Abstract
Glycerol-3-phosphate acyltransferase (GPAT) catalyzes the initial and rate-limiting step in glycerolipid synthesis. Several mammalian GPAT activities have been recognized, including N-ethylmaleimide (NEM)-sensitive isoforms in microsomes and mitochondria and an NEM-resistant form in mitochondrial outer membrane (GPAT1). We have now cloned a second mitochondrial isoform, GPAT2 from mouse testis. The open reading frame encodes a protein of 798 amino acids with a calculated mass of 88.8 kDa and 27% amino acid identity to GPAT1. Testis mRNA expression was 50-fold higher than in liver or brown adipose tissue, but the specific activity of NEM-sensitive GPAT in testis mitochondria was similar to that in liver. When Cos-7 cells were transiently transfected with GPAT2, NEM-sensitive GPAT activity increased 30%. Confocal microscopy confirmed a mitochondrial location. Incubation of GPAT2-transfected Cos-7 cells with trace (3 μM; 0.25μCi) [1-14C]oleate for 6 h increased incorporation of [14C]oleate into TAG 84%. In contrast, incorporation into phospholipid species was lower than in control cells. Although a polyclonal antibody raised against full-length GPAT1 detected an ∼89 kDa band in liver and testis from GPAT1 null mice and both 89 and 80 kDa bands in BAT from the knockout animals, the GPAT2 protein expressed in Cos-7 cells was only 80 kDa. In vitro translation showed a single product of 89 kDa. Unlike GPAT1, GPAT2 mRNA abundance in liver was not altered by fasting or refeeding. GPAT2 is likely to have a specialized function in testis.
The synthesis of triacylglycerol and all glycerophospholipids begins with the acylation of glycerol-3-phosphate with long-chain fatty acyl-CoA to produce 1-acylglycerol-3-phosphate. This reaction is catalyzed by glycerol-3-phosphate acyltransferase (GPAT1; EC 3.1.3.9) which exhibits the lowest specific activity of all enzymes in the glycerol-3-phosphate pathway, suggesting that it is the rate limiting step [1].
Three mammalian GPAT activities have been differentiated based on their subcellular location and biochemical properties [2, 3]. GPAT activity is present in microsomal and mitochondrial fractions. In most tissues, 90% of the activity is attributed to a microsomal GPAT which is sensitive to inactivation by sulfhydryl reagents such as NEM. Microsomal GPAT3, prominent in adipose tissue, has been cloned [4]. Microsomal GPAT activity does not appear to be regulated in liver. In contrast, the mitochondrial isoform (GPAT1)2 that has been purified [5] and cloned [6, 7], is highly regulated by insulin and SREBP1c under conditions in which fatty acid and triacylglycerol synthesis are enhanced [1]. GPAT1, which comprises 30−50% of total GPAT activity in rodent liver is NEM-resistant. In our studies of Gpat1−/− mouse liver, we identified a second mitochondrial GPAT activity [3]. This isoform, GPAT2, was first recognized because a polyclonal antibody raised to full-length GPAT1 identified a protein of similar molecular mass in liver from GPAT1 null mice. Characterization of GPAT2 activity in liver mitochondria from Gpat1−/− mice showed that, unlike GPAT1, GPAT2 is sensitive to NEM inactivation, has no preference for palmitoyl-CoA compared with oleoyl-CoA, is inhibited by dihydroxyacetone-phosphate and polymixin B, is unaffected by incubation with acetone, and is more sensitive to thermal inactivation [3].
In order to further characterize the expression and functionality of this second mitochondrial GPAT isoform, we endeavored to clone and express GPAT2. A search of the mouse and human databases with a sequence encompassing the GPAT1 active site region [8] identified a putative glycerolipid acyltransferase with significant homology to GPAT1 and a similar molecular mass. Here we report the cloning and expression of GPAT2 and demonstrate that overexpression of GPAT2 in Cos-7 cells increases [1-14C]oleate incorporation into triacylglycerol.
Experimental Procedures
Chemicals.
All chemicals were purchased from Sigma unless otherwise indicated. Lipid standards were from Avanti Polar Lipids.
Animals.
Animal protocols were approved by the University of North Carolina at Chapel Hill Institutional Animal Care and Use Committee. Male Gpat1−/− (backcrossed 6 times to C57BL/6) and wild type C57BL/6 mice were genotyped by PCR as described previously [9]. Mice were housed on a 12-h/12-h light/dark cycle with free access to water and Prolab RMH 3000 SP76 chow. Male Sprague-Dawley rats (150 g) were housed on a 12-h/12-h light/dark cycle with free access to water and food (Dyets #611000). For a study of response to fasting and refeeding, livers were obtained from rats that had been fasted for 48 h or fasted for 24 h, then refed a 69% sucrose diet (Dyets #111780) for 24 h.
Gene cloning and construction of the recombinant pcDNA3.1-GPAT2-Flag plasmid.
Mouse testis cDNA was synthesized from a GPAT1 null mouse using the Superscript First strand synthesis Kit for RT-PCR (Invitrogen) according to the manufacturer's protocol. Touch-down PCR was performed to clone the GPAT2 cDNA. Primers were synthesized by UNC's oligonucleotide core facility. Forward primer for GPAT2: 5'-TCA CGG ATC CAT GTT GAA ATC CAA CCC CCA AAC CCA GCA-3'. Reverse primer: 5'-CTG CAG ATG ACC TGT CGG ATA AAT TGT TCC AGC TTG-3'. The PCR reaction was performed using the Pfx DNA polymerase kit (Invitrogen) The reaction contained 1X PCR buffer, 200 μM dNTPs, 0.3 μM primers, 1.25 units Pfx DNA polymerase, cDNA template and 2X amplification buffer. The mixture was heated to 94 °C for 5 min to activate the Pfx DNA polymerase. The reaction was incubated for 30s at 94 °C. Primers annealed to the template DNA after reducing the temperature to 60 °C for 30s. The annealing temperature was reduced 0.5 °C /cycle for 20 cycles followed by 15 cycles at 50 °C. Extension was performed for 2.3 min at 68 °C, resulting in a 2.4 kb cDNA product. The PCR product was cloned into TOPO vector (Invitrogen) and sequenced by the UNC Genome Analysis Facility. 5'RACE was performed to ensure that the complete cDNA was obtained using the BD SMART RACE cDNA Amplification Kit (BDBioSciences Clonetech) according to the manufacturer's instructions. This analysis verified that the there were no other upstream transcription initiation start sites and identified a 135 bp untranslated region (5'-ACGCGGGGCCTGCCCTCCATCTTGGTTTTGGCGAGAACCTTCCTTGCAGGCGTAGGG GTATTTGCTGCAGCTGAGAACTTTTTAAAGGTCAGGTCTGTTGGAGAGAGACACAGTGCT TGATTCACTGATCTTACAGACCTTC-3').
