Abstract
Hybrid zones between closely related species or subspecies provide useful settings for studying the genetic architecture of speciation. Using markers distributed throughout the mouse genome, we use a hybrid zone between two recently diverged species of house mice (Mus musculus and Mus domesticus) as a natural mapping experiment to identify genomic regions that may be involved in reproductive isolation. Using cline analysis we document a nearly 50-fold variation in level of introgression among markers. Some markers have extremely narrow cline widths; these genomic regions may contribute to reproductive isolation. Biological processes associated with these narrow clines include physiological and immune responses to the environment as well as physiological and behavioral aspects of reproduction. Other autosomal markers exhibit asymmetrically broad clines, usually with high frequencies of M. domesticus alleles on the M. musculus side of the hybrid zone. These markers identify genome regions likely housing genes with alleles that are spreading from one species to the other. Biological processes associated with these wide clines include cell signaling, olfaction, and pheromone response. These processes play important roles in survival and reproduction, and associated genes are likely targets of selection. Patterns of linkage disequilibrium in the center of the hybrid zone suggest that isolation may be caused by multiple epistatic interactions between sets of genes. These data highlight the complex genetic architecture underlying speciation even at early stages of divergence and point to some of the biological processes that may govern this architecture.
The genetic basis of speciation is a central problem in evolutionary biology. Because reproductive isolation delineates the point at which emerging species will no longer freely exchange genes, considerable speciation research has focused on the genetics of reproductive isolation. In principle, reproductive isolation between taxa might be due to premating isolation, postmating but prezygotic isolation, or postzygotic isolation. Postzygotic isolation may be due to either inviability or sterility of hybrid individuals (either F1’s or subsequent generations), or to some combination of these factors. Moreover, these fitness effects could be due to intrinsic genetic incompatibilities and/or they might depend heavily on ecological context (Coyne and Orr 2004). Historically, there have been two major approaches to studying the genetic basis of reproductive isolation. The first approach relies on crosses in the laboratory between species that display partial prezygotic or postzygotic reproductive isolation. The second approach utilizes naturally occurring hybrid populations to make inferences about the genetics of reproductive isolation.
Hybrid zone studies offer several advantages not inherent in studies based on laboratory crosses. They consider all aspects of hybrid fitness, including intrinsic genetic incompatibilities, in addition to extrinsic ecological effects. They often provide many more generations of recombination than are obtainable in the lab, making it possible to pinpoint more precisely the chromosomal regions that are involved in reproductive isolation. In addition, even in cases where the specific phenotype involved in reproductive isolation is unknown or unclear, it may still be possible to detect the effects of selection through geographic patterns of changes in allele frequencies. For example, genes contributing to reproductive isolation are expected to introgress less than neutral markers (Hunt and Selander 1973; Harrison 1990; Tucker et al. 1992; Rieseberg et al. 1999; Buerkle and Rieseberg 2001). Thus, with reasonable coverage of markers throughout the genome, a hybrid zone can be used as a natural mapping experiment to identify the number and location of genomic regions that may be important in maintaining isolation between recently diverged taxa. Conversely, hybridization may be a source of beneficial alleles (Arnold et al. 1991; Rieseberg 1991, 1997). Advantageous alleles that are introduced from one species into another are expected to introgress faster and thus further (due to selection) than neutral genes.
House mice belonging to the Mus musculus species complex provide an excellent system for speciation research: They show intermediate levels of reproductive isolation, they can be crossed in the lab, they hybridize in nature, and a wealth of genetic and genomic tools are available. House mice are variably referred to in the literature as either distinct species or subspecies of Mus musculus and include M. domesticus (M. musculus domesticus) of Western Europe, North Africa, and the Middle East; M. musculus (M. m. musculus) of Eastern Europe and northern Asia; and M. castaneus (M. m. castaneus) of southeastern Asia. These three taxa are thought to have diverged from a common ancestor in west central Asia (Prager et al. 1998) or further east (Boursot et al. 1993, 1996) 0.35–0.9 million years ago (Mya) (She et al. 1990; Boursot et al. 1996; Suzuki et al. 2004). Through laboratory crosses between M. musculus, M. domesticus, and M. castaneus or strains derived from these taxa, some components of reproductive isolation have been identified. Most notably, there is clear evidence of hybrid male sterility from multiple studies (Forejt and Ivanyi 1975; Forejt 1996; Alibert et al. 1997; Oka et al. 2004, 2007; Storchova et al. 2004; Britton-Davidian et al. 2005; Trachtulec et al. 2005; Vyskocilova et al. 2005). There is evidence for limited female sterility in some crosses (Britton-Davidian et al. 2005) but not in others (Forejt and Ivanyi 1975; Britton-Davidian et al. 2005). There is also evidence for weak premating isolation in some crosses (Laukaitis et al. 1997; Smadja and Ganem 2002; Smadja et al. 2004) but not in others (Smadja and Ganem 2005). Finally, several studies have revealed higher parasite loads in hybrid mice in nature (Sage et al. 1986a) and in the lab (Moulia et al. 1993).
A hybrid zone between M. musculus and M. domesticus formed after the movement of M. domesticus into Western Europe within the last 3000 yr (Cucchi et al. 2005). This hybrid zone has been studied in six transects along its 2400-km length from Denmark to the Transcaucasus (Ursin 1952; Hunt and Selander 1973; van Zegeren and van Oortmerssen 1981; Schnell and Selander 1981; Sage et al. 1986a, b; Vanlerberghe et al. 1986, 1988a, b; Nance et al. 1990; Tucker et al. 1992; Prager et al. 1993; Munclinger et al. 2002; Macholán et al. 2003, 2007; Payseur et al. 2004; Bozikova et al. 2005; Britton-Davidian et al. 2005; Dod et al. 2005; Raufaste et al. 2005). Using several phenotypic traits, allozymes, and other molecular markers, these studies have documented clinal patterns of variation and have shown that the majority of mice in the hybrid zone have recombinant genotypes.
Previous studies have demonstrated limited introgression of markers on the X chromosome (Tucker et al. 1992; Dod et al. 1993; Payseur et al. 2004; Macholán et al. 2007), which supports the hypothesis that the X chromosome harbors a large number of genes causing hybrid sterility or inviability (Muller 1940, 1942; Hagen and Scriber 1989; Turelli and Orr 1995; Orr 1997; Turelli and Begun 1997; Presgraves and Orr 1998; Saetre et al. 2003). However, there are several reasons for expecting autosomal genes to play an important role in reproductive isolation as well. First, the autosomes contain 94.3% of the mouse genome. Previous studies have suggested that many genes may contribute to reproductive isolation in these taxa (Raufaste et al. 2005; Macholán et al. 2007); thus, it seems likely that at least some of these genes will be autosomal. Second, Haldane’s Rule, the sterility or inviability of the heterogametic sex (Haldane 1922), seems to be due to epistatic interactions between recessive X-linked genes and autosomal dominant genes (Turelli and Orr 1995). Crosses between Mus musculus and Mus domesticus obey Haldane’s Rule (males preferentially show reduced fertility), and thus hybrids are expected to have autosomal loci involved in incompatibilities. Finally, several laboratory crosses using inbred lines have implicated a few autosomal regions in reproductive isolation between these taxa (Forejt and Ivanyi 1975; Forejt 1996; Britton-Davidian et al. 2005; Oka et al. 2007).
Despite the expected importance of autosomal genes in speciation, there have been no comprehensive studies of reproductive isolation in wild hybrid mice using markers across the genome. Here, we report on the differential introgression of loci across a hybrid zone in Bavaria, Germany using markers located on all mouse autosomes. Our goals are to (1) identify the number and location of autosomal regions showing reduced introgression as a means of finding genomic regions contributing to reproductive isolation, (2) identify genomic regions showing unusually high levels of introgression as a means of finding genomic regions that may contain beneficial alleles, and (3) use functional annotations of genes in regions of broad versus limited introgression to get a first look at biological processes associated with both patterns.
