Abstract
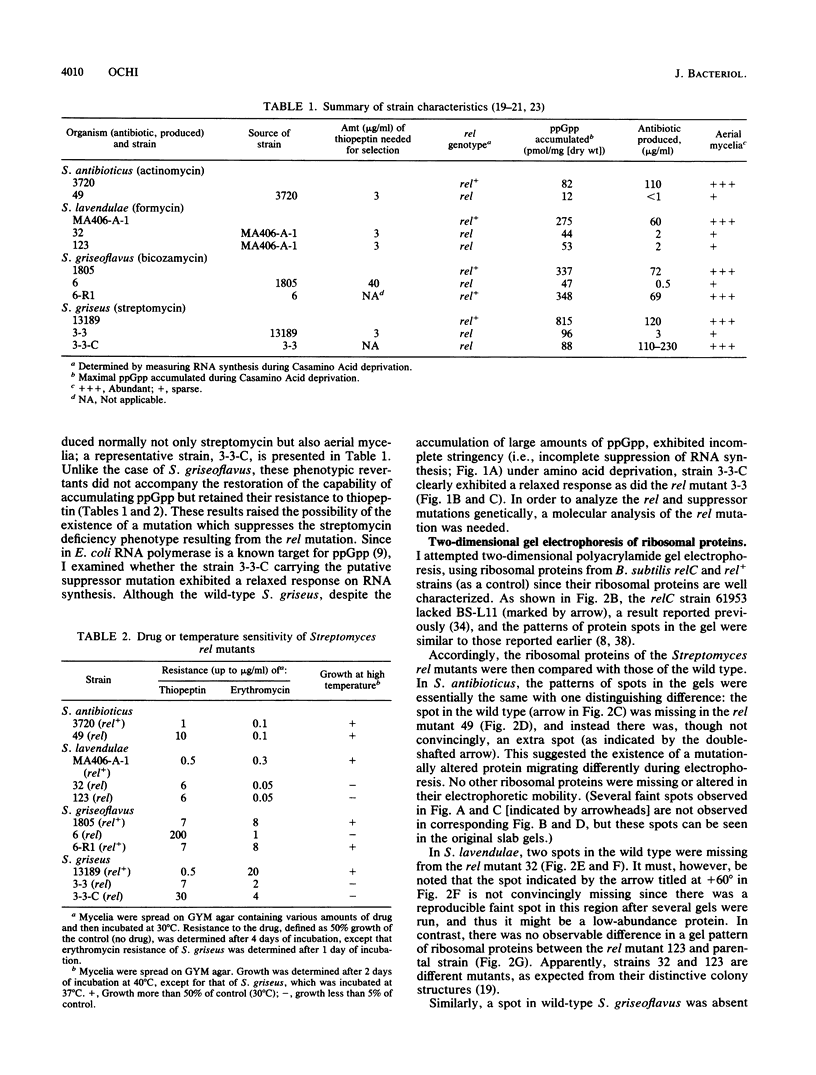
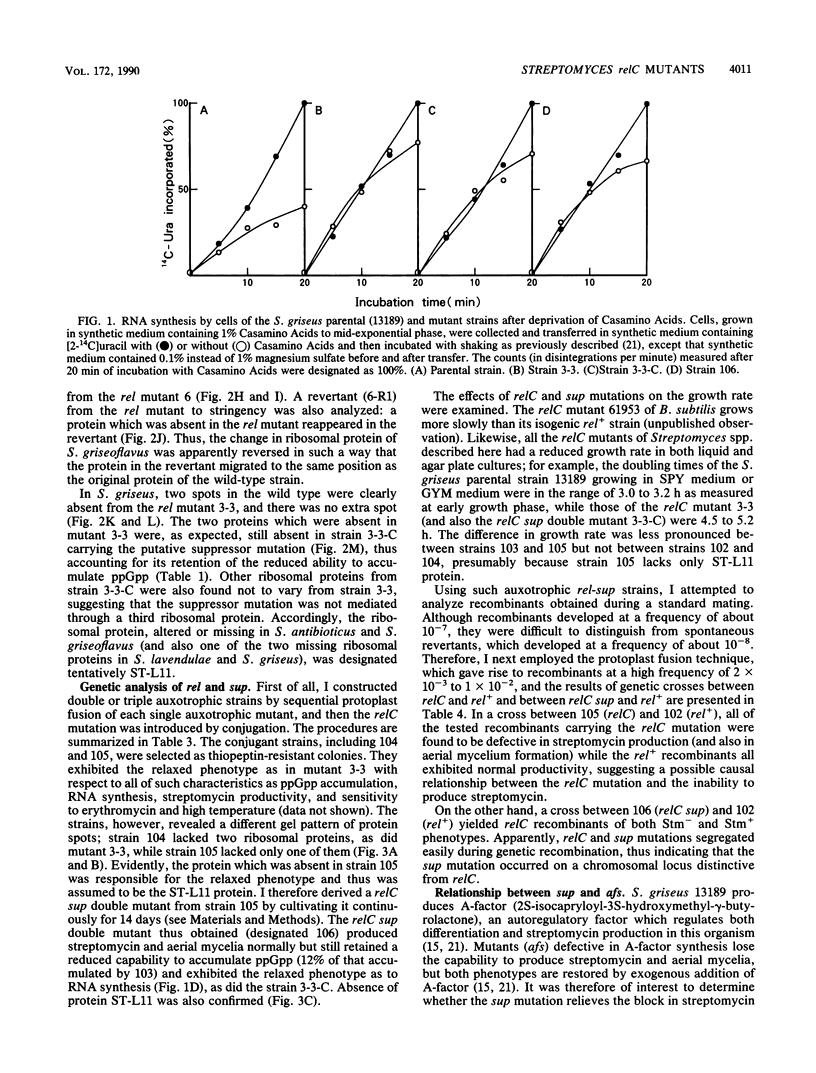
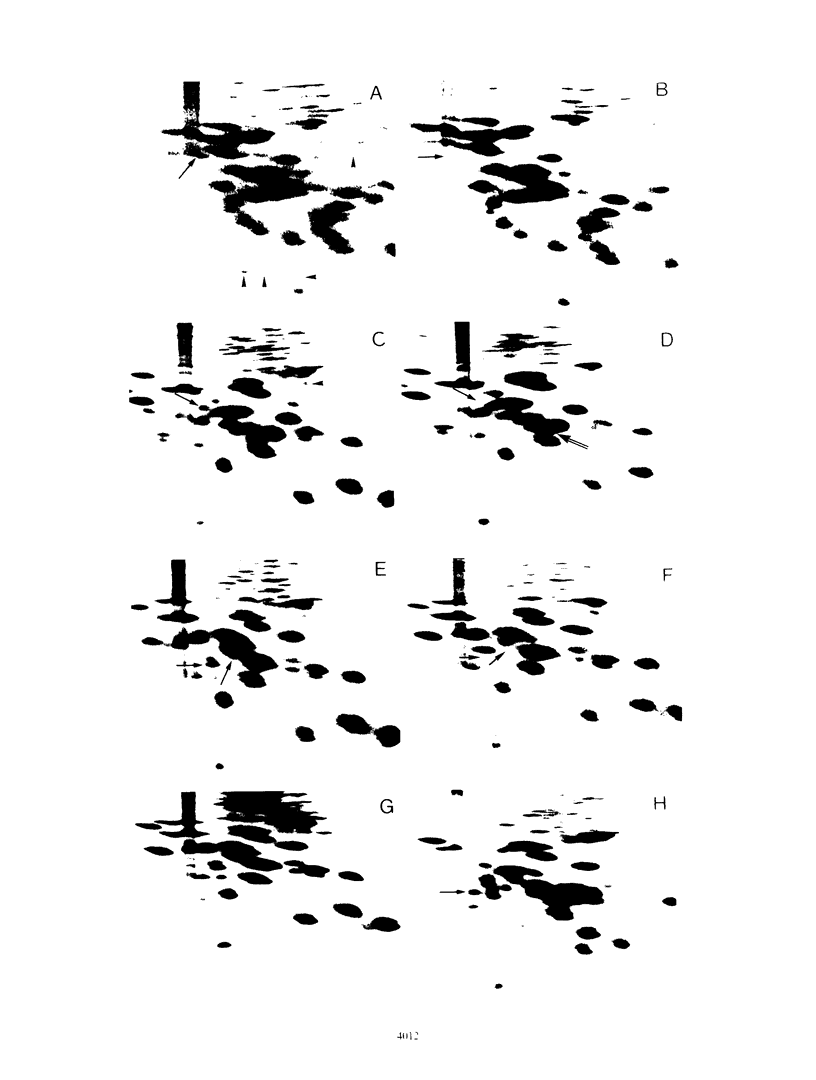
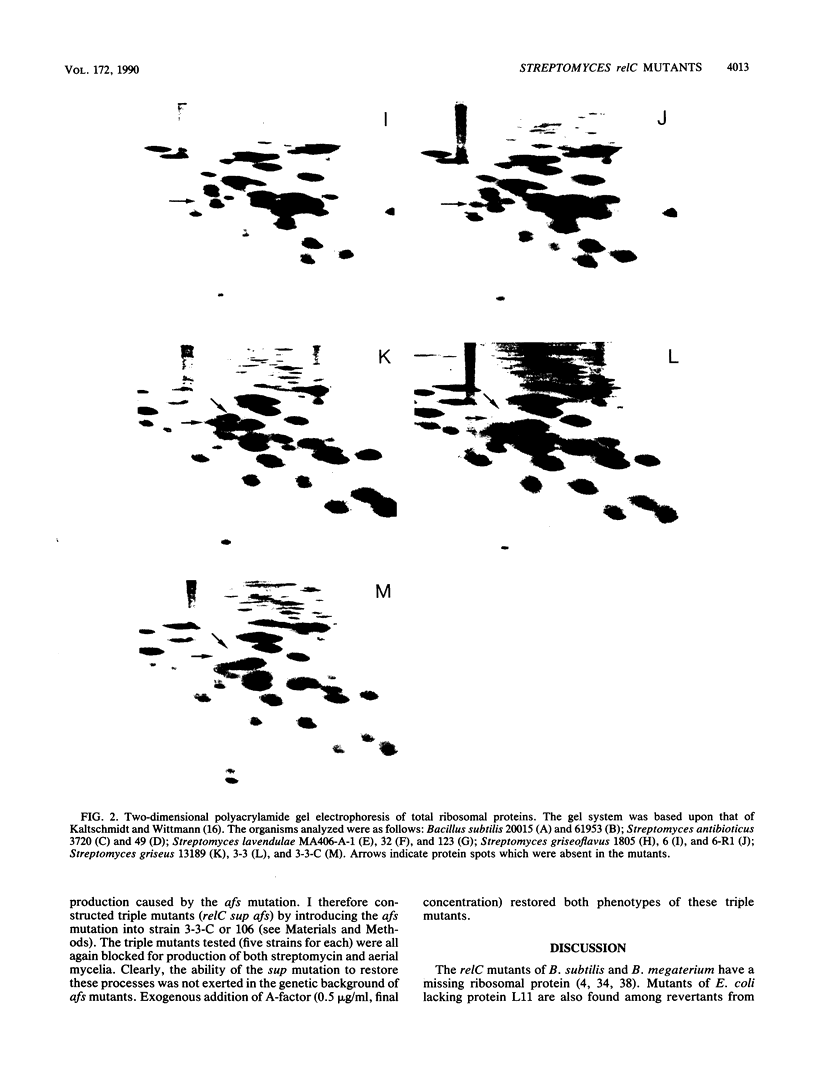
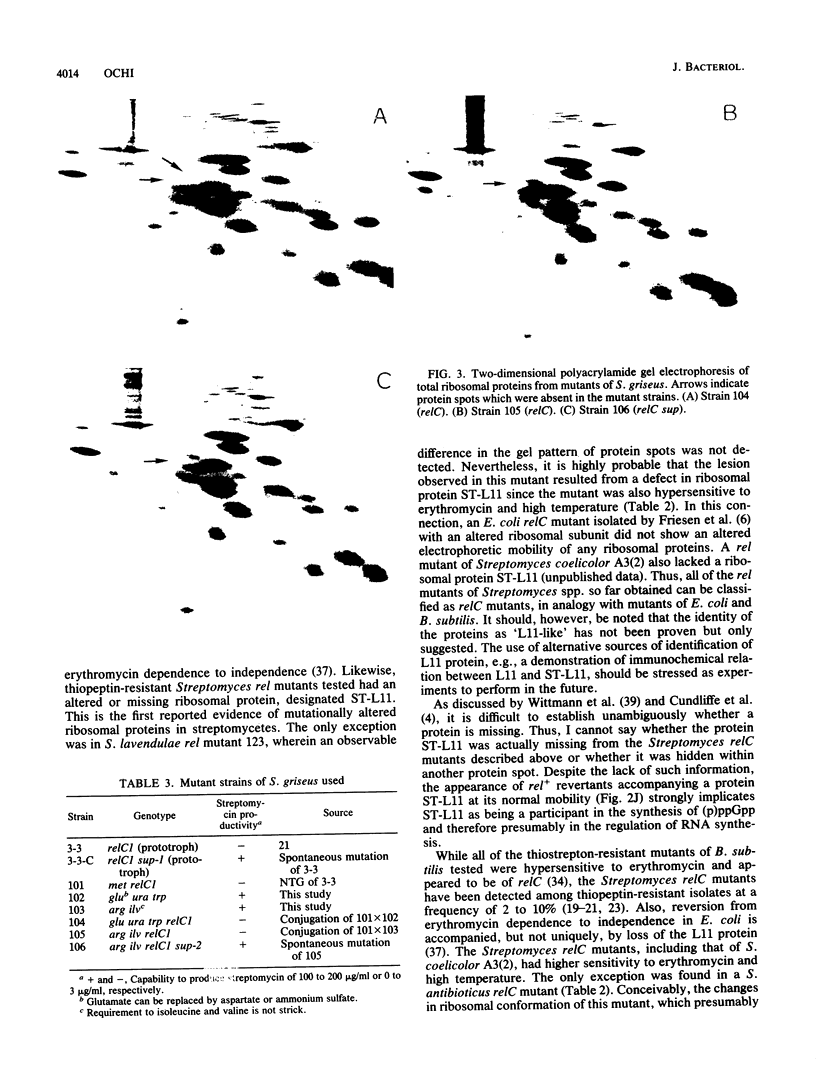
Several relaxed (rel) mutants have been obtained from Streptomyces species by selecting colonies resistant to thiopeptin, an analogue of thiostrepton. Using two-dimensional gel electrophoresis, I compared the ribosomal proteins from rel and rel+ pairs of S. antibioticus, S. lavendulae, S. griseoflavus, and S. griseus. It was found that all of the Streptomyces rel mutants thus examined had an altered or missing ribosomal protein, designated tentatively ST-L11. These rel mutants therefore could be classified as relC mutants and were highly sensitive to erythromycin or high temperature. A relC mutant of S. griseus was defective in streptomycin production, but phenotypic reversion of this defect to normal productivity was found at high incidence among progeny of the relC mutant. This phenotypic reversion did not accompany a reappearance of ribosomal protein ST-L11, and furthermore the ability of accumulating ppGpp still remained at a low level, thus suggesting existence of a mutation (named sup) which suppresses the streptomycin deficiency phenotype exhibited by the relC mutant. Genetic analysis revealed that there is a correlation between the rel mutation and the inability to produce streptomycin or aerial mycelia. The sup mutation was found to lie at a chromosomal locus distinct from that of the relC mutation. It was therefore concluded that the dependence of streptomycin production on the normal function of the relC gene could be entirely bypassed by a mutation at the suppressor locus (sup). The