Abstract
Facio-scapulo-humeral dystrophy (FSHD), a muscular hereditary disease with a prevalence of 1 in 20,000, is caused by a partial deletion of a subtelomeric repeat array on chromosome 4q. Earlier, we demonstrated the existence in the vicinity of the D4Z4 repeat of a nuclear matrix attachment site, FR-MAR, efficient in normal human myoblasts and nonmuscular human cells but much weaker in muscle cells from FSHD patients. We now report that the D4Z4 repeat contains an exceptionally strong transcriptional enhancer at its 5′-end. This enhancer up-regulates transcription from the promoter of the neighboring FRG1 gene. However, an enhancer blocking activity was found present in FR-MAR that in vitro could protect transcription from the enhancer activity of the D4Z4 array. In vivo, transcription from the FRG1 and FRG2 genes could be down- or up-regulated depending on whether or not FR-MAR is associated with the nuclear matrix. We propose a model for an etiological role of the delocalization of FR-MAR in the genesis of FSHD.
Facio-scapulo-humeral muscular dystrophy (FSHD) is an autosomal dominant neuromuscular disease with a prevalence of 1 in 20,000 (Lunt and Harper 1991). FSHD is characterized by progressive weakness and atrophy of muscles of the face, upper arms, and shoulder girdle. The disorder is associated with a shortened repeat array that remains present at a subtelomeric position on chromosome 4q after deletion of an integral number of 3.3-kb tandem repeats. The size of the D4Z4 polymorphic locus varies in normal individuals from 35 to 300 kb but is consistently less than 35 kb in length in FSHD patients (van Deutekom et al. 1993). D4Z4 elements have been shown to contain a cryptic DUX4 gene potentially coding for a double homeodomain protein (van Geel et al. 1999). An overall perturbation of mRNA expression profiles and protein content has been observed in FSHD patients (Tupler et al. 1999; Winokur et al. 2003; Wohlgemuth et al. 2003; Laoudj-Chenivesse et al. 2005). However, since no gene has been found altered by the intrachromosomal deletion, the mechanism leading to FSHD remains unexplained and alternative hypotheses are clearly needed.
In eukaryotic nuclei and metaphase chromosomes, the DNA is organized into loop domains (for review, see Vassetzky et al. 2000), which are anchored to the nuclear skeleton or matrix via specific sequences called S/MARs, for scaffold/matrix-associated regions (Mirkovitch et al. 1984; Cockerill and Garrard 1986). Generally A/T-rich, S/MARs are DNA fragments between 200 and 1000 bp in length. Some S/MARs are present in nontranscribed regions or within introns. Others found in the vicinity of enhancers, insulators, replication origins, or transcribed genes are defined as function-related (Vassetzky et al. 2000).
Previously, we demonstrated the existence of a nuclear matrix attachment site (S/MAR) in the immediate vicinity of the D4Z4 repeat (Fig. 1). We then demonstrated that the S/MAR adjacent to the D4Z4 array was prominent in normal human myoblasts and nonmuscular human cells but much weaker in muscle cells derived from FSHD patients. We also reported that the D4Z4 repeat array and upstream genes reside in a single loop in FSHD myoblasts but are located in two distinct loops in nonmuscular cells and normal human myoblasts (Petrov et al. 2006). In FSHD muscle, the decreased number of D4Z4 repeats results in an inappropriate up-regulation of adjacent 4q35 genes, including FRG1, FRG2, and SLC25A4 (previously known as ANT1) (Gabellini et al. 2002; Rijkers et al. 2004; Laoudj-Chenivesse et al. 2005). Interestingly, overexpressing FRG1 in transgenic mice provokes an FSHD-like phenotype (Gabellini et al. 2005). It has also been proposed that overexpression would result from the absence of a D4Z4-binding transcriptional repressor complex (Gabellini et al. 2002). Other hypotheses include the existence within D4Z4 of a transcriptional enhancer, which could up-regulate the expression of FRG2 (Petrov et al. 2003; Rijkers et al. 2004). The presence of a S/MAR (referred to as FR-MAR) between the D4Z4 array and proximal genes raises the question as to whether it functions as a border element insulating adjacent genes from the effect of D4Z4 in normal human cells. Indeed, within a given chromatin domain, border elements have been shown to protect genes from stimulatory and repressive effects exerted by flanking genomic regions (for review, see Gaszner and Felsenfeld 2006), and S/MARs have previously been characterized as implicated in bordering functions (Bode et al. 2000).
Figure 1.
