Abstract
smad genes constitute a family of nine members whose products serve as intracellular mediators of transforming growth factor β signals. SMAD2, which is a tumor suppressor involved in colorectal and lung cancer, has been shown to induce dorsal mesoderm in Xenopus laevis in response to transforming growth factor β and activins. The smad2 gene is expressed ubiquitously during murine embryogenesis and in many adult mouse tissues. Animals that lacked smad2 died before 8.5 days of development (E8.5). E6.5 homozygous mutants were smaller than controls, lacked the extraembryonic portion of the egg cylinder, and appeared strikingly similar to E6.5 smad4 mutants. This similarity was no longer evident at E7.5, however, because the smad2 mutants contained embryonic ectoderm within their interiors. Molecular analysis showed that smad2 mutant embryos did not undergo gastrulation or make mesoderm. The results demonstrate that smad2 is required for egg cylinder elongation, gastrulation, and mesoderm induction.
Keywords: transforming growth factor β/smad4/gastrulation/extraembryonic membranes
The transforming growth factor β (TGFβ) superfamily consists of numerous related cytokines, including TGFβ itself, activins, bone morphogenic proteins (BMPs), and others (reviewed in refs. 1 and 2), which perform a variety of biological functions. The ligands of the TGFβ superfamily transmit their signal through a number of related transmembrane serine/threonine kinases. These receptors are heteromeric, with a type I and a type II subunit required for signal transduction. Upon ligand binding the type II receptor phosphorylates the type I receptor, which then transmits the signal to downstream targets (3–5).
The recent discovery of the highly conserved vertebrate smad genes, smad1–9 (6–16), has advanced our understanding of the downstream signaling cascade utilized by the type I receptors. The products of the smad genes are phosphorylated by the type I receptors and transmit the TGFβ signal to the nucleus, where they participate in the activation of downstream genes (reviewed in refs. 17 and 18).
The SMAD proteins consist of a highly conserved amino-terminal MH1 domain and a carboxyl-terminal MH2 domain, which are separated by a proline-rich linker region (6) and are found as homotrimers in solution (19). The type I receptors phosphorylate the SMAD proteins at a highly conserved SS(V/M)S motif located at the carboxyl terminus of the MH2 domain (20, 21). However, SMAD1–SMAD5 are pathway specific, in that they cannot be phosphorylated by every receptor. SMAD1 and SMAD5 mediate signals in the BMP pathway (6, 8, 21), whereas SMAD2 and SMAD3 convey signals originated by TGFβ and activins (7, 8, 11, 14, 20). Upon phosphorylation these four SMAD proteins all form a stable complex with SMAD4, with which they are translocated into the nucleus (21, 22). In addition to the pathway-specific and common SMADs there are also inhibitory SMAD proteins, SMAD6 and SMAD7 (12, 13, 23, 24). Both smad2 and smad4 are on chromosome 18q21 in humans, and both have been identified as tumor suppressor genes.
A great deal has been learned about the function of smad2 in Xenopus, where it has been shown to induce dorsal mesoderm (8, 11, 25). To learn more about the functions of smad2 in mammals we have disrupted it by gene targeting in mice. Removal of the carboxyl-terminal half of the MH2 domain results in recessive embryonic lethality at day 6.5 (E6.5) of development because of a failure to form the extraembryonic portion of the egg cylinder. The embryonic ectoderm is able to proliferate, however, and fills the interior of the embryo at later stages. At this time point they resemble embryos deficient in the TGFβ homologue NODAL (26–28).
MATERIALS AND METHODS
Targeting Vector.
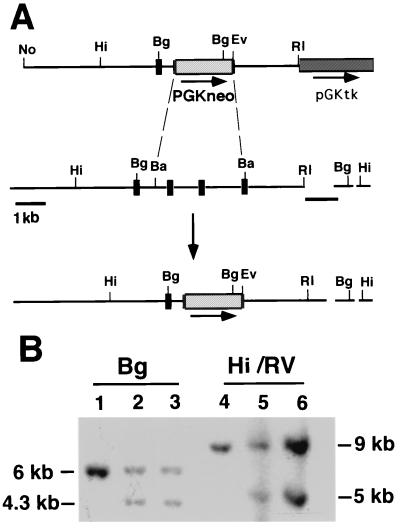
An 1130-bp smad2 cDNA sequence was amplified from brain cDNA by using primers (5′-GTGTGGATTGTTACCTTTGG-3′) and (5′-CATGAATACTACGACGGAGG-3′) and used to screen a mouse genomic library (Stratagene). A 2.5-kb EcoRI/BamHI fragment containing the most 3′ exon of smad2 was subcloned into the EcoRI site of ploxP (29) to create pMW110. A 7-kb AspI/BamHI fragment containing upstream smad2 sequences was subcloned into pBSKS (Stratagene). This subclone was digested with AspI and treated with the Klenow fragment of DNA polymerase, and the smad2 sequences were released with NotI. This fragment was cloned into NotI/HpaI-digested pMW110 to create psmad2neo (Fig. 1A).