Results
Cline analysis
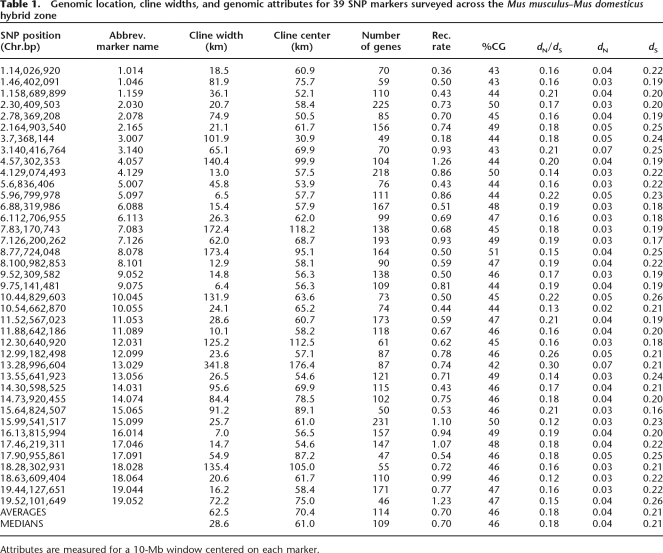
Scatter plots for all single nucleotide polymorphism (SNP) markers are shown in Supplemental Figures 1 and 2. Most changes in allele frequency occurred over a short distance; the median cline width was only 28.6 km (Table 1). Despite this small median cline width, there was enormous variation among loci in the level of introgression (Table 1; Supplemental Table 1). Three autosomal markers on three different chromosomes had cline widths <10 km: marker 5.097 (width 6.5 km), marker 9.075 (width 6.4 km), and marker 16.014 (width 7.0 km) (Table 1). These were the lowest autosomal cline widths observed. They are similar to the two-parameter estimates of cline widths for all but one of the 13 X-linked markers studied in Payseur et al. (2004). (The average two-parameter estimate of cline width for 12 X-linked markers [excluding marker X.100, with a cline width of 270.8 km] is 9.3 [range = 3.4–42.0 km, data not shown].) At the other extreme, eight markers had cline widths >100 km: markers 3.007, 4.057, 7.083, 8.078, 10.045, 12.031, 13.029, and 18.028. The average cline width for these eight markers was 165 km, or more than five times the median value for all markers. As extreme outliers, they are reasonable candidates for harboring genes under positive selection.
Table 1.
Genomic location, cline widths, and genomic attributes for 39 SNP markers surveyed across the Mus musculus–Mus domesticus hybrid zone
Attributes are measured for a 10-Mb window centered on each marker.
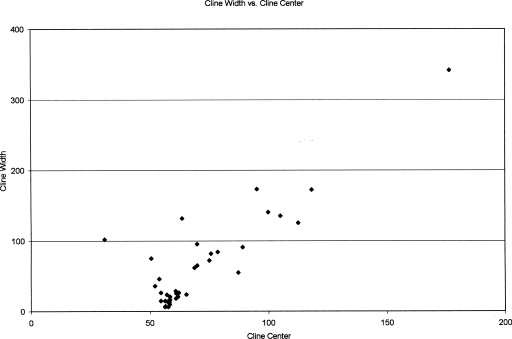
There was a strong positive correlation between cline center and cline width (P < 0.001 Spearman’s rank correlation test; Fig. 1). Wider clines had centers more to the east along the transect, indicating a pattern of asymmetric introgression, from M. domesticus to M. musculus. Evidence for introgression from M. musculus to M. domesticus was minimal and mainly limited to mice collected from the Augsburg Zoo. The presence of M. musculus alleles in this westernmost locality is likely the result of passive (long-distance) transport with zoo animals.
Figure 1.
Plot of cline width vs. cline center. These data were generated from two-parameter models of cline shape for 39 autosomal markers.
Genomic attributes of marker regions
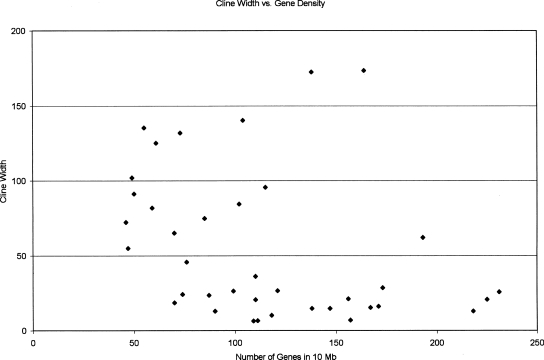
We investigated several genomic features to identify attributes of regions showing high or low levels of introgression. The number of genes in the 10-Mb windows centered on each marker ranged from 46 to 231, and, excluding marker 13.029, there was a significant negative correlation between gene density and cline width (Fig. 2; R2 = 0.102, P = 0.050). We also calculated the mean value of dN/dS between mouse and rat for all genes in each 10-Mb window. There was a significant positive correlation between mean dN/dS and cline width, although this pattern was driven by a single marker (13.029) with a window showing an unusually high mean dN/dS value. When this marker was removed from the analysis, the regression was no longer significant. The correlation between GC content and cline width was not significant, even including the marker 13.029 (R2 = 0.099, P = 0.051). Surprisingly, no significant correlation was observed between the local rate of recombination and cline width, although this may be due partly to the fact that recombination rates vary relatively little in the mouse (Shifman et al. 2006).
Figure 2.
Plot of cline width versus gene density.
Linkage disequilibrium (LD)
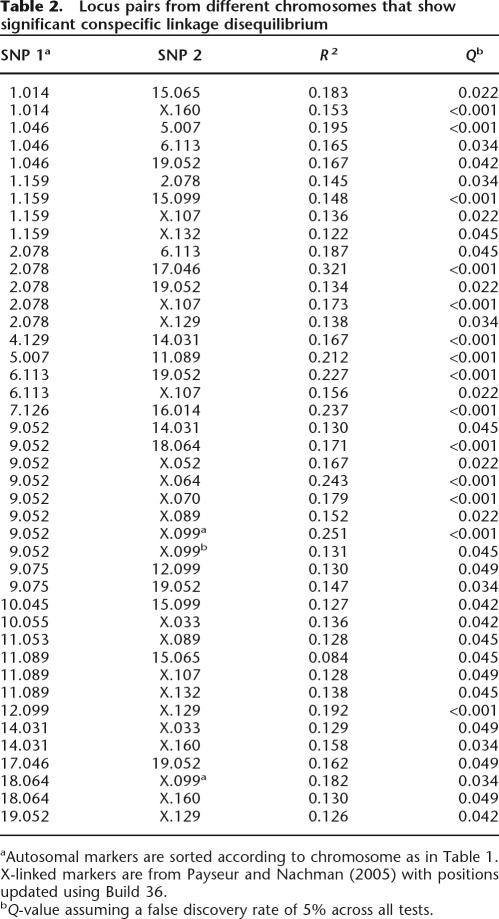
Theory predicts that reproductive isolation is most likely to be due to epistatic interactions (Dobzhansky–Muller incompatibilities), rather than to the effects of single underdominant genes (Coyne and Orr 2004). We studied nonrandom associations of alleles at different loci in one hybrid population near the middle of the hybrid zone (Neufahrn) to identify genomic regions that might be involved in such epistatic interactions. Observed values of R2 from the Neufahrn population ranged from 0 to 0.321, with a mean of 0.036. In the absence of selection and in a randomly mating population at equilibrium, the expected value of R2 is near 0. Slightly but significantly more than half (603) of the 1127 tests showed associations among conspecific alleles (P = <10−6; binomial test), indicating a bias toward conspecific, two-locus genotypes. After accounting for multiple tests, 68 SNP pairs showed significant linkage disequilibrium, and conspecific alleles were associated in 42 of these cases (Table 2). Since different chromosomes assort independently each generation, and since the average R2 in this population is near 0, these exceptions might be the result of epistatic incompatibilities. Some SNPs showed LD with several genomic regions (Table 2), suggesting the possibility of complex interactions involving multiple loci. However, the 68 locus pairs showing significant LD did not show a greater proportion of conspecific/heterospecific associations when compared with the 1059 nonsignificant locus pairs (P = 0.528; Fisher’s exact test). Additionally, there was no evidence for a bias toward X–autosome pairs (vs. autosome–autosome pairs) among significant tests (P = 0.451; Fisher’s exact test).
Table 2.
Locus pairs from different chromosomes that show significant conspecific linkage disequilibrium
aAutosomal markers are sorted according to chromosome as in Table 1. X-linked markers are from Payseur and Nachman (2005) with positions updated using Build 36.
bQ-value assuming a false discovery rate of 5% across all tests.
