Abstract
Animal cells contain a single centrosome that nucleates and organizes a polarized array of microtubules which functions in many cellular processes. In most cells the centrosome is composed of two centrioles surrounded by an ill-defined “cloud” of pericentriolar material. Recently, γ-tubulin-containing 25-nm diameter ring structures have been identified as likely microtubule nucleation sites within the pericentriolar material of isolated centrosomes. Here we demonstrate that when Spisula centrosomes are extracted with 1.0 M KI they lose their microtubule nucleation potential and appear by three-dimensional electron microscopy as a complex lattice, built from 12- to 15-nm thick elementary fiber(s), that lack centrioles and 25-nm rings. Importantly, when these remnants are incubated in extracts prepared from Spisula oocytes they recover their 25-nm rings, γ-tubulin, and microtubule nucleation potential. This recovery process occurs in the absence of microtubules, divalent cations, and nucleotides. Thus, in animals the centrosome is structurally organized around a KI-insoluble filament-based “centromatrix” that serves as a scaffold to which those proteins required for microtubule nucleation bind, either directly or indirectly, in a divalent cation and nucleotide independent manner.
The centrosome is the major microtubule-organizing center of animal cells (1). This structure is responsible for nucleating and orienting microtubules, which are major components of the cytoskeletal framework, but indirectly it has a more general influence on cellular organization. For example, microtubules have been shown to be involved in the positioning of organelles (2, 3), in transportation of cellular components by way of microtubule-dependent motors (4), and in the formation of the spindle apparatus in mitotic cells (5). Thus, as a result of its microtubule-organizing center function, the centrosome plays a major role in the organization of the entire cell cytoplasm.
During specific phases of the cell cycle (e.g., mitosis; ref. 1), or after fertilization (6, 7), centrosomes undergo a dramatic maturation process that is characterized by an increase in their microtubule nucleation potential (MNP). When isolated from a variety of sources, centrosomes retain the ability to nucleate radial (astral) arrays of microtubules in defined media (8–10). This MNP can be removed by treatment with various reagents including salts, chaotropic agents, and proteases (9, 11–14). Interestingly, under certain conditions, removal of the MNP does not inhibit the ability of isolated centrosomes to support parthenogenetic development of frog embryos when injected into frog oocytes (12). This indicates that some chaotrope-insoluble centrosome remnant-structure, capable of recovering complete centrosome function, must survive such treatments.
Typical animal cell centrosomes are composed of two centrioles surrounded by an amorphous cloud of pericentriolar material (PCM). Electron microscopy (EM) studies indicate that microtubules originate from within the PCM and are not continuous with the short microtubules that make up the centriole (15). Therefore, the centrosome’s ability to nucleate microtubules resides within the relatively undefined PCM. Although the molecular basis of microtubule nucleation remains unresolved, recent evidence indicates that γ-tubulin, a member of the tubulin protein family, is involved (16–20). Importantly, 25-nm-diameter ring-shaped structures that are the same diameter as microtubules have been identified by three-dimensional EM tomography within the PCM of Drosophila centrosomes (21) and contain γ-tubulin (22). Similar ring structures have been identified in isolated Spisula centrosomes (8). Further, an oligomeric protein complex composed of γ-tubulin and a number of other proteins [γ-tubulin ring complexes (γ-TuRCs)] that speeds the rate of microtubule polymerization in vitro has been isolated from Xenopus oocyte lysates (23). These isolated γ-TuRCs are also 25 nm in diameter, and thus it has been proposed that γ-TuRCs located within the PCM of centrosomes serve as the templates for the nucleation of microtubules (8, 21–24).
If this hypothesis is correct, the ability of a centrosome to nucleate microtubules should correlate with the presence of γ-TuRCs within the PCM. Thus, the removal of γ-tubulin and 25-nm-diameter rings from the PCM should result in a loss of MNP, and their replacement should result in the recovery of MNP. Moreover, because treatment of centrosomes with chaotropic agents removes MNP, then the molecular and structural definition of the chaotrope-insoluble centrosome remnant becomes an important objective for understanding centrosome assembly and function.
To approach these questions we have focused our attention on a model system that offers numerous advantages for a biochemical approach to understanding centrosome composition and function, namely, oocytes of the Atlantic surf clam, Spisula solidissima (8, 25, 26). These oocytes are arrested at the G2/M transition of meiosis I, and can be parthenogenetically activated to complete meiosis by adding KCl to oocyte sea water suspensions. Thus, an entire population of oocytes can be induced to proceed through the meiotic cell cycle in synchrony (6). Importantly, methods have been developed for the isolation and storage of milligram quantities of homogeneous centrosomes from one specific time point in the meiotic cell cycle, 4 min after oocyte activation (8, 26).