Figure 1.
(A) Disruption of smad2 through homologous recombination in embryonic stem (ES) cells. The targeting vector psmad2neo deletes the carboxyl-terminal portion of smad2. The top line is the targeting vector, the middle the genomic locus before recombination, and the bottom the locus after recombination. 5′ is on the left. The size and position of exons are not drawn to scale. The arrow represents the direction of neo transcription, the dashed lines the portion deleted, and the short black lines the 3′ flanking and 5′ internal probes used to detect homologous recombination. (B) Southern blot of ES DNAs. The 3′ flanking probe recognizes a 6-kb BglII fragment from the wild-type allele; this fragment is reduced to 4.3 kb in the targeted allele because of the presence of a BglII site on the neo gene. Correct targeting was confirmed with a HindIII/EcoRV digest, which releases a 9-kb band from the wild type and a 5-kb band from the targeted allele. The 5′ internal probe was used to further confirm correct targeting (data not shown). Both correctly targeted clones are shown. Hi, HindIII; No, NotI; Bg, BglII; Ba, BamHI; RI, EcoRI; and RV, EcoRV.
Electroporation of ES Cells and Generation of Germ-Line Chimeras.
psmad2neo was digested with NotI and electroporated into TC1 ES cells as described (30). Genomic DNAs were isolated, digested with BglII, and electrophoresed for Southern blotting. Targeted ES cells were microinjected into C57BL/6 blastulae to generate chimeras, which were mated to NIH Black Swiss females (Taconic Farms). Half of the agouti progeny from these matings carried the smad2ΔC allele.
Genotype Analysis.
Mice were genotyped either by Southern blotting as above or by PCR. For PCR analysis the mutant allele was amplified by using a primer from the smad2 sequences that were not deleted in the targeting (5′-CATGAATACTACGACGGAGG-3′) and a primer from neo (5′-ATCGCCTTCTATCGCCTTCTTGACGAGTTC-3′), which amplify a 600-bp product from the mutant allele. Mutant alleles were also identified by primers that amplified an internal segment of the neo gene. For the wild-type allele, the smad2 primer from above was used in conjunction with primers from the region deleted in the smad2ΔC mutation. These included (5′-CTCCTTGATGGATGAACTTC-3′), which amplifies a 150-bp product, and (5′-GGACCAGACTCACTAGTTCA-3′), which amplifies a 300-bp product.
Histological Analysis and in Situ Hybridization.
Histology and in situ hybridization were carried out by using standard procedures. The probes used included T (30), lim1 (31), fgf8 (32), bmp4 (a kind gift of C. Chang, Vanderbilt Univ., Nashville, TN), and H19 (33).
RESULTS
Disruption of smad2 by Gene Targeting.
The targeting vector psmad2neo (Fig. 1A) contains 9.5 kb of genomic smad2 sequences with a 2.5-kb deletion, into which we have placed a PGKneo cassette (34). This deletion removes the carboxyl-terminal 86 amino acids from the SMAD2 protein, which itself contains 487 amino acids. Homologous recombination between the targeting vector and the endogenous locus will result in loss of the phosphorylation site and approximately half of the MH2 domain. Phosphorylation of SMAD2 by the type I TGFβ receptors has been shown to be required for nuclear localization and signal transduction activity (11, 20), and phosphorylation-deficient mutants of smad2 have been found associated with colorectal tumors (11). The deletion of the smad2 carboxyl-terminal domain is therefore expected to create a null allele, which we refer to as smad2ΔC.
The psmad2neo vector was electroporated into TC1 ES (35), and 2 of the 179 G418/FIAU-resistant clones analyzed were found to be correctly targeted (Fig. 1B) [FIAU is 1-(2′-deoxy-2′fluoro-β-d-arabinofuranosyl)-5-iodouracil]. Both clones were injected into blastocysts and both resulted in germ-line transmission of the smad2ΔC allele. Southern blots and PCR analysis showed that the mutation was passed on to agouti offspring of the chimeras (data not shown).
The smad2ΔC Mutation Is a Recessive Embryonic Lethal.
Mice heterozygous for the smad2ΔC mutation are viable, healthy, and fertile. They are indistinguishable from their wild-type siblings in growth rates and litter sizes. They have not shown any tendency to develop tumors during a 9-month study period, although an increased rate of cancer could require more time to become evident.
