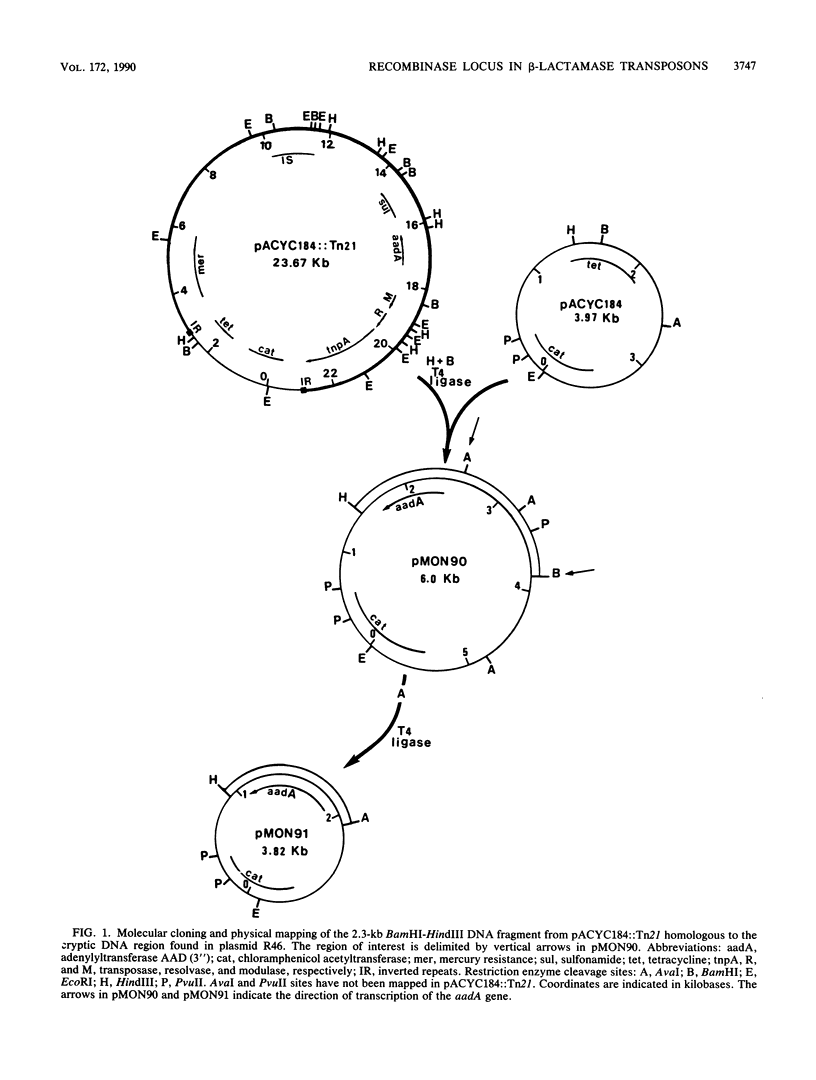
Abstract
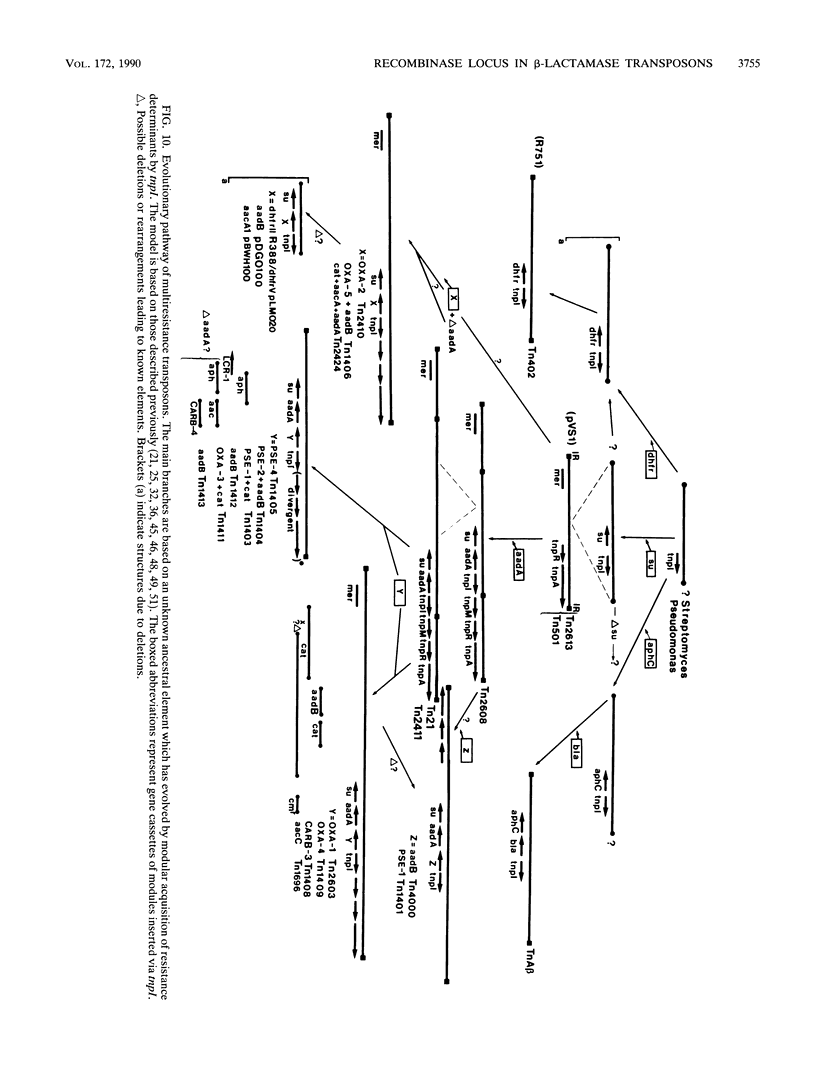
A novel discrete mobile DNA element from Tn21 from the plasmid R100.1 is described, and its mobilization function was confirmed experimentally. In addition, the element behaves as a recombinase-active locus (tnpI) which facilitates insertions of antibiotic resistance genes as modules or cassettes at defined hot spots or integration sites. A similar tnpI sequence was detected by DNA hybridization in a series of beta-lactamase transposons and plasmids and localized on their physical maps. The genetic function of the locus cloned from Tn21 into pACYC184 was tested for conduction and integration into the plasmids R388 and pOX38Km, and the results suggested recombinase-integrase activity and recA independence. DNA sequence analysis of the tnpI locus revealed no inverted or direct terminal repeats or transposition features of class I and class II transposons. The coding capacity revealed three putative open reading frames encoding 131, 134, and 337 amino acids. Orf3 encoded a putative polypeptide product of 337 amino acids that shared highly significant identity with the carboxyl region of integrase proteins. A comparison and an alignment of the tnpI locus from Tn21 and its flanking sequences identified similar sequences in plasmids and in transposons. The alignment revealed discrete nucleotide changes in these tnpI-like loci and a conserved 3' and 5' GTTA/G hot spot as a duplicated target site. Our data confirm the remarkable ubiquity of tnpI associated with antibiotic resistance genes. We present a model of transposon modular evolution into more complex multiresistant units via tnpI and site-specific insertions, deletions, and DNA rearrangements at this locus.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altenbuchner J., Schmid K., Schmitt R. Tn1721-encoded tetracycline resistance: mapping of structural and regulatory genes mediating resistance. J Bacteriol. 1983 Jan;153(1):116–123. doi: 10.1128/jb.153.1.116-123.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos P., Landy A., Abremski K., Egan J. B., Haggard-Ljungquist E., Hoess R. H., Kahn M. L., Kalionis B., Narayana S. V., Pierson L. S., 3rd The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986 Feb;5(2):433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett P. M., Grinsted J., Choi C. L., Richmond M. H. Characterisation of Tn501, a transposon determining resistance to mercuric ions. Mol Gen Genet. 1978 Feb 7;159(1):101–106. doi: 10.1007/BF00401753. [DOI] [PubMed] [Google Scholar]
