Abstract
Saccharomyces cerevisiae cells lacking the MDM12 gene product display temperature-sensitive growth and possess abnormally large, round mitochondria that are defective for inheritance by daughter buds. Analysis of the wild-type MDM12 gene revealed its product to be a 31-kD polypeptide that is homologous to a protein of the fission yeast Schizosaccharomyces pombe. When expressed in S. cerevisiae, the S. pombe Mdm12p homolog conferred a dominant-negative phenotype of giant mitochondria and aberrant mitochondrial distribution, suggesting partial functional conservation of Mdm12p activity between budding and fission yeast. The S. cerevisiae Mdm12p was localized by indirect immunofluorescence microscopy and by subcellular fractionation and immunodetection to the mitochondrial outer membrane and displayed biochemical properties of an integral membrane protein. Mdm12p is the third mitochondrial outer membrane protein required for normal mitochondrial morphology and distribution to be identified in S. cerevisiae and the first such mitochondrial component that is conserved between two different species.
Mitochondria are essential organelles that arise only by growth and division of preexisting mitochondria (Attardi and Schatz, 1988). Before completion of cytokinesis, a daughter cell must therefore receive a mitochondrial mass sufficient for viability. The molecular mechanisms and cellular components that mediate this mitochondrial inheritance are beginning to be elucidated through the analysis of Saccharomyces cerevisiae mutants exhibiting specific defects in mitochondrial distribution. These mitochondrial distribution and morphology (mdm)1 mutants were isolated by screening collections of temperature-sensitive strains by fluorescence microscopy to identify cells that failed to transfer mitochondria into daughter buds at the nonpermissive temperature (McConnell et al., 1990). Characterization of some of the mdm mutants has indicated that mitochondrial inheritance is a specific, active process that depends on a number of novel cellular components (Yaffe, 1996). One of these components, the Mdm1p protein, is a cytoskeletal element that exhibits structural similarities to the intermediate filament proteins of animal cells (McConnell and Yaffe, 1992, 1993). In addition, two different proteins of the mitochondrial outer membrane, Mdm10p and Mmm1p, were shown to be required for maintenance of the normal mitochondrial reticular network as well as for mitochondrial transmission to daughter cells (Sogo and Yaffe, 1994; Burgess et al., 1994). This report describes a third protein of the mitochondrial outer membrane, Mdm12p, which is essential for normal mitochondrial morphology and inheritance and which possesses a conserved homolog in the fission yeast Schizosaccharomyces pombe.
Materials and Methods
Strains and Genetic Techniques
S. cerevisiae strains used in this study were derived from wild-type strains MYY290 (MAT a, leu2, his3, ura3), MYY291 (MATα, leu2, his3, ura3), or MYY298 (MAT a/α, leu2, his3, ura3) (Smith and Yaffe, 1991; McConnell and Yaffe, 1992). mdm12 mutant strains MYY620 (MAT a, leu2, his3, ura3, mdm12-1), MYY621 (MATα, leu2, his3, ura3, mdm12-1), MYY623 (MAT a, leu2, his3, ura3, mdm12::URA3), and MYY624 (MATα, leu2, his3, ura3, mdm12::URA3) are described below. mdm10 mutant strains included MYY503 (MAT a, leu2, his3, ura3, mdm10::URA3) (Sogo and Yaffe, 1994) and MYY505 (MAT a, leu2, his3, ura3, mdm10::LEU2) (Berger, K., and M. Yaffe, unpublished results). SOT1 strains MYY626 (MAT a, leu2, his3, ura3, SOT1), MYY627 (MAT a, leu2, his3, ura3, mdm10::URA3, SOT1), MYY628 (MAT a, leu2, his3, ura3, mdm12-1, SOT1), and MYY629 (MATα, leu2, his3, ura3, mdm12::URA3, SOT1) are described below. Strain MYY461 (MAT a, leu2, his3, ura3, top1::URA3) was isolated as a Ura+ transformant generated by transforming strain MYY290 with a top1::URA3 disruption cassette, which was a gift from C. Holm (University of California, San Diego). Growth conditions and media for S. cerevisiae were essentially as described (Rose et al., 1990). Yeast were transformed using lithium acetate (Ito et al., 1983). Escherichia coli strains DH5α and MH6 were used to amplify plasmid DNA. DNA manipulations were as described (Sambrook et al., 1989).
Identification of the mdm12-1 Mutant
The mdm12-1 mutant was isolated from a collection of temperature-sensitive strains by microscopic screening as previously described (Yaffe, 1995). The original mutant isolate was backcrossed three times to the wild-type parental strain to yield strain MYY620, and meiotic progeny from the final backcross displayed 2:2 cosegregation of temperature-sensitive growth and defects in mitochondrial distribution and morphology.
Cloning and Sequence Analysis of MDM12
The MDM12 gene was isolated by complementation of the temperaturesensitive phenotype of the mdm12-1 mutant. mdm12-1 cells were transformed with a yeast genomic DNA library in centromere vector p366 (obtained from M. Hoekstra, ICOS Inc., Bothell, WA). Leu+ transformants were selected at 23°C and were replica plated to 37°C to identify temperature-resistant colonies. Six different clones were isolated, and restriction analysis revealed that these plasmids contained overlapping DNA inserts. Complementing activity was localized to a 1.3-kb KpnI–XbaI DNA fragment by subcloning and transformation of mdm12-1 cells.
The 1.3-kb fragment that complemented mdm12-1 was subcloned into plasmid pBluescript KS(+) (Stratagene Inc., La Jolla, CA) and digested with Exonuclease S1 to generate a set of nested deletions to use as sequencing templates. Nucleotide sequence of both strands of the complementing DNA region was determined by dideoxynucleotide sequencing (Sanger et al., 1977). Oligonucleotide primers used for sequencing and PCR amplification (described below) were purchased from Operon Technologies Inc. (Alameda, CA). Subsequent to nucleotide sequencing of MDM12 DNA in our laboratory, the DNA sequence for the region including MDM12 was made available by the Saccharomyces Genome Database (SGD). Our DNA sequence data are largely in agreement with those provided by the SGD and are available from EMBL/GenBank/ DDBJ under accession number U62252.
Mapping of MDM12 and mdm12-1
MDM12 was physically mapped by hybridization of a 32P-labeled DNA fragment containing the cloned MDM12 gene to a set of filters containing a mapped set of genomic clones provided by Dr. Linda Riles (Washington University, St. Louis, MO). The cloned sequences hybridized to two overlapping genomic clones from a region near the centromere of chromosome XV.