suppressing effect of the sup mutation on the relC mutation was blocked when the afs mutation (defective in A-factor synthesis) was introduced into a relC sup double mutant. It is proposed that the sup gene or its product can be direct or indirect target for ppGpp.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambulos N. P., Jr, Rogers E. J., Alexieva Z., Lovett P. S. Induction of cat-86 by chloramphenicol and amino acid starvation in relaxed mutants of Bacillus subtilis. J Bacteriol. 1988 Dec;170(12):5642–5646. doi: 10.1128/jb.170.12.5642-5646.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundliffe E., Dixon P., Stark M., Stöffler G., Ehrlich R., Stöffler-Meilicke M., Cannon M. Ribosomes in thiostrepton-resistant mutants of Bacillus megaterium lacking a single 50 S subunit protein. J Mol Biol. 1979 Aug 5;132(2):235–252. doi: 10.1016/0022-2836(79)90393-0. [DOI] [PubMed] [Google Scholar]
- Dixon P. G., Beven J. E., Cundliffe E. Properties of the ribosomes of antibiotic producers: effects thiostrepton and micrococcin on the organisms which produce them. Antimicrob Agents Chemother. 1975 Jun;7(6):850–855. doi: 10.1128/aac.7.6.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen J. D., Fiil N. P., Parker J. M., Haseltine W. A. A new relaxed mutant of Escherichia coli with an altered 50S ribosomal subunit. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3465–3469. doi: 10.1073/pnas.71.9.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant J. A. Stringent control in E. coli. Annu Rev Genet. 1979;13:393–415. doi: 10.1146/annurev.ge.13.120179.002141. [DOI] [PubMed] [Google Scholar]