Using isolated Spisula centrosomes, we demonstrate that their MNP can be dissociated by treatment with 1.0 M KI. Importantly, the loss of MNP corresponds with the removal of γ-tubulin and 25-nm ring structures from the PCM. We further show that the KI-insoluble Spisula centrosome remnant is composed of a structural network of 12- to 15-nm diameter filaments. Most importantly, the KI-insoluble remnant recovers MNP, γ-tubulin, and 25-nm rings when treated with high-speed Spisula oocyte extracts, and recovery of MNP is independent of microtubules, nucleotides, and divalent cations.
METHODS
Preparation of Spisula Oocyte Extracts.
Adult Spisula solidissima were obtained from the Marine Biological Laboratory (Woods Hole, MA). Gonads were dissected, and oocytes were collected, washed and parthenogenetically activated with KCl as described (6, 8, 25–27). Lysates that contain centrosomes were prepared from oocytes 4 min postactivation, frozen, and stored at −80°C (8, 27).
To make high-speed centrosome-free extracts, frozen-stored lysates were thawed on ice, diluted 1:1 in PEM (5 mM Pipes/1 mM EGTA/1 mM MgSO4, pH 7.2) and centrifuged three times at 39,000 × g at 4°C to clarify. Supernatants were collected and this process repeated three times. The third supernatant was centrifuged at 100,000 × g for 60 min at 4°C and the final supernatant/extract collected and used in the assays for recovery of MNP. When needed, these extracts were diluted 1/20 in PEM containing 20 μM colchicine. In some experiments, samples were supplemented with 5–50 mM 6-dimethylaminopurine (6-DMAP). For experiments involving EDTA, the 100,000 × g extract was diluted in PE buffer (5 mM Pipes/1 mM EGTA, pH 7.2) containing 20 μM colchicine and 5–100 mM EDTA.
Isolation of 3×-Cycled Sea Urchin Tubulin.
Sea urchin tubulin was isolated from Strongylocentrotus purpuratus oocytes by three cycles of microtubule polymerization and depolymerization (8, 28). After the third cycle, microtubules were depolymerized, centrifuged to clarify, and the resulting supernatant adjusted to a concentration of 0.7 mg/ml protein. Samples were aliquoted and stored at −80°C.
Disassembly and Reassembly of Centrosome MNP.
Centrosomes were isolated from frozen-stored Spisula oocyte lysates using sucrose density-gradient fractionation methods previously described (8, 26). Centrosomes were treated with either PEM or PEM containing 1.0 M KI for 15 min at room temperature and immobilized onto glass coverslips by centrifugation through a 10% sucrose cushion at 12,000 × g for 15 min at 4°C (9, 10, 26, 27). Coverslips were then washed three times with PEM, incubated for 10 min in either PEM or high-speed oocyte extract, supplemented with colchicine, EDTA, or 6-DMAP as described above. Coverslips were washed in PEM and incubated in tubulin reassembly buffer (100 mM Pipes/1 mM EGTA/5 mM MgSO4, pH 6.9) containing 0.35 mg/ml sea-urchin tubulin (8, 28) for 15 min to allow microtubule nucleation and aster formation. Samples were fixed by adding an equal volume of 2% glutaraldehyde in reassembly buffer, incubated for 15 min, postfixed in −20°C methanol, and processed for immunofluorescence analysis.
Immunofluorescence.
Fixed samples on coverslips were washed in PBS, treated with 10 mg/ml sodium borohydride, washed, and blocked in PBS containing 5% nonfat dry milk and 5 mg/ml BSA for 30 min. Coverslips are then incubated in primary rat anti-tubulin (Serotec) and affinity-purified rabbit polyclonal antibody prepared against the C-terminal sequence CAATRPDYISWGTQDK of Xenopus γ-tubulin (23). In some experiments we also used a mouse mAb that recognizes a Spisula centrosome protein, SpiCen300 (X. Wu, G. Pens, T. Ohta, J. Vogel, R. Kuriyama, and R.E.P., unpublished work). Coverslips were incubated in primary antibodies for 15 min, washed, and incubated in FITC-conjugated anti-rat and rhodamine-conjugated anti-rabbit secondary antibodies (Jackson ImmunoResearch) for γ-tubulin staining, or rhodamine-conjugated anti-mouse secondary antibodies for SpiCen300 staining, respectively. All antibody incubations were for 30 min at room temperature. After incubations in secondary antibodies, coverslips were washed and permanently mounted as described (8, 26, 27). Images were collected using a Bio-Rad MRC 1000 laser-scanning confocal system, in conjunction with a Nikon Diaphot 200 microscope equipped with a 60×/1.4 NA objective lens. In addition, some images were taken with a Zeiss Axioplan epifluorescence microscope equipped with a 63×/1.3 NA objective lens and a Hamamatsu SIT-video camera that was coupled to a Metamorph image processing system (Universal Imaging, Media, PA).
SDS/PAGE and Immunoblot Analyses.
