Abstract
L-selectin, a lectin-like receptor, mediates rolling of lymphocytes on high endothelial venules (HEVs) in secondary lymphoid organs by interacting with HEV ligands. These ligands consist of a complex of sialomucins, candidates for which are glycosylation- dependent cell adhesion molecule 1 (GlyCAM-1), CD34, and podocalyxin. The ligands must be sialylated, fucosylated, and sulfated for optimal recognition by L-selectin. Our previous structural characterization of GlyCAM-1 has demonstrated two sulfation modifications, Gal-6-sulfate and GlcNAc-6-sulfate in the context of sialyl Lewis x. We now report the cloning of a Gal-6-sulfotransferase and a GlcNAc-6-sulfotransferase, which can modify GlyCAM-1 and CD34. The Gal-6-sulfotransferase shows a wide tissue distribution. In contrast, the GlcNAc-6-sulfotransferase is highly restricted to HEVs, as revealed by Northern analysis and in situ hybridization. Expression of either enzyme in Chinese hamster ovary cells, along with CD34 and fucosyltransferase VII, results in ligand activity, as detected by binding of an L-selectin/IgM chimera. When coexpressed, the two sulfotransferases synergize to produce strongly enhanced chimera binding.
Keywords: sulfotransferase, carbohydrate, L-selectin, high endothelial venule, endothelium
Sulfation of carbohydrates attached to glycoproteins, glycolipids, and proteoglycans confers highly specific functions onto these macromolecules (Nelson et al., 1995; Hooper et al., 1996; Rosenberg et al., 1997). Examples of sulfation-dependent recognition phenomena are seen in diverse biological systems, including blood clotting, cytokine sequestration and receptor binding, control of the circulatory half-life of pituitary hormones, and plant–bacterial symbiosis. Therefore, the sulfotransferases that catalyze the sulfation of carbohydrates have become the focus of increasing research interest.
In recent years, the role of sulfated carbohydrates in leukocyte–endothelial interactions has been established (Kansas, 1996; Rosen and Bertozzi, 1996). During the process of normal lymphocyte recirculation, L-selectin on lymphocytes interacts with a set of sulfated glycoprotein ligands on the endothelial cells of high endothelial venules (HEVs).1 This interaction leads to lymphocyte rolling, which constitutes the first step in the recruitment cascade that culminates in the migration of the lymphocyte into the underlying tissue (Springer, 1994; Butcher and Picker, 1996). The involvement of L-selectin in leukocyte recruitment to certain sites of acute and chronic inflammation has also been demonstrated (Lewinsohn et al., 1987; Ley et al., 1991; Michie et al., 1993). The high endothelial cells (HECs) of HEVs within lymphoid organs and of HEV-like vessels that develop at sites of chronic inflammation have long been known to incorporate large amounts of [35S]sulfate in several species (Andrews et al., 1982; Girard and Springer, 1995a). The functional significance of this extensive sulfation was not understood until the ligands for L-selectin were elucidated.
To date, four discrete HEV-associated glycoprotein ligands for L-selectin have been identified at the molecular level: glycosylation-dependent cell adhesion molecule 1 (GlyCAM-1) (Lasky et al., 1992), CD34 (Baumhueter et al., 1993; Puri et al., 1995; Shailubhai et al., 1997), mucosal addressin cell adhesion molecule 1 (MAdCAM-1) (Berg et al., 1993), and podocalyxin (Sassetti et al., 1998). At least one other component, Sgp200, remains to be cloned (Berg et al., 1991; Hemmerich et al., 1994b; Hoke et al., 1995). These glycoproteins all contain mucin-like domains and are heavily O-glycosylated (Puri et al., 1995). It is well established that optimal recognition of these ligands by the C-type lectin domain of L-selectin requires sialylation (Imai et al., 1991), fucosylation (Maly et al., 1996), and sulfation (Imai et al., 1993). The protein scaffolds underlying these ligands are also expressed in tissues other than HEVs, such as mammary gland, non-HEV endothelium, hematopoietic precursors, and kidney podocytes (Dowbenko et al., 1993; Baumhueter et al., 1994; Kershaw et al., 1997). However, the L-selectin–reactive glycoforms of these proteins are highly restricted to HECs (Dowbenko et al., 1993; Baumhueter et al., 1994; Sassetti et al., 1998).
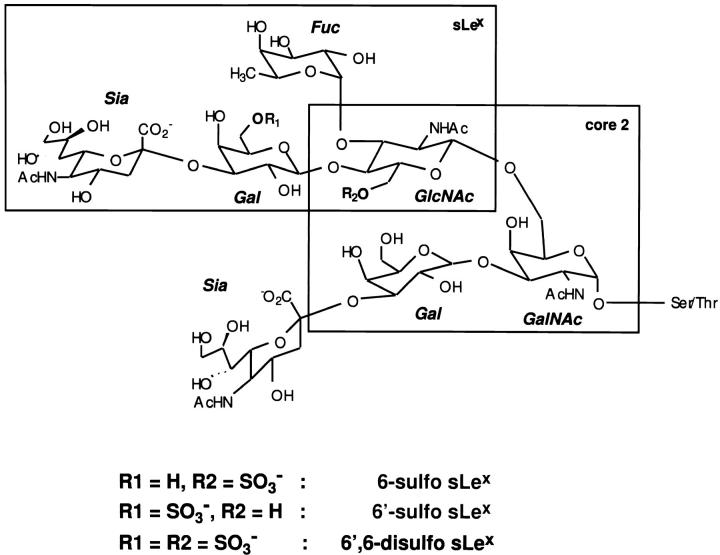
To understand the posttranslational modifications that underlie recognition of ligands by L-selectin, we previously undertook the analysis of the oligosaccharides of GlyCAM-1 which was affinity purified with recombinant L-selectin (Hemmerich and Rosen, 1994; Hemmerich et al., 1994a, 1995). We detected two specific sulfated monosaccharides: N-acetylglucosamine-6-sulfate (GlcNAc-6-sulfate) and galactose-6-sulfate (Gal-6-sulfate), which were present in equal amounts. These sulfate modifications are found, respectively, within two capping structures based on sialyl Lewis x (sLex): sialyl 6-sulfo Lex and sialyl 6′-sulfo Lex (see Fig. 1 and Table I). The simplest chains of GlyCAM-1 consist of sialyl 6′-sulfo Lex and sialyl 6-sulfo Lex as extensions from an internal trisaccharide known as core 2 (see Fig. 1 and Table I). More complex structures with increased chain size and multiple sulfate esters comprise the majority of the chains and remain to be analyzed in detail.
Figure 1.
Sulfated O-linked carbohydrate chains of GlyCAM-1. Oligosaccharides bearing the 6-sulfo sLex and the sialyl 6′-sulfo Lex motif, which extend from the core 2 structure, are shown. The presence of the sialyl 6′,6-disulfo Lex motif is strongly suspected. Structures of the more complex O-linked chains of GlyCAM-1 remain to be determined.
Table I.
Nomenclature and Structure of Oligosaccharides
| Name | Structure | |
|---|---|---|
| sLex | Siaα2→ 3Galβ1→ 4[Fucα1→ 3]GlcNAc | |
| Sialyl 6′-sulfo Lex | Siaα2→ 3[SO3→ 6]Galβ1→ 4[Fucα1→ 3]GlcNAc | |
| Sialyl 6-sulfo Lex | Siaα2→ 3Galβ1→ 4[Fucα1→ 3][SO3→ 6]GlcNAc | |
| Sialyl 6′,6-disulfo Lex | Siaα2→ 3[SO3→ 6]Galβ1→ 4[Fucα1→ 3] [SO3→ 6]GlcNAc | |
| Core 2 | Galβ1→ 3[GlcNAcβ1→ 6]GalNAc | |
| 6′,6-disulfo lactose | [SO3→ 6]Galβ1→ 4[SO3→ 6]Glc |
The sulfate dependence for L-selectin binding of HEV ligands was first established in pharmacological experiments with metabolic inhibitors of sulfation, i.e., chlorate and brefeldin A (Imai et al., 1993; Hemmerich et al., 1994b; Crommie et al., 1995; Shailubhai et al., 1997). Additional evidence for the importance of sulfation has emerged from studies with two mAbs, MECA-79 and G72. MECA-79 stains lymph node HEVs and blocks L-selectin– dependent lymphocyte attachment to HEVs in vitro and in vivo (Streeter et al., 1988). This antibody also stains HEV-like vessels that develop at sites of chronic inflammation (Michie et al., 1993; Lee and Sarvetnick, 1994; Girard and Springer, 1995a; Onrust et al., 1996). MECA-79 reacts with all the L-selectin ligands described above (Imai et al., 1991; Berg et al., 1993; Hemmerich et al., 1994b; Sassetti et al., 1998). This broad reactivity is thought to reflect its binding to a posttranslational modification that is common to many L-selectin ligands (Berg et al., 1993; Hemmerich et al., 1994b). The presence of the MECA-79 epitope is currently accepted as a predictor of L-selectin ligand activity (Girard and Springer, 1995a). However, endothelial and leukocyte ligands for L-selectin exist that lack this epitope (Fuhlbrigge et al., 1996; Clark et al., 1998). Although the precise structure of the MECA-79 epitope has not been elucidated, it is critically dependent on sulfation but does not require sialic acid or fucose (Hemmerich et al., 1994b; Maly et al., 1996; Shailubhai et al., 1997). The mAb G72 appears to have a more complex epitope in that it reacts with synthetic sialyl 6-sulfo Lex in a sialic acid– and sulfate-dependent manner (Mitsuoka et al., 1998). G72 stains HEVs in human peripheral lymph node intensely and blocks the binding of a human L-selectin/IgG chimera to HEV in vitro. Taken together, these results strongly implicate sulfated structures, including sialyl 6-sulfo Lex, as important recognition motifs for L-selectin binding to HEVs.
The contribution of individual sulfate esters to L-selectin binding affinity has been examined in several studies using sulfated sLex structures or analogues thereof. Direct binding measurements and competition studies have yielded evidence for enhanced binding (compared with sLex) from the Gal-6-sulfate modification (Tsuboi et al., 1996; Koenig et al., 1997), the GlcNAc-6-sulfate modification (Scudder et al., 1994; Saunders et al., 1996; Galustian et al., 1997), and from both (Yoshino et al., 1997). Significantly, Bertozzi et al. (1995) found that several sulfated variants of lactose were equal to or more potent than sLex in inhibiting the binding of L-selectin to GlyCAM-1 in an ELISA. For example, 6′,6-disulfo lactose (see Table I) competed twofold better than sLex, demonstrating that the relevant sulfate modifications by themselves, in the absence of any contribution from sialic acid or fucose, can confer a significant degree of binding to L-selectin.
