Abstract
A critical step in the signal-induced activation of the transcription factor NF-κB is the site-specific phosphorylation of its inhibitor, IκB, that targets the latter for degradation by the ubiquitin–proteasome pathway. We have previously shown that mitogen-activated protein kinase/ERK kinase kinase 1 (MEKK1) can induce both this site-specific phosphorylation of IκBα at Ser-32 and Ser-36 in vivo and the activity of a high molecular weight IκB kinase complex in vitro. Subsequently, others have identified two proteins, IκB kinase α (IKK-α) and IκB kinase β (IKK-β), that are present in a tumor necrosis factor α-inducible, high molecular weight IκB kinase complex. These kinases are believed to directly phosphorylate IκB based on the examination of the kinase activities of IKK immunoprecipitates, but more rigorous proof of this has yet to be demonstrated. We show herein that recombinant IKK-α and IKK-β can, in fact, directly phosphorylate IκBα at Ser-32 and Ser-36, as well as homologous residues in IκBβ in vitro, and thus are bona fide IκB kinases. We also show that MEKK1 can induce the activation of both IKK-α and IKK-β in vivo. Finally, we show that IKK-α is present in the MEKK1-inducible, high molecular weight IκB kinase complex and treatment of this complex with MEKK1 induces phosphorylation of IKK-α in vitro. We conclude that IKK-α and IKK-β can mediate the NF-κB-inducing activity of MEKK1.
The transcriptional activator protein NF-κB plays a critical role in immune and inflammatory responses (for recent reviews, see refs. 1 and 2). NF-κB is sequestered in the cytoplasm of most cell types by virtue of its association with the IκB family of inhibitor proteins, which include IκBα, IκBβ, and IκBɛ. Upon exposure to a wide variety of stimuli, including the proinflammatory cytokines tumor necrosis factor α (TNF-α) and interleukin 1, UV-irradiation, virus infection, bacterial lipopolysaccharide, and oxidative stress, the IκB protein is phosphorylated at its N terminus. In the case of IκBα, this occurs at Ser-32 and Ser-36; the corresponding residues in IκBβ are Ser-19 and Ser-23. This phosphorylation then targets IκB for degradation by the ubiquitin–proteasome pathway, thereby liberating NF-κB, which is then free to translocate to the nucleus and bind to DNA (3).
The mechanism by which the phosphorylation of the N terminus of IκB occurs has recently been the subject of intense investigation (4, 5). A large, approximately 700-kDa multiprotein complex isolated from uninduced HeLa cells was originally demonstrated to phosphorylate IκBα at Ser-32 and -36 (6). This complex could be activated in vitro by the mitogen-activated protein kinase kinase kinase (MAP3K) MEKK1, and the latter was shown to induce the site-specific phosphorylation of IκBα in vivo (7). A model was proposed in which MEKK1 activates IκBα kinase activity, which in turn leads to the phosphorylation of IκBα. Inherent in this model was the possibility that other MAP3Ks might also activate IκBα kinase activity (7).
Subsequently, the MAP3K NF-κB inducing kinase (NIK) was identified in a yeast two-hybrid screen based on its interaction with the TRAF2 component of the TNF-α receptor complex (8). The use of NIK as a bait in another yeast two-hybrid screen resulted in the cloning of the cDNA for IκB kinase α (IKK-α) (9). IKK-β was then identified in a DNA database search by virtue of its homology to IKK-α (10). IKK-α and IKK-β cDNAs were independently cloned by using peptide sequences obtained from purified proteins (11–13). These proteins were isolated from high molecular weight complexes purified from TNF-α-treated HeLa cells. Immunoprecipitates of both IKK-α and IKK-β can phosphorylate IκBα and IκBβ at the regulatory N-terminal Ser residues, and both IKKs can be activated by NIK (9–13). In addition, dominant negative versions of NIK, IKK-α, and IKK-β can inhibit TNF-α- and interleukin 1-induced NF-κB activation (8–13).
These recent reports raise two important issues. (i) MEKK1 and NIK are MAP3Ks that share significant homology in their catalytic domains (8, 14). Thus, one issue is whether MEKK1, like NIK, can activate IKK-α and/or IKK-β. (ii) Previous reports did not demonstrate whether IKK-α and IKK-β can directly phosphorylate IκB, because they used immunoprecipitates (as opposed to purified versions) of IKK-α and IKK-β to examine their kinase activity (9–13). Thus, IKK-α and IKK-β have not yet been shown to directly phosphorylate IκB. Herein, we show that MEKK1 can activate both IKK-α and IKK-β in vivo and that recombinant versions of the latter two proteins can directly phosphorylate IκBα and IκBβ in vitro. These findings lend support to a general model for NF-κB activation that involves the sequential involvement of a MAP3K, an IKK, and an IκB.
MATERIALS AND METHODS
Plasmids.
pcDNA3-FlagJNK1, pcDNA3-NIK and pcDNA3-FlagNIK, and pRK-FlagIKK-α and -β and their dominant negative mutants, were gifts from Roger Davis, David Wallach, and David Goeddel, respectively, and have been described (8–10, 15). pCMV4-IκBβ and mutants were gifts from Dean Ballard (16). pBS-IκBα (S32T/S36T) was constructed by overlapping PCR using pBS-IκBα as a template (17). The sources of pCMV5-MEKK1 (which encodes the C-terminal 672 residues of MEKK1), pcDNA3-FlagMEKK1Δ (which encodes the C-terminal 321 residues), (PRDII)2CAT, (PRDIV)6CAT, pCMV-lacZ, pBS-IκBα, pBS-IκBα (S32A), pBS-IκBα (S36A), pBS-IκBα (S32A/S36A), and pRSET-IκBα have been described (3, 7).
