS-BSA, 1% solution P at 22°C for 1 h. After one wash with PBS-BSA, and three with PBS, the samples were fixed with 2.5% glutaraldehyde (biological grade; Polysciences Inc.) in 40 mM potassium phosphate, pH 6.5, 0.5 mM MgCl2 for 2 h at 22°C. The glutaraldehyde was removed by washing once with buffer, then incubating the samples three times with 50 mM MES, 200 mM sucrose, pH 6.0, at 4°C for 5 min each. The Nanogold particles were then silver enhanced in the dark for 3–5 min at 22°C with NPG silver enhancement solution (Gilerovitch et al., 1995). After enhancement the samples were washed three times with cold 200 mM MES, pH 6.15, in the dark over 10 min, removed from the dark room, and washed twice with 0.1 M sodium acetate, pH 6.1. After postfixing with 2% osmium tetroxide the samples were processed for serial thin section EM.
Other Methods
EM, flow cytometry, elutriation, and GAL induction were as before (Kilmartin and Goh, 1996), as was immunofluorescence with anti-GFP (Kilmartin et al., 1993) and anti-Tub4p (Wigge et al., 1998). The anti-GFP antibodies used were affinity-purified rabbit polyclonals kindly given by K. Sawin (ICRF) and P. Silver (Dana-Farber Cancer Institute, Boston, MA), and mouse monoclonal 3E6 (Quantum Biotechnology). Cells (107/ml) were treated with α-factor (10 μg/ml, 20 μg/ml for tetraploid cells) for 105 min (or 3 h for the prolonged treatment) at 30°C, or with DAPI (5 μg/ml) for 20 min.
Results
EM of SPB Duplication
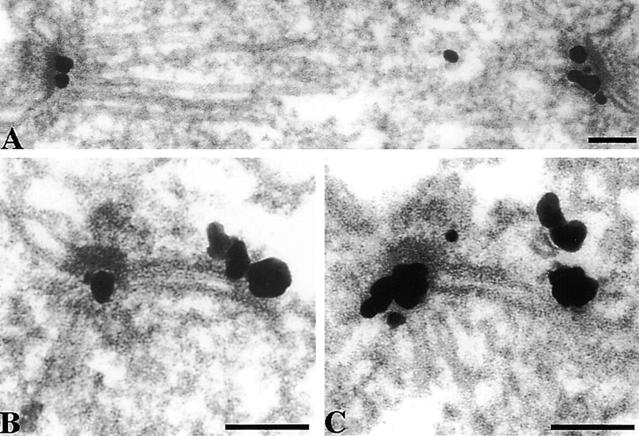
At present little is known about the process of SPB duplication because intermediates have not been observed by EM of vegetatively growing cells (Byers, 1981a) due presumably to the rapidity of the process. It has been known for some time that the process probably starts with the assembly of the satellite structure at the distal end of the half-bridge (Fig. 1 A, arrow). (In all of the EM figures in this paper, except for spindles, the cytoplasm is at the top, the nucleoplasm at the bottom, and the new SPB on the right.) Assembly of the satellite occurs between the end of anaphase and Start (Byers and Goetsch, 1974, 1975). There are some characteristic features of the half-bridge evident from these and other (Schutz and Winey, 1998) micrographs and those in Fig. 1: the lipid bilayers are more densely stained and are continuous with the bilayers outside of the half-bridge (Fig. 1 A). The bilayer on the inner nuclear side has a very thin layer of electron dense material closely associated with it whereas the cytoplasmic outer bilayer has a thicker layer which we call the half-bridge outer layer, it runs parallel to the bilayer in the cytoplasm with a gap of ∼15–20 nm and appears to intersect with the central plaque and the satellite (Fig. 1, A and B, and see Fig. 9). We looked for intermediate structures after the satellite stage by examining cells synchronized with α-factor and released for 30 min to increase the fraction of cells undergoing SPB duplication. In some cells at this time point the half-bridge structure appeared elongated and the satellite was replaced by a large plaque-like structure on the cytoplasmic side of the half-bridge (serial sections in Fig. 1, B1 and B2, and single section in Fig. 1 C), which we call the duplication plaque. This plaque, which was similar in diameter to the existing central plaque, appeared to have a more direct association with the cytoplasmic lipid bilayer of the half-bridge than the half-bridge outer layer. The duplication plaque is very similar in structure to the partly assembled SPB attached to the cytoplasmic side of the half-bridge observed at the arrest phenotype of mps2-1 (Winey et al., 1991), ndc1-1 (Winey et al., 1993), and mps1-737 (Schutz and Winey, 1998). There was a characteristic bend in the half-bridge associated with the side of the duplication plaque proximal to the existing SPB. At this time point the distal end of the half-bridge was often fused (Fig. 1, B1 and C) with usually a pore structure immediately adjacent to the fused half-bridge (Fig. 1 C, arrow). This pore, which was not morphologically distinguishable from a nuclear pore complex, may aid in fusion of the half-bridge. A proposed further stage in the duplication process was found in some cells where the duplication plaque was partly inserted into the nuclear membrane so that it appeared to be in direct contact with the nucleoplasm. This insertion was often at an angle to the existing SPB, and the plaque did not appear to have nuclear microtubules attached, though material, presumably outer plaque, was attached to the cytoplasmic side (Fig. 1, D and E). Finally at 45 min after α-factor release at 30°C all cells examined had side-by-side (Fig. 1 F) or separated SPBs.
Figure 1.
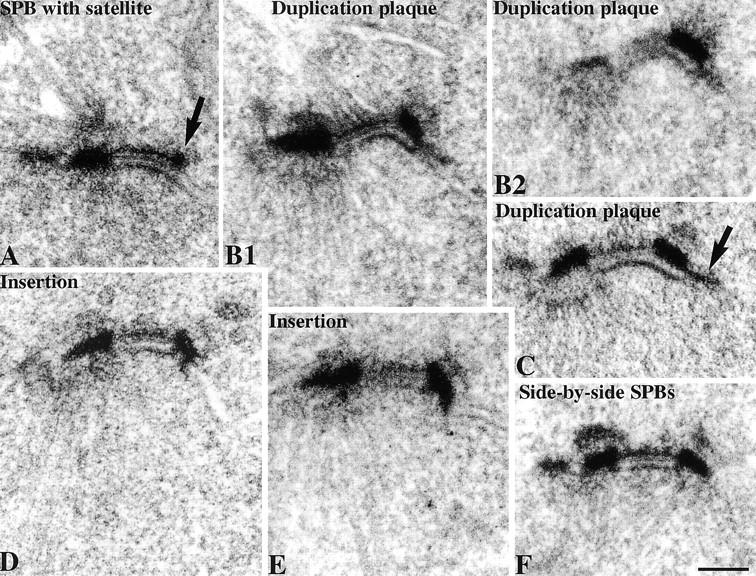
Stages during SPB duplication as shown by EM of thin sections of the SPB region of cells arrested in α-factor (A), released for 30 min (B1–E), or 45 min (F). Arrows in A show the satellite and in C a pore structure attached to the fused half-bridge. Serial thin sections are shown in B1 and B2. In all of the EM figures in this paper except spindles, cytoplasm is in the top part of each panel and the existing SPB is on the left. Bar, 0.1 μm.
Figure 9.
