Abstract
In early development of Xenopus laevis, it is known that activities of polypeptide growth factors are negatively regulated by their binding proteins. In this study, follistatin, originally known as an activin-binding protein, was shown to inhibit all aspects of bone morphogenetic protein (BMP) activity in early Xenopus embryos. Furthermore, using a surface plasmon resonance biosensor, we demonstrated that follistatin can directly interact with multiple BMPs at significantly high affinities. Interestingly, follistatin was found to be noncompetitive with the BMP receptor for ligand binding and to form a trimeric complex with BMP and its receptor. The results suggest that follistatin acts as an organizer factor in early amphibian embryogenesis by inhibiting BMP activities by a different mechanism from that used by chordin and noggin.
TGF-β family ligands are known to regulate a variety of cell differentiation processes that lead to morphogenesis in early vertebrate development (1). In Xenopus laevis, two classes of the TGF-β family ligands are believed to determine the dorsoventral pattern of the mesoderm in early gastrula embryos (2). The first class of ligands includes those that are related to activin and Vg1. This group has been shown to induce formation of the dorsal mesoderm, which gives rise to a variety of tissues including muscle and the notochord (3–5). The second class includes the BMP family of ligands, which inhibit dorsal mesoderm formation and induce cells to take on ventral fates, such as that of blood cells (6). BMP also is involved in the dorsoventral specification of ectodermal cell fate by inducing the epidermis and inhibiting neural cell differentiation (7). These biological activities of TGF-β ligands are known to be negatively regulated by their specific binding proteins (8, 9). For example, follistatin is capable of binding activin extracellularly, and this binding inhibits the activation of activin receptors (10). Therefore, follistatin has been considered to be an important component in the regulation of mesoderm induction (11). Similarly, among BMPs, at least BMP-4 is known to be strictly regulated by its binding proteins, chordin (12), and noggin (13). Chordin and noggin, both of which function as neural inducing agents, have been shown to bind BMP-4 directly thus interfering with its ability to bind its receptor (14, 15). Interestingly, the expression of both chordin and noggin is restricted to the Spemann’s organizer of early Xenopus gastrula (12, 13), suggesting that they may act as organizer factors inhibiting the antineural activity of BMP-4. These observations led to the notion that the inhibition of polypeptide growth factor activity by binding proteins is a key step in the control of early developmental events (16).
However, the specificity of ligand recognition by binding proteins is not fully understood. For example, recent studies on follistatin by using Xenopus embryos demonstrate that the binding specificity of follistatin is a complicated issue. Follistatin induces the secondary body axis when overexpressed in ventral blastomeres (17), and in ectoderm, it can induce neural tissue (18). These observations indicate that follistatin might inhibit not only activin but also BMPs through direct binding because many of the phenotypes caused by the overexpression of follistatin in early Xenopus embryos are similar to those obtained by overexpression of dominant-negative BMP receptors. Although the coprecipitation of follistatin with BMP-4 recently has been shown in Xenopus (19), no extensive studies that clarify the mechanism by which follistatin inhibits BMP family members have been reported. These questions led us to investigate, first, whether follistatin inhibits all BMP activities in Xenopus embryos and second, whether such inhibition is caused by the direct binding of follistatin to BMPs. Finally, we further explored the mechanism by which follistatin inhibits BMP activity.
MATERIALS AND METHODS
Embryo Manipulations.
Xenopus embryos were obtained by artificial fertilization, and 2- or 4-cell stage embryos were microinjected with synthetic RNAs as previously described (20). For evaluation of mesodermal markers, dorsal or ventral marginal region was excised when the injected embryo reached stage 10. For animal cap assay, presumptive ectoderm fragments were collected when the injected embryos reached stage 8.5.
Reverse Transcription–PCR (RT-PCR).
For the detection of molecular marker expression, total RNA was isolated by using TRIzol (GIBCO/BRL) according to manufacturer’s instructions and analyzed by RT-PCR. Primers were as follows: as a dorsal organizer marker goosecoid, upstream, 5′-ACTACTATGGACAGTTGCACG-3′ and downstream, 5′-TTCTGATTCCTCTGATGAAGATC-3′; a ventrolateral mesoderm marker Xvent1, upstream, 5′-TTCCCTTCAGCATGGTTCAAC-3′ and downstream, 5′-GCATCTCCTTGGCATATTTGG-3′; a definitive ventral mesoderm marker αT1 globin, upstream, 5′-TTGCTGTCTCACACCATC-3′ and downstream, 5′-TCTGTACTTGGAGGTGAG-3′; an internal input control histoneH4, upstream, 5′-ATAACATCCAGGGCATCACC-3′ and downstream, 5′-ACATCCATAGCGGTGACGGT-3′.
Recombinant Proteins.
Recombinant Xenopus BMP-4 (BMP-4) and the soluble form of the mouse BMP type I receptor (sBMPR), the extracellular domain of the receptor, were obtained by using a silkworm expression system (21), and recombinant human follistatin (FS-288) was obtained by using a Chinese hamster ovary cell expression system (22). Recombinant activin A was a gift from Y. Eto (Ajinomoto, Tokyo), Xenopus BMP-4/7 heterodimer (BMP-4/7) and Xenopus BMP-7 (BMP-7) were gifts from Y. Fujisawa (Takeda, Osaka), and Xenopus chordin was a gift from S. Piccolo (14). Recombinant human TGF-β1 was purchased (King Jyohzo, Hyogo).