The mdm12-1 mutation was tested for linkage to TOP1, a locus on the right arm and near the centromere of chromosome XV, by meiotic mapping. The meiotic progeny of a cross of strain MYY621 (mdm12-1 TOP1) and strain MYY461 (MDM12 top1::URA3) yielded a total of 45/45 tetrads of the parental ditype (2 Ura+ Ts+, 2 Ura− Ts−), indicating linkage of the MDM12 and TOP1 loci to within 1 centiMorgan (Sherman and Wakem, 1991).
MDM12 Gene Replacement
A 1.9-kb KpnI–ClaI DNA fragment containing MDM12 was subcloned into pBluescript KS(+) to yield plasmid pKB34. The URA3 gene was isolated on a HindIII fragment from plasmid pFL1 (Chevallier et al., 1980), the fragment ends were filled with Klenow, and the fragment was used to replace most of the MDM12 gene by ligation into the unique SnaBI and MscI sites in plasmid pKB34. The resulting mdm12::URA3 disruption cassette was excised from the vector by digestion with KpnI and ClaI and transformed into diploid strain MYY298. Replacement of one of two chromosomal copies of MDM12 coding sequences with URA3 was confirmed by Southern blot analysis. The mdm12::URA3 (null) haploid strains (MYY623 and MYY624) were recovered by sporulation of the heterozygous diploid.
Analysis of Loss of Respiratory Function
Haploid meiotic progeny were obtained by sporulation of diploid cells heterozygous for either the mdm10::LEU2 mutation or the mdm12::URA3 mutation (obtained by crossing MYY291 to MYY505 or to MYY623, respectively), and the genotype of haploid segregants was determined. For each strain, individual cultures of four different haploid segregants were grown overnight in YPD liquid medium at 23°C, and dilutions were plated onto YPD agar medium. Strains of three different genotypes, MDM10 MDM12 (strain MYY290), mdm12::URA3, and mdm10::LEU2, were tested. Plates were incubated until small colonies were visible, and colonies were replica plated onto YPG medium. Colonies that grew on YPD but not on YPG were scored as having lost respiratory function (rho− or rhoo). For each strain a total of at least 243 colonies was counted.
Cloning and Analysis of the S. pombe MDM12 Homolog
The DNA region containing the S. pombe Mdm12p homolog, which was designated mdm12p, was amplified by PCR from genomic DNA of S. pombe strain 975 using oligonucleotide primers flanking the open reading frame. Primer sequences were 5′-GTACTAATTCGAAGATGAAAGAGTGAATG-3′ (PMT-U01) and 5′-ATGCTGCAATAGTGCTCGCCAAGA-3′ (PMT-L01). PCR amplification was performed with Taq polymerase (Fisher Scientific, Pittsburgh, PA) in Taq reaction buffer supplemented with 2.5 mM MgCl2 and 0.2 mM dNTPs (Boehringer Mannheim Corp., Indianapolis, IN) using an ERICOMP thermal cycler. The PCR product was cloned into the vector pCR2.1 (Invitrogen, San Diego, CA) to generate plasmid pPM1. Nucleotide sequencing of the cloned mdm12p was performed to verify the sequence reported in the database and to resolve ambiguities in this sequence. The DNA sequence obtained is available from GenBank under accession number U64674.
To generate a vector for expression of mdm12p in S. cerevisiae, the putative open reading frame was amplified from genomic DNA of S. pombe strain 975 by PCR using a second upstream primer immediately 5′ of and including the predicted translational start site. Primers used were PMTU02 (5′-GGGAATTCCAGAACAATGTCTATTGACTTTGATTGG-3′) and PMT-L01 (described above). The primer PMT-U02 introduced an EcoRI site (underlined) just upstream of the translational start site (bold). The PCR product was cloned into the pCR2.1 vector to generate plasmid pPM2. An EcoRI fragment containing mdm12p coding sequences was isolated from pPM2 and ligated into EcoRI-digested DNA of the S. cerevisiae centromere-based expression plasmid pAC1 (Hurt et al., 1985). The orientation of the mdm12p fragment in pAC1 was verified by restriction analysis. The resulting construct, plasmid pPM3, contains the S. pombe mdm12p gene under transcriptional control of the constitutive S. cerevisiae ADH1 promoter.
To construct a vector for the constitutive, high-level expression of S. cerevisiae MDM12, primers MDM12-U01 (5′-GGGTCGACAAATGTCTTTTGATATTAATTGGAGTA-3′) and MDM12-L01 (5′-GGGGATCCTTTAACTCTTTTGGGTCATCTAA-3′) were used for PCR amplification of S. cerevisiae genomic DNA from strain MYY290. The PCR product was cloned into plasmid PCR2.1, and a 1-kb EcoRI fragment containing MDM12 was subcloned into the EcoRI site of plasmid pAC1 to yield plasmid pKB37.
Isolation of the SOT1 Mutation
Haploid cells harboring the mdm10::URA3 mutation (strain MYY503) were grown overnight in YPD medium at 23°C to a density of ∼108 cells/ml. A total of 1.5 × 1010 cells was plated onto 40 YPD agar plates (4 × 108 cells/plate). Plates were incubated at 37°C, and a single temperature-resistant colony was obtained. Genetic analysis determined that temperatureresistance was caused by a single nuclear mutation unlinked to MDM10. Subsequent analysis showed that the suppressor mutation, designated SOT1, also suppressed defects caused by either the mdm12-1 or the mdm12-null mutation as well as those of the mdm10-null mdm12-null double mutant. In addition, a mutation that was determined genetically to be a second allele of SOT1 with similar suppressing activity was isolated independently in a similar selection for suppressors of the mdm12-null mutation.
Preparation of Antibodies to Mdm12p
The peptide (KGSWINLDFNDDDE), comprising a two–amino acid linker followed by 12 residues corresponding to the COOH terminus of the predicted MDM12 product, was synthesized, conjugated to keyhole limpet hemocyanin, and used to immunize rabbits (Research Genetics, Huntsville, AL). For use in indirect immunofluorescence microscopy experiments, antibodies were purified on an affinity column prepared by coupling the Mdm12p COOH-terminal peptide to Affi-Gel 15 (Bio Rad Laboratories, Hercules, CA) per the manufacturer's instructions. Affinity-purified antibodies were subsequently preabsorbed against fixed mdm12-null cells (Pringle et al., 1991) before use. For immunoblot analysis, antibodies were purified on an affinity column prepared by coupling a β-galactosidase– Mdm12p fusion protein consisting of LacZ fused to all but the first three amino acids of Mdm12p (Berger, K., unpublished results) to Affi-Gel 10. Antibodies prepared by affinity purification against the LacZ–Mdm12p fusion protein recognized the same Mdm12p species as those purified against the COOH-terminal peptide but recognized fewer cross-reacting species (data not shown). Binding of anti-Mdm12p antibodies to both types of affinity columns and elution with 0.1 M glycine, pH 2.2, were essentially as described (Harlow and Lane, 1988), except that the column was washed with a solution of 0.14 M NaCl, 2.7 mM KCl, 5.4 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.2 (PBS), instead of Tris buffer. Preimmune sera subjected to the affinity purification protocol failed to yield antibodies with strong reaction to any of the protein species recognized by the affinity-purified immune antibodies.