Isolated centrosomes (8) were diluted in PEM buffer, pelleted at 120,000 × g for 30 min at 4°C, and resuspended in PEM buffer or PEM containing 1.0 M KI, incubated for 15 min at room temperature and centrifuged at 120,000 × g for 30 min at 4°C. Samples were aspirated dry, pellets were resuspended in 25 mM Tris/0.5% SDS (pH 7.2), and the protein concentrations were determined by the bicinchoninic acid (BCA) protein assay method (Pierce). Samples were diluted into sample buffer and equal protein was loaded onto four SDS/20% polyacrylamide gels (29). Gels were run at 100 V using a mini-PROTEAN II gel system (Bio-Rad) and stained with colloidal Coomassie (30). For immunoblots, proteins separated on SDS/polyacrylamide gels were transferred to nitrocellulose at 100 V for 2 hr on ice (31). Blots were blocked with 5% nonfat dry milk in TBST (Tris-buffered saline containing 0.05% Tween-20, pH 7.5) (8) for 1 hr, incubated in affinity-purified polyclonal γ-tubulin antibody for 1 hr, washed, incubated with horseradish peroxidase-conjugated anti-rabbit IgG secondary antibodies (Promega) for 1 hr, washed and developed with Super-Signal Substrate (Pierce). All washes and antibody incubations were at room temperature.
Intermediate Voltage EM (IVEM) Tomography.
Isolated centrosomes, KI-insoluble centrosome remnants (KICRs), and KICRs incubated in 100,000 × g centrosome-free oocyte supernatants were prepared and immobilized onto glass coverslips as described above. They were then washed in buffer, fixed with 0.5% glutaraldehyde for 15 min, washed, postfixed with 1% OsO4 for 15 min on ice, washed, and stained with 1% uranyl acetate for 3 hr. After washing, the samples were dehydrated in increasing concentrations of ethanol, transferred to acetone, and flat-embedded on the glass coverslip (8). After removing the coverslip with hydrofluoric acid, 10–15 serial 0.25-μm-thick sections were cut from the embedment surface that interfaced with the glass coverslip. After mounting in the center of Formvar-coated slot grids that had been lightly decorated with 15-nm-diameter colloidal gold, the sections were stained in uranyl acetate and lead citrate. Double-tilt IVEM tomography (±60o) was then conducted as described elsewhere (8, 32). IVEM images were originally recorded on film at ×30,000 and then digitized so that the size of each pixel was 3.1 nm. For this particular study we reconstructed samples from three separate experiments; three controls, three KI-extracted, and two reconstituted centrosomes.
RESULTS
Salt Extraction Removes MNP from the Centrosome.
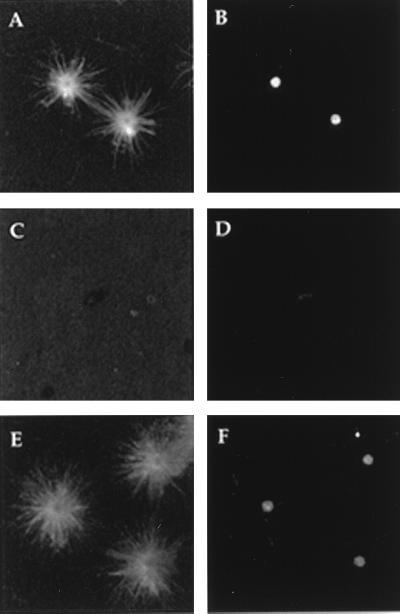
To explore the relationship between the structure, MNP, and composition of centrosomes, we developed an assay for the disassembly and reassembly of MNP using centrosomes isolated from S. solidissima oocytes (8, 26). Centrosomes were treated with PEM buffer (control) or buffer containing 1.0 M KI, sedimented onto glass coverslips (9), washed, and incubated in tubulin-containing media to allow microtubule nucleation and aster formation. Centrosome MNP and γ-tubulin content was then determined by double-label indirect immunofluorescence analysis using primary antibodies against microtubules and γ-tubulin. Coverslips with bound centrosomes that had been treated with PEM buffer contained an abundance of astral microtubule arrays (Fig. 1A), and each aster displayed a dense γ-tubulin staining spot at the center (Fig. 1B). Thus, treatment with buffer and immobilization of centrosomes onto glass coverslips had little or no effect on the centrosomes MNP. However, when centrosomes were treated with 1.0 M KI, no γ-tubulin staining (Fig. 1D) and no asters (Fig. 1C) were found associated with these coverslips. Importantly, EM studies revealed that KI-insoluble centrosome remnants (KICRs; see below) were indeed bound to these coverslips, but their ability to nucleate microtubules had been removed (Fig. 1C). To determine if these centrosome remnants could recover MNP, coverslips containing immobilized KICRs were incubated in high-speed centrosome-free Spisula oocyte extracts (27) containing 20 μM colchicine for 10 min at room temperature, washed, challenged with tubulin-containing medium, and screened for aster content. Asters were easily found on these coverslips, indicating that the KICRs had indeed recovered their MNP (Fig. 1E). Further, the center of each aster contained a dense γ-tubulin-stained spot (Fig. 1F), indicating that γ-tubulin that was present in the high-speed oocyte extract bound to the KICRs. As a control, blank coverslips were treated with oocyte extract followed by tubulin, and no asters were observed (data not shown). Together, these data reveal that KI-treatment removes the γ-tubulin and the MNP from isolated Spisula centrosomes, leaving a KICR that can be sedimented onto glass and that can recover its MNP when incubated in soluble oocyte extract.