Protein–Protein Interaction Analysis by Surface Plasmon Resonance Biosensor.
Binding experiments were performed using the BIACORE2000 (Biacore AB). The basic principles and its use have been previously documented (23). Purified samples were injected over the surfaces at a flow rate of 20 μl/min at 25°C for 120 s. For the blank runs, all samples were injected over mock-coupled sensor chip surfaces containing no protein simultaneously with each experimental run. All curves were corrected for background by subtracting the blank run, using biaevaluation software version 2.1 (Biacore AB). The buffer for sample dilution and running buffer was Hepes buffered saline (HBS, 10 mM Hepes/150 mM NaCl/3.4 mM EDTA/0.005% Tween 20, pH 7.4). BMPs, activin A, TGF-β1, and FS-288 were immobilized on the sensor chip surface (CM5, certified grade, Biacore AB) by the amine-coupling method (24). sBMPR that was biotinylated using sulfo-NHS-biotin (Pierce) was immobilized on the sensor chip surface (SA5, research grade, Biacore AB), which had been preimmobilized with streptavidin. The immobilization level of sBMPR was 684 resonance units. The sequential experiment was performed by using the coinjection method (Biacore AB). Arrowheads represent the initiation and termination of injections.
Chemical Cross-Linking and Two-Dimensional (2D) Electrophoresis.
FS-288 (450 ng) and BMP-4 (450 ng) were incubated for 1 hr at room temperature in 100 μl of HBS containing 0.02% Tween 20. Dithiobis (sulfosuccinimidylpropionate) (Pierce) was added to a final concentration of 0.15 mM, incubated for 30 min at room temperature, and stopped by adding Tris⋅HCl (pH 8.0) to a final concentration of 50 mM. Diagonal SDS/PAGE analysis was performed as described (25). In brief, samples (10 μl) were electrophoresed in 0.2 × 7 cm strips of 1 mm-thick 12.5% polyacrylamide gel without reducing agents. Each gel strip was removed after the first electrophoresis and incubated in a solution containing 5% 2-mercaptoethanol for 1 hr at 37°C. After incubation, the gel strip was placed onto a 15% SDS slab gel (7.5 × 9 cm) and then electrophoresed in the second dimension under reducing condition. Visualization of the proteins was performed by Western blotting as described below.
Western Blotting Analysis.
Protein samples, resolved by 2D electrophoresis, were electroblotted onto poly(vinylidene difluoride) membranes (Millipore). The membrane was blocked with 20 mM Tris (pH 7.5), 150 mM NaCl, and 0.5% Tween 20 containing 5% nonfat dry milk and reacted with the antibodies mixture of BMP-4 antibody (Ab97) and follistatin antiserum (Rb32) at 4°C overnight (26, 22). The membrane was then reacted with a horseradish peroxidase-conjugated antibody for 1 hr at room temperature. Western blot was developed with the chemiluminescent ECL plus kit (Amersham) as described by the manufacturer.
RESULTS AND DISCUSSION
Inhibition of BMP-Induced Ventralization by Follistatin.
One of the major roles of embryonic BMPs in Xenopus is to cause the ventralization of the mesoderm during gastrulation (27, 28). All three BMP subtypes identified so far, namely BMP-2, -4, and -7 (29) cause a ventralized embryo that lacks the anterior head structure and notochord when they are dorsally overexpressed by mRNA microinjection. Therefore, BMP activity is easily examined by microinjecting mRNA, and the potency can be evaluated by observing the extent of ventralization (20). As previously reported (30), the dorsal injection of 200 pg of BMP-2 (data not shown) or BMP-4 mRNA ventralized embryos to an average dorsoanterior index of 0–1 (Fig. 1Ab). In contrast, 500 pg of BMP-7 mRNA was required to cause a similar amount of ventralization (Fig. 1Ac), which is consistent with the previous observation that the ventralizing activity of BMP-7 mRNA is relatively weak compared with that of BMP-4 mRNA (20).
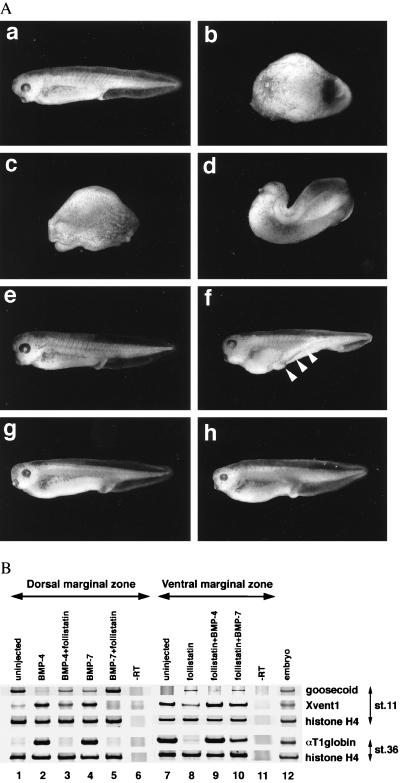
Figure 1.