- Boccard F., Smokvina T., Pernodet J. L., Friedmann A., Guérineau M. The integrated conjugative plasmid pSAM2 of Streptomyces ambofaciens is related to temperate bacteriophages. EMBO J. 1989 Mar;8(3):973–980. doi: 10.1002/j.1460-2075.1989.tb03460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot M., Mercier J., Levesque R. C. Development of natural and synthetic DNA probes for OXA-2 and TEM-1 beta-lactamases. Antimicrob Agents Chemother. 1987 May;31(5):728–734. doi: 10.1128/aac.31.5.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Cameron F. H., Groot Obbink D. J., Ackerman V. P., Hall R. M. Nucleotide sequence of the AAD(2'') aminoglycoside adenylyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538-1 and dhfrII in R388. Nucleic Acids Res. 1986 Nov 11;14(21):8625–8635. doi: 10.1093/nar/14.21.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M., Galas D. J. Cointegrate formation mediated by Tn9. II. Activity of IS1 is modulated by external DNA sequences. J Mol Biol. 1983 Oct 15;170(1):61–91. doi: 10.1016/s0022-2836(83)80227-7. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman C. J., Higgins C. F. Fimbrial phase variation in Escherichia coli: dependence on integration host factor and homologies with other site-specific recombinases. J Bacteriol. 1987 Aug;169(8):3840–3843. doi: 10.1128/jb.169.8.3840-3843.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R. M., Vockler C. The region of the IncN plasmid R46 coding for resistance to beta-lactam antibiotics, streptomycin/spectinomycin and sulphonamides is closely related to antibiotic resistance segments found in IncW plasmids and in Tn21-like transposons. Nucleic Acids Res. 1987 Sep 25;15(18):7491–7501. doi: 10.1093/nar/15.18.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch P. R., Wang C. L., Woodward M. J. Construction of a Tn5 derivative determining resistance to gentamicin and spectinomycin using a fragment cloned from R1033. Gene. 1986;48(2-3):203–209. doi: 10.1016/0378-1119(86)90078-8. [DOI] [PubMed] [Google Scholar]
- Humphreys G. O., Willshaw G. A., Anderson E. S. A simple method for the preparation of large quantities of pure plasmid DNA. Biochim Biophys Acta. 1975 Apr 2;383(4):457–463. doi: 10.1016/0005-2787(75)90318-4. [DOI] [PubMed] [Google Scholar]
- Huovinen P., Huovinen S., Jacoby G. A. Sequence of PSE-2 beta-lactamase. Antimicrob Agents Chemother. 1988 Jan;32(1):134–136. doi: 10.1128/aac.32.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y., Watson J. M., Haas D., Leisinger T. Genetic and molecular characterization of the Pseudomonas plasmid pVS1. Plasmid. 1984 May;11(3):206–220. doi: 10.1016/0147-619x(84)90027-1. [DOI] [PubMed] [Google Scholar]
