Abstract
Many human diseases, including Fanconi anemia, hemophilia B, neurofibromatosis, and phenylketonuria, can be caused by 5′-splice-site (5′ss) mutations that are not predicted to disrupt splicing, according to position weight matrices. By using comparative genomics, we identify pairwise dependencies between 5′ss nucleotides as a conserved feature of the entire set of 5′ss. These dependencies are also conserved in human–mouse pairs of orthologous 5′ss. Many disease-associated 5′ss mutations disrupt these dependencies, as can some human SNPs that appear to alter splicing. The consistency of the evidence signifies the relevance of this approach and suggests that 5′ss SNPs play a role in complex diseases.
The sequenced genomes of a wide range of organisms allow global, comparative analyses of regulatory sequences. The genomic set of splice-site sequences corresponds to a large-scale splicing experiment performed by nature under evolutionary constraints. Here we focus on 5′-splice-site (5′ss) sequences of the U2-type GT-AG class, which comprise over 98% of all splice sites, and use disease-causing mutations, human single nucleotide polymorphisms (SNPs), and variations in natural splice sites in the genome (within and between species) to infer properties inherent to 5′ss, with important implications for human genetics.
Splice sites are conserved sequences at both ends of an intron that are recognized during the initial steps of splicing (Hastings and Krainer 2001; Brow 2002; Jurica and Moore 2003). Both the 5′ss and the 3′ splice site (3′ss) conform to degenerate motifs that are recognized by specific splicing factors. The U2-type GT-AG 5′ss, spanning 3 nucleotides (nt) at the 3′ end of the exon and 6 nt at the 5′ end of the intron, is initially recognized via base pairing to the 5′ end of the U1 snRNA (Fig. 1; Zhuang and Weiner 1986; Séraphin et al. 1988; Siliciano and Guthrie 1988). Later in the reaction, U5 and U6 snRNAs base pair to exonic or intronic 5′ss positions, respectively (Newman and Norman 1992; Wassarman and Steitz 1992; Kandels-Lewis and Séraphin 1993; Lesser and Guthrie 1993; Crotti et al. 2007). Additional elements influence splice-site selection, such as exonic or intronic splicing enhancers (ESE, ISE) or silencers (ESS, ISS) (Cartegni et al. 2002; Ladd and Cooper 2002).
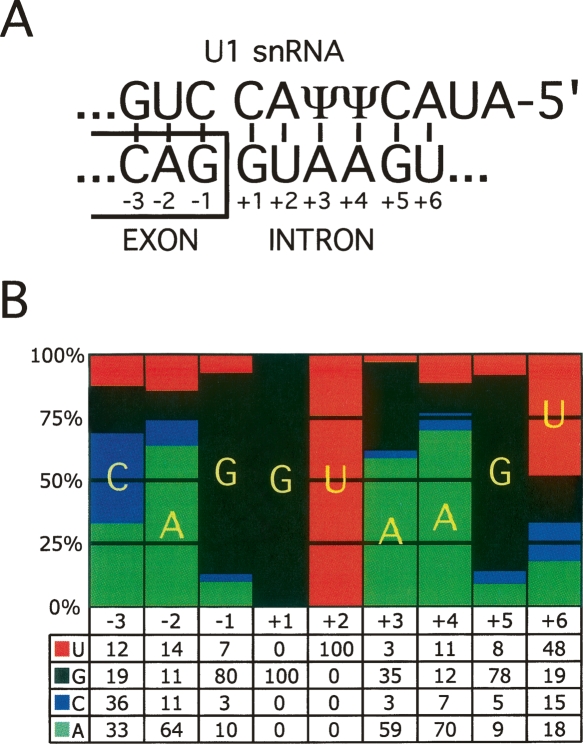
Figure 1.
Features of the 5′ splice site (5′ss). (A) Base pairing between the 5′ end of the U1 snRNA and the consensus 5′ss sequence. ψ denotes pseudo-uridine, which is a modified uridine nucleotide capable of base pairing to both A and G nucleotides. The conventional numbering of positions relative to the exon–intron boundary is indicated. For example, in the text −1G refers to nucleotide G at position −1. (B) A pictorial representation of the position weight matrix (PWM) of the human U2-type GT-AG 5′ss; the bars represent the percentages of nucleotides at each position of the 5′ss. The actual percentages are shown below the representation.
Even though the mammalian 5′ss consensus sequence (CAG|GTAAGT) is perfectly complementary to the 5′ end of U1 snRNA, individual 5′ss exhibit considerable variation at different positions, indicating a tolerance for mismatches in U1 base pairing. The free energy of the 5′ss/U1 base pairing is not always a good predictor of 5′ss efficiency (Roca et al. 2005), suggesting the existence of other factors that influence 5′ss selection. It is known that proteins such as the U1-C polypeptide (Du and Rosbash 2002), which is a component of the U1 snRNP, PRPF8 (also known as PRP8) (Maroney et al. 2000), and members of the SR and hnRNP protein families (Mayeda and Krainer 1992; Cáceres et al. 1994; Buratti et al. 2004) are also involved in 5′ss selection. Most methods for estimating 5′ss efficiency are based on position weight matrices (PWMs) that are calculated from collections of splice sites (for a depiction of a PWM, see Methods and Fig. 1) (Shapiro and Senapathy 1987; Senapathy et al. 1990). Compared with PWMs, which assume independence between positions within the 5′ss, methods that have considered the dependencies between these positions (Brunak et al. 1991; Yeo and Burge 2004) have provided some improvements in predicting 5′ss efficiency (Roca et al. 2003, 2005; Buratti et al. 2007). Such analyses have been previously carried out on smaller data sets (Burge and Karlin 1997; Carmel et al. 2004) or implicitly considered in various 5′ss scoring algorithms (Brunak et al. 1991; Yeo and Burge 2004; Krawczak et al. 2007).
Mutations at 5′ss are frequent among mutations that cause human disease, from genetic disorders to cancer (Krawczak et al. 1992; Nakai and Sakamoto 1994; Teraoka et al. 1999; Stenson et al. 2003; Buratti et al. 2007). In many cases, the mutations cause aberrant splicing by affecting the invariant GT dinucleotide at positions +1 and +2 of the 5′ss. However, the effects of other disease-causing 5′ss mutations are less clear-cut: Nucleotide substitutions affecting the less conserved positions can cause splicing defects in some but not all 5′ss, suggesting that the remaining 5′ss positions and/or the overall context dictate the extent to which splicing is disrupted.
We present a comprehensive analysis of the pairwise associations between nucleotides at different 5′ss positions, using the splice-site compilation from SpliceRack (Sheth et al. 2006), which is a collection of more than 500,000 splice sites from five genomes. We explicitly and exhaustively identify combinations that appear to be significant. We show that there are constraints on combinations of nucleotides at different positions of the 5′ss. We further show that these constraints are indeed operational in the evolutionary process using four data sets: disease-causing mutations at 5′ss, orthologous pairs of 5′ss between mouse and human, SNPs at 5′ss, and simulated SNPs. It is remarkable that all these disparate sources of information seem to be consistent with a single explanation: that the pairwise associations between positions of the 5′ss are a determinant of 5′ss efficiency.
Results
The primary determinant of the efficiency of a 5′ss is its match to the PWM (Zhuang and Weiner 1986; Séraphin et al. 1988; Siliciano and Guthrie 1988) given by the log-odds score (Methods), which reflects, at least in part, the stability of base pairing to the U1 snRNA 5′ end (Fig. 1). However, many 5′ss with similar PWM scores show differences in splicing efficiency, suggesting secondary effects that are ignored by the PWM. To study additional determinants of 5′ss efficiency, we have analyzed the associations between pairs of nucleotides at different positions of the U2-type GT-AG 5′ss in five species: Homo sapiens, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, and Arabidopsis thaliana.