PANTHER analysis of genes
We classified genes into functional categories in order to search for functions that were overrepresented in genomic regions of high or low introgression. In total, 12,437 genes found within 15 Mb of each marker used in the study were classified into 240 different categories of biological processes. Of these genes, 2315 (18.6%) were not recognized by PANTHER, 6133 (49.3%) were classified into one or more biological process categories, and the remaining 3989 (32.1%) were unclassified.
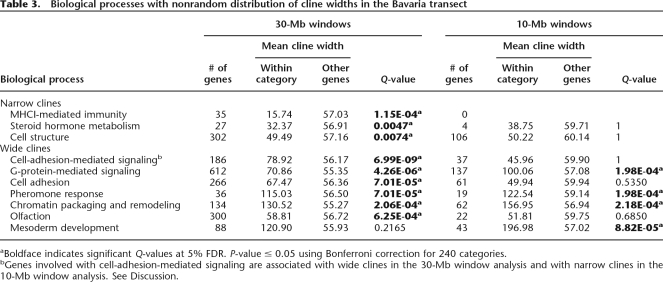
In the analysis of genes in 10-Mb windows, four biological processes were significantly associated with wide clines: G-protein-mediated signaling, pheromone response, chromatin packaging and remodeling, and mesoderm development (Table 3). In the analysis of genes in the 30-Mb windows, the first three of the biological processes listed above were associated with wide clines, as well as three additional biological processes: cell-adhesion-mediated signaling, cell adhesion, and olfaction. Cell adhesion, however, showed an opposite but nonsignificant trend in the analysis of the 10-Mb windows. There is a cluster of 60 protocadherin genes located 5–15 Mb upstream of marker 18.028, which has a cline width of 135.4 km. These genes are not in the 10-Mb windows, which accounts for the difference in the trends between the 10-Mb and 30-Mb analyses. Finally, mesoderm development showed a nonsignificant trend toward a wide cline in the 30-Mb analysis. Thus, there is good agreement between 10-Mb and 30-Mb windows in the identification of processes significantly associated with wide clines.
Table 3.
Biological processes with nonrandom distribution of cline widths in the Bavaria transect
aBoldface indicates significant Q-values at 5% FDR. P-value ≤ 0.05 using Bonferroni correction for 240 categories.
bGenes involved with cell-adhesion-mediated signaling are associated with wide clines in the 30-Mb window analysis and with narrow clines in the 10-Mb window analysis. See Discussion.
In the analysis of biological processes that were significantly associated with narrow clines, three processes were significant using 30-Mb windows: MHCI-mediated immunity, steroid hormone metabolism, and cell structure. The latter two processes showed similar but nonsignificant trends in the analysis of 10-Mb windows.
Discussion
We documented patterns of introgression across the mouse genome and documented regions of both extensive and restricted gene flow between recently diverged species of house mice. An earlier study of this same transect (Payseur et al. 2004) identified a region in the center of the X chromosome with an especially narrow cline. This same region was also identified as harboring a locus involved in hybrid male sterility between M. musculus and M. domesticus mice (Oka et al. 2004; Storchova et al. 2004), suggesting that cline shape can be used to pinpoint regions of the genome involved in reproductive isolation. Here, we find similarly narrow clines for multiple autosomal markers, and these are also likely found in genomic regions involved in reproductive isolation. In fact, one marker, 11.089, with a cline width of 10.1 km, is in close proximity to a recently identified QTL interacting with the center of the X chromosome to produce hybrid male sterility in these taxa (Oka et al. 2007). The fact that cline width is negatively associated with gene density suggests that selection, rather than random effects, determines the degree of introgression for each marker. It also suggests that the number of loci underlying reproductive isolation is rather large (Payseur and Nachman 2005).
Harr (2006) identified ten genomic regions, two on the X and eight on autosomes, of elevated differentiation between wild derived strains of M. musculus and M. domesticus and suggested they are likely to harbor genes associated with hybrid unfitness. Most of the strains used by Harr came from within the ranges of musculus and domesticus, not from near the hybrid zone. Four of our markers, 1.159, 8.078, 10.045, and 10.055, fall within the regions of elevated differentiation identified by Harr. Two of these markers, 1.159 and 10.055, have relatively narrow cline widths of 36.1 km and 24.1 km, respectively. However, the other two markers, 8.078 and 10.045, have relatively wide cline widths of 173.4 km and 131.9 km, respectively. In addition, both chromosome 10 markers, with very different patterns of introgression despite being located ∼10 Mb apart, fall within a single region of elevated differentiation that is 31 Mb in size (Harr 2006). Thus, while there is some concordance between the results presented here and those of Harr (2006), it is clear that regions showing reduced introgression in the hybrid zone are not always the same as those showing increased Fst between samples taken outside of the hybrid zone.
Reasoning that selection against heterospecific combinations of alleles that cause inviability or sterility can maintain nonrandom associations between loci in hybrid populations (Gardner et al. 2002; Payseur and Hoekstra 2005), LD among SNP pairs was measured. Several patterns emerge that provide evidence for natural selection. First, despite the fact that there is little overall LD in the middle of the hybrid zone, a number of SNP pairs show significant associations after correcting for multiple tests. Many of these significant associations show a bias against heterospecific genotypes, suggesting that selection tends to remove these interlocus combinations in the center of the hybrid zone. Second, these SNP pairs map to different chromosomes, demonstrating that linkage is not responsible for the maintenance of these associations. Third, several SNPs show strong associations with multiple, unlinked genomic regions. Because chance (and false-positive) associations are not expected to involve the same loci, this pattern also suggests the presence of functional interactions. Furthermore, these associations involving the same marker with two or more other markers suggest that selection may be acting on complex epistatic interactions (i.e., involving three or more genes), even in early stages of speciation (Coyne and Orr 2004). It is also worth noting that some markers with narrow cline widths show strong associations with each other. For example, 9.052, which has a cline width of 14.8 km, is in linkage disequilibrium with X.089, a marker with very little introgression (Payseur et al. 2004). Because patterns of LD and introgression provide independent ways of identifying incompatibility candidates, these marker pairs seem worthy of further attention.
Hybrid populations of laboratory mice have also been surveyed for linkage disequilibrium between unlinked SNPs (Payseur and Hoekstra 2005; Payseur and Place 2007). The regions surveyed in these studies are generally not the same as those surveyed in the present study. Nevertheless, of the 23 individual autosomal SNP markers involved in extreme associations between conspecific alleles in laboratory mice (Payseur and Hoekstra 2005), 10 were <10 Mb from SNP markers surveyed in our analysis. Interestingly, eight of these markers show narrow cline widths ranging in size from 6.4 to 26.3 km. The remaining two markers were furthest from the SNPs identified in Payseur and Hoekstra (2005) and had the widest cline widths, 45.8 and 62 km, respectively. Agreement between these data sets in the identification of regions potentially involved in reproductive isolation reinforces the notion that reproductive isolation between these taxa involves disrupted interactions between multiple autosomal loci, “Dobzhansky–Muller incompatibilities” (Dobzhansky 1937; Muller 1942; Coyne and Orr 2004).
The only study to explicitly map multiple partners of incompatibilities that underlie reproductive isolation phenotypes in mice is Oka et al. (2007). In this study, a region from the middle of the X chromosome of M. molossinus (a mixture of Asian M. musculus and M. castaneus genomes) caused hybrid male sterility when combined with regions on chromosomes 1, 9, and 11 from C57BL6 (an inbred strain primarily descended from M. domesticus). The central part of the X chromosome also shows significant conspecific associations with SNPs from chromosomes 1, 9, and 11 in the hybrid zone (Table 2). Although the large size of the mapped regions and low density of markers surveyed in the hybrid zone preclude strong conclusions, the agreement between these two studies suggests that some loci showing strong linkage disequilibrium are linked to incompatibilities associated with hybrid male sterility.