To address the effect of a loss of smad2 function on murine development, smad2ΔC/+ animals were bred inter se to yield offspring homozygous for the disrupted allele. However, no homozygotes were found in the 50 offspring genotyped at weaning (Table 1), suggesting that the mutation resulted in recessive embryonic lethality.
Table 1.
Genotypic and phenotypic analysis of embryos derived from smad2ΔC/+ crosses
| Age | No. with phenotype
|
No. with genotype
|
||||
|---|---|---|---|---|---|---|
| Normal | Abnormal | Resorbtions* | +/+ | +/− | −/− | |
| E6.5 | 17 (17) | 3 (3) | 1 | 6 | 11 | 3 |
| E7.5 | 40 (34) | 5 (4) | 5 | 8 | 26 | 4 |
| E8.5 | 30 (30) | 5 (5) | 8 | 4 | 26 | 5 |
| E9.5 | 15 (15) | 0 | 3 | 7 | 8 | 0 |
| Total | 102 | 13 | 17 | 25 | 71 | 12 |
| P21 | 50 | 0 | — | 20 | 30 | 0 |
Numbers inside parentheses are the number of embryos genotyped. P21, postnatal day 21 (weaning).
Resorbtions were not subjected to genotypic analysis.
The timing of this lethality was determined by examining embryos at several stages of development, as shown in Table 1. Abnormal or resorbed embryos were found between E6.5 and E8.5, and PCR analysis showed all were homozygous for the deleted smad2 sequences (data not shown). This suggests that the deletion of the smad2 carboxyl-terminal domain caused abnormal development and embryonic death.
E6.5 smad2ΔC/ΔC embryos were much smaller than their sibling controls (Fig. 2A) with a pronounced reduction in the extraembryonic portion of the egg cylinders. Indeed they bore a superficial resemblance to embryos lacking smad4 (29, 36).
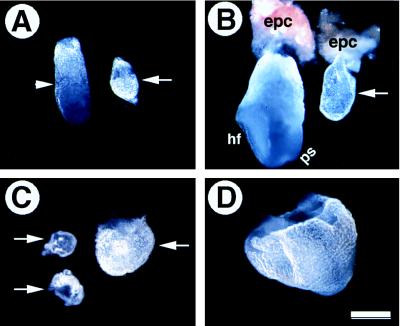
Figure 2.
Morphological analysis of smad2ΔC/ΔC embryos (arrows) and their littermate controls. (A) E6.5 embryos. Arrowhead points to the boundary between extraembryonic and embryonic portions of the control embryo. The mutant embryo is round without such a boundary. (B) E7.5 embryos. Mutant embryos remain small and do not contain any normal structures such as headfold (hf) and primitive streak (ps). The ectoplacental cone (epc) is normal in the mutants. (C and D) E8.5 mutant (C) and control (D) embryos. Note that the mutant embryos exhibit some variation in size. However, even the largest mutant embryo (right in C) is much smaller than its littermate control (D). (Bar, 20 μm for A and B and 40 μm for C and D.)
By E7.5 normal embryos had developed readily recognizable structures, such as the ectoplacental cone, primitive streak, and headfold (Fig. 2B). smad2ΔC/ΔC embryos either were resorbed (Table 1) or did not form a distinguishable headfold or primitive streak. They did exhibit an increase in size over E6.5 smad2ΔC/ΔC embryos, although they were still diminutive compared with their normal siblings. Interestingly, the ectoplacental cone appeared normal in E7.5 smad2ΔC/ΔC embryos (Fig. 2B), as it did in embryos lacking smad4 (29, 36).
By E8.5 many of the smad2ΔC/ΔC embryos were either resorbed or in the process of resorption (Table 1). The remainder were poorly organized and extremely small (Fig. 2C). Some of the smad2ΔC/ΔC mutants formed small embryos within larger membranous sacs (embryo on the right in Fig. 2C), whereas the wild-type and smad2ΔC/+ embryos have formed a visible embryonic axis, a defined head, somites, and other embryonic structures (Fig. 2D and not shown). These results clearly demonstrate that smad2 is required for early embryonic development, and its loss results in early postimplantation lethality.