The D4Z4 repeat contains a strong transcriptional enhancer. (A) Schematic representation of the D4Z4 repeat in its chromosome 4q35 environment. The SLC25A4 (previously known as ANT1), FRG1, and FRG2 genes are shown as gray rectangles. Telomeric and D4Z4 repeats are shown as circles and triangles, respectively. A scaffold/matrix attachment region depicted as FR-MAR (dark gray) is located between the genes and the D4Z4 array. (B) The D4Z4 repeat unit contains a strong transcriptional enhancer. The transcriptional effect of the D4Z4 repeat unit and its deletion fragments was tested 48 h after transfection. The enhancer strength is quantified relative to the luciferase activity generated with the SV40 enhancer. The numbers refer to the nucleotide positions starting from the upstream KpnI restriction site. (C) The minimal D4Z4 enhancer is neither species nor tissue specific. The effect of the minimal D4Z4 enhancer was tested in human primary myoblasts, HeLa cells, the human rhabdomyosarcoma cell line RMS, and the murine myoblast cell line C2C12. Luciferase activity is shown relative to that obtained with the SV40 enhancer.
Here, we have studied in detail the ability of the D4Z4 repeat to regulate transcriptional activity. We have found evidence of an exceptionally strong transcriptional enhancer at the 5′-end of the repeat. This enhancer could up-regulate transcription from the FRG1 gene promoter. We have also found that the S/MAR located in the vicinity of the D4Z4 array exerts an enhancer blocking activity in vivo, thus insulating the FRG1 and FRG2 genes from the effect of D4Z4. We propose a model whereby this S/MAR regulates chromatin accessibility and expression of genes implicated in the genesis of FSHD.
Results and Discussion
The D4Z4 repeat contains a strong enhancer
A partial deletion of the D4Z4 repeat array on human chromosome 4q affects transcription of neighboring genes (Tupler et al. 1999; Winokur et al. 2003; Wohlgemuth et al. 2003; Laoudj-Chenivesse et al. 2005). To test the hypothesis that the repeat array could directly regulate gene transcription, we tested the transcription modulating activity of a D4Z4 repeat unit using a pGL3 luciferase reporter system. Constructs and control plasmids were transfected into human primary myoblasts where reporter gene expression was measured 48 h post-transfection. As seen in Figure 1B, the presence of the SV40 enhancer increased transcription fourfold in comparison with the enhancer-less pGL3-promoter plasmid. The D4Z4 repeat cloned into the pGL3–promoter plasmid stimulated luciferase synthesis with efficiency similar to that observed with the SV40 enhancer used as a positive control. Thus, the D4Z4 repeat exhibits properties of a transcriptional enhancer.
Transcriptional activators, or enhancers, are short DNA sequences composed of elements ranging in size from 10 to several hundreds of nucleotide pairs that promote transcription irrespective of their location in relation with the regulated gene. Enhancers can act at very long distances, greater than 100 kb. They can be located either at the 5′-end or 3′-end of coding sequences as well as within introns. To characterize the D4Z4 enhancer, we submitted a repeat unit to progressive deletion. This translated into an increasing enhancing effect. The strongest transcriptional enhancing activity was obtained with the two smallest fragments produced, which were 319 and 170 bp in length. Located at the 5′-end of the D4Z4 repeat, these two fragments increased transcription 12-fold as compared with the basal promoter and proved to be threefold more powerful than the SV40 enhancer (Fig. 1B). The increased activity of the minimal enhancer as compared with the whole D4Z4 repeat unit may be explained by the presence of a transcriptional repressor element, D4Z4 binding element (DBE) (Gabellini et al. 2002), within the D4Z4. Removal of this element increases the transcriptional activity of the remaining part of D4Z4. However, it is important to note that the D4Z4 repeat as a whole has an overall strong enhancer activity comparable with that of the SV40 enhancer (Fig. 1B).
We then tested the tissue and species specificity of the minimal 170-bp-long enhancer by transfecting reporter plasmids into the murine myoblast cell line C2C12, the human rhabdomyosarcoma (RMS) cell line, and HeLa cells. Similar to the SV40 enhancer, this minimal D4Z4 enhancer was found to lack any strong tissue or species specificity (Fig. 1C). When further tested in stable transfectants, the enhancer produced similar results (data not shown).
The D4Z4 enhancer up-regulates transcription from the FRG1 promoter
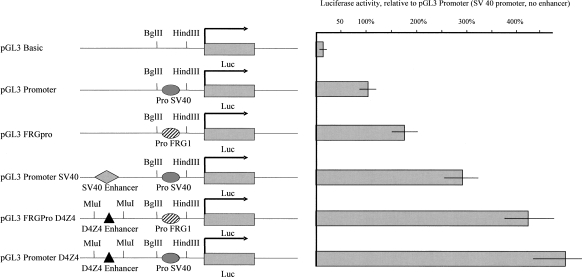
Next, we studied the minimal D4Z4 enhancer for its ability to up-regulate the activity of the promoter of FRG1, a neighboring gene which encodes a putative splicing factor (Gabellini et al. 2005; van Koningsbruggen et al. 2006) found overexpressed in myoblasts from FSHD patients (Gabellini et al. 2002; Rijkers et al. 2004; Laoudj-Chenivesse et al. 2005). A 577-bp fragment from the FRG1 gene promoter region was transferred into the promoterless pGL3-Basic plasmid for transfection into human primary myoblasts and HeLa cells. We used a luciferase assay to evaluate the enhancer activity of D4Z4 in comparison with the well-studied SV40 enhancer. As seen in Figure 2, both the SV40 promoter (pGL3 Promoter) and the FRG1 promoter (PGL3 FRGpro) constructs efficiently induced transcription as compared with the control promoterless plasmid (pGL3 Basic). When the D4Z4 minimal enhancer was transfected along with the FRG1 (pGL3FRGProD4Z4) or SV40 (pGL3PromoterD4Z4) promoters into myoblasts, transcription was up-regulated two- to threefold (Fig. 2). This was more efficient than with a plasmid that contained the SV40 enhancer in addition to the SV40 promoter (pGL3 Promoter SV40). Similar results were obtained in stable HeLa cell transfectants (data not shown). Together, these data suggested that D4Z4 could play a direct role in the transcriptional regulation of the FRG1 gene on chromosome 4q.
