Abstract
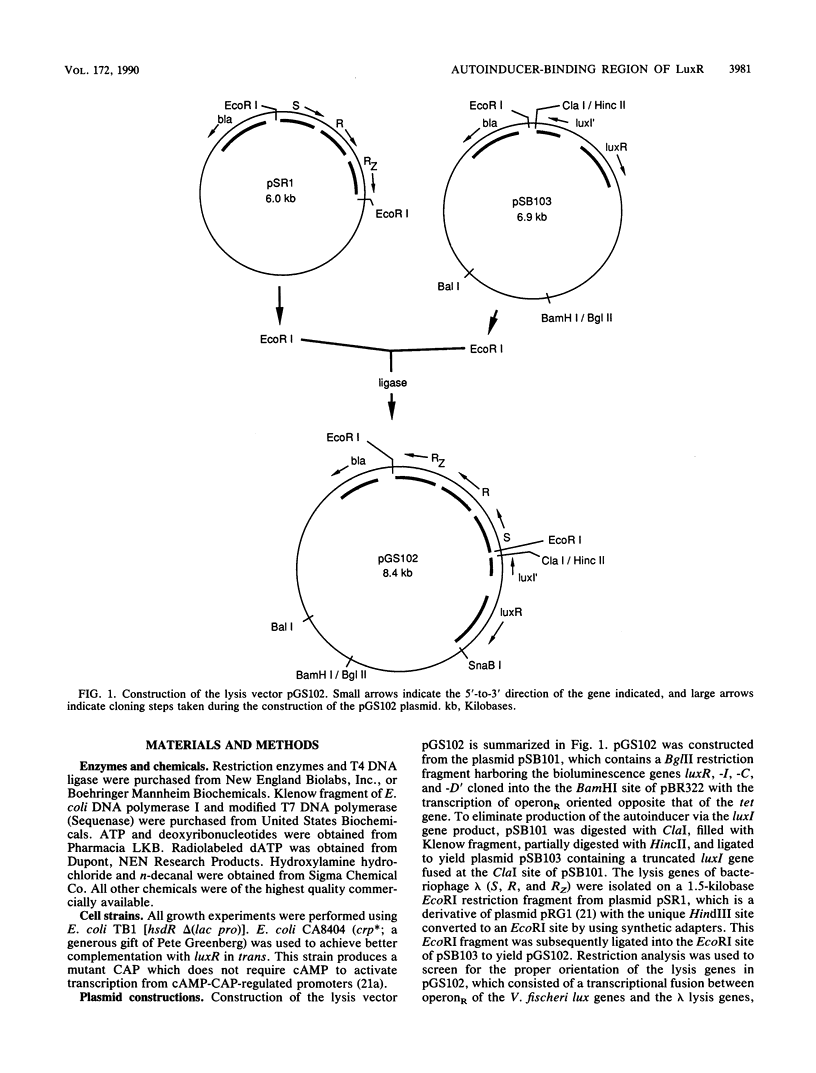
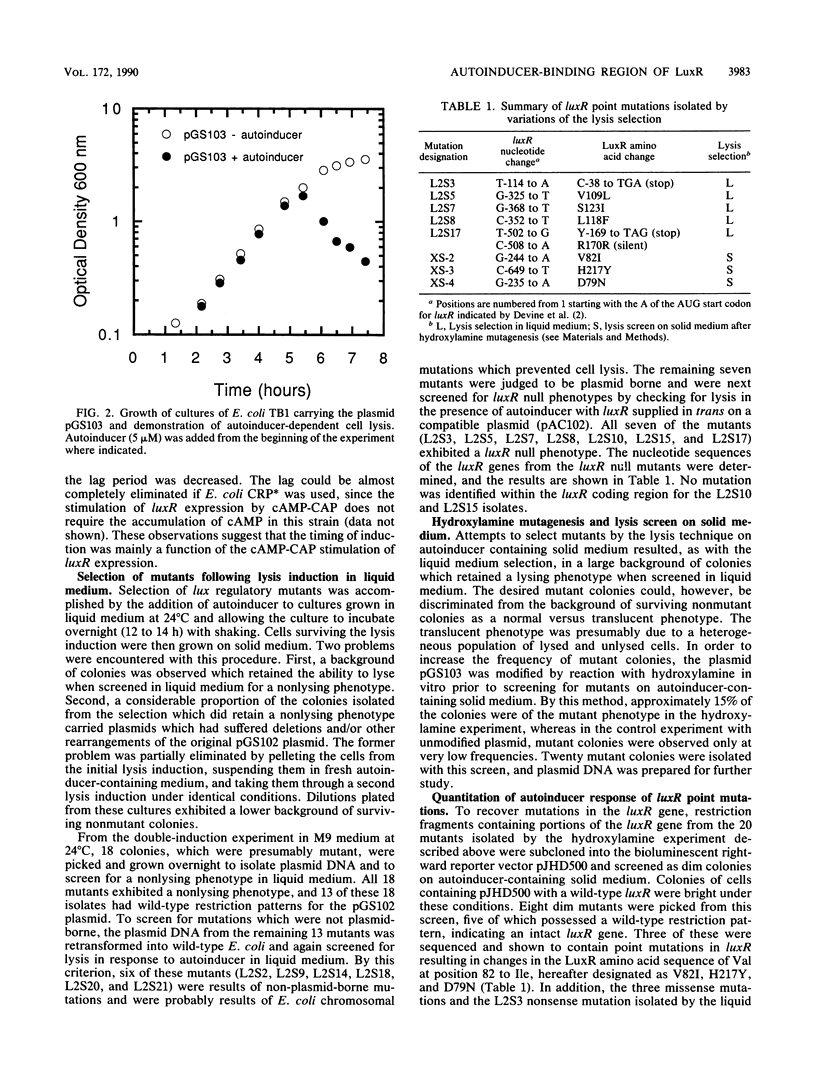
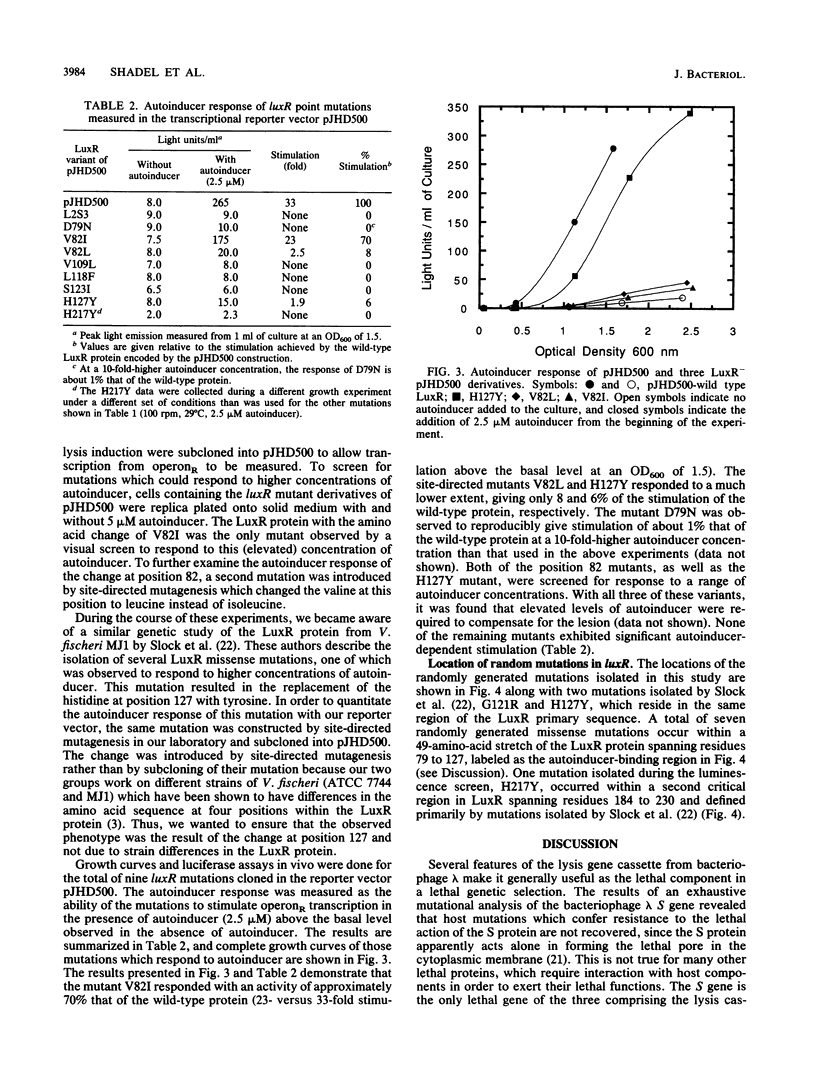
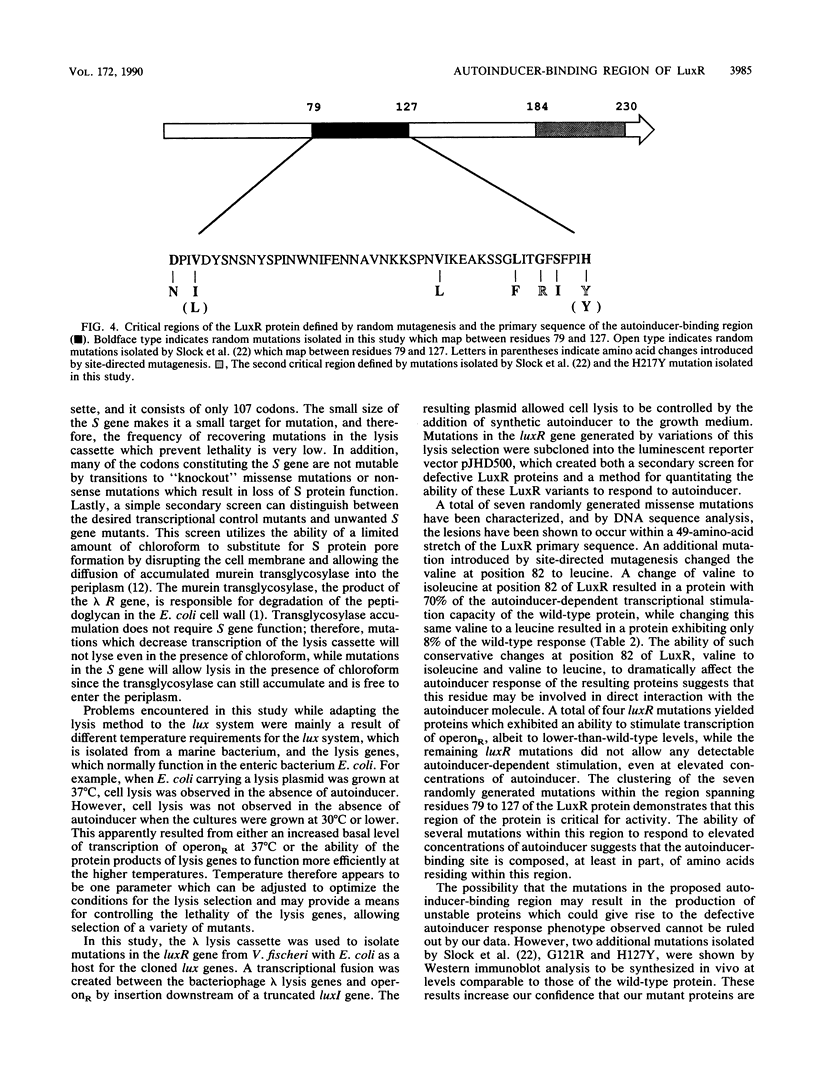
A lethal genetic selection utilizing the bacteriophage lambda lysis genes (S, R, RZ) has been developed and used in conjunction with a luminescence screen to allow the isolation and characterization of six missense mutations and two nonsense mutations in the luxR gene from Vibrio fischeri ATCC 7744. A transcriptional fusion of the lysis genes in operonR downstream of a truncated luxI gene allows control of cell lysis by the addition of synthetic autoinducer to the growth medium. The six missense mutations isolated resulted in changes in the LuxR protein of Asp at position 79 to Asn (hereafter designated as D79N), V82I, V109L, L118F, S123I, and H217Y. Variant LuxR proteins with amino acid changes of D79N, V82I, V82L, and H127Y were shown to require higher concentrations of autoinducer to elicit a certain amplitude response than is required by the wild-type protein. We believe that the clustering of a total of seven randomly generated missense mutations in a 49-amino-acid region of the LuxR primary sequence defines a critical portion of the LuxR protein. The observation that proteins with lesions in this region responded to elevated levels of autoinducer suggests that the autoinducer-binding site is constructed, at least in part, from several amino acid residues within the 79-to-127 region of the LuxR protein.