- Kratz J., Schmidt F., Wiedemann B. Characterization of Tn2411 and Tn2410, two transposons derived from R-plasmid R1767 and related to Tn2603 and Tn21. J Bacteriol. 1983 Sep;155(3):1333–1342. doi: 10.1128/jb.155.3.1333-1342.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhstoss S., Richardson M. A., Rao R. N. Site-specific integration in Streptomyces ambofaciens: localization of integration functions in S. ambofaciens plasmid pSAM2. J Bacteriol. 1989 Jan;171(1):16–23. doi: 10.1128/jb.171.1.16-23.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafond M., Couture F., Vézina G., Levesque R. C. Evolutionary perspectives on multiresistance beta-lactamase transposons. J Bacteriol. 1989 Dec;171(12):6423–6429. doi: 10.1128/jb.171.12.6423-6429.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque R. C., Jacoby G. A. Molecular structure and interrelationships of multiresistance beta-lactamase transposons. Plasmid. 1988 Jan;19(1):21–29. doi: 10.1016/0147-619x(88)90059-5. [DOI] [PubMed] [Google Scholar]
- Levesque R. C., Medeiros A. A., Jacoby G. A. Molecular cloning and DNA homology of plasmid-mediated beta-lactamase genes. Mol Gen Genet. 1987 Feb;206(2):252–258. doi: 10.1007/BF00333581. [DOI] [PubMed] [Google Scholar]
- Levesque R., Roy P. H. Mapping of the plasmid (pLQ3) from Achromobacter and cloning of its cephalosporinase gene in Escherichia coli. Gene. 1982 Apr;18(1):69–75. doi: 10.1016/0378-1119(82)90057-9. [DOI] [PubMed] [Google Scholar]
- Macrina F. L., Kopecko D. J., Jones K. R., Ayers D. J., McCowen S. M. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid. 1978 Jun;1(3):417–420. doi: 10.1016/0147-619x(78)90056-2. [DOI] [PubMed] [Google Scholar]
- Mahillon J., Lereclus D. Structural and functional analysis of Tn4430: identification of an integrase-like protein involved in the co-integrate-resolution process. EMBO J. 1988 May;7(5):1515–1526. doi: 10.1002/j.1460-2075.1988.tb02971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E., de la Cruz F. Transposon Tn21 encodes a RecA-independent site-specific integration system. Mol Gen Genet. 1988 Feb;211(2):320–325. doi: 10.1007/BF00330610. [DOI] [PubMed] [Google Scholar]
- Meyer J. F., Nies B. A., Wiedemann B. Amikacin resistance mediated by multiresistance transposon Tn2424. J Bacteriol. 1983 Aug;155(2):755–760. doi: 10.1128/jb.155.2.755-760.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossakowska D., Ali N. A., Dale J. W. Oxacillin-hydrolysing beta-lactamases. A comparative analysis at nucleotide and amino acid sequence levels. Eur J Biochem. 1989 Mar 15;180(2):309–318. doi: 10.1111/j.1432-1033.1989.tb14649.x. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Nobuta K., Tolmasky M. E., Crosa L. M., Crosa J. H. Sequencing and expression of the 6'-N-acetyltransferase gene of transposon Tn1331 from Klebsiella pneumoniae. J Bacteriol. 1988 Aug;170(8):3769–3773. doi: 10.1128/jb.170.8.3769-3773.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nücken E. J., Henschke R. B., Schmidt F. R. Nucleotide sequence of an OXA-2 beta-lactamase gene from the R-plasmid R1767 derived plasmid pBP11 and comparison to closely related resistance determinants found in R46 and Tn2603. J Gen Microbiol. 1989 Apr;135(4):761–765. doi: 10.1099/00221287-135-4-761. [DOI] [PubMed] [Google Scholar]
- Olson E. R., Chung S. T. Transposon Tn4556 of Streptomyces fradiae: nucleotide sequence of the ends and the target sites. J Bacteriol. 1988 Apr;170(4):1955–1957. doi: 10.1128/jb.170.4.1955-1957.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette M., Bissonnette L., Roy P. H. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 beta-lactamase gene. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7378–7382. doi: 10.1073/pnas.84.21.7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Lanka E. Conservation of a common 'backbone' in the genetic organization of the IncP plasmids RP4 and R751. Nucleic Acids Res. 1987 Mar 11;15(5):2385–2385. doi: 10.1093/nar/15.5.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pargellis C. A., Nunes-Düby S. E., de Vargas L. M., Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988 Jun 5;263(16):7678–7685. [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A convenient and adaptable package of computer programs for DNA and protein sequence management, analysis and homology determination. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):643–655. doi: 10.1093/nar/12.1part2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M., Ekaterinaki N., Nimmo E., Sherratt D. Analysis of Tn7 transposition. Mol Gen Genet. 1986 Dec;205(3):550–556. doi: 10.1007/BF00338097. [DOI] [PubMed] [Google Scholar]
- Rubens C. E., McNeill W. F., Farrar W. E., Jr Transposable plasmid deoxyribonucleic acid sequence in Pseudomonas aeruginosa which mediates resistance to gentamicin and four other antimicrobial agents. J Bacteriol. 1979 Sep;139(3):877–882. doi: 10.1128/jb.139.3.877-882.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F. R., Nücken E. J., Henschke R. B. Structure and function of hot spots providing signals for site-directed specific recombination and gene expression in Tn21 transposons. Mol Microbiol. 1989 Nov;3(11):1545–1555. doi: 10.1111/j.1365-2958.1989.tb00140.x. [DOI] [PubMed] [Google Scholar]
