Abstract
Whereas short-term regulation of insulin biosynthesis at the level of translation is well accepted, glucose-dependent transcriptional control is still believed to be a long-term effect occurring after more than 2 hr of glucose stimulation. Because pancreatic β cells are exposed to elevated glucose levels for minutes rather than hours after food uptake, we hypothesized the existence of a short-term transcriptional control. By studying the dynamics of newly synthesized (prepro)insulin RNA and by employing on-line monitoring of gene expression in single, insulin-producing cells, we were able to provide convincing evidence that insulin gene transcription indeed is affected by glucose within minutes. Exposure of insulinoma cells and isolated pancreatic islets to elevated glucose for only 15 min resulted in a 2- to 5-fold elevation in (prepro)insulin mRNA levels within 60–90 min. Similarly, insulin promoter-driven green fluorescent protein expression in single insulin-producing cells was significantly enhanced after transient glucose stimulation. Thus, short-term signaling, such as that involved in insulin secretion, also may regulate insulin gene transcription.
Insulin is of vital importance in maintaining glucose homeostasis in mammals. This, and its unique role as the only anabolic peptide hormone, necessitates strict regulation and fast-acting mechanisms guaranteeing efficient insulin biosynthesis and secretion. Although pancreatic beta cells secrete only part of their stored hormone in response to elevated glucose concentrations, biosynthesis of (prepro)insulin (PPI) is started immediately to rapidly replenish the insulin stores. It is commonly believed that glucose exhibits its immediate, or short-term, effect on insulin biosynthesis at the posttranscriptional and translational levels rather than at the level of transcription. Elevated glucose concentrations have been shown to enhance the stability of PPI mRNA (1), translation initiation (2), and translation elongation (2, 3). The effect of glucose on the generation of the PPI mRNA is commonly accepted to be a long-term effect, occurring after more than 2 hr of exposure to elevated glucose levels (4). To this end, the molecular mechanisms underlying glucose-stimulated insulin gene transcription mainly have been studied after incubation with high glucose concentrations over several hours or even days (5–15). According to this view, mechanisms regulating PPI gene transcription are dissociated from the immediate glucose effect and from short-term signaling associated with the regulation of translation or insulin secretion. However, from the physiological point of view pancreatic beta cells are exposed to elevated glucose levels for minutes rather than hours after food-uptake. Therefore, we hypothesized the existence of a short-term control, allowing glucose to exert physiological regulation at the level of insulin gene transcription.
The aim of the present study was to evaluate whether glucose stimulation for only 15 min, thus mimicking the physiological situation, affects insulin gene transcription. By employing the nuclear run-off technique to study insulin gene transcription initiation, RNase-protection analysis and comparative reverse transcription–PCR (RT-PCR) to determine endogenous PPI mRNA levels, together with the methodology of on-line monitoring of insulin gene promoter-driven green fluorescent protein (GFP) expression in isolated rat pancreatic islets, islet cells, and the hamster insulinoma-derived cell line HIT-T15 (16), we were able to demonstrate convincingly the existence of short-term transcriptional control of the insulin gene regulated by glucose.
MATERIALS AND METHODS
Expression Constructs.
prIns1GFP was generated by subcloning the rat insulin I promoter sequence (−411 bp to +1 bp) into p0GFP (provided by K. Michelsen, Karolinska Institute, Sweden). pRcCMVGFP was provided by M. Vogel and H. H. Niller, University Regensburg, Germany. Plasmid pc-fosGFP was constructed by exchanging the chloramphenicol acetyltransferase versus the GFP gene in pFC700 (17). pFC700 was kindly provided by J. P. Loeffler (Institut de Physiologie et de Chimie Biologique, Strasbourg, France). pTREGFP was generated by subcloning the GFP cDNA downstream the Tet-On promoter of pTRE (Invitrogen).
Cell Culture and Transfection.
HIT-T15 cells were obtained from ATCC (Manassas, VA). HIT cells were reported to show glucose responsiveness at subphysiological concentrations, i.e., between 0.1 and 2 mM of the sugar (14). HIT cells were grown in RPMI 1640 medium supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mM glutamine, and 10% fetal calf serum at 5% CO2 and 37°C. Pancreatic islets were isolated from normally fed male Wistar rats (150–200 g body weight) by collagenase digestion. Isolated islets and cells of disaggregated islets were incubated overnight at 5% CO2 and 37°C in RPMI 1640 medium containing 5.6 mM glucose and supplemented as described above.
Transfection of HIT-T15 and islet cells was performed overnight by the lipofectamine technique in RPMI 1640 medium containing 11.1 or 5.6 mM glucose, respectively, without serum and antibiotics. After transfection, HIT cells were split and cultured for a further 16 hr on 24-mm glass coverslips in RPMI 1640 medium containing 0.1 mM glucose, 10% fetal calf serum, 100 μg/ml streptomycin, and 100 units penicillin, at 5% CO2 and 37°C.
Cells were stimulated for 15 min with 16.7 mM glucose in RPMI 1640 medium, supplemented as above, or for 15 min with 100 ng/ml PMA in supplemented RPMI 1640 medium, when studying c-fos promoter-driven GFP expression.