In our search for the relevant sulfotransferases responsible for the synthesis of the specific sulfated carbohydrates on L-selectin ligands, Bowman et al. (1998) recently identified a GlcNAc-6-sulfate sulfotransferase activity in porcine organs. This activity is highly expressed in lymph node and spleen, relative to other tissues. Strikingly, it is greatly enriched in HECs, with only scant activity detectable in lymph node lymphocytes or HEC-depleted stromal cells. To identify candidate sulfotransferases at the molecular level, we probed expressed sequence tag (EST) databases for human sequences with homology to the cDNA sequence for a chicken Gal-6-sulfotransferase (Fukuta et al., 1995; Habuchi et al., 1996, 1997). Here, we describe the cloning of two cDNAs encoding carbohydrate sulfotransferases that can modify GlyCAM-1 and CD34. One enzyme is a Gal-6-sulfotransferase, which has been independently cloned by another group. The other is a novel enzyme exhibiting GlcNAc-6-sulfotransferase activity. The Gal-6-sulfotransferase has a wide tissue distribution, whereas the GlcNAc-6-sulfotransferase is highly restricted to HECs. Finally, we show that both enzymes can contribute to the generation of functional ligands for L-selectin and do so in a synergistic manner.
Materials and Methods
Semiquantitative Reverse Transcription PCR Analysis
HECs were purified from human tonsils as described previously (Sassetti et al., 1998). Human umbilical vein endothelial cells (HUVECs) were purchased from Clonetics. Total RNA was isolated from HECs, HUVECs, and human tonsillar lymphocytes by lysis and extraction with RNAZol (Tel-Test, Inc.). Approximately 45 μg RNA was obtained from 7 × 106 cells. First strand cDNA was made from 2 μg total RNA primed with random hexamers using AMV reverse transcriptase (RT; Life Technologies, Inc.). PCR reactions were carried out in a total volume of 10 μl of 1× KlenTaq buffer (Clontech) containing 400 nM primers, 200 μM dNTPs, 0.2 μl KlenTaq Advantage DNA polymerase mix (KlenTaq polymerase; Clontech), and 1.0 μl of twofold serially diluted cDNA as template. Cycling conditions were as follows: 1 min at 94°C; 30 cycles of 30 s at 92°C followed by 1 min 15 s at 68°C; one cycle of 10 min at 68°C. The following primers were used: for HEC-GlcNAc6ST, 5′-AAACTCAAGAAGGAGGACCAACCCTACTATGTGATGC-3′ and 5′-GTGGATTTGCTCAGGGACAGTCCAGCTAGACAGAAGAT-3′, which amplify a 456-bp fragment corresponding to nucleotides (nt) 884–1339 in Fig 3 a; for hypoxanthine phosphoribosyltransferase (HPRT), 5'-CCTGCTGGATTACATCAAAGCACTG-3′ and 5′-TCCAACACTTCGTGGGGTCCT-3′. The resulting amplified DNA was electrophoresed and visualized by ethidium bromide.
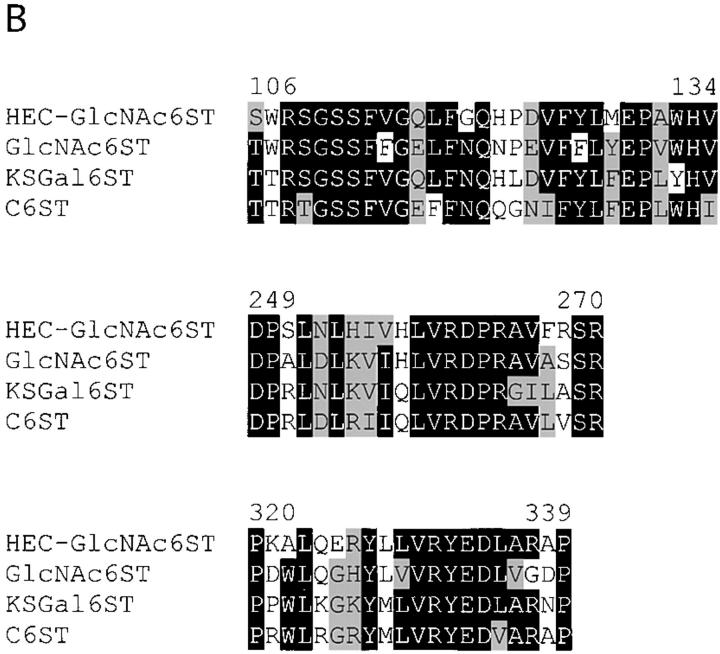
Figure 3.

Molecular features of human and mouse HEC-GlcNAc6ST. (A) cDNA sequence for human HEC-GlcNAc6ST and predicted protein sequences of human (h) and mouse (m) HEC-GlcNAc6STs (EMBL/GenBank/DDBJ accession nos. AF131325 and AF131236, respectively). The open reading frame is denoted by capital letters, and the predicted amino acid sequences are indicated below the nucleotide sequence. The putative transmembrane domains are underlined, and three potential N-linked glycosylation sites (for each sequence) are indicated by asterisks. (B) Alignment of regions of high conservation among human carbohydrate 6-sulfotransferases (Gal-6, GlcNAc-6, and GalNAc-6). Protein sequences were aligned using the ClustalW algorithm (Thompson et al., 1994). Black shading indicates identity at that residue among at least three of the sequences; grey shading indicates similarity among the shaded residues.
Preparation of an HEC cDNA Expression Library
A cDNA expression library was prepared from human HECs using the SMART cDNA technology (Clontech), which incorporates a long-distance PCR amplification step of first strand cDNA. 1 μg of total HEC RNA prepared as above was mixed with a modified oligo (dT) primer (1 μM) containing a NotI site and a universal site for 3′ priming of the PCR reaction and the SMART oligonucleotide (1 μM), which provides a universal site for 5′ priming of the PCR reaction. This mixture was heated at 72°C for 2 min to disrupt RNA secondary structure, and first strand cDNA was synthesized by Moloney murine leukemia virus (MMLV) RT in a total volume of 10 μl. 2 μl of this reaction mixture was subjected to 18 cycles of long-distance PCR primed by the universal primers using KlenTaq polymerase (Clontech). The PCR reaction mixture was incubated in the presence of proteinase K for 1 h at 45°C to destroy the KlenTaq polymerase activity, followed by heat inactivation at 90°C for 10 min. The double-stranded cDNA was polished by treatment with T4 DNA polymerase at 16°C for 30 min, followed by ligation to EcoRI/BstXI adaptors (Invitrogen) overnight at 16°C in the presence of T4 DNA ligase. Adaptor-ligated cDNAs were digested with NotI and then phosphorylated. The double-stranded cDNA was purified by size fractionation, ligated into EcoRI/ NotI-digested pCDNA1.1 (Invitrogen), and introduced into Escherichia coli MC1061/p3 (Invitrogen) by electroporation. The library contained 500,000 independent clones with an average insert size of 1.1 kb.
Molecular Cloning of HEC-GlcNAc6ST
HEC-GlcNAc6ST (human) was cloned from the HEC cDNA library by modification of a pool selection procedure (Kolodkin et al., 1997). In brief, an aliquot (comprising 400,000 colony forming units) of the amplified bacterial stock of the HEC cDNA library was plated onto 200 LB plates and grown for ∼18 h at 37°C. Each pool of 2,000 colonies was harvested and grown for an additional 2 h at 37°C, and glycerol stocks were made. PCR analysis was performed, using the HEC-GlcNAc6ST–specific primers described above, to identify positive pools. 1 of the 9 positive pools was titered and plated onto 40 plates to yield 100 colonies per plate. These pools were expanded and analyzed as in the first round. A single positive subpool was titered and plated onto 20 plates of 10 colonies each. Analysis of individual colonies by PCR resulted in a single positive clone, which was sequenced (Sanger et al., 1977). To clone the murine HEC-GlcNAc6ST, a 241-bp probe (nt 26–267) was amplified from the EST clone (AA522184; Research Genetics Inc., Huntsville, AL) and used as probe for screening a bacterial artificial chromosome (BAC) library from the C57BL/6 mouse (Genome Systems, Inc.). From the single positive clone, DNA was purified and sequenced directly, using primers derived from EST AA522184 (forward: 5′-TGGGTCAGCATGCCTTCCATACTAAC-3′; reverse: 5′-TTCTAAGATTCCGGTTGCTTCTCCGTGGAC-3′) and then obtaining sequence upstream (1559 nt) and downstream (582 nt). The resulting 1926 nt sequence was confirmed by resequencing in both directions.
Molecular Cloning of KSGal6ST
A human fetal brain library (λ ZAP; Stratagene) was the kind gift of Dr. Marc Tessier-Lavigne at the University of California, San Francisco. Approximately 106 plaques were screened with a probe consisting of a 730-bp HindIII/BamHI restriction fragment from IMAGE Consortium clone 40604 (EMBL/GenBank/DDBJ accession no. R55609) using standard techniques (Sambrook et al., 1989). 18 independent positive plaques were identified after the second round of hybridization. Cloned fragments were excised and sequenced as above.
Northern Blot Analysis
The probe for HEC-GlcNAc6ST consisted of a 496-bp fragment from LifeSeq clone no. 2620445, corresponding to nt 1021–1516 of the cloned cDNA (see Fig. 3 A). The probe was labeled with [α-32P]dATP (Amersham Pharmacia Biotech) by the random decamer priming method (Strip-EZ DNA kit; Ambion, Inc.). Multiple Tissue Northern blots (Clontech) containing poly(A)+ RNA from various human tissues were hybridized at 60°C overnight in Rapid-Hyb (Amersham Pharmacia Biotech) and then washed twice at room temperature for 15 min in 2× SSC/0.1% SDS followed by two 15-min washes at 60°C in 0.1× SSC/0.1% SDS and autoradiography. For the Northern blot to establish expression in HECs, poly(A)+ RNA was prepared from 1.5 × 107 HECs and 2.0 × 107 HUVECs, respectively. Isolation of the poly(A)+ RNA with oligo(dT) latex beads was performed according to the manufacturer's protocol (Oligotex Direct kit; QIAGEN, Inc.). Approximately 2 μg poly(A)+ RNA was loaded per lane. The RNA was separated by electrophoresis in a 1% denaturing agarose-formaldehyde gel and transferred to positively charged nylon filters (Hybond N+). The filters were hybridized and washed as for the Multiple Tissue blots. Blots were stripped using the Strip-EZ DNA kit (Ambion, Inc.), according to the manufacturer's protocol.
In Situ Hybridization
Paraffin sections (5 μM) from C57BL/6 mice were deparaffinized, fixed in 4% paraformaldehyde, and treated with proteinase K. After washing in 0.5× SSC, the sections were covered with hybridization solution (50% formamide, 300 mM NaCl, 20 mM Tris, pH 8.0, 5 mM EDTA, 1× Denhardt's, 10% dextran sulfate, 10 mM DTT), prehybridized for 1–3 h at 55°C, and hybridized overnight with sense or antisense 35S-labeled riboprobe transcribed from the IMAGE consortium clone 851801 (EMBL/ GenBank/DDBJ accession no. AA522184; Research Genetics, Inc.) which had been modified by digestion with SacI followed by religation. After hybridization, sections were washed at high stringency, dehydrated, dipped in photographic emulsion NTB2 (Eastman Kodak Co.), stored at 4°C for 2–8 wk, developed, and counterstained with hematoxylin and eosin.
