Abstract
Members of the polo subfamily of protein kinases play pivotal roles in cell proliferation. In addition to the kinase domain, polo kinases have a strikingly conserved sequence in the noncatalytic domain, termed the polo-box. The function of the polo-box is currently undefined. The mammalian polo-like kinase Plk is a functional homologue of Saccharomyces cerevisiae Cdc5. Here, we show that Plk localizes at the spindle poles and cytokinetic neck filaments. Without impairing kinase activity, a conservative mutation in the polo-box disrupts the capacity of Plk to complement the defect associated with a cdc5–1 temperature-sensitive mutation and to localize to these subcellular structures. Our data provide evidence that the polo-box plays a critical role in Plk function, likely by directing its subcellular localization.
The polo subfamily of protein kinases has been identified in various eukaryotic organisms and plays pivotal roles in cell division and proliferation. A feature of the polo subfamily members is the presence of a distinct region of homology in the C-terminal noncatalytic domain, termed the polo-box (1) (Fig. 1 A and B). Members of this subfamily include mammalian Plk (1–5), Snk (6), Fnk/Prk (7, 8), Xenopus laevis Plx1 (9), Drosophila melanogaster polo (10), Schizosaccharomyces pombe Plo1 (11), and Saccharomyces cerevisiae Cdc5 (12).
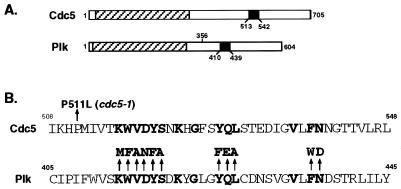
Figure 1.
(A) Structures of Cdc5 and Plk. A hatched box denotes the kinase domain, and a closed box denotes the polo-box. Plk lacking the C-terminal domain (PlkΔC) has lost amino acid residues 356 to 604. (B) Identification of the cdc5–1 mutation site and diagram showing the polo-box mutants generated in Plk. Yeast genomic DNAs prepared from the cdc5–1 mutant (H5C1A1) and its parental wild-type strain (H4939–1b) (a gift of L. Hartwell, University of Washington) were used as templates to amplify full length cdc5–1 and CDC5 genes, respectively, by using the PCR. Restriction and complementation analyses confirmed the cloned genes (data not shown). DNA sequence analysis revealed a point mutation (conversion of Pro511 to Leu) in the cdc5–1 allele. Introduction of the P511L mutation into the wild-type Cdc5 was sufficient to abolish its capacity to complement the cdc5–1 defect (data not shown). Conserved amino acids among all of the polo family members known to date are in bold letters; arrows point to amino acids changed in the point mutations.
The M phase of the cell cycle is a highly regulated process that requires a series of coordinated biochemical and cellular events to ensure the faithful partitioning of genetic and cytoplasmic components. It is apparent from genetic and biochemical analyses that many polo kinases regulate diverse cellular events at various stages of M phase (for reviews, see refs. 13 and 14), such as centrosome maturation (15) and bipolar spindle formation (10, 11, 15). Polo kinases also appear to regulate important biochemical steps in G2/M phase, such as activation of Cdc2 through Cdc25C phosphatase (9), DNA damage checkpoint adaptation (16), and regulation of the anaphase-promoting complex (17, 18).
One additional function attributed to polo kinases is the induction of cytokinesis-associated septal structures (11, 19). In an otherwise wild-type yeast genetic background, ectopic expression of a mutationally activated form of Plk induces a class of cells with unusually elongated buds that develop multiple septal structures (19). In addition, cells depleted of Cdc5 protein fail to complete cytokinesis and have a dumbbell-shaped terminal morphology (12). In fission yeast, loss of plo1+ function leads to mitotic arrest, with failure of both formation of the F-actin ring and deposition of septal components, whereas overexpression of Plo1 induces multiple septa at any phase of the cell cycle (11), suggesting that Plo1 is both essential and sufficient for septum and actin ring formation. In animal cells, current models of cytokinesis emphasize the potential of either the centrosomes or the midzone to provide a signal for cleavage furrow induction at the equatorial cortex (20–24). Studies (25, 26) have shown that Plk localizes at both the centrosomes and the midzone/midbody during the late stage of the mammalian cell cycle. Thus, the subcellular localization of Plk supports its proposed role in cytokinesis (27).
