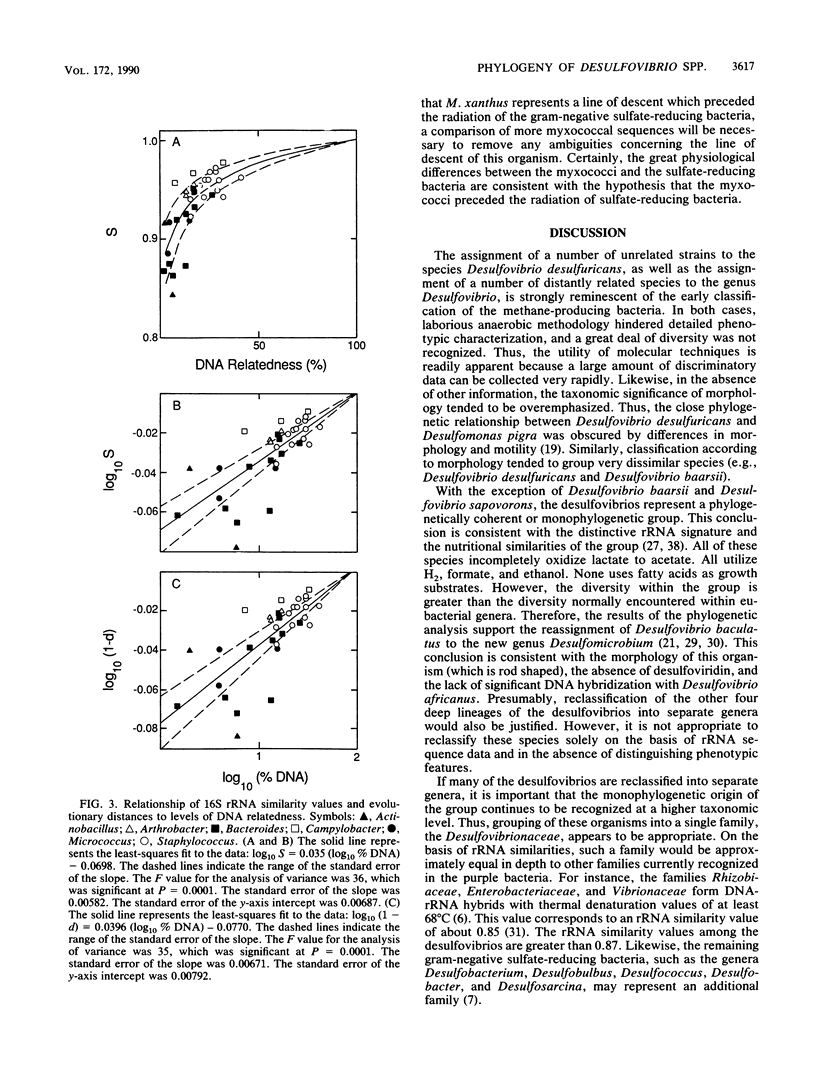
Abstract
The different nutritional properties of several Desulfovibrio desulfuricans strains suggest that either the strains are misclassified or there is a high degree of phenotypic diversity within the genus Desulfovibrio. The results of partial 16S rRNA and 23S rRNA sequence determinations demonstrated that Desulfovibrio desulfuricans ATCC 27774 and "Desulfovibrio multispirans" are closely related to the type strain (strain Essex 6) and that strains ATCC 7757, Norway 4, and El Agheila Z are not. Therefore, these latter three strains of Desulfovibrio desulfuricans are apparently misclassified. A comparative analysis of nearly complete 16S rRNA sequences in which we used a least-squares analysis method for evolutionary distances, an unweighted pair group method, a signature analysis method, and maximum parsimony was undertaken to further investigate the phylogeny of Desulfovibrio species. The species analyzed were resolved into two branches with origins deep within the delta subdivision of the purple photosynthetic bacteria. One branch contained five deep lineages, which were represented by (i) Desulfovibrio salexigens and Desulfovibrio desulfuricans El Agheila Z; (ii) Desulfovibrio africanus; (iii) Desulfovibrio desulfuricans ATCC 27774, Desulfomonas pigra, and Desulfovibrio vulgaris; (iv) Desulfovibrio gigas; and (v) Desulfomicrobium baculatus (Desulfovibrio baculatus) and Desulfovibrio desulfuricans Norway 4. A correlation between 16S rRNA sequence similarity and percentage of DNA relatedness showed that these five deep lineages are related at levels below the minimum genus level suggested by Johnson (in Bergey's Manual of Systematic Bacteriology, vol. 1, 1984). We propose that this branch should be grouped into a single family, the Desulfovibrionaceae. The other branch includes other genera of sulfate-reducing bacteria (e.g., Desulfobacter and Desulfococcus) and contains Desulfovibrio sapovorans and Desulfovibrio baarsii as separate, distantly related lineages.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdollahi H., Nedwell D. B. Serological characteristics within the genus Desulfovibrio. Antonie Van Leeuwenhoek. 1980;46(1):73–83. doi: 10.1007/BF00422231. [DOI] [PubMed] [Google Scholar]
- Badziong W., Thauer R. K. Growth yields and growth rates of Desulfovibrio vulgaris (Marburg) growing on hydrogen plus sulfate and hydrogen plus thiosulfate as the sole energy sources. Arch Microbiol. 1978 May 30;117(2):209–214. doi: 10.1007/BF00402310. [DOI] [PubMed] [Google Scholar]
- Belland R. J., Trust T. J. Deoxyribonucleic acid sequence relatedness between thermophilic members of the genus Campylobacter. J Gen Microbiol. 1982 Nov;128(11):2515–2522. doi: 10.1099/00221287-128-11-2515. [DOI] [PubMed] [Google Scholar]
- Chuba P. J., Bock R., Graf G., Adam T., Göbel U. Comparison of 16S rRNA sequences from the family Pasteurellaceae: phylogenetic relatedness by cluster analysis. J Gen Microbiol. 1988 Jul;134(7):1923–1930. doi: 10.1099/00221287-134-7-1923. [DOI] [PubMed] [Google Scholar]