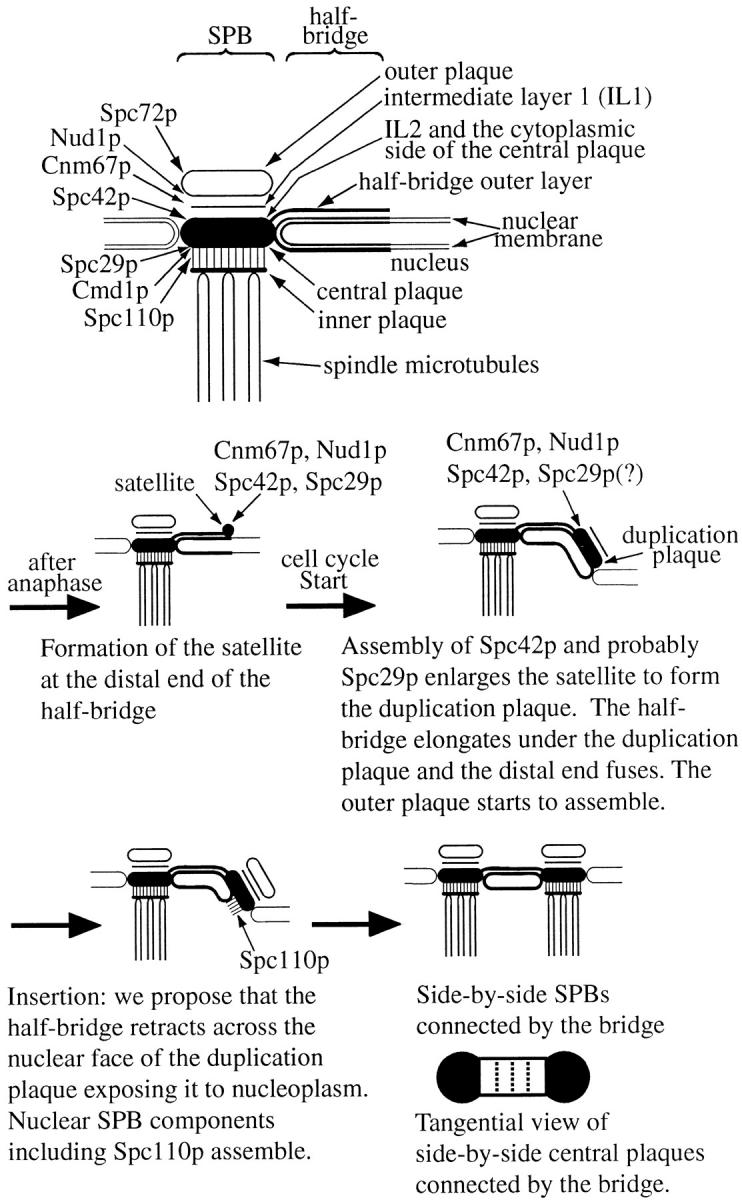
Diagram of the main core SPB components during the different stages of SPB duplication. Cytoplasmic microtubules which can grow from both the outer plaque and the half-bridge during SPB duplication are not shown.
A simple interpretation of these images (shown schematically in Fig. 9) is that the duplication plaque is a precursor of the central plaque and its assembly is initiated from the satellite. Towards the end of this assembly the distal end of the half-bridge fuses to allow insertion of the duplication plaque into the nuclear envelope, followed by the formation of nuclear microtubules. These results suggest that SPB duplication is initially a cytoplasmic event with the later stages taking place in the nucleus.
Main Components of the SPB Core
We now wished to examine the role of the main structural components of the SPB in the cytological events of SPB duplication described above. Since the initial events in SPB duplication mainly involve the central part of the SPB, we analyzed an SPB core fraction by MALDI mass spectrometry analysis (Wigge et al., 1998) to identify the main components concerned. These heparin-extracted SPB cores (Rout and Kilmartin, 1990) appeared to contain a partly depleted outer plaque, intermediate layer 1 (IL1), and the central plaque, which in conventional EM thin sections includes IL2 (Bullitt et al., 1997); the inner plaque appeared to be absent (Fig. 2 B). A thin section through the whole pellet of this material shows only profiles of SPB cores sectioned at various angles, suggesting that they are highly enriched (Fig. 2 A). Coomassie-stained SDS gels of SPB cores (Fig. 2, C and D) show only a few main bands when compared with intact spindle poles. Alkaline phosphatase was used to sharpen the bands of the phosphoproteins (Wigge et al., 1998), and gels from two preparations are presented to show the bands consistent between different preparations (Fig. 2, C and D). In addition we show λ phosphatase–digested material to show that there are no main bands concealed by the alkaline phosphatase (Fig. 2 E). The main SPB core bands were identified by MALDI mass spectrometry as Pom152p, Spc110p, Nud1p, Cnm67p, Spc42p, Spc29p, Sec53p (phosphomannomutase), and Cmd1p (calmodulin). All of these proteins had been identified previously in our preparation of spindle poles (Wigge et al., 1998). It seems likely that Pom152p and Sec53p are contaminants as they were in the earlier preparation (Wigge et al., 1998). Spc110p, Nud1p, Cnm67p, Spc42p, and Cmd1p are all SPB components and have been shown to localize to the central and outer plaques (Rout and Kilmartin, 1990, 1991; Spang et al., 1996b; Sundberg et al., 1996; Wigge et al., 1998), but Spc29p has not yet been localized within the SPB. We have now localized GFP-Spc29p by immunoEM to the nuclear side of the central plaque in 90% of the SPBs examined (Fig. 3 A). The other 10% showed additional cytoplasmic staining of the central plaque. However in ∼25% of the SPBs, which serial sectioning showed were single SPBs, there was an additional focus of GFP-Spc29p staining to one side. When the half-bridge was correctly oriented in the section, this staining was clearly located at the distal cytoplasmic end of the half-bridge (Fig. 3, B and C). Thus, GFP-Spc29p has both a nuclear and cytoplasmic localization like some other SPB components such as Spc98p (Rout and Kilmartin, 1990; Geissler et al., 1996). When synchronized cells were examined, staining of GFP-Spc29p at the end of the half-bridge was absent in cells with small buds and detected again when large-budded and unbudded cells were present. This suggests an association between the staining and the presence of the satellite, and indeed later we show localization of GFP-Spc29p to the satellite in synchronized cells (see Fig. 6, E1 and E2). We also examined Nud1p-GFP and Spc42p-GFP log phase cells and found a similar proportion of SPBs showing cytoplasmic staining at the distal end of the half-bridge (data not shown). The proportion of single SPBs showing distal half-bridge staining (20–25%) is higher than the proportion of satellite-bearing SPBs in a log phase culture in rich medium (∼8% in the diploid K842, data not shown). This suggests that in addition to satellite staining, a precursor to the satellite might also be stained. Some of these SPBs did appear to lack satellites; however, these could have been obscured by silver deposition. In addition, during processing for immunoEM satellite-bearing SPBs could have been enriched and the satellite partly removed. Therefore, a more definite conclusion as to whether a satellite precursor is being stained should come from immunoEM of GFP-tagged SPB components in some of the cell cycle mutants.
Figure 2.
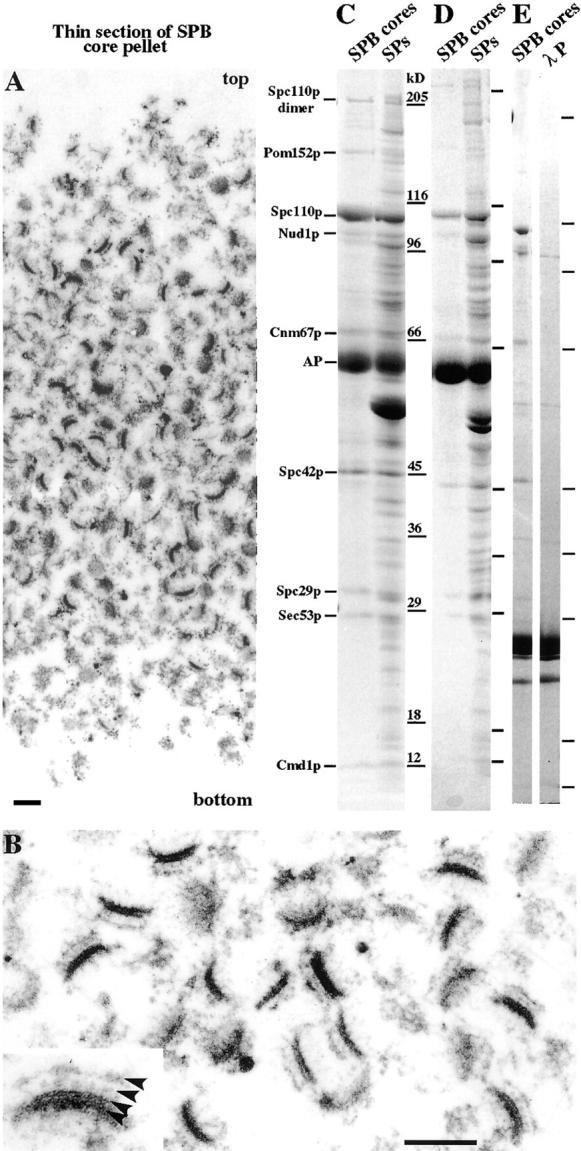
Analysis of heparin-extracted SPB cores. (A) Thin section of the entire pellet. B is a higher magnification, and the inset in B shows the structural features of the lower right SPB core with arrowheads showing from the top down: the depleted outer plaque, IL1, IL2, and the central plaque. In SPBs from whole cells the IL2 and central plaque layers appear fused and together are called the central plaque. (C and D) Coomassie-stained SDS gradient gels of alkaline phosphatase (AP) or (E) λ phosphatase (λP)-digested SPB cores together with λ phosphatase alone. Identifications of the main bands by MALDI mass spectrometry are to the left of C. The SPB cores in C were isolated from ∼1011 cells. Lines beside the gel lanes in D and E show the positions of the same molecular weight standards used in C. Bars, 0.2 μm.