Sulfation of GlyCAM-1/IgG in COS Cells
For generation of recombinant GlyCAM-1/IgG fusion protein, COS-7 cells were grown to 80% confluency in a T162 culture flask (Corning-Costar) and transfected with 8 μg of a plasmid encoding GlyCAM-1/IgG and 8 μg of plasmid encoding either HEC-GlcNAc6ST (pCDNA1.1), KSGal6ST (pCDNA3.1), or the empty vector (pCDNA1.1) using Lipofectamine (Life Technologies) in Opti-MEM (Life Technologies). The cDNA encoding the GlyCAM-1/IgG chimera was constructed by amplifying the entire coding sequence of murine GlyCAM-1 (Lasky et al., 1992) by PCR and cloning the resulting fragment into the pIG1 vector (Simmons, 1993). Cells were grown for 12 h after transfection in DME containing 10% FBS, then cell layers were washed once with PBS and cultured for 72 h in serum-free medium (Endothelial SFM, 16 ml per flask; GIBCO BRL) supplemented with Na2 35SO4 (0.25 mCi/ml, 1,400 Ci/ mmol; ICN). Recombinant GlyCAM-1/IgG fusion protein was isolated from the conditioned medium (CM) by affinity chromatography on protein A–agarose (Watson et al., 1990). The protein was then transferred into 50 mM ammonium bicarbonate on a Centricon 30 concentrator (Amicon, Inc.). 1% of each sample was analyzed by SDS-PAGE, and the remaining samples were lyophilized for subsequent acid hydrolysis and positional analysis.
Analysis of Sulfated GlyCAM-1/IgG Carbohydrates
The lyophilized recombinant GlyCAM-1/IgG samples were hydrolyzed in 0.2 M H2SO4, and the hydrolysates were prepared for analysis by high pH anion exchange chromatography (HPAEC) essentially as described previously (Hemmerich et al., 1994a), with the following modifications: (a) after 30 min hydrolysis in 0.2 M H2SO4 at 96°C, and before the initial gel filtration step, excess sulfate was precipitated by addition of an equivalent amount of Ba(AcO)2; (b) the second ion exchange column (DEAE-Sepharose) was equilibrated in 2 mM pyridine-acetate, pH 5.0, and eluted with a gradient of 2–500 mM pyridine-acetate. The monosulfated oligosaccharides eluted from this column between 50 and 250 mM pyridine acetate. The eluate was lyophilized, redissolved in 100 μl of water, and 30-μl samples of the resulting solution were subjected directly to HPAEC. HPAEC was performed using a Dionex DX 500 chromatographic analysis system and a Carbopac 1 column (Dionex Corp.) essentially as described (Hemmerich et al., 1994a, 1995), with column elution performed isocractically with 150 mM NaOH in 400 mM NaOAc at 1 ml/min. The authentic carbohydrate standards used in this analysis were obtained as described (Hemmerich et al., 1994a, 1995).
Flow Cytometry
CHO/FTVII/C2GnT cells were grown to 80% confluency in T75 flasks (6 × 106 cells per flask; Nalge Nunc) and then transfected with plasmids encoding C2GnT (1 μg, pCDNA1.1), fucosyltransferase VII (FTVII; 1 μg, pCDNA3.1), human CD34 (2 μg, pRK5), sulfotransferases (at the concentrations indicated for each experiment), and vector plasmid (pCDNA3.1) to achieve 8 μg total DNA per flask, using Lipofectamine. 48 h after transfection, the cells were harvested in 0.6 mM EDTA in PBS without calcium and magnesium, washed once in assay buffer (0.1% BSA in PBS), and resuspended at 4 × 106/ml in assay buffer. 25 μl of this cell suspension was added to wells of 96-well plates containing 50 μl of L-selectin/IgM chimera, or assay buffer. Cells were incubated on ice for 30 min, washed twice in assay buffer, and resuspended in 50 μl assay buffer containing secondary staining reagents. Cells were incubated for another 30 min, washed twice in assay buffer, and then resuspended in 50 μl assay buffer containing tertiary staining reagents and/or directly conjugated primary mAbs. For the sialyl 6-sulfo Lex or overall sLex determinations, cells were incubated with G72 mAb or HECA 452 (PharMingen), respectively, diluted in assay buffer, and then reacted with rabbit anti– mouse IgG-FITC (Zymed Laboratories, Inc.) (for G72) or mouse anti–rat IgM-FITC (PharMingen) (for HECA 452). The cells were incubated for 30 min, washed twice, and fixed in 1.5% paraformaldehyde in PBS, pH 7, for 20 min before transfer into 300 μl of assay buffer. Cells were analyzed on a FACSort™ (Becton Dickinson) flow cytometer using CELLQuest software (Becton Dickinson). To produce the L-selectin/IgM chimera (Maly et al., 1996), COS cells (10-cm plates) were transfected with L-selectin/IgM cDNA in pCDM8 (8 μg cDNA, 50 μl Lipofectamine per plate) and incubated in Opti-MEM for 10 d, at which time the CM was harvested and clarified by centrifugation. The CM was concentrated to half its original volume, titered, and used neat. Biotinylated goat anti– human IgM (μ) and streptavidin-FITC were purchased from Caltag Laboratories. The L-selectin mAb Mel-14 (Gallatin et al., 1983) was purified by ammonium sulfate precipitation from hybridoma cell culture supernatant, followed by immunoaffinity purification with mouse L-selectin/IgG. The control for Mel-14 was a rat IgG2a myeloma (Zymed Laboratories, Inc.). For the two-color analysis, we used mouse anti-CD34 phycoerythrin (PE) (QBend10-PE; Coulter Corp.) and the isotype-matched control, mouse IgG1-PE (Caltag Laboratories).
Results
Identification of Two cDNAs Predicting Carbohydrate Sulfotransferases
Our previous structural analysis of the carbohydrate chains of GlyCAM-1 indicated that the Gal-6-sulfate and GlcNAc-6-sulfate modifications accounted for essentially all of the sulfation of GlyCAM-1 (Hemmerich et al., 1994a). The previously cloned chicken chondroitin/keratan sulfate sulfotransferase (C6/KSST) has been shown by Habuchi et al. (1997) to catalyze sulfation at C-6 of galactose in Siaα2→ 3Galβ1→ 4GlcNAc, which is a core structure within the capping groups shown in Fig. 1. We used the cDNA sequence encoding C6/KSST to probe the National Center for Biotechnology Information (NCBI) dbEST and LifeSeq (Incyte Pharmaceuticals, Inc.) human EST databases for related sequences. When we examined expression of transcripts corresponding to the ESTs by Northern analysis, one (corresponding to LifeSeq clone no. 2620445, derived from a human breast epithelial cell line) was of particular interest, because its tissue distribution was highly restricted (original finding not shown, but result demonstrated in Fig. 2 B). To determine whether this gene was expressed in HECs, a cell type in which L-selectin ligands are elaborated, we carried out a semiquantitative RT-PCR analysis on HEC cDNA with primers derived from this EST. HECs were purified, as described previously, from human tonsils using immunomagnetic separation with MECA-79 as the probe (Girard and Springer, 1995b; Sassetti et al., 1998). The resulting cells had a purity of >99% and represented <1 per 1,000 of the stromal cells in tonsils. In parallel, we prepared cDNA from HUVECs and tonsillar lymphocytes. As shown in Fig. 2 A, the primers specific for the novel gene amplified a fragment of 456 bp from HEC cDNA, but failed to amplify this product from the HUVEC or lymphocyte cDNA.
Figure 2.
Expression of HEC-GlcNAc6ST transcripts in HECs. (A) Semiquantitative RT-PCR analysis. Fragments of the HEC-GlcNAc6ST and HPRT sequences were amplified by PCR from serial dilutions of cDNA prepared from purified HECs, HUVECs, and tonsillar lymphocytes. The reaction products (456 and 300 bp, respectively) were analyzed by agarose electrophoresis and ethidium bromide staining. −RT, PCR reactions in which the template was generated by omission of RT. (B) Northern blotting. Northern blots containing poly(A)+ RNA from various human tissues (left and center) and from HECs and HUVECs (right) were probed with a 500-bp fragment from the HEC-GlcNAc6ST cDNA (top panels). The blots were stripped and reprobed with a 300-bp probe for β-actin (bottom panels).
Since we had a relatively small number of HECs available, we used a PCR-based technique (SMART technology; Clontech, Inc.) to produce cDNA from total RNA, from which we prepared a plasmid expression library. Using PCR amplification of the 456-bp fragment to identify positive pools, we isolated a full-length cDNA clone from the library by a pool selection procedure, as described in Materials and Methods. Initially, 200 pools of 2,000 colony-forming units/pool were screened. After two additional rounds of screening, a single positive clone was obtained. The cDNA corresponding to this clone contains a single open reading frame of 1,158 nucleotides. The cDNA is apparently full-length as indicated by the presence of an upstream stop codon and a Kozak sequence surrounding the first ATG, and a poly(A) tail at the 3′ end. This open reading frame predicts a type II transmembrane protein of 386 amino acids with 3 potential sites for N-linked glycosylation. The new cDNA sequence was used to probe the human databases for additional matching ESTs. One EST was identified in the LifeSeq database that mapped to the new gene at the 5′ end of its protein coding region. When the clone (no. 2617407, derived from gall bladder) corresponding to this EST was fully sequenced, its sequence completely matched the original cDNA sequence within the coding region. There were two base changes in the 3′ untranslated region and divergence in the 5′ untranslated region. We present the sequence corresponding to LifeSeq clone no. 2617407 (Fig. 3 A; EMBL/Genbank/DDBJ accession no. AF131235), since the library from which it was cloned was created without a PCR amplification step. The predicted amino acid sequence of this novel gene is 31% identical to C6/KSST (Fukuta et al., 1995), the chicken enzyme used in the original EST search. We have termed this novel gene HEC-specific GlcNAc-6-sulfotransferase (HEC-GlcNAc6ST) on the basis of the characterization described below.
Using the human HEC-GlcNAc6ST cDNA sequence as a probe, we identified a highly related mouse sequence in the NCBI dbEST database (EMBL/GenBank/DDBJ accession no. AA522184). A 241-bp probe based on this EST was used for screening of a BAC library from mouse (C57BL/6). The probe was found to hybridize to a single BAC. The genomic clone within this BAC contained an open reading frame of 1167 bp (EMBL/Genbank/DDBJ accession no. AF131236), which is 77% identical at the nt level to the coding region of the human HEC-GlcNAc6ST. No introns were detected. The sequence predicts a protein of 388 amino acids (Fig. 3 A), which is 73% identical to that of human HEC-GlcNAc6ST. We term this apparent mouse homologue mHEC-GlcNAc6ST.
The expression of HEC-GlcNAc6ST was confirmed to be highly restricted by further analysis. On a conventional Northern blot (human), transcripts corresponding to this gene were absent from most tissues (Fig. 2 B). Low levels of a 2.4-kb transcript were apparent in lymph node, liver (adult and fetal), and pancreas. A prominent band of the same size was detected at relatively high levels in mRNA from HECs, but was undetectable in HUVEC mRNA (Fig. 2 B). An additional transcript at ∼6 kb was expressed in liver and pancreas, and trace levels appeared to be present in lymph node, HECs, and HUVECs (Fig. 2 B). Using an antisense riboprobe based on the original mouse EST, we carried out in situ hybridization on mouse tissues. Strikingly, mHEC-GlcNAc6ST transcripts were detected only in the HEVs of lymph node (Fig. 4). No hybridizing signal was found in other cell types of the lymph node, or in several other organs, including spleen, thymus, liver, skeletal muscle, pancreas, stomach, and kidney (data not shown). A weak signal was detected in gut intestinal epithelium (data not shown). The sense control did not yield signal in any tissue.
