Abstract
Mitochondrial dysfunction is associated with many human diseases. There has not been a systematic genetic approach for identifying regulators of basal mitochondrial biogenesis and function in higher eukaryotes. We performed a genome-wide RNA interference (RNAi) screen in Drosophila cells using mitochondrial Citrate synthase (CS) activity as the primary readout. We screened 13,071 dsRNAs and identified 152 genes that modulate CS activity. These modulators are involved in a wide range of biological processes and pathways including mitochondrial-related functions, transcriptional and translational regulation, and signaling pathways. Selected hits among the 152 genes were further analyzed for their effect on mitochondrial CS activity in transgenic flies or fly mutants. We confirmed a number of gene hits including HDAC6, Rpd3(HDAC1), CG3249, vimar, Src42A, klumpfuss, barren, and smt3 which exert effects on mitochondrial CS activities in vivo, demonstrating the value of Drosophila genome-wide RNAi screens for identifying genes and pathways that modulate mitochondrial function.
Mitochondrial dysfunction is associated with a wide range of human diseases, including neurodegenerative diseases, cardiovascular disease, diabetes, aging, and cancer (Schapira 2006). This is due to diverse roles of mitochondria in ATP production, apoptosis regulation, thermogenesis, free radical production, and calcium homeostasis (Ryan and Hoogenraad 2007). The molecular mechanisms for regulating mitochondrial function and biogenesis are not fully understood. Two classes of transcriptional regulators implicated in mitochondrial biogenesis have been identified in mammalian cells in recent years (Scarpulla 2002, 2006). One class consists of transcriptional factors, including NRF1 (nuclear respiratory factor 1), NFE2L2 (also known as NRF2), and ESRRA (estrogen-related receptor alpha, also known as ERRα), that regulate basal mitochondrial biogenesis by binding to the promoters of a number of nuclear genes that specify mitochondrial functions. In vivo evidence from mice showed that homozygous deletion of NRF1 resulted in lethality between embryonic days 3.5 and 6.5 (Huo and Scarpulla 2001). These null blastocytes were defective in maintaining a mitochondrial membrane potential and had severely reduced mtDNA (mitochondrial DNA) levels (Huo and Scarpulla 2001). Moreover, NRF1 and NFE2L2 (NRF2) recognition sites have been found in a number of nuclear genes required for mitochondrial respiratory function (Scarpulla 2002). The second class of transcription regulators includes nuclear coactivators in the PPARGC1 (peroxisome proliferators-activated receptor γ, coactivator-1, formerly PGC1) family, such as PPARGC1A (formerly PGC-1α), PPARGC1B (formerly PGC-1β), and PPRC1 (peroxisome proliferators-activated receptor γ, coactivator related 1). The PPARGC1 (PGC1) family members do not bind DNA but regulate mitochondrial gene expression through their interaction with transcription factors and other coactivators, coordinating mitochondrial gene expression with signaling pathways and mediating adaptive regulation of mitochondrial biogenesis (Scarpulla 2006). Environmental signals, such as cold or exercise, induce the expression of the PPARGC1 (PGC1) family members, which in turn activate transcription factors (NRF1, NFE2L2, and ESRRA) to induce the expression of genes critical for mitochondrial function (Wu et al. 1999; Lehman et al. 2000; Lin et al. 2002; Meirhaeghe et al. 2003; Mootha et al. 2004; Schreiber et al. 2004). PPARGC1A (PGC-1α) also interacts with a number of nuclear hormone receptors involved in the regulation of metabolic pathways such as retinoic X receptors, thyroid hormone receptor, and the peroxisome proliferator-activated receptors (Knutti and Kralli 2001; Puigserver and Spiegelman 2003). Other regulators of mitochondrial biogenesis have also been identified, such as CAMK4 (calcium/calmodulin-dependent protein kinase IV), PRKAA (AMP-activated protein kinase, also known as AMPK), and NO (nitric oxide), and they appear to modulate mitochondrial biogenesis at least in part through PPARGC1A (PGC-1α) (Reznick and Shulman 2006).
Mitochondrial biogenesis has also been studied in the model organism Drosophila. Mitochondria are assembled by proteins encoded by either the mitochondrial genome or the nuclear genome. The entire mitochondrial DNA as well as nuclear genes encoding mitochondrial proteins are highly conserved between the mammalian and Drosophila system (Sanchez-Martinez et al. 2006). Thus, Drosophila has been successfully utilized as a model system to study various mitochondrial diseases (Sanchez-Martinez et al. 2006). A pathogenic mutation was identified in the Drosophila mitochondrial mt:ATPase6 gene, which encodes a subunit of complex V. This Drosophila mutant recapitulates key features of the human disorders caused by the same mutation and has been used as a model for mitochondrial encephalomyopathy (Celotto et al. 2006). The Drosophila technical knockout (tko) gene encodes the mitochondrial ribosomal protein S12. tko mutants exhibit various defects including defective response to sound, which has been used as a model for mitochondrial deafness (Toivonen et al. 2001; Jacobs et al. 2004). Drosophila mutants for tamas, encoding the mtDNA polymerase catalytic subunit, die in the late larval stage and show defects in locomotion and in the development of the visual system (Iyengar et al. 1999). The phenotype of tamas mutants is similar to lopo (low power) mutants, which die in the late larval stage and show defects in the developing visual system (Maier et al. 2001). lopo mutant flies contain a mutation in mitochondrial single-strand DNA-binding protein (mtSSB), which is required for strand displacement during mtDNA replication. The Drosophila homolog of NRF1 is erect wing (ewg). It has been characterized and was shown to be essential for neurogenesis and myogenesis (DeSimone and White 1993; DeSimone et al. 1996; Fazio et al. 2001). These cases provide compelling evidence in support of using Drosophila as a model system to study mitochondrial function and biogenesis, even though, so far, a PPARGC1A (PGC-1α) homolog has not been identified in Drosophila. In addition, there has not been a reported study employing a systematic genetic approach, for which Drosophila is well suited, to identify regulators of basal mitochondrial biogenesis and function in higher eukaryotes.
To identify new regulators of mitochondrial biogenesis and function, we performed a whole-genome RNA interference (RNAi) screen in Drosophila cells by using the activity of the mitochondrial enzyme Citrate synthase (CS) as the primary readout. The Drosophila system was chosen for its high efficiency of RNAi, lower genetic redundancy, and long history as a good genetic model for in vivo validation. CS catalyzes the first reaction in the Krebs cycle (TCA cycle) to convert acetyl-CoA and oxaloacetate to citrate (Fig. 1A). CS activity has been used as a common marker for mitochondrial oxidative capacity in preclinical and clinical research (Bruce et al. 2004; Kelley et al. 2002). It has been shown that CS activity and mtDNA content are positively correlated in muscle (Moyes et al. 1997; Wang et al. 1999). CS activity was also induced by PPARGC1A (PGC-1α) overexpression (Mortensen et al. 2006). Recently, a mutation in Citrate synthase was identified in Drosophila, and the mutants develop increased seizure susceptibility and exhibit reduced ATP levels consistent with disruption of mitochondrial energy production (Fergestad et al. 2006). After an RNAi screen with 13,071 dsRNAs followed by a confirmation step, we identified 152 genes that affect CS activity in the S2 cell line. The molecular functions of these genes are quite diverse, including mitochondrial-related functions, transcriptional and translational regulation, and signaling pathways. We selected a subset of hits among the 152 genes to further analyze their effect on mitochondrial CS activity in mutant or transgenic flies. We have confirmed that HDAC6, Rpd3(HDAC1), CG3249, vimar, Src42A, klumpfuss, barren, and smt3 modulate mitochondrial CS activity in vivo. Candidate genes identified in our genome-wide RNAi screen serve as a starting point for the identification and characterization of novel genes and pathways that modulate mitochondrial function.
Figure 1.
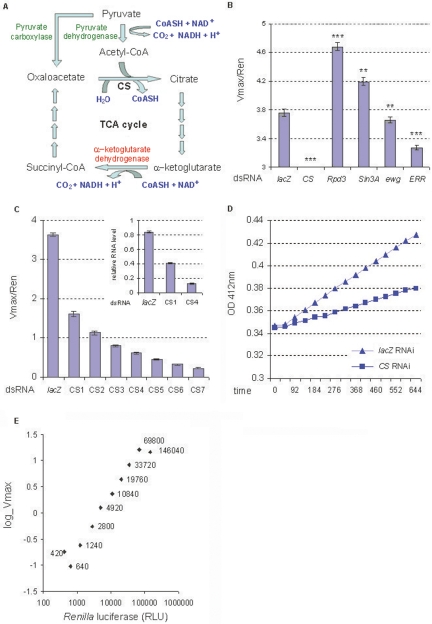
Development of a CS activity assay for genome-wide RNAi screen in S2 cells. (A) TCA cycle and pyruvate metabolism. The biochemical reactions catalyzed by Citrate synthase, Pyruvate dehydrogenase, and α-ketoglutarate dehydrogenase are shown. (B) The effects of dsRNAs of the known mitochondria regulators on the normalized CS activity (Vmax/Ren) in S2 cells (n = 48). The value of CS is 0.22; 5 μg/mL dsRNA was used. **P < 0.01, ***P < 0.001. Error bars indicate the standard errors. (C) Dose response of the CS activity to Citrate synthase RNAi (n = 48). The normalized CS activities were reduced by RNAi in a dose-dependent manner. Seven dosages of Citrate synthase dsRNA were used, from CS1 to CS7, being 0.5, 1, 2, 4, 8, 16, 32 μg/mL/106 cells. The relative Citrate synthase RNA expression levels after RNAi are shown (upper right). (D) The stability of the CS activity in S2 cells. The CS activity level was represented by the Vmax; 0.5 μg/mL dsRNA was used. Optical densities (OD) at 412 nm for cell lysates from lacZ RNAi and Citrate synthase RNAi are shown on the Y-axis. The X-axis shows time in seconds, and data were captured every 46 sec. (E) The linear range of the CS activity in S2 cells (n = 48). The Y-axis shows CS activity (Vmax). The X-axis shows the Renilla luciferase activity (RLU) of cell lysates. The Renilla luciferase activities are labeled next to each data point. Log scales are applied to both X- and Y-axes.
Results
Development of a cell-based assay
A cell-based assay was developed for an RNAi screen. We used a S2 cell line stably transfected with a Renilla luciferase reporter gene driven by a basal promoter. The CS activity is represented by Vmax of the CS-catalyzed citrate synthesis reaction from oxaloacetate and acetyl-CoA (see Methods). The Renilla luciferase activity is represented by the relative light unit (RLU) (see Methods). The Renilla luciferase activity was used to normalize the CS activity, correcting the effect of RNAi on cell growth or viability. To validate the CS assay, the known mitochondrial regulators were evaluated. As expected, CS activities were highly reduced in the cells transfected with Citrate synthase dsRNA (Fig. 1B). Sin3A, a cofactor of Rpd3 (Drosophila HDAC1), was reported to negatively regulate mitochondrial mass in Drosophila cells (Pile et al. 2003). CS activities were significantly elevated in the cells transfected with Rpd3 dsRNA and Sin3A dsRNA (Fig. 1B). In contrast, CS activities were significantly reduced in the cells transfected with dsRNA of ERR (estrogen related receptor) or ewg (Drosophila NRF1), two known positive regulators of mitochondrial biogenesis (Fig. 1B). These results demonstrated that CS activity positively correlates with mitochondrial density/function and can be modulated by the known modulators of this process. Thus, CS activity is an appropriate indicator of mitochondrial function in the Drosophila system. To quantitatively modulate CS activity, RNAi was performed with different doses of Citrate synthase dsRNA. CS activity was reduced by Citrate synthase RNAi in a dose-dependent manner (Fig. 1C). The reduction of CS activity correlated with the reduction of Citrate synthase RNA expression levels (Fig. 1C, upper right). We tested the stability and the linearity of the CS assay. CS enzyme activity was stable for at least 644 sec (Fig. 1D) under the assay conditions. This time window is long enough for 15 readings of 384 samples (one 384-well plate), which provides sufficient data points for calculating the Vmax. There was also a good linear range of CS activity when using the amounts of cell lysates having the Renilla luciferase activity ranging from 640 to 69,800 RLU (Fig. 1E).
