Abstract
We have developed a portable and easily configurable genome annotation pipeline called MAKER. Its purpose is to allow investigators to independently annotate eukaryotic genomes and create genome databases. MAKER identifies repeats, aligns ESTs and proteins to a genome, produces ab initio gene predictions, and automatically synthesizes these data into gene annotations having evidence-based quality indices. MAKER is also easily trainable: Outputs of preliminary runs are used to automatically retrain its gene-prediction algorithm, producing higher-quality gene-models on subsequent runs. MAKER’s inputs are minimal, and its outputs can be used to create a GMOD database. Its outputs can also be viewed in the Apollo Genome browser; this feature of MAKER provides an easy means to annotate, view, and edit individual contigs and BACs without the overhead of a database. As proof of principle, we have used MAKER to annotate the genome of the planarian Schmidtea mediterranea and to create a new genome database, SmedGD. We have also compared MAKER’s performance to other published annotation pipelines. Our results demonstrate that MAKER provides a simple and effective means to convert a genome sequence into a community-accessible genome database. MAKER should prove especially useful for emerging model organism genome projects for which extensive bioinformatics resources may not be readily available.
Genome annotation, not genome sequencing, is becoming the bottleneck in genomics today. New genomes are being sequenced at a far faster rate than they are being annotated. As of 2007, there are 126 completely sequenced, but unpublished genomes, and the backlog of unpublished and unannotated genomes continues to grow (Liolios et al. 2006). Eukaryotic genomes are particularly at risk as their large size and intron-containing genes make them difficult substrates for annotation. There are currently more than ∼800 Eukaryotic genome projects under way (Liolios et al. 2006). Many of them belong to emerging model organisms (http://grants.nih.gov/grants/guide/pa-files/PA-04-135.html), and are represented by relatively small research communities. Annotating these genomes and distributing the results for the benefit of the larger biomedical community is proving difficult for many of these communities, as they often lack bioinformatics experience. One solution to this problem is to outsource the annotation to one of the major annotation databases such as Ensembl (Stabenau et al. 2004) or VectorBase (Lawson et al. 2007). This has proven a fruitful strategy for several groups (c.f. VectorBase), but the numbers of sequenced genomes far exceeds the capacity and the stated purview of these projects; Ensembl, e.g., is restricted to vertebrate genomes and VectorBase to insect vectors of human disease.
In an attempt to ameliorate this problem, many sequencing centers, data repositories, and model organism databases make their annotation software available to the public (http://www.broad.mit.edu/tools/software.html; http://www.tigr.org/software/genefinding.shtml) (Stabenau et al. 2004). However this is not their primary mission, and they usually only make subsets of their internal systems available—and these generally require significant in-house bioinformatics support (Lawson et al. 2007). Thus, despite the best efforts of the bioinformatics community, large numbers of unannotated genomes continue to accumulate, underscoring an urgent need for simpler, more portable annotation pipelines.
Developing an easy-to-use annotation pipeline imposes several design constraints. First, it must be easy to configure and run, requiring minimal bioinformatics and computer resources. In other words, external executables and software need to be minimal, and installation must be routine, even for users with only rudimentary UNIX skills. Second, an easy-to-use pipeline must also provide both a compute and an annotation engine. In practical terms, it must be able to identify repeats, to align ESTs and proteins to the genome, and to automatically synthesize these data into feature-rich gene annotations, including alternative splicing and UTRs, as well as attributes such as evidence trails, and confidence measures. Third, every genome is different and an easy-to-use annotation pipeline must be, therefore, easily configurable and trainable. If not, the evidence gathered by the compute pipeline will be of poor quality, and the annotation process will be compromised.
Another essential feature of an easy-to-use annotation pipeline is that its output formats must be both comprehensive and database ready. This task has been simplified by the Generic Model Organism Database (GMOD) project (http://www.gmod.org), which provides a generic genome database schema and genome visualization tools. GMOD, however, does not provide a means to produce the contents of a database; these must be created by an external annotation pipeline. Therefore, to take advantage of GMOD tools, annotation pipelines must write their outputs in GMOD-compatible Generic Feature Format (GFF3; www.sequenceontology.org/gff3.shtml). However, creating GFF3 files containing all of the information necessary to populate a GMOD database is a complex task. These files must contain descriptions of EST and protein alignments, repeats, and gene predictions. They must also include EST and protein alignments not associated with any annotation, so that false negatives can be identified. Without such data, downstream automated and manual annotation management is seriously compromised.
Finally, to qualify as truly user-friendly, an annotation pipeline should provide an easy means to annotate, view, and edit individual contigs and BACs. This allows users to analyze partial genome assemblies and to independently annotate regions of interest using their own data sets, ideally without the overhead of a database and with only minimal compute resources such as a laptop computer.
We have designed an easy-to-use annotation tool called MAKER in an attempt to meet all of these design criteria. Our goal was to provide emerging genome projects with the means to independently annotate protein-coding genes and to create a GMOD database. MAKER identifies repeats, aligns ESTs and proteins to a genome, makes gene predictions, and integrates these data into protein-coding gene annotations. Moreover, its outputs can be loaded directly into GMOD browsers and databases with no post-processing. As proof of principle, we have used MAKER to annotate the genome of the planarian Schmidtea mediterranea and to create a new genome database, SmedGD (http://smedgd.neuro.utah.edu). We have also compared MAKER’s performance to other published annotation pipelines as part of the nGASP contest hosted by WormBase (http://www.wormbase.org/wiki/index.php/NGASP). Our results demonstrate that MAKER provides a simple-to-use, yet effective means to annotate an individual contig or BAC or to convert an entire genome sequence into a community-accessible genome database. MAKER is not exhaustive: it does not identify noncoding RNA genes, nor is it intended as a comprehensive solution to every problem in genome annotation. Rather, MAKER is designed to jump-start genomics in emerging model organisms by providing a robust first round of database-ready protein-coding gene annotations.
Results
Benchmarking MAKER on Caenorhabditis elegans
In order to obtain a performance benchmark, we ran MAKER on a 10-megabase (Mb) portion of the C. elegans genome, as part of the nGASP competition (http://www.wormbase.org/wiki/index.php/NGASP). nGASP provided two annotated 10-Mb regions of the C. elegans genome, one for training, and the other for testing. We trained MAKER using the boot-strap procedure outlined in the Methods section and then compared MAKER’s performance on the testing region to three other nGASP participants: SNAP (Korf 2004), Augustus (Stanke et al. 2006), and Gramene—an Ensembl-based pipeline (Stabenau et al. 2004) managed by the Gramene group (www.gramene.org). SNAP was run in its ab initio gene prediction mode; Gramene is an evidence-based annotation pipeline that assembles its own computational evidence; and Augustus is a gene-prediction algorithm that can be used to produce either ab initio or evidence-based predictions when provided with an external GFF3 file of EST and protein alignment data. The evidence-based Augustus annotations summarized in Table 1 used GFF3 files of aligned ESTs and proteins provided by nGASP.