Construction of Epitope-tagged Mdm12p
The 1.3-kb KpnI–XbaI DNA fragment including MDM12 was cloned into plasmid pRS316 (Sikorski and Hieter, 1989) to generate plasmid pKB35. Plasmid pJR1265, the source of c-myc epitope, consists of six tandem repeats of the epitope sequences in the polylinker of plasmid pBluescript KS(+) and was a gift from R. Hampton (University of California, San Diego). Sequences corresponding to five tandem copies of the c-myc epitope were purified from plasmid pJR1265 after digestion with ClaI and NcoI and treatment with Klenow and were ligated into the unique SnaBI site within the MDM12 gene in pKB35 to yield pKB36. The correct orientation and fusion junctions of the c-myc–MDM12 construction were confirmed with restriction endonuclease and DNA sequence analysis.
Fluorescence Microscopy
Staining of live cells with the mitochondria-specific dye DASPMI (2-[4dimethylaminostryl]-1-methylpyridinium iodide) and fluorescence microscopy were as previously described (Yaffe, 1995). Indirect immunofluorescence microscopy was performed as described previously (McConnell et al., 1990). The 9E10 antibodies (Evan et al., 1985) used for immunodetection of c-myc–Mdm12p were purchased from Berkeley Antibody Co. (Richmond, CA).
Cell Fractionation
Yeast cells were grown in semisynthetic lactate medium (Daum et al., 1982), homogenized, and subjected to differential centrifugation to isolate mitochondria and other subcellular fractions as previously described (Yaffe, 1991; Schauer et al., 1985). Mitochondrial subfractions were isolated as described by Daum et al. (1982). Extraction of mitochondria with sodium carbonate solution was performed as described (Sogo and Yaffe, 1994). SDS-PAGE and immunoblotting were performed by standard procedures (Douglas and Butow, 1976; Towbin et al., 1979).
Results
The mdm12-1 Mutation Causes Abnormal Mitochondrial Morphology and Distribution
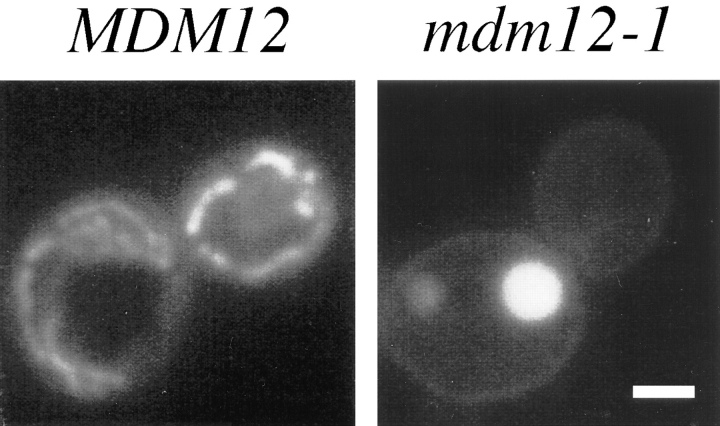
The mdm12-1 mutant was identified in a collection of S. cerevisiae temperature-sensitive strains by microscopic screening of cells stained with the mitochondria-specific vital dye DASPMI (McConnell et al., 1990; Sogo and Yaffe, 1994). When visualized by fluorescence microscopy, wildtype mitochondria appeared as extended tubules and tubular networks peripherally distributed in both mother and bud portions of the cell (Fig. 1). In contrast, mdm12-1 mutant cells typically displayed one or two large round mitochondria per cell (Fig. 1). These giant mitochondria were frequently localized exclusively to the mother portion of cells, with mitochondrial staining largely absent from daughter buds. The mdm12-1 mutant displayed giant mitochondria and defective mitochondrial distribution at both permissive (23°C) and nonpermissive (37°C) temperatures (see Fig. 2), and mdm12-1 cells grew slowly at the permissive temperature. Large round mitochondria were present in mdm12-1 cells cultured on both fermentable (glucose) and nonfermentable (glycerol) carbon sources. Despite their grossly altered morphology, the mdm12-1 mutant mitochondria appeared to be competent for respiration because mdm12-1 cells grew, although very slowly, at 23°C on media containing glycerol. In addition, DASPMI uptake by mdm12-1 mitochondria indicated the presence of a significant membrane potential (Bereiter-Hahn, 1976).
Figure 1.
mdm12 mutant cells display giant mitochondria that are defective for distribution to buds. MDM12 (wild type; MYY290) and mdm12-1 (MYY621) cells were grown at 23°C in YPD medium, stained with DASPMI, and viewed by fluorescence microscopy. Representative wild-type (left) and mdm12-1 cells (right) are shown. Bar, 2 μm.
Figure 2.
Indirect immunofluorescence confirms the mitochondrial inheritance and morphology defects of the mdm12 mutant. MDM12 (wild type; MYY290) and mdm12-1 (MYY621) cells were grown in YPD medium at 23°C, incubated at 37°C for 2 h, fixed with formaldehyde, and processed for immunofluorescence. Mitochondria were detected with antibodies specific for the mitochondrial outer membrane protein OM14 (B), and mitochondrial and nuclear DNAs were visualized by DAPI staining (C). Brightfield images of the corresponding cells are shown in A. Bar, 2 μm.
The defects in mitochondrial distribution and morphology in mdm12-1 mutant cells were confirmed by indirect immunofluorescence microscopy using antibodies specific for OM14, a major protein of the mitochondrial outer membrane (Fig. 2 B). Nuclear division and distribution appeared unaffected by the mdm12-1 mutation based on DAPI (4,6-diamidino-2-phenylindole) staining of nuclear DNA (Fig. 2 C).
The phenotype of the mdm12-1 mutant closely resembled that of another mutant, mdm10, previously characterized by our laboratory (Sogo and Yaffe, 1994), as well as that of a different mutant, mmm1 (Burgess et al., 1994). The mdm12-1 mutation was recessive and genetically distinct from mdm10 based on complementation analysis and allelism tests. In addition, the cloned MDM10 gene did not complement the mdm12-1 mutation. Haploid mdm10-null mdm12-1 double mutants were viable at 23°C and displayed temperature-sensitive growth, mitochondrial distribution, and mitochondrial morphology phenotypes similar to those of the single mutant (parental) strains (data not shown). The MDM12 gene was not identical to MMM1 (see below), and the mdm12-null mmm1-null double mutant displayed no synthetic phenotypes (data not shown). The mdm12 mutant was also complemented by other available mdm mutants tested. The mdm12-1 mutation therefore appeared to define a new MDM gene.