Whole genome RNAi screen
A genome-wide RNAi screen was then carried out in S2 cells using the validated assay described earlier, followed by a confirmation screen. The RNAi screen was carried out with a library of 13,071 dsRNAs. Duplicates were used for each dsRNA and normalized Z-scores (NZ, see Methods) were calculated. Primary hits were selected if NZ scores of both replicates were larger than 2 or smaller than −2. Additional hits with only one replica having NZ larger than 3 or smaller than −3 were also selected for the confirmation screen. A total of 821 primary hits, ∼6% of the collection, were selected from the primary screen (Fig. 2A). Among them, 573 of the 821 selected genes have human homologs defined as having BLASTP E-value ≤−10. A confirmation screen was performed using six replicas for each dsRNA. Citrate synthase and lacZ dsRNAs were included as the positive and negative controls, respectively. The P-value of the CS activity in cells transfected with individual dsRNA was calculated relative to the lacZ dsRNA controls. As expected, CS activities were reduced by 75%–85% in the cells transfected with Citrate synthase dsRNA compared with cells transfected with lacZ dsRNA (Fig. 2B). From the confirmation screen, a total of 153 primary hits were verified. Among them, there are 76 hits whose dsRNAs led to up-regulation of CS activity (P < 0.05). The fold change of CS activities in cells transfected with these dsRNAs over the lacZ control ranged from 1.15 to twofold (Fig. 2B). There are two hits (dsRNA fragments) corresponding to one gene CG4849 (Supplemental Table S1). For those hits whose dsRNAs led to down-regulation of CS activities, 77 hits had a P-value <0.01, their fold change of CS activities over the control ranged from 0.13 to 0.8 (Fig. 2B). Here, a P-value of 0.01 was chosen as the cutoff since there were three lacZ RNAi controls having P-values between 0.01 and 0.05 (Fig. 2C).
Figure 2.
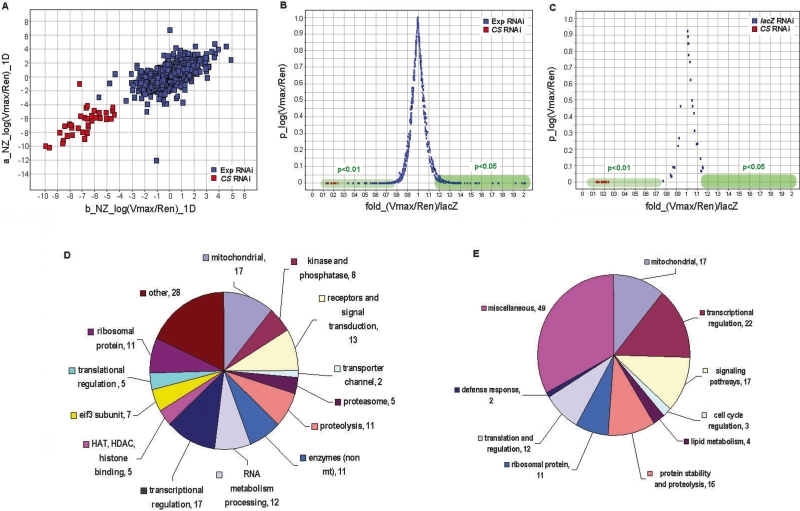
Whole genome RNAi screen. (A) The scatter plot shows the data from the first 12 duplicated plates of the primary screen. The X- and Y-axes represent NZ1D for replica b and replica a, respectively. Each spot represents the result of one dsRNA. The Citrate synthase dsRNA concentration was used as 5 μg/mL. “Exp RNAi” indicates experimental RNAi, “CS RNAi” indicates control Citrate synthase RNAi. (B) The result of confirmation screen for the primary hits. The X-axis represents average fold change of normalized CS activity induced by a dsRNA (six replicates) against the lacZ RNAi controls. The Y-axis represents P-value. Hits were selected based on their P-values against lacZ RNAi controls. The hits that negatively affect CS activities (their RNAi result in an increase in CS activity) were selected if their P < 0.05. The hits that positively affect CS activities (their RNAi result in a decrease in CS activity) were selected if their P < 0.01. The Citrate synthase dsRNA concentration was the same as in A. (C) The distribution of the lacZ RNAi controls in the confirmation screen as shown in B. (D) The classification of the confirmed hits by their molecular functions. (E) The classification of the confirmed hits by their biological processes and pathways.
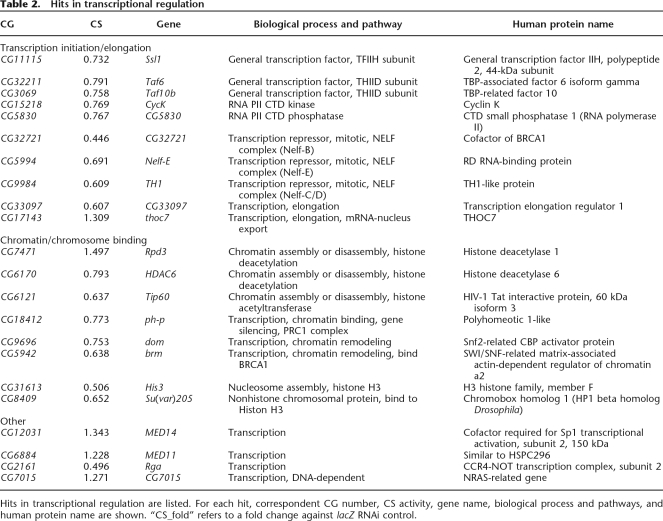
A total of 153 hits were identified from the whole-genome RNAi screen (see Supplemental Table S1). To provide a global view of what classes and pathways these gene hits are in, the hits were classified by their associated Gene Ontology annotation or InterPro protein domain (Fig. 2D). The hits fall into diverse classes of proteins such as kinases, phosphatases, receptors, other enzymes, and transcription regulators. They are involved in a number of pathways and biological processes (Fig. 2E). Among the 153 hits, 17 genes have mitochondria-related functions (Table 1), 22 genes have functions in transcriptional regulation (Table 2), and 17 genes have functions in signaling pathways (Table 3). These are the top three categories containing relatively more hits than other categories. The discovery of 17 genes with mitochondrial functions demonstrates a statistically significant enrichment of hits in this functional category (P < e−5, hypergeometric test), since there are only 2%–3% genes encoding proteins associated with mitochondrial function in the whole genome (Sardiello et al. 2003; Catalano et al. 2006). With the exception of the mitochondrial function category, genes in other categories do not seem to be overrepresented, indicating the specificity of our screen. Moreover, for several protein complexes, multiple subunits were identified, indicating the validity of the screen. These included two subunits of Pyruvate dehydrogenase, two subunits for α-ketoglutarate dehydrogenase, two subunits of ATP synthase, three subunits of NELF (negative elongation factor) complex, and two subunits of TFIID (Tables 1, 2).
Table 1.
Hits in mitochondrial function
Hits with mitochondrial function are listed. For each hit, correspondent CG number, CS activity, gene name, biological process and pathways, and human protein name are shown. “CS_fold” refers to a fold change against lacZ RNAi control. “mt” refers to mitochondrial.
Table 2.
Hits in transcriptional regulation
Hits in transcriptional regulation are listed. For each hit, correspondent CG number, CS activity, gene name, biological process and pathways, and human protein name are shown. “CS_fold” refers to a fold change against lacZ RNAi control.
Table 3.
Hits in signaling pathways and major biological processes
Hits in signaling pathways and major biological processes are listed. For each hit, correspondent CG number, CS activity, gene name, biological process and pathways, and human protein name are shown. “CS_fold” refers to a fold change against lacZ RNAi control.
Hits encoding mitochondrial proteins
Among the 17 identified hits in the mitochondrial function category, five of them are involved in the TCA cycle and pyruvate metabolism (Table 1). These include E1β (CG11876) and E3 (CG7430) subunits of Pyruvate dehydrogenase (PDH), and E1 (CG11661) and E3 (CG7430) subunits of α-ketoglutarate dehydrogenase. These enzyme complexes have similar compositions, consisting of E1, E2, and shared E3 subunits. They also catalyze very similar chemical reactions, utilizing CoASH and NAD+ and yielding CO2 and NADH (Fig. 1A). The reaction catalyzed by CS is the first rate-limiting step in the TCA cycle which is regulated by substrate availability and product inhibition. RNAi against PDH subunits resulted in an up-regulation of CS activity, which could be due to the compensation caused by the reduction of acetyl-CoA, a substrate for CS (Fig. 1A). Similarly, RNAi treatment of Pyruvate carboxylase resulted in an up-regulation of CS activity (Table 1). Pyruvate carboxylase catalyzes the conversion of pyruvate to oxaloacetate which is also a substrate for CS (Fig. 1A). On the other hand, RNAi against α-ketoglutarate dehydrogenase resulted in a reduction of CS activity (Table 1). This could be due to product inhibition. α-ketoglutarate dehydrogenase catalyzes a biochemical reaction downstream from CS in the TCA cycle (Fig. 1A). The reduction of α-ketoglutarate dehydrogenase will indirectly lead to accumulation of citrate, a product of CS, which will in turn inhibit CS activity (Fig. 1A). The identification of genes involved in both mechanisms of CS regulation, substrate availability, and product inhibition indicates that the screen was sensitive enough to uncover the enzymes that regulate CS activity through metabolic intermediates.
Among the rest of the identified hits associated with mitochondrial function, a number of them have been previously characterized in the Drosophila system. shibire was identified as a positive modulator of CS activity. shibire is a homolog of yeast dynamin 1 and human dynamin 1-like, which is one of the major regulators for mitochondrial fission (Chen and Chan 2005). Drosophila homolog of mouse Slc25a4 (also known as ANT), sesB (stress sensitive B), was also identified as a positive modulator of CS activity. Slc25a4 (ANT) exchanges matrix ATP for cytosolic ADP across the inner mitochondrial membrane, providing mitochondrial energy to the cytosol (Klingenberg 1992). Homozygous Slc25a4 knockout mice develop mitochondrial myopathy and hypertrophic cardiomyopathy with increased levels of blood lactate and compensatory mitochondrial proliferation (Graham et al. 1997). In agreement with its mitochondrial function in mice, temperature-sensitive sesB mutants show complete loss of synaptic transmission in the visual system (Zhang et al. 1999; Rikhy et al. 2003). Moreover, we have identified tamas, mtSSB, and mitochondrial ribosomal protein S18C as modulators of CS activity. The Drosophila mutants of these genes have been characterized and shown to be good mitochondrial disease models (Sanchez-Martinez et al. 2006).
Rpd3(HDAC1) and HDAC6 modulate mitochondrial functions
Among the hits in the transcriptional regulation category, Rpd3(HDAC1) and HDAC6 were identified as positive and negative modulators of CS activity, respectively. Compared with the lacZ RNAi controls, CS activity was increased by 50% in the cells treated with Rpd3 RNAi and was reduced by 20% in the cells treated with HDAC6 RNAi (Fig. 3A). To evaluate the significance of Rpd3 and HDAC6 in regulating mitochondrial function, transgenic flies bearing the inducible RNAi constructs for Rpd3 or HDAC6 were used to knock down Rpd3 or HDAC6 expression in vivo. Global knockdown of Rpd3 or HDAC6 in these transgenic Drosophila leads to developmental lethality; however, muscle-specific knockdown of either Rpd3 or HDAC6 gives rise to viable and fertile adults. The knockdown of Rpd3 and HDAC6 was efficient, as seen by highly reduced Rpd3 and HDAC6 protein levels in these transgenic flies (Fig. 3B,C).
Figure 3.