Although it is apparent that polo kinases play multiple roles during M-phase progression and cytokinesis, the role of the polo-box, a strikingly conserved amino acid sequence present in all the members of polo kinase subfamily, is not yet known. We reported that murine Plk is a functional homolog of S. cerevisiae Cdc5 (19). In this communication, we used Plk expression in yeast to provide evidence that the polo-box is required for the capacity of Plk to complement functionally the cdc5–1 defect. The data reported here suggest that the polo-box serves to target the catalytic activity of Plk to the spindle poles and cytokinetic neck filaments.
MATERIALS AND METHODS
Strains, Growth Conditions, and Transformations.
Yeast strains used in this study are 1788 [isogenic diploid of EG123, MATα leu2–3, 112 ura3–52 trp1–1 his4 can1r (28) and KKY921–2B (MATa cdc5–1 leu2 trp1 ura1) (12)]. Yeast cells were cultured in yeast extract/peptone (1% yeast extract/2% Bacto-peptone) supplemented with 2% glucose, 2% raffinose (Sigma), or 2% galactose (J.T. Baker) as required. Synthetic minimal medium (29) supplemented with the appropriate nutrients was employed to select for plasmid maintenance. Yeast transformation was carried out by the lithium acetate method (30).
Generation of Plk Mutants and Enhanced Green Fluorescent Protein (EGFP)-Plk Fusion Constructs.
Site-directed mutagenesis in murine PLK cDNA was carried out by using the Sculptor in vitro mutagenesis system (Amersham). A 1.4-kb SacI fragment isolated from each partially digested mutant clone was ligated into YCplac111-GAL1-HA-PLK digested with SacI (19). All of the mutant clones were sequenced to confirm the mutations introduced into the PLK coding sequence. YCplac111-GAL1-HA-PLK and its C-terminal deletion mutant were constructed as described (19). The C-terminal deletion mutant lacks amino acid residues 356 to 604. YCplac111-GAL1-EGFP-PLK fusion constructs were generated by inserting a 700-bp XhoI fragment of the EGFP coding sequence into the XhoI site present at the N-terminal PLK coding sequence. The EGFP coding sequence was amplified by PCR by using the pEGFP-N1 plasmid (CLONTECH) as a template.
Expression of Plk in S. cerevisiae.
To express wild-type and mutant forms of Plk in S. cerevisiae under the control of the GAL1 promoter, yeast transformants harboring each construct were grown in yeast extract/peptone/raffinose to an OD600 of 0.8 at 30°C. Cultures then were washed twice with water and were resuspended in yeast extract/peptone/galactose at an OD600 of 0.05 and were cultured continuously. All the cdc5–1 transformants were cultured at 23°C. To examine the complementation of the cdc5–1 defect by Plk expression, cells were cultured at 37°C, however.
Flow Cytometry Analyses.
Flow cytometry analyses were carried out as described (19). In brief, cells were harvested and were fixed with 70% ethanol, then were treated with RNase A (1 mg/ml) in PBS for 30 min at 37°C. After disrupting the cells for 1 min with a sonicator (model W-375, Heat Systems/Ultrasonics), cells were stained with propidium iodide (50 μg/ml) in PBS. Flow cytometry analyses were performed with a cellquest program (Becton Dickinson).
Immunoprecipitation, Kinase Assays, and Western Analyses.
Yeast cells were lysed with an equal volume of glass beads (Sigma) as described (19). The obtained lysates were spun at 2,000 × g for 2 min to remove unbroken cells and beads. The resulting supernatants were considered as total cellular lysates. For immunoprecipitation, total cellular lysates were subjected to further centrifugation at 15,000 × g for 30 min to clarify heavy cellular materials. Before incubation with affinity-purified anti-Plk antibody, the resulting supernatants (S15) were diluted to 1 ml with TBSN buffer (20 mM Tris⋅Cl, pH8.0/150 mM NaCl/0.5% Nonidet P-40/5 mM EGTA/1.5 mM EDTA/0.5 mM Na3VO4/20 mM p-nitrophenyl phosphate) supplemented with protease inhibitors. Protein A-Sepharose 4B (Zymed) was added and incubated for an additional 1 hr to precipitate the antibody.