Figure 1.
Fluorescence micrographs of asters formed by centrosomes and centrosome remnants after indirect immunofluorescent labeling for both tubulin (A, C, and E) and γ-tubulin (B, D, and F). The MNP (A) and γ-tubulin (B) present in centrosomes are removed by treatment with 1.0 M KI (C and D). The MNP recovers (E) and γ-tubulin returns (F) when KI-insoluble centrosome remnants are incubated in oocyte extracts.
Protein Analysis of the Centrosome and KICR.
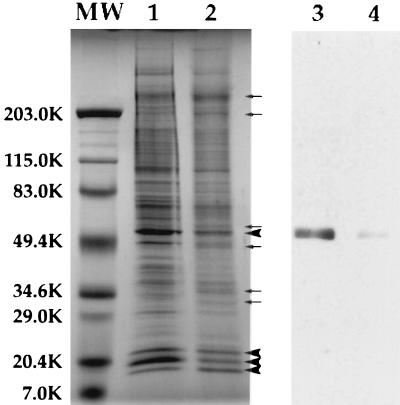
The composition of centrosomes and KICRs was assessed by gel electrophoresis and Western blot analyses. Centrosomes were treated with PEM or PEM containing 1.0 M KI, the resultant soluble and insoluble fractions were separated by centrifugation, and the protein content of both fractions determined. Protein analysis revealed that the KICR represents roughly 10% of the total centrosome protein (data not shown). Comparison of centrosome (Fig. 2, lane 1) and KICR (Fig. 2, lane 2) protein profiles revealed that the KICR was enriched in a number of proteins. The major proteins found in the KICR were three proteins of ≈20 kDa, and a protein of ≈50 kDa (Fig. 2, lane 2), which also copurified with isolated centrosomes (ref. 8; Fig. 2, lane 1). In addition, a number of other proteins were enriched in the KICR fraction (Fig. 2, lane 2).
Figure 2.
SDS/PAGE and immunoblot analyses of centrosomes and KICRs. Some of the proteins present in isolated centrosome fractions (lane 1) are removed by KI treatment (lane 2). Three proteins of ≈20 kDa, which copurify with centrosomes (lane 1), along with a protein of ≈50 kDa are the most abundant proteins in KICR fractions (large arrowheads, lane 2). Additionally, at least six proteins are enriched in the KICR fraction compared with control centrosomes (small arrows, lane 2). Immunoblots of centrosomes (lane 3) and KICRs (lane 4) with affinity-purified polyclonal anti-γ-tubulin antibody reveals that γ-tubulin that is present in isolated centrosome fractions (lane 3), is removed by KI treatment and KICR fractions (lane 4) contain negligible γ-tubulin.
Importantly, Western blot analysis revealed that although centrosomes contain an abundance of γ-tubulin (Fig. 2, lane 3), the KICR fraction contained a negligible amount of γ-tubulin (Fig. 2, lane 4). This confirms the indirect immunofluorescence observations that suggested that KI-treatment dissociated γ-tubulin from centrosomes, and little if any was left associated with KICRs (Fig. 1D and Fig. 2, lane 4).
Ultrastructural Analysis of Centrosomes, KI-Insoluble Centrosome Remnants, and Recovered KI-Insoluble Centrosome Remnants.