Tissue Culture and Transfection.
HeLa cells were maintained as described (7). Transfections, performed in 3.5-cm-diameter wells, were conducted as described (18). Protein concentration, chloramphenicol acetyltransferase (CAT), and β-galactosidase measurements on cellular extracts were performed as described (7).
Western Blotting.
Immunoprecipitates or aliquots of whole cell lysates were mixed with 2× SDS/PAGE loading buffer and then subjected to SDS/PAGE in 8% or 10% gels. Western blotting was then performed as described (7) by using anti-Flag, anti-JNK1 (C-17), or anti-IKK-α (H-744) polyclonal antibodies (Santa Cruz Biotechnology), anti-rabbit IgG-horseradish peroxidase conjugates (Santa Cruz Biotechnology), and Enhanced Chemiluminescence substrate (Amersham).
Preparation of Recombinant Proteins and Peptides.
pFastBacHT-IKK-α and pFastBacHT-IKK-β were constructed by subcloning the coding sequence fragments of pRK-FlagIKK-α or pRK-FlagIKK-β, respectively, into pFastBacHTb (Life Technologies). Recombinant bacmids and baculovirus were prepared according to the manufacturer’s instructions. (His)6IKK-α and (His)6IKK-β were isolated by using Ni-nitrilotriacetic acid agarose from baculovirus-infected Sf9 cells. (His)6IKK-β was further purified by applying it to a Mono Q column equilibrated with 25 mM Bis-Tris, pH 6.3/10% glycerol and eluting it with a 0–1 M NaCl gradient in the same buffer. The concentrations of (His)6IKK-α and (His)6IKK-β were determined by SDS/PAGE followed by staining with Coomassie blue or silver and comparison with BSA standards. (His)6MEKK1Δ was prepared as described (7).
pGEX-IκBα-(5–55), pGEX-IκBα-(5–55) (S32A/S36A), and pRSET-IκBα (S32A/S36A) were gifts from Jeremiah Hagler in the laboratory of T.M. These plasmids as well as pGEX-IκBα-(5–55) (S32A), pGEX-IκBα-(5–55) (S36A), and pGEX-IκBα-(5–55) (S32T/S36T) were constructed by subcloning restriction enzyme fragments from appropriate pBS-IκBα wild-type or mutant constructs into pGEX-5X-1 (Pharmacia) or pRSET (Invitrogen). pGEX-IκBβ-(1–46), pGEX-IκBβ-(1–46) (S19A), pGEX-IκBβ-(1–46) (S23A), pGEX-IκBβ-(1–46) (S19A/S23A), pGEX-IκBβ-(1–46) (S19T/S23T), and pRSET-IκBβ and pRSET-IκBβ (S19A/S23A) were constructed by subcloning restriction enzyme fragments from appropriate pCMV4-IκBβ wild-type or mutant constructs as above.
Glutathione S-transferase (GST) fusion proteins were purified from Escherichia coli HB101 transformed with the corresponding pGEX expression vectors, by using glutathione-agarose affinity chromatography (19). (His)6-tagged proteins were purified by using Ni-NTA agarose from E. coli BL21(DE3)LysS transformed with the corresponding pRSET expression vectors. The concentrations of the various IκB substrate proteins were determined by the Bradford method.
The following peptides were synthesized, isolated in greater than 95% purity (BioSynthesis), and dissolved in water: IKK-β-(171–187), KELDQGSLCTSFVGTLQ; IKK-β-(171–187) (S177A/S181A), KELDQGALCTAFVGTLQ.
Protein Kinase Assays.
When purified recombinant proteins were used as both enzyme and substrate, kinase assays were performed in 10 μl of buffer A (20 mM Hepes, pH 7.6/20 mM β-glycerophosphate/0.1 mM sodium orthovanadate/10 mM MgCl2/50 mM NaCl/1 mM DTT) containing 0.5 μg of IκBα or IκBα fusion protein substrate, 50 μM ATP, and 5 μCi of [γ-32P]ATP (1 Ci = 37 GBq), and conducted at 30°C for 30 min. Reaction products were subjected to SDS/PAGE in 9 or 10% gels. When peptide substrates were used, the assay conditions were as above, except that 2 μg of peptide was used, and reactions were performed for 60 min. Peptide reaction products were subjected to Tris-Tricine 10–20% gradient SDS/PAGE (Bio-Rad).
Immunocomplex kinase assays for IKK and c-Jun N-terminal kinase (JNK) were performed by the methods of DiDonato et al. (11) and Derijard et al. (15), respectively, with minor modifications. Whole cell extracts were prepared by lysing cells in buffer B (20 mM Tris⋅HCl, pH 7.6/150 mM NaCl/25 mM β-glycerophosphate/2 mM EDTA/2 mM pyrophosphate/1 mM sodium orthovanadate/10% glycerol/1% Triton X-100/1 mM DTT) containing leupeptin (10 μg/ml) and 1 mM phenylmethylsulfonyl fluoride. After centrifugation of the lysate at 16,000 × g for 10 min at 4°C, the supernatant was incubated with 10 μl of M2-agarose with end over end rotation for 1–2 hr at 4°C. For IKK assays, the resin was then washed once with buffer B, three times with buffer A containing 3 M urea, and then twice with buffer A. IKK reactions were initiated by the addition of 10 μl of buffer A containing 0.5 μg of GST-IκBα-(5–55), 200 μM ATP, and 5 μCi of [γ-32P]ATP and performed at 30°C for 15–45 min. For JNK assays, the resin was washed three times with buffer B then once with buffer A. JNK reactions were initiated by the addition of 10 μl of buffer A containing 0.36 μg of GST-cJun-(1–79) (GST fused to residues 1–79 of c-Jun; Santa Cruz Biotechnology), 50 μM ATP, and 5 μCi of [γ-32P]ATP, and performed at 30°C for 20 min. Kinase reaction products were subjected to SDS/PAGE in 8 or 10% gels. Kinase activities were quantified by using a phosphorimager.