The PANTHER analysis of 10- and 30-Mb windows provides preliminary information on the biological processes of genes that are nonrandomly associated with regions of both reduced and extensive introgression. The 30-Mb analysis has greater statistical power than the 10-Mb analysis due to the larger number of genes, resulting in a larger number of significant associations. However, the biological meaning of these associations is less clear at the larger window size. Nevertheless, these data suggest that reproductive isolation may be driven by multiple kinds of interactions. These include ecological responses such as immune function. Interestingly, higher loads of intestinal parasites have been documented in mice from the hybrid zone, indicating possible reduced viability of hybrids (Sage et al. 1986a; Moulia et al. 1991, 1993). In addition, genes associated with physiological and behavioral aspects of reproduction involving steroid hormone metabolism appear to be associated with reduced introgression. Inspection of the expression patterns (SymAtlas v1.2.4; Su et al. 2002) for cell structure genes associated with the nine narrowest clines suggests involvement of a wide range of tissues, but with the majority being associated with sensory apparatuses such as the tongue, snout epidermis, digits, and epidermis.
Across the full set of markers, introgression is found to be strongly asymmetric. If a marker has extensive introgression across the hybrid zone, it most often occurs only from M. domesticus to M. musculus. With the exception of the Czech transect (Macholán et al. 2007), this same pattern has been documented in other studies of the Mus hybrid zone (Vanlerberghe et al. 1986; Tucker et al. 1992; Dod et al. 1993, 2005; Payseur et al. 2004; Raufaste et al. 2005). There are four possible explanations for this pattern, which are not mutually exclusive. The first possibility is that there are intrinsic genomic incompatibilities between M. domesticus alleles and M. musculus alleles, with many more M. musculus alleles being incompatible with a M. domesticus genetic background than the converse. The second possibility is that mate preference is asymmetric between the two species. The third possibility is the dispersal patterns differ between the two species, and the fourth possibility is that the hybrid zone has shifted over time.
There is evidence for asymmetric mating preferences between M. domesticus and M. musculus. A preference for conspecific urine signals has been shown in M. musculus, but no such preference is apparent in M. domesticus (Smadja and Ganem 2002; Smadja et al. 2004). There also seems to be a trend toward conspecific preferences in M. musculus for salivary androgen-binding proteins (Bimova et al. 2005). A lack of conspecific mate preference in M. domesticus could contribute to the observed asymmetric pattern of introgression, assuming that M. domesticus simply are less choosy about their mates, and more likely to mate with hybrid mice and M. musculus.
There is also behavioral evidence that M. domesticus is dominant to M. musculus in male–male competition (van Zegeren and van Oortmerssen 1981), and that the M. domesticus males tend to be more aggressive. The behavioral dominance of M. domesticus could result in a shift of the position of the hybrid zone to the east (moving from M. domesticus toward M. musculus).
Preliminary analyses based on partial annotation of the mouse genome indirectly corroborate the laboratory studies on aggression and mating behavior, as genes involved in pheromone response are nonrandomly associated with genome regions of extensive introgression. These include regions housing vomeronasal receptor genes and genes coding for mouse urinary protein (Mup). There are five Mup genes located within 5 Mb of marker 4.057, with a cline width of 140 km. They bind a variety of pheromones affecting mouse physiology and behavior including estrus, puberty, and intermale aggression (Novotny 2003). Quite possibly, M. domesticus alleles at these loci confer a fitness advantage over M. musculus alleles.
The other processes nonrandomly associated with wide clines include genes broadly associated with cell signaling and with olfaction. The latter process includes a large number (n = 300) of olfactory receptor loci that, upon closer inspection, are associated with either narrow or wide clines. These rapidly evolving loci are obvious targets of selection as they play important roles in survival and reproduction and likely reflect responses to the environment (Lane et al. 2001).
Conclusion
This study represents the most detailed genetic survey of a vertebrate hybrid zone. With markers situated on all mouse autosomes, we have used patterns of introgression to map genomic regions contributing to the maintenance of genetic isolation between recently diverged species. Patterns of LD corroborate that isolation is likely caused by epistatic interactions between sets of parental alleles. We find multiple regions of the genome with narrow clines, and these regions probably house genes involved in reproductive isolation. They include genes associated with a variety of biological processes including reproductive physiology and behavior and physiological and sensory responses to the environment. Autosomal regions with wide asymmetric clines have different effects on the fitness of hybrid mice. Interestingly, these regions also include genes involved in reproductive behavior and sensory responses to the environment. A more fully annotated genome coupled with denser sampling of the genome for patterns of introgression along with comparisons of introgression across multiple transects will provide a more complete understanding of the genetic underpinnings of reproductive isolation in this system.
Methods
Sampling
Four hundred forty-nine mice used in this study were collected by R.D. Sage from a transect through the hybrid zone in the German state of Bavaria, and western Austria. Collecting for this transect was performed by R.D. Sage in 1984, 1985, and 1992. The location of the hybrid zone and the transect are shown in Payseur et al. (2004), and information on collecting localities and numbers of mice from each locality is found in Supplemental Table 2. Sampling was performed in a roughly linear, east–west manner, and transect distances (in kilometers) were calculated from the western end of the transect.
Development and scoring of molecular markers
Single nucleotide markers for autosomes were identified using the SNP database described by Lindblad-Toh et al. (2000), http://www.broad.mit.edu/snp/mouse/, and the later SNP database from the mouse genome project, http://www.ncbi.nlm.nih.gov/projects/SNP/MouseSNP.cgi. To identify SNPs with fixed differences between M. domesticus and M. musculus, markers were sequenced in 10 allopatrically distributed mice from each species (Supplemental Table 3). The genomic location of markers was originally checked against the UCSC Mouse Genome database (build 33, May 2004, http://genome.ucsc.edu). Markers were chosen with the goal of having a relatively even marker density throughout the genome. Three markers were developed for the largest chromosomes, 1 and 2, and two markers per chromosome were developed for the remaining autosomes with the exception of chromosome 16. Markers fixed for alternate alleles in M. domesticus and M. musculus were used to design probes for TaqMan genotyping, through Applied Biosystems Assays-By-Design service. Approximately 500 bp of sequence centered on the SNP was checked for repeat regions using RepeatMasker (http://www.repeatmasker.org/). Reactions for TaqMan genotyping were set up using 5 μL of TaqMan master mix, 40 μL of the custom fluorescently labeled assay mix, ∼0.01 μg of DNA in a 3-μL volume, and 1.75 μL of sterile dH2O in 96-well plates. Genotyping plates were run on a real-time PCR machine. Thirteen X-linked markers were taken from Payseur et al. (2004) and reanalyzed here to obtain two-parameter estimates of cline shape. Genotyping data for all markers is found in Supplemental Table 4.
Cline analysis
Cline shape was estimated individually for each marker. As the goal of these analyses was to allow comparison between markers, a relatively simple two-parameter model was used to estimate the center and width of the cline for each marker, similar to the analyses in Bozikova et al. (2005). This model describes the relationship between allele frequency and geographic distance for individuals along a transect. The position of the cline center and the cline width were used in comparisons between markers. This two-parameter model consists of a hyperbolic tangent function that describes a sinusoidal curve (Equation 1):
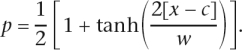 |
In this equation, p = allele frequency, x = geographic distance along a transect, c = the center of the hybrid zone, defined as the point of steepest slope, and w = 1/slope of the cline at the center. All cline analyses were performed using ClineFit software, available from Adam Porter (Porter et al. 1997) at http://www-unix.oit.umass.edu/~aporter/software/index.html. ClineFit estimates are based on data from individuals, using a Metropolis algorithm (Metropolis et al. 1953) to numerically estimate the parameters that describe the cline shape. Because the estimates are based on individuals, rather than populations, populations with small samples are weighted less than populations with large samples. We conducted some analyses excluding the populations with smallest samples sizes, and the results were unchanged. Two-unit support limits are derived from the likelihood searches for each parameter. These support limits are roughly analogous to 95% confidence intervals (Edwards 1992).
The data set presented here was also analyzed using six-parameter models (see Supplemental Methods). Spearman nonparametric rank correlation tests were used to detect correlations between the estimated cline parameters. Correlation tests were performed for two different types of comparisons: (1) comparing cline widths and centers for each marker, and (2) comparing the estimated cline widths with the local gene density. These tests were performed in SPSS 11.0 for Macintosh OS X.