Two-nucleotide associations
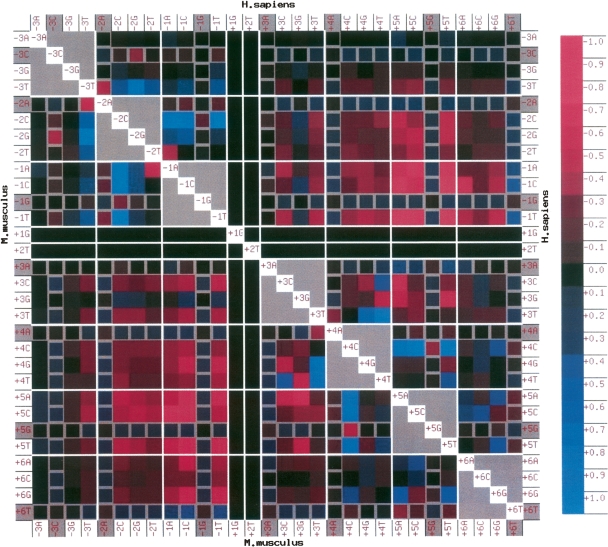
PWMs for each of the five species can be calculated from the genomic 5′ss data in SpliceRack (Sheth et al. 2006), consisting of 183,678 H. sapiens 5′ss, 174,671 M. musculus 5′ss, 40,367 D. melanogaster 5′ss, 93,699 C. elegans 5′ss, and 111,351 A. thaliana 5′ss. The PWMs are calculated by counting the occurrences of nucleotides at each position within the 5′ss. The expected occurrence of pairs of nucleotides at two positions, e.g., A at +4 and G at +5, can be calculated from the PWM frequencies by assuming independence between the two positions (for nomenclature of positions in 5′ss, see Fig. 1). The actual frequency of occurrence for such pairs can be measured from the genomic data, by counting the occurrence of pairs of nucleotides (16 possible combinations) at all possible combinations of positions (21 unique combinations, if the +1 and +2 invariant positions are excluded). The deviations of the actual count from the expected count are measured using a log-odds scoring scheme (see Methods), and for visualization, the results are scaled and shown on a colored matrix. A combined matrix showing data from M. musculus and H. sapiens is depicted in Figure 2; a matrix for C. elegans and A. thaliana is shown in Supplemental Figure S1; and the D. melanogaster matrix is shown in Supplemental Figure S2.
Figure 2.
Association matrix for H. sapiens (upper right triangle, above diagonal) and M. musculus (lower left triangle, below diagonal). Row and column labels identify each square; e.g., the square identified by row −3A and column +5T represents the pair −3A+5T in human, as it lies in the upper right triangle. The color of the squares indicates the bias for (blue) or against (maroon) the pair of nucleotides. The log-odds score for each pair (the log of the ratio of actual versus expected counts) is calculated and scaled so that the maximum positive value is assigned a score of +1 and the minimum negative value is assigned a score of −1. The scale on the right shows the values indicated by the color tone. The diagonal elements are colored white, and gray is used for associations within a position. Gray borders are used to mark the rows and columns containing consensus nucleotides at any given position (e.g., +5G). A black square is used for the invariant (GT) dinucleotide at +1 and +2, and white lines are used to demarcate each position at the 5′ss. Note that the association matrices for M. musculus and H. sapiens are strikingly similar.
The surprising observation is that the pattern of deviations of the actual counts from the expected counts is, to a great extent, conserved between species. This suggests that these patterns of deviation are a result of the mechanisms of 5′ss recognition by the splicing machinery (e.g., cf. upper and lower triangles in Fig. 2 as well as Supplemental Figs. S1, S2). The mouse and human patterns show remarkably close similarity. We show the variability for a few pairs within the human genome and across genomes in Table 1. There are some differences, but the overall patterns of deviation are clearly preserved and presumably reflect the pressures that arise from the conservation of the splicing machinery.
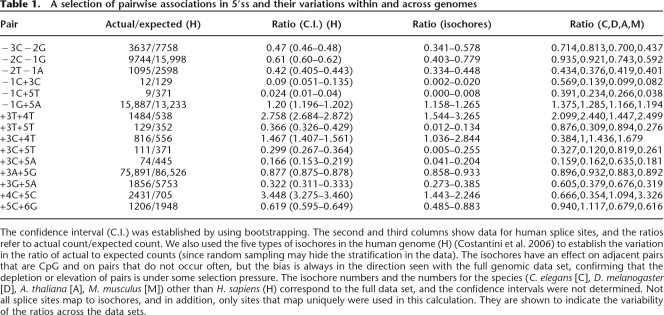
Table 1.
A selection of pairwise associations in 5′ss and their variations within and across genomes
The confidence interval (C.I.) was established by using bootstrapping. The second and third columns show data for human splice sites, and the ratios refer to actual count/expected count. We also used the five types of isochores in the human genome (H) (Costantini et al. 2006) to establish the variation in the ratio of actual to expected counts (since random sampling may hide the stratification in the data). The isochores have an effect on adjacent pairs that are CpG and on pairs that do not occur often, but the bias is always in the direction seen with the full genomic data set, confirming that the depletion or elevation of pairs is under some selection pressure. The isochore numbers and the numbers for the species (C. elegans [C], D. melanogaster [D], A. thaliana [A], M. musculus [M]) other than H. sapiens (H) correspond to the full data set, and the confidence intervals were not determined. Not all splice sites map to isochores, and in addition, only sites that map uniquely were used in this calculation. They are shown to indicate the variability of the ratios across the data sets.
The depletion (maroon coloration) in the parts of the matrix that connect the exonic and intronic portions of the 5′ss is a striking feature in all species (Figs. 2, S1, S2; see associations between positions −2 or −1 and positions +3, +4, +5, or +6). This implies that having a nonconsensus nucleotide on the exonic side causes a depletion of nonconsensus nucleotides on the intronic side, and vice versa, which is consistent with the proposal of a seesaw linkage pattern (Burge and Karlin 1997; Carmel et al. 2004). Some combinations are surprising in the distance they span, such as the enhancement of the pair −1T+6T. The pair −1C+3C shows a strong depletion in all species, for reasons unknown to us. Another striking pattern is the association between positions −1 and +5 wherein a G (consensus) at −1 allows any nucleotide at position +5; similarly, +5G (consensus) allows any nucleotide at −1. We show some of the conserved pairwise associations in Table 1.
Some features in the pairwise association matrix might be the result of constraints other than the splicing mechanism. For example, the combinations −3T−2A and −2T−1A are severely depleted in the five species, probably because these combinations can be part of two of the three stop codons (TAG and TAA). The combination +5C+6C is enriched, probably reflecting the gradual conversion of U12-type GT-AG introns into U2-type GT-AG introns (Burge et al. 1998; Sheth et al. 2006). From the pairwise association matrix, it can be seen that the CpG dinucleotide is depleted in higher species, whereas D. melanogaster and C. elegans do not show such depletion, consistent with the fact that in higher organisms CpG is underrepresented, due to methylation of the C followed by de-amination (Tweedie et al. 1997). The spliceosome could have evolved to use patterns that might be species-specific, even though the drive to maintain the patterns might come from other processes.
Another possible explanation for some of the pairwise associations could lie in nucleotide biases in error-prone DNA repair. However, the dependence of these mechanisms on the neighboring nucleotides is not very pronounced (Krawczak et al. 1998), arguing against a large contribution to the associations.
There are some species-specific features in the pairwise associations. For example, there are tighter associations (brighter blues) between the exonic nucleotides at positions −2 and −1 in mammals and A. thaliana than in the two invertebrates (Figs. 2, S1, S2). In contrast, D. melanogaster and C. elegans have more biases between intronic nucleotides (positions +3 to +6 show brighter blues).
The human genome is organized into isochores, i.e., regions with relatively stable GC content compared with the variation of the GC content across the genome (Costantini et al. 2006). The isochores have been classified into five groups (H3 [GC > 53.5%], H2 [46.6–53.5%], H1 [41.6–46.5%], L2 [37.6–41.5%], and L1 [<37.5%]). In order to use this stratification to study variations in the associations, the 5′ss were classified into the five isochore categories, and the expected and actual counts for pairs within each set were calculated. In Table 1 we show the variation of the ratios of actual to expected counts for a selection of pairs. Overall, the associations are preserved, both within and across species.
Implications for the splicing machinery
A striking feature that is conserved across species is the depletion of U1-noncomplementary pairs across the exon–intron boundary (e.g., −1C and +5T in Fig. 2). This implies that U1 snRNA requires complementarity on at least one side of the invariant GT.