Figure 3.
Localization of GFP-Spc29p by immunoEM in log phase cells. (A) Short spindle. (B and C) Single SPBs with half-bridges. About 200 SPBs were examined. Bars, 0.1 μm.
Figure 6.
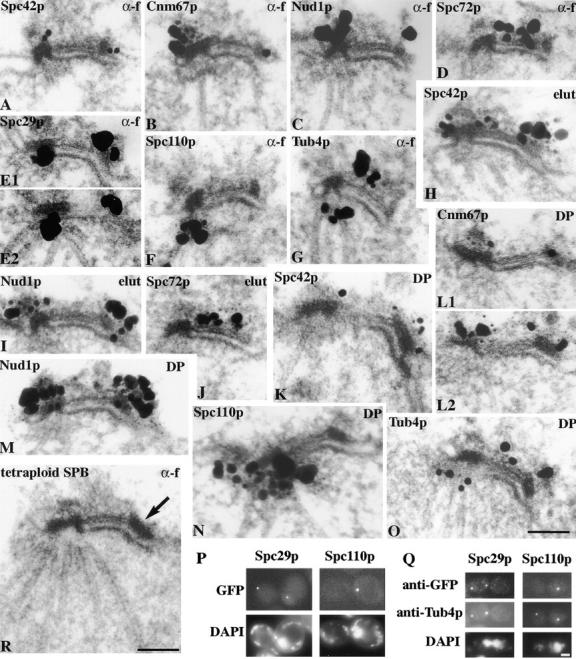
ImmunoEM localization of GFP-labeled Spc42p, Cnm67p, Nud1p, Spc72p, Spc29p, and Spc110p in cells arrested in G1 with α-factor (G1 α-f), G1 elutriated cells grown for 25 min (elut), and cells released from α-factor for 30 min to observe the duplication plaque (DP). Tub4p was detected with affinity-purified polyclonal antibodies. Adjacent serial sections have the same panel letter and are numbered. (P) Fluorescence of GFP-Spc29p and GFP-Spc110p in unfixed mps2 cells after 3 h at 37°C then treated with DAPI for 0.5 h at 37°C; (Q) immunofluorescence of the same cells with anti-GFP and anti-Tub4p; (R) EM of a thin section of a tetraploid MATa cell arrested with α-factor. The arrow indicates the satellite. Bars, 0.1 μm for the immunoEM and EM and 2 μm for the light microscopy.
In summary we have characterized an SPB core fraction and shown that it is composed mainly of six SPB components all of which localize to the central and outer plaques.
Approximate Arrangement of SPB Components
Since the initial stages of SPB duplication are cytoplasmic (see above) we were particularly interested in the main SPB components which had a cytoplasmic location in the SPB and how they were arranged, since components close to the central plaque are more likely to be involved in the initial events of SPB duplication. First we considered the outer plaque components Cnm67p, Nud1p (Wigge et al., 1998), and Spc72p (Knop and Schiebel, 1998; Wigge et al., 1998). Spc72p is depleted from the SPB core preparation suggesting it has a peripheral location, consistent with the finding that it interacts with the Tub4p complex (Knop and Schiebel, 1998). Cells containing a deletion of CNM67 have depleted outer plaques but intact central plaques (Brachat et al., 1998); thus, we have used this strain to establish which of these outer plaque components are still present at the SPB. We present most of the data as the GFP and DAPI staining given by unfixed cells as this is probably free from fixation artifacts such as the absence of staining caused by over-fixation (Kilmartin et al., 1993). In unfixed anaphase, B cells' SPB position can be predicted without the need for tubulin staining as close to the two front edges of the separating chromatin (Kilmartin and Adams, 1984). For fixed cells, we mostly used immunoEM instead of immunofluorescence because of its greater resolution.
As expected, we found that Spc42p-GFP localizes to the SPB region in a strain containing a deletion of Cnm67p (Fig. 4, A and B) which has a depleted outer plaque. However, to our surprise we found that the outer plaque component Spc72p-GFP also localized to the SPB region (Fig. 4, C and D) in the same background. The reason for this became clear on immunoEM where, in contrast to Spc42p-GFP which localized as normal to the central plaque (data not shown), Spc72p-GFP localized to the half-bridge in all the SPBs examined (Fig. 4 E), including those in short spindles. Cytoplasmic microtubules are attached to the half-bridge throughout the cell cycle in cells deleted for Cnm67p (Brachat et al., 1998), and Spc72p is presumably associated with the Tub4p complex (Knop and Schiebel, 1998) at this site. When this strain was rescued by transformation with CNM67, Spc72p-GFP now localized normally to the outer plaque (Fig. 4 F). When Nud1p-GFP was localized in the strain deleted for Cnm67p we found two types of staining pattern. Most cells with buds had a prominent dot which was probably not associated with the SPB since it was not coincident with DAPI staining (Fig. 4, G and H). However, in unbudded cells and cells with small buds we found a weakly staining dot which was DAPI-associated (data not shown) and immunofluorescence showed this was SPB-associated (Fig. 4, I and J). ImmunoEM of these cells showed that most of the SPBs were unstained, but ∼10% had Nud1p-GFP localized to the half-bridge (Fig. 4 K), or with an unstructured particle associated with the half-bridge (data not shown). Most of the stained SPBs appeared to have a satellite or were side-by-side SPBs showing that they had come from unbudded or small-budded cells. When these cells are rescued with CNM67 we found that in all cells Nud1p-GFP now localized to the SPB region (Fig. 4, L and M) and immunoEM showed it had returned to the outer plaque in all the SPBs examined (Fig. 4 N).
Figure 4.
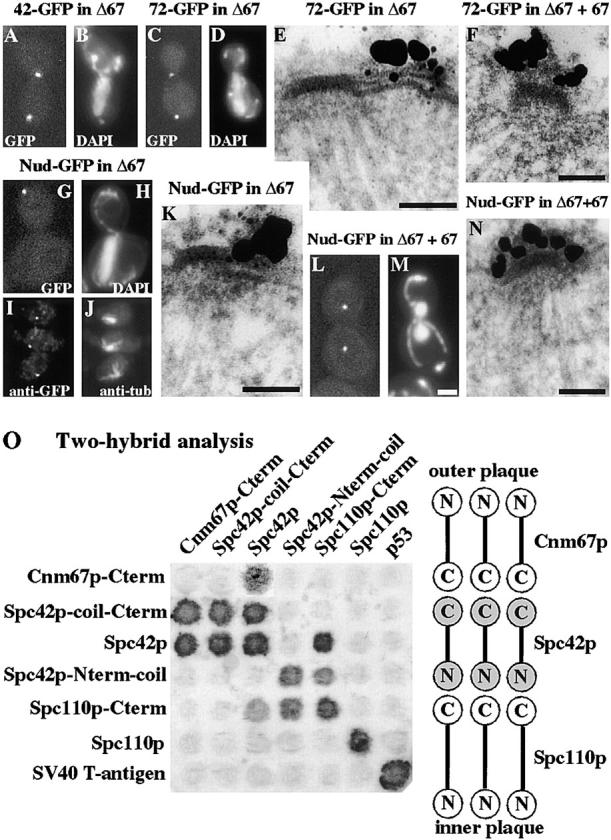
Localization of Spc42p-GFP (42-GFP), Spc72p-GFP (72-GFP), and Nud1p-GFP (Nud-GFP) in cells containing a deletion of CNM67 (Δ67) with or without a rescuing plasmid carrying CNM67 (67), using either fluorescence of unfixed cells treated with DAPI (A–D, G, H, L, and M), immunofluorescence with anti-GFP (I) and anti-tubulin (J), or immunoEM (E, F, K, and N). About 200 SPBs were examined for E and K, and ∼30 SPBs for F and N. (O) Two-hybrid analysis of the interactions between Cnm67p, Spc42p, and Spc110p. The activation domain is on the left and the DNA-binding domain at the top. Bars, 0.1 μm for the immunoEM and 2 μm for the light microscopy.
In conclusion these results show that while the localization of Spc42p-GFP to the cytoplasmic side of the central plaque is independent of Cnm67p, the localization of Spc72p-GFP and Nud1p-GFP to the outer plaque requires Cnm67p. This, coupled with the depletion of Spc72p in the SPB cores, suggests that the approximate order of cytoplasmic SPB components from the nuclear membrane out is Spc42p, Cnm67p, Nud1p, and Spc72p.
The finding that Spc42p is the closest cytoplasmic SPB component to the nucleus suggests that it should interact in some way with the nuclear SPB components Spc110p, Cmd1p, and Spc29p. We tested for interactions between Spc42p and Spc110p since both have been detected in an enriched complex (Knop and Schiebel, 1997), and we also sought to confirm possible interactions between Spc42p and Cnm67p using both a genetic and a two-hybrid approach.