IκBα mobility shift assays were performed as described (7), except that creatine phosphokinase and phosphocreatine were omitted.
Purification of IκBα Kinase Complex.
IκBα kinase complex was purified as described (6) with minor modifications. Cytoplasmic extracts (S100) from uninduced HeLa cells were subjected to precipitation with 30% ammonium sulfate, resuspended, dialyzed against buffer C (50 mM Tris⋅HCl, pH 7.5/5% glycerol/0.5 mM DTT/0.1 mM phenylmethylsulfonyl fluoride), and applied to a Mono Q HiTrap column (Pharmacia). The column was washed with buffer C containing 0.2 M KCl and material was eluted with buffer C containing 0.3 M KCl. Fractions containing IκBα kinase activity were then pooled, concentrated, and applied first to a Superdex-200 column (Pharmacia) equilibrated in buffer C containing 150 mM NaCl and then to a BioQ2 column (Bio-Rad), and material was eluted with a linear gradient of 150–405 mM NaCl in buffer C. Fractions containing activity were dialyzed against buffer D (20 mM Tris⋅HCl, pH 7.5/20 mM NaCl/50% glycerol/0.5 mM DTT/0.1 mM phenylmethylsulfonyl fluoride/leupeptin at 10 μg/ml).
IKK-α Immunoprecipitations.
IκBα kinase complex was incubated in 110 μl of buffer D containing 20 mM β-glycerolphosphate, 0.1 mM sodium orthovanadate, 2.5 μM okadaic acid, 10 mM MgCl2, 10 μM ATP, and 25 μCi of [γ-32P]ATP at 30°C for 30 min in the absence or presence of 40 ng of MEKK1Δ. Reactions were then incubated with either anti-Stat1 (E-23) or anti-IKK-α (H-744) polyclonal antibodies (Santa Cruz Biotechnology) prebound to protein G-agarose in 600 μl of buffer E (50 mM Tris⋅HCl, pH 8.0/0.5% sodium deoxycholate/1% Nonidet P-40/0.1% SDS) with rocking for 1 hr at 4°C. Resins were washed twice with buffer E, twice with buffer A containing 3 M urea, and then twice with buffer E. Immunoprecipitated material was eluted with 2× SDS loading buffer and subjected to SDS/PAGE in 8% gels. Reactions performed in the presence of [γ-32P]ATP were analyzed by autoradiography, and parallel reactions performed in its absence were analyzed by Western blotting.
RESULTS
Phosphorylation of IκBα and IκBβ by IKK-α and IKK-β in Vitro.
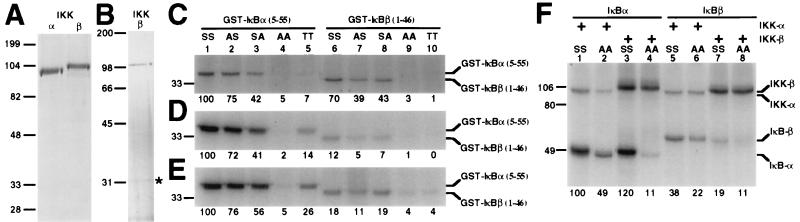
To examine whether IKK-α and IKK-β can directly phosphorylate the critical Ser residues at the N terminus of IκB, polyhistidine-tagged IKK-α and IKK-β were purified as recombinant proteins from baculovirus-infected insect cells by using Ni-agarose chromatography (Fig. 1A). When IKK-α (Fig. 1C) or IKK-β (Fig. 1D) were incubated with GST fused to residues 5–55 of IκBα [GST-IκBα-(5–55)] or residues 1–46 of IκBβ [GST-IκBβ-(1–46)] in the presence of [γ-32P]ATP, phosphorylation of the IκB fusion proteins was readily detected (Fig. 1 C and D, lanes 1 and 6). A double mutation of either the IκBα Ser-32 and -36 to Ala (lane 4) or the IκBβ Ser-19 and -23 to Ala (lane 9) virtually abolished phosphorylation, indicating that phosphorylation of the wild-type proteins occurred at these residues.
Figure 1.
Phosphorylation of IκBα and IκBβ by recombinant IKK-α and IKK-β. (His)6-tagged IKK-α (A) and IKK-β (A and B) were purified from baculovirus-infected Sf9 cells with Ni-NTA agarose chromatography (A) and further by Mono Q chromatography (B), and 500 (A) or 20 ng (B) was subjected to SDS/PAGE in 8 or 10% gels and stained with Coomassie blue (A) or silver (B). An asterisk in B denotes a polypeptide copurifying with IKK-β. (C–E) GST-IκBα-(5–55) (0.5 μg) or GST-IκBβ-(1–46) (0.5 μg) bearing the indicated residues was incubated in the presence of [γ-32P]ATP with recombinant IKK-α (10 ng) (C) or IKK-β (1 ng) (D and E) purified by Ni-agarose chromatography (C and D) or further by Mono Q chromatography (E). The first and second residues correspond to positions 32 or 36 of IκBα, respectively, or to positions 19 and 23 of IκBβ, respectively. Reaction products were subjected to SDS/PAGE in 10% gels and analyzed by autoradiography. (F) (His)6IκBα (0.5 μg), (His)6IκBα (S32A/S36A) (0.5 μg), (His)6IκBβ (0.5 μg), or (His)6IκBβ (S19A/S23A) (0.5 μg) was incubated in the presence of [γ-32P]ATP with 10 ng of recombinant IKK-α or IKK-β purified by Ni-agarose chromatography. Reaction products were subjected to SDS/PAGE in 9% gels and analyzed by autoradiography. The slower migration of wild-type as compared with S32A/S36A (His)6IκBα reflects phosphorylation at Ser-32 and -36 (6). (A–F) Molecular mass markers (in kDa) are indicated to the left, and the positions of IKK-α, IKK-β, and the IκBα and IκBβ substrates are indicated to the right. The relative levels of 32P incorporation into the IκB substrates are indicated below the gels.