To test GFP as a quantitative measure we employed the Tet-On system (Invitrogen). pTREGFP and pTet-On were cotransfected into HIT cells in a ratio of 1:1. After transfection, HIT cells were split and cultured on glass coverslips as described above in supplemented RPMI 1640 medium. Quantitative induction of GFP expression was performed by addition of doxycycline to the medium for 24 hr to final concentrations of 0, 0.25, 0.5, 1.0, 2.0, and 2,000 ng/ml, according to the instruction manual.
Nuclear Run-Off Analysis.
HIT cells were preincubated overnight at substimulatory glucose concentrations (0.1 mM) in RPMI 1640 medium supplemented as above. After stimulation with 16.7 mM glucose for 15 min, cells were washed and incubated further, until isolation of nuclei in supplemented RPMI 1640 medium, containing 0.1 mM glucose and 10% fetal calf serum. Nuclei were isolated and run-off reactions were performed as described (18). Labeled RNA was hybridized to 2.5 μg cDNA of insulin, β-actin, and control pBluescript DNA, which were immobilized on nitrocellulose filters. Hybridization was performed as described (18), with an equal number of cpm (2.5 × 106 cpm) to each sample from all experimental conditions. After hybridization, the excess of RNA probe was digested by treatment with RNase A as described (18). Filters were dried and analyzed by phosphorimaging. Values obtained for PPI mRNA were normalized by β-actin-mRNA values.
Quantification of PPI mRNA Amounts.
Levels of PPI mRNA were analyzed by RNase-protection analysis and comparative RT-PCR. For RNase-protection analysis radiolabeled cRNA were generated on the respective linearized cDNA-containing plasmids by employing the SP6/T7 in vitro transcription kit (Boehringer Mannheim) and [α-32P]CTP. After purification by polyacrylamide gel electrophoresis (6% acrylamide/7 M urea in 1× TBE), equal cpm of the labeled cRNA probes (8 × 104 cpm/μl, final activity) were mixed with the total RNA in hybridization solution, incubated for 5 min at 90°C, and hybridized at 45°C overnight. RNase protection was performed by using the RPA II kit (Ambion). Quantification of protected complexes was performed by phosphorimaging. Values obtained for PPI mRNA were normalized by β-actin-mRNA values. Levels of PPI mRNA in HIT cells were analyzed by RT-PCR using primers 5′-AGAAGCCATCAGCAAGCAGG-3′ and 5′-GGTGCAGCACTGATCCACAA-3′ and primers 5′-TGCCCAGGCTTTTGTCAAAC-3′ and 5′-CTCCAGTGCCAAGGTCTGAA-3′ for PPI mRNA in rat pancreatic islets. Levels of β-actin mRNA were analyzed by using primers 5′-AACTGGAACGGTGAAGGCGA-3′ and 5′-AACGGTCTCACGTCAGTGTA-3′. Total RNA obtained from either 106 HIT cells or 10 pancreatic islets was reverse-transcribed by using moloney murine leukemia virus revertase. Aliquots of the generated cDNA were used for PCR-mediated amplification using the RT-PCR Kit (Stratagene) and [α-32P]dCTP. PCR conditions were chosen that guaranteed the amplification of insulin and actin fragments within the linear range (Fig. 1C). PCR was performed in an AutogeneII thermocycler (Grant, U.K.) using a linked program (1 cycle: 5 min at 94°C, 5 min at 54°C, and 2 min at 72°C, and 25 cycles: 1 min at 94°C, 2 min at 54°C, and 2 min at 72°C). Labeled PCR products were separated on a 6% polyacrylamide sequencing gel and analyzed by phosphorimaging. Quantification was performed with tina 2.07d software (Raytest), using coamplified RT-PCR products for β-actin as the internal standard.
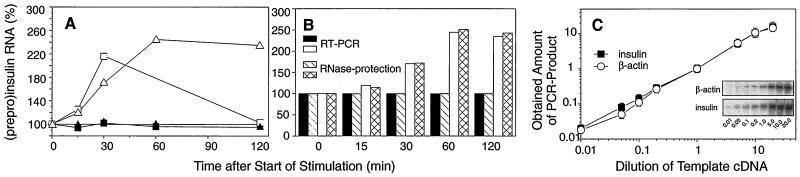
Figure 1.