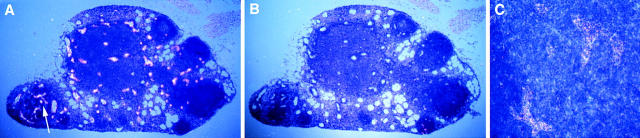
Figure 4.
In situ hybridization to detect HEC-GlcNAc6ST transcripts in mouse lymph node. Sections of C56BL/6 mouse lymph node were hybridized with 35S-labeled sense or antisense riboprobes based on the clone corresponding to the mouse homologue of HEC-GlcNAc6ST. Dark field micrographs of the sections are shown. Signal is seen as bright dots. (A) Hybridization with antisense probe, whole lymph node shown. The only source of signal are HEVs, seen as distinctive high-walled vessels in the cortex of the node. (B) Hybridization with sense probe of section adjacent to that in A. (C) Higher magnification view of area indicated by arrow in A. Two large HEVs are evident.
Our screening of the human EST databases with the chicken C6/KSST led to the identification of multiple ESTs that mapped to another gene. We cloned a cDNA for this gene by conventional techniques from a fetal brain library as described in Materials and Methods. While this work was in progress, the same cDNA (with only two base changes in untranslated regions) was published independently by Fukuta et al. (1997). The expressed protein was characterized as a keratan sulfate sulfotransferase with Gal-6-sulfotransferase activity (KSGal6ST) (Fukuta et al., 1997). The predicted protein is a type II transmembrane protein of 411 amino acids with 32% identity to HEC-GlcNAc6ST. Fukuta et al. (1997) reported the expression of KSGal6ST in many tissues, including lymph node. We confirmed these results; in addition, we detected its expression in HECs, albeit at apparently low levels, by performing RT-PCR with HEC cDNA and the HEC-cDNA library (data not shown).
Sulfation of GlyCAM-1 by KSGal6ST and HEC-GlcNAc6ST
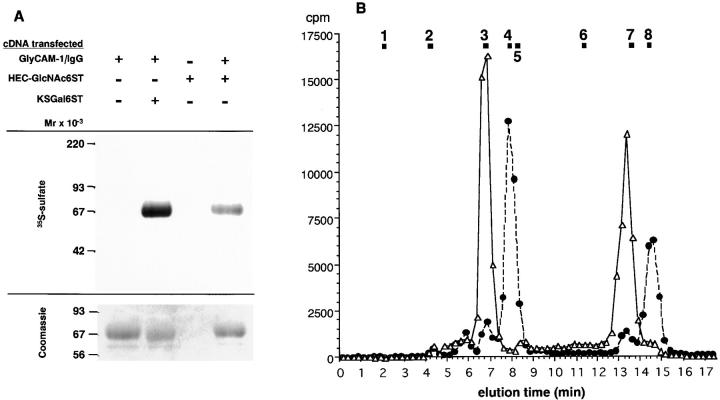
To test whether KSGal6ST and HEC-GlcNAc6ST (human) were capable of sulfating an L-selectin ligand, we transfected COS cells with a cDNA encoding a GlyCAM-1/IgG chimera and a cDNA encoding one or the other sulfotransferase. The transfected cells were cultured in the presence of [35S]sulfate, and radiolabeled GlyCAM-1/IgG was purified from the CM on protein A–agarose. As shown in Fig. 5 A, there was substantial incorporation of radioactivity into GlyCAM-1/IgG when either KSGal6ST or HEC-GlcNAc6ST cDNA (but not empty vector) was included in the cotransfection.
Figure 5.
Sulfation of GlyCAM-1/IgG by sulfotransferases. COS cells were transfected with combinations of plasmids encoding GlyCAM-1/IgG, KSGal6ST, and HEC-GlcNAc6ST, as indicated. Transfected cells were cultured in the presence of [35S]sulfate, and recombinant GlyCAM-1/IgG was isolated from the CM. 1% of the captured material was analyzed by SDS-PAGE, and the remainder was subjected to hydrolysis and compositional analysis. (A) Autoradiograph (top) and Coomassie Blue staining (bottom) of SDS gel. Densitometric quantification of the Coomassie Blue–stained bands showed that each lane, except the control lane without GlyCAM-1/IgG plasmid, contained approximately equal amounts of GlyCAM-1/IgG. (B) Sulfated carbohydrates produced in GlyCAM-1 by transfection with HEC-GlcNAc6ST (▵) or KSGal6ST (•) were analyzed by Dionex chromatography after acid hydrolysis. The following standards are indicated: 1, GlcNAc-3SO3 −; 2, [35S]SO4 2−; 3, Galβ1→ 4[SO3 –→ 6]GlcNAc; 4, [SO3 –→ 6]Galβ1→ 4GlcNAc; 5, Gal-4SO3 –; 6, Gal-3SO3 –; 7, GlcNAc-6SO3 –; 8, Gal-6SO3 –.
To establish the regiochemistry of sulfation on radiolabeled GlyCAM-1/IgG, samples resulting from the two sulfotransferase transfections were subjected to hydrolysis and Dionex HPLC analysis according to our previously established procedures (Hemmerich et al., 1994a). As shown in Fig. 5 B, transfection with KSGal6ST resulted in sulfated mono- and disaccharides that comigrated with [SO3→ 6]Gal and [SO3→ 6]Galβ1→ 4GlcNAc, confirming this enzyme as a Gal-6-sulfotransferase (Fukuta et al. 1997). In contrast, transfection with HEC-GlcNAc6ST resulted in products that corresponded to [SO3→ 6]GlcNAc and Galβ1→ 4[SO3→ 6]GlcNAc. Thus, this enzyme was established to be a novel GlcNAc-6-sulfotransferase.
Generation of Sialyl 6-sulfo Lex with HEC-GlcNAc6ST
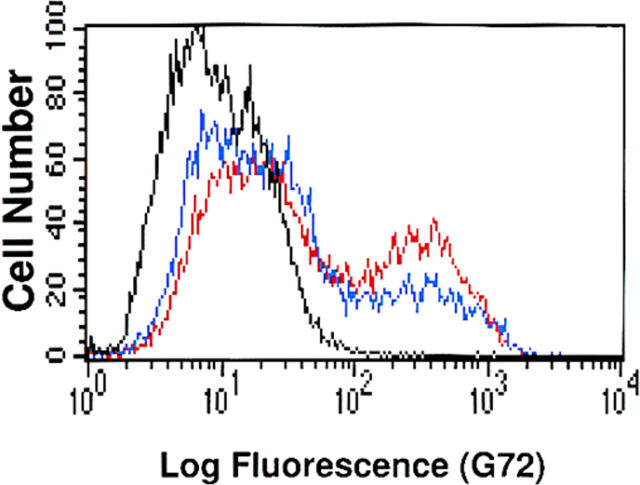
To determine whether the HEC-GlcNAc6ST could contribute to the generation of sialyl 6-sulfo Lex (Table I), we took advantage of the G72 mAb, which recognizes this structure in a sulfate-dependent manner (Mitsuoka et al., 1998). We carried out transfection experiments with CHO cells, termed CHO/FTVII/C2GnT, which had been stably transfected with (a) fucosyltransferase VII (FTVII) to provide α1→ 3 Fuc, and (b) core 2 β1→ 6 N-acetylglucosaminyltransferase (C2GnT) (Bierhuizen and Fukuda, 1992) to provide a core structure for O-linked glycans (Fig. 1) upon which extended chains with sLex capping groups can be elaborated (Li et al., 1996). The modified CHO cells were transiently transfected with HEC-GlcNAc6ST or KSGal6ST cDNA as a control. Transfection of the CHO cells with HEC-GlcNAc6ST cDNA resulted in significant staining by G72 as measured by flow cytometry (Fig. 6), whereas transfection with KSGal6ST did not yield staining (not shown). Similar results were obtained with G152 (Mitsuoka et al., 1998), a related mAb (data not shown). Inclusion of a CD34 cDNA in the transfection did not significantly enhance staining by G72, indicating that an exogenous mucin scaffold was not required for the expression of this epitope.
Figure 6.
Generation of sialyl 6-sulfo Lex by transfection with HEC-GlcNAc6ST cDNA. CHO/FTVII/C2GnT cells were transfected with a cDNA encoding HEC-GlcNAc6ST, with or without CD34 cDNA. Cells were stained with the G72 mAb to detect the presence of sialyl 6-sulfo Lex. Histogram shows G72 staining for the transfections with (red) or without (blue) CD34 cDNA, or staining of the isotype control antibody for the transfection with CD34 cDNA (black).
Contribution of KSGal6ST and HEC-GlcNAc6ST to the Generation of L-selectin Ligand Activity
Since HEC-GlcNAc6ST and KSGal6ST are both expressed in HECs and are capable of sulfating GlyCAM-1/IgG in transfected cells, they are candidates to participate in the biosynthesis of L-selectin ligands. Our previous structural analysis indicated that Gal-6-sulfate and GlcNAc-6-sulfate are present equally in native GlyCAM-1 oligosaccharides. However, as reviewed above, the relative contribution of the two sulfate esters to L-selectin binding affinity has been a matter of considerable uncertainty. To test whether these enzymes can contribute to the generation of ligand activity, we carried out further flow cytometry experiments with an L-selectin/IgM chimera as a probe. The CHO/FTVII/C2GnT cells were transiently transfected with cDNAs for the two sulfotransferases (singly or in combination) plus a CD34 cDNA. As shown in Fig. 7 A, no binding of the chimera was detected in the absence of the sulfotransferases. Transfection with either sulfotransferase yielded positive staining, both in terms of the proportion of positive cells and their mean fluorescence intensity (MFI). Strikingly, the combination of KSGal6ST and HEC-GlcNAc6ST cDNAs produced strongly enhanced binding of L-selectin/IgM, which greatly exceeded the sum of the signals from the individual sulfotransferases (Fig. 7 A). This apparent synergistic effect was evident over a considerable range of cDNA concentrations in the transfection mixtures (Tables II and III). The binding of the L-selectin/IgM chimera induced by the sulfotransferases was confirmed to be specific as indicated by its calcium dependence (data not shown) and complete inhibition binding by a function-blocking anti–L-selectin mAb (MEL-14; Fig. 7 B). The effects of the sulfotransferases on the L-selectin/ IgM chimera were not due to differences in transfection efficiency, as the CD34 expression levels were essentially constant, irrespective of the combinations of sulfotransferase cDNAs in the transfection mixtures (Table II). Furthermore, to control for the possibility that the sulfotransferases might cause changes in essential glycosylation parameters that could affect L-selectin binding, we stained the transfected cells with the HECA 452 mAb. This antibody recognizes sLex-related structures and is widely used as a reporter for glycosylation modifications (sialylation and fucosylation) pertinent to selectin ligands (Wagers et al., 1997; Tu et al., 1999). Because this antibody reacts equally well with sulfated (at Gal-6, GlcNAc-6, or both) and nonsulfated sLex structures (Mitsuoka et al., 1998), it was of particular utility for detecting the overall presence of sLex on the cell surface of the transfectants. As shown in Table III, the expression of the HECA 452 epitope was not significantly altered (<25% variation) by transfection with the sulfotransferase cDNAs in any combination. The final issue we investigated was the contribution of CD34 to the expression of L-selectin ligand activity. In striking contrast to the results with G72, the binding of L-selectin/IgM was dependent on the presence of the CD34 protein scaffold, as there was a nearly complete loss of staining when the CD34 cDNA was omitted from the transfection (Fig. 7 C). Interestingly, among the CD34-positive population, only those cells expressing the highest levels stained with the L-selectin/IgM chimera (Fig. 7 A).