Genomic attributes of marker regions
Genomic attributes were calculated for 10-Mb windows centered on each marker using Build 36 of the ENSEMBL mouse sequence. Previous work in the hybrid zone suggested that linkage disequilibrium extends beyond this distance (Payseur et al. 2004), so that windows of this size are reasonable for tracking the effects of linked sites. Gene density was measured by counting the number of known and predicted genes. The mean of dN (number of nonsynonymous substitutions per nonsynonymous site), dS (number of synonymous substitutions per synonymous site), and dN/dS for all genes in each window were calculated by comparing mouse and rat sequences (M. Dean and J. Good, unpubl.). Outliers were excluded after the examination of scatter plots between dN, dS, dN/dS, and gene position in each window. Recombination rates were estimated based on the relationship between the genetic and physical maps of the Mus musculus genome, using data from 2293 heterogeneous stock (HS) mice (http://gscan.well.ox.ac.uk/#genetic_map; Shifman et al. 2006). Genetic positions are for 9904 single nucleotide polymorphisms (SNPs) typed in 2293 HS animals. Recombination rates were calculated as the slope of a linear regression comparing the genetic positions of SNPs (in centiMorgans, cM) against their physical position (in megabase pairs, Mb) for each 10-Mb window.
Linkage disequilibrium analysis
Linkage disequilibrium was measured in 63 mice from a single locality, Neufahrn bei Freising, in the center of the transect. Linkage disequilibrium was estimated using two-locus genotype counts (Weir et al. 2004). Specifically, genotypes at each locus were recoded as 0, 1, or 2 for autosomal loci and X-linked loci in females, and 0 or 2 for X-linked loci in males. Linkage disequilibrium was estimated as the squared Pearson’s correlation coefficient (genotypic R2) between these recoded genotypes. The probability of each observed R2 value was estimated by comparison to 1000 R2 values obtained by randomizing genotypes across individuals. We adjusted statistical significance to achieve a 5% false discovery rate (FDR) (Storey and Tibshirani 2003) across all tests.
Two markers, 17.091 (which was fixed for the M. domesticus allele in this locality) and 3.007 (which harbored a single M. domesticus allele), were not included in these analyses. To ensure independent assortment among loci in each generation, linkage disequilibrium was measured for pairs of loci residing on different chromosomes. Because the X chromosome is involved in reproductive isolation between house mouse species (Oka et al. 2004; Storchova et al. 2004), we included 13 X-linked markers (Payseur et al. 2004) in our survey of linkage disequilibrium.
PANTHER analysis of genes
Using the PANTHER database (Thomas et al. 2003; Mi et al. 2005), genes found within 30 Mb of each marker used in the study were classified into different categories of biological processes. As biological process groups are not mutually exclusive, some genes belong to more than one group. In addition, some biological process groups are subsets of others (e.g., “Pheromone response” is a subset of “Sensory perception”).
For each of the biological processes identified, genes with PANTHER classifications were divided into two groups: (1) those associated with the biological process in question and (2) all others. Genes unclassified or unrecognized by PANTHER were excluded from this analysis, but including these genes does not significantly change the results (not shown). Genes within 10-Mb and 30-Mb windows were ranked according to the cline widths for the Bavaria transect. Genes associated with more than one marker were included once for each associated marker. A nonrandom distribution of cline widths for genes in a particular biological process was identified by comparing groups 1 and 2. If genes for a particular biological process tend to be associated with narrow (or wide) clines, this tendency can be identified by a significant P-value from a two-tailed Mann-Whitney U-test. Because multiple biological processes were tested, it is necessary to correct for multiple testing. Q-values were calculated (Storey and Tibshirani 2003), and those values significant at a 5% false discovery rate (FDR) are reported. A Bonferroni correction for multiple tests was also used. This correction is especially conservative because some biological processes are subsets of others.
Acknowledgments
Richard Sage collected all of the animals from the Bavaria transect and generously provided DNA samples and tissues to both the Tucker and Nachman laboratories. Michael Hammer provided some DNAs from M. musculus. Matt Dean, Milos Macholán, and Jaroslav Piálek provided helpful comments on an earlier draft. Research was supported by NSF DEB0212667 to P.K.T., NSF DEB0213013 to M.W.N., and MEC/Fulbright grant from Secretaría de Estado de Universidades e Investigación from the Spanish Ministerio de Educación y Ciencia to M.A.S.-F.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6757907
References
- Alibert P., Fel-Clair F., Manolakou K., Britton-Davidian J., Auffray J.C., Fel-Clair F., Manolakou K., Britton-Davidian J., Auffray J.C., Manolakou K., Britton-Davidian J., Auffray J.C., Britton-Davidian J., Auffray J.C., Auffray J.C. Developmental stability, fitness, and trait size in laboratory hybrids between European subspecies of the house mouse. Evolution Int. J. Org. Evolution. 1997;51:1284–1295. doi: 10.1111/j.1558-5646.1997.tb03975.x. [DOI] [PubMed] [Google Scholar]
- Arnold S.J., Buckner C.M., Robinson J.J., Buckner C.M., Robinson J.J., Robinson J.J. Pollen-mediated introgression and hybrid speciation in Louisiana irises. Proc. Natl. Acad. Sci. 1991;88:1398–1402. doi: 10.1073/pnas.88.4.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bimova B., Karn R.C., Pialek J., Karn R.C., Pialek J., Pialek J. The role of salivary androgen-binding protein in reproductive isolation between two species of house mouse: Mus musculus musculus and Mus musculus domesticus. Biol. J. Linn. Soc. Lond. 2005;84:349–361. [Google Scholar]
- Boursot P., Auffray J.C., Britton-Davidian J., Bonhomme F., Auffray J.C., Britton-Davidian J., Bonhomme F., Britton-Davidian J., Bonhomme F., Bonhomme F. The evolution of house mice. Annu. Rev. Ecol. Syst. 1993;24:119–152. [Google Scholar]
- Boursot P., Din W., Anand R., Darviche D., Dod B., Von Deimling F., Talwar G.P., Bonhomme F., Din W., Anand R., Darviche D., Dod B., Von Deimling F., Talwar G.P., Bonhomme F., Anand R., Darviche D., Dod B., Von Deimling F., Talwar G.P., Bonhomme F., Darviche D., Dod B., Von Deimling F., Talwar G.P., Bonhomme F., Dod B., Von Deimling F., Talwar G.P., Bonhomme F., Von Deimling F., Talwar G.P., Bonhomme F., Talwar G.P., Bonhomme F., Bonhomme F. Origin and radiation of the house mouse: Mitochondrial DNA phylogeny. J. Evol. Biol. 1996;9:391–415. [Google Scholar]
- Bozikova E., Munclinger P., Teeter K.C., Tucker P.K., Macholán M., Piálek J., Munclinger P., Teeter K.C., Tucker P.K., Macholán M., Piálek J., Teeter K.C., Tucker P.K., Macholán M., Piálek J., Tucker P.K., Macholán M., Piálek J., Macholán M., Piálek J., Piálek J. Mitochondrial DNA in the hybrid zone between Mus musculus musculus and Mus musculus domesticus: A comparison of two transects. Biol. J. Linn. Soc. Lond. 2005;84:363–378. [Google Scholar]
