Abstract
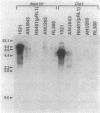
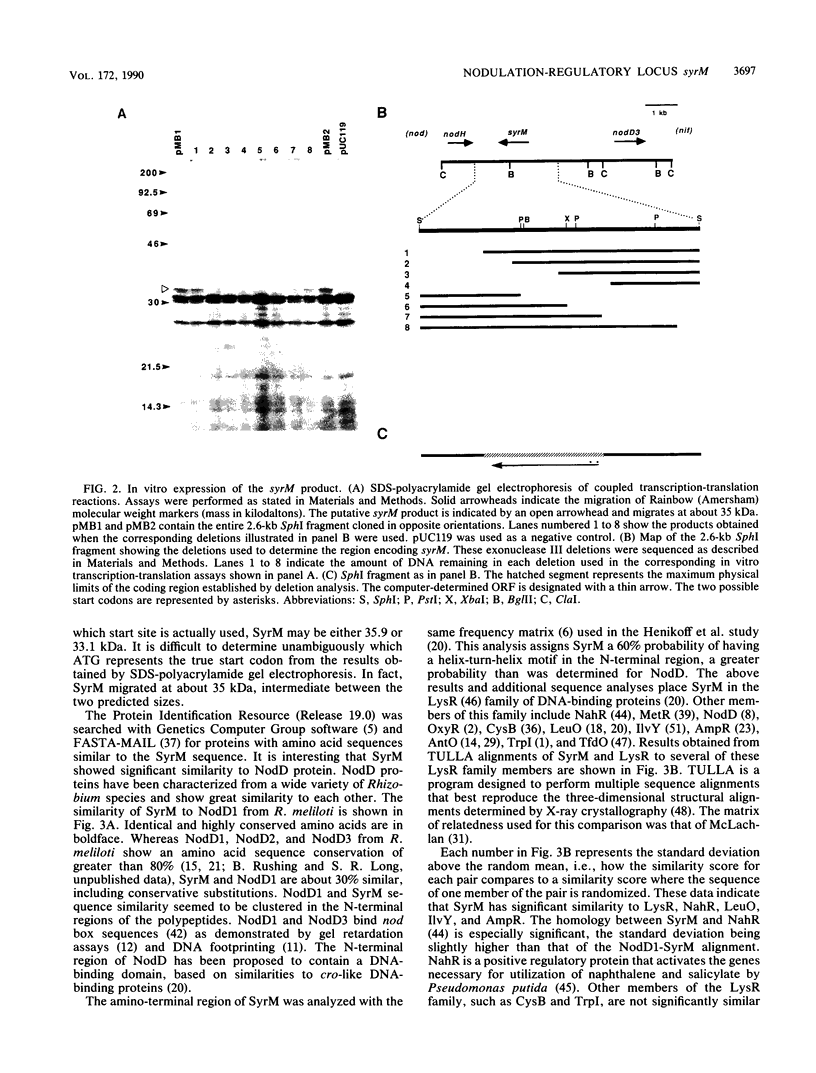
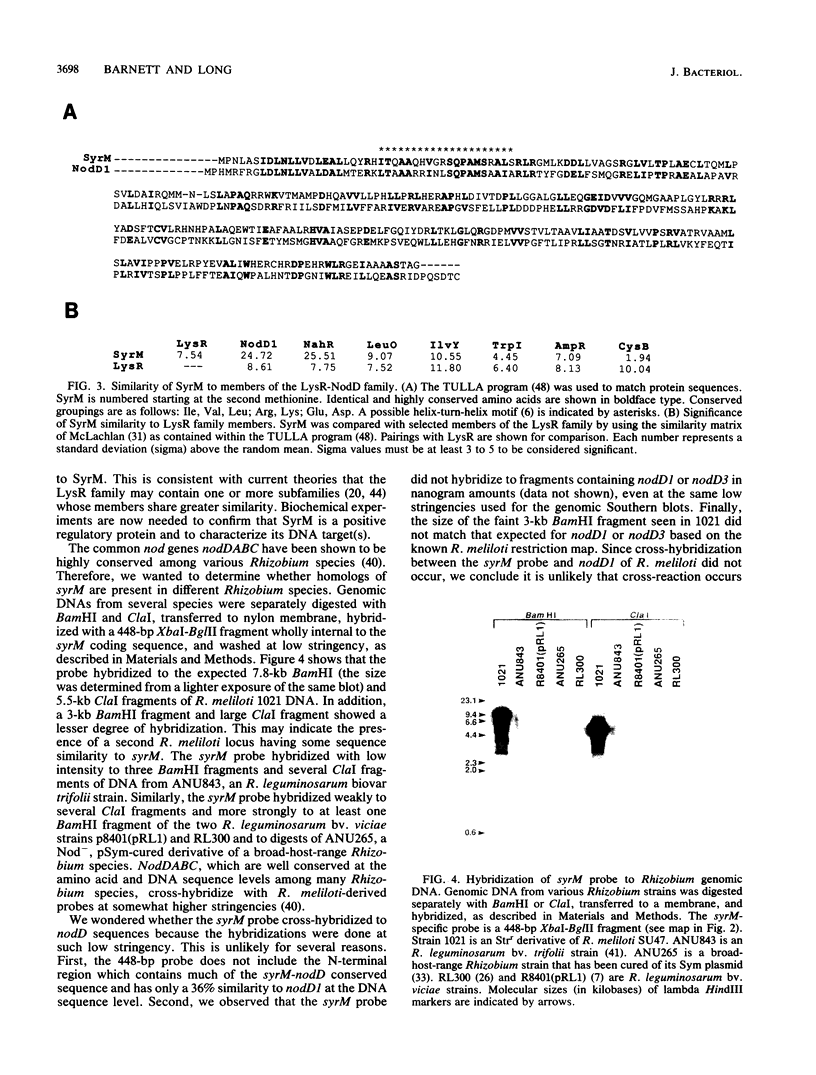
Rhizobium meliloti nodulation (nod) genes are expressed when activated by trans-acting proteins in the NodD family. The nodD1 and nodD2 gene products activate nod promoters when cells are exposed to plant-synthesized signal molecules. Alternatively, the same nod promoters are activated by the nodD3 gene when nodD3 is carried in trans along with a closely linked global regulatory locus, syrM (symbiotic regulator) (J. T. Mulligan and S. R. Long, Genetics 122:7-18, 1989). In this article we report the nucleotide sequence of a 2.6-kilobase SphI fragment from R. meliloti SU47 containing syrM. Expression from this locus was confirmed by using in vitro transcription-translation assays. The open reading frame encoded a protein of either 33 or 36 kilodaltons whose sequence shows similarity to NodD regulatory proteins.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chang M., Hadero A., Crawford I. P. Sequence of the Pseudomonas aeruginosa trpI activator gene and relatedness of trpI to other procaryotic regulatory genes. J Bacteriol. 1989 Jan;171(1):172–183. doi: 10.1128/jb.171.1.172-183.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman M. F., Storz G., Ames B. N. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci U S A. 1989 May;86(10):3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debellé F., Maillet F., Vasse J., Rosenberg C., de Billy F., Truchet G., Dénarié J., Ausubel F. M. Interference between Rhizobium meliloti and Rhizobium trifolii nodulation genes: genetic basis of R. meliloti dominance. J Bacteriol. 1988 Dec;170(12):5718–5727. doi: 10.1128/jb.170.12.5718-5727.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Systematic method for the detection of potential lambda Cro-like DNA-binding regions in proteins. J Mol Biol. 1987 Apr 5;194(3):557–564. doi: 10.1016/0022-2836(87)90681-4. [DOI] [PubMed] [Google Scholar]
- Egelhoff T. T., Fisher R. F., Jacobs T. W., Mulligan J. T., Long S. R. Nucleotide sequence of Rhizobium meliloti 1021 nodulation genes: nodD is read divergently from nodABC. DNA. 1985 Jun;4(3):241–248. doi: 10.1089/dna.1985.4.241. [DOI] [PubMed] [Google Scholar]