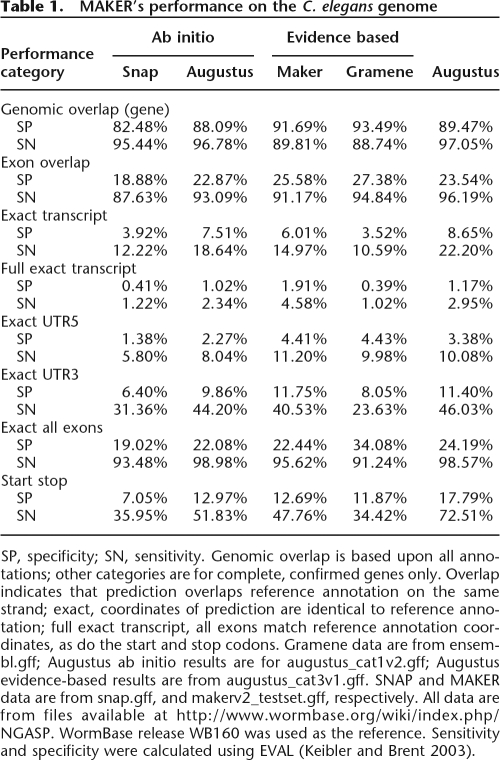
Table 1.
MAKER’s performance on the C. elegans genome
SP, specificity; SN, sensitivity. Genomic overlap is based upon all annotations; other categories are for complete, confirmed genes only. Overlap indicates that prediction overlaps reference annotation on the same strand; exact, coordinates of prediction are identical to reference annotation; full exact transcript, all exons match reference annotation coordinates, as do the start and stop codons. Gramene data are from ensembl.gff; Augustus ab initio results are for augustus_cat1v2.gff; Augustus evidence-based results are from augustus_cat3v1.gff. SNAP and MAKER data are from snap.gff, and makerv2_testset.gff, respectively. All data are from files available at http://www.wormbase.org/wiki/index.php/NGASP. WormBase release WB160 was used as the reference. Sensitivity and specificity were calculated using EVAL (Keibler and Brent 2003).
Overall, MAKER’s performance on the C. elegans genome was comparable to that of Gramene and to Augustus when run in the evidence-based mode. All three programs had very similar sensitivity and specificity values for genomic overlap—a measure of the percentage of genes overlapped by an annotation. MAKER’s genomic overlap sensitivity (89.81%) was greater than that of Gramene’s (88.74%) and less than that of Augustus’ (97.05%), indicating that ∼90% of annotated C. elegans genes were overlapped by at least a portion of a MAKER annotation. MAKER’s genomic overlap specificity (91.69%) was also intermediate between those of Augustus (89.47%) and Gramene (93.49%).
When considering the remaining categories in Table 1, it should be kept in mind that these refer to the subset of annotations (32%) that WormBase denoted as complete and confirmed WB160 genes. The low specificities reported for all three programs reflect this fact.
MAKER’s weakest performance was in the exon nucleotide accuracy and exon overlap and categories. For all genes, its exon level nucleotide accuracy is 61.82%, Gramene’s is 70.8%, and Augustus’ is 70.83% and 77.62% (evidence-based). For confirmed genes, MAKER’s exon level overlap specificity is similar to that of the other programs (Table 1), but its sensitivity is still 3.67% less than that of Gramene and 5.02% less than that of Augustus when run in its evidence-based mode. On confirmed genes, MAKER’s exact all exon (Table 1) accuracy is similar to those of the other two evidence-based programs. MAKER fell squarely between Gramene and Augustus in correctly annotating entire transcripts (Exact Transcript) but outperformed the other two programs when the start and stop of translation is also taken into account (Full Exact transcript). MAKER also outperformed the other two programs in accurately identifying 5′ UTRs (Exact UTR5). MAKER was more effective at precisely identifying 3′ UTRs (Exact UTR3) than was Gramene and was only slightly less accurate than Augustus. The last category in Table 1, Start Stop, provides a measure of how well MAKER did at identifying start and stop codons. Once, again, MAKER’s performance is comparable to the other programs. MAKER outperformed Gramene in this category, though Augustus was the clear winner. In total then, the data in Table 1 demonstrate that MAKER’s overall performance on the C. elegans genome is in most instances comparable to that of Gramene and Augustus.
A proof-of-principle collaboration
In order to demonstrate MAKER’s suitability as an annotation tool for the genomes of emerging model organisms, we partnered with the S. mediterranea genome project to annotate its genome and create a GMOD-based genome database. S. mediterranea is a model planarian species, known for its ability to regenerate complete animals from miniscule fragments of its body (Randolph 1897; Morgan 1898). S. mediterranea is an emerging model organism for regeneration studies following demonstrations that it is amenable to modern cell (Robb and Sánchez Alvarado 2002), molecular (Sánchez Alvarado et al. 2002), and RNAi (Sánchez Alvarado and Newmark 1999) techniques. Its annotated genome will provide a central resource for the planarian and regenerative medicine research community.
The S. mediterranea genome and assembly
The S. mediterranea genome was sequenced and assembled by the Washington University Genome Sequencing Center (St. Louis, MO). The final assembly is 902,775,852 nucleotides in length, consistent with Cot and nuclear volume analyses carried out prior to sequencing, which place the S. mediterranea genome at ∼850 Mb (http://genome.wustl.edu/ancillary/data/whitepapers/Schmidtea_mediterranea_WP.pdf). S. mediterranea was sequenced to a depth of ∼10×. The assembly’s contig length distributions are similar to those of the human and Drosophila genomes (data not shown). Its super-contigs, however, are shorter, as technical issues precluded the construction of a BAC library for this organism; thus, no BAC end reads were available during the assembly process; 89.30% of the genome is in super-contigs 10 kb or longer and 44.62% is in super-contigs longer than 50 kb. The final assembly contains 43,673 contigs with a median length of 11,260 bp. The genome has a high AT content (67%).
ESTs
At time of compute, there were 78,101 ESTs from S. mediterranea. These were derived from a variety of libraries (see Methods), and consist of both 5′ and 3′ reads. As the S. mediterranea EST collection was quite redundant, we collapsed the ESTs into contigs using the CAP3 program (Huang and Madan 1999). This process yielded 15,011 contigs. MAKER aligned 13,026 (88%) of the EST contigs to the genome, using the splice-site aware Exonerate algorithm (Slater and Birney 2005). Of the remainder, about half were not found in the assembly, and low sequence complexity prohibited alignment of the other half. These numbers provide an estimate of 90% for the overall completeness of the assembly, a finding consistent with the experimental estimates of genome size (http://genome.wustl.edu/ancillary/data/whitepapers/Schmidtea_mediterranea_WP.pdf) and the size of the assembly.
S. mediterranea repeats
In total, RepeatMasker (http://repeatmasker.org) flagged 22% of the S. mediterranea genome as low-complexity sequence. MAKER also uses BLASTX together with an internal library of transposon and virally encoding proteins to identify mobile-elements (see Architecture of MAKER section). This process masked an additional 4.18% of the genome. Finally, we used Muscle (Edgar 2004) and PILER (Edgar and Myers 2005) to identify additional S. mediterranea specific and highly divergent repeats, missed by the previous processes. MAKER used these as a RepeatMasker library. This masked another 1.2% of the genome. In total, 27.4% of the genome was identified as repetitive. The creation of a custom library for use with MAKER is optional but recommended.