A mammalian expression plasmid coding full-length mouse GPAT2 was engineered by unidirectional subcloning of the 2,394 bp cDNA fragment from the TOPO vector described above into the BamH1 and Xho1 sites of pcDNA3.1(+) mammalian expression vector (Invitrogen). The resulting GPAT2 clone also contained the 9 amino acid Flag epitope (DYKDDDDK) at the C-terminus.
Transient transfection.
Cos-7 cells were routinely cultured in DMEM at 37 °C with 10% heat-inactivated FBS plus 100 U/ml penicillin and 100 μg/ml streptomycin, 5% CO2, and 100% humidity. The medium was changed every other day. Cos-7 cells were plated at a cell density of 1.2 × 106 per 60 mm dish and were transfected 8 h after plating. Cells were transfected with either 4 μg pcDNA3.1(+) vector, 4 μg pcDNA3.1(+)-GPAT1-Flag, or 4 μg pcDNA3.1(+)-GPAT2-Flag recombinant constructs using a 3:1 (μL:μg) Fugene 6:DNA complex (Roche Diagnostics Corp.). Transfection efficiency was monitored by transfecting a separate dish of cells with 4 μg pBi-tet vector and 12 μg pTetoff to express GFP, which was visualized using a fluorescent microscope. The highest GPAT2-Flag expression was achieved 18−36 h after transfection as determined by immunoblot analysis against the Flag (M2) epitope (data not shown).
Immunofluorescence.
Cells were transiently transfected as described above. Twenty-four h after transfection the cells were trypsinized and seeded into an 8 well chamber slide at 40,000 cells per well (Nalge Nunc International), and returned to the 37 °C incubator. After 12 h the cells were washed with PBS and incubated for 45 min with 100 nM Mitotracker Red CMXROS (Molecular Probes) in culture medium, rinsed with PBS, and fixed with ice cold acetone: methanol (1:1; v/v) at room temperature for 10 min. After washing with PBS, cells were permeabilized with 0.18% Triton X-100. The fixed cells were blocked for 15 min with 5% NGS in PBS, pH 7.8 that contained 0.05% Triton X-100. Cells were then incubated with a 1/1000 dilution of anti-FLAG M2 monoclonal antibody (Sigma-Aldrich) in 1% NGS, 0.05% Triton X-100 in PBS for 60 min. Cells were washed with PBS plus 0.05% Triton X-100 and incubated for 30 min with a 1/500 dilution of Alexafluor 488-conjugated secondary antibody: goat anti-mouse IgG (Molecular Probes) in PBS, pH 7.2 with 1% BSA. Cells were then washed in PBS plus 1% NGS, and the cover slips were mounted with Gelmount (Biomeda Corp). Confocal microscopy was performed on a Zeiss Confocal LSM Pascal 5 equipped with an excitation argon laser 488/515 to visualize cells stained with Mitotracker and Alexa 488. Co-localization analysis was performed using the manufacturer's included quantitative co-localization software.
Immunoblotting.
100 μg of protein was electrophoresed on an 8% polyacrylamide gel containing 1% SDS and transferred to a PVDF membrane. The expression of GPAT1-FLAG or GPAT2- FLAG was analyzed by blotting with anti-FLAG monoclonal antibody or anti-GPAT antibody. The rabbit polyclonal anti-GPAT antibody was raised to gel-purified full-length rat GPAT1 expressed in E. coli [10]. The membrane was washed extensively, and then probed with horseradish peroxidase-conjugated goat anti-mouse IgG or goat anti-rabbit IgG (Pierce). GPAT-Flag and mtGPAT proteins were visualized by a reaction with Supersignal chemiluminescent substrate (Pierce) following exposure to x-ray film.
Glycerol 3-Phosphate Synthesis and GPAT Assay.
sn-[2-3H]Glycerol 3-phosphate was synthesized enzymatically from [2-3H]glycerol (1 mCi/ml) (PerkinElmer Life Sciences) as described [11]. GPAT activity was assayed in a 200 μl mixture containing 75 mM Tris-HCl, pH 7.5, 4 mM MgCl2, 1 mg/ml BSA (essentially fatty acid-free), 1 mM dithiothreitol, 8 mM NaF, 800 μM [3H]glycerol 3-phosphate, and 80 μM palmitoyl-CoA [11]. The reaction was initiated by adding 1–30 μg of protein to the assay mix. For NEM inhibition studies, protein was pre-incubated with 1 mM NEM for 15 min on ice before addition to the assay mix. All assays were performed for 10 min at 37 °C unless otherwise indicated, and measured initial rates. Products were identified by thin layer chromatography with CHCl3/pyridine/88% formic acid (50/30/7; v/v).
Quantitative
RT-PCR-Rat liver and mouse tissue samples stored in RNALater (Qiagen) were homogenized with a rotor-stator homogenizer and RNA was isolated with the RNeasy kit (Qiagen). Samples were analyzed in triplicate on an ABI Prism 7700 Sequence Detection System and data were analyzed using the relative standard curve method as described in User Bulletin #2 (Applied Biosystems) at the UNC Animal Clinical Chemistry and Gene Expression Facility. Primer and probe sequences for rat and mouse genes are shown in Table 1.
Table 1.
Primers and probes for qRT-PCR of rat GPAT1, GPAT2, and β-actin.
| GPAT1 |
| Forward Primer: 5′ -ACAGTGGCAGAGCAAGATGA |
| Reverse Primer: 5′ -ACTTCTCCAGTTCAGAGGCT |
| Probe: 5′ - AGTCCTGGCCTTGCAGAGCAGCAG |
| GPAT2 |
| Forward Primer: 5′ - CTCCTGGTTGCAGAGGAGA |
| Reverse Primer: 5′ - |
| AGCAGCTTTGCACTCAGATG |
| Probe: 5′ - CGGCCAGCCTGTGACACAGGAA |
| β-actin |
| Forward Primer: 5′ - TGCCTGACGGTCAGGTCA |
| Reverse Primer: 5′ - CAGGAAGGAAGGCTGGAAG |
| Probe: 5′ - CACTATTGGCAACGAGCGGTTCCG |
Primers were designed to target the GPAT1 and GPAT2 exon 14/15 splice junction and to be compatible with a single qRT-PCR thermal profile (48 °C for 30 min, 95 °C for 10 min, and 40 cycles of 95 °C for 15 s and 60 °C for 1 min) such that multiple transcripts could be analyzed simultaneously (Table 1). The reaction mixture contained forward and reverse primers (50 nM each), sample RNA (100 ng), 1x AmpliTaq Gold® PCR Master Mix (Applied Biosystems) (containing AmpliTaq Gold® DNA Polymerase, dNTPs, Gold Buffer and MgCl2) in a final volume of 30 μL. For each set of primers, a no-template control and a no-reverse amplification control were included. Fold changes in gene expression were determined using the △ Ct method with normalization to ß-actin expression.