After U1 snRNA is displaced from the 5′ss, U5 snRNA binds to the exonic 5′ss positions via a U-rich sequence in the invariant loop 1 (Newman and Norman 1992). However, we found that combinations of A nucleotides at exonic positions are not enhanced, consistent with the finding that the U5 loop 1 is dispensable in vivo in yeast (O’Keefe et al. 1996).
We observed that the pair +3C+4T is enhanced in the five species. This association likely reflects base pairing to the U6 ACAGAG box (the nucleotides involved in base pairing to positions +3 and +4 are in italics) (Wassarman and Steitz 1992; Kandels-Lewis and Séraphin 1993; Lesser and Guthrie 1993). The remaining nucleotides that would base pair to U6 but not to U1 do not show an association that is phylogenetically conserved.
There are a number of biased combinations that cannot be explained by base pairing to any of these snRNAs, such as an enhancement of +4C+5C in all species, or −2C−1T in the vertebrates. Most importantly, these tend to be less conserved than the previous associations. It is possible that these combinations are part of binding sites for proteins that influence 5′ss selection, such as U1-C, PRPF8, SR proteins, or hnRNP proteins (Mayeda and Krainer 1992; Cáceres et al. 1994; Maroney et al. 2000; Du and Rosbash 2002; Buratti et al. 2004). Whether these dinucleotide associations reflect a portion of a protein binding site and whether the binding specificity of some of these proteins is slightly different between species remain to be elucidated.
Scoring the pairwise associations within 5′ss
We used the pairwise association matrix and a log-odds scoring scheme to score 5′ss for the level of association (see Methods). This allowed us to quantify the effect of 5′ss changes on splicing efficiency. We used the data sets from disease-causing mutations, SNPs at 5′ss, orthologous mouse–human 5′ss pairs, and simulated SNPs at 5′ss to estimate the magnitude of change in associations that can significantly alter splice-site efficiency, and then to predict SNPs at 5′ss that can affect splicing (see below). On the Web site accompanying this article (the URL is listed at the end of the article), we present an interactive picture that allows exploration of this pairwise-association matrix, which can be used to study novel SNPs and mutations at 5′ss for their predicted effects on splicing.
In Table 1 we show the variations for a few pairs across isochores within the human genome. This establishes that, overall, the compositional variations can change the ratios, but the trend remains the same; depleted (enhanced) pairs are depleted (enhanced) in all sets, irrespective of the origin of the 5′ss. Most patterns are maintained across species, with the CpG motifs being prominent exceptions. The justification for the scoring scheme lies in the conserved nature of these depletions and enhancements and in the fact that the scoring scheme is an indicator of the effects of disrupting the pairwise association patterns.
Disease-causing mutations at 5′ss
We expected disease-causing mutations at 5′ss to be disruptive of the PWM scores as well as the pairwise-association scores. For this study, we used 595 disease-causing 5′ss mutations from the HGMD database (Stenson et al. 2003) plus another independent set (Madsen et al. 2006), excluding mutations that affect the nearly invariant +1 and +2 positions.
The reduction in PWM scores caused by many of the mutations cannot, by itself, explain the severe effects on splicing, because there are other pairs of functional 5′ss in the human genome that have the same nucleotide change. Indeed, we observed that the natural 5′ss tend to have better pairwise-association scores than the mutant 5′ss (Fig. 3).
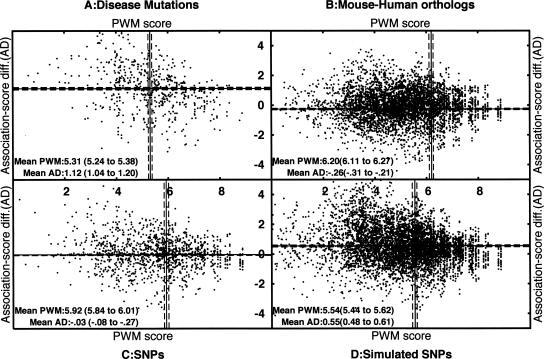
Figure 3.
Scatter plots showing combinations of PWM scores (X-axis) and association-score differences (Y-axis) for 5′ss from four data sets. The four data sets are disease-causing mutations (A, 595 cases), orthologous mouse–human pairs of 5′ss (B, 19,940 pairs), SNPs at 5′ss (C, 1260 SNPs), and simulated SNPs at 5′ss (D, 8074 cases). Each spot in the data set corresponds to a 5′ss and a single-nucleotide change to the 5′ss. In each data set, along the X-axis, we plotted the original 5′ss PWM score; in the case of SNPs and orthologous 5′ss, the 5′ss with the higher score is considered the original 5′ss. Along the Y-axis, we plotted the difference in association scores due to the change (score of original pair − score of new pair). A change to a pair with reduced association score will give a positive score difference, and an increase in association score will give a negative value. The solid lines show the averages along the X- and Y-axes, whereas the dashed lines show the confidence intervals for the means (calculated by a bootstrap method). The numbers are also given as text within the plots, with the confidence interval shown in parentheses. The disease panel is clearly biased toward low PWM scores and greater disruption of associations, compared with the other data sets. The averages for the four data sets are well separated on the basis of the confidence intervals.
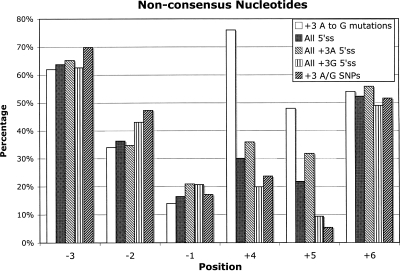
We picked a well-studied set among disease-causing 5′ss mutations, i.e., a subclass that consists of A-to-G transitions at position +3, for further theoretical and experimental analyses. A (59%) and G (35%) are both conserved at position +3 (Sheth et al. 2006), probably because both can base pair to a pseudo-uridine nucleotide (ψ), a post-transcriptionally modified uridine isomer (Reddy et al. 1981) in the 5′ arm of the U1 snRNA (Fig. 1). Thus, it is not obvious why the A to G mutations at +3 in genes such as ACADSB (also named SBCAD), BRCA1, APC, and NF1 can cause genetic diseases (Madsen et al. 2006). It has been shown that 5′ss with disease-causing +3 A-to-G mutations exhibit two distinct features: (1) They are intrinsically weak (Madsen et al. 2006); and (2) they frequently have nonconsensus nucleotides at positions +4 and +5 (for confirmation, see Fig. 4; Ohno et al. 1999; Madsen et al. 2006). Figure 4 also shows that 5′ss with G at position +3 and SNPs with A/G alleles at position +3 prefer consensus nucleotides at +4 and +5, in agreement with the second observation.
Figure 4.
Percentage of nonconsensus nucleotides at positions (X-axis) of the 5′ss for five classes of 5′ss: all 5′ss in human (186,630 cases), 5′ss with A at +3 (110,598), 5′ss with G at +3 (64,837), 5′ss with SNPs having alleles A/G at position +3 (93), and 5′ss with disease-causing A-to-G mutations at +3 (50 cases). 5′ss with disease-associated A-to-G mutations at position +3 tend to have nonconsensus nucleotides at positions +4 and +5 more frequently than the other sets, which suggests their disruptive nature, based on the association matrix (Fig. 2).
These observations can be explained by the dependencies between position +3 (A/G) and the nucleotides at positions +4 and +5 (Fig. 2). The association of +3A to nonconsensus nucleotides at +4 (C, G, T) and +5 (A, C, T) is blue (enhanced), whereas the association of +3G to nonconsensus nucleotides at +4 and +5 is maroon (depleted). An interesting prediction is that if both +4 and +5 are nonconsensus, then the splicing defect in the mutant can be fixed by converting either +4 or +5 independently to the consensus (see below).
Experimental tests of the associations
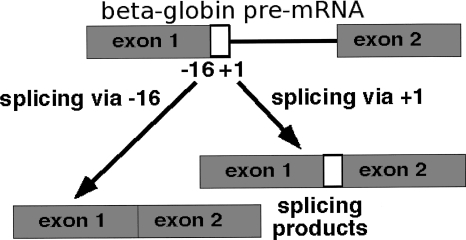
We carried out in vitro splicing experiments in order to confirm our prediction for the +3 A-to-G mutations. We used a 5′ss-competition assay described in Figure 5. We inserted variants of the 5′ss (GGG/GUACAU) from exon 3 of the ACADSB gene into a beta-globin (HBB) minigene, which allows direct comparison of the efficiencies of any two 5′ss (Roca et al. 2005). An A-to-G mutation at position +3 of this 5′ss causes a rare metabolic disease, ACADSB deficiency (Madsen et al. 2006).