Inhibitory interaction between follistatin and BMPs. (A) Phenotypes of Xenopus embryos dorsally coinjected with BMP mRNA and follistatin mRNA. BMP-4 mRNA (200 pg), either alone (b) or with 100 pg of follistatin mRNA (d), and BMP-7 mRNA (500 pg), either alone (c) or with 50 pg of follistatin mRNA (e), were injected into the equatorial region of the two dorsal blastomeres at the four-cell stage as described (28). In contrast, equatorial regions of two ventral blastomeres at the four-cell stage were injected with follistatin mRNA (60 pg), either alone (f), with 500 pg of BMP-4 mRNA (g), or with 2 ng of BMP-7 mRNA (h). Embryos were evaluated and photos were taken at the tadpole stage. (a) Uninjected embryo. (B) Expression of molecular markers in dorsal and ventral marginal explants. Embryos were either uninjected (lane 1, 6, 7, 11, and 12) or injected with the mRNAs indicated on the top of each lane into the equatorial region of two dorsal (lane 2–5) or ventral blastomeres (lane 8–10) at the four-cell stage. The amounts of mRNA for dorsal injection were 200 pg of BMP-4 (lane 2), 200 pg of BMP-4 plus 200 pg of follistatin (lane 3), 500 pg of BMP-7 (lane 4), and 500 pg of BMP-7 plus 50 pg of follistatin (lane 5). For ventral injection, 60 pg of follistatin mRNA, 500 pg of BMP-4 mRNA, and 2 ng of BMP-7 mRNA were used. Lane 12 shows the expression of markers in whole embryos and lane 6 and 11 show the control reactions with no RT step. Dorsal or ventral marginal zones were excised and explanted at the early gastrula stage and incubated until sibling embryos reached stages 11 and 36.
To examine the antagonistic role of follistatin vis-à-vis BMP-4 and BMP-7, 200 pg of BMP-4 and 500 pg of BMP-7 mRNA, both of which cause nearly complete ventralization (average dorsoanterior index of 0–1), were used. The antagonistic effect was tested by coinjecting BMP mRNAs with follistatin mRNA into the two dorsal blastomeres of four-cell embryos. Coinjection of 100 pg follistatin mRNA with 200 pg of BMP-4 partially rescued the ventralized phenotype to average dorsoanterior index of 4 (Fig. 1Ad). Complete rescue, however, was not achieved even at 300 pg of follistatin mRNA (data not shown). In contrast, only 10 pg follistatin mRNA was needed to significantly rescue the ventralized phenotype of embryos treated with 500 pg of BMP-7 mRNA (data not shown), and 50 pg effected almost complete rescue (Fig. 1Ae). These results suggest that follistatin can antagonize BMP-7 more efficiently than BMP-4. This antagonistic effect was further confirmed using molecular markers expressed in the dorsal marginal (equatorial) region using RT-PCR (Fig. 1B). Expression of the dorsal mesoderm marker goosecoid, which is suppressed by BMP-4 and BMP-7 mRNA overexpression, was restored by the coinjection of follistatin mRNA. In particular, follistatin effectively returned the BMP-7-suppressed goosecoid expression to normal levels. Conversely, a ventral mesoderm marker, Xvent1, ectopically induced in the dorsal mesoderm by BMPs, was suppressed by follistatin to normal levels and a definitive ventral mesoderm marker, αT1 globin, induced by BMP, also was down-regulated by the overexpression of follistatin (lane 2–5). Thus, by both criteria, i.e., morphology and molecular markers, the preferential inhibition of BMP-7 by follistatin in dorsal mesoderm was confirmed.
BMP Can Rescue Dorsalization and Neuralization Induced by Follistatin.
It has been shown that follistatin induces the dorsal mesoderm and a secondary body axis in Xenopus when it is ventrally overexpressed (17). These effects are indistinguishable from those caused by the ventral overexpression of a dominant-negative BMP receptor that inhibits both BMP-2 and BMP-4 (30, 31). Therefore, it has been speculated that follistatin might inhibit the BMP-signaling pathway (9, 17). In our experiments, 60 pg of follistatin mRNA ventrally injected was sufficient to induce a secondary axis although it lacks anterior structures such as a brain and eyes (Fig. 1Af, arrowheads). To examine whether this secondary axis formation is caused by the inhibitory effects of follistatin on BMP, we tested whether BMP could rescue the secondary axis formation. Coinjection of 50 pg of BMP-4 mRNA with 60 pg of follistatin mRNA partially suppressed the formation of secondary axis (data not shown), and 500 pg of the mRNA almost completely suppressed it (Fig. 1Ag). In contrast, no significant inhibitory effect of BMP-7 mRNA was observed with 500 pg (data not shown), and 2 ng was required to suppress the secondary axis to a level similar to that of embryos rescued by 500 pg of BMP-4 mRNA (Fig. 1Ah). Analysis of molecular marker expression further demonstrated that follistatin-induced dorsal marker gene induction and ventral marker suppression were reciprocally returned to normal levels by the coinjection of BMP mRNAs (Fig. 1B), although the rescuing effect of BMP-7 (lane 10) appeared to be weak compared with that of BMP-4 (lane 9).