The S. mediterranea high-confidence gene set
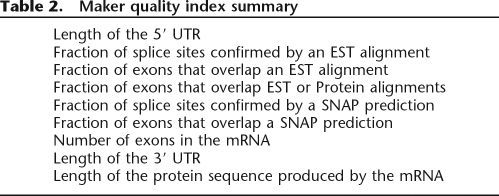
In order to produce a maximally inclusive set of compute data and annotations for our downstream analyses, we ran MAKER over every contig in the S. mediterranea genome assembly regardless of size. The resulting data are summarized in Supplemental Table 1. Following procedures similar to those used to annotate other eukaryotic genomes (Rubin et al. 2000; Venter et al. 2001), we next sought to assemble a high-confidence (HC) gene set from among the 65,563 MAKER genes. To do so, we took advantage of the MAKER Quality Indices generated for each transcript, which document the number of exons confirmed by EST, and/or protein evidence (see Methods; Table 2). We included in the HC set every gene having at least one transcript confirmed by an EST alignment with at least one canonical splice site. In total, there were 12,620 genes that met this criteria. We also included in the HC gene set any MAKER annotation with protein homology (BLASTX E < 1 × 10−6) to the Swiss-Prot database (Bairoch and Apweiler 2000); 24,209 MAKER genes met this criterion. We then used RPS-BLAST (http://web.csb.ias.edu/blast/rpsblast.txt) to screen the annotations for Pfam (Bateman et al. 2004) domains (E < 1 × 10−3; minimum coverage >40%). This identified 15,702 domain-containing annotations. We also screened the 128,339 SNAP predictions not overlapping a MAKER annotation for protein homology with SWISS-PROT (Bairoch and Apweiler 2000) and for Pfam (Bateman et al. 2004) domains using the same significance thresholds; 378 of them were homologous to SWISS-PROT proteins, and 1633 had one or more domains. In total, this gave us a set of 31,955 protein-coding genes supported by combinations of EST, protein, and domain homology.
Table 2.
Maker quality index summary
Protein-coding gene numbers
Our purpose in assembling the HC gene set was to produce a set of gene models suitable for comparison to other annotated eukaryotic genomes. Gene number is one such comparison. Though protein-coding gene numbers have been a subject of controversy, most annotated model Eukaryotes contain on the order of 15,000–25,000 protein-coding genes (for discussion, see Yandell et al. 2005). Drosophila, e.g., is believed to contain fewer than 15,000 protein coding genes (Yandell et al. 2005), and the WS160 WormBase release puts the number of C. elegans genes at slightly less than 20,000. The latest Ensembl (Stabenau et al. 2004) release of the human genome contains 21,724 known protein-coding genes.
Although there is no a priori reason to assume that S. mediterranea might not contain 31,955 protein-coding genes (the number of genes in the HC set), this possibility is not well supported by available experimental evidence. We therefore sought to determine what percentage of the annotations might be split across the short super-contigs characteristic of the S. mediterranea genome assembly. To do so, we cloned and sequenced 31 high-molecular-weight S. mediterranea mRNAs without recourse to the MAKER annotations. We aligned each mRNA to the genome assembly (30 were found in the assembly) and found that 28 corresponded to MAKER annotations, nine of these (30%) were split across multiple contigs, and four (14%) were annotated as multiple genes on a single S. mediterranea contig. By comparison, only 12 of the mRNAs were overlapped by SNAP ab initio predictions, and three of these were split. Though these are small numbers, they suggest that 90.3% of S. mediterranea genes correspond to at least one MAKER annotation, 30% of S. mediterranea genes are split among multiple contigs, and MAKER has incorrectly split ∼14% of mediterranea genes into two or more annotations. Taking these percentages as indicative of the genome as a whole would place the S. mediterranea protein-coding gene number at 15,570, a number in good agreement with the annotated gene numbers in other model animals.
Evaluating MAKER’s performance on S. mediterranea
The absence of a large corpus of known S. mediterranea genes and mRNAs makes it difficult to assess MAKER’s performance by comparison to known S. mediterranea gene structures. Instead we have used the protein domains to gain a rough indication of overall annotation completeness and quality. Domain data also provide a measure of how much MAKER’s synthesis procedure improved upon SNAP’s ab initio predictions for this genome.
We used RPS-BLAST (http://web.csb.ias.edu/blast/rpsblast.txt) and Pfam (Bateman et al. 2004) to identify protein domains in S. mediterranea predicted and annotated proteins. In total, 21.54% of MAKER annotations and 38% of HC annotations contain at least one known domain. We used the same procedure to identify domains in the annotated proteomes of other animals and found that 35.5% of Drosophila melanogaster, 31.9% of C. elegans, and 36.4% human annotated proteins contain known domains. Thus, the percentage of protein domains in the HC set is comparable to those of other annotated animal proteomes. We further categorized the annotations using the Gene Ontology (http://www.geneontology.org) classifications of the domains they encode and used these data to compare the S. mediterranea annotations to other annotated animal genomes. These data are shown in Supplemental Table 2 and demonstrate that the HC genes contain an unbiased, diverse, and comprehensive sampling of the S. mediterranea proteome.
MAKER improves upon SNAP’s ab initio predictions
As a control, we ran a version of SNAP trained for the AT-rich genome of C. elegans over the S. mediterranea genome: Only 3.49% of those predictions contained domains, whereas when trained for S. mediterranea using the universal gene-based procedure (for details, see Methods), 5.17% of SNAP ab initio predictions contained domains. By comparison, 21.54% of MAKER S. mediterranea annotations and 38% of HC annotations contain at least one domain (Supplemental Table 1). Of the 128,339 ab initio SNAP predictions not overlapping MAKER annotations, only 1.27% contained domains.
In contrast to S. mediterranea, MAKER’s training and synthesis procedures produced only modest increases in gene-level specificity for C. elegans, with most of the improvements to accuracy coming from refinements to transcript structures. In C. elegans, MAKER’s overall genomic overlap and exon overlap accuracies were similar to SNAP’s (Table 1). Real gains, however, were observed in the other categories. MAKER’s exact transcript sensitivity was 6.01% compared with SNAP’s 3.92%; exact transcript specificity also showed gains, rising from SNAP’s 12.22% to 14.97%. Likewise, full exact transcript sensitivity and specificity increased by a factor of four. The synthesis process also improved the accuracy of UTR annotation; in fact, Table 1 somewhat misrepresents the nature of the improvement, as EVAL (Keibler and Brent 2003) considers stop codons to be part of the 3′ UTR; likewise, it also considers an incomplete codon preceding the annotated translation to be 5′ UTR. Excluding UTRs of less than four nucleotides in length, the Exact UTR5 and UTR3 values are 0% for SNAP. MAKER’s synthesis procedures also improved upon SNAP’s ability to correctly identify start and stop codons; for these features, specificity rose from 7.05% to 12.69% and sensitivity from 35.95% to 47.76%.