(A) Dynamics of transcription initiation (squares) and cytoplasmic PPI mRNA amounts (triangles) in response to short-term glucose stimulation in HIT cells. Transcription initiation was studied by nuclear run-off analysis in nuclei of nonstimulated cells (■) cultured at 0.1 mM glucose and cells stimulated for 15 min with 16.7 mM glucose (□). Amounts of cytoplasmic PPI mRNA were determined by RNase-protection analysis. The mRNA levels of nonstimulated cells are shown as ▴ and of stimulated cells as ▵. The values of PPI mRNA were normalized to amounts of β-actin mRNA. Elevation of RNA levels in stimulated cells is shown as percentage of RNA levels of the nonstimulated control (given as 100%). Data are from a representative experiment. This experiment has been performed three times with similar results. Maximum elevation of transcription initiation at 30 min varied from 2.1- to 2.7-fold; maximum elevation of cytoplasmic PPI mRNA at 60 min varied from 1.9- to 2.8-fold. (B) Dynamics of cytoplasmic PPI mRNA levels obtained from the run-off experiment were measured by RNase-protection analysis and by comparative RT-PCR. mRNA levels of nonstimulated cells are shown as hatched bars (RNase-protection) and solid bars (RT-PCR). mRNA levels of stimulated cells are presented as cross-hatched bars (RNase-protection) and open bars (RT-PCR). Values of PPI mRNA were normalized to amounts of β-actin mRNA. Elevation of PPI mRNA levels in stimulated cells is presented as percentage of mRNA levels of the nonstimulated control (100%). (C) To prove that the amplification of both the PPI-mRNA-derived product (■) as well as the β-actin mRNA-product (○) remained in the linear range of PCR, the following dilutions of the template cDNA were used (0.01, 0.05, 0.1, 0.2, 1.0, 5.0, 10.0, and 20.0). Template cDNA was prepared from nonstimulated islets cultured at 5.6 mM glucose. The working dilution used for mRNA quantification is presented as 1.0. Shown values represent the average values of three independent experiments in arbitrary units. Inset shows data from a representative RT-PCR, performed three times.
On-Line Monitoring of Insulin Promoter-Driven GFP Expression in HIT-T15 and Pancreatic Islet Cells by Digital Imaging Fluorescence Microscopy and Laser-Scanning Confocal Microscopy.
For digital imaging fluorescence microscopy cells were grown on 24-mm glass coverslips. After glucose stimulation the coverslips were placed into a perifusion chamber, which was mounted on an inverted microscope (Zeiss Axiovert 135TV). Temperature was kept at 37°C. The objective lens used was a Zeiss Plan-NEOFLUAR ×25/0.8 Imm Korr. Fluorescence imaging was performed with a cooled charge-coupled device camera (CH250 with KAF 1400, Photometrics, Tucson, AZ) connected to an imaging system (Inovision, Durham, NC) with fluorescence excitation from a SPEX fluorolog-2 CM1T11I spectrofluorimeter (Spex Industries, Edison, NJ). The autofluorescence was minimized by using the following wavelength settings: excitation at 485 nm, a 505-nm dichroic mirror, and an emission band-pass filter of 500–530 nm. Cells were exposed every 20 min to the excitation light for 1 min, and fluorescence was measured by digital imaging. The position of cells on the coverslip was checked by overlaying the fluorescence and phase-contrast images. Fluorescence intensity was calculated by using the isee software for UNIX (Inovision). Laser-scanning confocal microscopy was performed by using the Leica CLSM (Leica Lasertechnik GmbH, Heidelberg, Germany). Cells were prepared and treated as described for fluorescence digital imaging. For the confocal microscope the following settings were used: ×40/1.30 oil Leitz Fluotar objective lens, excitation wavelength 488 nm (argon/krypton laser), and a long-pass 515-nm emission filter.
RESULTS
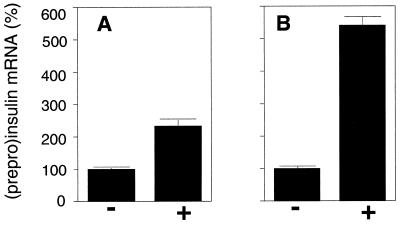
When studying the effect of short-term glucose stimulation on insulin gene expression, two major processes have to be taken into account, namely the renewed synthesis of PPI mRNA and the degradation of PPI mRNA. The impact of both mechanisms dictates the steady-state levels of PPI mRNA. To analyze the effect of short-term glucose stimulation on the renewed synthesis of PPI mRNA we performed nuclear run-off analysis. For this purpose, insulin-producing HIT-T15 cells were precultured overnight at substimulatory glucose concentrations (0.1 mM glucose), stimulated for 15 min with 16.7 mM glucose, and cultured further at 0.1 mM glucose until harvest of cells and isolation of nuclei, i.e., 0, 15, 30, and 120 min after start of stimulation. As shown in Fig. 1A, elevated levels of insulin gene transcripts were seen as early as 15 min after start of glucose stimulation. Transcription activity was maximal at 30 min, but was markedly decreased at 120 min after start of glucose stimulation. To study steady-state PPI mRNA levels under the same conditions, we prepared cytoplasmic mRNA from the samples of the same experiment and quantified PPI mRNA levels by two independent methods, RNase-protection analysis and comparative RT-PCR (Fig. 1B). The latter approach was established to analyze mRNA levels in very limited amounts of tissue, such as pancreatic islets and islet cells. Therefore, PCR conditions were chosen in which the amplification of both the PPI-mRNA-derived product as well as the β-actin mRNA-product (control) remained in the linear range of the PCR (Fig. 1C). As shown in Fig. 1B, both methods revealed almost identical results and demonstrated that in the cytoplasm PPI mRNA levels also increased in response to glucose as early as 15 min after start of stimulation and reached a maximum, i.e., a more than 2-fold elevation, at 60 min. When islets had been preincubated at substimulatory glucose concentrations, i.e., 5.6 mM, stimulation for only 15 min with 16.7 mM glucose resulted in a more than 5-fold increase in the endogenous PPI mRNA level at 60 min after start of stimulation (Fig. 2).