Intact decidua of E6.5–E8.5 embryos were subjected to histological sectioning to better understand the defect present in the smad2ΔC/ΔC mutants. At E6.5 normal embryos undergo a process of rapid cell division and elongation to form the egg cylinder (Fig. 3A). A clear boundary was seen between the embryonic and extraembryonic portions (arrow in Fig. 3A), and we often detected mesoderm involuting at this stage. Cuboidal cells of the visceral endoderm could be seen encompassing the entire structure, although they flattened out at the distal end of the egg cylinder (Fig. 3A). In contrast, the E6.5 smad2ΔC/ΔC embryos lacked extraembryonic ectoderm and extraembryonic endoderm, and the ectoplacental cone was found directly adjacent to the epiblast (Fig. 3B). At this stage smad2ΔC/ΔC and smad4ex8/ex8 embryos bore a striking similarity (29, 36), suggesting that SMAD2 and SMAD4 are required to function together for egg cylinder elongation. In contrast to what was found in embryos lacking smad4 (36), smad2ΔC/ΔC embryos appear to have a well developed visceral endoderm surrounding the epiblast, although the cuboidal cell architecture is maintained around the circumference of the embryo. This result suggests that other SMAD proteins may synergize with SMAD4 to direct the development of this lineage.
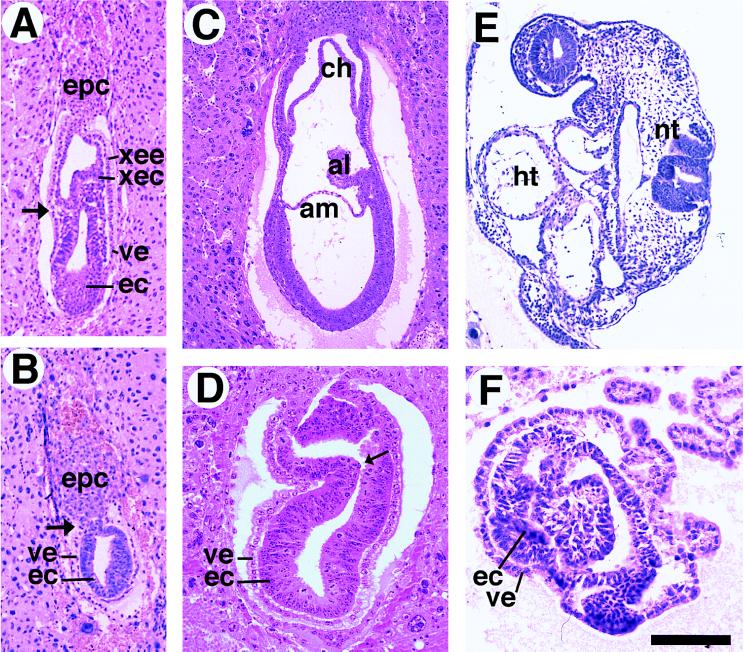
Figure 3.
Histological sections of embryos generated from crosses between smad2ΔC/+ mice. (A and B) Sagittal section of E6.5 control (A) and mutant (B) embryos. Arrows point to the boundary between extraembryonic and embryonic portions of the embryo. epc, ectoplacental cone; ec, embryonic ectoderm; ve, visceral endoderm; xec, extraembryonic ectoderm; xee, extraembryonic endoderm. (C and D) E7.5 control (C) and mutant (D) embryos. The embryonic ectoderm of the mutant embryo grew significantly, whereas no normal structures were visible. al, allantois; am, amnion; ch, chorion. The arrow in D points to an infolding of embryonic ectoderm. (E and F). E8.5 control (E) and mutant (F) embryos. In mutant embryos, the presumptive embryonic ectoderm continues to grow without forming any structures seen in normal embryos such as heart (ht) and neural tube (nt). (Bar, 200 μm for A and B, 260 μm for C, 130 μm for D, 970 μm for E, and 110 μm for F.)
At E7.5 normal embryos had a well defined mesodermal layer. The allantois, chorion, and amnion were clearly visible (Fig. 3C). Those smad2ΔC/ΔC embryos that were not resorbed at this stage either lacked morphologically distinguishable chorion, amnion, and allantois (Fig. 3D) or exhibited a diminished extaembryonic portion (not shown). The visceral endoderm and ectoderm were visible, but a mesodermal cell layer could not be seen in the smad2ΔC/ΔC embryos (Fig. 3D). The smad2ΔC/ΔC embryos were enlarged compared with their counterparts at E6.5, however, and in sharp contrast to embryos lacking smad4, smad2ΔC/ΔC embryos exhibited infolding of the embryonic ectoderm (arrow in Fig. 3D) that in most embryos appeared as aggregates of cells filling the proamniotic cavity (not shown). In this regard embryos resemble those deficient in NODAL, a TGFβ homologue that itself is necessary for proper gastrulation (26–28). Reichert’s membrane and parietal endoderm also appeared normal in the smad2ΔC/ΔC embryos (not shown), despite the other defects seen. E8.5 animals had many structures recognizable in sections, such as the heart and the neural tube (Fig. 3E). However, E8.5 smad2ΔC/ΔC embryos lacked any recognizable structure or organization and closely resembled E7.5 smad2ΔC/ΔC embryos (Fig. 3F). Endodermal and ectodermal layers are evident, cells fill the embryonic interior, and no mesoderm can be seen. Reichert’s membrane and the parietal endoderm still appeared normal in a number of cases, however. We therefore believe that the smad2ΔC/ΔC animals arrest at E7.5 without undergoing gastrulation.
smad2ΔC/ΔCEmbryos Exhibited a Defect in the Extraembryonic Portion of the Egg Cylinder.