One intriguing observation shown in the above experiments (Fig. 1) is that the antagonism between BMPs and follistatin is related to the specificity of follistatin to BMP subtypes. In dorsal blastomeres, BMP-7 mRNA is inhibited by the overexpression of follistatin more efficiently than BMP-4 mRNA is, suggesting that follistatin may have a higher affinity for BMP-7 than BMP-4. It further suggests that BMP-7 may be the biological target of follistatin. However, this possibility is unlikely because, first, in ventral blastomeres and animal cap cells, BMP-4 mRNA appeared to be more efficient in blocking follistatin and, second, BMP subunits tend to form heterodimers, thus it cannot be assumed that follistatin blocked BMP-7 homodimers. Namely, it is very likely that exogenously introduced BMP mRNA contributed to the formation of heterodimers with related BMP subunits translated by endogenous mRNAs. Supporting this possibility, a cleavage mutant of BMP-7 inhibits the processing of, not only BMP-7, but also BMP-4, presumably by heterodimerization (32). It is therefore possible that follistatin efficiently blocked BMP-2/7 or BMP-4/7 heterodimer whose ventral mesoderm-inducing activity is significantly higher than that of BMP homodimers (20). This suggestion is consistent with the observation that follistatin binds to BMP-4/7 heterodimer more efficiently than to homodimers. Alternatively, the amount of BMP-4/7 heterodimer generated by BMP-7 mRNA injection may be rather low in dorsal blastomeres because the amount of BMP-4 mRNA is limited, particularly on the dorsal side of the gastrula embryo (33). This heterodimerization may account for the observation that a smaller amount of follistatin mRNA could overcome the ventralizing activity and rescue the ventralized phenotype caused by BMP-7 mRNA injection, whereas higher doses were required to rescue the phenotype caused by BMP-4 mRNA injection. In the latter case, the injection of BMP-4 mRNA may lead to the efficient formation of heterodimer with BMP-7, whose transcript is uniformly distributed and abundant in early embryos (20). In ventral blastomeres, on the other hand, BMP-4 mRNA injection may result in the formation of BMP-4/7 heterodimer, which efficiently cancels the antagonistic effect of follistatin, whereas BMP-7 mRNA injection generates mostly BMP-7 homodimers and only trace amounts of BMP-4/7 heterodimer, both of which are less effective at rescuing embryos dorsalized by follistatin.
In the animal cap, follistatin acts as an inducer of neural differentiation (18) as does a dominant-negative BMP receptor. We confirmed that NCAM induction by follistatin was completely inhibited by BMP-4 and partially inhibited by BMP-7 (data not shown), consistent with a previous observation for BMP-4 (17). Conversely, the suppression of epidermal keratin was reversed partially by the coinjection of BMP-4 or BMP-7 mRNA (data not shown). Thus, follistatin appears to inhibit all the known BMP activities in early Xenopus embryogenesis.
Follistatin Interacts with BMP Extracellularly.
We next tested whether follistatin antagonizes BMPs extracellularly or whether it acts on the BMP pathway intracellularly. Although it is generally accepted that follistatin binds activin extracellularly, there remains a possibility that it interacts with the BMP-signaling pathway intracellularly, acting through a specific membrane receptor that has not yet been identified. To address this possibility, we used constitutively activated forms of BMP receptors (34, 35). Two types of Ser/Thr kinase receptors are known to mediate TGF-β family ligands. Type I receptors harbor a unique sequence called a GS box or type I box, which is rich in Gly and Ser, and is believed to play essential roles in signaling in association with type II receptors. Type II receptors trigger signaling by phosphorylating the GS box on type I receptors in a ligand-dependent manner. Substitution of Gln to Thr in the GS box is known to lead to constitutive activation of the TGF-β receptor (36). By using type I receptors mutated in a similar way, at least two have been shown to mediate the effects of BMP on ventralizing activity in early Xenopus embryos in a ligand-independent manner. They are constitutively active forms of ActRI (CA-ALK2) (34) and of BMPRIA (CA-ALK3) (35). Using such constitutively active receptors, it is possible to determine whether follistatin interacts with BMP extracellularly or intracellularly, i.e., in theory, the extracellular interaction of follistatin with BMPs should not inhibit the ligand-independent ventralizing signal generated by the constitutively active receptors. As shown in Fig. 2, the dorsal overexpression of constitutively activated forms of both ActRI (34) and BMPRIA (data not shown) resulted in partially ventralized embryos without head formation as previously reported (Fig. 2Aa). As we expected, ventralization by the constitutively active receptor overexpression was not inhibited by follistatin coexpression. (Fig. 2Ab). Molecular marker expression also demonstrated that the effects of constitutively active receptors were not rescued by the coinjection of follistatin mRNA (Fig. 2B). These results suggest that the interaction between follistatin and BMP occurs extracellularly.