Figure 2.
Elevation of endogenous PPI mRNA levels after glucose stimulation in HIT-T15 cells (A) and in isolated rat pancreatic islets (B). RNA was prepared 90 min and 60 min after start of glucose stimulation of HIT cells and pancreatic islets, respectively. Amounts of PPI and β-actin mRNA were determined by RNase-protection analysis. The values of PPI mRNA were normalized to amounts of β-actin mRNA. Elevation of endogenous PPI mRNA levels in stimulated cells (+) is presented as percentage of mRNA levels of the nonstimulated control (−), the average value of which is given as 100%. All data are shown as mean values ± SD (n = 3).
To our surprise PPI mRNA levels in HIT cells started to decrease 120 min after start of glucose stimulation and reached almost base-line level at 180 min (Fig. 3A). Because these data implied an increase in degradation of PPI mRNA rather than a stabilization in response to the short-term glucose stimulus, we next analyzed the effect of the glucose stimulus on the stability of both total, i.e., preexisting plus newly synthesized, and preexisting PPI mRNA pools.
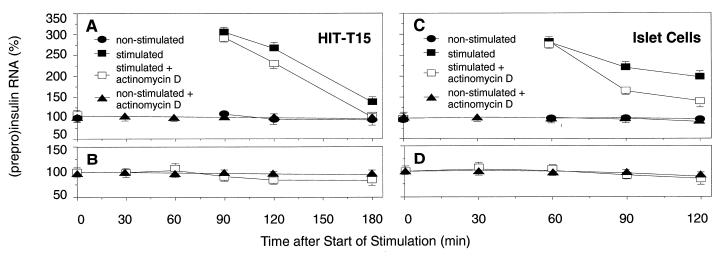
Figure 3.
The effect of short-term glucose stimulation on PPI mRNA stability in HIT-T15 cells (A and B) and pancreatic islet cells (C and D). (A and C) Analysis of the stability of the total PPI mRNA pool. (B and D) Analysis of the stability of the preexisting PPI mRNA pool. Values of PPI mRNA were normalized to amounts of β-actin mRNA. Elevation of RNA levels in stimulated cells is shown as percentage of RNA levels of the nonstimulated control, the average value of which is given as 100%. Data are shown as mean values ± SD (n = 3).
To study the effect of short-term glucose stimulus on the total PPI mRNA pool, we stimulated HIT cells and cultured islet cells for 15 min with 16.7 mM glucose, incubated the cells further at substimulatory glucose levels, 0.1 mM for HIT cells, and 5.6 mM for islet cells, to allow enforced insulin gene transcription, mRNA processing, and nuclear–cytoplasmic transport. Transcription was stopped 90 min after start of stimulation in HIT cells and 60 min after start of stimulation in pancreatic islet cells, by adding 5 μg/ml actinomycin D to the culture medium until harvest of cells. We analyzed PPI mRNA levels in HIT cells 0, 90, 120, and 180 min (Fig. 3A) and in islet cells 0, 60, 90, and 120 min (Fig. 3C) after start of stimulation. In both insulinoma HIT cells and in isolated pancreatic islet cells, the total PPI mRNA pool was degraded faster after glucose stimulation when renewed mRNA synthesis was abolished by actinomycin D.
To study the effect of the glucose stimulus on the stability of the preexisting PPI mRNA pool, HIT cells and pancreatic islets cells were preincubated with actinomycin D for 30 min before the 15-min glucose stimulation and throughout stimulation. Results from these experiments (Fig. 3 B and D) show very little changes in the stability of the preexisting PPI mRNA. These data led us to suggest that short-term glucose stimulation preferentially initiates degradation of newly synthesized PPI mRNA compared with preexisting mRNA.
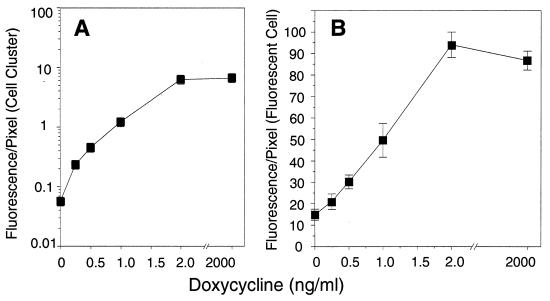
To prove that short-term, stimulated insulin promoter-driven transcription can be monitored at the single-cell level, we fused the rat insulin gene I promoter to the cDNA coding for the “humanized” form of the GFP mutant S65T (19) and measured on-line glucose-stimulated GFP expression in transfected HIT and islet cells by either digital imaging fluorescence microscopy or confocal microscopy. The use of GFP as a reporter gene to monitor gene expression was suggested by Chalfie et al. (20), and the promoter of the rat insulin I gene has been shown to contain glucose-stimulatory elements (5–7, 10). To show that GFP can be used as a quantitative measure we employed the well accepted Tet-On system. Therefore, the GFP gene was expressed under control of the inducible Tet-On promoter in transfected HIT cells and GFP expression was measured by digital imaging fluorescence microscopy. Using increasing amounts of doxycycline (0–2,000 ng/ml) as the inducer, GFP expression was elevated more than 100-fold in the whole population of transfected cells, i.e., when measured as “fluorescence/pixel of cell cluster” (Fig. 4A), where the “cell cluster” reflects the sum of both GFP-expressing and nonexpressing cells, and more than 4-fold when measured as “fluorescence/pixel of GFP-expressing cell” (Fig. 4B).