We found synthetic lethal effects between SPC110 and SPC42. An enhanced ts phenotype (by 5°C) was found in the double spc110-2i spc42-10i mutant. Synthetic lethal effects were also detected between CNM67 and SPC42 but not SPC110. Here cells containing a deletion of CNM67 maintained by CNM67 on a CEN URA3 plasmid were spotted at various dilutions onto 5-fluoroorotic acid media at 23°C which selects against the URA3 plasmid. When the spc42-10i mutation was present, the plasmid loss rate was reduced 1,000-fold as judged by the absence of colony growth at high dilutions, but was unaffected by the presence of the spc110-1i or spc110-2i mutations.
We also looked at the interaction between Spc42p, Spc110p, and Cnm67p by two-hybrid analysis (Fig. 4 O). Here we found an interaction between the NH2 terminus of Spc42p and the COOH terminus of Spc110p and between the COOH terminus of Spc42p and the COOH terminus of Cnm67p. These interactions may not be direct and may also involve other SPB components, particularly the nuclear components Cmd1p (calmodulin) and Spc29p (we were able to detect a very weak two-hybrid interaction between full-length Spc29p and the NH2 terminus of Spc42p, data not shown). Both Spc29p and Cmd1p localize close to the nuclear edge of the central plaque (Fig. 3; Spang et al., 1996b; Sundberg et al., 1996); this is where the COOH terminus of Spc110p probably resides since Cmd1p has been shown to interact with this part of Spc110p (Geiser et al., 1993; Stirling et al., 1994). The localizations of these SPB components are shown schematically in Fig. 9. The finding that Spc42p has different interactions with its NH2 and COOH termini suggests that it exists as a single layer in the SPB rather than the double layer which is its probable arrangement in the overexpressed polymer (Bullitt et al., 1997).
Phenotypes of ts Mutants in NUD1 and SPC29
Of the six core SPB components described in the above section, five are essential (Kilmartin et al., 1993; Donaldson and Kilmartin, 1996; Wigge et al., 1998) and one, Cnm67p (Brachat et al., 1998), is non-essential. The cytoplasmic peripheral component Spc72p is also non-essential (Souès and Adams, 1998), although this depends on the genetic background (Chen et al., 1998; Knop and Schiebel, 1998). Of the five essential genes, EM phenotypes have been described for CMD1 (Sun et al., 1992), SPC42 (Donaldson and Kilmartin, 1996), and SPC110 (Kilmartin and Goh, 1996; Sundberg et al., 1996; Sundberg and Davis, 1997). spc42 mutants have a defect in SPB duplication, whereas spc110 and cmd1-101 mutants appear to duplicate most of the SPB normally but have a defect in nuclear microtubule attachment; thus, the phenotype of these alleles is compatible with the location of the gene product in the SPB. We wished to examine the phenotypes of the two remaining essential SPB core components to see whether they were also compatible with their location in the SPB.
Elutriated G1 nud1-44 cells released at 37°C passed through mitosis at a slightly slower rate than wild-type cells (Donaldson and Kilmartin, 1996). Of 20 cells examined by EM at 2 h, 3 showed satellite-bearing SPBs, 4 side-by-side SPBs, 12 short spindles, and 1 a postanaphase spindle. At 4 h cells (>80% had large buds) contained postanaphase spindles (Fig. 5, A–C). Eight of these cells were examined by EM and SPBs were found at both ends of the spindle (Fig. 5 D). This phenotype suggests that the cells are unable to exit mitosis. The SPBs appeared to have a depleted outer plaque (Fig. 5 D) and cytoplasmic microtubules were often found to terminate at the half-bridge (Fig. 5, E1 and E2). This part of the phenotype is similar to that of cells lacking Cnm67p which also has a depleted outer plaque (Brachat et al., 1998).
Figure 5.
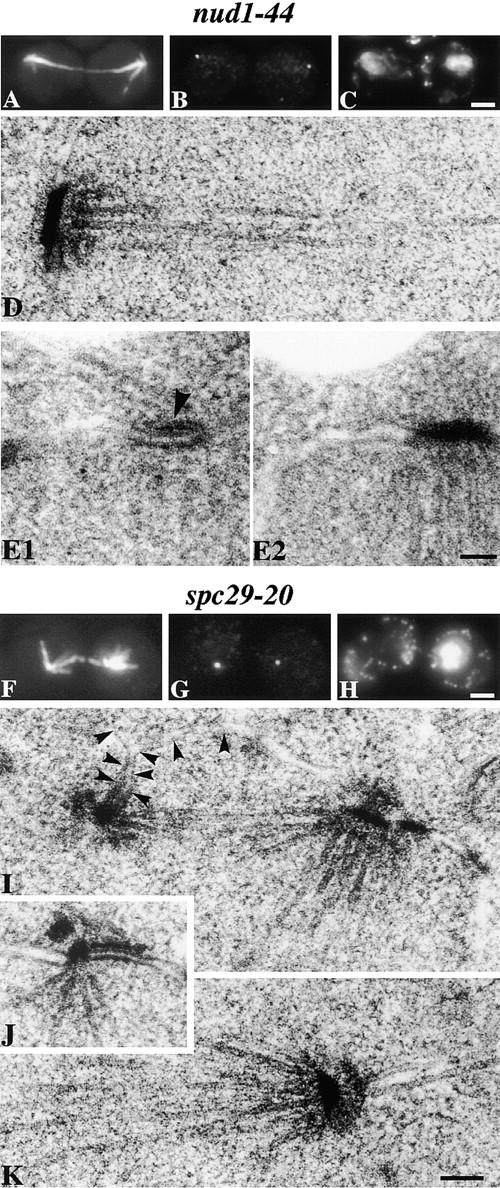
ts phenotype of nud1-44 and spc29-20 alleles. nud1-44 cells were synchronized by elutriation and spc29-20 cells synchronized by α-factor (90 min at 23°C followed by 60 min at 37°C). Immunofluorescence of a synchronized nud1-44 cell (A–C) and an spc29-20 cell (F–H) after 4 h at 37°C with anti-tubulin (A and F), anti-Tub4p (B and G), and DAPI (C and H). (D) EM of a synchronized nud1-44 cell after 4 h at 37°C showing an SPB at the end of a postanaphase spindle. Adjacent serial sections of a synchronized nud1-44 cell containing a short spindle after 2 h at 37°C (E1 and E2) showing that a cytoplasmic microtubule is still interacting with the half-bridge (arrowhead). EM of synchronized spc29-20 cells after 2 h (I and J) and 4 h (K) at 37°C. Arrowheads in I indicate the inner nuclear membrane and show the small SPB is still inserted in the nuclear membrane. Bars, 0.1 μm for the EM and 2 μm for the immunofluorescence.
spc29-20 cells synchronized with α-factor and released at 37°C completed DNA replication at 1 h, and immunofluorescence at 1.5 h suggested that ∼25% of the cells contained spindles (data not shown). However, EM analysis of nine spindles (and three side-by-side SPBs) between 1 and 2 h showed an abnormality: one apparently normal-sized SPB and one small SPB (Fig. 5 I). The small SPB, which is presumably the new one, remains inserted in the nuclear envelope at these time points and appeared to have a normal half-bridge (Fig. 5, I and J). Thus, although SPB duplication can occur in spc29-10 cells, SPB assembly is apparently defective. At 4 h nearly 80% of the cells had large or multiple buds, and immunofluorescence showed that two-thirds had one focus of microtubule staining while the other third showed an additional much weaker focus of microtubule staining which was not apparently associated with nuclear DNA (Fig. 5, F–H). Complete serial sectioning of whole nuclei from seven cells showed one apparently normal SPB (Fig. 5 K), but the nuclear microtubules from these SPBs were not apparently connected to another structure. We presume that the weaker focus of microtubule staining seen by immunofluorescence represents the smaller SPB which has become separated from the main body of the nucleus. We did not find these SPBs by EM, possibly due to partial breakdown of the structure, as was found in some spc110 alleles (Sundberg and Davis, 1997). Flow cytometry of both nud1-44 and spc29-20 showed they progressed normally through the first round of DNA replication, but did not appear to arrest with a G2 DNA content. However, we could not reach a definite conclusion on this because the peaks at later time points were often not distinct and could be caused by increased background due to the increased cell size (data not shown). Two other alleles, nud1-52 and spc29-10, were examined by immunofluorescence after an asynchronous block and showed similar phenotypes to nud1-44 and spc29-20, respectively (data not shown).