To further define the targets of phosphorylation, IκBα and IκBβ fusion proteins bearing single point mutations at these residues were examined as substrates. Both IKK-α and IKK-β efficiently phosphorylated all of these substrates (Fig. 1 C and D, lanes 2, 3, 7, and 8), implying that both IKKs phosphorylate both Ser-32 and -36 of IκBα and Ser-19 and -23 of IκBβ. Mutation of these residues to Thr resulted in substrates that are poorer than the wild-type version (Fig. 1 C and D, lanes 5 and 10); overexpressed IκBαs bearing the same mutations at either site displayed impaired TNF-α-induced degradation in vivo (20). The marked preference reported for IKK-α for Ser-23 over Ser-19 of IκBβ (9) was not observed here (Fig. 1C, compare lanes 7 and 8). Whether the discrepancy in results is due to differences in the source of IKK-α [transfected mammalian 293 cells (9) vs. baculovirus-infected Sf9 cells (present report)] or differences in the substrates used [IκBβ-(1–311) vs. GST-IκBβ-(1–46)] is not clear.
In addition to the GST fusion proteins containing the N terminus of IκB, we also examined full-length IκBα and IκBβ as substrates. As with the GST fusion proteins, the full-length proteins were both substrates for both IKK-α and IKK-β (Fig. 1F, lower bands of odd numbered lanes). In contrast to the GST fusion proteins, however, double mutations of IκBα Ser-32 and -36 to Ala or IκBβ Ser-19 and -23 to Ala only partially diminished 32P incorporation into the substrates (lower bands of even numbered lanes). This result indicates that at least in vitro, IKK-α and IKK-β can phosphorylate IκB residues distinct from these N-terminal residues. Studies conducted with IKK-α immunoprecipitates suggest that additional IκBα phosphoacceptor sites reside between residues 263 and 315 (11). Both IKK-α and IKK-β displayed autophosphorylating activity (Fig. 1F, upper bands).
To address the possibility that the purified IKK-β is contaminated with an insect cell kinase activity, the recombinant IKK-β was further purified by Mono Q chromatography (Fig. 1B) and assayed for kinase activity (Fig. 1E). This IKK-β phosphorylated the wild-type and mutant IκBα and IκBβ fusion proteins with a substrate specificity essentially the same as that of the less-purified IKK-β (Fig. 1, compare D and E). A 30-kDa polypeptide still copurified with IKK-β (Fig. 1B, asterisk). However, assay of other fractions containing this polypeptide but not IKK-β revealed no detectable IκB kinase activity (data not shown). We conclude that the observed activity derives from the purified recombinant IKK-β.
Activation of IKK-α and IKK-β by MEKK1 in Vivo.
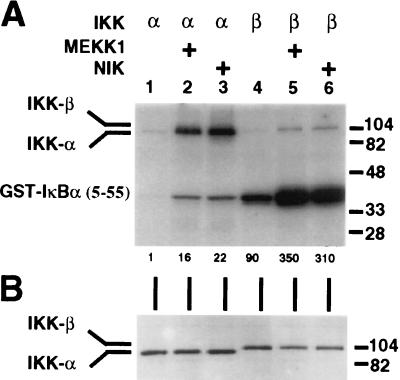
To address whether MEKK1 can activate either IKK-α or IKK-β activity, HeLa cells were cotransfected with expression vectors for either IKK-α or IKK-β and expression vectors for MEKK1, NIK, or expression vector alone. The IKK was then immunoprecipitated and assayed for kinase activity against GST-IκBα-(5–55). As shown in Fig. 2A, overexpressed and immunoprecipitated IKK-α displays barely detectable IκBα kinase activity under the conditions used (Fig. 2A, lane 1, lower band). This activity can be augmented by coexpression with NIK (Fig. 2A, lane 3) (9) and, importantly, also by coexpression with MEKK1 (Fig. 2A, lane 2). Overexpressed and immunoprecipitated IKK-β displays a substantially higher basal IκBα kinase activity (Fig. 2A, lane 4, lower band) relative to IKK-α, as noted by others (10, 12, 13). Significantly, this activity can also be augmented by coexpression with either MEKK1 or NIK (Fig. 2A, lanes 5 and 6). In all cases, IKK expression levels were not significantly affected by this coexpression (Fig. 2B). Immunoprecipitated IKK-α has relatively weak IκBα kinase activity with the GST-IκBα-(5–55) substrate, whereas immunoprecipitated IKK-β has relatively strong activity (Fig. 2A, compare lower bands of lanes 1 and 4). In addition, both immunoprecipitated proteins display autophosphorylating activity (Fig. 2A, upper bands).
Figure 2.