Improvements were greater for the emerging genome
MAKER’s synthesis procedure resulted in a far greater enrichment for protein-domain containing annotations in S. mediterranea than it did in C. elegans. In total, only 5.17% of S. mediterranea ab initio SNAP predictions encode proteins with domains, whereas 21.54% of the MAKER annotations do. In C. elegans, by comparison, 38.65% of ab initio SNAP predictions and 44.81% of MAKER annotations have domains. Hence, the enrichment (16%) of MAKER annotations compared with SNAP predictions for domains in C. elegans is modest compared with 315% enrichment seen in S. mediterranea. The difference appears to be due primarily to the lower specificity of SNAP on the S. mediterranea compared with the C. elegans genome. Three lines of evidence support this conclusion. First, the ratio of SNAP to MAKER transcripts is much higher in S. mediterranea than C. elegans, 2.5× compared with 1.3×, respectively. Second, there is a greater correspondence between SNAP predictions and MAKER annotations in C. elegans than there is in S. mediterranea—only 22.39% of SNAP predictions do not overlap a MAKER annotation in C. elegans, whereas 76% fail to do so in S. mediterranea. Third, of the SNAP predictions not overlapping an S. mediterranea MAKER annotation, only 1.27% contain domains (RPS blast to PFAM E < 1 × 10−3; percent coverage > 40%), again suggesting that many are false positives. Despite these differences, 38% of genes in the S. mediterranea HC gene-set encode protein domains, a value quite similar to the 32% in the WB160 release of the C. elegans proteome. These facts demonstrate that the ability of MAKER to screen and improve upon SNAP’s ab initio predictions is greatest in emerging model organisms such as S. mediterranea for which there is limited training data.
SmedGD
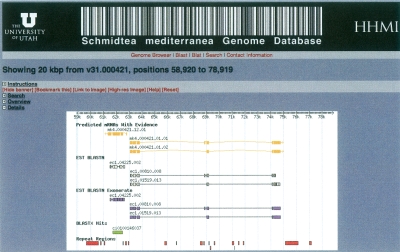
We used the GFF3 output from MAKER to jump-start SmedGD, a publicly available resource for the planarian and regeneration research communities. SmedGD houses the S. mediterranea genome assembly, its MAKER annotations, and their associated computational evidence (Robb et al. 2007). Because SmedGD conforms to GMOD specifications (http://www.gmod.org), its contents can be queried and viewed over the Web using GMOD tools such as GBrowse (http://www.gmod.org). The use of GMOD schemas ensures interoperability of database contents, allowing them to be shared and compared with the contents of other GMOD databases such as FlyBase, WormBase, and SGD. Figure 1 shows a screen shot from SmedGD, showing a MAKER annotation and its accompanying compute data. SmedGD is located at http://smedgd.neuro.utah.edu.
Figure 1.
SmedGD, the GMOD-based S. mediterranea genome database constructed directly from MAKER’s outputs (http://smedgd.neuro.utah.edu).
Discussion
We used MAKER on the genomes of both an established and an emerging model organism. Our results for the C. elegans genome demonstrate that the accuracy of MAKER on a model organism genome is comparable to that of other annotation pipelines, whereas our work on the S. mediterranea genome shows that MAKER provides an effective means to annotate an emerging genome and to create a genome database.
MAKER’s performance on established genomes
We compared MAKER’s sensitivity and specificity to those of Augustus (Stanke et al. 2006) and Gramene (www.gramene.org)—an Ensembl-based pipeline (Stabeneau et al. 2004) in eight different categories using the EVAL program (see Table 1) (Keibler and Brent 2003). Two categories, genomic overlap and exon overlap, give an indication of what percentage of annotated C. elegans genes were overlapped by a MAKER annotation. MAKER’s performance in the first category was similar to the other two programs; its accuracy was 90.75% compared with 91.12% and 93.26% for Gramene and Augustus, respectively. However, MAKER faired worse in the Exon overlap category, exhibiting a slight tendency to drop exons, resulting in a 3.67% and 5.02% underperformance for sensitivity in this category relative to Gramene and Augustus, respectively. MAKER was the most effective of the three annotation pipelines at calling entire transcripts, including start and stop codons (full exact transcript; Table 1). Its performance in the remaining categories was comparable to Gramene and Augustus.
MAKER’s performance on emerging genomes
Emerging genomes place particular demands on annotation pipelines. The differences in the C. elegans and S. mediterranea annotations illustrate many of the challenges unique to annotating an emerging genome. Our results make it clear that good performance on an established genome is no guarantee of similar performance on an emerging genome. Poor ab initio gene-finder performance—even when retrained—makes an evidence-based process to inform and filter gene predictions absolutely crucial. As judged by domain content, MAKER was only able to improve upon SNAP’s C. elegans ab initio predictions by 16%, whereas in S. mediterranea MAKER’s annotations are enriched 315% for protein domains compared with the SNAP ab initio predictions. The differences are due to ab initio gene finder performance. In C. elegans, the ab initio predictors did well, and the evidence assembled by the compute pipeline usually did little more than confirm their structure. In S. mediterranea, the situation was very different, and MAKER’s synthesis procedure played a much greater role. These facts demonstrate the necessity of a trainable, evidence-based process to inform and filter gene predictions when annotating emerging genomes; MAKER’s quality indices proved instrumental in this regard, both for training and for assembling the HC gene set.
MAKER outputs are GMOD and Apollo compliant
The S. mediterranea genome project used MAKER’s GFF3 outputs to create SmedGD, a GMOD database (http://www.gmod.org) for the S. mediterranea genome. SmedGD is available at http://smedgd.neuro.utah.edu and is intended to provide a basic resource for planarian functional and comparative genomics. In total, less than 60 d were required to convert the S. mediterranea genome assembly into a genome database. SmedGD thus demonstrates the power of MAKER to jump-start genomics in emerging model organisms by providing a first round of database-ready, protein-coding gene annotations.
MAKER is ideal for smaller projects
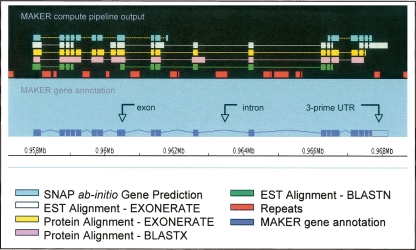
MAKER can also be used to annotate individual contigs and BACs. For S. mediterranea, MAKER ran on a single-core MAC laptop (2 GHz CPU with 2 GB RAM) at a rate of 4.1 h/Mb of sequence; this means that a 100 KB BAC can be annotated on a laptop computer in less than half an hour. Furthermore, the outputs can be immediately viewed and edited in Apollo (see Fig. 3) (Lewis et al. 2002) without the added overhead of a genome database. These features make MAKER ideal for small-scale applications and will prove useful for researchers working in emerging model organisms for which only partial assemblies are available.
Figure 3.
Apollo view of a MAKER gene annotation and its associated evidence. Evidence gathered by MAKER’s compute pipeline (upper panel) is synthesized into the resulting MAKER annotation (lower panel).