- Britton-Davidian J., Fel-Clair F., Lopez J., Alibert P., Boursot P., Fel-Clair F., Lopez J., Alibert P., Boursot P., Lopez J., Alibert P., Boursot P., Alibert P., Boursot P., Boursot P. Postzygotic isolation between the two European subspecies of the house mouse: Estimates from fertility patterns in wild and laboratory-bred hybrids. Biol. J. Linn. Soc. Lond. 2005;84:379–393. [Google Scholar]
- Buerkle C.A., Rieseberg L.H., Rieseberg L.H. Low intraspecific variation for genomic isolation between hybridizing sunflower species. Evolution Int. J. Org. Evolution. 2001;55:684–691. doi: 10.1554/0014-3820(2001)055[0684:livfgi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Coyne J.A., Orr H.A., Orr H.A. Speciation. Sinauer Associates, Inc.; Sunderland, MA: 2004. p. 545. [Google Scholar]
- Cucchi T., Vigne J.D., Auffray J.C., Vigne J.D., Auffray J.C., Auffray J.C. First occurrence of the house mouse (Mus musculus domesticus Schwarz and Schwarz, 1943) in the Western Mediterranean: A zooarcheological revision of subfossil occurrences. Biol. J. Linn. Soc. Lond. 2005;84:429–445. [Google Scholar]
- Dobzhansky T. Genetics and the origin of species. Columbia University Press; New York: 1937. p. 364. [Google Scholar]
- Dod B., Jermiin L.S., Boursot P., Chapman V.H., Nielsen J.T., Bonhomme F., Jermiin L.S., Boursot P., Chapman V.H., Nielsen J.T., Bonhomme F., Boursot P., Chapman V.H., Nielsen J.T., Bonhomme F., Chapman V.H., Nielsen J.T., Bonhomme F., Nielsen J.T., Bonhomme F., Bonhomme F. Counterselection on sex-chromosomes in the Mus musculus European hybrid zone. J. Evol. Biol. 1993;6:529–546. [Google Scholar]
- Dod B., Smadja C., Karn R.C., Boursot P., Smadja C., Karn R.C., Boursot P., Karn R.C., Boursot P., Boursot P. Testing for selection on the androgen-binding protein in the Danish mouse hybrid zone. Biol. J. Linn. Soc. Lond. 2005;84:447–459. [Google Scholar]
- Edwards A.W.F. Likelihood. Johns Hopkins University Press; Baltimore, MD: 1992. p. 275. [Google Scholar]
- Forejt J. Hybrid sterility in the mouse. Trends Genet. 1996;12:412–417. doi: 10.1016/0168-9525(96)10040-8. [DOI] [PubMed] [Google Scholar]
- Forejt J., Ivanyi P., Ivanyi P. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.) Genet. Res. 1975;24:189–206. doi: 10.1017/s0016672300015214. [DOI] [PubMed] [Google Scholar]
- Gardner K., Buerkle A., Whitton J., Rieseberg L.H., Buerkle A., Whitton J., Rieseberg L.H., Whitton J., Rieseberg L.H., Rieseberg L.H. Inferring epistasis in wild sunflower hybrid zones. In: Wolf J.B., et al., editors. Epistasis and the evolutionary process. Oxford University Press; New York: 2002. pp. 264–279. [Google Scholar]
- Hagen R.H., Scriber J.M., Scriber J.M. Sex-linked diapause, color, and allozyme loci in Papilio glaucus: Linkage analysis and significance in a hybrid zone. J. Hered. 1989;80:179–185. [Google Scholar]
- Haldane J.B.S. Sex ratio and unisexual sterility in animal hybrids. J. Genet. 1922;12:101–109. [Google Scholar]
- Harr B. Genomic islands of differentiation between house mouse subspecies. Genome Res. 2006;16:730–737. doi: 10.1101/gr.5045006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R.G. Hybrid zones: Windows on evolutionary process. Oxf. Surv. Evol. Biol. 1990;7:69–128. [Google Scholar]
- Hunt W.G., Selander R.K., Selander R.K. Biochemical genetics of hybridisation in European house mice. Heredity. 1973;31:11–33. doi: 10.1038/hdy.1973.56. [DOI] [PubMed] [Google Scholar]
- Lane R.P., Cutforth T., Young J., Athanasiou M., Friedman C., Rowen L., Evans G., Axel R., Hood L., Trask B.J., Cutforth T., Young J., Athanasiou M., Friedman C., Rowen L., Evans G., Axel R., Hood L., Trask B.J., Young J., Athanasiou M., Friedman C., Rowen L., Evans G., Axel R., Hood L., Trask B.J., Athanasiou M., Friedman C., Rowen L., Evans G., Axel R., Hood L., Trask B.J., Friedman C., Rowen L., Evans G., Axel R., Hood L., Trask B.J., Rowen L., Evans G., Axel R., Hood L., Trask B.J., Evans G., Axel R., Hood L., Trask B.J., Axel R., Hood L., Trask B.J., Hood L., Trask B.J., Trask B.J. Genomic analysis of orthologous mouse and human olfactory receptor loci. Proc. Natl. Acad. Sci. 2001;98:7390–7395. doi: 10.1073/pnas.131215398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laukaitis C.M., Critser E.S., Karn R.C., Critser E.S., Karn R.C., Karn R.C. Salivary androgen-binding protein (ABP) mediates sexual isolation in Mus musculus. Evolution Int. J. Org. Evolution. 1997;51:2000–2005. doi: 10.1111/j.1558-5646.1997.tb05121.x. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Winchester E., Daly M.J., Wang D.G., Hirschhorn J.N., Laviolette J.P., Ardlie K., Reich D.E., Robinson E., Sklar P., Winchester E., Daly M.J., Wang D.G., Hirschhorn J.N., Laviolette J.P., Ardlie K., Reich D.E., Robinson E., Sklar P., Daly M.J., Wang D.G., Hirschhorn J.N., Laviolette J.P., Ardlie K., Reich D.E., Robinson E., Sklar P., Wang D.G., Hirschhorn J.N., Laviolette J.P., Ardlie K., Reich D.E., Robinson E., Sklar P., Hirschhorn J.N., Laviolette J.P., Ardlie K., Reich D.E., Robinson E., Sklar P., Laviolette J.P., Ardlie K., Reich D.E., Robinson E., Sklar P., Ardlie K., Reich D.E., Robinson E., Sklar P., Reich D.E., Robinson E., Sklar P., Robinson E., Sklar P., Sklar P., et al. Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat. Genet. 2000;24:381–386. doi: 10.1038/74215. [DOI] [PubMed] [Google Scholar]
- Macholán M., Krytufek E., Vohralík V., Krytufek E., Vohralík V., Vohralík V. The location of the Mus musculus / M. domesticus hybrid zone in the Balkans: Clues from morphology. Acta Theriol. (Warsz.) 2003;48:177–188. [Google Scholar]
- Macholán M., Munclinger P., Sugerkova M., Dufkova P., Bimova B., Zima J., Piálek J., Munclinger P., Sugerkova M., Dufkova P., Bimova B., Zima J., Piálek J., Sugerkova M., Dufkova P., Bimova B., Zima J., Piálek J., Dufkova P., Bimova B., Zima J., Piálek J., Bimova B., Zima J., Piálek J., Zima J., Piálek J., Piálek J. Genetic analysis of autosomal and X-linked markers across a mouse hybrid zone. Evolution Int. J. Org. Evolution. 2007;61:746–771. doi: 10.1111/j.1558-5646.2007.00065.x. [DOI] [PubMed] [Google Scholar]
- Metropolis N., Rosenbluth A., Rosenbluth M., Teller A., Teller E., Rosenbluth A., Rosenbluth M., Teller A., Teller E., Rosenbluth M., Teller A., Teller E., Teller A., Teller E., Teller E. Equation of state calculations by fast computing machines. J. Chem. Phys. 1953;21:1087–1092. [Google Scholar]
- Mi H., Lazareva-Ulitsky B., Loo R., Kejariwal A., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., Lazareva-Ulitsky B., Loo R., Kejariwal A., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., Loo R., Kejariwal A., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., Kejariwal A., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., Guo N., Muruganujan A., Doremieux O., Campbell M.J., Muruganujan A., Doremieux O., Campbell M.J., Doremieux O., Campbell M.J., Campbell M.J., et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;31:334–341. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulia C.J., Aussel P., Bonhomme F., Boursot P., Nielsen J.T., Renaud F., Aussel P., Bonhomme F., Boursot P., Nielsen J.T., Renaud F., Bonhomme F., Boursot P., Nielsen J.T., Renaud F., Boursot P., Nielsen J.T., Renaud F., Nielsen J.T., Renaud F., Renaud F. Wormy mice in a hybrid zone: A genetic control of susceptibility to parasite infection. J. Evol. Biol. 1991;4:679–687. [Google Scholar]