MDM12 Encodes a Novel Protein with a Homolog in Fission Yeast
To identify the molecular basis of the mdm12 mutant defects, the MDM12 gene was isolated from a wild-type S. cerevisiae genomic DNA library by complementation of the mdm12-1 temperature-sensitive growth defect. Nucleotide sequence analysis of a 1.3-kb DNA region (Fig. 3), which fully complemented the mutant phenotype, revealed a single open reading frame of 813 bp encoding a putative polypeptide of ∼31 kD. An analysis of the predicted MDM12 product failed to reveal any known sequence motifs (Bairoch, 1992); however, the region from amino acid 24 to residue 40 is uncharged and of sufficient length to comprise a potential membrane-spanning domain. Subsequent to our analysis, the MDM12 open reading frame was independently identified as part of DNA sequence analysis by the Saccharomyces Genome Database and has been designated YOL009c.
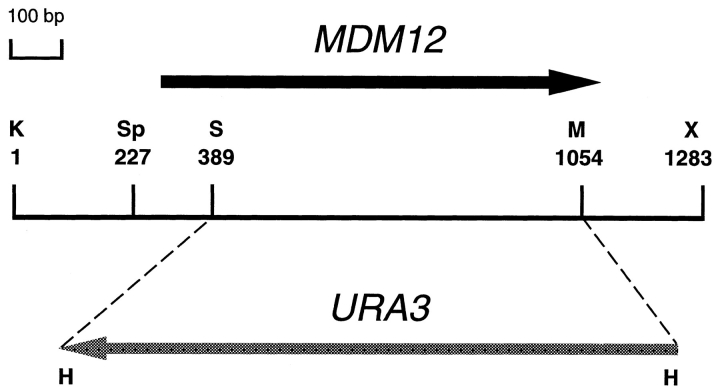
Figure 3.
Restriction map of MDM12. Represented is a 1,283-bp KpnI–XbaI DNA fragment containing MDM12 complementing activity. The MDM12 open reading frame (solid black arrow) extends from nucleotide 284 to nucleotide 1099. In the mdm12:: URA3 gene replacement, the portion of the MDM12 coding region between the SnaBI and MscI sites, corresponding to most of the MDM12 gene, is replaced with URA3 coding sequences (shaded gray arrow). Restriction sites: H, HindIII; K, KpnI; M, MscI; S, SnaBI; Sp, SphI; X, XbaI.
The cloned MDM12 gene was physically mapped to a region near the centromere on the left arm of chromosome XV (as described in Materials and Methods). The mdm12-1 mutation was mapped via genetic linkage analysis to the same region. These mapping data indicated that the DNA clone identified by complementation of the mdm12-1 mutant represented the authentic MDM12 gene rather than an extragenic suppressor.
The SWISSPROT and GENBANK data bases were searched for proteins homologous to Mdm12p using the BLAST sequence comparison program (Altschul et al., 1990). This search identified a single homologous protein: an uncharacterized, hypothetical gene product in the fission yeast S. pombe. Because the DNA sequence of the S. pombe gene reported in the database contained a number of ambiguities, the gene was cloned by PCR, and its complete nucleotide sequence was determined. This analysis revealed a longer open reading frame than that previously reported in the database, with the gene encoding a potential product of 273 amino acids (Fig. 4 A). Alignment of the predicted S. cerevisiae Mdm12p with its S. pombe homolog indicated that the proteins shared significant homology over their entire lengths, with 32% sequence identity and 50% sequence similarity (Fig. 4 A). In addition to homology with the S. pombe protein, Mdm12p exhibited one region of sequence similarity to another protein, Mmm1p (Fig. 4 B). Although the level of identity between these regions of Mdm12p and Mmm1p was low (23%), the resemblance was noteworthy because loss of Mmm1p function produces a phenotype similar to that caused by an mdm12 mutation (Burgess et al., 1994).
Figure 4.
Alignment of S. cerevisiae Mdm12p with its S. pombe homolog and with a region of Mmm1p. (A) Alignment of S. cerevisiae protein (S.c.) with S. pombe protein (S.p.). (B) Alignment of amino acid residues 123– 178 of Mdm12p with residues 278–333 of Mmm1p. Alignment was performed by the CLUSTALW program (Baylor College of Medicine, Waco, TX). Amino acid identities are indicated by a vertical bar, and similarities are indicated by :. Similarities include R and K, D and E, S and T, N and Q, and A, L, I, M, V, and F. The uncharged region of Mdm12p from amino acids 24 to 40, corresponding to a potential membrane-spanning domain, is underlined.
MDM12 Is Essential for Normal Mitochondrial Morphology, Mitochondrial Inheritance, and Sporulation
To investigate the phenotype of an mdm12-null mutation, the DNA region corresponding to most of the MDM12 coding sequences was replaced with the URA3 gene in one of two chromosomal copies in a diploid cell. Sporulation of the heterozygous diploid yielded four viable spores, of which two were wild type and two harbored the mdm12:: URA3 mutation. The mdm12-null cells displayed a phenotype very similar to the original mdm12-1 mutant: slow growth at 23°C, no growth at 37°C, and enlarged, spherical mitochondria, which were largely absent from buds at both 23°C and 37°C (see below). Transformation of mdm12-null cells with a centromere-based plasmid encoding the MDM12 gene corrected all of the mutant defects. These results indicate that MDM12 is required for the efficient transmission of mitochondria to daughter buds and for the maintenance of normal mitochondrial morphology.
The deficient mitochondrial inheritance caused by loss of MDM12 function was apparent at both permissive (23°C) and nonpermissive (37°C) temperatures. To quantify these inheritance defects, mitochondrial distribution in populations of mutant or wild-type cells was examined by fluorescence microscopy after incubation at 23°C or 37°C. At both temperatures, 100% of wild-type (MDM12) cells exhibited normal mitochondrial inheritance with mitochondria distributed in both mother and bud. In contrast, 84% (235/281) of mdm12-null cells grown at 23°C displayed a defect in mitochondrial inheritance. The fraction of mdm12-null cells with mitochondrial distribution defects increased to 89% (114/128) after incubation for 4 h at 37°C.
One difference between the mdm12-1 and mdm12-null mutants was that cells harboring the latter mutation were defective for sporulation. Diploid cells homozygous for the mdm12-null mutation did not form spores or asci when plated on sporulation medium, whereas mdm12-1/mdm12-1 homozygous diploids were capable of sporulation (data not shown).
Although mdm12 cells were able to grow (albeit very slowly) on nonfermentable carbon sources, mdm12 and mdm10 mutant cells readily produced respiration-deficient (rho− or rhoo) cells at rates much higher than that of the wild-type parental strain. During overnight growth on glucose-containing medium, 59 ± 4% of mdm12-null cells and 98 ± 2% of mdm10-null cells lost mitochondrial respiratory function, whereas only 4 ± 2% of wild-type cells became respiration deficient during the same treatment.