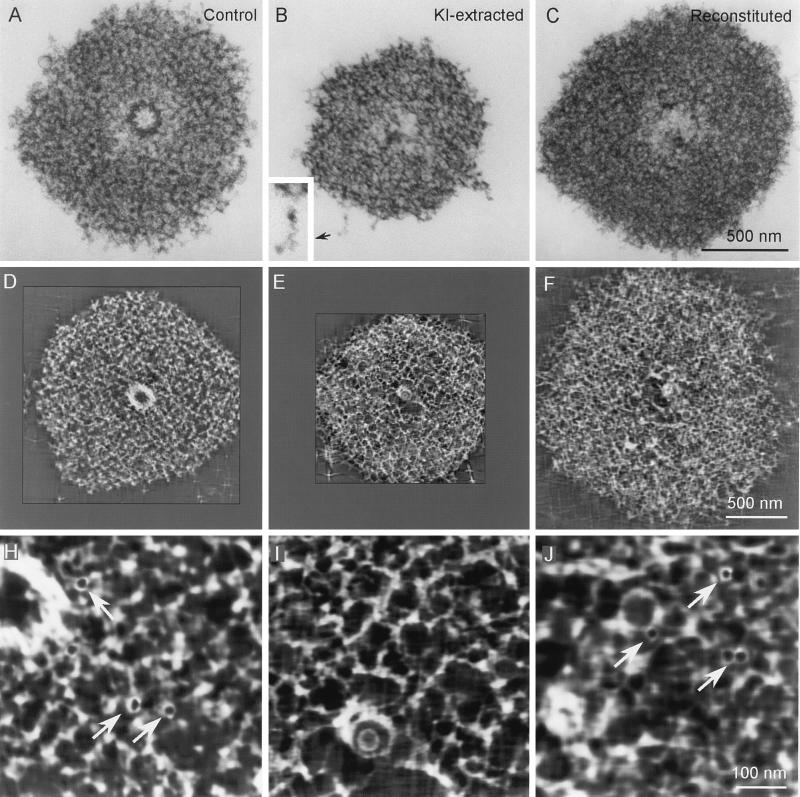
IVEM tomography (8, 21, 22) was used to analyze the structural features of centrosomes, KICRs, and KICRs that had recovered MNP when treated with high-speed oocyte extracts (recovered KICRs). As previously reported, numerous 25 nm-diameter rings were found within the PCM of isolated Spisula centrosomes when analyzed by IVEM tomography (Fig. 3H; see also ref. 8). However, these rings were absent from centrosomes that had been treated with KI (Fig. 3I). Thus, KICRs, which do not nucleate microtubules, do not contain 25-nm rings. Importantly, analyses of KICRs that had recovered MNP as a result of treatment with high-speed oocyte extracts revealed that these recovered KICRs contained an abundance of these 25-nm rings (Fig. 3J). Thus, as was the case for γ-tubulin content, the loss and recovery of the centrosome’s MNP correlates with the loss and recovery of 25-nm rings.
Figure 3.
IVEM analyses of control (A, D, and H), KI-extracted (B, E, and I), and reconstituted (C, F, and J) Spisula centrosomes. A–C are electron micrographs of 0.25-μm-thick sections cut through the centriole-containing region of the centrosome. D–F represent single 3-nm-thick slices through the tomographic volume computed from centrosomes similar to those in A–C. H–J are higher magnification images of the centrosome matrix pictured in D–F. Note that the 25-nm-diameter ring-shaped structures seen in control (arrows in H) and reconstituted (arrows in J) centrosomes are absent from the KI-extracted centrosome (I). Note also that the matrix of the KI-extracted centrosome is comprised of an interconnected latticework of 12- to 15-nm-diameter filaments (see D, E, H, and I) that can be resolved in the raw IVEM images at the peripheral edges of the matrix (B Inset).
In addition to removing the 25-nm rings and other matrix components, KI-treatment had several additional effects on the structure of isolated Spisula centrosomes. The most conspicuous effects were that KI significantly reduced the overall diameter of the centrosome and removed the centriole (Fig. 3 A–E). This latter finding is consistent with previous reports that KI disrupts the structure of centrioles in isolated mammalian centrosomes (12). Although centrosome diameter and MNP recovered when KICRs were treated with oocyte extract (Fig. 3 B, C, E, and F), the centrioles were not recovered (Fig. 3 C and F). Most importantly, however, was the consistent finding that the PCM of KI-extracted centrosomes appeared as a loosely organized but interconnected three-dimensional lattice of 12- to 15-nm-diameter fibrous polymers (Fig. 3 D, E, H, and I). The fibrous nature of the KICR was apparent throughout the volume of the reconstructions (Fig. 3 E and I) and was clearly evident even in electron micrographs of 0.25-μm-thick sections (Fig. 3B), especially at the thinner peripheral edges where individual fibers could be resolved (Fig. 3B, Inset).
Recovery of MNP.
Because all assays for the recovery of MNP by KICRs were conducted in the presence of 20 μM colchicine, it is reasonable to conclude that the recruitment and binding of molecules required for the nucleation of microtubules to the KICR does not require polymerized microtubules. To extend our analysis of what is required for KICRs to recover their MNP, we tested the effects of EDTA (a chelator of divalent cations) and 6-DMAP (an ATP analog and general protein kinase inhibitor) on the recovery process. KICRs were prepared, immobilized onto glass coverslips, and incubated in high-speed oocyte extracts containing EDTA (5–100 mM) or 6-DMAP (5–50 mM), washed, treated with tubulin media, fixed, and assayed by indirect immunofluorescence using antibodies to tubulin to visualize and count asters. To locate and count KICRs and centrosomes, we used a mAb that recognizes a centrosome specific protein, SpiCen300 (X. Wu, G. Peng, T. Ohta, J. Vogel, R. Kuriyama, and R.E.P., unpublished results), which we found recognizes both KICRs and centrosomes regardless of MNP (data not shown).