Rpd3(HDAC1) and HDAC6 modulate mitochondrial functions in vivo. (A) The CS activity in S2 cells transfected with Rpd3 and HDAC6 RNAi (n = 6). The lacZ RNAi acts as a negative control and Citrate synthase RNAi as a positive control. P-value was calculated against lacZ RNAi control group. (B) Rpd3 protein was highly reduced in the transgenic Rpd3 RNAi flies as shown by Western blot. Lysates from the Rpd3 RNAi flies and control siblings were blotted with the antibody against Rpd3 and Tubulin. The Tubulin levels were used as loading controls. (C) HDAC6 protein was highly reduced in the transgenic HDAC6 RNAi flies as shown by Western blot. Lysates from the transgenic HDAC6 RNAi and control siblings were blotted with the antibodies against HDAC6 and Tubulin. The Tubulin levels were used as loading controls. (D) The CS activity in the transgenic Rpd3 RNAi flies and control siblings (n = 8). (E) The CS activity in the transgenic HDAC6 RNAi flies and control siblings (n = 8). (F) The COX activity in the transgenic Rpd3 RNAi flies and control siblings (n = 8). (G) The COX activity in the transgenic HDAC6 RNAi flies and control siblings (n = 8). *P < 0.05, **P < 0.01, ***P < 0.001. Error bars indicate the standard errors.
In the transgenic flies with muscle-specific knockdown of HDAC6, a 19% reduction in CS activity was observed compared with their sibling controls (Fig. 3E). To determine whether HDAC6 inhibition affects the function of other components in mitochondria in addition to CS activity, cytochrome c oxidase (COX) activity, indicative of electron transport chain function, was measured. A 34% reduction of COX activity was observed in the transgenic flies with muscle-specific knockdown of HDAC6 compared to their sibling controls (Fig. 3G). These results indicate that HDAC6 positively modulates mitochondrial functions in vivo, consistent with the results obtained in S2 cells (Fig. 3A).
We initially analyzed the CS and COX activity in the adult transgenic flies with muscle-specific knockdown of Rpd3. Although COX activity was elevated by 47%, the change of the CS activity was inconclusive (data not shown). We then analyzed the larvae of the transgenic flies with global knockdown of Rpd3 which is adult-lethal. A 60% increase in CS activity and a threefold increase in COX activity were observed in these larvae compared to their sibling controls (Fig. 3D,F). In agreement with results from S2 cells (Fig. 3A), Rpd3 is seen to negatively modulate mitochondrial CS and COX activity in vivo.
Here, we provide the first in vivo evidence for HDAC1 and HDAC6 in modulating mitochondrial enzyme activities, although the identities of the HDAC1 or HDAC6 substrates that mediate this modulation in our system are not known.
In vivo validation for CG3249, vimar, Src42A, klumpfuss, smt3, and barren
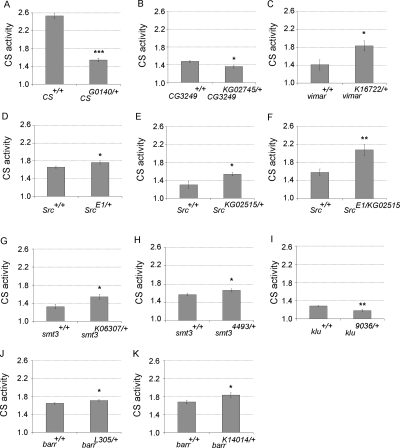
Among the identified hits in signaling pathways and major biological processes, 23 hits were selected to be analyzed in flies, taking advantage of the publicly available large collection of Drosophila mutants (Supplemental Table S2). Six of them were shown to have significant effects on CS activity, including CG3249, vimar, Src42A, klumpfuss, smt3, and barren (Fig. 4).
Figure 4.
The CS activities in heterozygous mutants (male flies used except for A) with mutation in Citrate synthase (A, female flies), CG3249 (B), vimar (C), Src42A (D–F), smt3 (G,H), klumpfuss (I), barren (J,K). n = 8. *P < 0.05, **P < 0.01, ***P < 0.001. Error bars indicate the standard errors.
Citrate synthase heterozygous mutants were first tested as a positive control. Average CS activity was reduced 39% in heterozygous Citrate synthase mutants compared to their wild-type siblings (Fig. 4A). Here, female adult flies were used since the Citrate synthase gene locus is on the X chromosome, while, in other experiments, male adult flies were used.
In the CG3249 heterozygous mutants, CS activity was reduced 8% (Fig. 4B). CG3249 encodes a homolog of the A kinase anchor protein (AKAP). A role for AKAP in regulating mitochondria function has been reported previously (Horbinski and Chu 2005; Livigni et al. 2006). AKAP anchors protein kinase A (PKA) to mitochondria where it increases PKA-dependent phosphorylation of the pro-apoptotic protein BAD and enhances cell survival (Harada et al. 1999; Affaitati et al. 2003). AKAP was also shown to target the complex containing PTPN21 (formerly PTPD1) and SRC tyrosine kinase to the mitochondria and enhance SRC-dependent phosphorylation of mitochondrial substrate proteins (Cardone et al. 2004; Livigni et al. 2006). We showed that CG3249 heterozygous mutants had reduced CS activity (Fig. 4B), consistent with a positive role for AKAP in regulating mitochondrial function.
In the vimar heterozygous mutants, a 30% increase in CS activity was observed (Fig. 4C). The human homolog of vimar is RAP1GDS1, a guanine exchange factor (GEF) that activates RAP1. RAP1 is a small G protein involved in the regulation of the actin cytoskeleton (Bos 2005). A number of GEFs have been identified for RAP1. RAPGEF3, or EPAC, was observed to localize to mitochondria and activate RAP1 by direct binding of cAMP in a PKA-independent manner (Qiao et al. 2002; Wang et al. 2006). Here, we identified another RAP1-GEF in modulating mitochondrial function, supporting a role for RAP1 in regulating mitochondrial function.
In two different alleles of heterozygous Src42A mutants, CS activities were increased 7% and 18% (Fig. 4D,E). CS activities were further increased 33% in Src42A mutants that are trans-heterozygous for these two mutant alleles compared to heterozygous siblings (Fig. 4F). Src42A encodes a tyrosine kinase and is a homolog of vertebrate SRC family tyrosine kinase. Drosophila Src42A was shown to regulate RTK (receptor tyrosine kinases) signaling and JUN kinase (JNK) activity (Lu and Li 1999; Tateno et al. 2000). The first evidence of Src mitochondrial localization was found in rat brain (Salvi et al. 2002). Under our experimental conditions, Src42A appears to negatively affect CS activity, supporting a role of Src kinase in regulating mitochondrial function.
In the klumpfuss heterozygous mutants, CS activity was reduced 8% (Fig. 4G). klumpfuss was shown to promote programmed cell death by down-regulation of cell survival signals mediated by the dEGFR/dRas1 signaling pathway (Rusconi et al. 2004). A connection between klumpfuss and mitochondrial function has not been reported previously.
In the smt3 heterozygous mutants with two different alleles, CS activities were increased by 17% and 6% (Fig. 4G,H). smt3 encodes the Drosophila member of SUMO (small ubiquitin-related modifier) proteins. There are four SUMO isoforms in mammals and one SUMO found in Drosophila. SUMO proteins conjugate to other proteins via a very similar mechanism to that of the ubiquitin system. SUMO modification appears to occur on diverse targets and plays important roles in protein stability, protein targeting, and other processes (Dohmen 2004). The first mitochondrial target identified for SUMO modification is DENR (also known as DRP1), which functions in mitochondrial fission. DENR Drosophila ortholog shibire was also identified in our screen (Table 1). DENR was shown to colocalize with SUMO1 and to be sumoylated in mammalian cells (Harder et al. 2004). Recently, SUMO protease SENP5 was shown to catalyze the cleavage of SUMO1 from DENR and to maintain mitochondrial morphology and function (Zunino et al. 2007). Here, we show that CS activity was increased in the smt3 heterozygous mutants, providing additional support for sumoylation in regulating mitochondrial function.
In two different barren heterozygous mutants, the CS activities were increased 4% and 9% (Fig. 4J,K), respectively. barren encodes a subunit of condensin, a complex required for chromosome condensation (Hirano et al. 1997; Legagneux et al. 2004). barren was first characterized in Drosophila, and homozygous barren mutants were embryonic lethal with a severe loss of PNS neurons (Kania et al. 1995; Bhat et al. 1996). The nonmitotic function has not yet been reported for barren, and further study is required to understand the specificity of barren on modulating CS activity.
Discussion
Described here is a genome-wide screen in higher eukaryotes to systematically search for modulators of basal mitochondrial function using CS activity as a readout. A total of 153 gene hits were identified from the screen. We also identified several pathways with multiple hits, including pathways with mitochondrial function, as well as pathways which have not been previously implicated in mitochondrial function.
Two genes (CG3249 and Pka-C2) in the PKA pathway were identified, consistent with the fact that this pathway is known to participate in mitochondrial biogenesis (Feliciello et al. 2005). The NF-κB family members have been shown to activate mitochondrial genes (Le Bras et al. 2006). We identified cactin (IκB) as a negative modulator of CS activity and CG7457 (NFκB) as a positive modulator of CS activity. This is in agreement with their known functions in which IκB proteins sequester NF-κB proteins and prevent NF-κB proteins from activating transcription of their targets. We also identified two genes in NO (nitric oxide)-mediated pathways, which have also been shown to participate in regulating mitochondrial biogenesis (Reznick and Shulman 2006). One Guanylyl cyclase (Gyc-89Da) was identified as a positive modulator of CS activity in the screen. Guanylyl cyclase was shown to mediate PPARGC1 (PGC-1α) activation of mitochondrial biogenesis and to be activated by NO (Nisoli et al. 2004, 2005). Three genes in the small GTPase-mediated signaling pathway were identified, and a link between the genes in this pathway and mitochondrial function has been reported (Qiao et al. 2002; Wang et al. 2006). Furthermore, a number of genes in metabolic pathways were identified such as Tre1 and Fas, which are known for their mitochondrial roles. Tre1 encodes the pineal hormone receptor with well-known mitochondrial-related functions such as stimulating antioxidant enzymes, increasing the efficiency of the electron transport chain, and promoting ATP synthesis (Martin et al. 2002; Leon et al. 2004). Fas encodes Fatty acid synthase, which is the only protein capable of synthesizing long-chain fatty acids. It utilizes acetyl-CoA, which is metabolized from citrate, a product of CS exported from mitochondria (Kuhajda 2006). Fas was identified as a positive modulator of CS activity. It is possible that the reduction of Fatty acid synthase by Fas RNAi results in an accumulation of acetyl-CoA and hence citrate, which will inhibit CS activity via product inhibition mechanism. We also identified three genes which are known to be involved in cell cycle regulation. It was proposed that a low-energy cell cycle checkpoint monitors the metabolic activity of the mitochondria before committing to another round of cell division (McBride et al. 2006). Other pathways or biological processes with multiple hits identified were protein stability and proteolysis, RNA metabolism, and defense responses (Table 3).
More importantly, we identified several novel modulators of CS activity and investigated their roles in modulating CS activity in vivo. These include CG3249, vimar, Src42A, klumpfuss, smt3, barren, and two HDACs, Rpd3(HDAC1) and HDAC6. HDAC1 has been reported in regulating mitochondrial function in vitro (Pile et al. 2003), while HDAC6 has not been shown previously to be involved in mitochondrial function. However, several HDAC6 substrates are known to play roles in mitochondrial function, including Hsp90 and tubulin. HDAC6 functions as an HSP90 deacetylase and is required for HSP90-dependent maturation of glucocorticoid receptor (GR) (Kovacs et al. 2005; Murphy et al. 2005). GR was shown to localize in mitochondria and stimulate mitochondrial biogenesis and function, including COX activity (Weber et al. 2002; Scheller and Sekeris 2003; Psarra et al. 2005). HDAC6 also deacetylates α-tubulin and destabilizes the microtubular network, but it does not affect tubulin protein level (Hubbert et al. 2002; Matsuyama et al. 2002; Haggarty et al. 2003; Zhang et al. 2003; Iwata et al. 2005; Kovacs et al. 2005). It is known that mitochondria require an intact microtubular network to function (Appaix et al. 2003). Previously, mammalian HDAC5 was shown to negatively regulate mitochondrial biogenesis in cells, and mammalian HDAC7 has been reported to have a mitochondrial localization (Czubryt et al. 2003; Bakin and Jung 2004). The Drosophila HDAC4 is the ortholog of mammalian HDAC5; however, this species does not have the ortholog of mammalian HDAC7. The absence of fly HDAC4 as a hit may be explained by the differences in genome complexity between the Drosophila and mammalian systems and by the potential overlapping functions of HDACs (Gallinari et al. 2007). Humans have a total of 11 HDACs for Class I and Class II. Among them, there are four HDACs for Class I (HDAC1, 2, 3, 8), six HDACs for Class II (HDAC4, 5, 6, 7, 9, 10), and HDAC11 shares similarity with both Class I and II (Dokmanovic and Marks 2005). In Drosophila, only five HDACs were found for Class I and Class II, with Rpd3(HDAC1) and Hdac3 for Class I, HDAC4 and HDAC6 for Class II, and HDAC11 for both Class I and II (Barlow et al. 2001; Gao et al. 2002).