Activation of IKK-α and IKK-β by MEKK1 and NIK. HeLa cells were transfected with 1 μg of pRK-FlagIKK-α or 1 μg of pRK-FlagIKK-β and 6 μg of pCMV5-MEKK1, 6 μg of pcDNA3-NIK, or 6 μg of pcDNA3. Cells were harvested 42 hr after transfection, and IKK-α or IKK-β was immunoprecipitated with anti-Flag M2-agarose. (A) IKK immunocomplexes were assayed for kinase activity by incubation with 0.5 μg of GST-IκBα-(5–55) in the presence of [γ-32P]ATP. Reaction products were subjected to SDS/PAGE in 10% gels and analyzed by autoradiography. The relative level of 32P incorporation into the GST-IκBα-(5–55) substrate is indicated below the gel. (B) IKK immunocomplexes were subjected to SDS/PAGE in 8% gels, transferred to nitrocellulose membrane, and probed with anti-Flag polyclonal antibodies. (A and B) The positions of IKK-α, IKK-β, and GST-IκBα-(5–55) are indicated to the left, and molecular mass markers (in kDa) are indicated to the right.
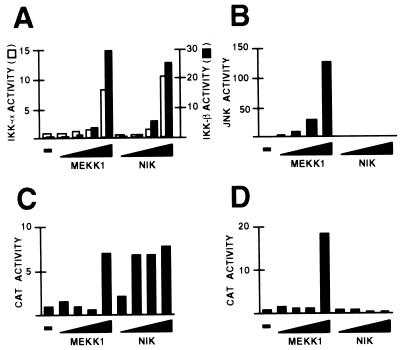
Stimuli such as proinflammatory cytokines and UV-irradiation not only activate the NF-κB pathway but also the JNK (also known as the stress-activated protein kinase) pathway (15, 21). Therefore, MEKK1 and NIK were further compared with respect to their activation of these pathways in HeLa cells. Two different assays were used. The first measured the kinase activity of coexpressed and immunoprecipitated IKK (as above) or JNK. The second measured the activity of a reporter gene for either the NF-κB or JNK pathways. The reporter gene for the NF-κB pathway is driven by two copies of the interferon β (IFN-β) enhancer PRDII element, which binds to NF-κB (22, 23). That for the JNK pathway is driven by six copies of the IFN-β enhancer PRDIV element, which binds to an ATF-2/c-Jun heterodimer (24); both ATF-2 and c-Jun contain transcriptional activation domains that can be phosphorylated and activated by the JNK pathway (25). As shown in Fig. 3A, MEKK1 and NIK display similar dose–response curves with respect to their activation of coexpressed IKK-α or IKK-β. However, NIK is a substantially more potent activator of the NF-κB reporter gene than MEKK1, with substantial activation seen with a NIK dose (50 ng) two orders of magnitude lower than that required to detect MEKK1-induced activation (5 μg) (Fig. 3C). We note, however, that increasing levels of reporter gene activity could be detected with intermediate levels of transfected MEKK1 DNA (between 0.5 and 5 μg; data not shown). NIK does not activate either coexpressed JNK or the JNK reporter gene, in contrast to MEKK1, which activates both (Fig. 3 B and D), consistent with previous reports (7, 26–28).
Figure 3.
Effects of MEKK1 and NIK on the NF-κB and JNK pathways. HeLa cells were transfected with 2 μg of pCMV-lacZ; 1 μg of pRK-FlagIKK-α (A), 1 μg of pRK-FlagIKK-β (A), 1 μg of pcDNA3-FlagJNK1 (B), 3 μg of (PRDII)2CAT (C), or 3 μg of (PRDIV)6CAT (D); and 5, 50, 500, or 5000 ng of either pCMV5-MEKK1 or pcDNA3-NIK. The total DNA dose was brought up to 8 (A and B) or 10 (C and D) μg with pcDNA3. Cells were harvested 40–42 hr after transfection. (A and B) Immunocomplex kinase assays. Whole cell extracts, the volumes of which were normalized for β-galactosidase activity, were subjected to immunoprecipitation with anti-Flag M2-agarose. Immunocomplexes were assayed for IKK (A) or JNK (B) activities by incubation with GST-IκBα-(5–55) or GST-cJun-(1–79), respectively, in the presence of [γ-32P]ATP. Western blots performed on aliquots of whole cell extracts revealed that comparable amounts of protein kinase were subjected to immunoprecipitation in each series of experiments (data not shown). Activities were normalized to that of IKK-α, IKK-β, or JNK alone, each of which was assigned a value of 1. (C and D) CAT assays. Activities were normalized to protein concentrations of cell extracts. Shown is a representative result from three experiments.
Inhibition of MEKK1-Induced NF-κB Activation in Vivo by Dominant Negative IKK-α and Dominant Negative IKK-β.
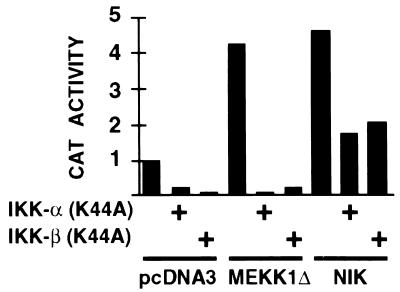
To examine whether IKK-α or IKK-β mediate MEKK1-induced activation of NF-κB, HeLa cells were cotransfected with the NF-κB reporter gene, expression vectors for MEKK1Δ (which contains the kinase domain of MEKK1), NIK, or the expression vector alone, and expression vectors for catalytically inactive IKK-α, IKK-β, or the expression vector alone. As shown in Fig. 4, both MEKK1Δ and NIK activate the NF-κB reporter gene, as expected. Under conditions where both dominant negative IKK-α and IKK-β inhibit NIK-induced activation of the NF-κB reporter gene, both mutants also potently inhibit MEKK1Δ-induced activation of this same reporter gene, providing further evidence that IKK-α and IKK-β can mediate MEKK1-induced NF-κB activation in vivo. Similar results were reported with 293 cells (29).