Kinase assays and Western analyses were carried out as described (19). In brief, the kinase activity of Plk was measured in a kinase mixture (50 mM Tris⋅Cl, pH 7.5/10 mM MgCl2/5 mM DTT/2 mM EGTA/0.5 mM Na3VO4/20 mM p-nitrophenyl phosphate) supplemented with 3 μg of dephosphorylated casein (Sigma) and 25 μM ATP (5 μCi of [γ-32P]ATP; 1 Ci = 37 Gbq). Western analyses were carried out with affinity-purified Plk antibody or HA antibody at a concentration of 0.5 μg/ml. Proteins that interact with antibodies were detected by the enhanced chemiluminescence (ECL) Western detection system (Amersham).
Cell Staining and Immunofluorescence Microscopy.
Indirect immunofluorescence was performed as described (19). Cdc10 was localized by using affinity-purified rabbit polyclonal anti-Cdc10 antibody (a gift of J. Chant, Harvard University, Cambridge, MA) and rhodamine-conjugated goat anti-rabbit IgG (Zymed). Microtubules were visualized by using YOL1/34 rat anti-tubulin antibody (Accurate Chemical and Scientific Corp., Westbury, NY) and goat anti-rat CY3 antibody (The Jackson Laboratory). DNA was visualized by using propidium iodide staining at 40 μg/ml. Stained cells were viewed under a Zeiss LSM410 confocal microscope equipped with a Krypton/Argon laser.
RESULTS
Requirement of both the Polo-Box and the Kinase Activity of Plk for Functional Complementation of the cdc5–1 Defect.
It has been reported (19) that Plk is a functional homolog of S. cerevisiae Cdc5. At the restrictive temperature, cells with a cdc5–1 mutation arrest late in mitosis as large budded cells (12, 16), with an accumulation of cells with 2N (G2/M) DNA content and a greatly diminished 1N (G1) population, as reflected in flow cytometry (Fig. 2A). We confirmed our previous observation that expression of wild-type Plk, but not the kinase-inactive PlkK82M, complements the cell division cycle defect of the cdc5–1 mutants, regenerating the 1N DNA-containing population to a similar extent as CDC5 expression (19) (Fig. 2 B–D).
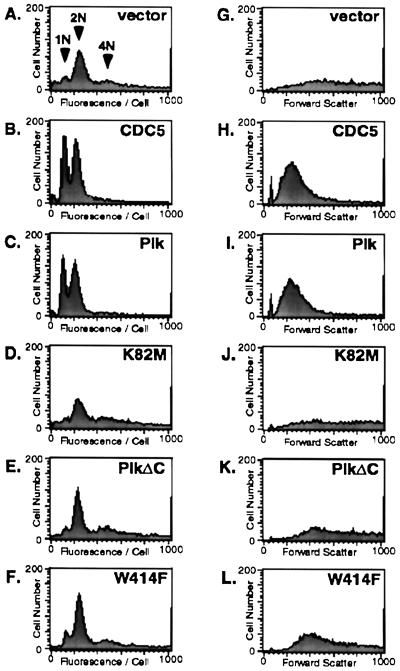
Figure 2.
Mutations in either the ATP-binding site or the polo-box abolish the capacity of Plk to complement the cdc5–1 defect. A haploid cdc5–1 mutant strain, KKY921–2B (MATa cdc5–1 leu2 trp1 ura1) (12) was transformed with various YCplac111-GAL1-HA-PLK constructs or with YCplac111-CDC5. To examine the ability of Plk constructs to complement the cdc5–1 defect, transformants were cultured at 37°C in yeast extract/peptone + 2% galactose medium for 10 hr as described (19) and were subjected to flow cytometry analyses. Vector, YCplac111-GAL1; CDC5, YCplac111-CDC5; PlK, YCplac111-GAL1-HA-Plk; K82M, YCplac111-GAL1-HA-PlkK82M; PlkΔC, YCplac111-GAL1-HA-PlkΔC; W414F, YCplac111-GAL1-HA-PlkW414F. (A–F) A G1 population (1N arrow) and G2/M population (2N arrow) are indicated in the vector panel. Cells with a DNA content >2N are indicated by the 4N arrow (a broad cell population with the third 4N arrow). (G–L) The increase in forward scatter at the x axis reflects an increase in cell size. The broad, spread out pattern observed in the cdc5–1 mutant transformed with vector (G) results from a heterogeneous population of enlarged cells whereas wild-type cells (data not shown) and the cdc5–1 mutant complemented with Cdc5 (H) or Plk (I) expression produce a distinct bell-shaped pattern of forward scatter.