Although the current screen was capable of yielding novel modulators of CS activity, there are a number of potential limitations that should be recognized. Some hits altered CS activity in the opposite direction of expectation based on previously characterized functions. For example, treatment of S2 cells with RNAi directed against either mtSSB, encoding the mitochondrial single-strand DNA-binding protein required for mtDNA replication (Maier et al. 2001), or mRpS18C, encoding a mitochondrial ribosomal protein required for synthesis of mitochondrial proteins, resulted in an increase, rather than a decrease, in CS activity. This could be due to a compensation or retrograde response from mitochondria, a mechanism that seems to be conserved from yeast to mammals. Yeast or mammalian cells, which have lost their mtDNA, compensate for mitochondrial dysfunction by up-regulation of a defined set of metabolic genes, including CS (Traven et al. 2001; Miceli and Jazwinski 2005). In addition, the CS RNAi screen may also miss some true modifiers due to knockdown efficiency and assay sensitivity. For practical reasons, fixed dsRNA dosage and assay time were used for the screen. However, it is known that the knockdown kinetics and dose requirements for dsRNA are not the same for all genes. This could be one of the reasons why the homologous genes of some of the best characterized mammalian regulators of mitochondrial biogenesis such as NRF1 and CAMK4 were not identified in the screen. For instance, Drosophila NRF1 homolog ewg (only one NRF homolog found in Drosophila) was not identified as a hit in the screen, but it behaved as a positive modulator of CS activity with relatively small effect in the assay development (Fig. 1B). Such a small change might be due to an indirect effect of NRF1 on the CS activity, since the NRF1-binding site was not found on the CS gene promoter (Kraft et al. 2006). Furthermore, even though the basal promoter-driven Renilla luciferase was used to normalize the cell growth or viability in the screen, it is entirely possible that an alteration of a gene in general transcription or translation machinery could have qualitatively similar but differential effects on the CS and Renilla luciferase activity, thus giving false positives. For this reason, we analyzed the Renilla luciferase activity separately, and the hits with 50% lower Renilla luciferase activity compared to lacZ RNAi controls were highlighted for potential false positives (Supplemental Table S1). Moreover, in our primary screen, CS activity, a single enzyme readout was used as an indicator for mitochondrial function. Thus, it is likely to miss some modifiers that do not affect CS activity but rather other components in mitochondria such as oxidative phosphorylation and fatty acid synthesis.
Finally, the in vivo validation for most of our hits was done with heterozygous mutants. This approach makes it feasible to test large numbers of mutations in parallel. However, heterozygosity is expected to reduce gene dosage by no more than 50% and will only validate those genes that are haplo-insufficient with respect to CS activity. For example, we observed that heterozygous Rpd3(HDAC1) flies do not have any associated developmental or adult phenotypes, and a change in CS activity was not observed (Supplemental Table S2). However, constitutive expression of Rpd3 RNAi leads to developmental lethality and a greater knockdown of Rpd3 expression; in this case a clear up-regulation of CS activity was observed. On the other hand, if a small effect in CS activity is seen in heterozygotes, a larger effect is expected in homozygotes or trans-heterozygotes. This was seen in the experiments with multiple alleles of Src42A. The Src42A trans-heterozygous mutants have 33% higher CS activity compared to the Src42A single-heterozygous mutants (Fig. 4F), while the single-heterozygous mutants have only 7% increase in CS activity compared to their wild-type control siblings (Fig. 4D). Despite the limitation in validating hits using heterozygous mutants, we observed reproducible effects on CS activity in the heterozygous mutant flies for the six gene hits (Fig. 4B–K). Haplo-insufficient genes often encode transcription factors, not signaling proteins. There are no haplo-insufficient phenotypes reported for these genes in the literature. The finding of haplo-insufficient phenotypes of these mutants on CS activity is likely attributable to the highly quantitative nature of the CS assay.
In summary, this is the first study that identifies novel modulators of basal mitochondrial function using an unbiased genomic approach in higher eukaryotes. The results from this study establish several new proteins and pathways that modulate mitochondrial function. This study also provides important new insights in the regulation of mitochondrial biogenesis and function as well as potential therapeutic targets for mitochondrial dysfunction diseases.
Methods
Whole genome RNAi screen
The dsRNA collection used for these studies was obtained from Ambion, Inc. This collection has normalized dsRNA concentration in each well and represents 13,071 annotated genes from the Drosophila genome, including all fly genes with human homologs. It was reformatted from the original 96-well plates to 384-well plates. The entire collection consists of thirty-six 384-well plates. RNAi was performed for 72 plates (36 plates, in duplicate). Each well contains 5 μL of dsRNA at a concentration of 50 ng/μL. The control dsRNAs were synthesized from the PCR amplicon collection by Heidelberg Fly Array (Hild et al. 2003) using the Megascript RNAi kit (Ambion). The amplicons selected were HFA18328 (Csyn), HFA08696 (HDAC1/Rpd3), HFA07552 (Sin3A), HFA18773 (ewg), and HFA10840 (ERR). Before the screen, 5 μL (50 ng/μL) of control dsRNA was loaded in each empty well; four replicates were used in each plate. In order to normalize for RNAi-mediated gene knockdown effects on cell growth and viability, a stable cell line bearing a Renilla luciferase reporter construct was used in the experiments. Renilla luciferase activity (Ren) was used to normalize CS activity. The Renilla luciferase reporter gene is driven by a basal promoter of the hsp70 gene of phRL–CMV plasmid. This plasmid was generated by replacing the CMV promoter with basal promoter of hsp70 gene.
For the RNAi experiment, S2 cells were harvested, washed once in 1× PBS and resuspended in serum free medium. Ten thousand cells in 10 μL serum free medium were loaded in each well containing dsRNA. After 1-h incubation, 30 μL complete medium was added to each well, the plates were sealed with gas permeable sheets (ABgene) and incubated at 25°C. At day 5, the cells were pelleted by centrifugation at 2000 rpm for 2 min, and medium was removed using a Biomek FX liquid handler. Plates were sealed and the cells were frozen at −80°C until use. In each well, cell pellet was lysed in 60 μL Renilla lysis buffer for 30 min. An aliquot of 20 μL cell lysate was used for the CS activity assay, 20 μL for the luciferase assay. To confirm the hits, dsRNAs of 573 hits were first cherry-picked into two 384-well plates, then aliquoted into six replicate plates with 5 μL (50 ng/μL) of dsRNA per well. The confirmation screen was performed in the same way as whole genome screen except that the cells were harvested at day 6 after RNAi treatment. The molecular information for 153 hits was batch-downloaded from flybase (http://flybase.bio.indiana.edu). Here, the GO term was only used as a classification tool.
CS activity assay
For the whole genome screen and the confirmation screen, cells (stable cell line) were lysed in Renilla lysis buffer (Promega) and assays were performed in 384-well plates (Falcon). CS activity was measured using 20 μL cell lysate in a total of 70 μL reaction solution containing 0.1 mM DTNB (Sigma), 0.3 mM acetyl-CoA (Roche), 1 mM oxaloacetate (Sigma), and 50 mM Tris pH 8.0 (USB). Oxaloacetate was added just prior to the measurement. The absorbance was measured at 412 nm every 46 sec for 15 time points using the Envision 2100 (PerkinElmer). Vmax (SLOPE) and R2 (RSQ) were calculated using Microsoft EXCEL. There were total 27,648 wells assayed for CS activity in the primary screen: 27,483 wells (99.4%) had R2 > 0.99, 79 wells had R2 between 0.98 and 0.99, 37 wells had R2 between 0.97 and 0.98, 49 wells had R2 < 0.97. For the 49 wells with R2 < 0.97, data were visually inspected, and the data points located in the range with best linearity were selected for estimating Vmax and calculating R2. After this manual process, the final R2 of the 49 wells ranged from 0.04 to 1.0. However, none of the 49 wells were included in the final primary hit list, due to their poor ranking of Vmax values.
Other than the primary and confirmation screens, CS assays were carried out in 96-well plates (Costar). Cells or single flies were lysed in extraction buffer containing 0.1% Triton X-100, 1 mM EDTA, and 20 mM HEPES, pH 7.2 (GIBCO). For cells in 96-well plates, 100 μL extraction buffer was used for each well. For individual flies, 250 μL extraction buffer was used for homogenization, and supernatant was transferred into one well of a 96-well plate. Eight individual flies were analyzed for each fly genotype, providing eight biological replicates. Ten microliters of cell or fly lysate was used for each CS assay reaction. Total reaction volume was 100 μL. Absorbance was measured at 412 nm every 15 sec for 10 min (at 25°C) using the kinetic mode of a Molecular Devices plate reading spectrometer. Three microliters of the lysate was used for total protein concentration measurement (Bio-Rad). The CS activity was normalized to total protein concentration.
Renilla luciferase assay
To measure Renilla luciferase activity, the Renilla luciferase assay system (Promega) was used for the primary screen. Dual-Glo Luciferase Assay system (Promega) was used for the confirmation screen. Experiments were performed according to manufacturer’s instructions. Luciferase activity was measured using an Envision 2100 (PerkinElmer) and presented as relative light unit (RLU).
Screen data analysis
From the primary screen, there were two data values for each well, the Vmax of CS activity and the Renilla luciferase activity (Ren). The Vmax was normalized by Ren such that the ratio of Vmax/Ren was calculated for each well. The Vmax/Ren value was normalized by a two-dimensional normalization scheme. The first dimension normalization (1D) was performed based on plate median of screened 384-well plates. Thus, the formula for calculating the 1D normalized value (denoted as x1D) is x1D = log x′1D, where x′1D = (Vmax/Ren)/medianplate(Vmax/Ren). The normalized Z score (NZ) was calculated for every x1D. The formula used for calculating NZ1D is: NZ1D = (x1D − median(x1D))/MAD(x1D). MAD(x1D) is the Median Absolute Deviation of x1D and is equal to 1.4826 × median (|x1D − median(x1D)|). After 1D normalization, the second dimension normalization (2D) was performed based on the median of a set of wells each from a different plate, but sharing the same relative location on a plate, for example, A1 wells of all screened plates. The formula for calculating the 2D normalized value (denoted as x2D) is x2D = log x′2D where x′2D = x′1D/medianwell(x′1D). For x2D of different plates and wells to be comparable, the normalized NZ was calculated for every x2D. The formula used for calculating NZ2D is: NZ2D = (x2D − median(x2D))/MAD(x2D). MAD(x2D) is the Median Absolute Deviation of x2D and equal to 1.4826 × median (|x2D − median(x2D)|).
Before 1D normalization, the mean of the plate medians of Vmax/Ren was 0.0033, and the standard deviation of the plate medians was 0.0028. This is indicative of plate-to-plate variation, likely due to factors such as different assay dates. After 1D normalization (based on plate median), the plate medians (plate medians of x′1D) are all set at 1, the mean of the well medians of x′1D is 1.0016 and the standard deviation of the well median is 0.0811. This indicates that there was no substantial well position-specific systematic error. After 2D normalization (based on well median), the well medians (well medians of x′2D) are all set at 1. Of the 36 duplicate plates, the Pearson product moment correlation coefficient (R) between the duplicates ranged from 0.32 to 0.87 with the first quartile at 0.52, the median at 0.63, and the third quartile at 0.72.
For confirmation of the primary screen hits, six replicates were used, and the CS activities (normalized by the Ren level) from the hit dsRNA-treated cells were compared to the lacZ dsRNA-treated controls by the t-test. The average CV of the CS assay was ∼3.5%, thus, on average, our confirmation assay is able to detect 5% or larger changes with P < 0.05 (t-test). The power of our confirmation assay was >0.8 when the change in mean CS activity was >7% (t-test).