Both the morphological and histological analyses suggested that the E6.5 smad2ΔC/ΔC embryos had a defect in the extraembryonic portion of the egg cylinder. We therefore characterized the extraembryonic lineages by examining expression of the H19 gene, which marks the extraembryonic endoderm and ectoderm, the ectoplacental cone, and the trophoblastic giant cells. A clear boundary can be seen between the epiblast and extraembryonic portion in normal E6.5 embryos (Fig. 4 A and C), and the extraembryonic endoderm and the extraembryonic ectoderm are well organized. In contrast the smad2ΔC/ΔC embryos exhibited abundant staining of H19 in the ectoplacental cone, but not in any extraembryonic ectoderm or extraembryonic endoderm (Fig. 4 B and D). The visceral endoderm was clearly of uniform thickness around the mutant embryo (arrowhead in Fig. 4D), unlike the normal embryos, in which the visceral endoderm flattened out at the distal tip of the egg cylinder (arrowhead in Fig. 4C). This observation may reflect a developmental delay, as the visceral endoderm is of uniform thickness at E5.5, or it could result from the failure of egg cylinder elongation. At E7.5 the smad2ΔC/ΔC embryos exhibited H19 staining in the ectoplacental cone and a small extraembryonic membrane, which itself was lacking in many mutants (not shown). The failure of egg cylinder elongation was also seen in embryos that lacked smad4; however, the defect in the smad2ΔC/ΔC embryos was considerably less severe, as the visceral endoderm appears less severely affected, and the ectoderm is capable of further development.
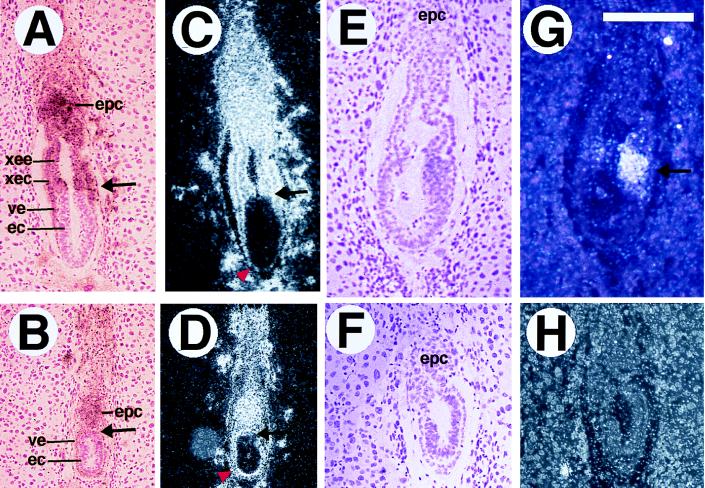
Figure 4.
In situ analysis of smad2ΔC/ΔC embryos at E6.5. A, B, E, and F are bright-field views, and the four others are dark-field views. (A–D) H19 expression in control (A and C) and mutant (B and D) embryos. H19 marks the ectoplacental cone (epc), extraembryonic endoderm (xee) and ectoderm (xec), and visceral endoderm (ve). The H19 expression domain in the E6.5 mutant is smaller than that of the control, and the extraembryonic ectoderm and extraembryonic endoderm appear to be missing. Arrows in A–D point to the boundary between extraembryonic and embryonic portions, and arrowheads in C and D point out the visceral endoderm, which is flattened at the distal tip of the control embryo (C), but remains thickened in the mutant (D). T expression is shown in normal (E and G) and mutant (F and H) embryos. T labels involuting mesodermal cells (arrow in G), which are not in the smad2ΔC/ΔC mutants. (Bar, 258 μm for A–D and 180 μm for E–H.)
smad2ΔC/ΔCEmbryos Fail to Form Mesoderm.