- Moulia C.J., LeBrun N., Dallas J., Orth A., Renaud F., LeBrun N., Dallas J., Orth A., Renaud F., Dallas J., Orth A., Renaud F., Orth A., Renaud F., Renaud F. Experimental evidence of genetic determinism in high susceptibility to intestinal pinworm infection in mice: A hybrid zone model. Parasitology. 1993;106:387–393. doi: 10.1017/s0031182000067135. [DOI] [PubMed] [Google Scholar]
- Muller H.J. Bearing of the Drosophila work on systematics. In: Huxley J.S., editor. The new systematics. Clarendon; Oxford, UK: 1940. pp. 185–268. [Google Scholar]
- Muller H.J. Isolating mechanisms, evolution, and temperature. Biol. Symp. 1942;6:71–125. [Google Scholar]
- Munclinger P., Bozíková E., Sugerková M., Piálek J., Macholán M., Bozíková E., Sugerková M., Piálek J., Macholán M., Sugerková M., Piálek J., Macholán M., Piálek J., Macholán M., Macholán M. Genetic variation in house mice (Mus, Muridae, Rodentia) from the Czech and Slovak Republics. Folia Zool. (Brno) 2002;51:81–92. [Google Scholar]
- Nance V., Vanlerberghe F., Nielsen J.T., Bonhomme F., Britton-Davidian J., Vanlerberghe F., Nielsen J.T., Bonhomme F., Britton-Davidian J., Nielsen J.T., Bonhomme F., Britton-Davidian J., Bonhomme F., Britton-Davidian J., Britton-Davidian J. Chromosomal Introgression in house mice from the hybrid zone between M. m. domesticus and M. m. musculus in Denmark. Biol. J. Linn. Soc. Lond. 1990;41:215–227. [Google Scholar]
- Novotny M.V. Phermones, binding proteins and receptor responses in rodents. Biochem. Soc. Trans. 2003;31:117–122. doi: 10.1042/bst0310117. [DOI] [PubMed] [Google Scholar]
- Oka A., Mita A., Sakurai-Yamatani N., Yamamoto H., Takagi N., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Mita A., Sakurai-Yamatani N., Yamamoto H., Takagi N., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Sakurai-Yamatani N., Yamamoto H., Takagi N., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Yamamoto H., Takagi N., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Takagi N., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Takano-Shimizu T., Toshimori K., Moriwaki K., Shiroishi T., Toshimori K., Moriwaki K., Shiroishi T., Moriwaki K., Shiroishi T., Shiroishi T. Hybrid breakdown caused by substitution of the X chromosome between two mouse subspecies. Genetics. 2004;166:913–924. doi: 10.1534/genetics.166.2.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Aoto T., Totsuka Y., Takahashi R., Ueda M., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Aoto T., Totsuka Y., Takahashi R., Ueda M., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Totsuka Y., Takahashi R., Ueda M., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Takahashi R., Ueda M., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Ueda M., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Mita A., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Sakurai-Yamatani N., Yamamoto H., Kuriki S., Takagi N., Yamamoto H., Kuriki S., Takagi N., Kuriki S., Takagi N., Takagi N., et al. Disruption of genetic interaction between two autosomal regions and the X chromosome causes reproductive isolation between mouse strains derived from different subspecies. Genetics. 2007;175:185–197. doi: 10.1534/genetics.106.062976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H.A. Haldane’s rule. Annu. Rev. Ecol. Syst. 1997;28:195–218. [Google Scholar]
- Payseur B.A., Hoekstra H.E., Hoekstra H.E. Signatures of reproductive isolation in patterns of single nucleotide diversity across inbred strains of mice. Genetics. 2005;171:1905–1916. doi: 10.1534/genetics.105.046193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur B.A., Nachman M.W., Nachman M.W. The genomics of speciation: Investigating the molecular correlates of X chromosome introgression across the hybrid zone between Mus domesticus and Mus musculus. Biol. J. Linn. Soc. Lond. 2005;84:523–534. [Google Scholar]
- Payseur B.A., Place M., Place M. Searching the genomes of inbred mouse strains for incompatibilities that reproductively isolate their wild relatives. J. Hered. 2007;98:115–122. doi: 10.1093/jhered/esl064. [DOI] [PubMed] [Google Scholar]
- Payseur B.A., Krenz J.G., Nachman M.W., Krenz J.G., Nachman M.W., Nachman M.W. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution Int. J. Org. Evolution. 2004;58:2064–2078. doi: 10.1111/j.0014-3820.2004.tb00490.x. [DOI] [PubMed] [Google Scholar]
- Porter A.H., Wenger R., Geiger H., Scholl A., Shapiro A.M., Wenger R., Geiger H., Scholl A., Shapiro A.M., Geiger H., Scholl A., Shapiro A.M., Scholl A., Shapiro A.M., Shapiro A.M. The Pontia daplidice-edusa hybrid zone in Northwestern Italy. Evolution Int. J. Org. Evolution. 1997;51:1561–1573. doi: 10.1111/j.1558-5646.1997.tb01479.x. [DOI] [PubMed] [Google Scholar]
- Prager E.M., Sage R.D., Gyllensten U., Thomas W.K., Hubner R., Jones C.S., Noble L., Searle J.B., Wilson A.C., Sage R.D., Gyllensten U., Thomas W.K., Hubner R., Jones C.S., Noble L., Searle J.B., Wilson A.C., Gyllensten U., Thomas W.K., Hubner R., Jones C.S., Noble L., Searle J.B., Wilson A.C., Thomas W.K., Hubner R., Jones C.S., Noble L., Searle J.B., Wilson A.C., Hubner R., Jones C.S., Noble L., Searle J.B., Wilson A.C., Jones C.S., Noble L., Searle J.B., Wilson A.C., Noble L., Searle J.B., Wilson A.C., Searle J.B., Wilson A.C., Wilson A.C. Mitochondrial DNA sequence diversity and the colonization of Scandinavia by house mice from East Holstein. Bio. J. Linn. Soc. Lond. 1993;50:85–122. [Google Scholar]
- Prager E.M., Orrego C., Sage R.D., Orrego C., Sage R.D., Sage R.D. Genetic variation and phylogeography of central Asian and other house mice, including a major new mitochondrial lineage in Yemen. Genetics. 1998;150:835–861. doi: 10.1093/genetics/150.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D.C., Orr H.A., Orr H.A. Haldane’s rule in taxa lacking a hemizygous X. Science. 1998;282:952–954. doi: 10.1126/science.282.5390.952. [DOI] [PubMed] [Google Scholar]
- Raufaste N., Orth A., Belkhir K., Senet D., Smadja C., Baird S.J.E., Bonhomme F., Dod B., Boursot P., Orth A., Belkhir K., Senet D., Smadja C., Baird S.J.E., Bonhomme F., Dod B., Boursot P., Belkhir K., Senet D., Smadja C., Baird S.J.E., Bonhomme F., Dod B., Boursot P., Senet D., Smadja C., Baird S.J.E., Bonhomme F., Dod B., Boursot P., Smadja C., Baird S.J.E., Bonhomme F., Dod B., Boursot P., Baird S.J.E., Bonhomme F., Dod B., Boursot P., Bonhomme F., Dod B., Boursot P., Dod B., Boursot P., Boursot P. Inferences of selection and migration in the Danish house mouse hybrid zone. Biol. J. Linn. Soc. Lond. 2005;84:593–616. [Google Scholar]
- Rieseberg L.H. Homoploid reticulate evolution in Helianthus (Asteraceae): Evidence from ribosomal genes. Am. J. Bot. 1991;78:1218– 1237. [Google Scholar]
- Rieseberg L.H. Hybrid origin of plant species. Annu. Rev. Ecol. Syst. 1997;28:359–389. [Google Scholar]
- Rieseberg L.H., Whitton J., Gardner K., Whitton J., Gardner K., Gardner K. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics. 1999;152:713–727. doi: 10.1093/genetics/152.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saetre G.P., Borge T., Lindroos K., Haavbie J., Sheldon B.C., Primmer C., Syvänen A.C., Borge T., Lindroos K., Haavbie J., Sheldon B.C., Primmer C., Syvänen A.C., Lindroos K., Haavbie J., Sheldon B.C., Primmer C., Syvänen A.C., Haavbie J., Sheldon B.C., Primmer C., Syvänen A.C., Sheldon B.C., Primmer C., Syvänen A.C., Primmer C., Syvänen A.C., Syvänen A.C. Sex chromosome evolution and speciation in flycatchers. Proc. R. Soc. Lond. B. Biol. Sci. 2003;270:53–59. doi: 10.1098/rspb.2002.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage R.D., Heyneman D., Lim K.C., Wilson A.C., Heyneman D., Lim K.C., Wilson A.C., Lim K.C., Wilson A.C., Wilson A.C. Wormy mice in a hybrid zone. Nature. 1986a;324:60–63. doi: 10.1038/324060a0. [DOI] [PubMed] [Google Scholar]