Figure 5.
Experimental system used to test 5′ss efficiency by a competition assay (Roca et al. 2005). The human beta-globin pre-mRNA has two 5′ss at positions −16 and +1, of which the +1 5′ss is always used because the −16 cryptic 5′ss is much weaker (Roca et al. 2003). Replacement of the −16 5′ss by the +1 5′ss sequence results in both 5′ss being used equally. This system can be used to test the relative efficiencies of 5′ss by a competition assay, wherein the −16 and/or +1 5′ss are replaced by the various 5′ss to be tested. When different 5′ss are compared, their relative positions are also swapped as the contexts are not equivalent, in that for 5′ss of comparable efficiency, either the +1 or the −16 site can get more efficiently used. Use of the two competing 5′ss results in products that differ in length, which is detected by gel electrophoresis.
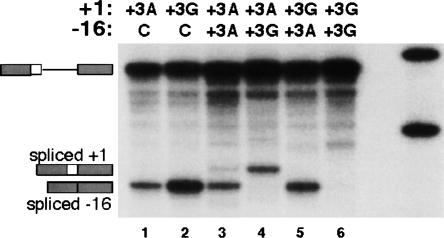
First, we found that the +3 A-to-G mutation in the ACADSB 5′ss severely reduces the 5′ss strength (Fig. 6). When the ACADSB wild-type 5′ss (+3A) or its +3 A-to-G mutant version (+3G) were tested in competition against the beta-globin cryptic 5′ss at −16 (Treisman et al. 1983), only the cryptic 5′ss was used (lanes 1, 2). Since the cryptic 5′ss is already a suboptimal 5′ss (Roca et al. 2003), we conclude that both the +3A and +3G 5′ss are weak. When the +3A and +3G 5′ss were competing against each other, splicing occurred only via the +3A 5′ss (lanes 4, 5). This finding indicates that the +3A 5′ss is much stronger than its +3G counterpart.
Figure 6.
The +3 A-to G-mutation in the ACADSB 5′ss has a severe effect on splicing efficiency. By using the competition strategy described in Figure 5, we performed three experiments. First, the ACADSB wild-type 5′ss (+3A, GGG/GUACAU) and the mutant (+3G, GGG/GUGCAU), which differs by having a G at +3, were placed in competition against the beta-globin cryptic (GUG/GUGAGG) 5′ss at −16 (Treisman et al. 1983). The cryptic 5′ss at −16 was the only 5′ss used (lanes 1,2), indicating that both +3A and +3G 5′ss are very weak. Second, a competition was done between +3A and +3G, which resulted in splicing only via the +3A 5′ss (lanes 4,5). Third, the same 5′ss was placed on the same substrate at both locations. Whereas the substrate with two +3A 5′ss resulted in use of both −16 and +1 5′ss (lane 3), two +3G 5′ss did not show any splicing (lane 6). Together, these results indicate that the +3A 5′ss is much stronger than the +3G 5′ss.
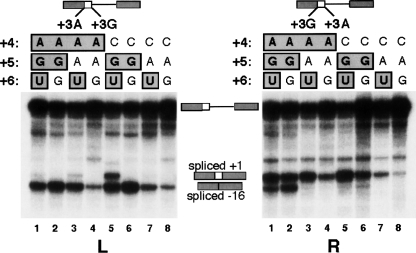
Second, we found that correcting positions +4 and/or +5 to match the consensus can alleviate the effects of the +3 A-to-G mutation, which is in concordance with our above-mentioned prediction derived from the pairwise associations. To test for the rescue of splicing by correcting these positions, we compared two ACADSB 5′ss with the same combination of nucleotides at positions +4, +5, and +6, but one having A at +3 and the other one G at +3 (Fig. 7). In general, we found that +3G 5′ss use was positively correlated with the number of consensus nucleotides at positions +4 to +6 (lanes 1–3, 5). However, the two reciprocal experiments shown in this figure do not match, in the sense that the activation of the +3G 5′ss is influenced by the relative position of the two competing 5′ss (lanes 1–6, cf. the +3G-spliced band between both panels). This difference is probably due to the influence of the sequences that flank the 5′ss at positions −16 and +1. Notwithstanding these positional effects, our conclusion that correction of positions +4 and/or +5 rescues splicing of the +3G 5′ss is still supported by the data.
Figure 7.
The deleterious effect of the +3 A-to-G mutation can be alleviated by correcting positions +4 and/or +5. In this competition assay, we created two sequences from the ACADSB exon 3 5′ss: one with +3A and another with +3G (for the experimental strategy, see Fig. 5; for the ACADSB 5′ss sequences, see Fig. 6). The same mutations were simultaneously introduced at positions +4, +5, and +6 at both 5′ss, to test their compensatory effects vis-à-vis the +3 A-to-G mutations. The nucleotides at positions +4, +5, and +6 are listed above the lanes, and the consensus nucleotides are boxed. The left and right panels are the same, except that the positions of the +3A and +3G 5′ss are swapped (indicated on the top diagram). With a few exceptions, the general trend is that the +3G 5′ss was used to a greater extent as the number of consensus nucleotides at positions +4 to +6 was increased (lanes 1–3, 5). The degree of use of the +3G 5′ss was not the same for identical mutants in the two panels (for details, see text), but despite these positional effects, the trend remains. The +3G 5′ss can be activated by correcting position +4 alone (lane 3) or +5 alone (lane 5). The rescue by +5 alone is surprising and indicates that associations between nonadjacent positions can affect splice-site efficiency.
Finally, the +3G 5′ss can be activated by correcting position +4 alone (Fig. 7, lane 3). This is not surprising, because a base pair at +4 with U1 might stabilize the wobble G-Ψ base pair at +3. Strikingly, correcting position +5 alone alleviated the severity of the A-to-G mutation at +3 (Fig. 7, lane 5). This indicates that nonadjacent interactions (such as those involving positions +3 and +5) at the 5′ss can affect splice-site efficiency.
Orthologous 5′ss between the mouse and human genomes
We expected that orthologous pairs of 5′ss between mouse and human genomes, which are very similar to each other according to the PWMs and the association matrices, should show patterns of nucleotide substitutions that do not substantially disrupt the PWM score or the association score. We used a published data set of 54,761 pairs of orthologous 5′ss from the human and mouse genomes (Carmel et al. 2004) to study the nature of differences between members of these pairs.
We found that 36.5% of these pairs have identical splice-site sequences and 36.4% show a single nucleotide change. The nucleotide change between orthologous pairs that differ at a single position does not usually disrupt the PWM score and the pairwise associations within 5′ss (Fig. 3). We also observed a compensatory trend: If one member of an orthologous 5′ss pair has a weaker PWM score, then its pairwise-association score tends to be higher (Fig. 8).
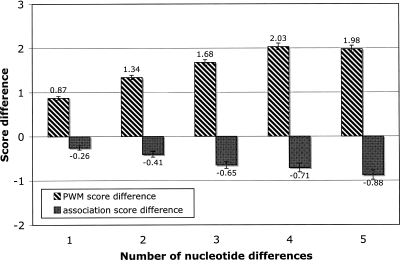
Figure 8.
Association improvements can compensate for reductions in PWM scores. In orthologous mouse–human pairs of 5′ss, association scores improve as PWM scores get weaker. The graph shows the average of the PWM-score differences (white bars) as well as association-score differences (black bars) for the orthologous 5′ss mouse–human pairs, organized by the number of nucleotide differences between the members of each pair (X-axis). A positive difference means a decrease in the score (and vice versa). The differences in PWM scores are always positive, as the 5′ss with the higher score within a pair is always considered first. Error bars, confidence intervals.