- Geisser M., Tischendorf G. W., Stöffler G. Comparative immunological and electrophoretic studies on ribosomal proteins of bacillaceae. Mol Gen Genet. 1973 Dec 20;127(2):129–145. doi: 10.1007/BF00333661. [DOI] [PubMed] [Google Scholar]
- Glass R. E., Jones S. T., Ishihama A. Genetic studies on the beta subunit of Escherichia coli RNA polymerase. VII. RNA polymerase is a target for ppGpp. Mol Gen Genet. 1986 May;203(2):265–268. doi: 10.1007/BF00333964. [DOI] [PubMed] [Google Scholar]
- Goldthwaite C., Smith I. Physiological characterization of antibiotic resistant mutants of Bacillus subtilis. Mol Gen Genet. 1972;114(3):190–204. doi: 10.1007/BF01788888. [DOI] [PubMed] [Google Scholar]
- Hara O., Horinouchi S., Uozumi T., Beppu T. Genetic analysis of A-factor synthesis in Streptomyces coelicolor A3(2) and Streptomyces griseus. J Gen Microbiol. 1983 Sep;129(9):2939–2944. doi: 10.1099/00221287-129-9-2939. [DOI] [PubMed] [Google Scholar]
- Highland J. H., Howard G. A., Ochsner E., Hasenbank R., Gordon J., Stöffler G. Identification of a ribosomal protein necessary for thiostrepton binding to Escherichia coli ribosomes. J Biol Chem. 1975 Feb 10;250(3):1141–1145. [PubMed] [Google Scholar]
- Hopwood D. A. Genetic analysis and genome structure in Streptomyces coelicolor. Bacteriol Rev. 1967 Dec;31(4):373–403. doi: 10.1128/br.31.4.373-403.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood D. A. The Leeuwenhoek lecture, 1987. Towards an understanding of gene switching in Streptomyces, the basis of sporulation and antibiotic production. Proc R Soc Lond B Biol Sci. 1988 Nov 22;235(1279):121–138. doi: 10.1098/rspb.1988.0067. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Hara O., Beppu T. Cloning of a pleiotropic gene that positively controls biosynthesis of A-factor, actinorhodin, and prodigiosin in Streptomyces coelicolor A3(2) and Streptomyces lividans. J Bacteriol. 