Figure 7.
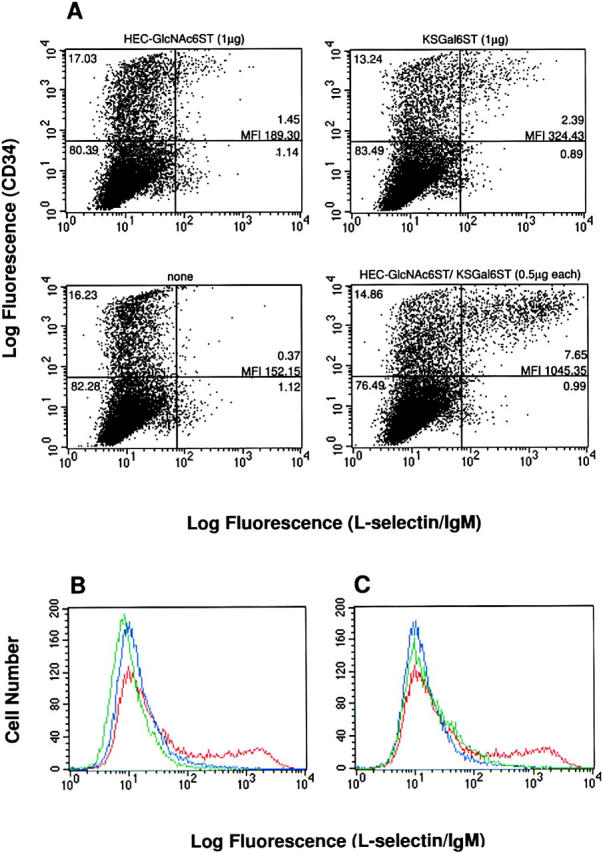
L-selectin reactivity conferred to CD34 by transfection with HEC-GlcNAc6ST and KSGal6ST cDNAs. CHO/FTVII/ C2GnT cells were transfected with different combinations of cDNAs encoding human CD34, HEC-GlcNAc6ST, and KSGal6ST. Cells were stained with the L-selectin/IgM chimera to detect ligand activity. (A) Two-color analysis showing CD34 expression (y-axis, staining with CD34-PE mAb) and L-selectin ligand activity (x-axis, FITC) in cells transfected with CD34 cDNA and cDNAs encoding HEC-GlcNAc6ST and KSGal6ST, alone or in combination as indicated. The horizontal bar is set such that all cells staining with the isotype-matched control for the CD34 mAb are included in the lower quadrants. The vertical bar is set to indicate the L-selectin/IgM staining of cells in which no sulfotransferase cDNA was included in the transfection mixture (bottom left panel). The fraction of positive cells (as a percentage of the total) in each quadrant is indicated. The mean fluorescence intensity (MFI) for the cells in the top right quadrants is indicated. The CD34 mAb did not interfere with staining by the L-selectin/IgM chimera. (B and C) Histograms showing L-selectin/IgM staining for the following transfections: CD34 cDNA only (blue, B and C); CD34/HEC-GlcNAc6ST/KSGal6ST cDNAs (red, B and C); CD34/HEC-GlcNAc6ST/KSGal6ST cDNAs with staining done in the presence of Mel-14 mAb (green, B); HEC-GlcNAc6/KSGal6ST cDNAs but no CD34 cDNA (green, C). The secondary staining reagent when used alone showed staining equal to that observed when Mel-14 mAb was used as an inhibitor. The isotype-matched control for Mel-14 showed no inhibitory effect.
Table II.
L-selectin/IgM and CD34 Staining of CHO Cells Transfected with Combinations of Sulfotransferase cDNAs
| cDNA transfected | MFI | |||||
|---|---|---|---|---|---|---|
| KSGal6ST | HEC-GlcNAc6ST | L-sel/IgM staining | CD34 staining | |||
| μg | ||||||
| 0 | 0 | 0 | 660 | |||
| 1.0 | 0 | 364 | 742 | |||
| 1.5 | 0 | 391 | 731 | |||
| 0 | 1.0 | 114 | 748 | |||
| 0 | 1.5 | 163 | 700 | |||
| 0.5 | 0.5 | 935 | 743 | |||
| 1.0 | 0.5 | 917 | 684 | |||
| 0.5 | 1.0 | 830 | 703 | |||
CHO/FTVII/C2GnT cells were cotransfected with plasmids encoding CD34 (2 μg) and each sulfotransferase singly or in combination in the indicated amounts. Data are expressed as the mean fluorescence intensity (MFI) of L-selectin/IgM staining in the L-sel–IgM+/CD34+ population with background signal (from transfectants with CD34 cDNA alone, value 139) subtracted. Also shown is the MFI for staining with a CD34 mAb (Qbend-10) for the population that was positive for CD34, as defined by staining with the class-matched control antibody.
Table III.
L-selectin/IgM and HECA 452 Staining of CHO Cells Transfected with Combinations of Sulfotransferase cDNAs
| cDNA transfected | MFI | |||||
|---|---|---|---|---|---|---|
| KSGal6ST | HEC-GlcNAc6ST | L-sel/IgM staining | HECA staining | |||
| μg | ||||||
| 0 | 0 | 0 | 665 | |||
| 1.0 | 0 | 197 | 697 | |||
| 0 | 1.0 | 60 | 496 | |||
| 0.5 | 0.5 | 836 | 561 | |||
CHO/FTVII/C2GnT cells were cotransfected with plasmids encoding CD34 (2 μg) and each sulfotransferase singly or in combination in the indicated amounts. Data are expressed as the MFI of L-selectin/IgM staining in the L-sel–IgM+/CD34+ population with background signal (from transfectants with CD34 cDNA alone, value 182) subtracted. Also shown is the MFI for staining with the HECA 452 mAb for the entire population. The MFI for staining with a control antibody was <10.
Discussion
Sulfation plays a central role in the interactions of L-selectin and P-selectin with their physiological ligands (Rosen and Bertozzi, 1996). PSGL-1, a leukocyte ligand for both P-selectin and L-selectin (McEver and Cummings, 1997), possesses a cluster of sulfated tyrosine residues, which facilitate binding of both selectins (Pouyani and Seed, 1995; Sako et al., 1995; Wilkins et al., 1995; Snapp et al., 1998). As reviewed above, L-selectin recognition of its cognate HEV ligands requires sulfation, as well as fucosylation and sialylation, for optimal binding. For the molecularly defined HEV ligands, sulfation is on sugar moieties rather than tyrosine residues (Rosen and Bertozzi, 1996). Our objective has been the molecular identification of the sulfotransferases that participate in the biosynthesis of these endothelial ligands. The specificities of the desired sulfotransferases were dictated by the analysis of the oligosaccharides of L-selectin–reactive GlyCAM-1, which revealed the presence of Gal-6-sulfate and GlcNAc-6-sulfate in the context of sLex (sialyl 6′-sulfo Lex and sialyl 6-sulfo Lex, respectively). Similarly, the functional glycoforms of CD34 also contain Gal-6-sulfate and GlcNAc-6-sulfate in approximately equal representations (Hemmerich and Rosen, unpublished observations). Histochemical evidence with carbohydrate-directed mAbs shows that sialyl 6-sulfo Lex is prominently displayed on the HEVs of human peripheral lymph node (Mitsuoka et al., 1998), further implicating a GlcNAc-6-sulfotransferase activity in the biosynthesis of L-selectin ligands. Bowman et al. (1998) recently characterized such an activity from porcine lymph node, which was highly enriched in HECs.
To identify the relevant sulfotransferases at the molecular level, we probed human EST databases for homologues of the chicken C6/KSST, a bona fide Gal-6-sulfotransferase. Here we report the cloning of two carbohydrate sulfotransferases. One is indeed a Gal-6-sulfotransferase, which was independently discovered by Fukuta et al. (1997) and named KSGal6ST. The second enzyme, which we have termed HEC-GlcNAc6ST, is a novel GlcNAc-6-sulfotransferase. These two sulfotransferases, together with the chicken C6/KSST (Fukuta et al., 1995), and the recently reported human chondroitin-6-sulfotransferase (C6ST, specificity for C-6 of GalNAc) (Fukuta et al., 1998) and GlcNAc-6-sulfotransferase (GlcNAc6ST) (Uchimura et al., 1998a,b) constitute a family of highly conserved enzymes. Overall, amino acid identities within the family range from 27 to 42%. These enzymes are type II transmembrane proteins with short cytoplasmic tails, features that are typical of glycosyltransferases and carbohydrate sulfotransferases.
Within this new family of carbohydrate sulfotransferases, there are three regions of amino acid sequence in which amino acid identity ranges from 45 to 54% and similarity from 80 to 90% (Fig. 3 C). Regions one and two contain elements that conform to the recently described consensus binding motifs for the high energy sulfate donor, 3′-phosphoadenosine 5′-phosphosulfate. These elements are found in all sulfotransferases characterized to date (Kakuta et al., 1998). In addition, regions one and three contain two stretches of sequence of 11 amino acids each (corresponding to amino acids 124–134 and 328–339, respectively, in the HEC-GlcNAc6ST sequence) that are highly conserved (>90% similarity). It is possible that these two elements contribute to a binding pocket that interacts with the 6-hydroxyl group of an appropriate oligosaccharide acceptor (Gal, GalNAc, or GlcNAc) to bring it into apposition with the donor phosphosulfate group.
Our characterization of the KSGal6ST and HEC-GlcNAc6ST supports their involvement in the synthesis of L-selectin ligands. KSGal6ST has a wide tissue distribution which includes HECs. HEC-GlcNAc6ST shows a highly restricted, although not absolute, localization to HECs. Second, the sulfotransferases catalyze the two specific sulfation modifications on a recombinant L-selectin ligand that have been established to occur on native ligands. In the case of HEC-GlcNAc6ST, we have used the G72 mAb to show that transfection with the cDNA leads to 6-sulfation of sLex on the cell surface of CHO/FTVII/ C2GnT cells. This finding establishes that recombinant HEC-GlcNAc6ST can be used to generate a highly specific sulfated structure that is present on lymph node HEVs (Mitsuoka et al., 1998).