Expression of the Fission Yeast Mdm12p in S. cerevisiae Mimics the mdm12 Mutant Phenotype
To investigate whether the S. pombe Mdm12p homolog might exhibit conserved function with the S. cerevisiae protein, the corresponding S. pombe gene, designated mdm12p, was cloned and expressed in S. cerevisiae under control of the constitutive ADH1 promoter. This construct did not complement the mdm12-1 mutant (data not shown). In wild-type (MDM12) cells, however, the cloned S. pombe gene conferred a dominant phenotype strikingly similar to that found in the mdm12 mutant: giant, round mitochondria and empty buds reflecting defective mitochondrial inheritance (Fig. 5 B). Mitochondria in a majority of wild-type cells harboring the cloned S. pombe Mdm12p homolog showed abnormal morphology: 70% of cells (355/507) had mitochondria that were either thickened and log shaped or spherical (Fig. 5 B), compared with only 1% of control cells (3/238) harboring vector alone (Fig. 5 A). These aberrant mitochondrial morphologies were apparent in cells harboring the cloned S. pombe gene at 23°C but were largely absent when cells were incubated at 37°C (data not shown). To determine whether high-level expression of S. cerevisiae MDM12 might produce a comparable mutant phenotype, the MDM12 gene was cloned behind the ADH1 promoter and expressed in S. cerevisiae cells. The ADH1–MDM12 construct fully complemented the mdm12-1 mutant and did not produce any apparent phenotype in wild-type cells. The similarity of the mitochondrial distribution and morphology defects caused by expression of the S. pombe homolog to the phenotype of mdm12 mutant cells suggests a partial conservation of function between the S. cerevisiae and S. pombe homologs of Mdm12p.
Figure 5.
Expression of the S. pombe Mdm12p homolog in S. cerevisiae produces abnormal mitochondrial morphology. Cells were grown overnight at 23°C in minimal synthetic medium lacking uracil to retain the plasmid and stained with DASPMI. Shown are two different wild-type yeast cells (strain MYY290) harboring either (A) pAC1 (vector) or (B) pAC1 containing mdm12p (plasmid pPM3, encoding the cloned S. pombe Mdm12p homolog). Bar, 2 μm.
Identification of SOT1, a Suppressor of Both mdm12 and mdm10
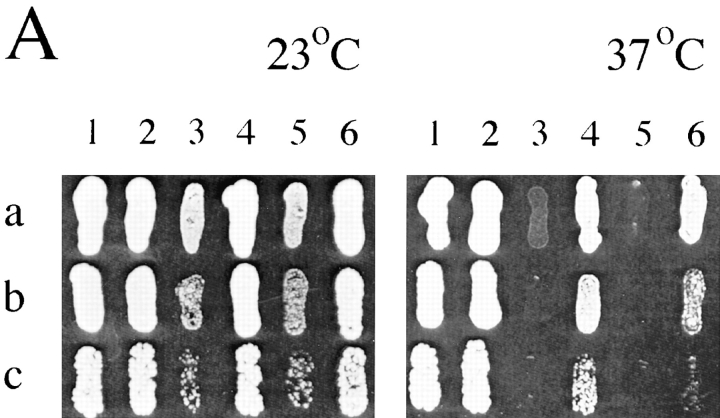
To characterize further the mdm10 and mdm12 mutant defects, mutations able to bypass the cellular requirement for MDM10 or MDM12 were identified by selecting for growth of the mutant cells at 37°C. One such mutation, designated SOT1 (suppressor of mdm10/mdm12), was initially identified as a suppressor of the mdm10-null mutation but was also found to suppress the temperature-sensitive growth defect of mdm12-null cells (Fig. 6 A). SOT1 also suppressed the growth defects of the mdm12-1 mutant and the mdm10 mdm12 double mutant (data not shown). Microscopic analysis of mdm10-null and mdm12-null cells harboring SOT1 revealed mitochondrial distribution and morphology at 23°C nearly identical to those found in wild-type cells (Fig. 6 B). Suppression of defects in growth (Fig. 6 A) and mitochondrial morphology (data not shown) was less effective at 37°C. In addition, although SOT1-suppressed haploid cells could grow at the elevated temperature, SOT1 failed to suppress many of the mdm12 mutant defects in diploid cells. mdm12/mdm12 cells that were either heterozygous (SOT1/sot1) or homozygous (SOT1/SOT1) for the SOT1 (mutant) allele failed to grow at 37°C and did not exhibit suppressed (normal) mitochondrial morphology at either 23°C or 37°C. This lack of suppression in diploid cells prevented a determination of the dominant or recessive character of SOT1 with respect to the mitochondrial morphology and high temperature growth traits. A single copy of SOT1 was able to suppress fully the sporulation defect of the mdm12-null homozygous diploid strain, which indicated that the SOT1 mutation was dominant (or codominant) to the wild-type allele (here designated sot1).
Figure 6.
SOT1 suppresses growth and mitochondrial morphology defects of mdm10 and mdm12 cells. (A) Dilutions of 10-fold (a), 100-fold (b), and 1,000-fold (c) of strains MYY290 (1), MYY626 (2), MYY503 (3), MYY627 (4), MYY624 (5), and MYY629 (6), were plated onto YPD agar medium and cultured for 80 h at 23°C or 37°C. The relevant genotype of each strain is: (1) MDM10 MDM12 sot1; (2) MDM10 MDM12 SOT1; (3) mdm10 MDM12 sot1; (4) mdm10 MDM12 SOT1; (5) MDM10 mdm12 sot1; (6) MDM10 mdm12 SOT1. (B) mdm10-null (MYY503), mdm12-null (MYY624), mdm10-null SOT1 (MYY627), or mdm12null SOT1 (MYY629) cells were grown overnight in YPD at 23°C and stained with DASPMI. (a) Unsuppressed cells (sot1). (b) Suppressed cells (SOT1). Bar, 2 μm.
In the absence of mdm10 or mdm12, the SOT1 mutation did not confer any apparent mutant phenotype. SOT1 cells grew normally at both low and high temperatures on a variety of media and displayed normal mitochondrial distribution and morphology. In addition, SOT1 was unable to suppress the defects in mitochondrial morphology caused by expression of the S. pombe Mdm12p homolog in S. cerevisiae cells (data not shown). SOT1 showed no genetic linkage to mdm12 or mdm10 and appears to define a novel gene.