In the orthologous 5′ss pairs with two nucleotide changes (19.2%), certain coordinated changes are preferred over others. For example, +5C+6C changes to +5T+6G more often than expected (72 vs. 28 expected; both combinations are strongly enriched), whereas −1T+6T to −1C+6A is strongly suppressed (occurs once vs. 20 expected; the first combination is enhanced, whereas the second combination is strongly depleted). The probability of a single change occurring, e.g., +5C to +5T, is calculated by counting the number of such transitions over the pairs of which at least one of them has a C at +5. The expected number of coordinated pairwise changes is calculated by assuming independence between the transitions at the two positions in the 5′ss. Evidently, the pairwise-association patterns reflect constraints on the evolution of 5′ss due to the conserved splicing machinery, and these association patterns could hold clues into mechanistic features of the splicing machinery.
SNPs at 5′ss
We expect SNPs that occur at splice sites to be neutral, i.e., not to affect splice-site efficiency in a majority of the cases. There have been a few reports of SNPs that do affect splicing (Shinmura et al. 2004; Field et al. 2005; Skarratt et al. 2005; ElSharawy et al. 2006; Kawase et al. 2007) but no comprehensive analysis of this phenomenon. In our study, we used the 1260 SNPs at 5′ss (dbSNP) (Smigielski et al. 2000) that are confirmed by population data and lie outside of positions +1 and +2.
We calculated the PWM scores and pairwise-association score differences for SNPs (Fig. 3C). The average PWM score for 5′ss with SNPs (5.92) is higher than that for simulated SNPs (5.54), which in turn is higher than the average score for disease-causing mutations (5.31). The PWM scores of orthologs that show one difference between human and mouse are higher on average than the SNP set (6.2 vs. 5.92), suggesting that strong 5′ss (PWM score >6.2) can tolerate such changes without affecting splicing.
From Figure 3, we can see that the disruption of the associations by the SNPs is smaller than the disruption by the simulated SNPs, which in turn is smaller than the disruption by the 5′ss disease-causing mutations, suggesting selective pressure on SNPs to preserve the associations. For instance, Figure 4 shows that SNPs with alleles A and G at position +3 tend to have consensus nucleotides at positions +4 and +5, as expected from the previous discussion on +3 A-to-G disease mutations.
By using these findings (see Fig. 3), we were able to establish criteria to identify SNPs that might disrupt splicing. We used the average score of the disease set to identify low PWM scores (≤5.31). The separation between disease-causing mutations and the SNPs and orthologous sets was used to identify high PWM-score differences (≥1). The average association-score difference for the disease data set was used to identify high association-score differences (≥1.1). We expect disruptive 5′ss SNPs to occur at 5′ss with low PWM scores, and for the SNPs to substantially disrupt either the PWM score or the association scores or both simultaneously.
Disruptive 5′ss SNP predictions and evidence for their effect on splicing
By using the criteria listed above, we created five sets from the 1260 SNPs in dbSNP that occur at 5′ss (Smigielski et al. 2000). The sets were picked to dissect the effects of the PWM scores, PWM-score differences between SNP alleles, and association-score differences between SNP alleles. The first set, designated LHH, consists of 35 SNPs at 5′ss that have low (L) PWM scores and alleles with high (H) association-score differences and high (H) PWM-score differences. The second set, LHL, consists of 35 SNPs at 5′ss with low PWM scores and alleles with high PWM-score differences and low association-score differences. The third set, LLH, consists of 43 SNPs at 5′ss with low PWM scores and alleles with low PWM-score differences and high-association score differences. The fourth set, HLL, consists of 58 SNPs at 5′ss with high PWM scores and alleles with low PWM-score differences and low association-score differences. Finally, the fifth set, LLL, consists of 111 SNPs at 5′ss with low PWM scores and alleles with low PWM-score differences and low association-score differences. The LHH, LLH, and LHL sets of SNPs are expected to be more disruptive of splicing, whereas the HLL and LLL sets are controls that should be less disruptive of splicing.
We used EST and mRNA alignments to the genome to establish disruption caused by SNPs at 5′ss, using the UCSC browser (Kent et al. 2002). The alignments were studied to pick unambiguous cases of intron retention (at least one intron besides the retained one was spanned by the EST/mRNA), exon skipping (the skipped exon is otherwise constitutively spliced), and cryptic splice-site utilization. A prediction was considered validated if alternative splicing was observed at the 5′ss in question, and when possible, the EST and mRNA data supporting the alternative event corresponded to the nonconsensus allele of the SNP. In the case of exon skipping, it is not possible to ascertain the allele at the SNP position, but by considering the proportion of internal exons among RefSeq mRNAs that are skipped in this manner (3734/166,622), we were able to infer the likely error rate in making the determination of exon skipping as being due to the SNP (one in 45). We estimate similar error rates for intron retention (one in 160) and cryptic-splice-site utilization (one in 70) based on previous work on the relative prevalence of the different types of alternatively spliced exons (Kim et al. 2007). Furthermore, we checked that a 5′ss following cassette exons (from the Alt Events track of the UCSC browser) was no more likely to contain a SNP than a randomly selected 5′ss.
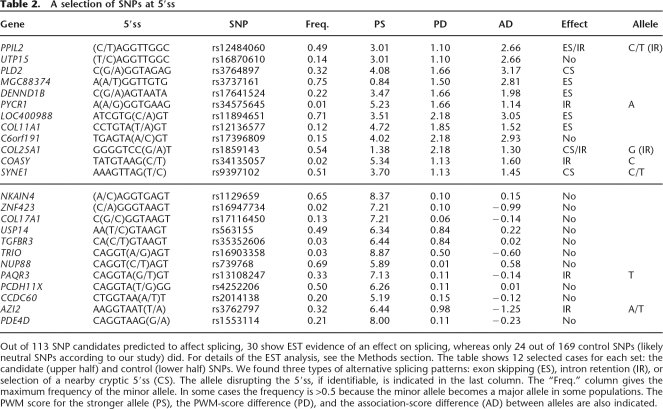
In agreement with our expectations, we found that the sets LHH (10 disruptive SNPs out of 35), LHL (eight disruptive SNPs out of 35), and LLH (12 disruptive SNPs out of 43) are more disruptive than the control sets HLL (five disruptive SNPs out of 58) and LLL (19 disruptive SNPs out 111). By using bootstrapping to estimate the variance of these numbers, we found that the LLL and LLH sets are significantly different from each other (P-value of 0.0003 using an unpaired t-test). The stronger disruption shown by the LLH set compared with the LLL set confirms the predictive nature of the association scores. A sampling of SNPs from these sets is given in Table 2.
Table 2.
A selection of SNPs at 5′ss
Out of 113 SNP candidates predicted to affect splicing, 30 show EST evidence of an effect on splicing, whereas only 24 out of 169 control SNPs (likely neutral SNPs according to our study) did. For details of the EST analysis, see the Methods section. The table shows 12 selected cases for each set: the candidate (upper half) and control (lower half) SNPs. We found three types of alternative splicing patterns: exon skipping (ES), intron retention (IR), or selection of a nearby cryptic 5′ss (CS). The allele disrupting the 5′ss, if identifiable, is indicated in the last column. The “Freq.” column gives the maximum frequency of the minor allele. In some cases the frequency is >0.5 because the minor allele becomes a major allele in some populations. The PWM score for the stronger allele (PS), the PWM-score difference (PD), and the association-score difference (AD) between alleles are also indicated.
This analysis is confounded by the fact that the ESTs might not have sampled the SNP alleles of interest, due to the genotype of the source. The intronic SNPs are also not likely to be sampled in the ESTs. In addition, some of the alternative spliced products might be degraded by NMD, due to the presence of a premature stop codon and thus would not be observed (Lejeune and Maquat 2005). In light of these limitations, it is encouraging that we still find such a high confirmation rate.
We expected the genotypes to show a preference for alleles of SNPs at 5′ss that do not disrupt splicing, but we did not detect a strong signal in the distribution of genotypes. The effect of SNPs at 5′ss might be alleviated by the sequence context. For example, in the ACADM (also called MCAD) gene (Nielsen et al. 2007), one allele of a SNP creates an ESS, and a mutation that disrupts a nearby ESE causes exon skipping only in individuals with the ESS-creating allele. Even if splicing is affected, there might be no phenotypic effects due to the following: (1) a small reduction in the correct mRNA levels; (2) compensation in the genetic network; (3) the function of the disrupted gene is required only under certain conditions; or (4) the alternate product due to the disruption of the particular 5′ss is also functional. Nevertheless, such SNPs can offer a window into the investigation of the genetic network. This analysis will allow investigation of disruptive SNPs and their role in complex diseases.