To extend our previous analyses and investigate whether the polo-box is required for the ability of Plk to complement the cdc5–1 defect, we created a Plk mutant lacking the C-terminal domain (PlkΔC), which includes the polo-box (amino acids 410–439 in Plk) (Fig. 1A). PlkΔC does not complement the cdc5–1 defect (Fig. 2E), despite a 3- to 4-fold increase in specific kinase activity (19). These results are consistent with the abilities of these Plk constructs to restore the cdc5–1 cell growth defect at the restrictive temperature when cultured on galactose-containing medium (data not shown) (19).
To examine specifically whether Plk requires its polo-box to complement the cdc5–1 defect, we generated 11 single point mutations in the polo-box (Fig. 1B). The ability of these mutants to complement the cdc5–1 defect was analyzed by flow cytometry. The most dramatic effect was seen with a W414F mutation, which essentially eliminated the capacity of Plk to complement the cdc5–1 defect (Fig. 2F). Mutations at V415A, L427A, and N437D also significantly reduced the capacity of Plk to complement this defect (data not shown). These results indicate that the polo-box is critical for the ability of Plk to complement the cdc5–1 defect. Consistent with the importance of the polo-box domain was our finding that the cdc5–1 allele possesses a single point mutation at the upstream boundary of the polo-box (Fig. 1B) that may alter the structural integrity of this region.
Microscopic observation revealed that the majority of cdc5–1 mutant cells at the restrictive temperature were enlarged greatly, with heterogeneous cell sizes and shapes (data not shown); in forward scatter, this phenotype results in a broad, spread out pattern (Fig. 2G). As with expression of Cdc5 (Fig. 2H), wild-type Plk expression, but not that of the kinase-inactive K82M mutant, restored the heterogeneously enlarged cell population to a uniform wild-type morphology (Fig. 2 I and J). Neither W414F nor PlkΔC expression was able to restore wild-type cell morphology (Fig. 2 K and L), consistent with its inability either to rescue growth at the restrictive temperature (data not shown) or to relieve the cdc5–1 cell cycle arrest. Taken together, the above data demonstrate the necessity of both the kinase activity and the polo-box for the ability of Plk to complement the cdc5–1 defect.
The W414F Mutation Does Not Influence the Expression Level or the Kinase Activity of Plk.
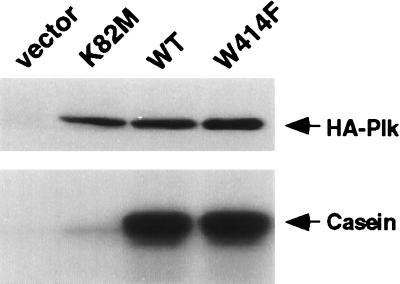
To eliminate the possibility that the inability of PlkW414F to complement the cdc5–1 defect is caused by a decrease in stability or kinase activity of W414F, we examined the expression level and the enzymatic activity of PlkW414F. Immune complex kinase assays revealed that both wild-type Plk and the PlkW414F mutant exhibit similar levels of specific activities, whereas PlkK82M does not possess a significant amount of activity (Fig. 3). In total cellular lysates, the expression levels of the wild-type and mutant forms of Plk were similar (data not shown). These results indicate that the inability of PlkW414F to complement the cdc5–1 defect is likely caused by the disrupted function of the polo-box by the introduction of the W414F mutation.
Figure 3.
Mutations in the polo-box do not impair Plk kinase activity in vitro. The cdc5–1 (KKY921–2B) cells bearing various YCplac111-GAL1-HA-PLK constructs were cultured under inducing conditions for 10 hr and were harvested. The lysates were centrifuged at 15,000 × g for 30 min to clarify heavy cellular materials. From 500 μg of cellular proteins present in the S15 fraction, the wild-type and mutant forms of HA-Plk were immunoprecipitated and subjected to in vitro kinase assays. (Upper) HA-tagged Plk was immunoprecipitated with affinity-purified anti-Plk antibody, was electrophoresed, and was detected on membranes by Western analysis with anti-HA antibody. (Lower) HA-tagged Plk was immunoprecipitated with anti-Plk antibody, and in vitro kinase assays were performed as described by using casein as a substrate (19). Vector, YCplac111-GAL1; K82M, YCplac111-GAL1-HA-PlkK82M; WT, YCplac111-GAL1-HA-Plk; W414F, YCplac111-GAL1-HA-PlkW414F.