- Sage R.D., Whitney J.B., Wilson A.C., Whitney J.B., Wilson A.C., Wilson A.C. Genetic analysis of a hybrid zone between domesticus and musculus mice (Mus musculus complex) hemoglobin polymorphisms. Curr. Top. Microbiol. Immunol. 1986b;127:75–85. doi: 10.1007/978-3-642-71304-0_9. [DOI] [PubMed] [Google Scholar]
- Schnell G.D., Selander R.K., Selander R.K. Environmental and morphological correlates of genetic variation in mammals. In: Joule J., Smith M.H., Smith M.H., editors. Mammalian population genetics. University of Georgia Press; Athens, GA: 1981. pp. 60–69. [Google Scholar]
- She J.X., Bonhomme F., Boursot P., Thaler L., Catzeflis F., Bonhomme F., Boursot P., Thaler L., Catzeflis F., Boursot P., Thaler L., Catzeflis F., Thaler L., Catzeflis F., Catzeflis F. Molecular phylogenies in the genus Mus: Comparative analysis of electrophoretic, scnDNA hybridization, and mtDNA RFLP data. Biol. J. Linn. Soc. Lond. 1990;41:83–103. [Google Scholar]
- Shifman S., Bell J.T., Copley R.R., Taylor M.S., Williams R.W., Mott R., Flint J., Bell J.T., Copley R.R., Taylor M.S., Williams R.W., Mott R., Flint J., Copley R.R., Taylor M.S., Williams R.W., Mott R., Flint J., Taylor M.S., Williams R.W., Mott R., Flint J., Williams R.W., Mott R., Flint J., Mott R., Flint J., Flint J. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 2006;4:e395. doi: 10.1371/journal.pbio.0040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smadja C., Ganem G., Ganem G. Subspecies recognition in the house mouse: A study of two populations from the border of a hybrid zone. Behav. Ecol. 2002;13:312–320. [Google Scholar]
- Smadja C., Ganem G., Ganem G. Asymmetrical reproductive character displacement in the house mouse. J. Evol. Biol. 2005;18:1485–1493. doi: 10.1111/j.1420-9101.2005.00944.x. [DOI] [PubMed] [Google Scholar]
- Smadja C., Catalan J., Ganem G., Catalan J., Ganem G., Ganem G. Strong premating divergence in a unimodal hybrid zone between two subspecies of the house mouse. J. Evol. Biol. 2004;17:165–176. doi: 10.1046/j.1420-9101.2003.00647.x. [DOI] [PubMed] [Google Scholar]
- Storchova R., Gregorova S., Buckiova D., Kyselova V., Divina P., Forejt J., Gregorova S., Buckiova D., Kyselova V., Divina P., Forejt J., Buckiova D., Kyselova V., Divina P., Forejt J., Kyselova V., Divina P., Forejt J., Divina P., Forejt J., Forejt J. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm. Genome. 2004;15:515–524. doi: 10.1007/s00335-004-2386-0. [DOI] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R., Tibshirani R. Statistical significance for genome-wide studies. Proc. Natl. Acad. Sci. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A.I., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinosa L.M., Moqrich A., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinosa L.M., Moqrich A., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinosa L.M., Moqrich A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinosa L.M., Moqrich A., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinosa L.M., Moqrich A., Wiltshire T., Orth A.P., Vega R.G., Sapinosa L.M., Moqrich A., Orth A.P., Vega R.G., Sapinosa L.M., Moqrich A., Vega R.G., Sapinosa L.M., Moqrich A., Sapinosa L.M., Moqrich A., Moqrich A., et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Shimada T., Terashima M., Tsuchiya K., Aplin K., Shimada T., Terashima M., Tsuchiya K., Aplin K., Terashima M., Tsuchiya K., Aplin K., Tsuchiya K., Aplin K., Aplin K. Temporal, spatial and ecological modes of evolution of Eurasian Mus based on mitochondrial and nuclear gene sequences. Mol. Phylogenet. Evol. 2004;33:626–646. doi: 10.1016/j.ympev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Kariak B., Daverman R., Diemer K., Muruganujan A., Narechania A., Campbell M.J., Kejariwal A., Mi H., Kariak B., Daverman R., Diemer K., Muruganujan A., Narechania A., Kejariwal A., Mi H., Kariak B., Daverman R., Diemer K., Muruganujan A., Narechania A., Mi H., Kariak B., Daverman R., Diemer K., Muruganujan A., Narechania A., Kariak B., Daverman R., Diemer K., Muruganujan A., Narechania A., Daverman R., Diemer K., Muruganujan A., Narechania A., Diemer K., Muruganujan A., Narechania A., Muruganujan A., Narechania A., Narechania A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtulec Z., Mihola O., Vlcek C., Himmelbauer H., Paces V., Forejt J., Mihola O., Vlcek C., Himmelbauer H., Paces V., Forejt J., Vlcek C., Himmelbauer H., Paces V., Forejt J., Himmelbauer H., Paces V., Forejt J., Paces V., Forejt J., Forejt J. Positional cloning of the Hybrid sterility 1 gene: Fine genetic mapping and evaluation of two candidate genes. Biol. J. Linn. Soc. Lond. 2005;84:637–641. [Google Scholar]
- Tucker P.K., Sage R.D., Warner J.H., Wilson A.C., Eicher E.M., Sage R.D., Warner J.H., Wilson A.C., Eicher E.M., Warner J.H., Wilson A.C., Eicher E.M., Wilson A.C., Eicher E.M., Eicher E.M. Abrupt cline for sex chromosomes in a hybrid zone between two species of mice. Evolution Int. J. Org. Evolution. 1992;46:1146–1163. doi: 10.1111/j.1558-5646.1992.tb00625.x. [DOI] [PubMed] [Google Scholar]
- Turelli M., Begun D.J., Begun D.J. Haldane’s rule and X-chromosome size in Drosophila. Genetics. 1997;147:1799–1815. doi: 10.1093/genetics/147.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M., Orr H.A., Orr H.A. The dominance theory of Haldane’s rule. Genetics. 1995;140:389–402. doi: 10.1093/genetics/140.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursin E. Occurrence of voles, mice, and rats (Muridae) in Denmark, with a special note on a zone of intergradation between two subspecies of the house mouse (Mus musculus L.) Vid. Medd. Dansk Naturhist. Foren. 1952;114:217–244. [Google Scholar]
- van Zegeren K., van Oortmerssen G.A., van Oortmerssen G.A. Frontier disputes between the West- and East-European house mouse in Schleswig-Holstein, West Germany. Z. Saugetierkd. 1981;46:363–369. [Google Scholar]
- Vanlerberghe F., Dod B., Boursot P., Bellis M., Bonhomme F., Dod B., Boursot P., Bellis M., Bonhomme F., Boursot P., Bellis M., Bonhomme F., Bellis M., Bonhomme F., Bonhomme F. Absence of Y-chromosome introgression across the hybrid zone between Mus musculus domesticus and Mus musculus musculus. Genet. Res. 1986;48:191–197. doi: 10.1017/s0016672300025003. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe F., Boursot P., Catalan J., Boursot P., Catalan J., Catalan J. Analyse genetique de la zone d’hybridation entre les deux sous-especes de souris Mus musculus domesticus et Mus musculus musculus en Bulgaire. Genome. 1988a;30:427–437. [PubMed] [Google Scholar]
- Vanlerberghe F., Boursot P., Nielsen J.T., Bonhomme F., Boursot P., Nielsen J.T., Bonhomme F., Nielsen J.T., Bonhomme F., Bonhomme F.1988b. . A steep cline for mitochondrial DNA in Danish mice Genet. Res. 52185–193. [DOI] [PubMed] [Google Scholar]
- Vyskocilova M., Trachtulec Z., Forejt J., Piálek J., Trachtulec Z., Forejt J., Piálek J., Forejt J., Piálek J., Piálek J. Does geography matter in hybrid sterility in mice? Biol. J. Linn. Soc. Lond. 2005;84:663–674. [Google Scholar]
- Weir B.S., Hill W.G., Cardon L.R., Hill W.G., Cardon L.R., Cardon L.R. Allelic association patterns for a dense SNP map. Genet. Epidemiol. 2004;27:442–450. doi: 10.1002/gepi.20038. [DOI] [PubMed] [Google Scholar]