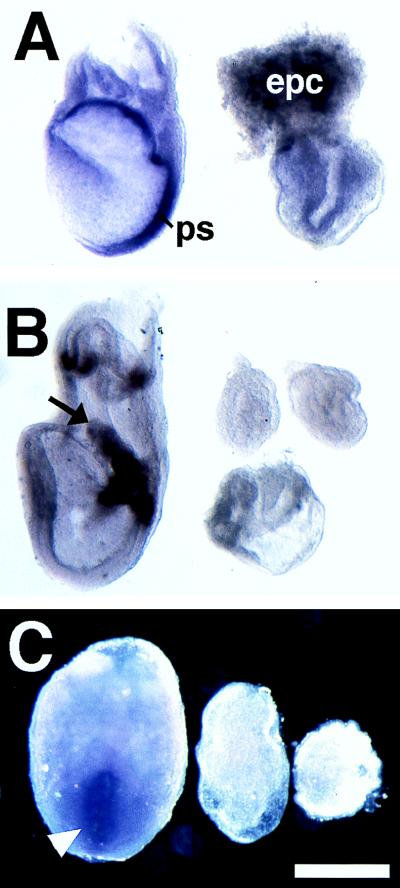
We used in situ hybridization to examine mesodermal induction in the smad2ΔC/ΔC mutants. Litters from smad2ΔC/+ crosses at E6.5 were sectioned serially and labeled with the widely used mesodermal marker T. Normal embryos exhibited involuting mesoderm at E6.5 (Fig. 4 E and G), whereas E6.5 smad2ΔC/ΔC embryos (n = 6) showed no detectable T staining (Fig. 4 F and H). We also performed serial section in situ hybridizations against E7.5 and E8.5 mutants, which similarly lacked T staining (n = 5, data not shown). In addition, we performed whole mount in situ hybridization against both early and late mesodermal markers (T and lim1, respectively). E7.0 and E7.5 smad2ΔC/ΔC embryos lacked staining for T (n = 6, Fig. 5A), and E8.5 embryos were not labeled with T or lim1 (data not shown). We also failed to detect expression of bmp4, a marker of extraembryonic mesoderm, in the E7.0 and E7.5 smad2ΔC/ΔC embryos (n = 6, Fig. 5B). Similarly, fgf8, which is found in the primitive streak, was not seen in the mutant embryos at E7.5 (n = 3, Fig. 5C).
Figure 5.
smad2ΔC/ΔC mutants do not make mesoderm. Whole mount in situ hybridization against markers of embryonic lineages. (A) T in E7.5 embryos. The primitive streak (ps) can be found in the normal embryo (Left) but not in the mutant embryo (Right). epc, ectoplacental cone. (B) bmp4 in E7.5 embryos. The arrow points to the allantois of the normal embryo (Left), whereas the chorion and amnion are less visible. The three mutant embryos shown on the Right fail to exhibit staining for this marker. (C) fgf8 in E7.5 embryos. The primitive streak (arrowhead) is stained in the normal embryo (Left). Note the lack of staining in the two mutant embryos on the Right. (Bar = 458 μm for A and C and 523 μm for B.)
DISCUSSION
smad2 Functions in Egg Cylinder Elongation and Mesoderm Induction.
SMAD2 is necessary for egg cylinder elongation, and in its absence the extraembryonic portion of the egg cylinder is not formed. Embryos that lack smad2 fail to undergo normal gastrulation, and they become arrested without inducing any mesoderm. Our present level of analysis is insufficient to determine which of these defects is primary. Failure of mesodermal induction could result from the absence of the extraembryonic endoderm and extraembryonic ectoderm; conversely, the failure of egg cylinder elongation may result from an absence of mesoderm. It is also possible that the two phenotypes result independently.
smad2 mutants have been recently observed with defects considerably different from those communicated here (37). Specifically, the embryos examined by Waldrip et al. (37) survived 1 day longer than the smad2ΔC/ΔC mutants, were able to form relatively normal extraembryonic membranes, and induced mesoderm, as determined by expression of T and bmp4. Interestingly, Waldrip et al. demonstrated an essential function of smad2 in the extraembryonic membranes for establishment of the anterioposterior embryonic axis.
We believe that the disruption introduced in Waldrip et al. (37) may not have resulted in a complete loss of function. Although neither our study nor that of Waldrip et al. has demonstrated the generation of a null mutation, the targeting reported here deleted the carboxyl terminus, which is known to be essential for SMAD2 activity (20, 21). We have also shown that homozygosity for this deletion results in embryonic lethality earlier than that seen by Waldrip et al. (37).
The mutation introduced by Waldrip et al. (37) abolished the translation start site; however, translation could have started from a downstream methionine codon in SMAD2, creating a product truncated in the amino-terminal domain. This truncation would actually be expected to increase the activity of the protein, as a similar protein exhibited an increased ability to induce mesoderm, morphogenetic cell movements, and axis duplication in Xenopus (25), probably because of the loss of an inhibitory domain in the amino terminus (38). The mutation examined by Waldrip et al. (37) may have left sufficient smad2 activity to allow for formation of the extraembryonic membranes, but not enough to pattern the embryo. Production of SMAD2 protein in the mutants was not analyzed by Waldrip et al.