Cell Labeling and Lipid Analysis.
Cos-7 cells were seeded in 60 mm dishes (1.5 × 106 cells/dish). Twelve h after the cells were seeded, they were transiently transfected with 4 μg of pcDNA3.1-GPAT2-Flag plasmid. Fourteen h after transfection, the cells were incubated with 2 ml DMEM/10% FBS with [14C]oleic acid at 0.25 μCi/dish (∼3 μM oleate), supplemented with 0.5% BSA and 1 mM carnitine. After a 6 h incubation the cells were washed with 0.1% BSA in ice-cold 0.9% NaCl, and scraped in ice-cold methanol and H2O. Cell lipids were extracted [12] and concentrated in a Savant SpeedVac concentrator. Neutral lipids and phospholipids were resolved and separated by TLC [13]. All samples were chromatographed in parallel with authentic lipid standards. The 14C-labeled lipids were detected using a Bioscan Image System.
In vitro Transcription-Translation Assays.
To determine the translated size of GPAT2, in vitro transcription and translation was performed using TNT-T7 Quick coupled transcription and non-radioactive translation system (Promega) with modifications. To initiate gene transcription, we generated a GPAT cDNA clone that was positioned behind a phage T7 promoter. The GPAT2 cDNA template, including T7 promoter, Kozak consensus sequence, 5' start codon, and oligo dT after the stop codon, was amplified by PCR using GPAT1 and GPAT2 recombinant plasmids. These PCR generated templates were used for GPAT1 and GPAT2 transcription to make mRNA templates for GPAT1 and GPAT2 proteins synthesized in vitro. The coupled transcription/translation reaction contained 20 μl TNT-T7 Quick master mixture, 20 mM methionine, 2 μl PCR-generated DNA template (∼150 ng), 1 μl Transcend Biotin-Lys-tRNA, and 1 μl pancreatic microsomal membranes. This reaction mixture was incubated at 37°C for 1 h. Aliquots (1.2 μl) of the biotin-labeled translation products with 15 μl SDS sample buffer were analyzed on an 8% polyacrylamide gel containing 1% SDS and visualized by detection of biotin/streptavidin-HRP.
Statistics.
Data were analyzed by Student's t-test and are shown as the means ± standard errors.
Results
Identification of the GPAT2 sequence.
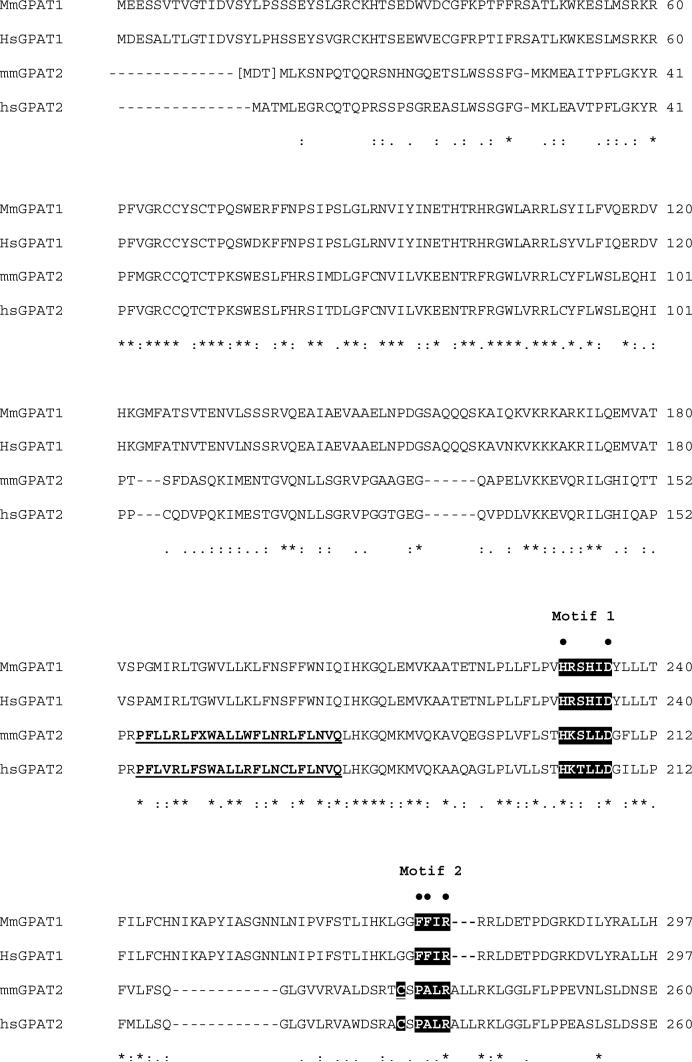
After finding that Gpat1−/− mice retained GPAT activity in liver mitochondria and that a polyclonal antibody raised to GPAT1 recognized another protein of approximately the same molecular mass [3], we used the sequence of mouse GPAT1 that delineates the active site region [8] (aa 229-aa357) to search mouse and human databases. Of several putative glycerolipid acyltransferases whose enzymatic function was unknown, only one had a predicted open reading frame that would encode a protein with a size similar to that of GPAT13. Sequences were identified that encoded 798 (NP_001074558) and 801 (BAF03614.1) amino acid mouse proteins of 88.8 or 89.1 kDa, respectively4 (Fig. 1A). Using the 798 GPAT2 amino acid sequence, the Blast alignment of GPAT1 and GPAT2 showed an amino acid sequence identity of 27% with GPAT1 and, in the active site region from amino acid 153 to 372, an amino acid identity of 24.5%. The active site region contained recognizable motifs 1 and 4 that had been identified in E. coli PlsB [8, 14] and mouse GPAT1 [15], and that are present in dihydroxyacetonephosphate acyltransferase [1], lysocardiolipin acyltransferase [16], and lysophosphatidylglycerol acyltransferase [17]. Motifs 2 and 3, however, differed in GPAT2. In mammals and bacteria, motif 2 normally contains the sequence FXXR, and the invariant arginine is essential for the activity of human tafazzin, human dihydroxyacetone-phosphate acyltransferase and E. coli PlsB, probably because it interacts with the phosphate portion of the glycerolphosphate substrates of these enzymes [1]. Motif 3 usually contains the consensus sequence FXEGXRXR in which the glutamic acid, glycine, and the first arginine are essential for activity [1, 15]. In GPAT2, however, the usual motif 2 and 3 sequences differ markedly from GPAT1 and are similar, instead, to sequences in several chloroplast GPATs (Fig. 1B). We suggest that the GPAT2 sequences PALR and GSPGPRLS comprise motifs 2 and 3, respectively, because they match the conserved plant sequences and because the assigned motif 3 contains one of the two arginines that was recognized as critical for enzymatic function of squash chloroplast GPAT [18].
Fig. 1. GPAT2 protein sequence analysis.