- Schmidt F. R., Nücken E. J., Henschke R. B. Structure and function of hot spots providing signals for site-directed specific recombination and gene expression in Tn21 transposons. Mol Microbiol. 1989 Nov;3(11):1545–1555. doi: 10.1111/j.1365-2958.1989.tb00140.x. [DOI] [PubMed] [Google Scholar]
- Schmidt F., Klopfer-Kaul I. Evolutionary relationship between Tn21-like elements and pBP201, a plasmid from Klebsiella pneumoniae mediating resistance to gentamicin and eight other drugs. Mol Gen Genet. 1984;197(1):109–119. doi: 10.1007/BF00327930. [DOI] [PubMed] [Google Scholar]
- Stokes H. W., Hall R. M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989 Dec;3(12):1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- Sundström L., Rådström P., Swedberg G., Sköld O. Site-specific recombination promotes linkage between trimethoprim- and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol Gen Genet. 1988 Aug;213(2-3):191–201. doi: 10.1007/BF00339581. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Matsushita K., Yamamoto T. Genesis of a complex transposon encoding the OXA-1 (type II) beta-lactamase gene. Antimicrob Agents Chemother. 1985 Aug;28(2):227–234. doi: 10.1128/aac.28.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Yamamoto T., Sawai T. Evolution of complex resistance transposons from an ancestral mercury transposon. J Bacteriol. 1983 Mar;153(3):1432–1438. doi: 10.1128/jb.153.3.1432-1438.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F. C., Phillips K. L., Gilbert T., Lockhart P., O'Hara P. J., Plorde J. J. Development of a DNA probe from the deoxyribonucleotide sequence of a 3-N-aminoglycoside acetyltransferase [AAC(3)-I] resistance gene. Antimicrob Agents Chemother. 1989 Apr;33(4):551–559. doi: 10.1128/aac.33.4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. M., Grinsted J. Physical and genetic analysis of the Inc-W group plasmids R388, Sa, and R7K. Plasmid. 1982 May;7(3):239–250. doi: 10.1016/0147-619x(82)90005-1. [DOI] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlleben W., Arnold W., Bissonnette L., Pelletier A., Tanguay A., Roy P. H., Gamboa G. C., Barry G. F., Aubert E., Davies J. On the evolution of Tn21-like multiresistance transposons: sequence analysis of the gene (aacC1) for gentamicin acetyltransferase-3-I(AAC(3)-I), another member of the Tn21-based expression cassette. Mol Gen Genet. 1989 Jun;217(2-3):202–208. doi: 10.1007/BF02464882. [DOI] [PubMed] [Google Scholar]
- de la Cruz F., Grinsted J. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol. 1982 Jul;151(1):222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]