Future improvements to MAKER
At present, MAKER uses only a single ab initio gene predictor and creates only protein-coding annotations. MAKER’s modular structure means that any gene predictor can be integrated into its architecture with minimal software development. To date, we have focused on integrating SNAP, as it was designed with easy trainability in mind (Korf 2004), but additional predictors could be integrated as well. Augustus is also trainable and comes with an optimization script that tries to find values for the meta-parameters, such as splice window sizes (Stanke and Morgenstern 2005). MAKER should be able to manufacture this information automatically as part of an extended training procedure, and we are currently exploring the feasibility of doing so. Extending MAKER to produce ncRNA annotations is another area of development. Tools for tRNA gene prediction exist (Lowe and Eddy 1997), as do ncRNA gene-finders (Holmes 2005; Rivas and Eddy 2001). These improvements will make for more complete genome databases and help end the annotation bottleneck.
Methods
Architecture of MAKER
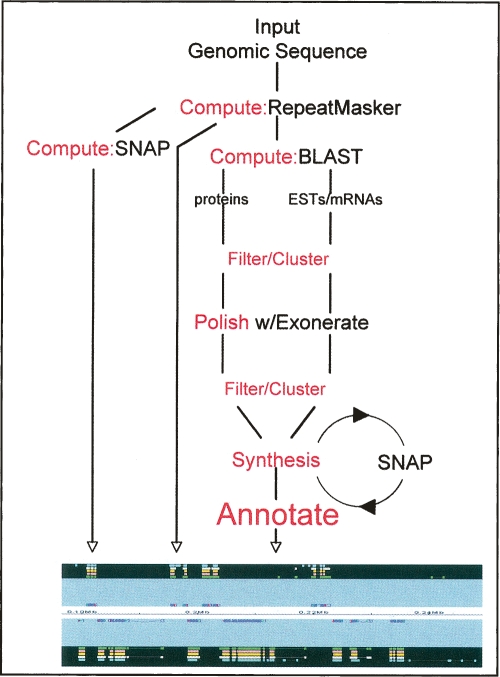
MAKER has a modular architecture that abstracts sequence analyses in a standardized object model. MAKER uses the CGL (Yandell et al. 2006) common object model, which extends the Bioperl (http://www.bioperl.org) GenericHit and GenericHSP classes with methods that facilitate comparative analyses and automatic annotation. MAKER’s modular construction allows it to break the annotation process down into a series of five discrete activities that are easily interoperable: compute, filter/cluster, polish, synthesis, and annotate (Fig. 2). MAKER performs these actions on sequences of any length by automatically cutting the input sequence into series of chunks (default is 100 kb), running each compute, and then merging the results.
Figure 2.
MAKER Overview. MAKER uses four external executables: RepeatMasker, BLAST, SNAP, and Exonerate. Actions corresponding to the five basic steps of automatic annotation are shown in red.
Step 1: Compute phase
A battery of sequence analysis programs is run on input genomic sequence. The purpose of these computes is to identify and Mask repeats and to assemble protein EST and mRNA alignments that will be used to inform MAKER’s gene-annotation process, which is outlined in steps 4 and 5 below. The default MAKER configuration uses four external programs: RepeatMasker (http://repeatmasker.org), BLAST (Altschul et al. 1990; Korf et al. 2003), Exonerate (Slater and Birney 2005), and SNAP (Korf 2004). Each is publicly available and free for academic use. All four programs are also easy to install and run on UNIX, Linux, and OS X.
Unless repeats are effectively masked, gene predictions and gene annotations will contain portions of transposons and viruses. MAKER uses a two-tier process to avoid this problem. First, RepeatMasker is used to screen the genome for low-complexity repeats; these are then “soft-masked,” e.g., transformed to lowercase letters rather than to Ns. Soft masking excludes these regions from nucleating BLAST alignments (Korf et al. 2003) but leaves them available for inclusion in annotations, as many protein-coding genes contain runs of low complexity sequence. MAKER also uses BLASTX together with an internal library of transposon and virally encoding proteins to identify mobile-elements. This process has been shown to substantially improve repeat masking as it identifies genome regions that are distantly related to the protein coding portions of transposons and viruses; these tend to be missed by RepeatMasker’s nucleotide-based alignment process, even when genome specific repeat libraries are available (Smith et al. 2007). Repeat regions identified in this process are masked to Ns. MAKER performs all of the actions automatically.
BLAST is used throughout the compute phase, first for repeat identification with RepeatMasker (as described above) and then to identify EST, mRNAs, and proteins with significant similarity to the input genomic sequence. Because BLAST does not take splice sites into account, its alignments are only rough approximations. MAKER therefore uses Exonerate (Slater and Birney 2005), a splice-site aware alignment algorithm to realign, or polish, sequences following filtering and clustering (see steps 2 and 3, below). Exonerate’s ability to align both protein and nucleotide sequences to the genome make it an economical choice for this task.
Step 2: Filter/cluster
Filtering consists of identifying and removing marginal predictions and sequence alignments on the basis of scores, percent identities, etc. Filtering criteria for each external executable are set by modifying the text-based maker_bopts.ctl file (see configuration README distributed with MAKER). New users are not expected to edit this file, but advanced users may do so to change the behavior of the program. After filtering, the remaining data are then clustered against the genomic sequence to identify overlapping alignments and predictions. Clustering has two purposes. First, it groups diverse computational results into a single cluster of data, all of which support the same gene or transcript. Second, it identifies redundant evidence. For example, highly expressed genes may be supported by hundreds if not thousands of identical ESTs. Clustering criteria are set in the maker_bopts.ctl file, which instructs MAKER to keep some maximum number of members within each cluster, sorted on some series of filtering attributes such as score or fraction of the hit-sequence aligned. The default parameters are appropriate for most applications but can be easily modified.
Step 3: Polish
This step realigns BLAST hits using a second alignment algorithm to obtain greater precision at exon boundaries. MAKER uses Exonerate (Slater and Birney 2005) to realign matching and highly similar ESTs, mRNAs, and proteins to the genomic input sequence. Because Exonerate takes splice-sites into account when generating its alignments, they provide MAKER with information about splice donors and acceptors. This information is especially useful in the synthesis and annotation steps (see below). The thresholds in the maker_bopts.ctl file earmark BLAST hits for polishing and are suitable for most applications but can be easily altered if desired (see configuration README distributed with MAKER).
Step 4: Synthesis
MAKER synthesizes information from the polished and clustered EST and protein alignments to produce evidence for annotations. To do so, it identifies ESTs that it judges correspond to the same alternatively splice transcript. This is accomplished by comparing the coordinates of each polished sequence alignment on the genomic sequence in the same way that a human annotator might, e.g., by looking for internal exons with differing boundaries. Next MAKER identifies those protein alignments whose coordinates are consistent with each of the EST splice forms. Once a set of EST and protein alignments—all consistent with the same spliced transcript—has been identified, positions on the genomic input sequence upstream and downstream of the alignments are labeled as possible intergenic regions. Those bases on the genomic sequence that fall between exons are labeled as putative introns, and bases overlapping the protein alignments are labeled as putative translated sequence. MAKER then calculates a score for each of these nucleotides on the query sequence based upon the percentage of similarity of the alignment, type of alignment, and a query nucleotide’s position within the alignment. These scores together with their putative sequence types, e.g., Intergenic, Coding, Intron, and UTR, are then passed to SNAP. Based upon this information, SNAP then modifies its internal Hidden Markov Model (HMM). In the absence of any supporting EST or protein alignments, MAKER uses SNAP’s ab initio prediction (for additional details, see Training SNAP).