A.) Alignment of mouse (NP_001074558) and human (AAH68596; BAD18392) GPAT2 and of mouse ( NP_032175) and human (NP_065969) GPAT1 amino acid sequences. Mouse GPAT2 is shown with the start site for the 801 amino acid protein in brackets.4 The conserved catalytic amino acid residues for GPAT (Motifs 1−4) are indicated by white lettering on black. Residues in motifs 1−4 that have been shown to be important for GPAT1 or E. coli plsB activity are indicated by a dot (•) above the residue [1]. Cysteines near active site motifs and present in GPAT2 only shown in white lettering on black and underlined. The transmembrane domains for GPAT1 [19] and the potential transmembrane domains for GPAT2 are bolded and underlined. Amino acid residues identical for all isoforms are indicated with an asterisk (*); conservation of strong groups is indicated with two dots (:); conservation of weak groups is indicated with one dot (.) B) Comparison of the active site regions of GPAT2, plant chloroplast GPATs, and Chlamydia GPAT. Q43307, Arabidopsis Thaliana; Q43869, Spinacia oleracea; Q42713, Carthamus tinctorius (safflower); P30706, Pisum sativum (pea); AAC68403, Chlamydia trachomatis. Glycerolipid acyltransferase motifs and identical and conserved amino acid residues in GPAT2, plant chloroplast GPATs, and Chlamydia GPAT are shown as above. The mmGPAT1 and hsGPAT1 amino acids in this region are shown for comparison.
GPAT2 is predicted to have at least two transmembrane domains.
Like GPAT1, GPAT2 does not contain known mitochondrial targeting sequences. Using algorithms based on the residue hydrophobicity, several potential transmembrane domains were identified. These transmembrane domains are also apparent in a Kyte-Doolittle hydrophobicity plot using a 21 residue window (data not shown). Nine different Internet programs (TMPred, HMMTOP, DAS, SOSUI, TMAP, MEMSAT 2, PRED-TMR2, Top-Pred II and Con Pred II) each strongly predicted the presence of two TMDs. One TMD was consistently predicted to be between amino acids 446 and 474. This predicted TMD coincides with the first TMD of mouse GPAT1 (Fig. 1A) [19]. All the algorithms predicted an additional TMD, either between residues 204−226 or 301−329. These two TMDs seem highly improbable because they lie within the region containing Motifs 1−4 of the active site of GPAT2 [8] and would cause portions of the active site to lie on opposite faces of the membrane. The TMD predicted to encompass residues 156–177 is possible because it lies between the N terminus and the active site domain (Fig. 1A). With transmembrane domains between 156 and 177 and between 446 and 474, it is predicted that GPAT2 would adopt a membrane topography in which both the N- and C-termini are located in the intermembrane space, allowing the active site to face the cytosol where it would have access to its glycerol-3-phosphate and acyl-CoA substrates.
Tissue distribution of mouse GPAT2.
To determine the relative expression of GPAT1 and GPAT2 in mouse tissues, we examined mRNA abundance by qRT-PCR (Fig. 2). We found highest expression of GPAT1 in brown and white adipose tissue, liver, soleus muscle, and heart. A previous study of GPAT2 (called xGPAT1) reported alternate exon use for GPAT2 that results in identical proteins but different tissue expression of the alternate transcripts [20]. Using primers that would identify both transcripts, we found that GPAT2 mRNA was expressed at very high levels in testis; expression was 50-fold lower in liver and brown adipose tissue, and even lower in other tissues. When mitochondria from liver and testis from Gpat1−/− mice were assayed for GPAT activity, a TLC analysis showed that 27% of the radiolabeled reaction products was lysophosphatidic acid, 67% was phosphatidic acid, and 6% was diacylglycerol. This product distribution is similar to that for GPAT1 and is consistent with the presence of an active AGPAT in the mitochondrial membranes.
Fig. 2. GPAT2 is highly expressed in testis.
Pieces of rat tissues stored in RNALater were homogenized with a rotor-stator homogenizer and RNA was isolated with the RNeasy kit. Samples were analyzed in triplicate on an ABI Prism 7700 Sequence Detection System and data were analyzed using the relative standard curve method. Expression was normalized to β-actin. BAT, brown adipose tissue; RET-Ad, retroperitoneal adipose; GON-Ad, gonadal adipose; ING-Ad, inguinal adipose; GAST, gastrocnemius, DM, duodenal mucosa.
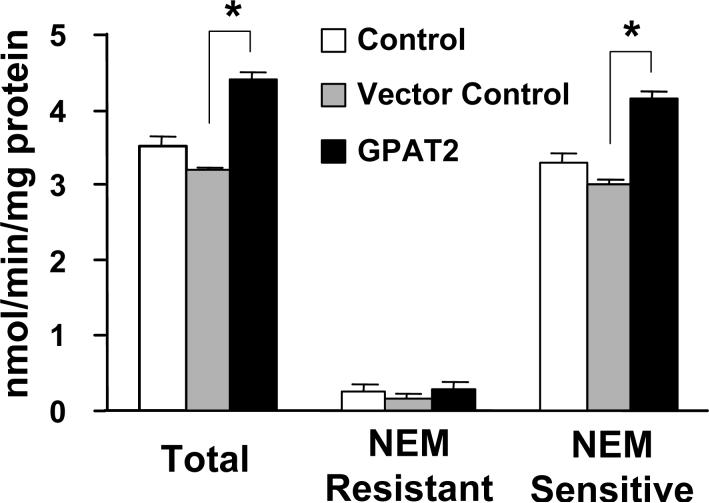
Recombinant GPAT2 shows NEM-sensitive activity and is recognized by anti-GPAT polyclonal antibody.
Cos-7 cells were transiently transfected with pcDNA3.1-GPAT2-Flag. In total cell membranes NEM-resistant GPAT activity was the same as the untransfected control, but NEM-sensitive GPAT activity was 30% higher (Fig. 3). When GPAT2 was transiently transfected into HEK293 cells, NEM-sensitive GPAT activity increased 24% and 57% at 18 and 36 h after transfection, respectively (18 h: 1.2 ± 0.1 vs. 1.49 ± 0.1 nmol/min/mg protein; 36 h: 1.2 ± 0.2 vs. 1.88 ± 0.04 nmol/min/mg protein; pcDNA3.1 vs. pcDNA3.1-GPAT2-Flag)
Fig. 3. Overexpression of GPAT2 increases NEM-sensitive GPAT activity.
Cos-7 cells were either not transfected (control), transfected with empty pcDNA3.1(+) (vector control), or transfected with pcDNA3.1(+)- GPAT2-Flag cDNA (GPAT2). Thirty-six h after transfection, cell homogenates were prepared and centrifuged at 100,000 x g to obtain total membranes. GPAT activity was assayed after incubation in the presence or absence of 2 mM NEM. Data are shown as means ± SE (n = 9); * p<0.05.