Using a flow cytometry assay based on the binding of an L-selectin/IgM chimera to transfected CHO/FTVII/C2GnT cells, we showed that each enzyme imparts L-selectin binding. Control experiments established that the effects of the sulfotransferases were not due to indirect effects on transfection efficiencies or to global changes in glycosylation parameters of the cells. In contrast to the generation of the G72 epitope, L-selectin/IgM binding required cotransfection of the cells with a CD34 cDNA. These results indicate that a specific protein scaffold, not present endogenously in CHO cells, is needed for optimal ligand activity, although at least one of the relevant sulfated carbohydrate epitopes (sialyl 6-sulfo Lex) can be formed without the provision of such a scaffold. This finding is analogous to the situation with P-selectin, in which sLex determinants can be formed by transfection of COS cells with an appropriate fucosyltransferase cDNA, but ligand activity requires the inclusion of PSGL-1 cDNA (Sako et al., 1993). As noted above, a shared feature of the molecularly defined HEV-associated ligands for L-selectin is the presence of a mucin region (Puri et al., 1995). This feature provides the potential for multivalent presentation of carbohydrate recognition determinants, which is thought to be important for enhancing the avidity of L-selectin interactions (Nicholson et al., 1998). Thus, the CD34 contribution to ligand activity seen in the present study may be primarily due to its mucin character. The ability of other mucins to perform this postulated scaffold function and their possible distinct usage of the different sulfotransferases are questions for future investigation.
While each sulfotransferase was capable of conferring L-selectin binding onto CD34 in the CHO/FTVII/C2GnT cells, the greatest effect was clearly produced by the combined action of the two sulfotransferases. The level of binding achieved in the cells transfected with both sulfotransferases could not be attained with either sulfotransferase alone, over a wide range of cDNA concentrations. These findings argue that optimal binding to L-selectin requires both the Gal-6-sulfate and GlcNAc-6-sulfate moieties. It is possible that this synergy arises through dual recognition of separate monosulfated chains, for example one chain capped by sialyl 6-sulfo Lex and the other by sialyl 6′-sulfo Lex. This mechanism would fit a model of selectin binding proposed by Varki (1994), in which a specific cluster of adjacent O-linked chains comprises the full recognition determinant. Alternatively, individual chains containing both modifications may underlie the synergistic effect. In this regard, it should be noted that most of the sulfated chains within GlyCAM-1 contain two or more sulfates, although the structure of these chains has not been solved (Hemmerich et al., 1995). It is not yet clear whether sialyl 6′,6-disulfo Lex (Table I) exists as a capping group on these multisulfated chains or whether there are extended chains containing sulfates on internal Gal or GlcNAc residues. Additional analysis of natural ligands will be required to resolve these important structural questions.
L-selectin normally functions in tethering and rolling of lymphocytes on HEVs under conditions of blood flow (Warnock et al., 1998). Future experiments must determine the impact of the two sulfation modifications on the kinetic parameters of L-selectin binding to ligands, as these parameters are the key determinants of the dynamic interactions of lymphocytes with HEVs (Alon et al., 1997). We are currently addressing these issues using a parallel plate flow chamber to recapitulate physiological flow dynamics within blood vessels (Lawrence et al., 1995).
On the basis of its activities and highly restricted expression pattern, HEC-GlcNAc6ST is likely to be specifically involved in the biosynthesis of the sialyl 6-sulfo Lex epitope and related structures within L-selectin ligands. Ultimately, it may be necessary to inactivate the gene in mice by gene knockout to define the biological roles of this enzyme. Experiments of this type were required to demonstrate the critical role for FTVII in the generation of selectin ligands, including the HEV ligands for L-selectin (Maly et al., 1996). We cannot rule out the possible involvement of another recently cloned GlcNAc-6-sulfotransferase identified in mice and humans (Uchimura et al., 1998a,b) in ligand biosynthesis. As revealed by in situ hybridization, transcripts encoding the latter enzyme were present in HECs of mouse lymph nodes, although the overall tissue expression pattern was extremely broad in both species, suggesting other functions as well. In this regard, it should be noted that 6-sulfated GlcNAc residues are present in the GAG chains of keratan sulfate and a variety of glycoproteins, including the N-linked oligosaccharides of thyroglobulin (Kamerling et al., 1988), gp120 of HIV (Shilatifard et al., 1993), and porcine zona pellucida (Noguchi and Nakano, 1992). Furthermore, the GlcNAc-6-sulfate motif in the context of the core-2 structure has been identified in respiratory mucins from cystic fibrosis patients (Lo-Guidice et al., 1994), and a GlcNAc-6-sulfotransferase activity, which can act on mucins, has been detected in human respiratory mucosa (Degroote et al., 1997).
With respect to the sialyl 6′-sulfo Lex epitope, our data on the acceptor specificity and expression pattern of KSGal6ST are consistent with a possible role for this enzyme in the generation of the epitope within HECs. However, this enzyme (Fukuta et al., 1997), like the GlcNAc-6-sulfotransferase described by Uchimura and colleagues (1998a,b), is broadly distributed, being expressed in all of the major organs. In analogy with HEC-GlcNAc6ST described in the present study, one might predict the existence of an HEC-specific, or more highly restricted, Gal-6-sulfotransferase that sulfates L-selectin ligands in vivo.
L-selectin–dependent leukocyte trafficking into lymphoid organs and inflammatory sites is likely to be subject to complex regulation, which may be affected at the level of L-selectin ligand biosynthesis through the availability of protein scaffolds or by posttranslational modification. Given the dramatic sulfate dependency of the interaction between L-selectin and its ligands, and the high [35S]sulfate incorporation seen in HEVs and HEV-like vessels (Girard and Springer, 1995a), it is tempting to speculate that regulation of the expression or activity of one or both of the relevant sulfotransferases represents critical control points for this process. It is further plausible that the differential expression of the sulfotransferases may contribute to the apparent variation in the posttranslational modifications of L-selectin ligands in different lymphoid organs (Berg et al., 1998; Mitsuoka et al., 1998). The expression of KSGal6ST and HEC-GlcNAc6ST, as well as other candidate sulfotransferases, should also be examined in HEV-like vessels that are induced at sites of chronic inflammation (Michie et al., 1993; Girard and Springer, 1995a; Onrust et al., 1996). Endothelial ligands for L-selectin are also inducible on flat-walled vessels in vivo, although the biochemical nature of these ligands is poorly understood (Ley and Tedder, 1995). In vitro models of endothelial activation suggest that sulfation is required for the activity of these ligands (Giuffrè et al., 1997; Zakrzewicz et al., 1997; Tu et al., 1999). Endothelial sulfotransferases that are implicated in inflammatory leukocyte trafficking would clearly assume importance as potential targets for antiinflammatory therapeutics.
Abbreviations used in this paper
- BAC
bacterial artificial chromosome
- C2GnT
core 2 β1→ 6 N-acetylglucosaminyltransferase
- C6/KSST
chicken chondroitin/keratan sulfate sulfotransferase
- C6ST
human chondroitin-6-sulfotransferase
- CM
conditioned medium
- EST
expressed sequence tag
- FTVII
fucosyltransferase VII
- Gal
galactose
- GlcNAc
N-acetylglucosamine
- GlyCAM-1
glycosylation-dependent cell adhesion molecule 1
- HEC
high endothelial cell
- HEC-GlcNAc6ST
- HEV
high endothelial venule
- HPAEC
high pH anion exchange chromatography
- HUVEC
human umbilical vein endothelial cell
- KSGal6ST
keratan sulfate Gal-6-sulfotransferase
- MFI
mean fluorescence intensity
- nt
nucleotide(s)
- PE
phycoerythrin
- RT
reverse transcriptase
- sLex
sialyl Lewis x
Footnotes
The Lifeseq EST database (Incyte Pharmaceuticals, Inc., Palo Alto) was accessed through a licensing agreement with Roche Bioscience. We thank Carmen Tam for assistance in the in situ hybridization experiments. We are grateful for the help of Ms. Katy Hoiles of Incyte Pharmaceuticals in recovering plasmids containing the LifeSeq ESTs used in this study. We are further indebted to Ms. Deborah McCarley for help in the experiments, to Mr. Steve Stoufer, Ms. Sophie Chow, and Dr. Chinh Bach for sequencing plasmids, and to Dr. Kurt Jarnagin for his support and advice during the course of the study. We thank Drs. Geoffrey Kansas for providing us with the CHO/FTVII/C2GnT cells, Minoru Fukuda for the C2GnT cDNA, John Lowe for the L-selectin/IgM and FTVII cDNAs, and David Simmons for the pIG1 plasmid. We thank Mark Singer for purifying the HECs, and Chris Sassetti for isolating the RNA from the HECs and preparing the cDNA. We are also grateful to Kirsten Tangemann, Carolyn Bertozzi, Chris Sassetti, and Kendra Bowman for many useful discussions.
The research was supported by grants to S.D. Rosen from the National Institutes of Health (R37 GM23547 and GM57411) and from Roche Bioscience. J.K. Lee is supported by a postdoctoral fellowship from the Arthritis Foundation.