Discussion
We have shown that the effects of disease-causing mutations are often a result of the disruption of conserved patterns in associations between nucleotides at different positions within the 5′ss, and SNPs that disrupt these pairwise associations tend to affect splicing. A majority of SNPs respect the associations, as expected from SNP neutrality. In addition, we have shown that orthologous 5′ss mouse–human pairs show changes that likewise respect the associations, which suggests the existence of selective pressure to maintain them. A set of simulated SNPs is more disruptive of associations than neutral SNPs but is better than the disease-causing mutations in this respect. This is expected from the lack of selection pressure on the simulated SNPs.
The conservation of the associations is indicative of selective pressures reflecting functional features of the splicing machinery, and allows inferences to be made about the underlying mechanisms. The pairwise associations confirm many known effects, but also suggest new areas for exploration. We found pairwise associations probably related to U1 base pairing, such as specific patterns involving consensus and nonconsensus nucleotides across the exon–intron boundary. We found one association probably related to base pairing to U6, but none related to U5. The long-range associations are likely related to protein–RNA interactions, and further experiments should shed light on them. Similar studies could be carried out using 3′ss. However, the longer span of this motif (Sheth et al. 2006) and its more plastic organization suggest that the pairwise associations at 3′ss will not reveal as many biases as the associations at 5′ss.
By using the data from disease-causing mutations in 5′ss, differences between orthologous pairs of mouse and human 5′ss, and genomic data for five species, we were able to generate criteria for prediction of splicing-disruptive SNPs at 5′ss. We have shown that circumstantial evidence from ESTs provides support for these predictions and encourages further experiments to study their effects in vivo.
The disruptive SNPs may provide insights into genetic networks. If a proven disruptive SNP is in Hardy-Weinberg equilibrium, it suggests that the genetic network is immune to the change, and this can be a starting point for investigating the reasons for this robustness of the network. Alternatively, such SNPs might be implicated in complex diseases, wherein their effects are apparent only under certain genetic and environmental conditions. For example, there has been work on SNPs affecting the p53 pathway, such as a SNP in the MDM2 gene that alters a transcription-factor binding site and hence the levels of p53 (Bond and Levine 2007). This results in phenotypes ranging from an effect on fertility, to the onset of tumor formation, and the response to various therapies, such as estrogen replacement. Our study is a first step in investigating the effect of SNPs at 5′ss on a genomic scale. The consequences of the disruptions could be stochastic in nature, leading to effects that might not be readily discernible in in vitro systems but might nevertheless affect the in vivo phenotypes.
This study illustrates the power of the convergence of different data sets for obtaining insights into mechanisms of gene expression and for understanding the neutral and disease-causing nucleotide changes found in human populations.
Methods
Scoring splice sites with PWM
PWMs reflect the frequencies of the 4 nt (A, C, G, and T) at each position of the splice site. PWMs can be used to score a site by converting the frequencies into a log-odds score (log of the ratio of the actual frequency and the expected frequency) (Sheth et al. 2006).
Scoring associations within splice sites
For the associations, we calculated the expected frequencies for each combination of a pair of positions and nucleotides, using the PWM and assuming independence of positions from each other. We then calculated a log-odds score for the combination, based on the actual numbers observed in the genomic data set. The total association score for a given 5′ss is the sum of the log-odds score for each nucleotide pair at the 5′ss. The change in association score measures the disruption (or enhancement) of the pairwise associations due to changes at the 5′ss caused by SNPs, mutations, or differences between pairs of orthologous 5′ss.
In order to avoid small statistical fluctuations affecting the scores (e.g., an actual occurrence of three versus an expected number of 45 would give the relevant pair a strong negative score), we excluded those pairs that are expected less than 50 times in the genome. This does not affect the scoring of most splice sites, as these pairs are by definition rare.
Collection of data sets
From the HGMD database (Stenson et al. 2003), we collected 1772 disease-causing mutations affecting U2-type GT-AG 5′ss sequences. After excluding mutations affecting +1 and +2 of the 5′ss, we were left with 582 mutations affecting the positions −3 to −1 and +3 to +6, which we used in our analysis. An additional 13 5′ss mutations changing a +3 A to G at 5′ss were added from a separate source (Madsen et al. 2006). We randomly selected 5′ss from SpliceRack (Sheth et al. 2006) and mutated a randomly selected position in the 5′ss to generate a list of simulated SNPs in 5′ss. From dbSNP (Smigielski et al. 2000), we extracted SNPs that have population data and mapped them to the genome to identify SNPs at 5′ss. For the orthologous set, we used a published data set (Carmel et al. 2004).
In vitro splicing
In the 5′ss competition assay, each test ACADSB 5′ss replaces the beta-globin 5′ss at positions −16 and/or +1 (Fig. 5). The competition between the two 5′ss provides an estimate of the difference in their intrinsic strengths.
The beta-globin wild-type 5′ss and the cryptic 5′ss at position −16 were replaced by various mutant ACADSB 5′ss by PCR mutagenesis. We inactivated a second cryptic 5′ss in beta-globin that is located 38 nt upstream of the authentic 5′ss (Treisman et al. 1983) to prevent competition with the various ACADSB 5′ss. The different mutant beta-globin minigenes, inserted into the pSP64 vector (Promega) were in vitro transcribed (Mayeda and Krainer 1999b). The pre-mRNA transcripts were then spliced in vitro using HeLa cell nuclear extract (Mayeda and Krainer 1999a), and the products were analyzed by electrophoresis in 5.5% poly-acrylamide/7M urea gels.
EST analysis
By using the UCSC genome browser (Kent et al. 2002), we located the relevant SNPs by entering the rs id numbers provided by dbSNP into the “position/search” input area. We used visual inspection to determine the nature of the alternative splicing event and tried to identify the allele that might be responsible for this splicing alteration. The browser was configured to display the “human mRNAs” and “spliced ESTs” tracks in “full” mode. Determining which allele was used in each EST was simplified by setting the “color track by codons or bases” option to “different mRNA bases” (follow the link on the track name to turn on this feature).