GPAT2 undergoes post-translational modification.
Although the calculated molecular mass of GPAT2 is 88.8 kDa (http://ca.expasy.org/tools/pi_tool.html), expression in Cos-7 cells or HEK293 cells (data not shown) produced a protein with an apparent molecular mass of about 80 kDa (Fig. 4). If methionine-65 acted as a second start site, it would generate a protein of about 80 kDa, so to determine the translated size of GPAT2, we performed in vitro transcription and translation of GPAT1 and GPAT2. Translation of the cDNAs produced proteins of ∼93 kDa for GPAT1 and ∼88 kDa for GPAT2. Both translation products had the predicted masses, thereby ruling out an alternate start site for GPAT2 (Fig. 4). We were unable to immunoprecipitate sufficient recombinant GPAT2 to sequence the N-terminus of the 80 kDa protein to determine where it had been truncated.
Fig. 4. Translated GPAT1 and 2 have similar molecular masses, whereas masses of GPAT1 and 2 expressed in cells differ.

Cos-7 cells were transfected with plasmids containing empty vector (EV), GPAT1-Flag or GPAT2-Flag for 36 h. Transcription and translation was performed in vitro as described in Experimental Procedures. The GPAT cDNA templates including the T7 promoter were used for transcription and translation in 25 μl reactions containing 20 mM methionine, 1 μl biotin-Lys-tRNA and 1 μl pancreatic microsomal membranes The reaction mixture was incubated at 37°C for 1 h. Lanes contained 60 μg of Cos-7 cell total particulate protein or 1.2 μl of GPAT1 or GPAT2 reaction aliquots. Proteins were analyzed on the same 8% gel by polyacrylamide gel electrophoresis. Proteins expressed in Cos-7 cells were visualized with anti-FLAG M2 antibodies and in vitro translated proteins were detected with biotin/streptavidin-HRP.
BAT in both wildtype and Gpat1−/− mice expresses both the ∼88 and the 80 kDa protein bands similar to that of GPAT2 transfected in Cos-7 cells (Fig. 5A). In contrast, in wildtype and Gpat1−/− mice, an anti-GPAT1 polyclonal antibody recognizes a ∼93 kDa band in both liver and testis that results from the expression of both GPAT1 and 2 (Fig. 5B) [3]. Neither liver nor testis express the 80 kDa protein.
Fig. 5. Protein expression of GPAT1 and GPAT2.
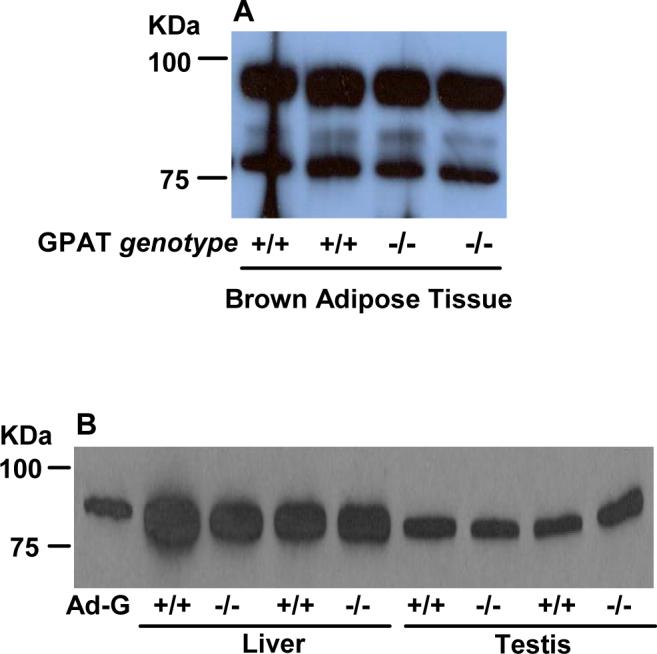
A) 40 μg brown adipose tissue total particulate protein or B) 50 μg liver or testis total particulate protein from WT (+/+) or Gpat1−/− mice. Proteins were separated by 8% SDS-PAGE and transferred to a PVDF membrane. Primary rabbit anti-GPAT1 polyclonal antibody was used to recognize GPAT isoforms. Ad-G, GPAT1 expressed in COS-7 cells with an adenovirus vector
Overexpressed GPAT2 localizes to mitochondria.
In Cos-7 cells that transiently express GPAT2, confocal microscopy showed a co-localization coefficient of ≥ 0.9 and Manders overlap coefficient=1, indicating that GPAT2-Flag colocalized with the mitochondrial marker to the mitochondria (Fig. 6). No GPAT2-Flag staining was observed in cells transfected with empty pcDNA3.1 or in cells transfected with pcDNA3.1-GPAT2-Flag but omitting the anti-Flag antibody during staining (data not shown).
Fig. 6. GPAT2 colocalizes with mitochondria.
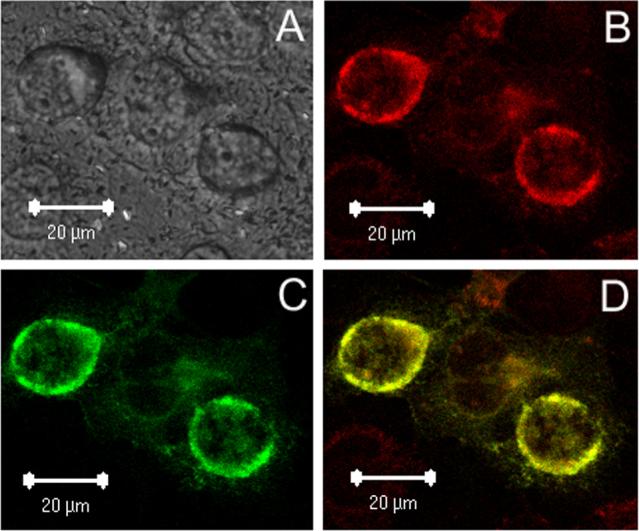
Cos-7 cells were transiently transfected with pcDNA3.1-GPAT2-Flag for 36 h and stained as described under Experimental Procedures. A) DIC; B) Mito-Tracker; C) anti-FLAG primary antibody and AlexaFluor 488-conjugated secondary antibody; D) merged image of B and C. The figure is representative of two independent transfections.