References
- Alon R, Chen S, Puri KD, Finger EB, Springer TA. The kinetics of L-selectin tethers and the mechanics of selectin-mediated rolling. J Cell Biol. 1997;138:1169–1180. doi: 10.1083/jcb.138.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews P, Milsom D, Ford W. Migration of lymphocytes across specialized vascular endothelium. V. Production of a sulphated macromolecule by high endothelial cells in lymph nodes. J Cell Sci. 1982;57:277–292. doi: 10.1242/jcs.57.1.277. [DOI] [PubMed] [Google Scholar]
- Baumhueter S, Dybdal N, Kyle C, Lasky LA. Global vascular expression of murine CD34, a sialomucin-like endothelial ligand for L-selectin. Blood. 1994;84:2554–2565. [PubMed] [Google Scholar]
- Baumhueter S, Singer MS, Henzel W, Hemmerich S, Renz M, Rosen SD, Lasky LA. Binding of L-selectin to the vascular sialomucin, CD34. Science. 1993;262:436–438. doi: 10.1126/science.7692600. [DOI] [PubMed] [Google Scholar]
- Berg EL, McEvoy LM, Berlin C, Bargatze RF, Butcher EC. L-selectin-mediated lymphocyte rolling on MAdCAM-1. Nature. 1993;366:695–698. doi: 10.1038/366695a0. [DOI] [PubMed] [Google Scholar]
- Berg EL, Robinson MK, Warnock RA, Butcher EC. The human peripheral lymph node vascular addressin is a ligand for LECAM-1, the peripheral lymph node homing receptor. J Cell Biol. 1991;114:343–349. doi: 10.1083/jcb.114.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg EL, Mullowney AT, Andrew DP, Goldberg JE, Butcher EC. Complexity and differential expression of carbohydrate epitopes associated with L-selectin recognition of high endothelial venules. Am J Pathol. 1998;152:469–477. [PMC free article] [PubMed] [Google Scholar]
- Bertozzi CR, Fukuda S, Rosen SD. Sulfated disaccharide inhibitors of L-selectin: deriving structural leads from a physiological selectin ligand. Biochemistry. 1995;34:14271–14278. doi: 10.1021/bi00044a001. [DOI] [PubMed] [Google Scholar]
- Bierhuizen MFA, Fukuda M. Expression cloning of a cDNA encoding UDP-GlcNAc:Galbeta1-3-GalNAc-R (GlcNAc to GalNAc) beta1-6GlcNAc transferase by gene transfer into CHO cells expressing polyoma large tumor antigen. Proc Natl Acad Sci USA. 1992;89:9326–9330. doi: 10.1073/pnas.89.19.9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman KG, Hemmerich S, Bakhta S, Singer MS, Bistrup A, Rosen SD, Bertozzi CR. Identification of an N-acetylglucamine-6-O-sulfotransferase activity specific to lymphoid tissue: an enzyme with a possible role in lymphocyte homing. Chem Biol. 1998;5:447–460. doi: 10.1016/s1074-5521(98)90161-2. [DOI] [PubMed] [Google Scholar]
- Butcher EC, Picker LJ. Lymphocyte homing and homeostasis. Science. 1996;272:60–66. doi: 10.1126/science.272.5258.60. [DOI] [PubMed] [Google Scholar]
- Clark RA, Fuhlbrigge RC, Springer TA. L-Selectin ligands that are O-glycoprotease resistant and distinct from MECA-79 antigen are sufficient for tethering and rolling of lymphocytes on human high endothelial venules. J Cell Biol. 1998;140:721–731. doi: 10.1083/jcb.140.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crommie D, Rosen SD. Biosynthesis of GlyCAM-1, a mucin-like ligand for L-selectin. J Biol Chem. 1995;270:22614–22624. doi: 10.1074/jbc.270.38.22614. [DOI] [PubMed] [Google Scholar]
- Degroote S, Lo-Guidice J-M, Strecker G, Ducourouble M-P, Roussel P, Lamblin G. Characterization of an N-acetylglucosamine-6-O-sulfotransferase from human respiratory mucosa active on mucin carbohydrate chains. J Biol Chem. 1997;272:29493–29501. doi: 10.1074/jbc.272.47.29493. [DOI] [PubMed] [Google Scholar]
- Dowbenko D, Kikuta A, Fennie C, Gillett N, Lasky LA. Glycosylation-dependent cell adhesion molecule 1 (GlyCAM-1) mucin is expressed by lactating mammary gland epithelial cells and is present in milk. J Clin Invest. 1993;92:952–960. doi: 10.1172/JCI116671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhlbrigge RC, Alon R, Puri KD, Lowe JB, Springer TA. Sialylated, fucosylated ligands for L-selectin expressed on leukocytes mediate tethering and rolling adhesions in physiologic flow conditions. J Cell Biol. 1996;135:837–848. doi: 10.1083/jcb.135.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuta M, Inazawa J, Torii T, Tsuzuki K, Shimada E, Habuchi O. Molecular cloning and characterization of human keratan sulfate Gal-6-sulfotransferase. J Biol Chem. 1997;272:32321–32328. doi: 10.1074/jbc.272.51.32321. [DOI] [PubMed] [Google Scholar]
- Fukuta M, Kobayashi Y, Uchimura K, Kimata K, Habuchi O. Molecular cloning and expression of human chondroitin 6-sulfotransferase. Biochim Biophys Acta. 1998;1399:57–61. doi: 10.1016/s0167-4781(98)00089-x. [DOI] [PubMed] [Google Scholar]
- Fukuta M, Uchimura K, Nakashima K, Kato M, Kimata K, Shinomura T, Habuchi O. Molecular cloning and expression of chick chondrocyte chondroitin 6-sulfotransferase. J Biol Chem. 1995;270:18575–18580. doi: 10.1074/jbc.270.31.18575. [DOI] [PubMed] [Google Scholar]
- Galustian C, Lawson AM, Komba S, Ishida H, Kiso M, Feizi T. Sialyl-lewis x sequence 6-O-sulfated at N-acetylglucosamine rather than at galactose is the preferred ligand for L-selectin and de-N-acetylation of the sialic acid enhances its binding strength. Biochem Biophys Res Commun. 1997;240:748–751. doi: 10.1006/bbrc.1997.7737. [DOI] [PubMed] [Google Scholar]
- Gallatin W, Weissman I, Butcher E. A cell-surface molecule involved in organ-specific homing of lymphocytes. Nature. 1983;304:30–34. doi: 10.1038/304030a0. [DOI] [PubMed] [Google Scholar]
- Girard J-P, Springer TA. High endothelial venules: specialized endothelium for lymphocyte migration. Immunol Today. 1995a;16:449–457. doi: 10.1016/0167-5699(95)80023-9. [DOI] [PubMed] [Google Scholar]
- Girard J-P, Springer TA. Cloning from purified high endothelial venule cells of hevin, a close relative of the antiadhesive extracellular matrix protein SPARC. Immunity. 1995b;2:113–123. doi: 10.1016/1074-7613(95)90083-7. [DOI] [PubMed] [Google Scholar]
- Giuffrè L, Cordey AS, Monai N, Tardy NY, Schapira M, Spertini O. Monocyte adhesion to activated aortic endothelium: role of L-selectin and heparan sulfate proteoglycans. J Cell Biol. 1997;136:945–956. doi: 10.1083/jcb.136.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habuchi O, Hirahara Y, Uchimura K, Fukuta M. Enzymatic sulfation of galactose residue of keratan sulfate by chondroitin 6-sulfotransferase. Glycobiology. 1996;6:51–57. doi: 10.1093/glycob/6.1.51. [DOI] [PubMed] [Google Scholar]
- Habuchi O, Suzuki Y, Fukuta M. Sulfation of sialyl lactosamine oligosaccharides by chondroitin 6-sulfotransferase. Glycobiology. 1997;7:405–412. doi: 10.1093/glycob/7.3.405. [DOI] [PubMed] [Google Scholar]
- Hemmerich S, Rosen SD. 6′-sulfated, sialyl Lewis X is a major capping group of GlyCAM-1. Biochemistry. 1994;33:4830–4835. doi: 10.1021/bi00182a011. [DOI] [PubMed] [Google Scholar]
- Hemmerich S, Bertozzi CR, Leffler H, Rosen SD. Identification of the sulfated monosaccharides of GlyCAM-1, an endothelial derived ligand for L-selectin. Biochemistry. 1994a;33:4820–4829. doi: 10.1021/bi00182a010. [DOI] [PubMed] [Google Scholar]
- Hemmerich S, Butcher EC, Rosen SD. Sulfation-dependent recognition of high endothelial venule (HEV)-ligands by L-selectin and MECA 79, an adhesion-blocking monoclonal antibody. J Exp Med. 1994b;180:2219–2226. doi: 10.1084/jem.180.6.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich S, Leffler H, Rosen SD. Structure of the O-glycans in GlyCAM-1, an endothelial-derived ligand for L-selectin. J Biol Chem. 1995;270:12035–12047. doi: 10.1074/jbc.270.20.12035. [DOI] [PubMed] [Google Scholar]
- Hoke D, Mebius RE, Dybdal N, Dowbenko D, Gribling P, Kyle C, Baumhueter S, Watson SR. Selective modulation of the expression of L-selectin ligands by an immune response. Curr Biol. 1995;5:670–678. doi: 10.1016/s0960-9822(95)00132-1. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Manzella SM, Baenziger JU. From legumes to leukocytes: biological roles for sulfated carbohydrates. FASEB J. 1996;10:1137–1146. doi: 10.1096/fasebj.10.10.8751716. [DOI] [PubMed] [Google Scholar]
- Imai Y, Lasky LA, Rosen SD. Sulphation requirement for GlyCAM-1, an endothelial ligand for L-selectin. Nature. 1993;361:555–557. doi: 10.1038/361555a0. [DOI] [PubMed] [Google Scholar]
- Imai Y, Singer MS, Fennie C, Lasky LA, Rosen SD. Identification of a carbohydrate-based endothelial ligand for a lymphocyte homing receptor. J Cell Biol. 1991;113:1213–1221. doi: 10.1083/jcb.113.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta Y, Pedersen LG, Pedersen LC, Negishi M. Conserved structural motifs in the sulfotransferase family. Trends Biochem Sci. 1998;23:129–130. doi: 10.1016/s0968-0004(98)01182-7. [DOI] [PubMed] [Google Scholar]
- Kamerling JP, Rijkse I, Maas AA, van Kuik JA, Vliegenthart JF. Sulfated N-linked carbohydrate chains in porcine thyroglobulin. FEBS Lett. 1988;241:246–250. doi: 10.1016/0014-5793(88)81070-6. [DOI] [PubMed] [Google Scholar]
- Kansas GS. Selectins and their ligands: current concepts and controversies. Blood. 1996;88:3259–3287. [PubMed] [Google Scholar]
- Kershaw DB, Beck SG, Wharram BL, Wiggins JE, Goyal M, Thomas P, Wiggins RC. Molecular cloning and characterization of human podocalyxin-like protein. Orthologous relationship to rabbit PCLP1 and rat podocalyxin. J Biol Chem. 1997;272:15708–15714. doi: 10.1074/jbc.272.25.15708. [DOI] [PubMed] [Google Scholar]
- Koenig A, Jain R, Vig R, Norgard-Sumnicht KE, Matta KL, Varki A. Selectin inhibition: synthesis and evaluation of novel sialylated, sulfated and fucosylated oligosaccharides, including the major capping group of GlyCAM-1. Glycobiology. 1997;7:79–93. doi: 10.1093/glycob/7.1.79. [DOI] [PubMed] [Google Scholar]
- Kolodkin AL, Levengood DV, Rowe EG, Tai YT, Giger RJ, Ginty D. Neuropilin is a semaphorin III receptor. Cell. 1997;90:753–762. doi: 10.1016/s0092-8674(00)80535-8. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lasky LA, Singer MS, Dowbenko D, Imai Y, Henzel WJ, Fennie C, Gillett N, Watson SR, Rosen SD. An endothelial ligand for L-selectin is a novel mucin-like molecule. Cell. 1992;69:927–938. doi: 10.1016/0092-8674(92)90612-g. [DOI] [PubMed] [Google Scholar]
- Lawrence MB, Berg EL, Butcher EC, Springer TA. Rolling of lymphocytes and neutrophils on peripheral node addressin and subsequent arrest on ICAM-1 in shear flow. Eur J Immunol. 1995;25:1025–1031. doi: 10.1002/eji.1830250425. [DOI] [PubMed] [Google Scholar]