Figure 4.
On-line monitoring of Tet-On promoter-driven GFP expression in HIT-T15 cells. GFP fluorescence of three independent series consisting of 20 individual HIT cell clusters was monitored by digital imaging fluorescence microscopy. Fluorescence was measured as “fluorescence/pixel of cell cluster,” where “cell cluster” reflects the sum of both GFP-expressing and nonexpressing cells (A) and as “fluorescence/pixel of GFP-expressing cell” (B). All data are shown as mean values ± SD.
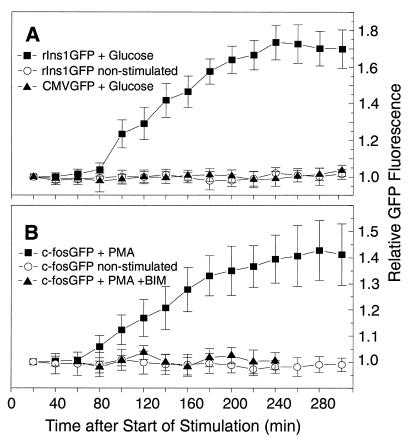
Because we expected the insulin promoter to respond to the glucose signal with similar dynamics as those reported for promoters of immediate early genes, we compared the glucose-stimulated expression of the insulin promoter-driven GFP gene (rIns1GFP) with the response of the c-fos promoter to phorbol 12-myristate 13-acetate (PMA) stimulation (c-fosGFP). The formation of the fluorophore of the GFPS65T protein has been reported to take at least 60 min after translation of the GFP mRNA (19). Considering this and the time necessary for transcription initiation, 10–120 min after start of stimulation (ref. 26 and our results), transcription elongation, transcript processing, nuclear–cytoplasmic transport, and translation, we expected a significant increase in GFP fluorescence between 80 and 120 min after start of stimulation. It is noteworthy that GFP expression by individual cells within a transfected cell population is heterogeneous, probably because of different copy numbers of GFP-expressing plasmids in different cells as a result of plasmid transfection. Therefore, we took the intensity of GFP fluorescence obtained at 20 min for each cell studied as the basal value 1.0, to which all other values were related. By employing digital imaging fluorescence microscopy, we were able to show that both glucose-stimulated insulin promoter-driven as well as PMA-stimulated c-fos promoter-driven GFP expression exhibit similar, if not identical, dynamics (Fig. 5 A and B), with an increased GFP fluorescence 80–100 min after start of stimulation. The specificity of the glucose effect on insulin promoter-driven GFP expression was shown by the observation that high glucose did not elevate GFP expression when driven by the human cytomegalovirus (CMV) promoter (Fig. 5A). An unspecific effect of PMA on fluorescence levels can be ruled out because PMA stimulation in combination with protein kinase C inhibitor bisindolylmaleimide failed to increase GFP fluorescence (Fig. 5B).
Figure 5.
On-line monitoring of insulin promoter-driven GFP expression (A) and c-fos promoter-driven GFP expression (B) in HIT-T15. On-line monitoring was started 20 min after the start of stimulation with glucose for rIns1GFP and with PMA for c-fosGFP. The 20-min value of GFP fluorescence was taken as 1.0. (A) On-line monitoring of individual HIT cells (n = 7) transfected with prIns1GFP and stimulated with 16.7 mM glucose for 15 min is shown as ■. Nonstimulated cells transfected with prIns1GFP (n = 10) are presented as ○. Cells transfected with pCMVGFP and stimulated with 16.7 mM glucose for 15 min are shown as ▴ (n = 10). (B) On-line monitoring of HIT cells transfected with pc-fosGFP. Cells stimulated for 15 min with 100 ng/ml PMA are shown as ■ (n = 7), and nonstimulated transfected cells are represented as ○ (n = 10). Transfected cells pretreated with 150 nM bisindolylmaleimide (BIM) and stimulated with PMA, shown as ▴ (n = 10), were monitored from minute 60 to 240 after start of stimulation. All data are shown as mean values ± SD.