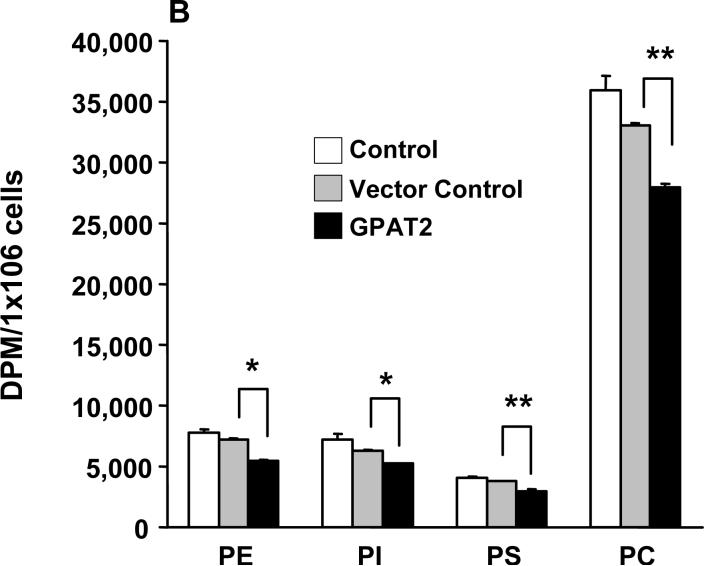
Overexpression of GPAT2 increases the incorporation of [14C]oleate into triacylglycerol.
To determine the function of GPAT2 in glycerolipid synthesis, we measured the incorporation of [1-14C]oleate into complex lipids. Cells were transiently transfected with pcDNA-GPAT2-Flag. Fourteen h after transfection, the cells were incubated with trace amounts (∼3 μM) of [14C]oleic acid for 6 h. In three independent experiments the incorporation of [1-14C]oleate into total phospholipid was 8 to 13% lower than in control cells (Fig. 7A), but 45% more [1-14C]oleate was incorporated into total neutral lipid compared to vector control cells (data not shown). The increased oleate incorporation into neutral lipid was entirely due to a 84% increase in [1-14C]oleate incorporation into triacylglycerol. Examination of the different phospholipid species in cells overexpressing GPAT2 showed diminished incorporation of [1-14C]oleate into phosphatidylethanolamine, phosphatidylinositol, phosphatidylserine, and phosphatidylcholine (Fig 7B).
Fig. 7. GPAT2 overexpression increases incorporation of [14C]oleic acid into triacylglycerol and decreases incorporation into phospholipids.
Cos-7 cells at 75% confluence were either untransfected (Control) or transfected with pcDNA3.1 (Vector control) or with pcDNA3.1-GPAT2-Flag plasmid (GPAT2). At 14 h after transfection the labeling media was replaced with DMEM supplemented with 10% FBS, 0.5% BSA, 1 mM carnitine, and 0.25μCi of [14C]oleic acid/dish and incubated for 6 h. Lipids were extracted and separated as described under Experimental Procedures (n=3). A. Neutral and phospholipids; B. Phospholipid species. The graphs are representative of an experiment that was repeated 3 times. * p< 0.05; ** p < 0.01 compared to vector control cells.
Physiological changes in GPAT2.
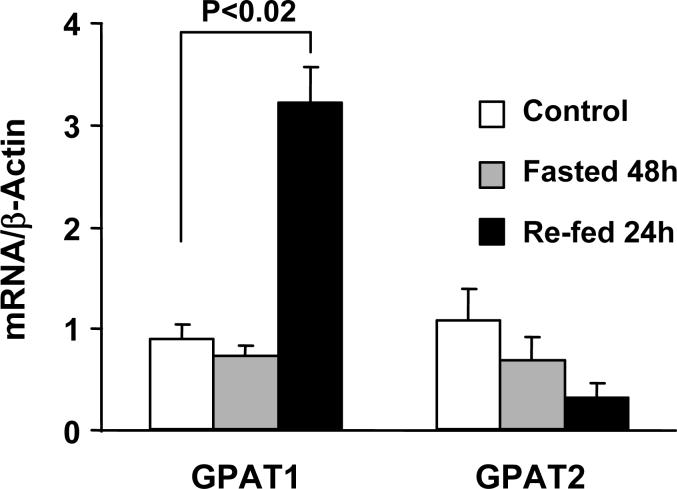
GPAT1 in liver is regulated by insulin and SREBP1c, and 8 h after fasted mice are refed, GPAT1 mRNA increases 20-fold [6]. To determine whether nutritional status regulates GPAT2 similarly, rats were fasted for 48 h or fasted and then refed a high sucrose diet. As expected, rat hepatic GPAT1 mRNA increased 3-fold 24 h after refeeding (Fig. 8). In contrast, hepatic GPAT2 mRNA expression did not change compared to the control animals.
Fig. 8. Regulation of GPAT1 and GPAT2 mRNA expression in rat liver.
GPAT1 and mtGPAT 2 expression in liver from untreated rats and from rats that were fasted for 48 h or fasted for 48 h and then refed for 24 h with a 69% sucrose diet. (n = 4 for each treatment)
Regulation of GPAT2.
High expression of rat GPAT2 in the testis prompted a search of the promoter region for potential androgen and estrogen response elements. The androgen receptor, progesterone receptor and glucocorticoid receptor all belong to the class I family of nuclear receptors that recognize and bind as homodimers to inverted repeats of 5'-TGTTCT- 3' [21-23]. In contrast, the estrogen receptor belongs to a separate class of nuclear receptors and recognizes the response element containing the sequence 5'– AGGTCA- 3' [22, 23]. The promoter region of rat GPAT2 was found by analyzing the known GPAT2 coding sequence with the NCBI high throughput gene sequence (htgs) BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/). A search for hormone response elements in 3.7 kb of this potential rat GPAT2 upstream sequence was carried out using the transcription factor binding site prediction program Alibaba2.1 [24]. Although Alibaba2.1 did not predict an androgen response element, the program predicted eleven potential glucocorticoid response elements, 4 estrogen response elements, and 2 progesterone response elements in this region. These preliminary findings suggest that corticosterone, estrogen and progesterone may activate expression of GPAT2 in the testis. Cloning the promoter region into a luciferase reporter and subsequent transactivation assays will be required to confirm this hypothesis.
Discussion
We first became aware of the existence of a second mitochondrial GPAT when a polyclonal antibody raised to GPAT1 recognized a protein of similar molecular mass in liver from mice null for GPAT1 [3]. It became apparent that a novel GPAT was present in mitochondria but, unlike GPAT1, this activity was inactivated by exposure to NEM. Using the active site region of GPAT1, we searched mouse and human databases, identified and cloned a cDNA that encoded a protein of ∼88 kDa, and expressed it.
The new GPAT2 is similar to GPAT1 in several ways. It is only 27 amino acids shorter, and it has similarly placed active site motifs in its N-terminal half. Like GPAT1 it has a long C-terminal region that differentiates it from AGPAT1, AGPAT2, GPAT3, and lysophospholipid acyltransferases [1, 4]. GPAT2 differs considerably, however, from other mammalian and bacterial glycerolipid acyltransferases in the position and amino acid sequence of the motifs that are essential for catalytic activity. A previously reported alignment of GPAT1 and GPAT2 [20] and a Clustal W analysis predict that motif 2 is comprised by the amino acid sequence FLPP and motif 3 by IFLEE. Because of the importance of the arginine in motif 2 (PALR), we believe that the correct alignment is as we show in Fig. 1B. The 38 amino acid gap found between motifs 1 and 2 in GPAT1 and the AGPAT isoforms is 12 residues shorter, the usual 35 amino acid gap between motifs 2 and 3 is 53 residues, and the amino acid sequences of both motif 2 and motif 3 are more similar to those of plant chloroplast GPATs than to E. coli plsB or to GPAT1. Firm identification of the hypothetical GPAT2 motifs as critical for catalysis will require mutational studies.