1983 Sep;155(3):1238–1248. doi: 10.1128/jb.155.3.1238-1248.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltschmidt E., Wittmann H. G. Ribosomal proteins. VII. Two-dimensional polyacrylamide gel electrophoresis for fingerprinting of ribosomal proteins. Anal Biochem. 1970 Aug;36(2):401–412. doi: 10.1016/0003-2697(70)90376-3. [DOI] [PubMed] [Google Scholar]
- Marahiel M. A., Zuber P., Czekay G., Losick R. Identification of the promoter for a peptide antibiotic biosynthesis gene from Bacillus brevis and its regulation in Bacillus subtilis. J Bacteriol. 1987 May;169(5):2215–2222. doi: 10.1128/jb.169.5.2215-2222.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi K. A decrease in GTP content is associated with aerial mycelium formation in Streptomyces MA406-A-1. J Gen Microbiol. 1986 Feb;132(2):299–305. doi: 10.1099/00221287-132-2-299. [DOI] [PubMed] [Google Scholar]
- Ochi K. Heterogeneity of ribosomal proteins among Streptomyces species and its application to identification. J Gen Microbiol. 1989 Oct;135(10):2635–2642. doi: 10.1099/00221287-135-10-2635. [DOI] [PubMed] [Google Scholar]
- Ochi K., Hitchcock M. J., Katz E. High-frequency fusion of Streptomyces parvulus or Streptomyces antibioticus protoplasts induced by polyethylene glycol. J Bacteriol. 1979 Sep;139(3):984–992. doi: 10.1128/jb.139.3.984-992.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi K. Metabolic initiation of differentiation and secondary metabolism by Streptomyces griseus: significance of the stringent response (ppGpp) and GTP content in relation to A factor. J Bacteriol. 1987 Aug;169(8):3608–3616. doi: 10.1128/jb.169.8.3608-3616.