The apparent depleted outer plaque and consequent attachment of cytoplasmic microtubules to the half-bridge in the nud1-44 spindles is in agreement with the localization of Nud1p to the outer plaque in wild-type cells. However, Nud1p must have an additional essential function as shown by the ts phenotype: it is required for cells to exit mitosis. This function is not dependent on its localization to the outer plaque since Nud1p remains an essential gene product in the absence of Cnm67p (data not shown) where it can localize to the half-bridge (Fig. 4 K). Possibly the requirement for cells to exit mitosis might depend on a localization of Nud1p to the half-bridge which we have not yet been able to detect in wild-type cells. Incidentally, while most SPB components are coiled-coil proteins (Wigge et al., 1998), Nud1p is a leucine-rich repeat protein (Kobe and Deisenhofer, 1994), it has at least six repeats with a consensus sequence of XLX2LNLSXNXaX2aX2aX2a where the second asparagine is at positions 510, 553, 576, 597, 630, and 652 (X is any amino acid and a is normally aliphatic). It is not clear what this homology means. Leucine-rich repeats are found in functionally diverse proteins and are involved in interactions with other proteins (Kobe and Deisenhofer, 1994).
The phenotype of spc29-20 shows a defect in spindle structure, probably caused by one SPB being smaller than the other. This could be due to partial dissociation of the new SPB as was found in some spc110 alleles (Sundberg and Davis, 1997). Alternatively, if preassembled Spc29p were stable at 37°C in the mutant, then a partial SPB might assemble from the satellite but not grow to full size due to defective soluble Spc29p. The primary defect in this mutant may be caused by the smaller SPB having insufficient microtubules to carry all the chromosomes.
In conclusion we show that the phenotypes of ts mutants in Nud1p and Spc29p are in part compatible with their localization to the SPB.
Components of the Satellite, Duplication Plaque, and Half-Bridge
In the scheme for SPB duplication presented earlier (Fig. 1) the simplest interpretation of the order of events is that the satellite is the precursor of the duplication plaque, although there are other possibilities such as the satellite dissociating from the half-bridge during duplication and production of a new SPB from fission of the parent (Byers, 1981a). It is difficult to distinguish between these alternatives, particularly if the events are very fast and thus only rarely represented even in synchronized cells. The precursor–product relationship between the satellite and duplication plaque would be more clearly established, though not proven, if these structures shared common components and had a similar morphology.
We looked for the presence of SPB components in both the satellite and the duplication plaque, starting with the cytoplasmic components since the initial steps in duplication occur in the cytoplasm. We consistently detected Spc42p-GFP and Nud1p-GFP in both the satellite and duplication plaque (Fig. 6, A, C, H, I, K, and M). For Spc42p-GFP it was essential to have multiple copies of SPC42-GFP integrated; this increased the size of the SPB and made it easier to distinguish the duplication plaque from the satellite. GFP-Cnm67p was also detected in both (Fig. 6, B, L1, and L2) but less consistently in the satellite; whether this is due to a lower concentration or greater susceptibility to fixation is not clear. Spc72p-GFP localized to the half-bridge in early G1 cells (Fig. 6, D and J). Tub4p stained the inner plaque of the existing SPB (Spang et al., 1996a) and the cytoplasmic surface of the half-bridge (Fig. 6, G and O), consistent with the growth of cytoplasmic microtubules from the half-bridge at this stage of the cell cycle (Byers and Goetsch, 1975). There was also some possible staining of the duplication plaque (Fig. 6 O), indicating that part of the periphery of the outer plaque has been assembled. We detected GFP-Spc29p in the satellite (Fig. 6, E1 and E2), but have not yet found a clear duplication plaque in this strain. Duplication plaques were seen infrequently in all the strains investigated because of their transient nature, but we presume that since GFP-Spc29p is present in both the satellite and the existing SPB, then it will also be present in the duplication plaque. We were concerned that the satellite may have an altered composition in α-factor–treated cells, so we repeated some of the results on G1 elutriated cells and were able to detect Spc42p-GFP and Nud1p-GFP in the satellite and Spc72p-GFP in the half-bridge (Fig. 6, H–J).
We also looked at the localization of a nuclear SPB component, GFP-Spc110p, and could detect it in the inner plaque but not in the satellite or duplication plaque (Fig. 6, F and N). This version of Spc110p had been tagged with GFP on the NH2 terminus which localizes to the inner plaque (Spang et al., 1996b). However, in case we were unable to detect Spc110p elsewhere because fixation reduced antibody accessibility (Kilmartin et al., 1993) we did an indirect experiment on unfixed cells. The mutant mps2-1 arrests during SPB duplication just before insertion with a structure on the cytoplasmic surface of the half-bridge very similar in morphology to the duplication plaque (Winey et al., 1991); the arrest phenotype looks very similar to Fig. 1 C. At later stages of the arrest the partly assembled SPB detaches from the existing SPB and migrates some distance away while remaining attached to the cytoplasmic face of the bridge or nuclear membrane, eventually coming to rest above an area of the nucleus containing little nuclear DNA (Winey et al., 1991). The two SPBs are thus cytologically distinguishable: the existing SPB is associated with nuclear DNA staining while the partly assembled SPB which resembles the duplication plaque is not. We tagged Spc110p, Spc29p, and Spc42p with GFP in this mutant to see whether these were detectable in the partly assembled SPB. Spc42p-GFP has been detected already in both SPBs in mps1-737 which has a similar phenotype (Schutz and Winey, 1998) and we found a similar result (data not shown). GFP-Spc29p gave the same result: two dots were detected and only one was associated with the DAPI staining (Fig. 6, P and Q). This is in agreement with the nuclear and cytoplasmic localization of Spc29p (Fig. 3) and indicative of its presence in the duplication plaque. However, with GFP-Spc110p we found only one dot, which was always associated with the nuclear DNA, suggesting that it corresponded to the existing SPB (Fig. 6 P). The partly assembled SPB could be detected with anti-Tub4p in this strain (Fig. 6 Q). Thus, these results indicate that Spc110p, and presumably calmodulin since it is bound to Spc110p (Geiser et al., 1993; Stirling et al., 1994), is assembled into the SPB after insertion into the nuclear membrane.
Since the satellite and duplication plaque contain common components, we might expect them to have a similar morphology. The morphology of the satellite cannot be readily observed by EM in haploid cells due to its small size. Since the diameter of the SPB increases with increasing ploidy (Byers and Goetsch, 1974), we thought the morphology of the satellite might be clearer in a tetraploid strain. When this strain was arrested in G1 with α-factor so that all the SPBs would be satellite-bearing, the satellites now appeared plaque-like (Fig. 6 R, arrow) with a more similar morphology to the duplication plaque.
In summary these results show that both the satellite and duplication plaque contain the same cytoplasmic SPB components and can have a similar morphology, all suggestive of a precursor–product relationship.
Polymorphic Structures Formed by Spc42p
The immunoEM results in the last section suggest that the satellite and duplication plaque contain common cytoplasmic SPB components. We wished to confirm this proposal by changing the structure of the cytoplasmic part of the existing SPB, since similar changes might also occur in the satellite and duplication plaque. We made use of polymorphic structures produced by Spc42p, these were found unexpectedly while attempting to investigate the function of the phosphorylation of Spc42p (Donaldson and Kilmartin, 1996), which occurs predominantly on serines (data not shown). To investigate the function of this phosphorylation we prepared a version of Spc42p in which all 34 serines were changed to alanine (S:A Spc42p). To our surprise S:A Spc42p was fully functional, although bilateral mating was lethal (data not shown). Although SPBs appear to have a normal morphology, there is an unusual structural polymorphism associated with S:A Spc42p. On GAL overexpression, instead of the normal dome-shaped structure (Donaldson and Kilmartin, 1996), a large electron-dense ball was associated with each central plaque (Fig. 7 A). ImmunoEM showed that this structure contained overexpressed S:A Spc42p (Fig. 7 B). In some cells, and in all cells treated with α-factor, the balls were associated with the central plaque and with the distal end of the cytoplasmic side of the half-bridge, where it presumably initiated from the satellite (Fig. 7, C1 and C2). Thus, perturbations introduced into the existing SPB are also evident in the satellite.
Figure 7.