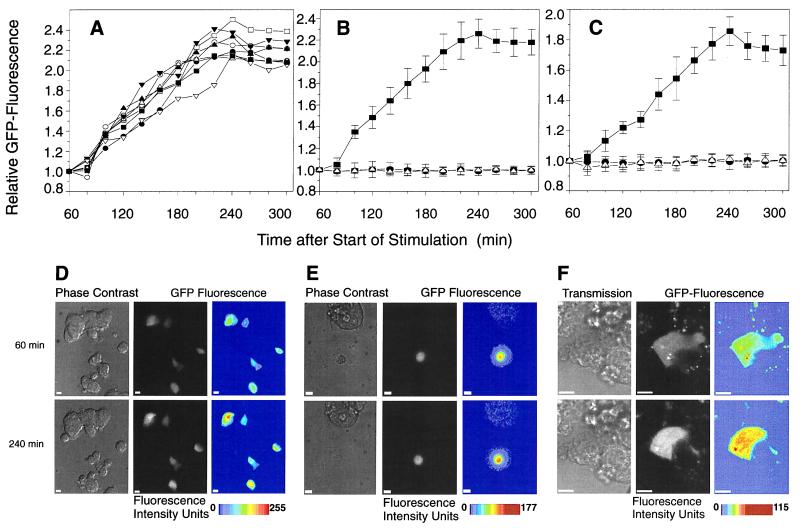
Both islet cells and HIT cells transfected with the insulin promoter-driven GFP construct (rIns1GFP) showed a similar increase in GFP fluorescence after the 15-min stimulation with 16.7 mM glucose and after preincubation at nonstimulatory glucose concentrations, i.e., 0.1 mM for HIT cells (Fig. 6 A and B) and 5.6 mM for islet cells (Fig. 6C). Again, the specificity of the glucose effect on insulin promoter-driven GFP expression was shown by the observation that high glucose did not elevate GFP expression when driven by the human CMV promoter (Fig. 6 B and C). Data obtained by either digital imaging or by confocal microscopy are shown in Fig. 6 A–E and F, respectively. Taken together, our results clearly demonstrate that the effect of glucose on insulin gene transcription has taken place within minutes rather than within hours and that glucose-stimulated insulin gene transcription reflects the transcriptional dynamics of an immediate early gene.
Figure 6.
On-line monitoring of insulin promoter-driven GFP expression in HIT-T15 cells and pancreatic islet cells. (A) On-line monitoring of eight individual HIT cells transfected with prIns1GFP and stimulated with 16.7 mM glucose for 15 min. (B) On-line monitoring of HIT cells transfected with prIns1GFP (■ and ▵) or pRcCMVGFP (•). Cells transfected with prIns1GFP (■, n = 8, summarized data from Fig. 2A) and with pRcCMVGFP (•, n = 10) were stimulated with 16.7 mM glucose. Nonstimulated cells transfected with prIns1GFP (n = 10) are shown as ▵. All data are shown as mean values ± SD. (C) On-line monitoring of pancreatic islet cells transfected with prIns1GFP (■ and ▵) or pRcCMVGFP (•). Cells transfected with prIns1GFP (■, n = 7) and with pRcCMVGFP (•, n = 10) were stimulated with 16.7 mM glucose. Nonstimulated cells transfected with prIns1GFP (n = 7) are shown as ▵. All data are shown as mean values ± SD. (D–F) Representative images (out of a total of 50) of cells are shown 60 and 240 min after the start of glucose stimulation. Images were obtained either by digital imaging fluorescence microscopy from transfected HIT (D) and primary islet cells (E) or from transfected HIT cells by laser-scanning confocal microscopy (F). Fluorescence images are shown as “gray scale” and “pseudo-color.” The “pseudo-color” images were created by converting the original “gray scale” data using tina 2.07d software (Raytest); the fluorescence signal increased from blue to red. The monitored cell cluster also is shown as a transmission/phase-contrast image (confocal microscopy/digital imaging, respectively). The scale bars represent 10 μm.
DISCUSSION
In the present study we show that stimulation of insulin-producing cells with high glucose concentrations for only 15 min results in transient elevation of insulin gene transcription, as verified by measurements of nuclear run-off, endogenous PPI mRNA levels, and insulin promoter-driven GFP expression. That insulin gene transcription can be triggered by glucose stimulation after incubation at a glucose concentration that is very close to euglycemic conditions, i.e., 5.6 mM glucose for isolated islets and islet cells (Figs. 2 and 3), makes it unlikely that the observed effect is solely a result of a recovery from hypoglycemic-to-normoglycemic conditions as has been suggested in refs. 21 and 22.
The possible existence of a short-term effect of glucose on insulin gene transcription was suggested indirectly, but as yet never clearly shown, by earlier findings (23–25). The strongest support comes from the findings of Efrat et al. (26), which clearly demonstrate that increased levels of nuclear PPI hn/mRNA can be detected in βTC-3 cells as early as 10 min after glucose stimulation. Similar dynamics obtained in the present study, when analyzing transcription initiation in HIT cells after a 15-min glucose stimulus on top of a 0.1 mM glucose background, support these earlier findings. These data clearly demonstrate a short-term regulation of insulin gene transcription by glucose. This new concept also is supported by the finding that increased binding of the pancreatic and duodenal homeobox factor 1, PDX-1 (27), to the insulin promoter occurs within 30 min of glucose stimulation (28). PDX-1 is identical or homologous to transcription factors described as IPF-1 (29), STF-1 (30), IDX-1 (31), GSF (32), and similar or identical to IUF-1 (33) and GSTF (34). Furthermore, PDX-1 is thought to be one of the transcription factors involved in glucose-dependent transcriptional control of PPI gene expression (9, 11, 28, 35, 36). The binding dynamics described for PDX-1, after glucose stimulation, are in agreement with data showing that maximal activity of insulin transcription initiation is reached within 30 min of glucose stimulation (ref. 26 and our data). Long-term exposure of pancreatic beta cells to high glucose levels, on the other hand, has been shown to lead to a desensitization of insulin-producing cells to the glucose signal, also at the level of transcription (34, 35). However, taking the relatively long half-life of the PPI mRNA into account, we expected a more than 2.5-fold elevation in transcription initiation. One possible explanation for this result would be that the actual half-life of PPI mRNA is much shorter than that obtained in actinomycin D experiments. This could be the case if an active PPI mRNA-degrading process is blocked by actinomycin D. A more likely explanation is that the run-off technique, which relies on the incorporation of radiolabeled nucleotides during the “run-off reaction” of initiated transcripts, is not quantitative when it comes to very short transcription units, such as that of the insulin gene. The stimulus may enhance both transcription initiation and transcript elongation, which here would result in an increased accumulation of “nearly finished” transcripts, which do allow only very little incorporation of radiolabeled nucleotides during the run-off reaction and thus would reflect transcription initiation in a distorted, nonquantitative manner.