We previously characterized GPAT2 in mitochondria from liver obtained from GPAT1 null mice [3]. Unlike GPAT1, GPAT2 was sensitive to NEM. The cysteines that confer sensitivity to NEM may be those near active site motifs 2 and 4 in GPAT2 (Cys232 and Cys337); these cysteines are not present in GPAT1 (Fig. 1A). GPAT2 also differs from GPAT1 because it exhibits no preference for 16:0-CoA compared to 18:1-CoA [3]. Interestingly, in light of the similarities between GPAT2 and chloroplast GPATs, chloroplast GPAT is believed to mediate chilling sensitivity in plants by altering the membrane content of unsaturated fatty acids in phosphatidylglycerol [25]. For example, Arabidopsis thaliana GPAT produces plants with a high content of cis-unsaturated fatty acids whereas the content of these fatty acids is low in plants containing squash GPAT. A natural mutation of L261F (which lies between motifs 3 and 4) in squash GPAT increases substrate selectivity for 18:1-ACP [18]. Whether the alterations in motifs 2 or 3 of GPAT1 or GPAT2 play a role in substrate specificity remains to be determined.
When we over-expressed GPAT2 in insect or mammalian cells, NEM-sensitive GPAT activity consistently increased by about 30%. GPAT2 mRNA is expressed at high levels in the testis and at much lower levels in all other tissues. In contrast, GPAT1 is expressed in several tissues prominent in triacylglycerol synthesis and metabolism, including adipose tissue, liver, heart and muscle. Despite the high mRNA expression in testis, the protein content of GPAT2 in Gpat1−/− mice and the specific activity of GPAT2 in mitochondria appeared to be similar in testis and liver. These data indicate a discrepancy between mRNA and protein expression, as was shown for GPAT1 [26]. Consistent with its tissue distribution, GPAT2 is not regulated in liver by fasting or refeeding. Regulation in testis via glucocorticoid, estrogen, or progesterone requires further study.
GPAT2 protein in tissues appears to have two different molecular masses, one of about 88 kDa that is consistent with its amino acid sequence, and a second of 80 kDa. In vitro translation produced a protein of about 88 kDa, suggesting that the lower molecular weight observed with transfected GPAT2-Flag in Cos-7 or HEK293 cells results from post-translational cleavage rather than from a second start site. Of interest in this regard is the fact that the plant chloroplast GPATs contain a cleaved N-terminal leader sequence [18]. Because recombinant GPAT2 is expressed only as the lower molecular mass, we do not know whether its relatively low activity is the result of the truncation or whether both sizes are equally active. The different sizes may also reflect differences in function or regulation. Interestingly, transfection of CHO-K1 cells with GPAT2 with a C-terminus myc epitope tag resulted in the expression of an 88 kDa protein, however the increase in NEM-sensitive GPAT activity (∼30%) was similar to that obtained with the 80 kDa protein product [20].
Although GPAT2 is present and active in Gpat1−/− mice, it does not complement the function of GPAT1. The Gpat1−/− mice have lower body weight, changes in the fatty acid composition of liver phospholipids [9] and resistance to the development of diet-induced fatty liver and hepatic insulin resistance [27]. Gpat1−/− mice also show marked increases in liver acyl-CoA levels and in β-oxidation [28]. Retention of the second mitochondrial GPAT activity does not prevent this phenotype.
The small amount of NEM-sensitive GPAT activity in mitochondrial membrane preparations has been previously attributed to microsomal contamination. This interpretation may not be valid in tissues which express GPAT2. The presence of a mitochondrial GPAT activity in testis that is regulated independently of GPAT1 suggests a novel function related to triacylglycerol synthesis. Further studies are warranted to determine which types of testicular cells express GPAT2 and whether GPAT2 is essential for sperm energy metabolism, membrane biogenesis, or another function specific to its testicular location.
Acknowledgments
This work was supported by grants DK56598 (RAC), TW06034 (RAC/MRG-B), and DK59931 (TML) from the NIH and 023032N (TML) from the American Heart Association.
Glossary
Abbreviations:
- AGPAT
1-acyl-sn-GPAT
- BAT
brown adipose tissue
- BSA
bovine serum albumin
- GPAT
glycerol-3-phosphate acyltransferase
- NEM
N-ethylmaleimide
- NGS
newborn goat serum
- PBS
phosphate buffered saline
- TMD
transmembrane domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Formerly called mtGPAT or mtGPAT1.
Human GPAT2 was also identified by Eva Rupp-Thuresson and Peter Lind, Biovitrum, Stockholm (personal communication).
Some of the protein sequences (e.g. NP_001074558) identified in GenBank begin with MLK and encode a protein of 798 amino acids. Others (e.g. BAF03614) begin with MDTMLK and encode a protein of 801 amino acids. We performed most of the studies reported here with the 797 amino acid protein, but transfection studies with the 801 amino acid form resulted in a similar increase in GPAT activity in Cos-7 cells and a truncated protein of about 80 kDa.