- Devereux R., Delaney M., Widdel F., Stahl D. A. Natural relationships among sulfate-reducing eubacteria. J Bacteriol. 1989 Dec;171(12):6689–6695. doi: 10.1128/jb.171.12.6689-6695.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W. M., Margoliash E. Construction of phylogenetic trees. Science. 1967 Jan 20;155(3760):279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. P., Yi C. S., LeGall J., Peck H. D., Jr Isolation of a new pigment, desulforubidin, from Desulfovibrio desulfuricans (Norway strain) and its role in sulfite reduction. J Bacteriol. 1973 Jul;115(1):453–455. doi: 10.1128/jb.115.1.453-455.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W., Schleifer K. H., Fox G. E., Seewaldt E., Stackebrandt E. A phylogenetic analysis of staphylococci, Peptococcus saccharolyticus and Micrococcus mucilaginosus. J Gen Microbiol. 1981 Aug;125(2):357–366. doi: 10.1099/00221287-125-2-357. [DOI] [PubMed] [Google Scholar]
- Moura I., Teixeira M., Huynh B. H., LeGall J., Moura J. J. Assignment of individual heme EPR signals of Desulfovibrio baculatus (strain 9974) tetraheme cytochrome c3. A redox equilibria study. Eur J Biochem. 1988 Sep 15;176(2):365–369. doi: 10.1111/j.1432-1033.1988.tb14290.x. [DOI] [PubMed] [Google Scholar]
- Nei M., Stephens J. C., Saitou N. Methods for computing the standard errors of branching points in an evolutionary tree and their application to molecular data from humans and apes. Mol Biol Evol. 1985 Jan;2(1):66–85. doi: 10.1093/oxfordjournals.molbev.a040333. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Lane D. J., Giovannoni S. J., Pace N. R., Stahl D. A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- Pace B., Campbell L. L. Homology of ribosomal ribonucleic acid of Desulfovibrio species with Desulfovibrio vulgaris. J Bacteriol. 1971 Jun;106(3):717–719. doi: 10.1128/jb.106.3.717-719.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk P. J., Zoltowska B., Trust T. J., Lane D. J., Olsen G. J., Pace N. R., Stahl D. A. Campylobacter pylori, the spiral bacterium associated with human gastritis, is not a true Campylobacter sp. J Bacteriol. 1987 May;169(5):2137–2141. doi: 10.1128/jb.169.5.2137-2141.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozanova E. P., Nazina T. N. Mezofil'naia palochkovidnaia bessporovaia bakteriia, vosstanavliviushchaia sul'faty. Mikrobiologiia. 1976 Sep-Oct;45(5):825–830. [PubMed] [Google Scholar]
- Schleifer K. H., Stackebrandt E. Molecular systematics of prokaryotes. Annu Rev Microbiol. 1983;37:143–187. doi: 10.1146/annurev.mi.37.100183.001043. [DOI] [PubMed] [Google Scholar]
- Stetter K. O., Lauerer G., Thomm M., Neuner A. Isolation of extremely thermophilic sulfate reducers: evidence for a novel branch of archaebacteria. Science. 1987 May 15;236(4803):822–824. doi: 10.1126/science.236.4803.822. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]