Surprisingly, short-term glucose stimulation led to a destabilization rather than stabilization of the PPI mRNA pool. Our data are based on measurements of PPI mRNA steady-state levels and on analysis of PPI mRNA stability employing actinomycin D. The data obtained led us to suggest that preferentially newly synthesized PPI mRNA is subjected to degradation, whereas the stability of the preexisting PPI mRNA remains unchanged. Very recently, Wang et al. (37) showed in an elegant way that glucose stimulates splicing of PPI II pre-mRNA and the occurrence of the matured transcript in the cytoplasm within minutes in mouse islets. This observation in combination with our data on insulin gene transcription and the dynamics of cytoplasmic PPI mRNA steady-state levels suggest a highly dynamic turnover of PPI mRNA.
In the present study we demonstrated that insulin gene transcription is initiated within minutes of glucose stimulation, suggesting that short-term rather than long-term glucose signaling triggers the transient elevation of cytoplasmic steady-state PPI mRNA levels. Hence, an elevation in the glucose concentration not only triggers translation of preexisting PPI mRNA, but in addition immediately enhances transcription of the insulin gene. This would allow the highly dynamic control of insulin biosynthesis at both the levels of transcription and translation in response to rapidly changing blood–glucose concentrations and would guarantee the appropriate level of insulin secretion to maintain glucose homeostasis. A further consequence of short-term rather than long-term control of PPI gene transcription is the possibility of the shared utilization of the signal-transduction pathway(s) employed by glucose. Whereas a long-term effect of glucose should uncouple insulin transcription from insulin secretion, a short-term regulation implies that PPI transcription, PPI mRNA translation, and insulin exocytosis can be mediated by the same messenger(s). This implies that mechanisms leading to impaired insulin secretion, as observed in cases of non-insulin-dependent diabetes mellitus, also impair insulin biosynthesis at the level of transcription, accelerating the pathophysiological processes associated with the disease.
Acknowledgments
We thank Jude T. Deeney for helpful discussions and comments on the manuscript. This work was supported by funds from the Karolinska Institute and by grants from the Swedish Diabetes Association, the Tore Nilsons Stiftelse för Medicinsk Forskning, Swedish Medical Research Council (03X-09890, 03XS-12708, 19X-00034, 03X-12549), The Nordic Insulin Foundation, Berth von Kantzows Foundation, and Juvenile Diabetes Foundation International (JDFI). B.L. is supported by a Postdoctoral Fellowship Award from JDFI.