The smad2 mutants reported here have shown no evidence of normal egg cylinder formation, and we have found no expression of either T or bmp4 at any embryonic stage. Moreover, deletions of either the amino-terminal or the carboxyl-terminal domains of smad2 generated in another laboratory resulted in a phenotype similar to that shown here (E. Li, personal communication).
smad2 and smad4 Function Cooperatively in Egg Cylinder Elongation.
We believe that SMAD2 and SMAD4 cooperate to convey signals necessary for egg cylinder elongation, as these two mutants both display this unusual phenotype. However the smad2ΔC/ΔC embryos are less affected than the smad4 mutants generated in our laboratory (29), as proliferating cell nuclear antigen (PCNA) staining and blastocyst culture experiments indicate they do not suffer the severe reduction in cell proliferation experienced by smad4 mutants (data not shown).
smad2 Is Necessary for Mesoderm Induction.
None of the markers used in this study, including T, lim1, bmp4, and fgf8, detected mesoderm in the smad2ΔC/ΔC embryos. These observations indicate that smad2 is essential for mesoderm induction, correlating well with earlier studies showing that SMAD2 was able to induce dorsal mesoderm in Xenopus (8, 11, 25). Data from our laboratory suggest that smad2ΔC/ΔC ES cells can form mesodermal derivatives (not shown), similar to cells that lack smad4 (36). SMAD2 may be needed to generate or receive inductive signals, or the failure of mesodermal induction may be a nonspecific consequence of the abnormal embryonic architecture.
smad2 May Mediate Signals from Several TGFβ Family Members.
E7.5 smad2ΔC/ΔC embryos do resemble nodal mutants, in that they both accumulate ectoderm within the embryonic interior due to folding of the ectoderm (26). nodal mutants are far less affected than smad2ΔC/ΔC embryos however, as the former have an overproliferation of embryonic ectoderm and trophectoderm cells, and they are able to make some mesodermal cells (27). The smad2ΔC/ΔC mutation may abrogate nodal signals, but there are other TGFβ signals that are terminated as well.
Several genes within the TGFβ signal transduction pathway function in the formation of the extraembryonic membranes. The smad4 gene has been alluded to earlier, but in addition mice lacking either the type I BMP receptor (BmpR1) or the type I activin receptor (ActRIB) also fail to make normal extraembryonic membranes or mesoderm (39, 40). It is unlikely that smad2 is involved in the BMP pathway, as it has been shown both biochemically and functionally to be specifically activated by the receptors for TGFβ and activin, and not by the BMP receptors (8, 11, 22). SMAD2 is expected to transmit the signal generated by ActRIB, however, and it is noteworthy that mutants lacking ActRIB do not form an organized extraembryonic epithelium, do not induce mesoderm, and exhibit marked similarities to smad2ΔC/ΔC mutants (40).
ABBREVIATIONS
- TGFβ
transforming growth factor β
- BMP
bone morphogenic protein
- En
day n of development
- ES
embryonic stem
References
- 1.Kingsley D M. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 2.Massague J. Cell. 1996;85:947–950. doi: 10.1016/s0092-8674(00)81296-9. [DOI] [PubMed] [Google Scholar]
- 3.Wrana J L, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang X F, Massague J. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 4.Letsou A, Arora K, Wrana J L, Simin K, Twombly V, Jamal J, Staehling-Hampton K, Hoffmann F M, Gelbart W M, Massague J, et al. Cell. 1995;80:899–908. doi: 10.1016/0092-8674(95)90293-7. [DOI] [PubMed] [Google Scholar]
- 5.Ruberte E, Marty T, Nellen D, Affolter M, Basler K. Cell. 1995;80:889–897. doi: 10.1016/0092-8674(95)90292-9. [DOI] [PubMed] [Google Scholar]
- 6.Liu F, Hata A, Baker J C, Doody J, Carcamo J, Harland R M, Massague J. Nature (London) 1996;381:620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Feng X, We R, Derynck R. Nature (London) 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 8.Graff J M, Bansal A, Melton D A. Cell. 1996;85:479–487. doi: 10.1016/s0092-8674(00)81249-0. [DOI] [PubMed] [Google Scholar]