Figure 4.
Inhibition of MEKK1- and NIK-induced NF-κB activation by dominant negative IKK-α or dominant negative IKK-β. HeLa cells were transfected with 3 μg of (PRDII)2CAT; 4 μg of pcDNA3-FlagMEKK1Δ, 0.1 μg of pcDNA3-FlagNIK, or 4 μg of pcDNA3; and 4 μg of pRK-FlagIKK-α (K44A), pRK-FlagIKK-β (K44A), or pcDNA3. The total DNA dose was brought up to 11 μg with pcDNA3. Cells were harvested 40 hr after transfection. CAT activities were normalized to protein concentrations of cell extracts. Shown is a representative result from three experiments.
Induced Phosphorylation of IKK by MEKK1.
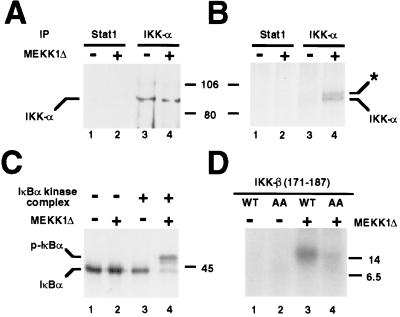
In previous studies, MEKK1 was shown to activate a 700-kDa IκB kinase complex isolated from uninduced cells (7), but these studies were carried out prior to the discovery of IKK-α and IKK-β. We therefore carried out Western blotting experiments with anti-IKK-α antibodies and found that IKK-α is indeed present in this complex (data not shown). In addition, the MEKK1-induced IκBα kinase activity could be immunoprecipitated with the anti-IKK-α antibodies (data not shown). Finally, a Western blot of the complex immunoprecipitated with anti-IKK-α antibodies revealed the presence of IKK-α (Fig. 5A, lane 3). To determine whether IKK-α in this complex is phosphorylated in response to MEKK1, the complex was incubated in the presence of [γ-32P]ATP with or without MEKK1Δ, subjected to immunoprecipitation with anti-IKK-α antibodies, and examined by autoradiography. In the absence of MEKK1Δ, there was marginal incorporation of 32P into IKK-α (Fig. 5B, lane 3). By contrast, in the presence of MEKK1Δ, this incorporation was markedly enhanced (lane 4). Importantly, the activity of the purified IκB kinase complex was dependent on MEKK1, as demonstrated by incubating the kinase complex with 35S-labeled Flag-IκBα in the absence or presence of MEKK1Δ and detecting the phosphorylation-dependent mobility shift of IκBα (Fig. 5C, compare lanes 3 and 4). This mobility shift was abolished by a S32A/S36A double mutation of IκBα (data not shown).
Figure 5.
MEKK1-induced phosphorylation of IKK. (A and B) IκBα kinase complex was incubated with or without 40 ng MEKK1Δ, in the absence (A) or presence (B) of [γ-32P]ATP. Reactions were then immunoprecipitated (IP) with either anti-Stat1 or anti-IKK-α antibodies and analyzed by Western blotting (A) using anti-IKK-α antibodies or by autoradiography (B). The position of IKK-α is as indicated and the position of a slower migrating species is denoted by an asterisk (see Results). (C) IκBα kinase complex was incubated with 35S-labeled in vitro-translated Flag-IκBα in the absence or presence of 40 ng MEKK1Δ. Reaction products were subjected to SDS/PAGE in 9% gels and detected by autoradiography. The positions of Flag-IκBα (IκBα) and phospho-Flag-IκBα (P-IκBα) are indicated to the left. (D) IKK-β-(171–187) (2 μg) or IKK-β-(171–187) (S177A/S181A) (2 μg) peptide was incubated with or without 100 ng of recombinant MEKK1Δ in the presence of [γ-32P]ATP. Reaction products were subjected to gradient SDS/PAGE in 10–20% gels and analyzed by autoradiography. The positions of molecular mass markers (in kDa) are indicated to the left (B) or right (A, C, and D).
An additional slower-migrating species that coimmunoprecipitates with IKK-α is detectable upon MEKK1Δ treatment (Fig. 5B, lane 4, asterisk). This species is most likely IKK-β because it has been previously shown to (i) display a slightly slower mobility than IKK-α by SDS/PAGE (10, 12, 13), (ii) copurify with IKK-α as a high molecular weight complex from TNF-α-treated HeLa cells (11–13), and (iii) interact with IKK-α in vitro and in vivo (10, 12, 13). Further studies are necessary to substantiate this proposal.
The experiment shown in Fig. 5B reveals MEKK1Δ-dependent 32P incorporation into IKK-α but does not assess the relative contributions to this by IKK-α autophosphorylation vs. phosphorylation by another kinase, such as MEKK1Δ. The latter is a distinct possibility, particularly because both IKK-α and IKK-β possess canonical MAP2K activation loop motifs (SXXXS) (13). Indeed, mutation of two IKK-β Ser residues (Ser-177 and Ser-181) in this motif to Glu results in a constitutively active kinase; conversely, their mutation to Ala results in a dominant negative inhibitor of NF-κB activation (13). To examine whether this activation loop might be directly phosphorylated by MEKK1, we synthesized a peptide that contains the activation loop of IKK-β, residues 171–187. As shown in Fig. 5D, when recombinant MEKK1Δ is incubated with this peptide in the presence of [γ-32P]ATP, phosphorylation of the peptide is observed (Fig. 5D, lane 3). Replacement of Ser-177 and Ser-181 with Ala substantially reduces phosphorylation (Fig. 5D, lane 4), indicating that phosphorylation of the wild-type peptide has occurred at either or both of these residues. Ser-176, the homologue of IKK-β Ser-177 in IKK-α, has been proposed to be the major site of IKK-α phosphorylation by NIK (30).