Acknowledgments
Jeremiah Faith and Susan Janicki gave insightful comments on the manuscript. The anonymous reviewers gave useful criticisms that helped improve the paper. A.J.O. and R.S. thank the DART Neurogenomics Alliance for support. X.R. and A.R.K. acknowledge support from NIH grant CA13106. A.R.R. acknowledges support from the Department of Biotechnology (India) grant BT/IN/BTOA/03/2005.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6859308
References
- Bond G.L., Levine A.J., Levine A.J. A single nucleotide polymorphism in the p53 pathway interacts with gender, environmental stresses and tumor genetics to influence cancer in humans. Oncogene. 2007;26:1317–1323. doi: 10.1038/sj.onc.1210199. [DOI] [PubMed] [Google Scholar]
- Brow D.A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- Brunak S., Engelbrecht J., Knudsen S., Engelbrecht J., Knudsen S., Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 1991;220:49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- Buratti E., Chivers M., Kralovicova J., Romano M., Baralle M., Krainer A.R., Vorechovsky I., Chivers M., Kralovicova J., Romano M., Baralle M., Krainer A.R., Vorechovsky I., Kralovicova J., Romano M., Baralle M., Krainer A.R., Vorechovsky I., Romano M., Baralle M., Krainer A.R., Vorechovsky I., Baralle M., Krainer A.R., Vorechovsky I., Krainer A.R., Vorechovsky I., Vorechovsky I. Aberrant 5′ splice sites in human disease genes: Mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2004;35:4250–4263. doi: 10.1093/nar/gkm402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E., Baralle M., Conti L.D., Baralle D., Romano M., Ayala Y.M., Baralle F.E., Baralle M., Conti L.D., Baralle D., Romano M., Ayala Y.M., Baralle F.E., Conti L.D., Baralle D., Romano M., Ayala Y.M., Baralle F.E., Baralle D., Romano M., Ayala Y.M., Baralle F.E., Romano M., Ayala Y.M., Baralle F.E., Ayala Y.M., Baralle F.E., Baralle F.E. hnRNP H binding at the 5′ splice site correlates with the pathological effect of two intronic mutations in the NF-1 and TSHb genes. Nucleic Acids Res. 2007;32:4224–4236. doi: 10.1093/nar/gkh752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C., Karlin S., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Burge C.B., Padgett R.A., Sharp P.A., Padgett R.A., Sharp P.A., Sharp P.A. Evolutionary fates and origins of U12-type introns. Mol. Cell. 1998;2:773–785. doi: 10.1016/s1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- Cáceres J.F., Stamm S., Helfman D.M., Krainer A.R., Stamm S., Helfman D.M., Krainer A.R., Helfman D.M., Krainer A.R., Krainer A.R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- Carmel I., Tal S., Vig I., Ast G., Tal S., Vig I., Ast G., Vig I., Ast G., Ast G. Comparative analysis detects dependencies among the 5′ splice-site positions. RNA. 2004;10:828–840. doi: 10.1261/rna.5196404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L., Chew S., Krainer A.R., Chew S., Krainer A.R., Krainer A.R. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Costantini M., Clay O., Auletta F., Bernardi G., Clay O., Auletta F., Bernardi G., Auletta F., Bernardi G., Bernardi G. An isochore map of human chromosomes. Genome Res. 2006;16:536–541. doi: 10.1101/gr.4910606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti L.B., Bacikova D., Horowitz D.S., Bacikova D., Horowitz D.S., Horowitz D.S. The PRP18 protein stabilizes the interaction of both exons with the U5 snRNA during the second step of pre-mRNA splicing. Genes & Dev. 2007;21:1204–1216. doi: 10.1101/gad.1538207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Rosbash M., Rosbash M. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature. 2002;419:86–90. doi: 10.1038/nature00947. [DOI] [PubMed] [Google Scholar]
- ElSharawy A., Manaster C., Teuber M., Rosenstiel P., Kwiatkowski R., Huse K., Platzer M., Becker A., Nurnberg P., Schreiber S., Manaster C., Teuber M., Rosenstiel P., Kwiatkowski R., Huse K., Platzer M., Becker A., Nurnberg P., Schreiber S., Teuber M., Rosenstiel P., Kwiatkowski R., Huse K., Platzer M., Becker A., Nurnberg P., Schreiber S., Rosenstiel P., Kwiatkowski R., Huse K., Platzer M., Becker A., Nurnberg P., Schreiber S., Kwiatkowski R., Huse K., Platzer M., Becker A., Nurnberg P., Schreiber S., Huse K., Platzer M., Becker A., Nurnberg P., Schreiber S., Platzer M., Becker A., Nurnberg P., Schreiber S., Becker A., Nurnberg P., Schreiber S., Nurnberg P., Schreiber S., Schreiber S., et al. SNPSplicer: systematic analysis of SNP-dependent splicing in genotyped cDNAs. Hum. Mutat. 2006;27:1129–1134. doi: 10.1002/humu.20377. [DOI] [PubMed] [Google Scholar]
- Field L.L., Bonnevie-Nielsen V., Pociot F., Lu S., Nielsen T.B., Beck-Nielsen H., Bonnevie-Nielsen V., Pociot F., Lu S., Nielsen T.B., Beck-Nielsen H., Pociot F., Lu S., Nielsen T.B., Beck-Nielsen H., Lu S., Nielsen T.B., Beck-Nielsen H., Nielsen T.B., Beck-Nielsen H., Beck-Nielsen H. OAS1 splice site polymorphism controlling antiviral enzyme activity influences susceptibility to type 1 diabetes. Diabetes. 2005;54:1588–1591. doi: 10.2337/diabetes.54.5.1588. [DOI] [PubMed] [Google Scholar]
- Hastings M.L., Krainer A.R., Krainer A.R. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 2001;13:302–309. doi: 10.1016/s0955-0674(00)00212-x. [DOI] [PubMed] [Google Scholar]
- Jurica M.S., Moore M.J., Moore M.J. Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Kandels-Lewis S., Séraphin B., Séraphin B. Involvement of U6 snRNA in 5′ splice site selection. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- Kawase T., Akatsuka Y., Torikai H., Morishima S., Oka A., Tsujimura A., Miyazaki M., Tsujimura K., Miyamura K., Ogawa S., Akatsuka Y., Torikai H., Morishima S., Oka A., Tsujimura A., Miyazaki M., Tsujimura K., Miyamura K., Ogawa S., Torikai H., Morishima S., Oka A., Tsujimura A., Miyazaki M., Tsujimura K., Miyamura K., Ogawa S., Morishima S., Oka A., Tsujimura A., Miyazaki M., Tsujimura K., Miyamura K., Ogawa S., Oka A., Tsujimura A., Miyazaki M., Tsujimura K., Miyamura K., Ogawa S., Tsujimura A., Miyazaki M., Tsujimura K., Miyamura K., Ogawa S., Miyazaki M., Tsujimura K., Miyamura K., Ogawa S., Tsujimura K., Miyamura K., Ogawa S., Miyamura K., Ogawa S., Ogawa S., et al. Alternative splicing due to an intronic SNP in HMSD generates a novel minor histocompatibility antigen. Blood. 2007;110:1055–1063. doi: 10.1182/blood-2007-02-075911. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The Human Genome Browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Magen A., Ast G., Magen A., Ast G., Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007;35:125–131. doi: 10.1093/nar/gkl924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak M., Reiss J., Cooper D.N., Reiss J., Cooper D.N., Cooper D.N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: Causes and consequences. Hum. Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- Krawczak M., Ball E.V., Cooper D.N., Ball E.V., Cooper D.N., Cooper D.N. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am. J. Hum. Genet. 1998;63:474–488. doi: 10.1086/301965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak M., Thomas N.S., Hundrieser B., Mort M., Wittig M., Hampe J., Cooper D.N., Thomas N.S., Hundrieser B., Mort M., Wittig M., Hampe J., Cooper D.N., Hundrieser B., Mort M., Wittig M., Hampe J., Cooper D.N., Mort M., Wittig M., Hampe J., Cooper D.N., Wittig M., Hampe J., Cooper D.N., Hampe J., Cooper D.N., Cooper D.N. Single base-pair substitutions in exon-intron junctions of human genes: Nature, distribution, and consequences for mRNA splicing. Hum. Mutat. 2007;28:150–158. doi: 10.1002/humu.20400. [DOI] [PubMed] [Google Scholar]
- Ladd A.N., Cooper T.A., Cooper T.A. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 2002;3:1–16. doi: 10.1186/gb-2002-3-11-reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F., Maquat L.E., Maquat L.E. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Lesser C.F., Guthrie C., Guthrie C. Mutations in U6 snRNA that alter splice site specificity: Implications for the active site. Science. 1993;6:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- Madsen P.P., Kibaek M., Roca X., Sachidanandam R., Krainer A.R., Christensen E., Steiner R.D., Gibson K.M., Corydon T.J., Knudsen I., Kibaek M., Roca X., Sachidanandam R., Krainer A.R., Christensen E., Steiner R.D., Gibson K.M., Corydon T.J., Knudsen I., Roca X., Sachidanandam R., Krainer A.R., Christensen E., Steiner R.D., Gibson K.M., Corydon T.J., Knudsen I., Sachidanandam R., Krainer A.R., Christensen E., Steiner R.D., Gibson K.M., Corydon T.J., Knudsen I., Krainer A.R., Christensen E., Steiner R.D., Gibson K.M., Corydon T.J., Knudsen I., Christensen E., Steiner R.D., Gibson K.M., Corydon T.J., Knudsen I., Steiner R.D., Gibson K.M., Corydon T.J., Knudsen I., Gibson K.M., Corydon T.J., Knudsen I., Corydon T.J., Knudsen I., Knudsen I., et al. Short/branched-chain acyl-CoA dehydrogenase deficiency due to an IVS3+3A>G mutation that causes exon skipping. Hum. Genet. 2006;118:680–690. doi: 10.1007/s00439-005-0070-4. [DOI] [PubMed] [Google Scholar]