Quantitative RT-PCR
RNA was extracted using the TurboCapture 8 mRNA kit (Qiagen) according to the manufacturer’s instruction. For cDNA synthesis, BD Sprint PowerScript PrePrimed 96 Plate with Oligo (dT) 18 primers was used (BD Bioscence). cDNA synthesis was performed by Thermal Cycle 9800 (Applied Biosystems), and then used for real-time PCR with SYBR green reagent (Applied Biosystems). Primers were designed from mRNA sequence using PrimerExpress software (Applied Biosystems). The primers used in the experiments are as following:
CS_F: 5′-AATGTGGGAGCCTATGTTCG,
CS_R: 5′-GCTTGCGGAAGTTCTTTACG,
rp49_F: 5′-GCTAAGCTGTCGCACAAATGG, and
rp49_R: 5′-ACTGTCCCTTGAAGCGTCGA.
RT-PCR was performed with the ABI 7000 sequence detection system (Applied Biosystems) according to the manufacturer’s protocol. The expression levels of CS gene were normalized by the expression level of rp49; three independent experiments were performed for RNAi treatments.
HDAC RNAi constructs and transgenic flies
The HDAC RNAi constructs were generated by cloning the HDAC1 and HDAC6 fragments into the pWIZ RNAi-transforming vector as described previously (Lee and Carthew 2003). The insertion fragments for HDAC1 (CG7471) and HDAC6 (CG6170) were amplified by PCR from the PCR amplicon collection by Heidelberg Fly Array (Hild et al. 2003). For HDAC1, the amplicon is HFA08696, primer-S: GACTAATGTGCAGCTTAAAATC, primer-R: GGAGGAGGCGTTCTATACC. For HDAC6, the amplicon is HFA20252, primer-S: GACGCACTGTATTCACATGG, primer-R: GGTGGAGACAGTGCCACT. Two insertion fragments with 520 bp of each were digested and inserted into the AvrII and NheI sites of the pWIZ vector in a 3′-to-5′ direction at 100 bp from the SV40 region as confirmed by sequencing (performed by SeqWright, Houston, TX). Transgenic fly lines were generated by microinjection of RNAi-transforming constructs into the germ lines of fly embryos (by C. Bayer, University of Central Florida, Orlando, FL).
Antibodies and Western blot
The polyclonal antibodies against HDAC1 and HDAC6 were produced in rabbits using a nonconserved peptide region of each HDAC (Covance, Denver, PA). The peptide used to generate antibody to HDAC6 is EHFERLKETSGIRDDERC, and for HDAC1 is CRKRPRLDKDTNSNK. Once the specificity for each HDAC was confirmed by Western blot, the final two bleeds were pooled and affinity-purified.
The wandering third-instar larvae were collected and stored at −80°C until use. The larvae were homogenized on ice in lysis buffer (1% non-idet P40 in PBS) containing protease inhibitors (Roche). The supernatants of the larval lysates were transferred to fresh tubes and heated in NuPAGE LDS (lithium dodecyl sulfate) sample buffer (Invitrogen) and resolved on SDS-PAGE (Invitrogen NuPAGE System). The proteins were transferred to nitrocellulose membranes, which were subjected to Western blot analysis. The signals were detected by chemiluminescence system (Amersham Biosciences). The final concentrations for the antibodies used for Western blots were 1:2000 for anti-HDAC1, 1:1000 for anti-HDAC6, 1:5000 for anti-tubulin (Sigma).
COX assay with single fly lysate
Cytochrome c oxidase Assay Kit (Sigma) was adapted to a 96-well plate format. Cytochrome c (Cyt C) solution was made at 5.4 mg/mL. For every 2.7 mg Cyt C, 10 μL DTT (0.1 mM) was added to reduce Cyt C until the ratio of absorbance 550 nm/565 nm is >10. Fifty microliters of lysate and 25 μL Cyt C were used for COX activity assays. Absorbance at 550 nm was acquired every 6 sec for 1 min. COX activity (Vmax) was calculated as the rate of reduction of absorbance with time (milli OD per minute). COX activity was normalized by total protein concentration; 250 μL extraction buffer was used to homogenize a single fly; 50 μL of the lysate was used for the COX assay, 3 μL for total protein concentration measurement. Eight replicates (one fly per replica) were used for each fly genotype.
Fly strains for in vivo assay
HDAC1 RNAi flies (UAS-HDACR15–17A3/tub-GAL4) and control flies (tub-GAL4/TM6B or UAS-HDAC1R15–17A3/TM6B) were siblings from the cross of UAS-HDAC1R15–17A3/TM6B and tub-GAL4/TM6B. The third-instar larvae under tubulin-GAL4 driver were used for Western blot, CS assay, and COX assay. HDAC6 RNAi flies (UAS-HDAC6R16–17A3/tub-GAL4) and control flies (TM6B/tub-GAL4 or TM6B/UAS-HDAC6R16–17A3) were siblings from the cross of UAS-HDAC6R16–17A3/TM6B and tub-GAL4/TM6B. The third-instar larvae under tubulin-GAL4 driver were used for Western blot. For CS and COX assay, adult male HDAC6 RNAi transgenic flies under mef-GAL4 driver were used. HDAC6 RNAi flies (UAS-HDAC6R16–9A3/mef-GAL4) and control flies (TM6B/mef-GAL4) were siblings from the cross UAS-HDAC6R16–9A3/TM6B and mef-GAL4.
Twenty-three selected genes tested in heterozygous mutants are l(1)G0030 (CS), tam, barr, brm, Rpd3, dom, Su(var)205, shi, Dyb, Src42A, mts, cdc2, S6K, smt3, CG3249, vimar, Tre1, Rab5, ph-p, klu, th, Rs1, CG8602. All strains were ordered from Bloomington public stock center. Stock numbers followed by genotypes are described below: 11936, w67c23 P{lacW}kdnG0140a P{lacW}G0140b, l(1)G0140G0140/FM7c, 3410, tam3 b1 Adhn4/CyO, 11117, y1 w67c23; P{lacW}barrk14014/CyO, 4402, y1 w67c23; barrL305/CyO, 4108, w1118; brmI21 P{white-un4}71F/TM3, Sb1, 3619, brm2 es ca1/TM6B, Sb1 Tb1 ca1, 3622, brm2 trxE2 ca1/TM6B, Tb1 ca1, 11633, P{PZ}Rpd304556 ry506/TM3, ryRK Sb1 Ser1, 10767, y1 w67c23; P{lacW}domk08108/CyO, 6234, In(1)wm4h; Su(var)2055/In(2L)Cy, In(2R)Cy, Cy1, 13253, y1 P{SUPor-P}shiKG03690/FM7a, 15494, y1 w67c23; P{EPgy2}DybEY01099, 13751, y1; P{SUPor-P}Src42AKG02515/CyO; ry506, 6408, w1118; Src42AE1/CyO, 11193, P{PZ}mts02496 cn1/CyO; ry506, 6629, cdc2E1–23 b1 pr1 cn1/CyO, 6643, w*; cdc2B47 cn1 bw1/CyO, P{ftz/lacB}E3, 11713, P{PZ}S6k07084 ry506/TM3, ryRK Sb1 Ser1, 11378, P{PZ}smt304493 cn1/CyO; ry506, 10419, y1 w67c23; P{lacW}smt3k06307/CyO, 12879, y1 P{SUPor-P}CG3249KG02745, 11199, y1 w67c23; P{lacW}vimark16722/CyO, 10089, w1118 P{EP}Tre1EP496, 10786, y1 w67c23; P{lacW}Rab5k08232/CyO, 5444, ph-d401 ph-p602 w1/FM7c, 14587, y1 w67c23; P{SUPor-P}blKG02524/CyO, 11733, P{PZ}klu09036 ry506/TM3, ryRK Sb1 Ser1, 12093, y1 w*; P{lacW}thj5C8/TM3, Sb1, 15310, y1 w67c23; P{EPgy2}thEY00710, 10885, y1 w67c23; P{lacW}Rs1k09514/CyO, 17262, w1118; P{EP}CG8602EP3102. Fly mutant stocks were crossed to wild-type strains Oregon R or w118. The sibling flies with or without mutations were compared for their mitochondrial CS activities. Age-matched wild-type strains were analyzed in parallel as additional controls for flies with balancers or markers.
Acknowledgments
We thank T. Hughes, M. Labow, and Y. Feng for their advice. We thank S. Zhao, B. Pak, J. Warner, S. An, J. Majercak, V. Iourgenko for their assistance in using the HTS equipments. We would also like to acknowledge the help of G. Chirn and C. Tao in data analysis. We thank M. Bowden and E. Petrella for critical reading of the manuscript. We are very grateful for the insightful comments of the anonymous reviewers that greatly improved the quality of the manuscript. We thank Bloomington Drosophila Stock Center at Indiana University (Bloomington, IN) for fly stocks.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6940108
References
- Affaitati A., Cardone L., De C.T., Carlucci A., Ginsberg M.D., Varrone S., Gottesman M.E., Avvedimento E.V., Feliciello A., Cardone L., De C.T., Carlucci A., Ginsberg M.D., Varrone S., Gottesman M.E., Avvedimento E.V., Feliciello A., De C.T., Carlucci A., Ginsberg M.D., Varrone S., Gottesman M.E., Avvedimento E.V., Feliciello A., Carlucci A., Ginsberg M.D., Varrone S., Gottesman M.E., Avvedimento E.V., Feliciello A., Ginsberg M.D., Varrone S., Gottesman M.E., Avvedimento E.V., Feliciello A., Varrone S., Gottesman M.E., Avvedimento E.V., Feliciello A., Gottesman M.E., Avvedimento E.V., Feliciello A., Avvedimento E.V., Feliciello A., Feliciello A. Essential role of A-kinase anchor protein 121 for cAMP signaling to mitochondria. J. Biol. Chem. 2003;278:4286–4294. doi: 10.1074/jbc.M209941200. [DOI] [PubMed] [Google Scholar]
- Appaix F., Kuznetsov A.V., Usson Y., Kay L., Andrienko T., Olivares J., Kaambre T., Sikk P., Margreiter R., Saks V., Kuznetsov A.V., Usson Y., Kay L., Andrienko T., Olivares J., Kaambre T., Sikk P., Margreiter R., Saks V., Usson Y., Kay L., Andrienko T., Olivares J., Kaambre T., Sikk P., Margreiter R., Saks V., Kay L., Andrienko T., Olivares J., Kaambre T., Sikk P., Margreiter R., Saks V., Andrienko T., Olivares J., Kaambre T., Sikk P., Margreiter R., Saks V., Olivares J., Kaambre T., Sikk P., Margreiter R., Saks V., Kaambre T., Sikk P., Margreiter R., Saks V., Sikk P., Margreiter R., Saks V., Margreiter R., Saks V., Saks V. Possible role of cytoskeleton in intracellular arrangement and regulation of mitochondria. Exp. Physiol. 2003;88:175–190. doi: 10.1113/eph8802511. [DOI] [PubMed] [Google Scholar]
- Bakin R.E., Jung M.O., Jung M.O. Cytoplasmic sequestration of HDAC7 from mitochondrial and nuclear compartments upon initiation of apoptosis. J. Biol. Chem. 2004;279:51218–51225. doi: 10.1074/jbc.M409271200. [DOI] [PubMed] [Google Scholar]
- Barlow A.L., van Drunen C.M., Johnson C.A., Tweedie S., Bird A., Turner B.M., van Drunen C.M., Johnson C.A., Tweedie S., Bird A., Turner B.M., Johnson C.A., Tweedie S., Bird A., Turner B.M., Tweedie S., Bird A., Turner B.M., Bird A., Turner B.M., Turner B.M. dSIR2 and dHDAC6: Two novel, inhibitor-resistant deacetylases in Drosophila melanogaster. Exp. Cell Res. 2001;265:90–103. doi: 10.1006/excr.2001.5162. [DOI] [PubMed] [Google Scholar]
- Bhat M.A., Philp A.V., Glover D.M., Bellen H.J., Philp A.V., Glover D.M., Bellen H.J., Glover D.M., Bellen H.J., Bellen H.J. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with Topoisomerase II. Cell. 1996;87:1103–1114. doi: 10.1016/s0092-8674(00)81804-8. [DOI] [PubMed] [Google Scholar]
- Bos J.L. Linking Rap to cell adhesion. Curr. Opin. Cell Biol. 2005;17:123–128. doi: 10.1016/j.ceb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Bruce C.R., Kriketos A.D., Cooney G.J., Hawley J.A., Kriketos A.D., Cooney G.J., Hawley J.A., Cooney G.J., Hawley J.A., Hawley J.A. Disassociation of muscle triglyceride content and insulin sensitivity after exercise training in patients with Type 2 diabetes. Diabetologia. 2004;47:23–30. doi: 10.1007/s00125-003-1265-7. [DOI] [PubMed] [Google Scholar]