DISCUSSION
Recombinant IKK-α and IKK-β were purified from baculovirus-infected insect cells (Fig. 1 A and B). Both proteins directly phosphorylate Ser-32 and -36 of IκBα and Ser-19 and -23 of IκBβ in vitro and are, therefore, bona fide IκB kinases. That being said, given the homology between the NF-κB pathway in mammals and the Dif immune response pathway in Drosophila (31), it is conceivable that in vivo these overexpressed IKKs (and by analogy, the insect cell IKK homologues) might exist as high molecular weight complexes in insect cells, just as they do in mammalian cells (11–13). Furthermore, association with other proteins might be necessary to achieve activation. Although the data imply that IKK-α/IKK-β heterodimerization is not obligatory for activity, it is conceivable that heterodimerization might modulate IKK-α or IKK-β activity and/or substrate specificity.
Recombinant IKK-α and IKK-β purified from baculovirus-infected Sf9 cells are active enzymes (Fig. 1). One possible reason for this is that overexpression of these kinases in insect cells, just as with the overexpression of certain kinases in mammalian cells, results in their constitutive activation. This, in turn, could be due to trans- or autophosphorylation or, perhaps, protein expression levels that exceed the capacity of endogenous negative regulatory factors. Another possibility is that the baculovirus infection itself might result in IKK activation, because virus infection is a well known activator of NF-κB in mammalian cells (22, 23, 32).
NIK is substantially more potent than MEKK1 in activating an NF-κB reporter gene (Fig. 3C), as observed (28). However, NIK and MEKK1 are equally effective in activating coexpressed IKK-α or IKK-β (Fig. 3A). The reason for this discrepancy is presently not clear. One possibility is that NIK might possess NF-κB inducing activities distinct from its IKK-inducing activity. However, it should be noted that catalytically inactive NIK does not activate NF-κB (8), implying that if this is indeed the case, then NIK phosphorylates a substrate in addition to IKK in the NF-κB pathway. Another possibility is that overexpressed IKK might respond less sensitively to coexpressed NIK (or MEKK1) than endogenous IKK. In support of the latter possibility, we find that overexpressed IKK-α or IKK-β can inhibit NIK- or MEKK1-induced activation of an NF-κB reporter gene (data not shown). Hence, overexpression of IKK itself might perturb the NF-κB signaling pathway.
These findings suggest limitations to experiments that examine the relative potency of overexpressed kinases such as NIK and MEKK1 in activating particular pathways. Just to highlight this general point, overexpressed IKK-α has substantially lower IκBα kinase activity than IKK-β (Fig. 2A) (10, 12, 13), yet antisense expression vectors and/or dominant negative versions of IKK-α and IKK-β indicate that both are essential for cytokine-induced activation of NF-κB (9–13). Thus, the potency of kinases observed in transfection experiments may not necessarily provide an accurate indication of their relative roles under physiological conditions.
We have previously proposed that MEKK1 is a coordinate activator of the NF-κB and JNK pathways (7). The fact that MEKK1 displays similar titration curves with the NF-κB and JNK reporter genes (Fig. 3 C and D) is consistent with this proposal; in both cases, significant activation of the reporter gene is observed at only the highest MEKK1 dose used (5 μg). We note that detectable JNK activation was observed at the lowest MEKK1 dose used (5 ng; Fig. 3B), raising the possibility that MEKK1 preferentially activates the JNK pathway (28). However, the activation observed at this dose is modest with respect to that seen at the highest dose (5 μg), and furthermore, no saturation of either IKK (Fig. 3A) or JNK (Fig. 3B) activities is seen with the MEKK1 dose range used. Thus, this provides further evidence that the two pathways can be coordinately activated by MEKK1.
A model for NF-κB activation (7) can now be extended to include the recently cloned NIK, IKK-α, and IKK-β. The essence of this model is a sequential involvement of a MAP3K (such as MEKK1 or NIK), an IKK (such as IKK-α or IKK-β), and an IκB. Although MEKK1 can directly phosphorylate a peptide containing the activation loop motif of IKK-β (Fig. 5D), it still remains to be rigorously determined whether MEKK1 and NIK directly activate IKK-α and IKK-β, and in fact, whether MEKK1 and NIK act in a functionally parallel manner, as would be predicted from the homology in their catalytic domains. Nonetheless, one implication of this model is that there might be additional MAP3Ks that activate IKK activity. These considerations suggest the possibility that at least some of the extremely large number of stimuli that activate NF-κB signal through distinct MAP3Ks.
This model, however, does not rule out other modes of regulation of IKK activity. Indeed, the 700-kDa IκBα kinase complex originally purified from uninduced HeLa cells can be activated in vitro by ubiquitination (6), suggesting that this particular IκBα kinase complex might serve to integrate phosphorylation- and ubiquitination-dependent signals leading to the activation of NF-κB. In addition, mutations in a helix–loop–helix domain of IKK-α severely impair its catalytic activity, suggesting that as yet to be identified protein–protein interactions might also regulate activity (12).