Step 5: Annotate
MAKER also post-processes the synthesis-generated SNAP predictions and recombines them with evidence to generate complete annotations. Each synthesis-generated SNAP prediction is checked against all ESTs and mRNAs, and 5′ and 3′ UTRs consistent with the prediction are identified based upon their coordinates relative to the predicted coding exons. The coordinates of the SNAP prediction are then altered to include these regions. This process is repeated for each of the synthesis-based predictions. Finally, compute evidence supporting each exon is added, and alternatively spliced forms are documented.
Additional details regarding MAKER’s architecture and implementation can be found in the release materials. All MAKER source code is publicly available; the current release along with installation instructions and documentation is available at http://www.yandell-lab.org/maker.
Inputs and outputs
The input to MAKER is a genomic sequence (of any length) in fasta format and three configuration files describing external executable, sequence database locations, and various compute parameters (see configuration README distributed with MAKER). MAKER also uses four sequence database files during the compute phase: a transposons file, an optional repeatmasker database file, a proteins file, and an ESTs/mRNAs file. Each file is in fasta format. The transposons file is bundled with MAKER and contains a selection of known transposon and virally encoded protein sequences. This file is used to identify and mask repeats missed by RepeatMasker, as this has been shown to substantially improve accuracy (Smith et al. 2007). In cases where no organism-specific repeat library is available, MAKER will automatically use the transposon file to mask mobile elements and the RepeatMasker program to identify and mask low-complexity sequences. The repeatmasker file is an optional fasta file containing organism specific repeat sequences, if available. The proteins file contains any proteins users would like aligned to the genome. We recommend they use the latest version of the SWISS-PROT database for this purpose (Bairoch and Apweiler 2000). Finally, users should also supply a file of ESTs and/or mRNAs sequences derived from the organism being annotated. Assembling these into contigs is helpful, but it is not required.
MAKER outputs GMOD-compliant annotations in GFF3 format (http://www.sequenceontology.org/gff3.shtml) containing alternatively spliced transcripts, UTRs, and evidence for each gene’s annotated transcript and protein sequences. This file can be directly imported into genome browsers and databases that adhere to Sequence Ontology (Eilbeck et al. 2005) and GMOD (http://www.gmod.org) standards. For convenience, MAKER also outputs multifasta files of transcripts and protein sequences for both annotations and ab initio SNAP predictions.
MAKER also writes a GAME XML file (http://www.fruitfly.org/annot/apollo/game.rng.txt) containing the same contents as the corresponding GFF3 file (http://www.sequenceontology.org/gff3.shtml); this file can be directly viewed in the Apollo genome browser (Figure 3) (Lewis et al. 2002). Apollo can also be used to directly edit annotations and to save them to GFF3 format, thus changes to MAKER annotations can be saved prior to uploading them into a GMOD browser or database. Apollo can also directly export the revised transcripts and protein sequences in fasta format. This is an especially useful feature for those seeking to annotate a single contig or BAC rather than an entire genome, as it circumvents the overhead associated with creating and maintaining a GMOD database. Figure 3 shows a portion of an annotated contig viewed in Apollo genome browser. Compute evidence assembled by MAKER is shown in the top panel; its resulting annotation, below. This figure demonstrates how MAKER synthesizes data gathered by its compute pipeline into evidence-informed gene annotations; while SNAP produced two ab initio predictions in this region, the EST and protein alignments clearly support a single gene. Note too the 3′ UTR on the MAKER annotation derived from the EST alignments.
The MAKER mRNA quality index
Compute data are essential for discriminating real genes from false positives. To simplify the quality evaluation process, each MAKER-annotated transcript has an associated quality index included in its GFF3 and GAME XML outputs. This is a nine-dimensional summary (Table 2) of a transcript’s key features and how they are supported by the data gathered by MAKER’s compute pipeline. The quality index associated with the mRNA shown in Figure 3 is QI:0|0.77|0.68|1|0.77|0.78|19|462|824.
Quality indices play a central role in training MAKER for a particular genome, where they are used to identify transcripts that are well supported by EST and protein evidence but poorly supported by ab initio SNAP predictions. These cases are used to retrain SNAP via the bootstrap procedure outlined below. MAKER’s quality indices also provide an easy means to sort and rank transcripts by key features such as number of exons, presence or absence of UTR, or degree of computational support. Quality indices were used to assemble the HC S. mediterranea genes described in the Results section.
Training MAKER
For optimal accuracy, a gene finder must be trained for a specific genome (Korf 2004), generally using several hundred existing gene-annotations drawn from a body of experimental data gathered over many years. Unfortunately, many emerging genomes do not have a history of experimental molecular biology. It has therefore become a common practice to use gene finders trained in one genome to predict genes in another—a far from optimal solution to the problem (for discussion, see Korf 2004). Information gathered from ab initio predictions is essential for the annotation process, even when other evidence is available. Moreover, in the absence of experimental evidence and sequence similarities, the probabilistic models produced by ab initio gene prediction programs offer the best guesses at gene structure. The SNAP (Korf 2004) gene finder was designed from the outset to be easily configured for any genome; hence its use in MAKER.
MAKER is trained for a genome using a two-step process. First, SNAP is trained by aligning a set of universal genes to the input genome (Parra et al. 2007). These universal genes are highly conserved in all eukaryotes and can be identified using pairwise and profile-HMM alignment methods. The resulting gene structures are used to create a first-pass version of SNAP for use in the next stage of the training process. This initial stage of the training procedure is automated, and complete details of the process can be found in the MAKER README. More extensive documentation is provided by Parra et al. (2007).
The genome-specific HMM produced in the first stage of SNAP training is further refined with a second stage of training. This is accomplished by running MAKER on a few megabases of genomic sequence (enough to result in a few hundred annotations). The resulting GFF3 outputs are then used as inputs to a script called maker2zff.pl, whose output is a ZFF file that can be used to automatically create a revised HMM. The maker2zff.pl script uses the quality index MAKER attaches to each annotation to identify a set of gene models with intron-exon structures that are unambiguously supported by EST alignments and protein homology. These genes are then used to further refine the SNAP HMM. The maker2fzff.pl script is bundled with MAKER, and programs necessary to create the HMM are included in the SNAP package. To train MAKER for the S. mediterranea genome, we first trained SNAP using the universal gene set as outlined above. We then ran MAKER on a randomly selected 100-Mb portion of the S. mediterranea genome (∼10% of the entire genome). The resulting GFF3 files were used as inputs to maker2zff.pl, and the refined SNAP-HMM was used in the final annotation run.