Surprisingly, neither EDTA nor 6-DMAP had any obvious effect on the KICRs ability to recover MNP (Table 1). The results indicate that 100% (50/50) of KICRs recovered MNP when incubated in oocyte extracts containing overwhelming concentrations of EDTA (5–100 mM). Similarly, 100% of KICRs recovered MNP when incubated in oocyte extracts containing 6-DMAP at concentrations as high as 50 mM (Table 1). These results suggested that the recovery of MNP by the KICR is independent of both ATP and divalent cations. To extend this analysis further, high-speed oocyte extract was subjected to two passes through Sephadex G-25 spin columns to deplete the extract of small molecules, including nucleotides and divalent cations. Surprisingly, 100% of the KICRs incubated in the Sephadex G-25 eluent recovered MNP (Table 1). Based on these results, we conclude that the recruitment and binding to the KICR of those centrosomal components that are necessary for microtubule nucleation is independent of nucleotides, divalent cations, and microtubules.
Table 1.
Recovery of MNP is independent of divalent cations, kinase activity, and nucleotides
| Sample | CEs (SpiCen300) | Asters (tubulin) |
|---|---|---|
| Centrosomes | 50 | 50 |
| KICR | 50 | 0 |
| KICR (extract) | 50 | 50 |
| KICR (extract + 5 mM EDTA) | 50 | 50 |
| KICR (extract + 100 mM EDTA) | 50 | 50 |
| KICR (extract + 5 mM DMAP) | 50 | 50 |
| KICR (extract + 50 mM DMAP) | 50 | 50 |
| KICR (extract: G-25 Spin column) | 50 | 50 |
Coverslips containing centrosomes (CEs) or KICRs were treated with PEM buffer or treated high-speed extracts, washed, incubated with tubulin, fixed, and processed for immunofluorescence using antibodies to SpiCen300 and tubulin. Coverslips were then analyzed by counting 50 SpiCen300 staining CEs or KICRs and determining how many of these had corresponding asters, judged by the tubulin staining.
DISCUSSION
Although centrosomes were first identified over 100 years ago (33, 34), their molecular composition, and the mechanism(s) by which they assemble, replicate, and nucleate microtubules remain mysterious. However, the discovery that γ-tubulin (16, 17) is necessary for centrosome-dependent microtubule nucleation (18–20), and that it is a component of 25-nm-diameter ring structures (22) residing within the PCM of isolated centrosomes (8, 21, 22) provides an attractive mechanism for how centrosomes nucleate microtubules (23). When isolated from Xenopus egg cytoplasm, these γ-TuRCs are the same diameter as the 13-protofilament microtubules found in cells (23) and appear to be composed of γ-tubulin and approximately six additional proteins. They also accelerate the rate of microtubule polymerization in vitro (23). Taken together, this evidence suggests that γ-TuRCs present within the centrosome’s PCM serve as templates for the polymerization of microtubules (23). If this hypothesis is correct, then the presence and absence of γ-tubulin and 25-nm-diameter rings within the centrosome’s PCM should correlate with the gain and loss of centrosome MNP. In addition, several other lines of investigation would then become important, including: (i) how are γ-TuRCs recruited to the centrosome, (ii) how are these structures bound and retained by the PCM, and (iii) what is the molecular nature of the PCM itself?
At this point, γ-TuRCs are under intensive investigation. However, with few exceptions, such as pericentrin (35), centrin (36), centrosomin (37), and ninein (38), other components of the PCM remain to be defined biochemically. As a result, the mechanism(s) by which γ-TuRCs are recruited and bound to the centrosome’s pericentriolar region remain to be resolved. The lack of progress in this area is due primarily to the lack of procedures for isolating sufficient quantities of material for analyses (39). As we have noted here and elsewhere (8, 26), one of the strengths of the Spisula system is that it allows significant quantities of homogeneous centrosomes to be isolated (8), which then makes a biochemical approach to understanding the mechansims that control centrosome assembly and function feasible.
In this study, we have shown that when isolated Spisula centrosomes are treated with 1.0 M KI their ability to nucleate microtubules is removed, which is consistent with the results of others using isolated mammalian centrosomes (9, 11–14). Importantly, we also found that KI treatment removes ≈90% of the total centrosome protein, including γ-tubulin and the 25-nm-diameter rings, both of which have been implicated in centrosome-dependent microtubule nucleation (8, 16–23). Treatment with KI also removes the centrioles. Therefore, KI reduces centrosomes to nonfunctional units that are stripped of centrioles, γ-tubulin, 25-nm rings, and their ability to nucleate microtubules. However, the effects of KI on the centrosome’s MNP are reversible because, as we have shown, KICRs recover their MNP when incubated in oocyte extracts. Thus, the approach described here allows one to reversibly remove and then add back to the centrosome those molecules and structural elements responsible for centrosome-dependent microtubule nucleation.