- Cardone L., Carlucci A., Affaitati A., Livigni A., DeCristofaro T., Garbi C., Varrone S., Ullrich A., Gottesman M.E., Avvedimento E.V., Carlucci A., Affaitati A., Livigni A., DeCristofaro T., Garbi C., Varrone S., Ullrich A., Gottesman M.E., Avvedimento E.V., Affaitati A., Livigni A., DeCristofaro T., Garbi C., Varrone S., Ullrich A., Gottesman M.E., Avvedimento E.V., Livigni A., DeCristofaro T., Garbi C., Varrone S., Ullrich A., Gottesman M.E., Avvedimento E.V., DeCristofaro T., Garbi C., Varrone S., Ullrich A., Gottesman M.E., Avvedimento E.V., Garbi C., Varrone S., Ullrich A., Gottesman M.E., Avvedimento E.V., Varrone S., Ullrich A., Gottesman M.E., Avvedimento E.V., Ullrich A., Gottesman M.E., Avvedimento E.V., Gottesman M.E., Avvedimento E.V., Avvedimento E.V., et al. Mitochondrial AKAP121 binds and targets protein tyrosine phosphatase D1, a novel positive regulator of src signaling. Mol. Cell. Biol. 2004;24:4613–4626. doi: 10.1128/MCB.24.11.4613-4626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano D., Licciulli F., Turi A., Grillo G., Saccone C., D'Elia D., Licciulli F., Turi A., Grillo G., Saccone C., D'Elia D., Turi A., Grillo G., Saccone C., D'Elia D., Grillo G., Saccone C., D'Elia D., Saccone C., D'Elia D., D'Elia D. MitoRes: A resource of nuclear-encoded mitochondrial genes and their products in Metazoa. BMC Bioinformatics. 2006;7:36. doi: 10.1186/1471-2105-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celotto A.M., Frank A.C., McGrath S.W., Fergestad T., Van Voorhies W.A., Buttle K.F., Mannella C.A., Palladino M.J., Frank A.C., McGrath S.W., Fergestad T., Van Voorhies W.A., Buttle K.F., Mannella C.A., Palladino M.J., McGrath S.W., Fergestad T., Van Voorhies W.A., Buttle K.F., Mannella C.A., Palladino M.J., Fergestad T., Van Voorhies W.A., Buttle K.F., Mannella C.A., Palladino M.J., Van Voorhies W.A., Buttle K.F., Mannella C.A., Palladino M.J., Buttle K.F., Mannella C.A., Palladino M.J., Mannella C.A., Palladino M.J., Palladino M.J. Mitochondrial encephalomyopathy in Drosophila. J. Neurosci. 2006;26:810–820. doi: 10.1523/JNEUROSCI.4162-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Chan D.C., Chan D.C. Emerging functions of mammalian mitochondrial fusion and fission. Hum. Mol. Genet. 2005;14:R283–R289. doi: 10.1093/hmg/ddi270. [DOI] [PubMed] [Google Scholar]
- Czubryt M.P., McAnally J., Fishman G.I., Olson E.N., McAnally J., Fishman G.I., Olson E.N., Fishman G.I., Olson E.N., Olson E.N. Regulation of peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) and mitochondrial function by MEF2 and HDAC5. Proc. Natl. Acad. Sci. 2003;100:1711–1716. doi: 10.1073/pnas.0337639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSimone S.M., White K., White K. The Drosophila erect wing gene, which is important for both neuronal and muscle development, encodes a protein which is similar to the sea urchin P3A2 DNA binding protein. Mol. Cell. Biol. 1993;13:3641–3649. doi: 10.1128/mcb.13.6.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSimone S., Coelho C., Roy S., Vijay Raghavan K., White K., Coelho C., Roy S., Vijay Raghavan K., White K., Roy S., Vijay Raghavan K., White K., Vijay Raghavan K., White K., White K. ERECT WING, the Drosophila member of a family of DNA binding proteins is required in imaginal myoblasts for flight muscle development. Development. 1996;122:31–39. doi: 10.1242/dev.122.1.31. [DOI] [PubMed] [Google Scholar]
- Dohmen R.J. SUMO protein modification. Biochim. Biophys. Acta. 2004;1695:113–131. doi: 10.1016/j.bbamcr.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Dokmanovic M., Marks P.A., Marks P.A. Prospects: Histone deacetylase inhibitors. J. Cell. Biochem. 2005;96:293–304. doi: 10.1002/jcb.20532. [DOI] [PubMed] [Google Scholar]
- Fazio I.K., Bolger T.A., Gill G., Bolger T.A., Gill G., Gill G. Conserved regions of the Drosophila erect wing protein contribute both positively and negatively to transcriptional activity. J. Biol. Chem. 2001;276:18710–18716. doi: 10.1074/jbc.M100080200. [DOI] [PubMed] [Google Scholar]
- Feliciello A., Gottesman M.E., Avvedimento E.V., Gottesman M.E., Avvedimento E.V., Avvedimento E.V. cAMP-PKA signaling to the mitochondria: Protein scaffolds, mRNA and phosphatases. Cell. Signal. 2005;17:279–287. doi: 10.1016/j.cellsig.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Fergestad T., Bostwick B., Ganetzky B., Bostwick B., Ganetzky B., Ganetzky B. Metabolic disruption in Drosophila bang-sensitive seizure mutants. Genetics. 2006;173:1357–1364. doi: 10.1534/genetics.106.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinari P., Di M.S., Jones P., Pallaoro M., Steinkuhler C., Di M.S., Jones P., Pallaoro M., Steinkuhler C., Jones P., Pallaoro M., Steinkuhler C., Pallaoro M., Steinkuhler C., Steinkuhler C. HDACs, histone deacetylation and gene transcription: From molecular biology to cancer therapeutics. Cell Res. 2007;17:195–211. doi: 10.1038/sj.cr.7310149. [DOI] [PubMed] [Google Scholar]
- Gao L., Cueto M.A., Asselbergs F., Atadja P., Cueto M.A., Asselbergs F., Atadja P., Asselbergs F., Atadja P., Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J. Biol. Chem. 2002;277:25748–25755. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- Graham B.H., Waymire K.G., Cottrell B., Trounce I.A., MacGregor G.R., Wallace D.C., Waymire K.G., Cottrell B., Trounce I.A., MacGregor G.R., Wallace D.C., Cottrell B., Trounce I.A., MacGregor G.R., Wallace D.C., Trounce I.A., MacGregor G.R., Wallace D.C., MacGregor G.R., Wallace D.C., Wallace D.C. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat. Genet. 1997;16:226–234. doi: 10.1038/ng0797-226. [DOI] [PubMed] [Google Scholar]
- Haggarty S.J., Koeller K.M., Wong J.C., Grozinger C.M., Schreiber S.L., Koeller K.M., Wong J.C., Grozinger C.M., Schreiber S.L., Wong J.C., Grozinger C.M., Schreiber S.L., Grozinger C.M., Schreiber S.L., Schreiber S.L. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc. Natl. Acad. Sci. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H., Becknell B., Wilm M., Mann M., Huang L.J., Taylor S.S., Scott J.D., Korsmeyer S.J., Becknell B., Wilm M., Mann M., Huang L.J., Taylor S.S., Scott J.D., Korsmeyer S.J., Wilm M., Mann M., Huang L.J., Taylor S.S., Scott J.D., Korsmeyer S.J., Mann M., Huang L.J., Taylor S.S., Scott J.D., Korsmeyer S.J., Huang L.J., Taylor S.S., Scott J.D., Korsmeyer S.J., Taylor S.S., Scott J.D., Korsmeyer S.J., Scott J.D., Korsmeyer S.J., Korsmeyer S.J. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol. Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- Harder Z., Zunino R., McBride H., Zunino R., McBride H., McBride H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr. Biol. 2004;14:340–345. doi: 10.1016/j.cub.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Hild M., Beckmann B., Haas S.A., Koch B., Solovyev V., Busold C., Fellenberg K., Boutros M., Vingron M., Sauer F., Beckmann B., Haas S.A., Koch B., Solovyev V., Busold C., Fellenberg K., Boutros M., Vingron M., Sauer F., Haas S.A., Koch B., Solovyev V., Busold C., Fellenberg K., Boutros M., Vingron M., Sauer F., Koch B., Solovyev V., Busold C., Fellenberg K., Boutros M., Vingron M., Sauer F., Solovyev V., Busold C., Fellenberg K., Boutros M., Vingron M., Sauer F., Busold C., Fellenberg K., Boutros M., Vingron M., Sauer F., Fellenberg K., Boutros M., Vingron M., Sauer F., Boutros M., Vingron M., Sauer F., Vingron M., Sauer F., Sauer F., et al. An integrated gene annotation and transcriptional profiling approach towards the full gene content of the Drosophila genome. Genome Biol. 2003;5:R3. doi: 10.1186/gb-2003-5-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Kobayashi R., Hirano M., Kobayashi R., Hirano M., Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Horbinski C., Chu C.T., Chu C.T. Kinase signaling cascades in the mitochondrion: A matter of life or death. Free Radic. Biol. Med. 2005;38:2–11. doi: 10.1016/j.freeradbiomed.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X.F., Yao T.P., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X.F., Yao T.P., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X.F., Yao T.P., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X.F., Yao T.P., Ito A., Nixon A., Yoshida M., Wang X.F., Yao T.P., Nixon A., Yoshida M., Wang X.F., Yao T.P., Yoshida M., Wang X.F., Yao T.P., Wang X.F., Yao T.P., Yao T.P. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Huo L., Scarpulla R.C., Scarpulla R.C. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol. Cell. Biol. 2001;21:644–654. doi: 10.1128/MCB.21.2.644-654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A., Riley B.E., Johnston J.A., Kopito R.R., Riley B.E., Johnston J.A., Kopito R.R., Johnston J.A., Kopito R.R., Kopito R.R. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- Iyengar B., Roote J., Campos A.R., Roote J., Campos A.R., Campos A.R. The tamas gene, identified as a mutation that disrupts larval behavior in Drosophila melanogaster, codes for the mitochondrial DNA polymerase catalytic subunit (DNApol-gamma125) Genetics. 1999;153:1809–1824. doi: 10.1093/genetics/153.4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs H.T., Fernández-Ayala D.J., Manjiry S., Kemppainen E., Toivonen J.M., O’Dell K.M., Fernández-Ayala D.J., Manjiry S., Kemppainen E., Toivonen J.M., O’Dell K.M., Manjiry S., Kemppainen E., Toivonen J.M., O’Dell K.M., Kemppainen E., Toivonen J.M., O’Dell K.M., Toivonen J.M., O’Dell K.M., O’Dell K.M. Mitochondrial disease in flies. Biochim. Biophys. Acta. 2004;1659:190–196. doi: 10.1016/j.bbabio.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Kania A., Salzberg A., Bhat M., D'Evelyn D., He Y., Kiss I., Bellen H.J., Salzberg A., Bhat M., D'Evelyn D., He Y., Kiss I., Bellen H.J., Bhat M., D'Evelyn D., He Y., Kiss I., Bellen H.J., D'Evelyn D., He Y., Kiss I., Bellen H.J., He Y., Kiss I., Bellen H.J., Kiss I., Bellen H.J., Bellen H.J. P-element mutations affecting embryonic peripheral nervous system development in Drosophila melanogaster. Genetics. 1995;139:1663–1678. doi: 10.1093/genetics/139.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D.E., He J., Menshikova E.V., Ritov V.B., He J., Menshikova E.V., Ritov V.B., Menshikova E.V., Ritov V.B., Ritov V.B. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51:2944–2950. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- Klingenberg M. Structure-function of the ADP/ATP carrier. Biochem. Soc. Trans. 1992;20:547–550. doi: 10.1042/bst0200547. [DOI] [PubMed] [Google Scholar]