Manufacturing an S. mediterranea specific RepeatMasker database
Repeat sequences were identified for the S. mediterranea genome by two methods. First, RepeatRunner (Smith et al. 2007) identified and masked sequences that had similarity to previously identified repeated elements. Second, PILER-DF (Edgar and Myers 2005) was used to find novel dispersed repeats. Settings for the various programs in the PILER suite are as follows: PALS was run with the parameter length = 150 (minimum hit length) and pctid = 94 (minimum percentage identity). PILER was run with the parameter famsize = 10 (minimum size of the repeat family). MUSCLE (Edgar 2004) was run with maxiters = 1 and diags = 1 as recommended in the documentation for PILER. There were 295 repeat families found by this method; most were helitrons (Kapitonov and Jurka 2001).
Manufacturing EST contigs from S. mediterranea ESTs
The 78,101 EST sequences from S. mediterranea were clustered into 15,011 contigs using CAP3 (Huang and Madan 1999).
Manufacturing the protein database
The reference proteins file consisted of proteome sequences from seven organisms and all known Platyhelminthes proteins. The C. elegans (W160), D. melanogaster (v4.3), Escherichia coli (NC_000913), Homo sapiens (v36.1), Mus musculus (v36.1), and Saccharomyces cerevisiae (08/2006) proteome sequences were downloaded from NCBI (http://ftp.ncbi.nih.gov/genomes). The Ciona intestinalis proteome (v1.0) was downloaded from the Joint Genome Institute downloads site (http://genome.jgi-psf.org/ciona4/ciona4.home.html). Platyhelminthes protein sequences were downloaded from NCBI’s Entrez in August 2006.
Compute times
We clocked MAKER on a 2.236-Mb sequence. On a 32 GB-RAM machine, with eight dual-core 2-GHz processors, the annotation took MAKER 549 min on one processor and 299 min using two processors. When external programs, such as BLAST are pre-run, the process time for MAKER on one processor was 31.33 min. For this test, MAKER produced a 3.7-Mb GFF3 file, a 60 MB GAME XML document, and four fasta files totaling 560 kilobytes. For S. mediterranea, MAKER ran on a single-core MAC laptop (2 GHz CPU with 2 GB RAM) at a rate of 4.1 h/Mb of sequence—this value includes all compute steps, e.g., compute phase, filter/cluster, polish, synthesis, and annotate.
Downloading and installing MAKER
MAKER is available for download from http://www.yandell-lab.org/downloads/maker/maker.tar.gz. Once downloaded, the MAKER package should be unzipped and untared. Full installation and usage instructions are available in the file called README.
Creating SmedGD
The GFF3 output files generated by MAKER were used to populate SmedGD. The files were uploaded into a mySQL database, using a standard Bioperl (http://www.bioperl.org) loading script, bp_seqfeature_load.pl. This script converts GFF3 formatted annotations to Bio∷SeqFeatureI objects, which are stored in the mySQL database. GBrowse, a tool distributed by GMOD (http://www.gmod.org) implementing a Bio∷DB∷SeqFeature∷Store database adaptor, accesses and displays rows of data or tracks that are mapped to specific locations in the genome. SmedGD consists of MAKER annotations as well as project specific features, such as additional protein homology, human curated genes, and RNA interference phenotype data. The database is available at http://smedgd.neuro.utah.edu.
Acknowledgments
We thank Bret Pearson for the cloning of the 31 S. mediterranea mRNAs discussed in the Results section. This work was supported in part by NIH grant K22-HG0064 to I.K. S.M.C.R. is supported by NIH Genetics Training Grant 5 T32 GM007464. A.S.A. is a Howard Hughes Medical Institute investigator.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6743907
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bairoch A., Apweiler R., Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Marshall M., Moxon S., Sonnhammer E.L., Moxon S., Sonnhammer E.L., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C., Myers E.W., Myers E.W. PILER: Identification and classification of genomic repeats. Bioinformatics. 2005;21:i152–i158. doi: 10.1093/bioinformatics/bti1003. [DOI] [PubMed] [Google Scholar]
- Eilbeck K., Lewis S.E., Mungall C.J., Yandell M., Stein L., Durbin R., Ashburner M., Lewis S.E., Mungall C.J., Yandell M., Stein L., Durbin R., Ashburner M., Mungall C.J., Yandell M., Stein L., Durbin R., Ashburner M., Yandell M., Stein L., Durbin R., Ashburner M., Stein L., Durbin R., Ashburner M., Durbin R., Ashburner M., Ashburner M. The Sequence Ontology: A tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes I. Accelerated probabilistic inference of RNA structure evolution. BMC Bioinformatics. 2005;6:73. doi: 10.1186/1471-2105-6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Madan A., Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov V.V., Jurka J., Jurka J. Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. 2001;98:8714–8719. doi: 10.1073/pnas.151269298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keibler E., Brent M.R., Brent M.R. Eval: A software package for analysis of genome annotations. BMC Bioinformatics. 2003;4:50. doi: 10.1186/1471-2105-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I., Yandell M., Bedell M., Yandell M., Bedell M., Bedell M. BLAST: An essential guide to the basic local alignment search tool. O’Reilly & Associates, Inc.; Sebastopol, CA: 2003. [Google Scholar]
- Lawson D., Arensburger P., Atkinson P., Besansky N.J., Bruggner R.V., Butler R., Campbell K.S., Christophides G.K., Christley S., Dialynas E., Arensburger P., Atkinson P., Besansky N.J., Bruggner R.V., Butler R., Campbell K.S., Christophides G.K., Christley S., Dialynas E., Atkinson P., Besansky N.J., Bruggner R.V., Butler R., Campbell K.S., Christophides G.K., Christley S., Dialynas E., Besansky N.J., Bruggner R.V., Butler R., Campbell K.S., Christophides G.K., Christley S., Dialynas E., Bruggner R.V., Butler R., Campbell K.S., Christophides G.K., Christley S., Dialynas E., Butler R., Campbell K.S., Christophides G.K., Christley S., Dialynas E., Campbell K.S., Christophides G.K., Christley S., Dialynas E., Christophides G.K., Christley S., Dialynas E., Christley S., Dialynas E., Dialynas E., et al. VectorBase: a home for invertebrate vectors of human pathogens. Nucleic Acids Res. 2007;35:D503–D505. doi: 10.1093/nar/gkl960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.E., Searle S.M., Harris N., Gibson M., Lyer V., Richter J., Wiel C., Bayraktaroglir L., Birney E., Crosby M.A., Searle S.M., Harris N., Gibson M., Lyer V., Richter J., Wiel C., Bayraktaroglir L., Birney E., Crosby M.A., Harris N., Gibson M., Lyer V., Richter J., Wiel C., Bayraktaroglir L., Birney E., Crosby M.A., Gibson M., Lyer V., Richter J., Wiel C., Bayraktaroglir L., Birney E., Crosby M.A., Lyer V., Richter J., Wiel C., Bayraktaroglir L., Birney E., Crosby M.A., Richter J., Wiel C., Bayraktaroglir L., Birney E., Crosby M.A., Wiel C., Bayraktaroglir L., Birney E., Crosby M.A., Bayraktaroglir L., Birney E., Crosby M.A., Birney E., Crosby M.A., Crosby M.A., et al. Genome Biol. Vol. 3. 