KI extraction leaves approximately 10% of the centrosome’s protein intact as a sedimentable structural unit, the KICR. We found that this KICR is composed of a number of proteins and that it appears structurally as a meshwork of 12–15-nm thick fibers that we term here the “centromatrix”. From these observations, we propose that the centromatrix defines the PCM by serving as a scaffold to which those components required for microtubule nucleation must bind. Because the centromatrix recovers its MNP when incubated in oocyte extracts, and because there is no significant loss in the recovery of MNP when these extracts are diluted 20-fold, the centrosomal components required for microtubule nucleation must be relatively abundant in the oocyte cytoplasm and ready for binding without further modifications. Together, these conclusions suggest that the rate-limiting step for increasing the MNP of a centrosome during its maturation, which occurs at the onset of mitosis and when the sperm aster forms after fertilization, is the assembly of the centromatrix and not the assembly or modification of soluble subunits such as γ-TuRCs. In this model, the number of microtubules that a centrosome can nucleate is regulated by the controlled assembly of the filaments comprising the KICR, and the more centromatrix assembled within the centrosome, the greater the number of docking sites available for structures such as γ-TuRCs.
The centromatrix as defined in this study is distinct from the “novel lattice” recently proposed by Dictenberg et al. (40) that contains both pericentrin and γ-tubulin as major structural components. Clearly, the filamentous centromatrix (the KICR) described here does not contain γ-tubulin, but must contain docking sites for the incorporation of γ-tubulin containing microtubule nucleation sites. Thus, during centrosome assembly and maturation, the assembly of the pericentrin/γ-tubulin lattice described by Dictenberg et al. (40) must follow and may be dependent on the assembly of the centromatrix scaffold described here. As yet we do not know whether pericentrin is part of the Spisula centromatrix.
In our experiments, the recovery of MNP coincides with the reappearance of γ-tubulin and 25-nm rings within the PCM of recovered KICRs. However, under these conditions the centriole was not reassembled within the KICR. We also found that the recovery of a KICR’s MNP occurs in the presence of 20 μM colchicine (an anti-microtubule drug), and thus in the absence of microtubules. Therefore, recruitment of those cytoplasmic subunits required for microtubule nucleation to the centromatrix is independent of microtubules and microtubule-dependent motor proteins. These results are consistent with the conclusions from studies on the mechanisms by which sperm asters assemble in Xenopus lysates (19, 20). However, the recovery of MNP by Spisula KICRs, and thus the binding of those components required for reinstating MNP to these remnants, is not inhibited by removing divalent cations from the extract, or by inhibiting extract kinase activity with 6-DMAP. This conclusion is further supported by our observation that passing oocyte extract through a Sephadex G-25 spin column, which removes all small molecules including nucleotides and divalent cations, had no effect on the recovery of the KICR’s MNP.
In general, our results are consistent with the report that urea-inactivated mammalian centrosomes recover their MNP when treated with Xenopus egg extracts (13). In those experiments, neither apyrase nor 6-DMAP inhibited centrosome recovery. However, our data appear to be in conflict with those reports implying that sperm aster assembly in Xenopus lysates requires ATP (19–20). A reasonable explanation for this apparent discrepancy in nucleotide requirements is that during fertilization the sperm brings naked centrioles into the egg that must first recruit and assemble a centromatrix (the KICR) around them before gaining the ability to nucleate microtubules, and that the assembly of this centromatrix is an energy dependent and rate-limiting step for centrosome maturation. The central theme of this hypothesis is that the assembly of the centromatrix is a necessary prerequisite for the recruitment of microtubule nucleation sites onto the centrosome, and that it is required for both centrosome assembly and increasing the centrosome’s MNP during centrosome maturation.
Acknowledgments
We acknowledge C. Lyddane and A. W. Suddith for their assistance in preparing the polyclonal anti-γ-tubulin antibodies. This work was supported by National Institutes of Health/General Medical Sciences 43264 to R.E.P., National Institutes of Health/General Medical Sciences 40198, and National Institutes of Health National Center For Research Resources P41 RR01219 that supports the Wadsworth Center Biological Microscopy and Image Reconstruction Resource facility as a National Biotechnological Resource.