The W414F Mutation Disrupts EGFP–Plk Localization at Spindle Poles and Septin Ring Structures.
The inability of the W414F mutant to complement the cdc5–1 defect led us to speculate that the polo-box may serve either to target Plk to particular subcellular locations or to mediate the interaction of Plk with essential substrates or activators. We examined the cellular localization of Plk that is fused to an EGFP and expressed under the GAL1 promoter. At the restrictive temperature, EGFP-fused wild-type Plk (EGFP–Plk) and PlkT210D (EGFP–PlkT210D) complemented the cdc5–1 defect to a similar degree as their parental unfused forms (data not shown). As expected, the introduction of the W414F mutation essentially eliminated the capacity of EGFP–Plk to complement the cdc5–1 defect, and the C-terminal deletion abolished complementation completely (data not shown). These results show that the EGFP-fused Plk proteins behave in the same manner as their nonfusion counterparts.
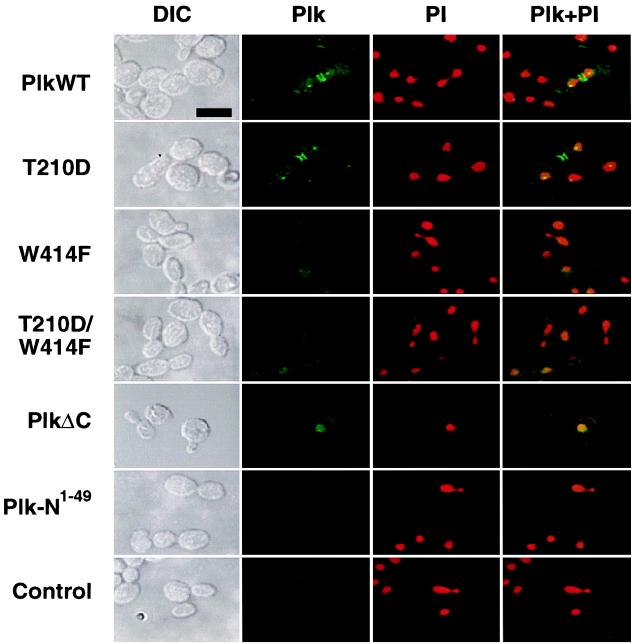
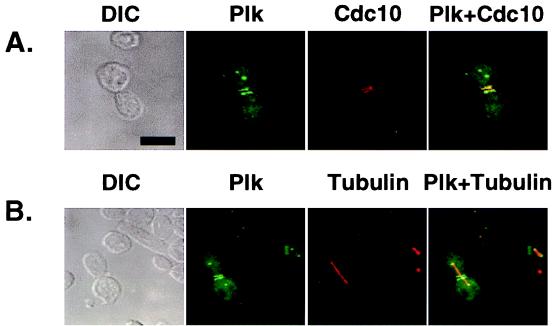
Ectopic expression of both wild-type EGFP–Plk and EGFP–PlkT210D in yeast yielded two distinct fluorescent dots in the cytoplasm as well as either one or two bright bands at the bud neck in budded cells (Fig. 4). To investigate whether the signals correspond to spindle poles and septin ring structures, respectively, we carried out subsequent immunostaining with antibodies against either tubulin or Cdc10, one of four septin components (Cdc3, Cdc10, Cdc11, and Cdc12) of the cytokinetic neck filaments in budding yeast (for review, see ref. 31). Our data revealed that the two dots of staining represented the spindle poles whereas the bands corresponded to the cytokinesis-associated septin ring structures (see Fig. 5 A and B). In contrast, the localization of Plk to these subcellular structures was either greatly diminished or absent in cells expressing EGFP–PlkW414F or the EGFP–PlkT210D/W414F double mutant. A Plk construct lacking the C-terminal domain (EGFP–PlkΔC) was diffusely localized to the cell periphery and chromatin whereas a construct lacking both the kinase and the C-terminal domains (EGFP-Plk-N1–49) yielded a diffuse signal (Fig. 4). These data show that the polo-box directs Plk localization to the spindle poles and the bud-neck filaments and that tryptophan 414 appears to play a critical role in targeting Plk to these structures. These observations indicate that the inability of the W414F mutant to complement the cdc5–1 defect is attributable to the loss of its specific cellular localization.
Figure 4.