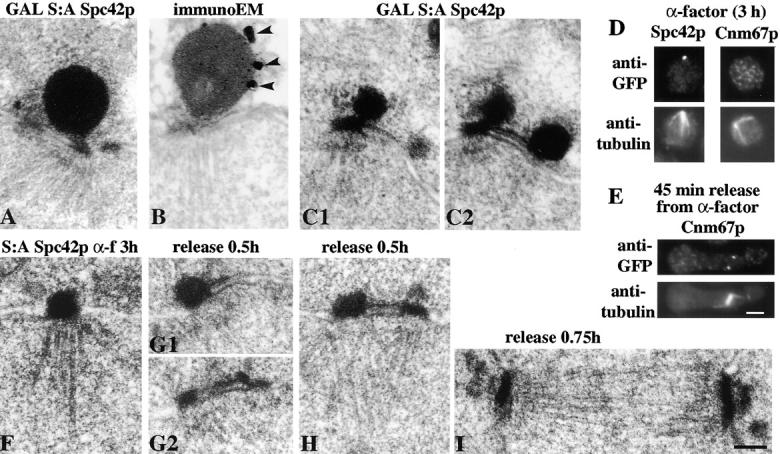
EM (A, C1, and C2) and immunoEM (B) of cells induced to overexpress S:A Spc42p under the control of the GAL promoter. C1 and C2 are adjacent serial sections. Arrowheads in B show the silver particles. (D) Immunofluorescence of Spc42p-GFP (left) and Cnm67p-GFP (right) in wild-type cells with anti-GFP (top) and anti-tubulin (bottom) after treatment with α-factor for 3 h at 30°C. (E) Immunofluorescence of Cnm67p-GFP 45 min after release from the α-factor treatment in D. (F) EM thin sections of S:A Spc42p cells treated with α-factor as in D and released for 0.5 h (G1, G2, and H) and 0.75 h (I). G1 and G2 are serial sections. These cells contained multiply integrated copies of S:A Spc42p because this increased the size of the balls and thus accentuated the morphological difference between the balls and plaques. In F–H, views were selected with the nuclear membrane bilayer in profile, thus excluding glancing sections of the central plaque. Bars, 0.1 μm for the EM and 2 μm for the immunofluorescence.
A similar structural transition was found on prolonged α-factor treatment (3 h) of the S:A Spc42p strain. We noticed that in wild-type cells prolonged α-factor treatment appears to deplete the outer plaque as shown by SPBs which are about to fuse during mating (Byers and Goetsch, 1975). Furthermore, we found that Cnm67p-GFP is not detectable at the SPB under these conditions in either unfixed (data not shown) or fixed cells, although it returns to the SPB on release from α-factor (Fig. 7, D and E). When we examined the morphology of SPBs in the S:A Spc42p strain after 3 h in α-factor, all the central plaques had been transformed into balls (Fig. 7 F). This structural transition may be due in part to the removal of a stabilizing interaction with Cnm67p and may also be responsible for the bilateral mating defect found in this strain. Cells remained viable on release from α-factor and all SPBs had reverted to the normal plaque-like morphology by the time of spindle formation (Fig. 7 I). It appeared that daughter SPBs formed with a normal plaque-like morphology (Fig. 7, G2 and H). We attempted to see if the duplication plaque could be made to form a ball in a strain carrying S:A Spc42p and a deletion of Cnm67p. This was not successful and the central plaque remains as a plaque in this strain (data not shown). Thus, it seems that both the absence of Cnm67p and some environmental effect of the α-factor–treated cell are necessary for ball formation.
In conclusion, we show that perturbations introduced into the central plaque are also evident in the satellite, suggesting they have similar structural features. In addition we show that changing all the serines in Spc42p to alanine can produce major reversible structural transitions in the central plaque. This shows the dominant role of Spc42p in central plaque structure and is consistent with the relatively small number of components found in this structure.
Inhibition of SPB Duplication by Overexpression of Spc110p Containing an Altered Potential NLS
A simple interpretation of the structure of the duplication plaque, given that it contains Spc42p which localizes to the cytoplasmic side of the central plaque (Rout and Kilmartin, 1991), but apparently not Spc110p which localizes to the nuclear side of the central plaque (Rout and Kilmartin, 1990), is that it consists of the cytoplasmic part of the central plaque attached to the half-bridge. Its orientation would be the same as the existing SPB since outer plaque components are able to assemble onto its cytoplasmic side. This model predicts that central plaque components such as Spc42p or Spc29p, instead of interacting with Spc110p as in the fully assembled SPB, would interact now either directly or indirectly with half-bridge proteins. This suggests that overexpressing Spc110p in the cytoplasm could compete with the interaction between the duplication plaque and the half-bridge, disrupt it, and inhibit SPB duplication.
Since Spc110p is a nuclear protein when overexpressed (Kilmartin and Goh, 1996), we altered a potential NLS so that the protein remained in the cytoplasm on overexpression. This version of Spc110p (ΔNLS-Spc110p) could not rescue a deletion of SPC110 unless the SV-40 T-antigen NLS was added to the NH2 terminus. This suggests that the lack of function of ΔNLS-Spc110p is due just to the removal of the potential NLS. When ΔNLS-Spc110p was overexpressed, immunofluorescent labeling was detected at the SPB but not in the nucleus (data not shown). EM showed that cells arrested during SPB duplication with a duplication plaque-like structure partly detached from the half-bridge (Fig. 8, A1 and A2), although one end remained attached to the distal end of the half-bridge at the site formerly occupied by the satellite. This suggested that ΔNLS-Spc110p had inhibited attachment of the plaque to the half-bridge possibly by forming a large polyhedral structure (Kilmartin and Goh, 1996). This would start to assemble between the half-bridge and the plaque and, if the plaque remained attached to the distal end of the half-bridge, cause it to swing up (clockwise in Fig. 8) to beyond a vertical position. Thus, in Fig. 8 A2, the outer plaque side of the displaced plaque (arrowhead) is on the right. ImmunoEM confirmed this possible arrangement, with labeling of ΔNLS-Spc110p on the side of the displaced plaque proximal to the SPB (Fig. 8, B1 and B2), the silver particles presumably staining the margins of the large polyhedral polymer, with internal staining of the polymer probably blocked by extensive cross-linking due to fixation. Spc42p-GFP was detected as expected on the distal side of the plaque (Fig. 8, C1 and C2). This result is consistent with the model that the duplication plaque contains Spc42p and probably Spc29p, that these proteins are probably involved in the attachment of the plaque to the half-bridge, and that this attachment is inhibited by inappropriate expression of Spc110p.
Figure 8.
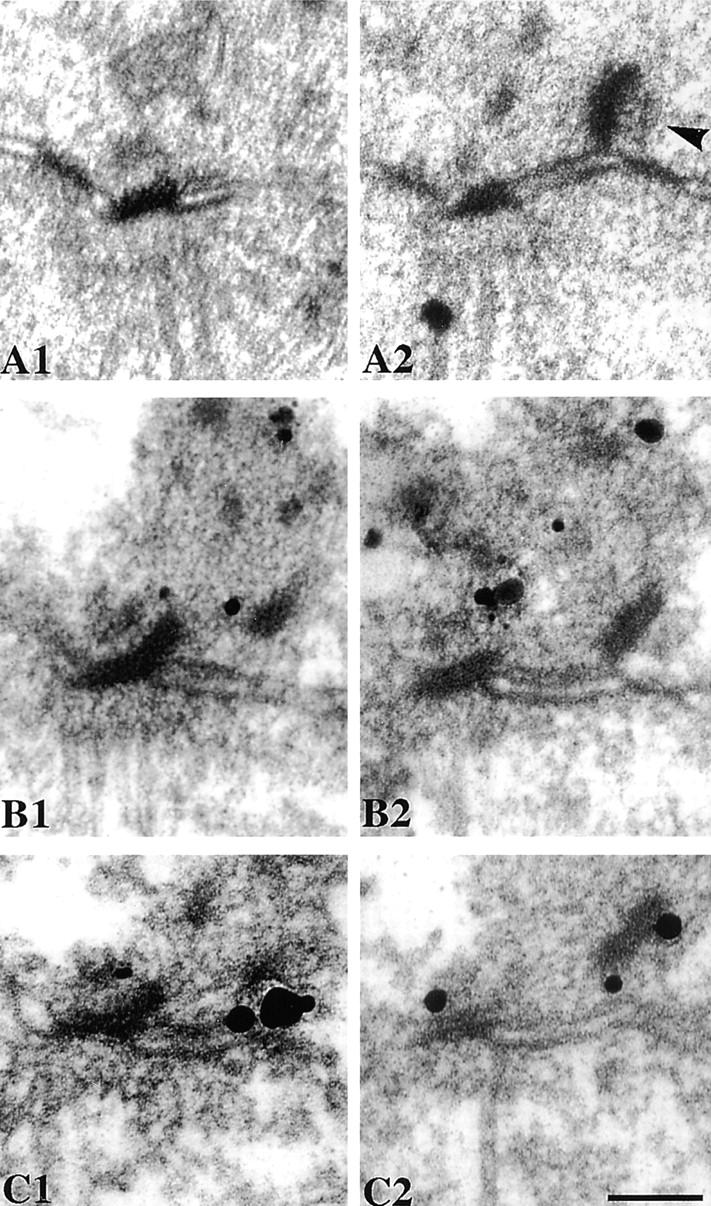
Thin section EM of cells overexpressing myc-ΔNLS-Spc110p for 2 h at 30°C under the control of the GAL promoter (A1 and A2), immunoEM with anti-myc to detect ΔNLS-Spc110p (B1 and B2), and with anti-GFP to detect Spc42p-GFP (C1 and C2). The numbers indicate serial sections. The arrowhead in A2 shows the outer plaque side of the displaced plaque. Bar, 0.1 μm.