- Faucher C., Maillet F., Vasse J., Rosenberg C., van Brussel A. A., Truchet G., Dénarié J. Rhizobium meliloti host range nodH gene determines production of an alfalfa-specific extracellular signal. J Bacteriol. 1988 Dec;170(12):5489–5499. doi: 10.1128/jb.170.12.5489-5499.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fisher R. F., Egelhoff T. T., Mulligan J. T., Long S. R. Specific binding of proteins from Rhizobium meliloti cell-free extracts containing NodD to DNA sequences upstream of inducible nodulation genes. Genes Dev. 1988 Mar;2(3):282–293. doi: 10.1101/gad.2.3.282. [DOI] [PubMed] [Google Scholar]
- Fisher R. F., Long S. R. DNA footprint analysis of the transcriptional activator proteins NodD1 and NodD3 on inducible nod gene promoters. J Bacteriol. 1989 Oct;171(10):5492–5502. doi: 10.1128/jb.171.10.5492-5502.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. F., Swanson J. A., Mulligan J. T., Long S. R. Extended Region of Nodulation Genes in Rhizobium meliloti 1021. II. Nucleotide Sequence, Transcription Start Sites and Protein Products. Genetics. 1987 Oct;117(2):191–201. doi: 10.1093/genetics/117.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg E. B., Arbel T., Chen J., Karpel R., Mackie G. A., Schuldiner S., Padan E. Characterization of a Na+/H+ antiporter gene of Escherichia coli. Proc Natl Acad Sci U S A. 1987 May;84(9):2615–2619. doi: 10.1073/pnas.84.9.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttfert M., Horvath B., Kondorosi E., Putnoky P., Rodriguez-Quiñones F., Kondorosi A. At least two nodD genes are necessary for efficient nodulation of alfalfa by Rhizobium meliloti. J Mol Biol. 1986 Oct 5;191(3):411–420. doi: 10.1016/0022-2836(86)90136-1. [DOI] [PubMed] [Google Scholar]
- Haughn G. W., Wessler S. R., Gemmill R. M., Calvo J. M. High A + T content conserved in DNA sequences upstream of leuABCD in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1986 Jun;166(3):1113–1117. doi: 10.1128/jb.166.3.1113-1117.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Haughn G. W., Calvo J. M., Wallace J. C. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Honma M. A., Asomaning M., Ausubel F. M. Rhizobium meliloti nodD genes mediate host-specific activation of nodABC. J Bacteriol. 1990 Feb;172(2):901–911. doi: 10.1128/jb.172.2.901-911.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma M. A., Ausubel F. M. Rhizobium meliloti has three functional copies of the nodD symbiotic regulatory gene. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8558–8562. doi: 10.1073/pnas.84.23.8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honoré N., Nicolas M. H., Cole S. T. Inducible cephalosporinase production in clinical isolates of Enterobacter cloacae is controlled by a regulatory gene that has been deleted from Escherichia coli. EMBO J. 1986 Dec 20;5(13):3709–3714. doi: 10.1002/j.1460-2075.1986.tb04704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath B., Kondorosi E., John M., Schmidt J., Török I., Györgypal Z., Barabas I., Wieneke U., Schell J., Kondorosi A. Organization, structure and symbiotic function of Rhizobium meliloti nodulation genes determining host specificity for alfalfa. Cell. 1986 Aug 1;46(3):335–343. doi: 10.1016/0092-8674(86)90654-9. [DOI] [PubMed] [Google Scholar]
- Jacobs T. W., Egelhoff T. T., Long S. R. Physical and genetic map of a Rhizobium meliloti nodulation gene region and nucleotide sequence of nodC. J Bacteriol. 1985 May;162(2):469–476. doi: 10.1128/jb.162.2.469-476.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston A. W., Beringer J. E. Identification of the rhizobium strains in pea root nodules using genetic markers. J Gen Microbiol. 1975 Apr;87(2):343–350. doi: 10.1099/00221287-87-2-343. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Long S. R. Rhizobium-legume nodulation: life together in the underground. Cell. 1989 Jan 27;56(2):203–214. doi: 10.1016/0092-8674(89)90893-3. [DOI] [PubMed] [Google Scholar]
- Mackie G. A. Structure of the DNA distal to the gene for ribosomal protein S20 in Escherichia coli K12: presence of a strong terminator and an IS1 element. Nucleic Acids Res. 1986 Sep 11;14(17):6965–6981. doi: 10.1093/nar/14.17.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan A. D. Tests for comparing related amino-acid sequences. Cytochrome c and cytochrome c 551 . J Mol Biol. 1971 Oct 28;61(2):409–424. doi: 10.1016/0022-2836(71)90390-1. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison N. A., Hau C. Y., Trinick M. J., Shine J., Rolfe B. G. Heat curing of a sym plasmid in a fast-growing Rhizobium sp. that is able to nodulate legumes and the nonlegume Parasponia sp. J Bacteriol. 1983 Jan;153(1):527–531. doi: 10.1128/jb.153.1.527-531.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan J. T., Long S. R. A family of activator genes regulates expression of Rhizobium meliloti nodulation genes. Genetics. 1989 May;122(1):7–18. doi: 10.1093/genetics/122.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan J. T., Long S. R. Induction of Rhizobium meliloti nodC expression by plant exudate requires nodD. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6609–6613. doi: 10.1073/pnas.82.19.