Mdm12p Is a Component of the Mitochondrial Outer Membrane
Two proteins previously shown to be required for normal mitochondrial morphology, Mdm10p and Mmm1p, are constituents of the mitochondrial outer membrane (Sogo and Yaffe, 1994; Burgess et al., 1994). To determine whether Mdm12p occupied a similar subcellular location, the protein was localized by indirect immunofluorescence microscopy and subcellular fractionation. To facilitate this analysis, antiserum was raised against a peptide whose sequence corresponded to the COOH terminus of Mdm12p, and antibodies specific to Mdm12p were purified on an affinity column (as described in Materials and Methods).
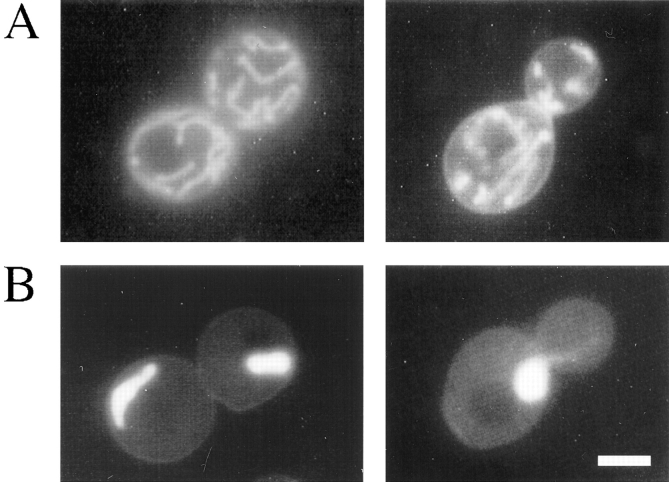
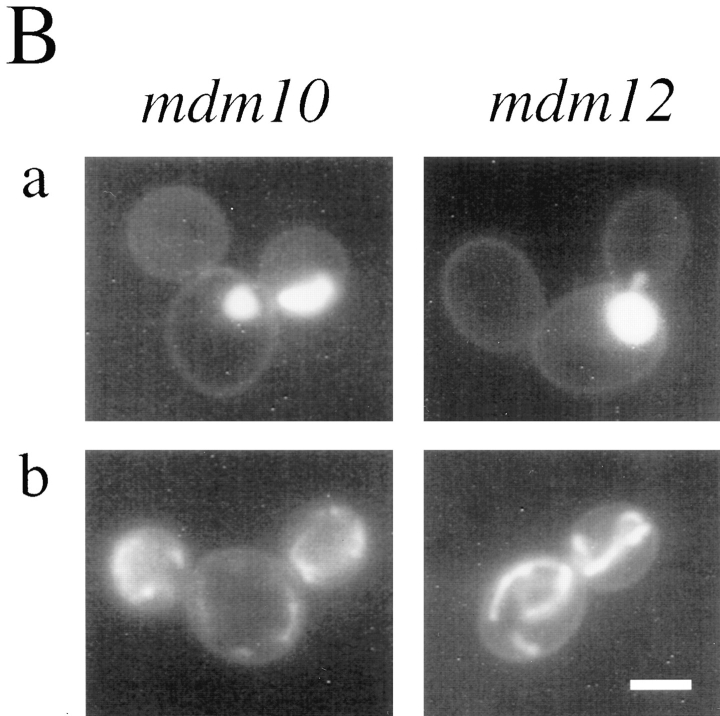
Indirect immunofluorescence microscopy of wild-type cells using affinity-purified anti-Mdm12p antibodies revealed that Mdm12p is associated with mitochondria (Fig. 7 A). The antibody failed to localize to mitochondria in mdm12-null cells (which harbored SOT1 to maintain normal mitochondrial morphology) (Fig. 7 B). Mdm12p was also detected in mitochondria in wild-type cells harboring the SOT1 mutation (Fig. 7 C) and in SOT1-suppressed mdm10-null cells (Fig. 7 D).
Figure 7.
Mdm12p is localized to mitochondria by indirect immunofluorescence. Cells were grown in YPD liquid medium at 23°C, fixed with formaldehyde, and processed for indirect immunofluorescence. (A) Wild type (MYY290); (B) mdm12-null SOT1 (MYY629); (C) wild type with SOT1 (MYY626); (D) mdm10-null SOT1 (MYY627). Upper panels show anti-Mdm12p indirect immunofluorescence and lower panels show DAPI staining of nuclear and mitochondrial DNAs. Bar, 2 μm.
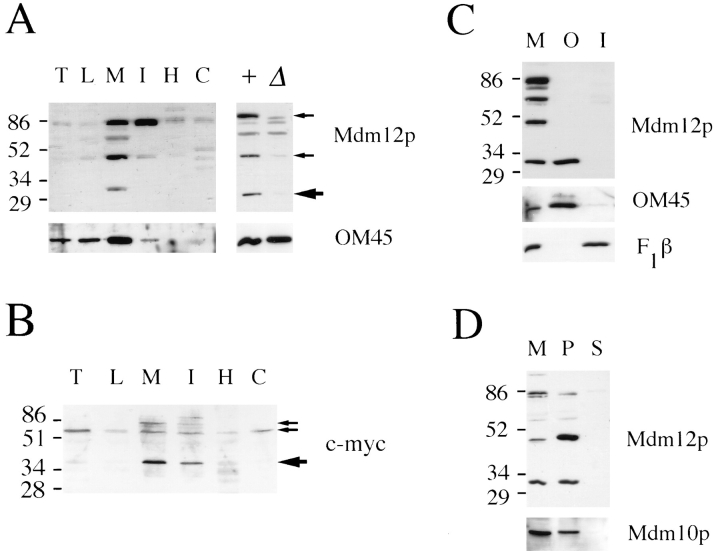
To identify the protein species corresponding to Mdm12p, subcellular fractions isolated from wild-type (MDM12) and mdm12-null cells were analyzed by immunoblotting. Both MDM12 and mdm12-null strains harbored the SOT1 suppressor mutation. A protein species of ∼31 kD, the predicted molecular size of Mdm12p, was detected in wildtype but not in mutant cell extracts (Fig. 8 A). This protein species was enriched in the mitochondrial fraction. In addition, the antibodies detected two additional protein species of ∼47 and 85 kD that appeared specific to MDM12 (Fig. 8 A). All three protein species were enriched in the mitochondrial fraction and deficient in fractions depleted of mitochondria (Fig. 8 A). Mdm12p showed a similar distribution in wild-type cells that did not carry the SOT1 mutation (data not shown). The relationship between the 31-kD protein and the two species of higher molecular mass is unclear. Various chemical treatments of mitochondria failed to change the relative amounts of the polypeptides (data not shown), and a specific consensus sequence for covalent modification (that might generate the higher molecular mass species) is absent from the predicted protein sequence of Mdm12p. The 47- and 85-kD species may represent Mdm12p-containing complexes or modified forms of Mdm12p. Alternatively, these species may be distinct from Mdm12p but share an antigenic epitope and, coincidentally, display decreased abundance in the absence of Mdm12p.