The requirement of the polo-box for Plk localization. To localize wild-type and mutant forms of Plk in a diploid wild-type strain, 1788, EGFP–Plk fusion constructs were generated and expressed under the control of the GAL1 promoter. Transformants expressing EGFP fusion constructs were stained with propidium iodide to visualize chromosomal DNA and were examined by confocal microscopy. PlkWT, YCplac111-GAL1-HA-EGFP-Plk; T210D, YCplac111-GAL1-HA-EGFP-PlkT210D; W414F, YCplac111-GAL1-HA-EGFP-PlkW414F; T210D/W414F, YCplac111-GAL1-HA-EGFP-PlkT210D/W414F; PlkΔC, YCplac111-GAL1-HA-EGFP-PlkΔC; Plk-N1–49, YCplac111-GAL1-HA-EGFP-Plk-N1–49; control, an irrelevant plasmid without EGFP. Plk-N1–49 contains only the N-terminal 49-aa residues of Plk fused to EGFP and serves as a background EGFP signal. DIC, differential interference contrast; Plk, EGFP–Plk expression; PI, propidium iodide staining of nuclei; Plk + PI, EGFP–Plk and propidium iodide images superimposed. (Bar = 5 μm.)
Figure 5.
Ectopically expressed Plk localizes at the spindle poles and bud neck filaments. EGFP–Plk fusion constructs were expressed under the control of the GAL1 promoter in a diploid wild-type strain, 1788. To enhance the signals present at the spindle poles and the cytokinetic septal structures, two tandem EGFPs were inserted into the N terminus of the Plk coding sequence. Transformants were cultured for subsequent immunostainings to examine Cdc10 and tubulin localizations. (A) Plk (green) and Cdc10 (red) localize at the neck filaments. Septin rings (red) are viewed edge on and therefore appear as lines. (B) Plk (green) localizes at the spindle poles. Spindles are visualized by microtubule staining (red). The spindles appear to emanate from the structures with which Plk associates. DIC, differential interference contrast; Plk, EGFP–Plk expression; Cdc10, Cdc10 staining; Tubulin, tubulin staining. Superimposed images are shown as Plk + Cdc10 and Plk + Tubulin. (Bar = 5 μm.)
DISCUSSION
The role of the polo-box, a strikingly conserved amino acid sequence present in the C-terminal noncatalytic domain of polo kinases, is not known. To shed light on the function of the polo-box, we used Plk expression in budding yeast. Either a conserved mutation in the polo-box, W414F, or a mutation inactivating the kinase activity of Plk, K82M, abolishes the capacity of Plk to functionally complement the cdc5–1 defect (Fig. 2). These data indicate that both the polo-box and the kinase activity are required for the mitotic functions of Plk. The importance of the polo-box is further supported by our recent observation that the introduction of the W414F mutation also completely abolished the capacity of Plk to induce abnormally elongated buds with ectopic septal structures (K.S.L., T.Z.G., and R.L.E., unpublished data). We have demonstrated that the polo-box is required for the localization of Plk to spindle poles and cytokinetic neck filaments (Figs. 4 and 5). Thus, the polo-box appears to function as an interaction domain to target the catalytic activity of the enzyme to specific subcellular locations, thereby allowing efficient interaction with its physiological substrates or activators.
We show that a single conserved amino acid change of tryptophan to phenylalanine at position 414 in the polo-box disrupts Plk localization at both the spindle poles and cytokinetic structures to a similar degree (Fig. 4). This observation suggests that the polo-box domain of Plk specifically recognizes a single binding protein or perhaps different binding partners with similar binding motifs at these distinct subcellular locations. Functional complementation assays for the cdc5–1 defect suggest that, among the examined amino acid residues conserved in the polo-box, W414 is the most critical amino acid residue for the function of Plk. However, additional amino acid residues (V415, L427, and N437) that are present in other conserved segments of the polo-box also appear to be important for Plk function (Fig. 2 and data not shown). Indeed, the critical interactions may extend well beyond the short module commonly referred to as the polo-box. In this regard, it should be noted that these data demonstrate that the polo-box is necessary for specific localization but do not demonstrate that it is sufficient. It is now of importance to determine the sequences that are sufficient to direct localization to the spindle poles and the bud neck.