ABBREVIATIONS
- MNP
microtubule organizing potential
- 6-DMAP
6-dimethylaminopurine
- KICR
KI insoluble centrosome remnant
- γ-TuRC
γ-tubulin ring complex
- EM
electron microscopy
- IVEM
intermediate voltage EM
- PCM
pericentriolar material
References
- 1.Kellogg D R, Moritz M, Alberts B M. Annu Rev Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- 2.Lee C M, Ferguson M, Chen L B. J Cell Biol. 1989;109:2045–2055. doi: 10.1083/jcb.109.5.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soltys B J, Gupta R S. Biochem Cell Biol. 1992;70:1174–1186. doi: 10.1139/o92-163. [DOI] [PubMed] [Google Scholar]
- 4.Schroer T A, Sheetz M P. Annu Rev Physiol. 1991;53:629–652. doi: 10.1146/annurev.ph.53.030191.003213. [DOI] [PubMed] [Google Scholar]
- 5.Sakai H. Cell Struct Funct. 1994;19:57–62. doi: 10.1247/csf.19.57. [DOI] [PubMed] [Google Scholar]
- 6.Allen R D. Biol Bull. 1953;105:213–239. [Google Scholar]
- 7.Schatten G. Dev Biol. 1994;165:299–335. doi: 10.1006/dbio.1994.1256. [DOI] [PubMed] [Google Scholar]
- 8.Vogel J M, Stearns T, Rieder C L, Palazzo R E. J Cell Biol. 1997;137:193–202. doi: 10.1083/jcb.137.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchison T, Kirschner M. Nature (London) 1984;312:232–237. doi: 10.1038/312232a0. [DOI] [PubMed] [Google Scholar]
- 10.Mitchison T, Kirschner M. Methods Enzymol. 1986;134:261–269. doi: 10.1016/0076-6879(86)34094-1. [DOI] [PubMed] [Google Scholar]
- 11.Kuriyama R. J Cell Sci. 1984;66:277–295. doi: 10.1242/jcs.66.1.277. [DOI] [PubMed] [Google Scholar]
- 12.Klotz C, Dabauvalle M-C, Paintrand M, Weber T, Bornens M, Karsenti E. J Cell Biol. 1990;110:405–415. doi: 10.1083/jcb.110.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buendia B, Draetta G, Karsenti E. J Cell Biol. 1992;116:1431–1442. doi: 10.1083/jcb.116.6.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohta K, Shiina N, Okumura E, Hisanaga S-I, Kishimoto T, Endo S, Gotoh Y, Nishida E, Sakai H. J Cell Sci. 1993;104:125–137. doi: 10.1242/jcs.104.1.125. [DOI] [PubMed] [Google Scholar]
- 15.Gould R R, Borisy G G. J Cell Biol. 1977;73:601–615. doi: 10.1083/jcb.73.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oakley C E, Oakley B R. Nature (London) 1989;338:662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- 17.Oakley B R, Oakley C E, Yoon Y, Jung M K. Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- 18.Joshi H C, Palacios M J, McNamara L, Cleveland D W. Nature (London) 1992;356:80–83. doi: 10.1038/356080a0. [DOI] [PubMed] [Google Scholar]
- 19.Felix M-A, Antony C, Wright M, Maro B. J Cell Biol. 1994;124:19–31. doi: 10.1083/jcb.124.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stearns T, Kirschner M W. Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- 21.Moritz M, Braunfeld J, Fung J C, Sedat J W, Alberts B M, Agard D A. J Cell Biol. 1995;130:1149–1159. doi: 10.1083/jcb.130.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moritz M, Braunfeld J, Sedat J W, Alberts B M, Agard D A. Nature (London) 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- 23.Zheng Y, Wong M L, Alberts B M, Mitchison T. Nature (London) 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- 24.Oakley B R. Nature (London) 1995;7:555–556. doi: 10.1038/378555a0. [DOI] [PubMed] [Google Scholar]
- 25.Palazzo R E, Brawley J B, Rebhun L I. Zool Sci (Tokyo) 1988;5:603–611. [Google Scholar]
- 26.Palazzo R E, Vogel J M. In: Mitosis and Meiosis, Methods in Cell Biology. Rieder C L, editor. Vol. 61. San Diego: Academic; 1998. , in press. [Google Scholar]
- 27.Palazzo R E, Vaisberg E, Cole R W, Rieder C L. Science. 1992;256:219–221. doi: 10.1126/science.1566068. [DOI] [PubMed] [Google Scholar]
- 28.Suprenant K A, Marsh J C. J Cell Sci. 1987;87:71–84. doi: 10.1242/jcs.87.1.71. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U K. Nature (London) 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Neuhoff V, Arold N, Taube D, Ehrhardt W. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 31.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Penczek P, Marko M, Buttle K, Frank J. Ultramicroscopy. 1995;60:393–410. doi: 10.1016/0304-3991(95)00078-x. [DOI] [PubMed] [Google Scholar]
- 33.Stearns T, Winey M. Cell. 1997;91:303–309. doi: 10.1016/s0092-8674(00)80414-6. [DOI] [PubMed] [Google Scholar]
- 34.Wilson E B. The Cell in Development and Heredity. New York: Macmillian; 1925. [Google Scholar]
- 35.Doxsey S J, Stein P, Evans L, Calarco P D, Kirschner M. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- 36.Baron A, Salisbury J. J Cell Biol. 1988;107:2669–2678. doi: 10.1083/jcb.107.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li K, Kaufman T C. Cell. 1996;85:585–596. doi: 10.1016/s0092-8674(00)81258-1. [DOI] [PubMed] [Google Scholar]
- 38.Bouckson-Castaing V, Moudjou M, Ferguson D J P, Mucklow S, Belkaid Y, Milon G, Crocker P R. J Cell Sci. 1996;109:179–190. doi: 10.1242/jcs.109.1.179. [DOI] [PubMed] [Google Scholar]
- 39.Brinkley B R. Annu Rev Cell Biol. 1985;1:145–172. doi: 10.1146/annurev.cb.01.110185.001045. [DOI] [PubMed] [Google Scholar]
- 40.Dictenberg J B, Zimmerman W, Sparks C A, Young A, Vidair C, Zheng Y, Carrington W, Fay F S, Doxsey S J. J Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]