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bienkowska-Szewczyk K., Lipinska B., Taylor A. The R gene product of bacteriophage lambda is the murein transglycosylase. Mol Gen Genet. 1981;184(1):111–114. doi: 10.1007/BF00271205. [DOI] [PubMed] [Google Scholar]
- Devine J. H., Shadel G. S., Baldwin T. O. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC7744. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5688–5692. doi: 10.1073/pnas.86.15.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap P. V., Greenberg E. P. Control of Vibrio fischeri luminescence gene expression in Escherichia coli by cyclic AMP and cyclic AMP receptor protein. J Bacteriol. 1985 Oct;164(1):45–50. doi: 10.1128/jb.164.1.45-50.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap P. V., Greenberg E. P. Control of Vibrio fischeri lux gene transcription by a cyclic AMP receptor protein-luxR protein regulatory circuit. J Bacteriol. 1988 Sep;170(9):4040–4046. doi: 10.1128/jb.170.9.4040-4046.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard A., Burlingame A. L., Eberhard C., Kenyon G. L., Nealson K. H., Oppenheimer N. J. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981 Apr 28;20(9):2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- Eberhard A. Inhibition and activation of bacterial luciferase synthesis. J Bacteriol. 1972 Mar;109(3):1101–1105. doi: 10.1128/jb.109.3.1101-1105.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J., Nealson K., Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983 Mar;32(3):773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- Engebrecht J., Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J., Silverman M. Nucleotide sequence of the regulatory locus controlling expression of bacterial genes for bioluminescence. Nucleic Acids Res. 1987 Dec 23;15(24):10455–10467. doi: 10.1093/nar/15.24.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. R., Howe M. New mutations in the S cistron of bacteriophage lambda affecting host cell lysis. Virology. 1969 May;38(1):200–202. doi: 10.1016/0042-6822(69)90148-2. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Wallace J. C., Brown J. P. Finding protein similarities with nucleotide sequence databases. Methods Enzymol. 1990;183:111–132. doi: 10.1016/0076-6879(90)83009-x. [DOI] [PubMed] [Google Scholar]
- Humphreys G. O., Willshaw G. A., Smith H. R., Anderson E. S. Mutagenesis of plasmid DNA with hydroxylamine: isolation of mutants of multi-copy plasmids. Mol Gen Genet. 1976 Apr 23;145(1):101–108. doi: 10.1007/BF00331564. [DOI] [PubMed] [Google Scholar]