- Maroney P.A., Romfo C.M., Nilsen T.W., Romfo C.M., Nilsen T.W., Nilsen T.W. Functional recognition of 5′ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol. Cell. 2000;6:317–328. doi: 10.1016/s1097-2765(00)00032-0. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Krainer A.R., Krainer A.R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Krainer A.R., Krainer A.R. Mammalian in vitro splicing assays. Methods Mol. Biol. 1999a;118:315–321. doi: 10.1385/1-59259-676-2:315. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Krainer A.R., Krainer A.R. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol. 1999b;118:309–314. doi: 10.1385/1-59259-676-2:309. [DOI] [PubMed] [Google Scholar]
- Nakai K., Sakamoto H., Sakamoto H. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene. 1994;141:171–177. doi: 10.1016/0378-1119(94)90567-3. [DOI] [PubMed] [Google Scholar]
- Newman A.J., Norman C., Norman C. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- Nielsen K.B., Sorensen S., Cartegni L., Corydon T.J., Doktor T.K., Schroeder L.D., Reinert L.S., Elpeleg O., Krainer A.R., Gregersen N., Sorensen S., Cartegni L., Corydon T.J., Doktor T.K., Schroeder L.D., Reinert L.S., Elpeleg O., Krainer A.R., Gregersen N., Cartegni L., Corydon T.J., Doktor T.K., Schroeder L.D., Reinert L.S., Elpeleg O., Krainer A.R., Gregersen N., Corydon T.J., Doktor T.K., Schroeder L.D., Reinert L.S., Elpeleg O., Krainer A.R., Gregersen N., Doktor T.K., Schroeder L.D., Reinert L.S., Elpeleg O., Krainer A.R., Gregersen N., Schroeder L.D., Reinert L.S., Elpeleg O., Krainer A.R., Gregersen N., Reinert L.S., Elpeleg O., Krainer A.R., Gregersen N., Elpeleg O., Krainer A.R., Gregersen N., Krainer A.R., Gregersen N., Gregersen N., et al. Seemingly neutral polymorphic variants may confer immunity to splicing-inactivating mutations: A synonymous SNP in exon 5 of MCAD protects from deleterious mutations in a flanking exonic splicing enhancer. Am. J. Hum. Genet. 2007;80:416–432. doi: 10.1086/511992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno K., Brengman J.M., Felice K.J., Cornblath D.R., Engel A.G., Brengman J.M., Felice K.J., Cornblath D.R., Engel A.G., Felice K.J., Cornblath D.R., Engel A.G., Cornblath D.R., Engel A.G., Engel A.G. Congenital end-plate acetylcholinesterase deficiency caused by a nonsense mutation and an A→G splice-donor-site mutation at position +3 of the collagenlike-tail-subunit gene (COLQ): How does G at position +3 result in aberrant splicing? Am. J. Hum. Genet. 1999;65:635–644. doi: 10.1086/302551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe R.T., Norman C., Newman A.J., Norman C., Newman A.J., Newman A.J. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell. 1996;86:679–689. doi: 10.1016/s0092-8674(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Reddy R., Henning D., Busch H., Henning D., Busch H., Busch H. Pseudouridine residues in the 5′-terminus of uridine-rich nuclear RNA I (U1 RNA) Biochem. Biophys. Res. Commun. 1981;98:1076–1078. doi: 10.1016/0006-291x(81)91221-3. [DOI] [PubMed] [Google Scholar]
- Roca X., Sachidanandam R., Krainer A.R., Sachidanandam R., Krainer A.R., Krainer A.R. Intrinsic differences between authentic and cryptic 5′ splice sites. Nucleic Acids Res. 2003;31:6321–6333. doi: 10.1093/nar/gkg830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca X., Sachidanandam R., Krainer A.R., Sachidanandam R., Krainer A.R., Krainer A.R. Determinants of the inherent strength of human 5′ splice sites. RNA. 2005;11:683–698. doi: 10.1261/rna.2040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy P., Shapiro M.B., Harris N.L., Shapiro M.B., Harris N.L., Harris N.L. Splice junctions, branch point sites, and exons: Sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Kretzner L., Rosbash M., Kretzner L., Rosbash M., Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 1988;183:2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M.B., Senapathy P., Senapathy P. RNA splice junctions of different classes of eukaryotes: Sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth N., Roca X., Hastings M.L., Roeder T., Krainer A.R., Sachidanandam R., Roca X., Hastings M.L., Roeder T., Krainer A.R., Sachidanandam R., Hastings M.L., Roeder T., Krainer A.R., Sachidanandam R., Roeder T., Krainer A.R., Sachidanandam R., Krainer A.R., Sachidanandam R., Sachidanandam R. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinmura K., Tao H., Yamada H., Kataoka H., Sanjar R., Wang J., Yoshimura K., Sugimura H., Tao H., Yamada H., Kataoka H., Sanjar R., Wang J., Yoshimura K., Sugimura H., Yamada H., Kataoka H., Sanjar R., Wang J., Yoshimura K., Sugimura H., Kataoka H., Sanjar R., Wang J., Yoshimura K., Sugimura H., Sanjar R., Wang J., Yoshimura K., Sugimura H., Wang J., Yoshimura K., Sugimura H., Yoshimura K., Sugimura H., Sugimura H. Splice-site genetic polymorphism of the human kallikrein 12 (KLK12) gene correlates with no substantial expression of KLK12 protein having serine protease activity. Hum. Mutat. 2004;24:273–274. doi: 10.1002/humu.9270. [DOI] [PubMed] [Google Scholar]
- Siliciano P.G., Guthrie C., Guthrie C. 5′ splice site selection in yeast: Genetic alterations in base pairing with U1 reveal additional requirements. Genes & Dev. 1988;2:1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- Skarratt K.K., Fuller S.J., Sluyter R., Dao-Ung L.P., Gu B.J., Wiley J.S., Fuller S.J., Sluyter R., Dao-Ung L.P., Gu B.J., Wiley J.S., Sluyter R., Dao-Ung L.P., Gu B.J., Wiley J.S., Dao-Ung L.P., Gu B.J., Wiley J.S., Gu B.J., Wiley J.S., Wiley J.S. A 5′ intronic splice site polymorphism leads to a null allele of the P2X7 gene in 1-2% of the Caucasian population. FEBS Lett. 2005;579:2675–2678. doi: 10.1016/j.febslet.2005.03.091. [DOI] [PubMed] [Google Scholar]
- Smigielski E.M., Sirotkin K., Ward M., Sherry S.T., Sirotkin K., Ward M., Sherry S.T., Ward M., Sherry S.T., Sherry S.T. dbSNP: A database of single nucleotide polymorphisms. Nucleic Acids Res. 2000;28:352–355. doi: 10.1093/nar/28.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson P., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N., Abeysinghe S., Krawczak M., Cooper D.N., Krawczak M., Cooper D.N., Cooper D.N. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- Teraoka S.N., Telatar M., Becker-Catania S., Liang T., Onengut S., Tolun A., Chessa L., Sanal O., Bernatowska E., Gatti R.A., Telatar M., Becker-Catania S., Liang T., Onengut S., Tolun A., Chessa L., Sanal O., Bernatowska E., Gatti R.A., Becker-Catania S., Liang T., Onengut S., Tolun A., Chessa L., Sanal O., Bernatowska E., Gatti R.A., Liang T., Onengut S., Tolun A., Chessa L., Sanal O., Bernatowska E., Gatti R.A., Onengut S., Tolun A., Chessa L., Sanal O., Bernatowska E., Gatti R.A., Tolun A., Chessa L., Sanal O., Bernatowska E., Gatti R.A., Chessa L., Sanal O., Bernatowska E., Gatti R.A., Sanal O., Bernatowska E., Gatti R.A., Bernatowska E., Gatti R.A., Gatti R.A., et al. Splicing defects in the ataxia-telangiectasia gene, ATM: Underlying mutations and consequences. Am. J. Hum. Genet. 1999;64:1617–1631. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Orkin S.H., Maniatis T., Orkin S.H., Maniatis T., Maniatis T. Specific transcription and RNA splicing defects in five cloned β-thalassaemia genes. Nature. 1983;302:591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Tweedie S., Charlton J., Clark V., Bird A., Charlton J., Clark V., Bird A., Clark V., Bird A., Bird A. Methylation of genomes and genes at the invertebrate-vertebrate boundary. Mol. Cell. Biol. 1997;17:1469–1475. doi: 10.1128/mcb.17.3.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman D.A., Steitz J.A., Steitz J.A. Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- Yeo G., Burge C.B., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A.M., Weiner A.M. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]