2002. Apollo: A sequence annotation editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liolios K., Tavernarakis N., Hugenholtz P., Kyrpides N.C., Tavernarakis N., Hugenholtz P., Kyrpides N.C., Hugenholtz P., Kyrpides N.C., Kyrpides N.C. The Genomes On Line Database (GOLD) v.2: A monitor of genome projects worldwide. Nucleic Acids Res. 2006;34:D332–D334. doi: 10.1093/nar/gkj145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T.M., Eddy S.R., Eddy S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T.H. Experimental studies of the regeneration of Planaria maculata. Arch. Entw. Mech. Org. 1898;7:364–397. [Google Scholar]
- Parra G., Bradnam K., Korf I., Bradnam K., Korf I., Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Randolph H. Observations and experiments on regeneration in planarians. Arch. Entw. Mech. Org. 1897;7:352–372. [Google Scholar]
- Rivas E., Eddy S.R., Eddy S.R. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb S.M., Sánchez Alvarado A., Sánchez Alvarado A. Identification of immunological reagents for use in the study of freshwater planarians by means of whole-mount immunofluorescence and confocal microscopy. Genesis. 2002;32:293–298. doi: 10.1002/gene.10087. [DOI] [PubMed] [Google Scholar]
- Robb S.M., Ross E., Sánchez Alvarado A., Ross E., Sánchez Alvarado A., Sánchez Alvarado A. SmedGD: The Schmidtea mediterranea genome database. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G.M., Yandell M.D., Wortman J.R., Gabor Miklos G.L., Nelson C.R., Hariharan I.K., Fortini M.E., Li P.W., Apweiler R., Fleischmann W., Yandell M.D., Wortman J.R., Gabor Miklos G.L., Nelson C.R., Hariharan I.K., Fortini M.E., Li P.W., Apweiler R., Fleischmann W., Wortman J.R., Gabor Miklos G.L., Nelson C.R., Hariharan I.K., Fortini M.E., Li P.W., Apweiler R., Fleischmann W., Gabor Miklos G.L., Nelson C.R., Hariharan I.K., Fortini M.E., Li P.W., Apweiler R., Fleischmann W., Nelson C.R., Hariharan I.K., Fortini M.E., Li P.W., Apweiler R., Fleischmann W., Hariharan I.K., Fortini M.E., Li P.W., Apweiler R., Fleischmann W., Fortini M.E., Li P.W., Apweiler R., Fleischmann W., Li P.W., Apweiler R., Fleischmann W., Apweiler R., Fleischmann W., Fleischmann W., et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A., Newmark P.A., Newmark P.A. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc. Natl. Acad. Sci. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A., Newmark P.A., Robb S.M., Juste R., Newmark P.A., Robb S.M., Juste R., Robb S.M., Juste R., Juste R. The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development. 2002;129:5659–5665. doi: 10.1242/dev.00167. [DOI] [PubMed] [Google Scholar]
- Slater G.S., Birney E., Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.D., Edgar R.C., Yandell M.D., Smith D.R., Celniker S.E., Myers E.W., Karpen G.H., Edgar R.C., Yandell M.D., Smith D.R., Celniker S.E., Myers E.W., Karpen G.H., Yandell M.D., Smith D.R., Celniker S.E., Myers E.W., Karpen G.H., Smith D.R., Celniker S.E., Myers E.W., Karpen G.H., Celniker S.E., Myers E.W., Karpen G.H., Myers E.W., Karpen G.H., Karpen G.H. Improved repeat identification and masking in Dipterans. Gene. 2007;389:1–9. doi: 10.1016/j.gene.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabenau A., McVicker G., Melsopp C., Proctor G., Clamp M., Birney E., McVicker G., Melsopp C., Proctor G., Clamp M., Birney E., Melsopp C., Proctor G., Clamp M., Birney E., Proctor G., Clamp M., Birney E., Clamp M., Birney E., Birney E. The Ensembl core software libraries. Genome Res. 2004;14:929–933. doi: 10.1101/gr.1857204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Morgenstern B., Morgenstern B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:W465–W467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Tzvetkova A., Morgenstern B., Tzvetkova A., Morgenstern B., Morgenstern B. AUGUSTUS at EGASP: using EST, protein and genomic alignments for improved gene prediction in the human genome. Genome Biol. 2006;7:S11–S18. doi: 10.1186/gb-2006-7-s1-s11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter J.C.M.D., Adams E.W., Myers P.W., Li R.J., Mural G.G., Sutton H.O., Smith M., Yandell C.A., Evans R.A., Holt J.D., Adams E.W., Myers P.W., Li R.J., Mural G.G., Sutton H.O., Smith M., Yandell C.A., Evans R.A., Holt J.D., Myers P.W., Li R.J., Mural G.G., Sutton H.O., Smith M., Yandell C.A., Evans R.A., Holt J.D., Li R.J., Mural G.G., Sutton H.O., Smith M., Yandell C.A., Evans R.A., Holt J.D., Mural G.G., Sutton H.O., Smith M., Yandell C.A., Evans R.A., Holt J.D., Sutton H.O., Smith M., Yandell C.A., Evans R.A., Holt J.D., Smith M., Yandell C.A., Evans R.A., Holt J.D., Yandell C.A., Evans R.A., Holt J.D., Evans R.A., Holt J.D., Holt J.D., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Yandell M., Bailey A.M., Misra S., Shu S., Wiel C., Evans-Holm M., Celniker S.E., Rubin G.M., Bailey A.M., Misra S., Shu S., Wiel C., Evans-Holm M., Celniker S.E., Rubin G.M., Misra S., Shu S., Wiel C., Evans-Holm M., Celniker S.E., Rubin G.M., Shu S., Wiel C., Evans-Holm M., Celniker S.E., Rubin G.M., Wiel C., Evans-Holm M., Celniker S.E., Rubin G.M., Evans-Holm M., Celniker S.E., Rubin G.M., Celniker S.E., Rubin G.M., Rubin G.M. A computational and experimental approach to validating annotations and gene predictions in the Drosophila melanogaster genome. Proc. Natl. Acad. Sci. 2005;102:1566–1571. doi: 10.1073/pnas.0409421102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell M., Mungall C.J., Smith C., Prochnik S., Kaminker J., Hartzell G., Lewis S., Rubin G.M., Mungall C.J., Smith C., Prochnik S., Kaminker J., Hartzell G., Lewis S., Rubin G.M., Smith C., Prochnik S., Kaminker J., Hartzell G., Lewis S., Rubin G.M., Prochnik S., Kaminker J., Hartzell G., Lewis S., Rubin G.M., Kaminker J., Hartzell G., Lewis S., Rubin G.M., Hartzell G., Lewis S., Rubin G.M., Lewis S., Rubin G.M., Rubin G.M. Large-scale trends in the evolution of gene structures within 11 animal genomes. PLoS Comput. Biol. 2006;2:e15. doi: 10.1371/journal.pcbi.0020015. [DOI] [PMC free article] [PubMed] [Google Scholar]