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi K. Occurrence of the stringent response in Streptomyces sp. and its significance for the initiation of morphological and physiological differentiation. J Gen Microbiol. 1986 Sep;132(9):2621–2631. doi: 10.1099/00221287-132-9-2621. [DOI] [PubMed] [Google Scholar]
- Ochi K. The tsr gene-coding plasmid pIJ702 prevents thiopeptin from inhibiting ppGpp synthesis in Streptomyces lividans. FEMS Microbiol Lett. 1989 Oct 1;52(1-2):219–223. doi: 10.1016/0378-1097(89)90200-0. [DOI] [PubMed] [Google Scholar]
- Ochi K., Tsurumi Y., Shigematsu N., Iwami M., Umehara K., Okuhara M. Physiological analysis of bicozamycin high-producing Streptomyces griseoflavus used at industrial level. J Antibiot (Tokyo) 1988 Aug;41(8):1106–1115. doi: 10.7164/antibiotics.41.1106. [DOI] [PubMed] [Google Scholar]
- Okanishi M., Suzuki K., Umezawa H. Formation and reversion of Streptomycete protoplasts: cultural condition and morphological study. J Gen Microbiol. 1974 Feb;80(2):389–400. doi: 10.1099/00221287-80-2-389. [DOI] [PubMed] [Google Scholar]
- Parker J., Watson R. J., Friesen J. D. A relaxed mutant with an altered ribosomal protein L11. Mol Gen Genet. 1976 Feb 27;144(1):111–114. doi: 10.1007/BF00277313. [DOI] [PubMed] [Google Scholar]
- Pestka S., Weiss D., Vince R., Wienen B., Stöffler G., Smith I. Thiostrepton-resistant mutants of Bacillus subtilis: localization of resistance to the 50S subunit. Mol Gen Genet. 1976 Mar 30;144(3):235–241. doi: 10.1007/BF00341721. [DOI] [PubMed] [Google Scholar]