Discussion
In this paper we have started to characterize the molecular events which occur during SPB duplication. EM of this process (Fig. 1) suggested that it starts with the assembly of the central part of the SPB, so to identify the more abundant proteins involved we prepared a highly enriched SPB core fraction. We found six main components by MALDI mass spectrometry, all of them previously identified (Wigge et al., 1998), which is a remarkably small number of main components for such a large organelle. We can estimate the mass of the SPB cores assuming each has ∼1,000 copies of Spc110p and Spc42p (Wigge et al., 1998) and assuming the other components (except for Nud1p) are in equivalent copy number as indicated from the Coomassie-stained gels (Fig. 2, C–E). This gives a mass of ∼0.3 GDa, which is at the lower end but compatible with the STEM mass analysis figure of 0.54 ± 0.23 GDa (Bullitt et al., 1997). We have presented an approximate model for the arrangement of some of these components in the SPB (Fig. 9). This has Spc110p, Cmd1p, and Spc29p on the nuclear side of the central plaque and Spc42p in the IL2 (Bullitt et al., 1997) on the cytoplasmic side of the central plaque. Note that in thin section EM of glutaraldehyde-fixed yeast cells the IL2 and central plaque layers appear merged, and we have called this merged structure the central plaque (Rout and Kilmartin, 1990). On the cytoplasmic side of the central plaque Cnm67p and Nud1p localize to the vicinity of IL1 and the outer plaque, and finally Spc72p is localized to the outer plaque itself since it is absent in SPB cores which have depleted outer plaques. However, it is clear that rearrangements of these components can occur in some circumstances, suggesting that the SPB is a dynamic organelle. Thus, Spc72p and Tub4p relocate from the outer plaque to the half-bridge in G1, presumably reflecting in part the assembly of cytoplasmic microtubules from the half-bridge in G1 (Byers and Goetsch, 1975; Knop and Schiebel, 1998). Nud1p, normally in the outer plaque (Wigge et al., 1998), can relocate to the half-bridge in G1/S in the absence of Cnm67p, although we do not know the function of this localization or whether this can happen in wild-type cells. In addition, Cnm67p and also possibly the entire outer plaque dissociates during prolonged α-factor treatment, perhaps facilitating SPB fusion during mating (Byers and Goetsch, 1975).
The combination of EM and immunoEM data presented in this paper suggests a number of stages in SPB duplication which are summarized in Fig. 9. One of the most interesting findings is that SPB components are present in the satellite, suggesting that duplication is already partly achieved at this stage. This finding indicates that there may be analogies between the satellite and the prereplicative complex for DNA replication (Diffley, 1996); both are assembled at the end of the preceding cell cycle and contain components which are primed to act once cells have passed through Start. The presence of Spc42p in the satellite and the apparent disruption of duplication plaque formation in spc42-10 mutants (Donaldson and Kilmartin, 1996) are consistent with the proposal that an early stage in SPB duplication is the crystalline lateral growth of Spc42p (Bullitt et al., 1997).
We presume that there are analogies between SPB duplication and centrosome duplication, since most processes in yeast, even the more specialized ones such as mating, use similar pathways to those found in higher eukaryotes. There are some common structural features between centrosome and SPB duplication (see Introduction), in particular the assembly of the new organelle at a set distance from the existing one. There may also be analogies between this process and the templated assembly of the new cytoskeleton in trypanosomes (Sherwin and Gull, 1989). Here new microtubules are nucleated by lateral microtubule-associated proteins in the preexisting microtubule cytoskeleton. This duplicates the exact pattern of the two dimensional microtubule array and can explain the earlier results showing cytoplasmic inheritance of the organization of the cell cortex in Paramecium (Beisson and Sonneborn, 1965). How might this templated assembly work in the case of the SPB? The bridge between side-by-side SPBs is a thin striated rectangular structure (Byers, 1981a) and is schematically shown in Fig. 9 with the two short edges adjacent to the central plaques of the two SPBs. One might envisage a protein complex localized specifically along the SPB-binding edges, or bridge components exposed at these two edges, which bind the central plaque components Spc29p and/or Spc42p. This binding always occurs at the cytoplasmic edge of the bridge and usually to the edge of the central plaque. Thus, the main function of the bridge is to provide two separate potential points of assembly along its SPB-binding edges. One of these is normally occupied by the existing SPB, the other is activated at some point in the preceding mitosis to bind cytoplasmic SPB components with the eventual formation of the satellite. There is a specialized attachment site for the distal end of the duplication plaque, and probably also for the satellite, at the cytoplasmic distal edge of the half-bridge. This site is different from the attachment of the nuclear face of the duplication plaque to the half-bridge, since overexpression of ΔNLS-Spc110p displaces the plaque from the half-bridge without detaching it from the distal cytoplasmic edge (Fig. 8). What happens during insertion is not clear, but a possible clue comes from images such as Fig. 1 D and Fig. 8 B1 here, and Figure 3 c in Byers and Goetsch (1974). These images suggest that the cytoplasmic edge of the half-bridge can bind to the nuclear face of the duplication or central plaque, in addition to the edge as described above. Thus, during insertion the half-bridge could retract with the cytoplasmic edge, presumably containing the satellite-binding site, withdrawing across the nuclear face of the duplication plaque (Fig. 9). The length of the half-bridge outer layer, or at least the distance between the near edges of the two plaques, appears to remain constant throughout duplication (Fig. 1), suggesting the retraction is confined to that part of the half-bridge immediately under the duplication plaque. The half-bridge outer layer may act as a rigid connecting strut between the existing SPB and the duplication plaque, perhaps stabilizing the latter during insertion. The retraction would expose the duplication plaque to nucleoplasm and thus allow assembly of nuclear components. This retraction may involve the half-bridge component Cdc31p (Spang et al., 1993), a member of the centrin family of proteins implicated in Ca2+-dependent contraction of basal body-associated fibers (Salisbury, 1995). Once SPB assembly is complete, the bridge would be severed in some way before spindle formation. This provides a new free SPB-binding edge associated with the half-bridge of each SPB for the assembly of the satellite during the next round of SPB duplication. A requirement for this model is that the self-nucleation rate for the assembly of SPB and half-bridge components is low, thereby directing assembly to preexisting templates or structures. This is certainly the case for structures assembled by Spc42p (Donaldson and Kilmartin, 1996), Spc110p (Kilmartin and Goh, 1996), and Spc72p (Souès and Adams, 1998). Thus, in this model the crucial element ensuring only two separate SPBs is the bridge and the specific binding of SPB components to either end of this structure. A problem for the future will be to characterize this binding further and also the role of the bridge in the other steps in SPB duplication, and the cell cycle controls which mediate them.
Abbreviations used in this paper
- bt
bis-tris
- GFP
green fluorescent protein
- IL
intermediate layer
- MALDI
matrix-assisted laser desorption/ ionization
- NLS
nuclear localization sequence
- SPB
spindle pole body
- ts
temperature-sensitive
Footnotes
We are particularly grateful to Sew Yeu Peak-Chew and Ian Fearnley for the mass spectrometry, Andy Kaja for technical help in strain and plasmid construction, Douglas Kershaw for cutting the serial thin sections, and Anne Donaldson for two of the EM pictures. Thanks are also due to M.S. Robinson for discussion. We apologize to the authors of references which we were unable to include because of length restrictions.
References
- Anderson RGW, Brenner RM. The formation of basal bodies (centrioles) in the Rhesus monkey oviduct. J Cell Biol. 1971;50:10–34. doi: 10.1083/jcb.50.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. . Nucl Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson J, Sonneborn TM. Cytoplasmic inheritance of the organization of the cell cortex in . Paramecium aurelia Proc Natl Acad Sci USA. 1965;53:275–282. doi: 10.1073/pnas.53.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Ivanovska I, Rose MD. Yeast ubiquitin-like genes are involved in the duplication of the microtubule organizing center. J Cell Biol. 1996;133:1331–1346. doi: 10.1083/jcb.133.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachat A, Kilmartin JV, Wach A, Philippsen P. Saccharomyces cerevisiaecells with defective spindle pole body outer plaques accomplish nuclear migration via half-bridge-organized microtubules. Mol Biol Cell. 1998;9:977–991. doi: 10.1091/mbc.9.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullitt E, Rout MP, Kilmartin JV, Akey CW. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell. 1997;89:1077–1086. doi: 10.1016/s0092-8674(00)80295-0. [DOI] [PubMed] [Google Scholar]
- Byers, B. 1981a. Cytology of the yeast cell cycle. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 59–96.