ABBREVIATIONS
- PPI
(prepro)insulin
- RT-PCR
reverse transcription–PCR
- GFP
green fluorescent protein
- PMA
phorbol 12-myristate 13-acetate
- CMV
cytomegalovirus
References
- 1.Welsh M, Nielsen D A, MacKrell A J, Steiner D F. J Biol Chem. 1985;250:13590–13594. [PubMed] [Google Scholar]
- 2.Welsh M, Scherberg N, Gilmore R, Steiner D F. Biochem J. 1986;235:459–467. doi: 10.1042/bj2350459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilligan M, Welsh G I, Flynn A, Bujalska I, Diggle T A, Denton R M, Proud C G, Docherty K. J Biol Chem. 1996;271:2121–2125. doi: 10.1074/jbc.271.4.2121. [DOI] [PubMed] [Google Scholar]
- 4.Bailyes E M, Guest P C, Hutton J C. In: Insulin: Molecular Biology to Pathology. Ashcroft F M, Ashcroft S J H, editors. Oxford: IRL; 1992. pp. 64–92. [Google Scholar]
- 5.Docherty K, Clark A. FASEB J. 1994;8:20–27. doi: 10.1096/fasebj.8.1.8299887. [DOI] [PubMed] [Google Scholar]
- 6.Stein R. Trends Endocrinol Metab. 1993;4:96–101. doi: 10.1016/1043-2760(93)90086-t. [DOI] [PubMed] [Google Scholar]
- 7.German M S, Moss L G, Rutter W J. J Biol Chem. 1990;265:22063–22066. [PubMed] [Google Scholar]
- 8.Goodison S, Kenna S, Ashcroft S J H. Biochem J. 1992;285:563–568. doi: 10.1042/bj2850563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melloul D, Ben-Neriah Y, Cerasi E. Proc Natl Acad Sci USA. 1993;90:3865–3869. doi: 10.1073/pnas.90.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.German M S, Wang J. Mol Cell Biol. 1994;14:4067–4075. doi: 10.1128/mcb.14.6.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen H V, Serup P, Leonard J, Michelsen B K, Madsen O D. Proc Natl Acad Sci USA. 1994;91:10465–10469. doi: 10.1073/pnas.91.22.10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma A, Stein R. Mol Cell Biol. 1994;14:871–879. doi: 10.1128/mcb.14.2.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redmon J B, Towle H C, Robertson R P. Diabetes. 1994;43:546–551. doi: 10.2337/diab.43.4.546. [DOI] [PubMed] [Google Scholar]
- 14.Sharma A, Fusco-DeMane D, Henderson E, Efrat S, Stein R. Mol Endocrinol. 1995;9:1468–1476. doi: 10.1210/mend.9.11.8584024. [DOI] [PubMed] [Google Scholar]
- 15.Odagiri H, Wang J, German M S. J Biol Chem. 1996;271:1909–1915. doi: 10.1074/jbc.271.4.1909. [DOI] [PubMed] [Google Scholar]
- 16.Santerre R F, Cook R A, Crisel R M, Sharp J D, Schmidt R J, Williams D C, Wilson C P. Proc Natl Acad Sci USA. 1981;78:4339–4343. doi: 10.1073/pnas.78.7.4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisch T M, Prywes R, Simon M C, Roeder R G. Genes Dev. 1989;3:198–211. doi: 10.1101/gad.3.2.198. [DOI] [PubMed] [Google Scholar]
- 18.Greenberg, M. E. & Bender, T. P. (1997) in Current Protocols in Molecular Biology, Unit 4.10. [DOI] [PubMed]
- 19.Heim R, Cubitt A B, Tsien R Y. Nature (London) 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 20.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 21.Giddings S J, Carnaghi L R, Shalwitz R A. Am J Physiol. 1993;265:E259–E266. doi: 10.1152/ajpendo.1993.265.2.E259. [DOI] [PubMed] [Google Scholar]
- 22.Philippe J, Pacheco I, Meda P. Diabetes. 1994;43:523–528. doi: 10.2337/diab.43.4.523. [DOI] [PubMed] [Google Scholar]
- 23.Permutt M A, Kipnis D M. J Biol Chem. 1972;247:1200–1207. [PubMed] [Google Scholar]
- 24.Okamoto H. Mol Cell Biochem. 1981;37:43–61. doi: 10.1007/BF02355886. [DOI] [PubMed] [Google Scholar]
- 25.Koh G, Seino Y, Takeda J, Fukumoto H, Kurose T, Tsuji K, Tsuda K, Taminato K, Imura H. Endocrinology. 1989;124:707–711. doi: 10.1210/endo-124-2-707. [DOI] [PubMed] [Google Scholar]
- 26.Efrat S, Surana M, Fleischer N. J Biol Chem. 1991;266:11141–11143. [PubMed] [Google Scholar]
- 27.Offield M F, Jetton T L, Labosky P A, Ray M, Stein R W, Magnuson M A, Hogan B L, Wright C V. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 28.MacFarlane W M, Read M L, Gilligan M, Bujalska I, Docherty K. Biochem J. 1994;303:625–631. doi: 10.1042/bj3030625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohlsson H, Karlsson K, Edlund T. EMBO J. 1993;12:1777–1788. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leonard J, Peers B, Johnson T, Ferreri K, Lee S, Montminy M R. Mol Endocrinol. 1993;7:1275–1283. doi: 10.1210/mend.7.10.7505393. [DOI] [PubMed] [Google Scholar]
- 31.Miller C P, McGehee R E, Jr, Habener J F. EMBO J. 1994;13:1145–1156. doi: 10.1002/j.1460-2075.1994.tb06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshak S, Totary H, Cerasi E, Melloul D. Proc Natl Acad Sci USA. 1996;93:15057–15062. doi: 10.1073/pnas.93.26.15057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boam D S, Docherty K. Biochem J. 1989;264:233–239. doi: 10.1042/bj2640233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson L K, Redmon J B, Towle H C, Robertson R P. J Clin Invest. 1993;92:514–519. doi: 10.1172/JCI116596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olson L K, Sharma A, Peshavaria M, Wright C V, Towle H C, Robertson R P, Stein R. Proc Natl Acad Sci USA. 1995;92:9127–9131. doi: 10.1073/pnas.92.20.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoffers D A, Thomas M K, Habener J F. Trends Endocrinol Metab. 1997;8:145–151. doi: 10.1016/s1043-2760(97)00008-8. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Shen L, Najafi H, Kolberg J, Matschinsky F M, Ureda M, German M. Proc Natl Acad Sci USA. 1997;94:4360–4365. doi: 10.1073/pnas.94.9.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]