- Knutti D., Kralli A., Kralli A. PGC-1, a versatile coactivator. Trends Endocrinol. Metab. 2001;12:360–365. doi: 10.1016/s1043-2760(01)00457-x. [DOI] [PubMed] [Google Scholar]
- Kovacs J.J., Murphy P.J., Gaillard S., Zhao X., Wu J.T., Nicchitta C.V., Yoshida M., Toft D.O., Pratt W.B., Yao T.P., Murphy P.J., Gaillard S., Zhao X., Wu J.T., Nicchitta C.V., Yoshida M., Toft D.O., Pratt W.B., Yao T.P., Gaillard S., Zhao X., Wu J.T., Nicchitta C.V., Yoshida M., Toft D.O., Pratt W.B., Yao T.P., Zhao X., Wu J.T., Nicchitta C.V., Yoshida M., Toft D.O., Pratt W.B., Yao T.P., Wu J.T., Nicchitta C.V., Yoshida M., Toft D.O., Pratt W.B., Yao T.P., Nicchitta C.V., Yoshida M., Toft D.O., Pratt W.B., Yao T.P., Yoshida M., Toft D.O., Pratt W.B., Yao T.P., Toft D.O., Pratt W.B., Yao T.P., Pratt W.B., Yao T.P., Yao T.P. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol. Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Kraft C.S., LeMoine C.M., Lyons C.N., Michaud D., Mueller C.R., Moyes C.D., LeMoine C.M., Lyons C.N., Michaud D., Mueller C.R., Moyes C.D., Lyons C.N., Michaud D., Mueller C.R., Moyes C.D., Michaud D., Mueller C.R., Moyes C.D., Mueller C.R., Moyes C.D., Moyes C.D. Control of mitochondrial biogenesis during myogenesis. Am. J. Physiol. Cell Physiol. 2006;290:C1119–C1127. doi: 10.1152/ajpcell.00463.2005. [DOI] [PubMed] [Google Scholar]
- Kuhajda F.P. Fatty acid synthase and cancer: New application of an old pathway. Cancer Res. 2006;66:5977–5980. doi: 10.1158/0008-5472.CAN-05-4673. [DOI] [PubMed] [Google Scholar]
- Le Bras B.M., Rouy I., Brenner C., Rouy I., Brenner C., Brenner C. The modulation of inter-organelle cross-talk to control apoptosis. Med. Chem. 2006;2:1–12. doi: 10.2174/157340606775197787. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Carthew R.W., Carthew R.W. Making a better RNAi vector for Drosophila: Use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Legagneux V., Cubizolles F., Watrin E., Cubizolles F., Watrin E., Watrin E. Multiple roles of Condensins: A complex story. Biol. Cell. 2004;96:201–213. doi: 10.1016/j.biolcel.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Lehman J.J., Barger P.M., Kovacs A., Saffitz J.E., Medeiros D.M., Kelly D.P., Barger P.M., Kovacs A., Saffitz J.E., Medeiros D.M., Kelly D.P., Kovacs A., Saffitz J.E., Medeiros D.M., Kelly D.P., Saffitz J.E., Medeiros D.M., Kelly D.P., Medeiros D.M., Kelly D.P., Kelly D.P. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon J., Cuna-Castroviejo D., Sainz R.M., Mayo J.C., Tan D.X., Reiter R.J., Cuna-Castroviejo D., Sainz R.M., Mayo J.C., Tan D.X., Reiter R.J., Sainz R.M., Mayo J.C., Tan D.X., Reiter R.J., Mayo J.C., Tan D.X., Reiter R.J., Tan D.X., Reiter R.J., Reiter R.J. Melatonin and mitochondrial function. Life Sci. 2004;75:765–790. doi: 10.1016/j.lfs.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Lin J., Puigserver P., Donovan J., Tarr P., Spiegelman B.M., Puigserver P., Donovan J., Tarr P., Spiegelman B.M., Donovan J., Tarr P., Spiegelman B.M., Tarr P., Spiegelman B.M., Spiegelman B.M. Peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- Livigni A., Scorziello A., Agnese S., Adornetto A., Carlucci A., Garbi C., Castaldo I., Annunziato L., Avvedimento E.V., Feliciello A., Scorziello A., Agnese S., Adornetto A., Carlucci A., Garbi C., Castaldo I., Annunziato L., Avvedimento E.V., Feliciello A., Agnese S., Adornetto A., Carlucci A., Garbi C., Castaldo I., Annunziato L., Avvedimento E.V., Feliciello A., Adornetto A., Carlucci A., Garbi C., Castaldo I., Annunziato L., Avvedimento E.V., Feliciello A., Carlucci A., Garbi C., Castaldo I., Annunziato L., Avvedimento E.V., Feliciello A., Garbi C., Castaldo I., Annunziato L., Avvedimento E.V., Feliciello A., Castaldo I., Annunziato L., Avvedimento E.V., Feliciello A., Annunziato L., Avvedimento E.V., Feliciello A., Avvedimento E.V., Feliciello A., Feliciello A.2006Mitochondrial AKAP121 links cAMP and src signaling to oxidative metabolism Mol. Biol. Cell 17263.– [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Li Y., Li Y. Drosophila Src42A is a negative regulator of RTK signaling. Dev. Biol. 1999;208:233–243. doi: 10.1006/dbio.1999.9196. [DOI] [PubMed] [Google Scholar]
- Maier D., Farr C.L., Poeck B., Alahari A., Vogel M., Fischer S., Kaguni L.S., Schneuwly S., Farr C.L., Poeck B., Alahari A., Vogel M., Fischer S., Kaguni L.S., Schneuwly S., Poeck B., Alahari A., Vogel M., Fischer S., Kaguni L.S., Schneuwly S., Alahari A., Vogel M., Fischer S., Kaguni L.S., Schneuwly S., Vogel M., Fischer S., Kaguni L.S., Schneuwly S., Fischer S., Kaguni L.S., Schneuwly S., Kaguni L.S., Schneuwly S., Schneuwly S. Mitochondrial single-stranded DNA-binding protein is required for mitochondrial DNA replication and development in Drosophila melanogaster. Mol. Biol. Cell. 2001;12:821–830. doi: 10.1091/mbc.12.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Macías M., León J., Escames G., Khaldy H., Acuña-Castroviejo D., Macías M., León J., Escames G., Khaldy H., Acuña-Castroviejo D., León J., Escames G., Khaldy H., Acuña-Castroviejo D., Escames G., Khaldy H., Acuña-Castroviejo D., Khaldy H., Acuña-Castroviejo D., Acuña-Castroviejo D. Melatonin increases the activity of the oxidative phosphorylation enzymes and the production of ATP in rat brain and liver mitochondria. Int. J. Biochem. Cell Biol. 2002;34:348–357. doi: 10.1016/s1357-2725(01)00138-8. [DOI] [PubMed] [Google Scholar]
- Matsuyama A., Shimazu T., Sumida Y., Saito A., Yoshimatsu Y., Seigneurin-Berny D., Osada H., Komatsu Y., Nishino N., Khochbin S., Shimazu T., Sumida Y., Saito A., Yoshimatsu Y., Seigneurin-Berny D., Osada H., Komatsu Y., Nishino N., Khochbin S., Sumida Y., Saito A., Yoshimatsu Y., Seigneurin-Berny D., Osada H., Komatsu Y., Nishino N., Khochbin S., Saito A., Yoshimatsu Y., Seigneurin-Berny D., Osada H., Komatsu Y., Nishino N., Khochbin S., Yoshimatsu Y., Seigneurin-Berny D., Osada H., Komatsu Y., Nishino N., Khochbin S., Seigneurin-Berny D., Osada H., Komatsu Y., Nishino N., Khochbin S., Osada H., Komatsu Y., Nishino N., Khochbin S., Komatsu Y., Nishino N., Khochbin S., Nishino N., Khochbin S., Khochbin S., et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21:6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride H.M., Neuspiel M., Wasiak S., Neuspiel M., Wasiak S., Wasiak S. Mitochondria: More than just a powerhouse. Curr. Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Meirhaeghe A., Crowley V., Lenaghan C., Lelliott C., Green K., Stewart A., Hart K., Schinner S., Sethi J.K., Yeo G., Crowley V., Lenaghan C., Lelliott C., Green K., Stewart A., Hart K., Schinner S., Sethi J.K., Yeo G., Lenaghan C., Lelliott C., Green K., Stewart A., Hart K., Schinner S., Sethi J.K., Yeo G., Lelliott C., Green K., Stewart A., Hart K., Schinner S., Sethi J.K., Yeo G., Green K., Stewart A., Hart K., Schinner S., Sethi J.K., Yeo G., Stewart A., Hart K., Schinner S., Sethi J.K., Yeo G., Hart K., Schinner S., Sethi J.K., Yeo G., Schinner S., Sethi J.K., Yeo G., Sethi J.K., Yeo G., Yeo G., et al. Characterization of the human, mouse and rat PGC1β (peroxisome-proliferator-activated receptor-γ co-activator 1β) gene in vitro and in vivo. Biochem. J. 2003;373:155–165. doi: 10.1042/BJ20030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miceli M.V., Jazwinski S.M., Jazwinski S.M. Nuclear gene expression changes due to mitochondrial dysfunction in ARPE-19 cells: Implications for age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2005;46:1765–1773. doi: 10.1167/iovs.04-1327. [DOI] [PubMed] [Google Scholar]
- Mootha V.K., Handschin C., Arlow D., Xie X., St P.J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., Handschin C., Arlow D., Xie X., St P.J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., Arlow D., Xie X., St P.J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., Xie X., St P.J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., St P.J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., Yang W., Altshuler D., Puigserver P., Patterson N., Altshuler D., Puigserver P., Patterson N., Puigserver P., Patterson N., Patterson N., et al. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen O.H., Frandsen L., Schjerling P., Nishimura E., Grunnet N., Frandsen L., Schjerling P., Nishimura E., Grunnet N., Schjerling P., Nishimura E., Grunnet N., Nishimura E., Grunnet N., Grunnet N. PGC-1α and PGC-1β have both similar and distinct effects on myofiber switching toward an oxidative phenotype. Am. J. Physiol. Endocrinol. Metab. 2006;291:E807–E816. doi: 10.1152/ajpendo.00591.2005. [DOI] [PubMed] [Google Scholar]
- Moyes C.D., Mathieu-Costello O.A., Tsuchiya N., Filburn C., Hansford R.G., Mathieu-Costello O.A., Tsuchiya N., Filburn C., Hansford R.G., Tsuchiya N., Filburn C., Hansford R.G., Filburn C., Hansford R.G., Hansford R.G. Mitochondrial biogenesis during cellular differentiation. Am. J. Phys. 1997;272:C1345–C1351. doi: 10.1152/ajpcell.1997.272.4.C1345. [DOI] [PubMed] [Google Scholar]
- Murphy P.J., Morishima Y., Kovacs J.J., Yao T.P., Pratt W.B., Morishima Y., Kovacs J.J., Yao T.P., Pratt W.B., Kovacs J.J., Yao T.P., Pratt W.B., Yao T.P., Pratt W.B., Pratt W.B. Regulation of the dynamics of hsp90 action on the glucocorticoid receptor by acetylation/deacetylation of the chaperone. J. Biol. Chem. 2005;280:33792–33799. doi: 10.1074/jbc.M506997200. [DOI] [PubMed] [Google Scholar]