Figure 8.
Mdm12p is localized to the mitochondrial outer membrane. The mobilities of molecular mass markers, indicated in kD, are shown at the left. (A) Subcellular fractions (left) were isolated from yeast strain MYY626 (wild type with SOT1) grown on semisynthetic lactate medium. Proteins were analyzed by SDSPAGE and Western blotting with antibodies specific for Mdm12p (top) or OM45 (bottom). Large arrow indicates protein species corresponding to the predicted size of Mdm12p, and small arrows indicate two additional species also specific to MDM12. T, total cell homogenate; L, low speed pellet; M, mitochondria; I, intermediate pellet; H, high speed pellet; C, cytosol; +, mitochondrial protein from MDM12 SOT1 cells; Δ, mitochondrial protein from mdm12-null SOT1 cells. (B) Immunoblot analysis of subcellular fractions from wild-type cells (strain MYY290) expressing c-myc–Mdm12p. Fractions are labeled as in A. Large arrow indicates species of predicted size of c-myc–Mdm12p, and small arrows indicate two additional protein species specific to cells harboring c-myc–MDM12. (C) Western blot of wild-type mitochondria (M), purified mitochondrial outer membrane (O), and inner membrane (I) fractions. 20 μg of protein was resolved in each lane. Fractions were analyzed using antibodies to Mdm12p (top), OM45 (middle), and F1β (bottom). (D) Mitochondria (M) were resuspended in 0.1 M Na2CO3. Supernatant (S) and membrane pellet (P) fractions were recovered by centrifugation for 1 h at 100,000 g. Protein samples were analyzed by immunoblotting with antibodies to Mdm12p or Mdm10p.
As an independent approach to investigating the MDM12 gene product, the localization of Mdm12p tagged with the c-myc epitope (described in Materials and Methods) was examined. Subcellular fractionation and Western blot analysis detected a polypeptide of ∼40 kD, corresponding to the expected size of c-myc–Mdm12p, which was enriched in the mitochondrial fraction (Fig. 8 B). Two other protein species of ∼60 and 83 kD, which were likewise specific to cells harboring the c-myc–tagged construct, were also detected. These larger species were reminiscent of the higher molecular mass bands detected with antibodies specific to the native Mdm12p (discussed above). The 83-kD species was enriched in the mitochondrial fraction, whereas the 60-kD species was more evenly distributed in all subcellular fractions and represented the major species detected in total cellular extracts (Fig. 8 B). One caveat to these results is that the c-myc–MDM12 construct only partially complemented the temperature-sensitive growth and mitochondrial morphology defects of mdm12 cells, and therefore the distribution and molecular forms of the epitope-tagged Mdm12p may not accurately reflect the properties of native Mdm12p.
Submitochondrial fractionation and immunoblot analysis of native Mdm12p revealed that the protein was associated with the mitochondrial outer membrane (Fig. 8 C). Only the 31-kD species was evident in the purified outer membrane fractions (Fig. 8 C). Separate analysis of isolated mitochondria suggested that the other MDM12-specific species were particularly protease sensitive (data not shown) and were likely to have been degraded during mitochondrial subfractionation. Mdm12p fractionated in a similar manner to another known mitochondrial outer membrane protein, OM45, and was not found in fractions enriched for mitochondrial inner membrane proteins such as F1β. Mdm12p was also absent from purified mitochondrial matrix and intermembrane space fractions (data not shown). These results demonstrate that Mdm12p is a protein of the mitochondrial outer membrane.
To characterize the association of Mdm12p with the mitochondrial outer membrane, peripheral membrane proteins were separated from integral membrane proteins by extraction of isolated mitochondria with 0.1 M sodium carbonate and centrifugation. The 31- and 47-kD species were recovered in the pellet fraction (Fig. 8 D), as was Mdm10p, which was previously shown to behave as an integral membrane protein (Sogo and Yaffe, 1994). These results suggest that Mdm12p is an integral protein of the mitochondrial outer membrane.
Discussion
Mdm12p is essential for normal mitochondrial morphology and inheritance. Loss of this protein leads to the appearance of giant mitochondria and defective transfer of mitochondria into daughter buds. This role for Mdm12p is very similar to that of Mdm10p, and several additional observations suggest that these two proteins function at the same step in mediating mitochondrial behavior. First, mdm10 mdm12 double mutant cells display a phenotype identical to that of either single mutant. Second, Mdm10p and Mdm12p share a similar subcellular and submitochondrial localization. Third, the SOT1 mutation suppresses loss of either Mdm10p or Mdm12p or the loss of both proteins. Finally, two recently identified multicopy suppressors each provide partial suppression (i.e., slow growth at 37°C and partial restoration of tubular mitochondrial morphology) of mdm10, mdm12, or the double mutant (Berger, K.H., and M.P. Yaffe, unpublished results).
Cells lacking Mdm10p, Mdm12p, or both proteins exhibited aberrant mitochondrial morphology at all temperatures, yet they were viable at 23°C. Even at this permissive temperature, the mdm12 mutant displayed defective mitochondrial inheritance and appeared to give rise to a large proportion of unbudded cells lacking mitochondria. This mitochondrial inheritance defect is likely to be responsible for the mutant's slow growth at permissive temperature because cells that failed to receive mitochondria would be inviable (Gbelska et al., 1983; Yaffe and Schatz, 1984). Growth of mdm12-deficient cells at permissive temperature suggests the existence of a backup or bypass pathway for mitochondrial distribution that operates at lower temperatures. At high temperatures, such a bypass pathway might fail to function or be insufficient for the metabolic needs of the cell. Although the molecular basis for a bypass pathway is unknown, the SOT1 mutation might define a component of this pathway and act to stimulate its activity.
Mdm12p is the third S. cerevisiae protein of the mitochondrial outer membrane to be identified whose loss causes temperature-sensitive growth and the appearance of enlarged, spherical mitochondria defective for distribution to buds at all temperatures. A potential role for these proteins—Mdm10p, Mmm1p, and Mdm12p—is as hooks or handles for attachment of mitochondria to cytoskeletal elements (Burgess et al., 1994; Sogo and Yaffe, 1994). Alternatively, these polypeptides might function primarily to alter properties of the mitochondrial outer membrane that are necessary for the maintenance of tubular mitochondrial morphology, and the mitochondrial inheritance defects might be a secondary consequence of the aberrant morphology. Given the similar phenotype caused by loss of Mdm12p, Mdm10p, or Mmm1p and the shared location of these three proteins, they may well function together in a complex.