- Kaplan H. B., Greenberg E. P. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol. 1985 Sep;163(3):1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab R., Neal G., Garrett J., Grimaila R., Fusselman R., Young R. Mutational analysis of bacteriophage lambda lysis gene S. J Bacteriol. 1986 Sep;167(3):1035–1042. doi: 10.1128/jb.167.3.1035-1042.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin D., Beckwith J. Deletion of the Escherichia coli crp gene. J Bacteriol. 1975 Apr;122(1):338–340. doi: 10.1128/jb.122.1.338-340.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slock J., VanRiet D., Kolibachuk D., Greenberg E. P. Critical regions of the Vibrio fischeri luxR protein defined by mutational analysis. J Bacteriol. 1990 Jul;172(7):3974–3979. doi: 10.1128/jb.172.7.3974-3979.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitzur S., Kuhn J. The transcription of bacterial luminescence is regulated by sigma 32. J Biolumin Chemilumin. 1988 Apr-Jun;2(2):81–93. doi: 10.1002/bio.1170020205. [DOI] [PubMed] [Google Scholar]
- Ulitzur S. The regulatory control of the bacterial luminescence system--a new view. J Biolumin Chemilumin. 1989 Jul;4(1):317–325. doi: 10.1002/bio.1170040144. [DOI] [PubMed] [Google Scholar]