- 9.Hoodless P A, Haerry T, Abdollah S, Stapleton M, O’Connor M B, Attisano L, Wrana J L. Cell. 1996;85:489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- 10.Hahn S A, Schutte M, Hoque A T, Moskaluk C A, da Costa L T, Rozenblum E, Weinstein C L, Fischer A, Yeo C J, Hruban R H, Kern S E. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 11.Eppert K, Scherer S W, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui L C, Bapat B, Gallinger S, Andrulis I L, et al. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 12.Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Nature (London) 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu Y Y, Grinnell B W, Richardson M A, Topper J N, Gimbrone M A, Jr, Wrana J L, Falb D. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 14.Nakao A, Roijer E, Imamura T, Souchelnytskyi S, Stenman G, Heldin C H, ten Dijke P. J Biol Chem. 1997;272:2896–2900. doi: 10.1074/jbc.272.5.2896. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Bhushan A, Vale W. Proc Natl Acad Sci USA. 1997;94:12938–12943. doi: 10.1073/pnas.94.24.12938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe T K, Suzuki M, Omori Y, Hishigaki H, Horie M, Kanemoto N, Fujiwara T, Nakamura Y, Takahashi E. Genomics. 1997;42:446–451. doi: 10.1006/geno.1997.4753. [DOI] [PubMed] [Google Scholar]
- 17.Heldin C H, Miyazono K, ten Dijke P. Nature (London) 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 18.Baker J C, Harland R M. Curr Opin Genet Dev. 1997;7:467–473. doi: 10.1016/s0959-437x(97)80072-x. [DOI] [PubMed] [Google Scholar]
- 19.Shi Y, Hata A, Lo R S, Massague J, Pavletich N P. Nature (London) 1997;388:87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- 20.Macias-Silva M, Abdollah S, Hoodless P A, Pirone R, Attisano L, Wrana J L. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 21.Kretzschmar M, Liu F, Hata A, Doody J, Massague J. Genes Dev. 1997;11:984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- 22.Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 23.Hata A, Lagna G, Massague J, Hemmati-Brivanlou A. Genes Dev. 1998;12:186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakao A, Afrakhte M, Moren A, Nakayama T, Christian J L, Heuchel R, Itoh S, Kawabata M, Heldin N E, Heldin C H, ten Dijke P. Nature (London) 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 25.Baker J C, Harland R M. Genes Dev. 1996;10:1880–1889. doi: 10.1101/gad.10.15.1880. [DOI] [PubMed] [Google Scholar]
- 26.Iannaconne P M, Zhou X, Khoka M, Boucher D, Kuehn M R. Dev Dyn. 1992;194:198–208. doi: 10.1002/aja.1001940305. [DOI] [PubMed] [Google Scholar]
- 27.Conlon F L, Lyons K M, Takaesu N, Barth K, Kispert A, Herrmann B, Robertson E J. Development (Cambridge, UK) 1994;120:1919–1928. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- 28.Varlet I, Collignon J, Robertson E J. Development (Cambridge, UK) 1997;124:1033–1044. doi: 10.1242/dev.124.5.1033. [DOI] [PubMed] [Google Scholar]
- 29.Yang X, Li C, Xu X, Deng C. Proc Natl Acad Sci USA. 1998;95:3667–3672. doi: 10.1073/pnas.95.7.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng C, Wynshaw-Boris A, Shen M M, Daugherty C, Ornitz D M, Leder P. Genes Dev. 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- 31.Barnes J D, Crosby J L, Jones C M, Wright C V E, Hogan B L M. Dev Biol. 1994;161:168–178. doi: 10.1006/dbio.1994.1018. [DOI] [PubMed] [Google Scholar]
- 32.Heikinheimo M, Lawshe A, Shackleford G M, Wilson D B, MacArthur C A. Mech Dev. 1994;48:129–138. doi: 10.1016/0925-4773(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 33.Poirier F, Chan C T, Timmons P M, Robertson E J, Evans M J, Rigby P W. Development (Cambridge, UK) 1991;113:1105–1114. doi: 10.1242/dev.113.4.1105. [DOI] [PubMed] [Google Scholar]
- 34.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 35.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 36.Sirard C, de la Pompa J L, Elia A, Itie A, Mirtsos C, Cheung A, Hahn S, Wakeham A, Schwartz L, Kern S E, et al. Genes Dev. 1998;12:107–119. doi: 10.1101/gad.12.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waldrip W R, Bikoff E K, Hoodless P A, Wrana J L, Robertson E J. Cell. 1998;92:797–808. doi: 10.1016/s0092-8674(00)81407-5. [DOI] [PubMed] [Google Scholar]
- 38.Hata A, Lo R S, Wotton D, Lagna G, Massague J. Nature (London) 1997;388:82–87. doi: 10.1038/40424. [DOI] [PubMed] [Google Scholar]
- 39.Mishina Y, Suzuki A, Ueno N, Behringer R R. Genes Dev. 1995;9:3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- 40.Gu Z, Nomura M, Simpson B B, Lei H, Feijen A, van den Eijnden-van Raaij J, Donahoe P K, Li E. Genes Dev. 1998;12:844–857. doi: 10.1101/gad.12.6.844. [DOI] [PMC free article] [PubMed] [Google Scholar]