- Lee MS, Sarvetnick N. Induction of vascular addressins and adhesion molecules in the pancreas of IFN-gamma transgenic mice. J Immunol. 1994;152:4597–4603. [PubMed] [Google Scholar]
- Lewinsohn DM, Bargatze RF, Butcher EC. Leukocyte-endothelial cell recognition: evidence of a common molecular mechanism shared by neutrophils, lymphocytes, and other leukocytes. J Immunol. 1987;138:4313–4321. [PubMed] [Google Scholar]
- Ley K, Tedder TF. Leukocyte interactions with vascular endothelium. New insights into selectin-mediated attachment and rolling. J Immunol. 1995;155:525–528. [PubMed] [Google Scholar]
- Ley K, Gaehtgens P, Fennie C, Singer MS, Lasky LA, Rosen SD. Lectin-like cell adhesion molecule 1 mediates leukocyte rolling in mesenteric venules in vivo. Blood. 1991;77:2553–2555. [PubMed] [Google Scholar]
- Li FG, Wilkins PP, Crawley S, Weinstein J, Cummings RD, McEver RP. Post-translational modifications of recombinant P-selectin glycoprotein ligand-1 required for binding to P- and E-selectin. J Biol Chem. 1996;271:3255–3264. [PubMed] [Google Scholar]
- Lo-Guidice JM, Wieruszeski JM, Lemoine J, Verbert A, Roussel P, Lamblin G. Sialylation and sulfation of the carbohydrate chains in respiratory mucins from a patient with cystic fibrosis. J Biol Chem. 1994;269:18794–18813. [PubMed] [Google Scholar]
- Maly P, Thall AD, Petryniak B, Rogers CE, Mith PL, Marks RM, Kelly RJ, Gersten KM, Cheng G, Saunders TL, et al. The alpha(1,3)fucosyltransferase Fuc-TVII controls lymphocyte homing, and blood leukocyte emigration through an essential role in L-, E- and P-selectin ligand biosynthesis. Cell. 1996;86:643–653. doi: 10.1016/s0092-8674(00)80137-3. [DOI] [PubMed] [Google Scholar]
- McEver RP, Cummings RD. Role of PSGL-1 binding to selectins in leukocyte recruitment. J Clin Invest. 1997;100:97–103. [PubMed] [Google Scholar]
- Michie SA, Streeter PR, Bolt PA, Butcher EC, Picker LJ. The human peripheral lymph node vascular addressin. Am J Pathol. 1993;143:1688–1698. [PMC free article] [PubMed] [Google Scholar]
- Mitsuoka C, Sawada-Kasugai M, Ando-Furui K, Izawa M, Nakanishi H, Nakamura S, Ishida H, Kiso M, Kannagi R. Identification of a major carbohydrate capping group of the L-selectin ligand on high endothelial venules in human lymph nodes as 6-sulfo sialyl Lewis X. J Biol Chem. 1998;273:11225–11233. doi: 10.1074/jbc.273.18.11225. [DOI] [PubMed] [Google Scholar]
- Nelson RM, Venot A, Bevilacqua MP, Linhardt RJ, Stamenkovic I. Carbohydrate-protein interactions in vascular biology. Annu Rev Cell Dev Biol. 1995;11:601–631. doi: 10.1146/annurev.cb.11.110195.003125. [DOI] [PubMed] [Google Scholar]
- Nicholson MW, Barclay AN, Singer MS, Rosen SD, van der Merwe PA. Affinity and kinetic analysis of L-selectin (CD62L) binding to GlyCAM-1. J Biol Chem. 1998;273:763–770. doi: 10.1074/jbc.273.2.763. [DOI] [PubMed] [Google Scholar]
- Noguchi S, Nakano M. Structure of the acidic N-linked carbohydrate chains of the 55-kDa glycoprotein family (PZP3) from porcine zona pellucida. Eur J Biochem. 1992;209:883–894. doi: 10.1111/j.1432-1033.1992.tb17361.x. [DOI] [PubMed] [Google Scholar]
- Onrust SV, Hartl PM, Rosen SD, Hanahan D. Modulation of L-selectin ligand expression during an immune response accompanying tumorigenesis in transgenic mice. J Clin Invest. 1996;97:54–64. doi: 10.1172/JCI118406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyani T, Seed B. PSGL-1 recognition of P-selectin is controlled by a tyrosine sulfation consensus at the PSGL-1 amino terminus. Cell. 1995;83:333–343. doi: 10.1016/0092-8674(95)90174-4. [DOI] [PubMed] [Google Scholar]
- Puri KD, Finger EB, Gaudernack G, Springer TA. Sialomucin CD34 is the major L-selectin ligand in human tonsil high endothelial venules. J Cell Biol. 1995;131:261–270. doi: 10.1083/jcb.131.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen SD, Bertozzi CB. Leukocyte adhesion: two selectins converge on sulphate. Curr Biol. 1996;6:261–264. doi: 10.1016/s0960-9822(02)00473-6. [DOI] [PubMed] [Google Scholar]
- Rosenberg RD, Shworak NW, Liu J, Schwartz IJ, Zhang L. Heparan sulfate proteoglycans of the cardiovascular system. Specific structures emerge but how is synthesis regulated? . J Clin Invest. 1997;100:S67–S75. [PubMed] [Google Scholar]
- Sako D, Chang XJ, Barone KM, Vachino G, White HM, Shaw G, Veldman GM, Bean KM, Ahern TJ, Furie B, et al. Expression cloning of a functional glycoprotein ligand for P-selectin. Cell. 1993;75:1179–1186. doi: 10.1016/0092-8674(93)90327-m. [DOI] [PubMed] [Google Scholar]
- Sako D, Comess KM, Barone KM, Camphausen RT, Cumming DA, Shaw GD. A sulfated peptide segment at the amino terminus of PSGL-1 is critical for P-selectin binding. Cell. 1995;83:323–331. doi: 10.1016/0092-8674(95)90173-6. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning, A Laboratory Manual. Cold Spring Harbor Laboratory Press. Plainview, New York.
- Sanger F, Nicklens S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti C, Tangemann K, Singer MS, Kershaw DE, Rosen SD. Identification of podocalyxin as a high endothelial venule ligand for L-selectin: parallels to CD34. J Exp Med. 1998;187:1965–1975. doi: 10.1084/jem.187.12.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WJ, Katsumoto TR, Bertozzi CR, Rosen SD, Kiessling LL. Selectin-carbohydrate interactions: an investigation into the relevant modifications of the Lewis x trisaccharides. Biochemistry. 1996;35:14862–14867. doi: 10.1021/bi9613640. [DOI] [PubMed] [Google Scholar]
- Scudder PR, Shailubhai K, Duffin KL, Streeter PR, Jacob GS. Enzymatic synthesis of a 6-sulfated sialyl-Lewisxwhich is an inhibitor of L-selectin binding to peripheral addressin. Glycobiology. 1994;4:929–933. doi: 10.1093/glycob/4.6.929. [DOI] [PubMed] [Google Scholar]
- Shailubhai K, Streeter P, Smith CE, Jacob GS. Sulfation and sialylation requirements for a glycoform of CD34, a major endothelial ligand for L-selectin in porcine peripheral lymph nodes. Glycobiology. 1997;7:305–314. doi: 10.1093/glycob/7.2.305. [DOI] [PubMed] [Google Scholar]
- Shilatifard A, Merkle RK, Helland DE, Welles JL, Haseltine WA, Cummings RD. Complex-type N-linked oligosaccharides of gp120 from human immunodeficiency virus type 1 contain sulfated N-acetylglucosamine. J Virol. 1993;67:943–952. doi: 10.1128/jvi.67.2.943-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, D.L. 1993. Cloning cell surface molecules by transient expression in mammalian cells. In Cellular Interactions in Development: A Practical Approach. D.A. Hartley, editor. IRL Press at Oxford University Press, Oxford. 93–127.
- Snapp KR, Ding H, Atkins K, Warnke R, Luscinskas FW, Kansas GS. A novel P-selectin glycoprotein ligand-1 monoclonal antibody recognizes an epitope within the tyrosine sulfate motif of human PSGL-1 and blocks recognition of both P- and L-selectin. Blood. 1998;91:154–164. [PubMed] [Google Scholar]
- Springer TA. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- Streeter PR, Rouse BTN, Butcher EC. Immunohistologic and functional characterization of a vascular addressin involved in lymphocyte homing into peripheral lymph nodes. J Cell Biol. 1988;107:1853–1862. doi: 10.1083/jcb.107.5.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi S, Isogai Y, Hada N, King JK, Hindsgaul O, Fukuda M. 6′-Sulfo sialyl Lex but not 6-sulfo sialyl Lex expressed on the cell surface supports L-selectin-mediated adhesion. J Biol Chem. 1996;271:27213–27216. doi: 10.1074/jbc.271.44.27213. [DOI] [PubMed] [Google Scholar]
- Tu L, Delahunty MD, Ding H, Luscinskas FW, Tedder TF. The cutaneous lymphocyte antigen is an essential component of the L-selectin ligand induced on human vascular endothelial cells. J Exp Med. 1999;189:241–252. doi: 10.1084/jem.189.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura K, Muramatsu H, Kadomatsu K, Fan QW, Kurosawa N, Mitsuoka C, Kannagi R, Habuchi O, Muramatsu T. Molecular cloning and characterization of an N-acetylglucosamine-6-O-sulfotransferase. J Biol Chem. 1998a;273:22577–22583. doi: 10.1074/jbc.273.35.22577. [DOI] [PubMed] [Google Scholar]
- Uchimura K, Muramatsu H, Kaname T, Ogawa H, Yamakawa T, Fan QW, Mitsuoka C, Kannagi R, Habuchi O, Yokoyama I, et al. Human N-acetylglucosamine-6-O-sulfotransferase involved in the biosynthesis of 6-sulfo sialyl Lewis X: molecular cloning, chromosomal mapping, and expression in various organs and tumor cells. J Biochem (Tokyo) 1998b;124:670–678. doi: 10.1093/oxfordjournals.jbchem.a022164. [DOI] [PubMed] [Google Scholar]
- Varki A. Selectin ligands. Proc Natl Acad Sci USA. 1994;91:7390–7397. doi: 10.1073/pnas.91.16.7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagers AJ, Stoolman LM, Kannagi R, Craig R, Kansas GS. Expression of leukocyte fucosyltransferases regulates binding to E-selectin: relationship to previously implicated carbohydrate epitopes. J Immunol. 1997;159:1917–1929. [PubMed] [Google Scholar]
- Warnock RA, Askari S, Butcher EC, von Andrian UH. Molecular mechanisms of lymphocyte homing to peripheral lymph nodes. J Exp Med. 1998;187:205–216. doi: 10.1084/jem.187.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson SR, Imai Y, Fennie C, Geoffroy JS, Rosen SD, Lasky LA. A homing receptor-IgG chimera as a probe for adhesive ligands of lymph node high endothelial venules. J Cell Biol. 1990;110:2221–2229. doi: 10.1083/jcb.110.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins PP, Moore KL, McEver RP, Cummings RD. Tyrosine sulfation of P-selectin glycoprotein ligand-1 is required for high affinity binding to P-selectin. J Biol Chem. 1995;270:22677–22680. doi: 10.1074/jbc.270.39.22677. [DOI] [PubMed] [Google Scholar]
- Yoshino K, Ohmoto H, Kondo N, Tsujishita H, Hiramatsu Y, Inoue Y, Kondo H. Studies on selectin blockers. 4. Structure-function relationships of sulfated sialyl Lewis X hexasaccharide ceramides toward E-, P-, and L-selectin binding. J Med Chem. 1997;40:455–462. doi: 10.1021/jm9605290. [DOI] [PubMed] [Google Scholar]
- Zakrzewicz A, Grafe M, Terbeek D, Bongrazio M, Auch-Schwelk W, Walzog B, Graf K, Fleck E, Ley K, Gaehtgens P. L-selectin-dependent leukocyte adhesion to microvascular but not to macrovascular endothelial cells of the human coronary system. Blood. 1997;89:3228–3235. [PubMed] [Google Scholar]