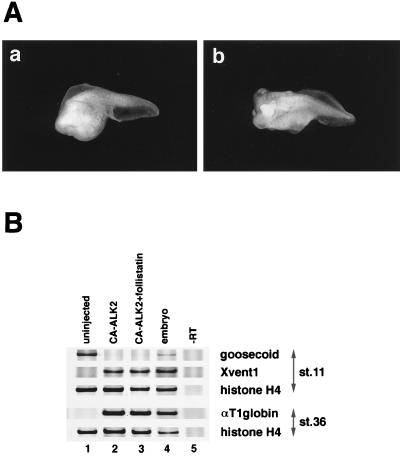
Figure 2.
Failure of follistatin to block intracellular BMP signaling. (A) Phenotype of ventralized embryos by the dorsal injection of mRNA for constitutively active BMP-signaling receptor, with or without follistatin mRNA. Fifty picograms of mRNA for a constitutively activated form of Xenopus ActRI (CA-ALK2) was injected dorsally with (b) or without (a) 200 pg of follistatin mRNA as described in the legend for Fig. 1. (B) Expression of molecular markers in dorsal marginal explants of embryos that were uninjected (lane 1), injected with 500 pg of mRNA for CA-ALK2 alone (lane 2), and coinjected with 500 pg of CA-ALK2 and 200 pg of follistatin (lane 3). Dorsal mRNA injection and RT-PCR analysis were performed as described in Fig. 1. Coinjection of follistatin mRNA did not reduce the effect of CA-ALK2 overexpression for both dorsal (goosecoid) and ventral (Xvent1, αT1 globin) markers. Similar results were obtained in the experiments using CA-ALK3 (data not shown).
Direct Binding of Follistatin to BMPs.
To ascertain that follistatin binds BMPs directly, we used a SPR biosensor (Biacore), which enables one to detect and follow protein–protein interactions in real time. First, the ability of follistatin to bind known TGF-β family proteins was tested. Purified activin A, TGF-β1, or BMP-4 was immobilized on a BIACORE sensor chip, and then FS-288 (5 μg/ml) was injected to flow over the sensor chips as an analyte. During the injection of FS-288, a rising slope of resonance signal was evident, indicating binding, and after injection, the resonance unit decreased slowly. However, this change in resonance signal was not detectable when the FS-288 was injected to flow over the TGF-β1-immobilized sensor chip surface. These results clearly suggest that FS-288 binds strongly to activin A but not to TGF-β1 (Fig. 3A). Interestingly, FS-288 showed significant binding to the BMP-4 homodimer although the level of response was relatively low compared with that obtained with activin A, even though the same amount of protein was fixed on the sensor chip. To test whether the interaction between BMP-4 and FS-288 is specific, we carried out a competition assay by using a biosensor. As shown in Fig. 3B, soluble FS-288 competed with the binding of BMP-4 to FS-288 immobilized on the sensor chip in a dose-dependent manner. We conclude from this result that the interaction between follistatin and BMP is specific. To further confirm this physical interaction by conventional biochemical way, we first performed immunoprecipitation assays. However, it failed to show the direct interaction, which is most likely to be caused by the fast dissociation rate of BMP-follistatin complex. Hence, we chemically cross-linked the follistatin-BMP complex. This complex was separated on 2D electrophoresis and visualized by Western blotting using specific antibodies for BMP and follistatin. As shown in Fig. 4B, BMP-4 bound to FS-288 migrated from 30 kDa to higher molecular masses (≈60, 90, 180, and 200 kDa<), whereas BMP-4 alone, without cross-linking (Fig. 4A) or with cross-linking (data not shown) did not shift significantly. This result supports that follistatin-BMP complex is indeed present in the reaction mixture.
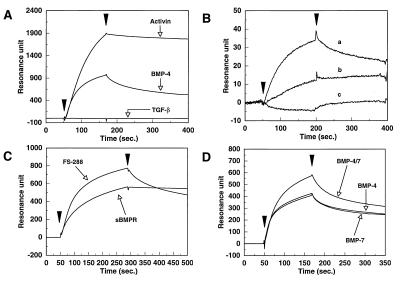
Figure 3.
Direct binding of follistatin to immobilized BMPs. (A) BMP-4, activin A, and TGF-β1 were immobilized on the sensor chip surface. The immobilization levels of BMP-4, activin A, and TGF-β1 were 2904, 2543, and 2681 resonance units, respectively. FS-288, at a concentration of 5 μg/ml, was injected over these surfaces. (B) FS-288 (807 resonance units) was immobilized on the sensor chip surface. (a) BMP-4 (1 μg/ml) was injected over the surface. (b) BMP-4 (1 μg/ml) and FS-288 (0.5 μg/ml) were incubated for 30 min at room temperature. The mixture was injected over the surface. (c) Mixture of BMP-4 (1 μg/ml) with FS-288 (2.5 μg/ml) was incubated and injected as described above. Excess soluble FS-288 almost completely abolished the binding activity on the FS-288 surface. (C) FS-288 and sBMPR, at the concentration of 5 μg/ml and 20 μg/ml, respectively, were injected over the surface of immobilized BMP-4, separately. (D) BMP-4, BMP-4/7, and BMP-7 were immobilized on the sensor chip surface. The immobilization levels of these ligands were 622, 768, and 697 resonance units, respectively. FS-288 at 5 μg/ml was injected over these surfaces.