The identification of a homolog of Mdm12p in fission yeast, together with the effect of the cloned homolog on mitochondrial morphology when expressed in S. cerevisiae, demonstrates that at least one component regulating mitochondrial distribution has been conserved between budding and fission yeast. This is a rather surprising finding because some of the central structures mediating mitochondrial distribution appear to be different in budding and fission yeast. In particular, recent studies have revealed that microtubules mediate mitochondrial distribution in S. pombe (Yaffe et al., 1996), whereas microtubules appear to play no role in mitochondrial inheritance in S. cerevisiae (Huffaker et al., 1988; Jacobs et al., 1988). Given the failure of the S. pombe mdm12p to complement the S. cerevisiae mdm12 mutant as well as the dominant-negative effect of expressing mdm12p in wild-type budding yeast, it appears that Mdm12p function may be only partially conserved. It is possible that similar components and structures on the mitochondrial surface are used to mediate the interaction of mitochondria with different cytoskeletal components in S. cerevisiae and S. pombe. An analysis of the function of the Mdm12p homolog in S. pombe may reveal details of its role in mitochondrial inheritance.
Acknowledgments
We thank Lena Best for expert technical assistance, Dr. R. Haselbeck for generation of strain MYY461, and Dr. R. Roberts for preparation of purified mitochondrial inner and outer membranes. We thank Dr. C. Holm for the top1::URA3 disruption cassette, R. Hampton for plasmid pJR1265, and Dr. L. Riles for the mapped λ prime clone grid filters. We greatly appreciate the encouragement and advice of past and present members of the Yaffe laboratory, and thank Harold Fisk, Mark Nickas, and Kelly Shepard for valuable comments on the manuscript.
This work was supported by grant GM44614 from the National Institutes of Health. K.H. Berger was supported by a National Institutes of Health postdoctoral fellowship (grant GM16173).
Abbreviations used in this paper
- DAPI
4,6-diamidino-2-phenylindole
- DASPMI
2-(4-dimethylaminostryl)-1-methylpyridinium iodide
- mdm
mitochondrial distribution and morphology
- YPD
yeast extract/peptone/ glucose
- YPG
yeast extract/peptone/glycerol
Footnotes
Please address all correspondence to Michael P. Yaffe, Department of Biology, 0347, University of California, San Diego, La Jolla, CA 92093-0347. Tel.: (619) 534-4769. Fax: (619) 534-4403. e-mail: myaffe@ucsd.edu
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1992;20:2013–2018. doi: 10.1093/nar/20.suppl.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereiter-Hahn J. Dimethylaminostyrylmethylpyridiniumiodine (DASPMI) as a fluorescent probe for mitochondria in situ. Biochim Biophys Acta. 1976;423:1–14. doi: 10.1016/0005-2728(76)90096-7. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Delannoy M, Jensen RE. MMM1encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J Cell Biol. 1994;126:1375–1391. doi: 10.1083/jcb.126.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallier M-R, Bloch JC, Lacroute F. Transcriptional and translational expression of a chimeric bacterial-yeast plasmid in yeast. Gene. 1980;11:11–19. doi: 10.1016/0378-1119(80)90082-7. [DOI] [PubMed] [Google Scholar]
- Daum G, Bohni PC, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- Douglas MG, Butow RA. Variant forms of mitochondrial translation products in yeast: evidence for location of determinants on mitochondrial DNA. Proc Natl Acad Sci USA. 1976;73:1083–1086. doi: 10.1073/pnas.73.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G, Lewis G, Ramsey G, Bishop JM. Isolation of monoclonal antibodies specific for human c-mycproto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gbelska Y, Subik J, Goffeau A, Kovac L. Intramitochondrial ATP and cell functions: yeast cells depleted of intramitochondrial ATP lose the ability to grow and multiply. Eur J Biochem. 1983;130:281–286. doi: 10.1111/j.1432-1033.1983.tb07148.x. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies. A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 726 pp.
- Huffaker TC, Thomas JH, Botstein D. Diverse effects of β-tubulin mutations on microtubule formation and function. J Cell Biol. 1988;106:1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt EC, Muller U, Schatz G. The first twelve amino acids of a yeast mitochondrial outer membrane protein can direct a nuclear-encoded cytochrome oxidase subunit to the mitochondrial inner membrane. EMBO (Eur Mol Biol Organ) J. 1985;4:3509–3518. doi: 10.1002/j.1460-2075.1985.tb04110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs CW, Adams AEM, Szaniszlo PJ, Pringle JR. Functions of microtubules in the Saccharomyces cerevisiaecell cycle. J Cell Biol. 1988;107:1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SJ, Yaffe MP. Nuclear and mitochondrial inheritance in yeast depends on novel cytoplasmic structures defined by the MDM1 protein. J Cell Biol. 1992;118:385–395. doi: 10.1083/jcb.118.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SJ, Yaffe MP. Intermediate filament formation by a yeast protein essential for organelle inheritance. Science (Wash DC) 1993;260:687–689. doi: 10.1126/science.8480179. [DOI] [PubMed] [Google Scholar]
- McConnell SJ, Stewart LC, Talin A, Yaffe MP. Temperaturesensitive yeast mutants defective in mitochondrial inheritance. J Cell Biol. 1990;111:967–976. doi: 10.1083/jcb.111.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR, Adams AEM, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–601. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., F. Winston, and P. Hieter. 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 198 pp.
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning. Cold Spring Harbor Laboratory, Plainview, NY. 545 pp.
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chainterminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer I, Emr S, Gross C, Schekman RW. Invertase signal and mature sequence substitutions that delay intercompartmental transport of active enzyme. J Cell Biol. 1985;100:1664–1675. doi: 10.1083/jcb.100.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Wakem P. Mapping yeast genes. Methods Enzymol. 1991;194:38–56. doi: 10.1016/0076-6879(91)94006-x. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BJ, Yaffe MP. A mutation in the yeast heat-shock factor gene causes temperature-sensitive defects in both mitochondrial protein import and the cell cycle. Mol Cell Biol. 1991;11:2647–2655. doi: 10.1128/mcb.11.5.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol. 1994;126:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehlin TH, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP. Analysis of mitochondrial function and assembly. Methods Enzymol. 1991;194:627–643. doi: 10.1016/0076-6879(91)94046-f. [DOI] [PubMed] [Google Scholar]
- Yaffe MP. Isolation and analysis of mitochondrial inheritance mutants from Saccharomyces cerevisiae. . Methods Enzymol. 1995;260:447–453. doi: 10.1016/0076-6879(95)60157-0. [DOI] [PubMed] [Google Scholar]
- Yaffe MP. The division and inheritance of mitochondria. Adv Mol Cell Biol. 1996;17:337–346. [Google Scholar]
- Yaffe MP, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP, Harata D, Verde F, Eddison M, Toda T, Nurse P. Microtubules mediate mitochondrial distribution in fission yeast. Proc Natl Acad Sci USA. 1996;93:11664–11668. doi: 10.1073/pnas.93.21.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]