- Byers, B. 1981b. Multiple roles of the spindle pole bodies in the life cycle of Saccharomyces cerevisiae. In Molecular Genetics in Yeast. Alfred Benzon Symposium 16. D. von Wettstein, J. Friis, M. Kielland-Brand, and A. Stenderup, editors. Munksgaard, Copenhagen. 119–131.
- Byers B, Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harbor Symp Quant Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. . J Bact. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XP, Yin H, Huffaker TC. The yeast spindle pole body component Spc72p interacts with Stu2p and is required for proper microtubule assembly. J Cell Biol. 1998;141:1169–1179. doi: 10.1083/jcb.141.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JFX. Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- Ding R, West RR, Morphew M, Oakley BR, McIntosh JR. The spindle pole body of Schizosaccharomyces pombeenters and leaves the nuclear envelope as the cell cycle proceeds. Mol Biol Cell. 1997;8:1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Kilmartin JV. Spc42p: a phosphorylated component of the S. cerevisiaespindle pole body (SPB) with an essential function during SPB duplication. J Cell Biol. 1996;132:887–901. doi: 10.1083/jcb.132.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. Centriole replication. J Biophys Biochem Cytol. 1961;10:163–193. doi: 10.1083/jcb.10.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, Sundberg HA, Chang BH, Muller EGD, Davis TN. The essential mitotic target of calmodulin is the 110-kilodalton component of the spindle pole body in Saccharomyces cerevisiae. . Mol Cell Biol. 1993;13:7913–7924. doi: 10.1128/mcb.13.12.7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Souès S, Kilmartin J, Schiebel E. The spindle pole body component Spc98p interacts with the γ-tubulin-like Tub4p of Saccharomyces cerevisiaeat the sites of microtubule attachment. EMBO (Eur Mol Biol Organ) J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Gilerovitch HG, Bishop GA, King JS, Burry RW. The use of electron microscopic immunocytochemistry and silver-enhanced 1.4-nm gold particles to localize GAD in the cerebellar nuclei. J Histochem Cytochem. 1995;43:337–343. doi: 10.1177/43.3.7868863. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and G.R. Fink. 1991. Guide to yeast genetics and molecular biology. In Methods in Enzymology. Academic Press, London. 933. [PubMed]
- Kilmartin JV, Adams AEM. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. . J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Dyos SL, Kershaw D, Finch JT. A spacer protein in the Saccharomyces cerevisiaespindle pole body whose transcript is cell cycle-regulated. J Cell Biol. 1993;123:1175–1184. doi: 10.1083/jcb.123.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Goh P-Y. Spc110p: assembly properties and role in the connection of nuclear microtubules to the yeast spindle pole body. EMBO (Eur Mol Biol Organ) J. 1996;15:4592–4602. [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Spc98p and Spc97p of the yeast γ-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO (Eur Mol Biol Organ) J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Receptors determine the cellular localization of a γ-tubulin complex and thereby the site of microtubule formation. EMBO (Eur Mol Biol Organ) J. 1998;17:3952–3967. doi: 10.1093/emboj/17.14.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the γ-tubulin of Saccharomyces cerevisiaeand functions in microtubule organisation and spindle pole body duplication. EMBO (Eur Mol Biol Organ) J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- Martin OC, Gunawardane RN, Iwamatsu A, Zheng Y. Xgrip109: a γ tubulin-associated protein with an essential role in γ tubulin ring complex (γTuRC) assembly and centrosome function. J Cell Biol. 1998;141:675–687. doi: 10.1083/jcb.141.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald HB, Byers B. A proteasome cap subunit required for spindle pole body duplication in yeast. J Cell Biol. 1997;137:539–553. doi: 10.1083/jcb.137.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Murphy SM, Urbani L, Stearns T. The mammalian γ-tubulin complex contains homologues of the yeast spindle pole body components Spc97p and Spc98p. J Cell Biol. 1998;141:663–674. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappin DJC, Hojrup P, Bleasby AJ. Protein identification by peptide mass fingerprinting. Curr Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- Rattner JB, Phillips SG. Independence of centriole formation and DNA synthesis. J Cell Biol. 1973;57:359–372. doi: 10.1083/jcb.57.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins E, Jentzsch G, Micali A. The centriole cycle in synchronized HeLa cells. J Cell Biol. 1968;36:329–339. doi: 10.1083/jcb.36.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Fink GR. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987;48:1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Rout MP, Kilmartin JV. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Kilmartin JV. Yeast spindle pole body components. Cold Spring Harbor Symp Quant Biol. 1991;56:687–692. doi: 10.1101/sqb.1991.056.01.077. [DOI] [PubMed] [Google Scholar]
- Salisbury JL. Centrin, centrosomes and mitotic spindle poles. Curr Opin Cell Biol. 1995;7:39–45. doi: 10.1016/0955-0674(95)80043-3. [DOI] [PubMed] [Google Scholar]
- Schutz AR, Giddings TH, Steiner E, Winey M. The yeast CDC37 gene interacts with MPS1and is required for proper execution of spindle pole body duplication. J Cell Biol. 1997;136:969–982. doi: 10.1083/jcb.136.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz AR, Winey M. New alleles of the yeast MPS1gene reveal multiple requirements in spindle pole body duplication. Mol Biol Cell. 1998;9:759–774. doi: 10.1091/mbc.9.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwin T, Gull K. Visualisation of detyrosination along single microtubules reveals novel mechanisms of assembly during cytoskeletal duplication in trypanosomes. Cell. 1989;57:211–221. doi: 10.1016/0092-8674(89)90959-8. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci. 1968;3:207–230. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- Souès S, Adams IR. SPC72: a spindle pole component required for spindle orientation in the yeast Saccharomyces cerevisiae. . J Cell Sci. 1998;111:2809–2818. doi: 10.1242/jcs.111.18.2809. [DOI] [PubMed] [Google Scholar]
- Spang A, Courtney I, Fackler U, Matzner M, Schiebel E. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiaeis a component of the half bridge of the spindle pole body. J Cell Biol. 1993;123:405–416. doi: 10.1083/jcb.123.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Geissler S, Grein K, Schiebel E. γ-Tubulin-like Tub4p of Saccharomyces cerevisiaeis associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J Cell Biol. 1996a;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Grein K, Schiebel E. The spacer protein Spc110p targets calmodulin to the central plaque of the yeast spindle pole body. J Cell Sci. 1996b;109:2229–2237. doi: 10.1242/jcs.109.9.2229. [DOI] [PubMed] [Google Scholar]
- Stirling DA, Welch KA, Stark MJR. Interaction with calmodulin is required for the function of Spc110p, an essential component of the yeast spindle pole body. EMBO (Eur Mol Biol Organ) J. 1994;13:4329–4342. doi: 10.1002/j.1460-2075.1994.tb06753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G-H, Hirata A, Ohya Y, Anraku Y. Mutations in yeast calmodulin cause defects in spindle pole body functions and nuclear integrity. J Cell Biol. 1992;119:1625–1639. doi: 10.1083/jcb.119.6.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg HA, Davis TN. A mutational analysis identifies three functional regions of the spindle pole component Spc110p in Saccharomyces cerevisiae. . Mol Biol Cell. 1997;8:2575–2590. doi: 10.1091/mbc.8.12.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg HA, Goetsch L, Byers B, Davis TN. Role of calmodulin and Spc110p interaction in the proper assembly of spindle pole body components. J Cell Biol. 1996;133:111–124. doi: 10.1083/jcb.133.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin A-M, Celati C, Moudjou M, Bornens M. Characterization of the human homologue of the yeast Spc98p and its association with γ-tubulin. J Cell Biol. 1998;141:689–701. doi: 10.1083/jcb.141.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiaeG1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO (Eur Mol Biol Organ) J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Souès S, Mann M, Kilmartin JV. Analysis of the Saccharomycesspindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Goetsch L, Baum P, Byers B. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Hoyt MA, Chan C, Goetsch L, Botstein D, Byers B. NDC1: a nuclear periphery component required for yeast spindle pole body duplication. J Cell Biol. 1993;122:743–751. doi: 10.1083/jcb.122.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]