Recent studies suggest that polo kinases have multiple roles during G2/M phase, such as activation of Cdc2 through Cdc25C phosphatase (9), maturation of mitotic centrosomes (15), DNA damage checkpoint adaptation (16), and regulation of the anaphase-promoting complex (17, 18, 32). It is not known whether all of the mitotic functions of Plk occur at the spindle poles and cytokinetic structures, therefore requiring Plk localization to these sites. Alternatively, it is possible that the polo-box interacts with other subcellular structures as well, and the strong EGFP–Plk signals at the spindle poles and cytokinetic neck filaments may simply reflect the enriched concentration of certain binding partners at these sites. Therefore, it will be interesting to investigate whether the polo-box is required for all of the proposed mitotic functions of polo kinases or for a subset of these events.
We have reported here that, in budding yeast, ectopically expressed EGFP–Plk localizes at the spindle poles and at the cytokinetic neck filaments. Consistent with our data, recent indirect immunofluorescence studies revealed that endogenous Cdc5 localizes at spindle poles (17). However, these studies did not detect Cdc5 at the bud neck filaments. Detection of Plk localization at the neck filaments could be attributable to the two different detection methods employed or to an additional property that Plk may have. Further experiments will be required to resolve this issue.
In mammalian cells, Plk localizes at the centrosomes and the midzone/midbody (2, 25), both of which have been implicated in directing the site of cytokinesis (20–24). Many eukaryotic cells rely on a mechanism in which the mitotic apparatus dictates the position of the cleavage plane during anaphase/telophase whereas in budding yeast, a future cleavage plane of septin ring structures is already present in late G1 in the absence of identifiable spindle structures. Although the morphological features and timing of certain events of cytokinesis in budding yeast and mammalian cells are strikingly different, it is likely that the molecular processes used for inducing cytokinesis have remained largely unchanged throughout evolution. Thus, the differences observed in Plk localization at the midzone/midbody as opposed to the cytokinetic neck filaments may reflect the difference in the temporal and spatial arrangement of mitotic events by which cytokinetic sites are specified in these evolutionarily distinct eukaryotic cells. In both systems, however, Plk localizes to the structures that are important for cytokinesis.
In this communication, we have demonstrated that complementation of the cdc5–1 defect in S. cerevisiae by mammalian Plk requires both the protein kinase activity and the polo-box domain and that the polo-box plays a crucial role to target the catalytic activity of Plk to the spindle poles and cytokinetic neck filaments. Further experiments are necessary to identify polo-box-interacting proteins and additional Plk substrates and to determine their roles in regulating cell division and cytokinesis.
Acknowledgments
We are grateful to J. Chant, D. P. Kiehart, R. Kuriyama, and B. Brott for helpful discussions and critical reading of this manuscript. We thank Laurie Scott for preparing the manuscript, J. Loconto for technical assistance, J. Chant for the provision of anti-Cdc10 antibody and anti-tubulin antibody, and L. Hartwell for yeast strains. This work is supported by a Postdoctoral/Postresidency Fellowship from the American Cancer Society, Massachusetts Division, Inc. (K.S.L.) and National Institutes of Health Grant CA42580 (R.L.E.). R.L.E. is the John F. Drum American Cancer Society Research Professor.
ABBREVIATION
- EGFP
enhanced green fluorescent protein
References
- 1.Clay F J, McEwen S J, Bertoncello I, Wilks A F, Dunn A R. Proc Natl Acad Sci USA. 1993;90:4882–4886. doi: 10.1073/pnas.90.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golsteyn R M, Schultz S J, Bartek J, Ziemiecki A, Ried T, Nigg E A. J Cell Sci. 1994;107:1509–1517. doi: 10.1242/jcs.107.6.1509. [DOI] [PubMed] [Google Scholar]