- Shand R. F., Blum P. H., Mueller R. D., Riggs D. L., Artz S. W. Correlation between histidine operon expression and guanosine 5'-diphosphate-3'-diphosphate levels during amino acid downshift in stringent and relaxed strains of Salmonella typhimurium. J Bacteriol. 1989 Feb;171(2):737–743. doi: 10.1128/jb.171.2.737-743.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I., Paress P., Cabane K., Dubnau E. Genetics and physiology of the rel system of Bacillus subtilis. Mol Gen Genet. 1980;178(2):271–279. doi: 10.1007/BF00270472. [DOI] [PubMed] [Google Scholar]
- Smith I., Paress P., Pestka S. Thiostrepton-resistant mutants exhibit relaxed synthesis of RNA. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5993–5997. doi: 10.1073/pnas.75.12.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M. J., Cundliffe E., Dijk J., Stöffler G. Functional homology between E. coli ribosomal protein L11 and B. megaterium protein BM-L11. Mol Gen Genet. 1980;180(1):11–15. doi: 10.1007/BF00267346. [DOI] [PubMed] [Google Scholar]
- Stark M. J., Cundliffe E. Requirement for ribosomal protein BM-L11 in stringent control of RNA synthesis in Bacillus megaterium. Eur J Biochem. 1979 Dec;102(1):101–105. doi: 10.1111/j.1432-1033.1979.tb06267.x. [DOI] [PubMed] [Google Scholar]
- Stöffler G., Cundliffe E., Stöffler-Meilicke M., Dabbs E. R. Mutants of Escherichia coli lacking ribosomal protein L11. J Biol Chem. 1980 Nov 10;255(21):10517–10522. [PubMed] [Google Scholar]
- Wienen B., Ehrlich R., Stöffler-Meilicke M., Stöffler G., Smith I., Weiss D., Vince R., Pestka S. Ribosomal protein alterations in thiostrepton- and Micrococcin-resistant mutants of Bacillus subtilis. J Biol Chem. 1979 Aug 25;254(16):8031–8041. [PubMed] [Google Scholar]
- Zuber P., Losick R. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J Bacteriol. 1987 May;169(5):2223–2230. doi: 10.1128/jb.169.5.2223-2230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]