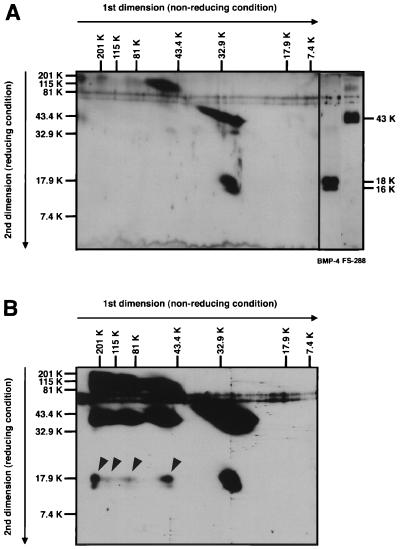
Figure 4.
Diagonal SDS/PAGE analysis of FS-288-BMP-4 complex. (A) For comparison of molecular mass, a mixture of FS-288 and BMP-4 was analyzed by 2D electrophoresis. Although FS-288 (32 kDa) tends to shift to higher molecular mass region (>60 kDa) on first dimension electrophoresis probably by self aggregation, a major band of BMP-4 shows the estimated molecular mass of dimeric form (30 kDa). FS-288 was detected as a 43-kDa protein, and BMP-4 was detected as 16- and 18-kDa proteins, which were generated because of difference of glycosylation, under reducing condition. (B) Cross-linked FS-288-BMP-4 complex was analyzed. Proteins were separated by 2D electrophoresis and subjected to Western blotting. At least, four-shifted bands (arrowheads) of BMP-4, which were not seen in A, were detected. Relative molecular mass is indicated horizontally for nonreducing, vertically for reducing condition.
Next, the kinetic parameters for the binding of FS-288 to BMP-4 were determined and compared with those for a soluble form of BMPRIA (sBMPR) that has previously been shown to bind BMP-2 and BMP-4 homodimers independently of the BMP type II receptor (21). The association rate constant for FS-288 (kass = 1.16 × 105) was faster than that for the soluble receptor binding to BMP-4 (kass = 3.81 × 104) (21). In contrast, following the injection, the dissociation of FS-288 was significantly faster than that of sBMPR (Fig. 3C). The dissociation rate constant of FS-288 was estimated from the resonance data (kdiss = 2.7 × 10−3). From the association and dissociation rate constants, the affinity of FS-288 for BMP-4 was calculated to be 23 nM, which is lower than that for sBMPR (9.6 nM) (21). Because BMP-4 is immobilized on the sensor surface by the amine-coupling method that may lead to random immobilization, we determined these values of rate constants from the sensorgrams at the low concentrations of FS-288 to obtain more reliable values (37). Although the association and dissociation rate constants are considered to be semi-quantitative, these data are thought to be valuable in the biological context. We then examined the specificity of FS-288 for each of the BMPs (Fig. 3D). FS-288 bound both BMP-2 (data not shown) and BMP-4 homodimers in an identical manner, as would be expected from the similarity of their primary sequences. It also bound BMP-7, whose primary structure is relatively distant from BMP-2 and -4, with almost the same binding profile as obtained with BMP-2 and -4. Interestingly, FS-288 showed a higher binding affinity for the BMP-4/7 heterodimer compared with other BMP homodimers (Fig. 3D).
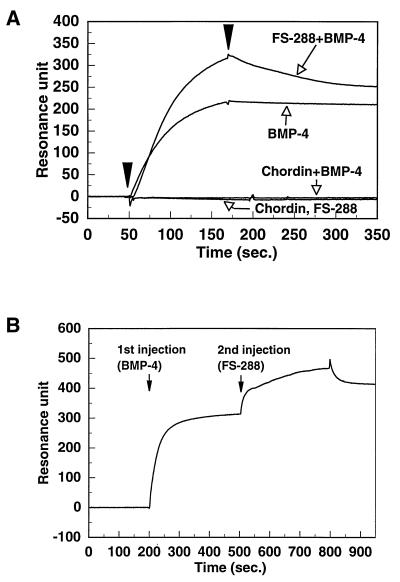
Next, we investigated the mechanism by which follistatin inhibits BMP activity. Chordin and noggin have been shown to interfere with the ability of BMP-4 to bind the BMP receptor and to inhibit BMP activity in a competitive manner (14, 15). In fact, as shown in Fig. 5A, a mixture of chordin and BMP-4 showed no detectable binding capacity for the BMPRIA receptor. Interestingly, FS-288 appears to bind BMP-4 in a noncompetitive manner; a mixture of FS-288 and BMP-4 was still able to bind soluble BMP receptor, suggesting that they form trimeric complex. To confirm this possibility, BMP-4 and FS-288 were sequentially added to the receptor by using the biosensor. After binding of BMP-4 to sBMPR immobilized on the sensor surface, a second increase of resonance signal was observed by sequential injection of FS-288 (Fig. 5B). This result suggests that the binding site of BMP-4 for its receptor may be distinct from that for follistatin and that follistatin and chordin inhibit the receptor activation with different mechanisms (Fig. 6).
Figure 5.