Acknowledgments
We are grateful to Drs. Roger Davis, David Wallach, David Goeddel, Dean Ballard, and Jeremiah Hagler for gifts of plasmids. We also thank Dr. Vito Palombella and members of the Maniatis laboratory for helpful discussions and critical readings of the manuscript. F.S.L. was supported by a postdoctoral research fellowship for physicians from the Howard Hughes Medical Institute. R.T.P. was supported by National Institutes of Health Biology Predoctoral Training Grant 5T32 GM07598. This work was supported by National Institutes of Health Grant AI20642 to T.M. and by ProScript, Inc.
ABBREVIATIONS
- CAT
chloramphenicol acetyltransferase
- GST
glutathione S-transferase
- IKK
IκB kinase
- JNK
c-Jun N-terminal kinase
- MAP3K
mitogen-activated protein kinase kinase kinase
- MEKK1
mitogen-activated protein kinase/ERK kinase kinase 1
- NIK
NF-κB inducing kinase
- TNF-α
tumor necrosis factor α
Note Added in Proof
Yin et al. (33) have recently shown that the human T cell leukemia virus type 1 (HTLV-1) transactivator protein Tax activates NF-κB through MEKK1.
Footnotes
A commentary on this article begins on page 9067.
References
- 1.Lee F S, Chen Z J, Maniatis T. In: Transcriptional Regulation of Endothelial Cell Adhesion Molecules. Collins T, editor. Austin, TX: Landes; 1998. , in press. [Google Scholar]
- 2.May M J, Ghosh S. Immunol Today. 1998;19:80–88. doi: 10.1016/s0167-5699(97)01197-3. [DOI] [PubMed] [Google Scholar]
- 3.Chen Z J, Hagler J, Palombella V, Melandri F, Scherer D, Ballard D, Maniatis T. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 4.Maniatis T. Science. 1997;278:818–819. doi: 10.1126/science.278.5339.818. [DOI] [PubMed] [Google Scholar]
- 5.Stankovski I, Baltimore D. Cell. 1997;91:299–302. doi: 10.1016/s0092-8674(00)80413-4. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z J, Parent L, Maniatis T. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 7.Lee F S, Hagler J, Chen Z J, Maniatis T. Cell. 1997;88:213–222. doi: 10.1016/s0092-8674(00)81842-5. [DOI] [PubMed] [Google Scholar]
- 8.Malinin N L, Boldin M P, Kovalenko A V, Wallach D. Nature (London) 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 9.Regnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 10.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 11.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. Nature (London) 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 12.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 13.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J, Young D B, Barbosa M, Mann M, Manning A, Rao A. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 14.Lange-Carter C A, Pleiman C M, Gardner A M, Blumer K J, Johnson G L. Science. 1993;260:315–319. doi: 10.1126/science.8385802. [DOI] [PubMed] [Google Scholar]
- 15.Derijard B, Hibi M, Wu I-H, Barrett T, Su B, Deng T, Karin M, Davis R J. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 16.McKinsey T A, Brockman J A, Scherer D C, Al-Murrani S W, Green P L, Ballard D W. Mol Cell Biol. 1996;16:2083–2090. doi: 10.1128/mcb.16.5.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1989. [Google Scholar]
- 18.Thanos D, Maniatis T. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 19.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 20.DiDonato J A, Mercurio F, Rosette C, Wu-Li J, Suyang H, Ghosh S, Karin M. Mol Cell Biol. 1996;16:1295–1304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyriakis J M, Banerjee P, Nikolaki E, Dai T, Ruble E A, Ahmad M F, Avruch J, Woodgett J R. Nature (London) 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 22.Visvanathan K V, Goodbourn S. EMBO J. 1989;8:1129–1138. doi: 10.1002/j.1460-2075.1989.tb03483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenardo M J, Fan C-M, Maniatis T, Baltimore D. Cell. 1989;57:287–294. doi: 10.1016/0092-8674(89)90966-5. [DOI] [PubMed] [Google Scholar]
- 24.Du W, Thanos D, Maniatis T. Cell. 1993;74:887–898. doi: 10.1016/0092-8674(93)90468-6. [DOI] [PubMed] [Google Scholar]
- 25.Gupta S, Campbell D, Derijard B, Davis R J. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 26.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis R J, Johnson G L, Karin M. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 27.Yan M, Dai T, Deak J C, Kyriakis J M, Zon L I, Woodgett J R, Templeton D J. Nature (London) 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]
- 28.Song H Y, Regnier C H, Kirschning C J, Ayres T M, Goeddel D V, Rothe M. Proc Natl Acad Sci USA. 1997;94:9792–9796. doi: 10.1073/pnas.94.18.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakano H, Shindo M, Sakon S, Nishinaka S, Mihara M, Yagita H, Okumura K. Proc Natl Acad Sci USA. 1998;95:3537–3542. doi: 10.1073/pnas.95.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ling L, Cao Z, Goeddel D V. Proc Natl Acad Sci USA. 1998;95:3792–3797. doi: 10.1073/pnas.95.7.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu L P, Anderson K V. Nature (London) 1998;392:93–97. doi: 10.1038/32195. [DOI] [PubMed] [Google Scholar]
- 32.Fujita T, Miyamoto M, Kimura Y, Hammer J, Taniguchi T. Nucleic Acids Res. 1989;17:3335–3346. doi: 10.1093/nar/17.9.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin M-J, Christerson L B, Yamamoto Y, Kwak Y-T, Xu S, Mercurio F, Barbosa M, Cobb M, Gaynor R. Cell. 1998;93:875–884. doi: 10.1016/s0092-8674(00)81447-6. [DOI] [PubMed] [Google Scholar]