- Nisoli E., Clementi E., Moncada S., Carruba M.O., Clementi E., Moncada S., Carruba M.O., Moncada S., Carruba M.O., Carruba M.O. Mitochondrial biogenesis as a cellular signaling framework. Biochem. Pharmacol. 2004;67:1–15. doi: 10.1016/j.bcp.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Nisoli E., Tonello C., Cardile A., Cozzi V., Bracale R., Tedesco L., Falcone S., Valerio A., Cantoni O., Clementi E., Tonello C., Cardile A., Cozzi V., Bracale R., Tedesco L., Falcone S., Valerio A., Cantoni O., Clementi E., Cardile A., Cozzi V., Bracale R., Tedesco L., Falcone S., Valerio A., Cantoni O., Clementi E., Cozzi V., Bracale R., Tedesco L., Falcone S., Valerio A., Cantoni O., Clementi E., Bracale R., Tedesco L., Falcone S., Valerio A., Cantoni O., Clementi E., Tedesco L., Falcone S., Valerio A., Cantoni O., Clementi E., Falcone S., Valerio A., Cantoni O., Clementi E., Valerio A., Cantoni O., Clementi E., Cantoni O., Clementi E., Clementi E., et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- Pile L.A., Spellman P.T., Katzenberger R.J., Wassarman D.A., Spellman P.T., Katzenberger R.J., Wassarman D.A., Katzenberger R.J., Wassarman D.A., Wassarman D.A. The SIN3 deacetylase complex represses genes encoding mitochondrial proteins: Implications for the regulation of energy metabolism. J. Biol. Chem. 2003;278:37840–37848. doi: 10.1074/jbc.M305996200. [DOI] [PubMed] [Google Scholar]
- Psarra A.M., Solakidi S., Trougakos I.P., Margaritis L.H., Spyrou G., Sekeris C.E., Solakidi S., Trougakos I.P., Margaritis L.H., Spyrou G., Sekeris C.E., Trougakos I.P., Margaritis L.H., Spyrou G., Sekeris C.E., Margaritis L.H., Spyrou G., Sekeris C.E., Spyrou G., Sekeris C.E., Sekeris C.E. Glucocorticoid receptor isoforms in human hepatocarcinoma HepG2 and SaOS-2 osteosarcoma cells: Presence of glucocorticoid receptor alpha in mitochondria and of glucocorticoid receptor beta in nucleoli. Int. J. Biochem. Cell Biol. 2005;37:2544–2558. doi: 10.1016/j.biocel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Puigserver P., Spiegelman B.M., Spiegelman B.M. Peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α): Transcriptional coactivator and metabolic regulator. Endocr. Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- Qiao J., Mei F.C., Popov V.L., Vergara L.A., Cheng X., Mei F.C., Popov V.L., Vergara L.A., Cheng X., Popov V.L., Vergara L.A., Cheng X., Vergara L.A., Cheng X., Cheng X. Cell cycle-dependent subcellular localization of exchange factor directly activated by cAMP. J. Biol. Chem. 2002;277:26581–26586. doi: 10.1074/jbc.M203571200. [DOI] [PubMed] [Google Scholar]
- Reznick R.M., Shulman G.I., Shulman G.I. The role of AMP-activated protein kinase in mitochondrial biogenesis. J. Physiol. 2006;574:33–39. doi: 10.1113/jphysiol.2006.109512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikhy R., Ramaswami M., Krishnan K.S., Ramaswami M., Krishnan K.S., Krishnan K.S. A temperature-sensitive allele of Drosophila sesB reveals acute functions for the mitochondrial adenine nucleotide translocase in synaptic transmission and dynamin regulation. Genetics. 2003;165:1243–1253. doi: 10.1093/genetics/165.3.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi J.C., Fink J.L., Cagan R., Fink J.L., Cagan R., Cagan R. klumpfuss regulates cell death in the Drosophila retina. Mech. Dev. 2004;121:537–546. doi: 10.1016/j.mod.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Ryan M.T., Hoogenraad N.J., Hoogenraad N.J. Mitochondrial-nuclear communications. Annu. Rev. Biochem. 2007;76:701–722. doi: 10.1146/annurev.biochem.76.052305.091720. [DOI] [PubMed] [Google Scholar]
- Salvi M., Brunati A.M., Bordin L., La R.N., Clari G., Toninello A., Brunati A.M., Bordin L., La R.N., Clari G., Toninello A., Bordin L., La R.N., Clari G., Toninello A., La R.N., Clari G., Toninello A., Clari G., Toninello A., Toninello A. Characterization and location of Src-dependent tyrosine phosphorylation in rat brain mitochondria. Biochim. Biophys. Acta. 2002;1589:181–195. doi: 10.1016/s0167-4889(02)00174-x. [DOI] [PubMed] [Google Scholar]
- Sanchez-Martinez A., Luo N., Clemente P., Adan C., Hernandez-Sierra R., Ochoa P., Fernandez-Moreno M.A., Kaguni L.S., Garesse R., Luo N., Clemente P., Adan C., Hernandez-Sierra R., Ochoa P., Fernandez-Moreno M.A., Kaguni L.S., Garesse R., Clemente P., Adan C., Hernandez-Sierra R., Ochoa P., Fernandez-Moreno M.A., Kaguni L.S., Garesse R., Adan C., Hernandez-Sierra R., Ochoa P., Fernandez-Moreno M.A., Kaguni L.S., Garesse R., Hernandez-Sierra R., Ochoa P., Fernandez-Moreno M.A., Kaguni L.S., Garesse R., Ochoa P., Fernandez-Moreno M.A., Kaguni L.S., Garesse R., Fernandez-Moreno M.A., Kaguni L.S., Garesse R., Kaguni L.S., Garesse R., Garesse R. Modeling human mitochondrial diseases in flies. Biochim. Biophys. Acta. 2006;1757:1190–1198. doi: 10.1016/j.bbabio.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M., Licciulli F., Catalano D., Attimonelli M., Caggese C., Licciulli F., Catalano D., Attimonelli M., Caggese C., Catalano D., Attimonelli M., Caggese C., Attimonelli M., Caggese C., Caggese C. MitoDrome: A database of Drosophila melanogaster nuclear genes encoding proteins targeted to the mitochondrion. Nucleic Acids Res. 2003;31:322–324. doi: 10.1093/nar/gkg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R.C. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim. Biophys. Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- Scarpulla R.C. Nuclear control of respiratory gene expression in mammalian cells. J. Cell. Biochem. 2006;97:673–683. doi: 10.1002/jcb.20743. [DOI] [PubMed] [Google Scholar]
- Schapira A.H. Mitochondrial disease. Lancet. 2006;368:70–82. doi: 10.1016/S0140-6736(06)68970-8. [DOI] [PubMed] [Google Scholar]
- Scheller K., Sekeris C.E., Sekeris C.E. The effects of steroid hormones on the transcription of genes encoding enzymes of oxidative phosphorylation. Exp. Physiol. 2003;88:129–140. doi: 10.1113/eph8802507. [DOI] [PubMed] [Google Scholar]
- Schreiber S.N., Emter R., Hock M.B., Knutti D., Cardenas J., Podvinec M., Oakeley E.J., Kralli A., Emter R., Hock M.B., Knutti D., Cardenas J., Podvinec M., Oakeley E.J., Kralli A., Hock M.B., Knutti D., Cardenas J., Podvinec M., Oakeley E.J., Kralli A., Knutti D., Cardenas J., Podvinec M., Oakeley E.J., Kralli A., Cardenas J., Podvinec M., Oakeley E.J., Kralli A., Podvinec M., Oakeley E.J., Kralli A., Oakeley E.J., Kralli A., Kralli A. The estrogen-related receptor α (ERRα) functions in PPARγ coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno M., Nishida Y., Chi-Yamada T., Nishida Y., Chi-Yamada T., Chi-Yamada T. Regulation of JNK by Src during Drosophiladevelopment. Science. 2000;287:324–327. doi: 10.1126/science.287.5451.324. [DOI] [PubMed] [Google Scholar]
- Toivonen J.M., Odell K.M., Petit N., Irvine S.C., Knight G.K., Lehtonen M., Longmuir M., Luoto K., Touraille S., Wang Z., Odell K.M., Petit N., Irvine S.C., Knight G.K., Lehtonen M., Longmuir M., Luoto K., Touraille S., Wang Z., Petit N., Irvine S.C., Knight G.K., Lehtonen M., Longmuir M., Luoto K., Touraille S., Wang Z., Irvine S.C., Knight G.K., Lehtonen M., Longmuir M., Luoto K., Touraille S., Wang Z., Knight G.K., Lehtonen M., Longmuir M., Luoto K., Touraille S., Wang Z., Lehtonen M., Longmuir M., Luoto K., Touraille S., Wang Z., Longmuir M., Luoto K., Touraille S., Wang Z., Luoto K., Touraille S., Wang Z., Touraille S., Wang Z., Wang Z., et al. Technical knockout, a Drosophila model of mitochondrial deafness. Genetics. 2001;159:241–254. doi: 10.1093/genetics/159.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven A., Wong J.M., Xu D., Sopta M., Ingles C.J., Wong J.M., Xu D., Sopta M., Ingles C.J., Xu D., Sopta M., Ingles C.J., Sopta M., Ingles C.J., Ingles C.J. Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial dna mutant. J. Biol. Chem. 2001;276:4020–4027. doi: 10.1074/jbc.M006807200. [DOI] [PubMed] [Google Scholar]
- Wang H., Hiatt W.R., Barstow T.J., Brass E.P., Hiatt W.R., Barstow T.J., Brass E.P., Barstow T.J., Brass E.P., Brass E.P. Relationships between muscle mitochondrial DNA content, mitochondrial enzyme activity and oxidative capacity in man: Alterations with disease. Eur. J. Appl. Physiol. Occup. Physiol. 1999;80:22–27. doi: 10.1007/s004210050553. [DOI] [PubMed] [Google Scholar]
- Wang Z., Dillon T.J., Pokala V., Mishra S., Labudda K., Hunter B., Stork P.J., Dillon T.J., Pokala V., Mishra S., Labudda K., Hunter B., Stork P.J., Pokala V., Mishra S., Labudda K., Hunter B., Stork P.J., Mishra S., Labudda K., Hunter B., Stork P.J., Labudda K., Hunter B., Stork P.J., Hunter B., Stork P.J., Stork P.J. Rap1-mediated activation of extracellular signal-regulated kinases by cyclic AMP is dependent on the mode of Rap1 activation. Mol. Cell. Biol. 2006;26:2130–2145. doi: 10.1128/MCB.26.6.2130-2145.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K., Bruck P., Mikes Z., Kupper J.H., Klingenspor M., Wiesner R.J., Bruck P., Mikes Z., Kupper J.H., Klingenspor M., Wiesner R.J., Mikes Z., Kupper J.H., Klingenspor M., Wiesner R.J., Kupper J.H., Klingenspor M., Wiesner R.J., Klingenspor M., Wiesner R.J., Wiesner R.J. Glucocorticoid hormone stimulates mitochondrial biogenesis specifically in skeletal muscle. Endocrinology. 2002;143:177–184. doi: 10.1210/endo.143.1.8600. [DOI] [PubMed] [Google Scholar]
- Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., Troy A., Cinti S., Lowell B., Scarpulla R.C., Cinti S., Lowell B., Scarpulla R.C., Lowell B., Scarpulla R.C., Scarpulla R.C., et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Zhang Y.Q., Roote J., Brogna S., Davis A.W., Barbash D.A., Nash D., Ashburner M., Roote J., Brogna S., Davis A.W., Barbash D.A., Nash D., Ashburner M., Brogna S., Davis A.W., Barbash D.A., Nash D., Ashburner M., Davis A.W., Barbash D.A., Nash D., Ashburner M., Barbash D.A., Nash D., Ashburner M., Nash D., Ashburner M., Ashburner M. stress sensitive B encodes an adenine nucleotide translocase in Drosophila melanogaster. Genetics. 1999;153:891–903. doi: 10.1093/genetics/153.2.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li N., Caron C., Matthias G., Hess D., Khochbin S., Matthias P., Li N., Caron C., Matthias G., Hess D., Khochbin S., Matthias P., Caron C., Matthias G., Hess D., Khochbin S., Matthias P., Matthias G., Hess D., Khochbin S., Matthias P., Hess D., Khochbin S., Matthias P., Khochbin S., Matthias P., Matthias P. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 2003;22:1168–1179. doi: 10.1093/emboj/cdg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunino R., Schauss A., Rippstein P., Ndrade-Navarro M., McBride H.M., Schauss A., Rippstein P., Ndrade-Navarro M., McBride H.M., Rippstein P., Ndrade-Navarro M., McBride H.M., Ndrade-Navarro M., McBride H.M., McBride H.M. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J. Cell Sci. 2007;120:1178–1188. doi: 10.1242/jcs.03418. [DOI] [PubMed] [Google Scholar]