Different competitive mechanisms of two BMP-4 antagonists. (A) BMP-4, chordin, and FS-288, at the concentration of 10 μg/ml, 2 μg/ml, 20 μg/ml, respectively, were injected over the surface immobilized sBMPR, separately. For the competition assay, a mixture of BMP-4 with chordin or FS-288 was injected. (B) FS-288 was injected after the injection of BMP-4 over the sBMPR-immobilized surface by using of the coinjection method. The concentrations of BMP-4 and FS-288 were 10 μg/ml and 5 μg/ml, respectively. Both proteins were injected at a flow rate of 10 μl/min for 300 s.
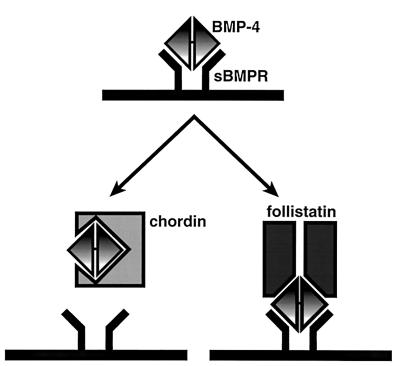
Figure 6.
Possible inhibitory mechanisms of the BMP action by follistatin and chordin. Chordin inhibits the binding of BMP-4 to its type I receptor (sBMPR). On the other hand, follistatin can bind to sBMPR through BMP-4, forming a trimeric complex.
The present study demonstrated that follistatin can inhibit the effects of all three BMP subunits: BMP-2, -4, and -7. Follistatin was previously shown to inhibit the growth inhibitory effect of OP-1 (BMP-7 homodimer) on Mv1Lu cells (38) at high concentrations. The present report is the first to reveal the negative effect of follistatin on BMP family proteins by direct binding. The restricted expression of follistatin mRNA in the Spemann’s organizer and its ability to block BMPs by binding to them support the idea that follistatin also acts as an organizer factor in Xenopus embryogenesis, but in a manner different from the other known organizer factors, noggin and chordin. Although we suggest the involvement of heterodimer formation, the identification of endogenous BMP heterodimers and negative regulation by their binding proteins awaits further studies.
We would like to thank Drs. Fujisawa and Hazama, Takeda Chemical Industries Ltd. for supplying us with recombinant BMP proteins, Dr. Eto, Ajinomoto Inc. for activin A, Drs. Piccolo and De Robertis for chordin-containing medium, Dr. Shibuya for the constitutively active BMPRIA cDNA, and Dr. Hemmati-Brivanlou for Xenopus follistatin plasmid. This work is supported by the “Research for the Future” Program of the Japan Society for the Promotion of Science (JSPS–RFTF96L00406).
ABBREVIATIONS
- BMP
bone morphogenetic protein
- FS-288
follistatin-288
- TGF-β
transforming growth factor-β
- sBMPR
soluble form of BMP type IA receptor
- RT
reverse transcription
- 2D
two-dimensional
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Kingsley D M. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 2.Sive H L. Genes Dev. 1993;7:1–12. doi: 10.1101/gad.7.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Asashima M, Nakano H, Shimada K, Kinoshita K, Ishii K, Shibai H, Ueno N. Roux’s Arch Dev Biol. 1990;198:330–335. doi: 10.1007/BF00383771. [DOI] [PubMed] [Google Scholar]
- 4.Smith J C, Price B M, Van Nimmen K, Huylebroeck D. Nature (London) 1990;345:729–731. doi: 10.1038/345729a0. [DOI] [PubMed] [Google Scholar]
- 5.Thomsen G H, Melton D A. Cell. 1993;74:433–441. doi: 10.1016/0092-8674(93)80045-g. [DOI] [PubMed] [Google Scholar]
- 6.Maeno M, Ong R C, Xue Y, Nishimatsu S, Ueno N, Kung H F. Dev Biol. 1994;161:522–529. doi: 10.1006/dbio.1994.1050. [DOI] [PubMed] [Google Scholar]
- 7.Wilson P A, Hemmati-Brivanlou A. Nature (London) 1995;376:331–333. doi: 10.1038/376331a0. [DOI] [PubMed] [Google Scholar]
- 8.Graff J M. Cell. 1997;89:171–174. doi: 10.1016/s0092-8674(00)80196-8. [DOI] [PubMed] [Google Scholar]
- 9.Thomsen G H. Trends Genet. 1997;13:209–211. doi: 10.1016/S0168-9525(97)01117-7. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura T, Takio K, Eto Y, Shibai H, Titani K, Sugino H. Science. 1990;247:836–838. doi: 10.1126/science.2106159. [DOI] [PubMed] [Google Scholar]
- 11.Fukui A, Nakamura T, Sugino K, Takio K, Uchiyama H, Asashima M, Sugino H. Dev Biol. 1993;159:131–139. doi: 10.1006/dbio.1993.1227. [DOI] [PubMed] [Google Scholar]
- 12.Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont L K, De Robertis E M. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith W C, Harland R M. Cell. 1992;70:829–840. doi: 10.1016/0092-8674(92)90316-5. [DOI] [PubMed] [Google Scholar]