- 3.Hamanaka R, Maloid S, Smith M R, O’Connell C D, Longo D L, Ferris D K. Cell Growth Differ. 1994;5:249–257. [PubMed] [Google Scholar]
- 4.Holtrich U, Wolf G, Bräuninger A, Karn T, Böhme B, Rübsamen-Waigmann H, Strebhardt K. Proc Natl Acad Sci USA. 1994;91:1736–1740. doi: 10.1073/pnas.91.5.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lake R J, Jelinek W R. Mol Cell Biol. 1993;13:7793–7801. doi: 10.1128/mcb.13.12.7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simmons D L, Neel B G, Stevens R, Evett G, Erikson R L. Mol Cell Biol. 1992;12:4164–4169. doi: 10.1128/mcb.12.9.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donohue P J, Alberts G F, Guo Y, Winkles J A. J Biol Chem. 1995;270:10351–10357. doi: 10.1074/jbc.270.17.10351. [DOI] [PubMed] [Google Scholar]
- 8.Li B, Ouyang B, Pan H, Reissmann P T, Slamon D J, Arceci R, Lu L, Dai W. J Biol Chem. 1996;271:19402–19408. doi: 10.1074/jbc.271.32.19402. [DOI] [PubMed] [Google Scholar]
- 9.Kumagai A, Dunphy W G. Science. 1996;273:1377–1380. doi: 10.1126/science.273.5280.1377. [DOI] [PubMed] [Google Scholar]
- 10.Llamazares S, Moreira A, Tavares A, Girdham C, Spruce B A, Gonzalez C, Karess R E, Glover D M, Sunkel C E. Genes Dev. 1991;5:2153–2165. doi: 10.1101/gad.5.12a.2153. [DOI] [PubMed] [Google Scholar]
- 11.Ohkura H, Hagan I M, Glover D M. Genes Dev. 1995;9:1059–1073. doi: 10.1101/gad.9.9.1059. [DOI] [PubMed] [Google Scholar]
- 12.Kitada K, Johnson A L, Johnston L H, Sugino A. Mol Cell Biol. 1993;13:4445–4457. doi: 10.1128/mcb.13.7.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glover D M, Ohkura H, Tavares A. J Cell Biol. 1996;135:1681–1684. doi: 10.1083/jcb.135.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane H, Nigg E A. Trends Cell Biol. 1997;7:63–68. doi: 10.1016/S0962-8924(96)10051-9. [DOI] [PubMed] [Google Scholar]
- 15.Lane H A, Nigg E A. J Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toczyski D P, Galgoczy D J, Hartwell L H. Cell. 1997;90:1097–1106. doi: 10.1016/s0092-8674(00)80375-x. [DOI] [PubMed] [Google Scholar]
- 17.Shirayama M, Zachariae W, Ciosk R, Nasmyth K. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Descombes P, Nigg E A. EMBO J. 1998;17:1328–1335. doi: 10.1093/emboj/17.5.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee K S, Erikson R L. Mol Cell Biol. 1997;17:3408–3417. doi: 10.1128/mcb.17.6.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devore J J, Conrad G W, Rappaport R. J Cell Biol. 1989;109:2225–2232. doi: 10.1083/jcb.109.5.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oegema K, Mitchison T J. Proc Natl Acad Sci USA. 1997;94:4817–4820. doi: 10.1073/pnas.94.10.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satterwhite L L, Pollard T D. Curr Opin Cell Biol. 1992;4:43–52. doi: 10.1016/0955-0674(92)90057-j. [DOI] [PubMed] [Google Scholar]
- 23.Rappaport R. Int Rev Cytol. 1986;105:245–281. doi: 10.1016/s0074-7696(08)61065-7. [DOI] [PubMed] [Google Scholar]
- 24.White J G, Borisy G G. J Theor Biol. 1983;101:289–316. doi: 10.1016/0022-5193(83)90342-9. [DOI] [PubMed] [Google Scholar]
- 25.Lee K S, Yuan Y-L, Kuriyama R, Erikson R L. Mol Cell Biol. 1995;15:7143–7151. doi: 10.1128/mcb.15.12.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golsteyn R M, Mundt K E, Fry A M, Nigg E A. J Cell Biol. 1995;129:1617–1628. doi: 10.1083/jcb.129.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mundt K E, Golsteyn R M, Lane H A, Nigg E A. Biochem Biophys Res Commun. 1997;239:377–385. doi: 10.1006/bbrc.1997.7378. [DOI] [PubMed] [Google Scholar]
- 28.Siliciano P G, Tatchell K. Cell. 1984;37:969–978. doi: 10.1016/0092-8674(84)90431-8. [DOI] [PubMed] [Google Scholar]
- 29.Sherman F, Fink G R, Hicks J B. Methods in Yeast Genetics. Plainview, N. Y.: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 30.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chant J. Cell. 1996;84:187–190. doi: 10.1016/s0092-8674(00)80972-1. [DOI] [PubMed] [Google Scholar]
- 32.Kotani S, Tugendreich S, Fujii M, Jorgensen P-M, Watanabe N, Hoog C, Hieter P, Todokoro K. Mol Cell. 1998;1:371–380. doi: 10.1016/s1097-2765(00)80037-4. [DOI] [PubMed] [Google Scholar]