References
- 1.Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 2004;43:134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 2.Saggerson E, Carpenter C, Cheng C, Sooranna SR. Subcellular distribution and some properties of N-ethylmaleimide-sensitive and-insensitive forms of glycerol phosphate acyltransferase in rat adipocytes. Biochem. J. 1980;190:183–189. doi: 10.1042/bj1900183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewin TM, Schwerbrock NMJ, Lee DP, Coleman RA. Identification of a new glycerol-3-phosphate acyltransferase isoenzyme, mtGPAT2, in mitochondria. J. Biol. Chem. 2004;279:13488–13495. doi: 10.1074/jbc.M314032200. [DOI] [PubMed] [Google Scholar]
- 4.Cao J, Li JL, Li D, Tobin JF, Gimeno RE. Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA. 2006;103:19695–19700. doi: 10.1073/pnas.0609140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vancura A, Haldar D. Purification and characterization of glycerophosphate acyltransferase from rat liver mitochondria. J. Biol. Chem. 1994;269:27209–27215. [PubMed] [Google Scholar]
- 6.Shin D-H, Paulauskis JD, Moustaid N, Sul HS. Transcriptional regulation of p90 with sequence homology to Escherichia coli glycerol-3-phosphate acyltransferase. J. Biol. Chem. 1991;266:23834–23839. [PubMed] [Google Scholar]
- 7.Bhat BG, Wang P, Kim J-H, Black TM, Lewin TM, Fiedorek TF, Coleman RA. Rat hepatic sn-glycerol-3-phosphate acyltransferase: Molecular cloning and characterization of the cDNA and expressed protein. Biochim. Biophys. Acta. 1999;1439:415–423. doi: 10.1016/s1388-1981(99)00103-1. [DOI] [PubMed] [Google Scholar]
- 8.Lewin TM, Wang P, Coleman RA. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 1999;38:5764–5771. doi: 10.1021/bi982805d. [DOI] [PubMed] [Google Scholar]
- 9.Hammond LE, Gallagher PA, Wang S, Posey-Marcos E, Hiller S, Kluckman K, Maeda N, Coleman RA. Mitochondrial glycerol-3-phosphate acyltransferase-deficient mice have reduced weight and liver triacylglycerol content and altered glycerolipid fatty acid composition. Mol. Cell. Biol. 2002;22:8204–8214. doi: 10.1128/MCB.22.23.8204-8214.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewin TM, Kim J-H, Granger DA, Vance JE, Coleman RA. Acyl-CoA synthetase isoforms 1, 4, and 5 are present in different subcellular membranes in rat liver and can be inhibited independently. J. Biol. Chem. 2001;276:24674–24679. doi: 10.1074/jbc.M102036200. [DOI] [PubMed] [Google Scholar]
- 11.Chang Y-Y, Kennedy EP. Biosynthesis of phosphatidyl glycerophosphate in Escherichia coli. J. Lipid Res. 1967;8:447–455. [PubMed] [Google Scholar]
- 12.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 13.Igal RA, Coleman RA. Acylglycerol recycling from triacylglycerol to phospholipid, not lipase activity, is defective in Neutral Lipid Storage Disease fibroblasts. J. Biol. Chem. 1996;271:16644–16651. doi: 10.1074/jbc.271.28.16644. [DOI] [PubMed] [Google Scholar]
- 14.Heath RJ, Rock CO. A conserved histidine is essential for glycerolipid acyltransferase catalysis. J. Bacteriol. 1998;180:1425–1430. doi: 10.1128/jb.180.6.1425-1430.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dircks LK, Ke J, Sul HS. A conserved seven amino acid stretch important for murine mitochondrial glycerol-3-phosphate acyltransferase activity: significance of arginine 318 in catalysis. J. Biol. Chem. 1999;274:34728–34734. doi: 10.1074/jbc.274.49.34728. [DOI] [PubMed] [Google Scholar]
- 16.Cao J, Liu Y, Lockwood J, Burn P, Shi Y. A novel cardiolipin-remodeling pathway revealed by a gene encoding an endoplasmic reticulum-associated Acyl-CoA:lysocardiolipin acyltransferase (ALCAT1) in mouse. J. Biol. Chem. 2004;279:31727–31734. doi: 10.1074/jbc.M402930200. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y, Cao J, Shi Y. Identification and characterization of a gene encoding human LPGAT1, an endoplasmic reticulum-associated lysophosphatidylglycerol acyltransferase. J. Biol. Chem. 2004;279:55866–55874. doi: 10.1074/jbc.M406710200. [DOI] [PubMed] [Google Scholar]
- 18.Slabas AR, Kroon JT, Scheirer TP, Gilroy JS, Hayman M, Rice DW, Turnbull AP, Rafferty JB, Fawcett T, Simon WJ. Squash glycerol-3-phosphate (1)-acyltransferase. Alteration of substrate selectivity and identification of arginine and lysine residues important in catalytic activity. J. Biol. Chem. 2002;277:43918–43923. doi: 10.1074/jbc.M206429200. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez-Baró MR, Granger DA, Coleman RA. Mitochondrial glycerol phosphate acyltransferase contains two transmembrane domains with the active site in the N-terminus, facing the cytosol. J. Biol. Chem. 2001;276:43182–43188. doi: 10.1074/jbc.M107885200. [DOI] [PubMed] [Google Scholar]
- 20.Harada N, Hara S, Yoshida M, Zenitani T, Mawatari K, Nakano M, Takahashi A, Hosaka T, Yoshimoto K, Nakaya Y. Molecular cloning of a murine glycerol-3-phosphate acyltransferase-like protein 1 (xGPAT1) Mol. Cell. Biochem. 2007;297:41–51. doi: 10.1007/s11010-006-9321-5. [DOI] [PubMed] [Google Scholar]
- 21.Beato M. Gene regulation by steroid hormones. Cell. 1989;56:335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 22.Glass CK. Differential recognition of target genes by nuclear receptor monomers, dimmers and heterodimers. Endocrine Rev. 1994;15:391–407. doi: 10.1210/edrv-15-3-391. [DOI] [PubMed] [Google Scholar]
- 23.Claessens F, Verrijdt G, Schoenmakers E, Haelens A, Peeters B, Verhoeven G, Rombauts W. Selective DNA binding by the androgen receptor as a mechanism for hormone-specific gene regulation. Journal. Steroid Biochem. Mol. Biol. 2001;76:23–30. doi: 10.1016/s0960-0760(00)00154-0. [DOI] [PubMed] [Google Scholar]
- 24.Grabe N. Alibaba2: Context Specific Identification of Transcription Factor Binding Sites. In Silico Biology. 2000. p. 19. [PubMed]
- 25.Murata N, Tasaka Y. Glycerol-3-phosphate acyltransferase in plants. Biochim. Biophys. Acta. 1997;1348:10–16. doi: 10.1016/s0005-2760(97)00115-x. [DOI] [PubMed] [Google Scholar]
- 26.Lewin TM, Granger DA, Kim J-H, Coleman RA. Regulation of mitochondrial snglycerol-3-phophate acyltransferase activity: response to feeding status is unique in various rat tissues and is discordant with protein expression. Arch. Biochem. Biophys. 2001;396:119–127. doi: 10.1006/abbi.2001.2604. [DOI] [PubMed] [Google Scholar]
- 27.Neschen S, Morino K, Hammond LE, Zhang D, Liu ZX, Romanelli AJ, Cline GW, Pongratz RL, Zhang XM, Choi CS, Coleman RA, Shulman GI. Prevention of hepatic steatosis and hepatic insulin resistance in mitochondrial acyl-CoA:glycerol-sn-3-phosphate acyltransferase 1 knock out mice. Cell Metab. 2005;2:55–65. doi: 10.1016/j.cmet.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Hammond LE, Neschen S, Romanelli AJ, Cline GW, Ilkayeva OR, Shulman GI, Muoio DM, Coleman RA. Mitochondrial glycerol-3-phosphate acyltransferase-1 is essential in liver for the metabolism of excess acyl-CoAs. J. Biol. Chem. 2005;280:25629–25636. doi: 10.1074/jbc.M503181200. [DOI] [PubMed] [Google Scholar]