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski J., Jagura-Burdzy G., Kredich N. M. DNA sequences of the cysB regions of Salmonella typhimurium and Escherichia coli. J Biol Chem. 1987 May 5;262(13):5999–6005. [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann L. S., Stauffer G. V. Nucleotide sequence of the Salmonella typhimurium metR gene and the metR-metE control region. J Bacteriol. 1987 Sep;169(9):3932–3937. doi: 10.1128/jb.169.9.3932-3937.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostas K., Kondorosi E., Horvath B., Simoncsits A., Kondorosi A. Conservation of extended promoter regions of nodulation genes in Rhizobium. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1757–1761. doi: 10.1073/pnas.83.6.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A., Sukordhaman M. Evidence that the transcription activator encoded by the Pseudomonas putida nahR gene is evolutionarily related to the transcription activators encoded by the Rhizobium nodD genes. J Bacteriol. 1989 Apr;171(4):1952–1959. doi: 10.1128/jb.171.4.1952-1959.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A., Wender P. E. Identification of the nahR gene product and nucleotide sequences required for its activation of the sal operon. J Bacteriol. 1986 Apr;166(1):9–14. doi: 10.1128/jb.166.1.9-14.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P., Patte J. C. Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. III. Nucleotide sequence and regulation of the lysR gene. J Mol Biol. 1983 Aug 5;168(2):333–350. doi: 10.1016/s0022-2836(83)80022-9. [DOI] [PubMed] [Google Scholar]
- Streber W. R., Timmis K. N., Zenk M. H. Analysis, cloning, and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J Bacteriol. 1987 Jul;169(7):2950–2955. doi: 10.1128/jb.169.7.2950-2955.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbiah S., Harrison S. C. A method for multiple sequence alignment with gaps. J Mol Biol. 1989 Oct 20;209(4):539–548. doi: 10.1016/0022-2836(89)90592-5. [DOI] [PubMed] [Google Scholar]
- Swanson J. A., Tu J. K., Ogawa J., Sanga R., Fisher R. F., Long S. R. Extended Region of Nodulation Genes in Rhizobium meliloti 1021. I. Phenotypes of Tn5 Insertion Mutants. Genetics. 1987 Oct;117(2):181–189. doi: 10.1093/genetics/117.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wek R. C., Hatfield G. W. Nucleotide sequence and in vivo expression of the ilvY and ilvC genes in Escherichia coli K12. Transcription from divergent overlapping promoters. J Biol Chem. 1986 Feb 15;261(5):2441–2450. [PubMed] [Google Scholar]