- 14.Piccolo S, Sasai Y, Lu B, De Robertis E M. Cell. 1996;86:589–598. doi: 10.1016/s0092-8674(00)80132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmerman L B, De Jesus-Escobar J M, Harland R M. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- 16.Moon R T, Brown J D, Yang-Snyder J A, Miller J R. Cell. 1997;88:725–728. doi: 10.1016/s0092-8674(00)81915-7. [DOI] [PubMed] [Google Scholar]
- 17.Sasai Y, Lu B, Steinbeisser H, De Robertis E M. Nature (London) 1995;376:333–336. doi: 10.1038/376333a0. [DOI] [PubMed] [Google Scholar]
- 18.Hemmati-Brivanlou A, Kelly O G, Melton D A. Cell. 1994;77:283–295. doi: 10.1016/0092-8674(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 19.Fainsod A, Deissler K, Yelin R, Marom K, Epstein M, Pillemer G, Steinbeisser H, Blum M. Mech Dev. 1997;63:39–50. doi: 10.1016/s0925-4773(97)00673-4. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki A, Kaneko E, Maeda J, Ueno N. Biochem Biophys Res Commun. 1997;232:153–156. doi: 10.1006/bbrc.1997.6219. [DOI] [PubMed] [Google Scholar]
- 21.Natsume T, Tomita S, Iemura S, Kinto N, Yamaguchi A, Ueno N. J Biol Chem. 1997;272:11535–11540. doi: 10.1074/jbc.272.17.11535. [DOI] [PubMed] [Google Scholar]
- 22.Inouye S, Guo Y, DePaolo L, Shimonaka M, Ling N, Shimasaki S. Endocrinology. 1991;129:815–822. doi: 10.1210/endo-129-2-815. [DOI] [PubMed] [Google Scholar]
- 23.Johnsson B, Löfás S, Lindquist G. Anal Biochem. 1991;198:268–277. doi: 10.1016/0003-2697(91)90424-r. [DOI] [PubMed] [Google Scholar]
- 24.Karlsson R, Roos H, Färgestam L, Persson B. Methods (Orlando) 1994;6:99–110. [Google Scholar]
- 25.Ueno N, Shoda A, Takebayashi K, Suzuki A, Nishimatsu S, Kikuchi T, Wakimasu M, Fujino M, Murakami K. Growth Factors. 1992;7:233–240. doi: 10.3109/08977199209046927. [DOI] [PubMed] [Google Scholar]
- 26.Nishimatsu S, Takebayashi K, Suzuki A, Murakami K, Ueno N. Growth Factors. 1993;8:173–176. doi: 10.3109/08977199309011020. [DOI] [PubMed] [Google Scholar]
- 27.Dale L, Howes G, Price B M, Smith J C. Development (Cambridge, UK) 1992;115:573–585. doi: 10.1242/dev.115.2.573. [DOI] [PubMed] [Google Scholar]
- 28.Jones C M, Lyons K M, Lapan P M, Wright C V, Hogan B L. Development (Cambridge, UK) 1992;115:639–647. doi: 10.1242/dev.115.2.639. [DOI] [PubMed] [Google Scholar]
- 29.Nishimatsu S, Suzuki A, Shoda A, Murakami K, Ueno N. Biochem Biophys Res Commun. 1992;186:1487–1495. doi: 10.1016/s0006-291x(05)81574-8. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki A, Thies R S, Yamaji N, Song J J, Wozney J M, Murakami K, Ueno N. Proc Natl Acad Sci USA. 1994;91:10255–10259. doi: 10.1073/pnas.91.22.10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graff J M, Thies R S, Song J J, Celeste A J, Melton D A. Cell. 1994;79:169–179. doi: 10.1016/0092-8674(94)90409-x. [DOI] [PubMed] [Google Scholar]
- 32.Hawley S H, Wunnenberg-Stapleton K, Hashimoto C, Laurent M N, Watabe T, Blumberg B W, Cho K W. Genes Dev. 1995;9:2923–2935. doi: 10.1101/gad.9.23.2923. [DOI] [PubMed] [Google Scholar]
- 33.Hemmati-Brivanlou A, Thomsen G H. Dev Genet. 1995;17:78–89. doi: 10.1002/dvg.1020170109. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki A, Kaneko E, Ueno N, Hemmati-Brivanlou A. Dev Biol. 1997;189:112–122. doi: 10.1006/dbio.1997.8652. [DOI] [PubMed] [Google Scholar]
- 35.Akiyama S, Katagiri T, Namiki M, Yamaji N, Yamamoto N, Miyama K, Shibuya H, Ueno N, Wozney J M, Suda T. Exp Cell Res. 1997;235:362–369. doi: 10.1006/excr.1997.3680. [DOI] [PubMed] [Google Scholar]
- 36.Wieser R, Wrana J L, Massague J. EMBO J. 1995;14:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuck P. Annu Rev Biophys Biomol Struct. 1997;26:541–566. doi: 10.1146/annurev.biophys.26.1.541. [DOI] [PubMed] [Google Scholar]
- 38.Yamashita H, ten Dijke P, Huylebroeck D, Sampath T K, Andries M, Smith J C, Heldin C H, Miyazono K. J Cell Biol. 1995;130:217–226. doi: 10.1083/jcb.130.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]