Figure 2.
The D4Z4 minimal enhancer up-regulates transcription from the FRG1 promoter. The effect of various promoter/enhancer combinations was observed 48 h after transfection. The luciferase activity is shown relative to that obtained with the pGL3 promoter plasmid carrying the SV40 promoter. The SV40 and FRG1 gene promoters are shown as plain and streaked gray ovals, respectively. The SV40 and D4Z4 enhancers are represented with a diamond and triangle, respectively. Luc stands for luciferase reporter gene.
FR-MAR exhibits an enhancer-blocking activity in vivo
Earlier studies have shown that gene expression profiles in the vicinity of the D4Z4 array are different in normal versus FSHD myoblasts (Tupler et al. 1999; Winokur et al. 2003; Wohlgemuth et al. 2003; Laoudj-Chenivesse et al. 2005). This may be the result of the reduction of the D4Z4 array and subsequent changes in chromatin organization (for review, see van der Maarel et al. 2006). Interestingly, it has been suggested that the D4Z4 repeat unit could possess both repressor and enhancer activities (Gabellini et al. 2002; Petrov et al. 2003; Rijkers et al. 2004). We have recently reported that the D4Z4 array was separated from neighboring genes by a nuclear matrix attachment site (FSHD-related S/MAR or FR-MAR), which does not function in the context of the retracted D4Z4 array in FSHD primary myoblasts (Petrov et al. 2006). Since S/MARs can increase gene transcription via an insulator-like effect (for review, see Bode et al. 2000), we decided to test the hypothesis that FR-MAR could similarly serve as an insulator protecting genes from the effect of neighbor sequences.
A series of pNEO plasmids were constructed based on the pGL3 series where the luciferase reporter gene was replaced by a neomycin resistance gene. First, the above-characterized 170-bp D4Z4 minimal enhancer was inserted in place of the SV40 enhancer in the pNEO plasmid. After transfection into HeLa cells and selection in the presence of G418, the number of resistant colonies was counted. As expected, the number of resistant colonies increased as compared with controls transfected with the enhancer-less plasmid (Fig. 3B). The FR-MAR and adjacent sequences (a 1263-bp HindIII fragment excised from the pGEM42 plasmid, see Fig. 3A) were then inserted, resulting in the number of G418-resistant colonies being reduced from 52 ± 7 to 22 ± 5 (Fig. 3B, cf. pNeo01 and pNeo02). As a further control, the 368-bp FR-MAR-containing fragment was then deleted from pNeo03 by HincII deletion and religation. This reconstituted the original ability of colonies to grow in the presence of G418 (45 ± 3 colonies), thus reinforcing the notion that FR-MAR indeed confers enhancer-blocking activity to transgenes (Fig. 3B, pNeo03). However, similar results would also have been obtained if FR-MAR acted as a silencer. To test this hypothesis, we used pNeo04 where the FR-MAR and adjacent sequences had been inserted in a plasmid containing only the SV40 promoter. Here, no inhibitory effect was obtained as compared with basal transcription (pNeo04, 15 ± 1 colonies vs. 17 ± 5 colonies for pNeo05, Fig. 3B). In our conditions, the construct integration efficacy was similar for all constructs, with an average of 1.5 copies per genome (Fig. 3C). The neomycin gene transcription has also been verified by Northern blot, and the data were very similar to those found by the colony counting method (Fig. 3D). From these data, we conclude that the FR-MAR indeed has enhancer-blocking activity.
Figure 3.
The FR-MAR has enhancer-blocking activity. (A) Restriction map of the pGEM42 plasmid used for FR-MAR cloning. (E) EcoRI; (Hd) HindIII; (Hc) HincII; (K) KpnI restriction sites. (B) For transfection into HeLa cells, the various plasmid constructs were linearized. Transformants were grown in the presence of 1.2 mg/mL G418 and viable colonies counted at day 10. (C) The number of inserted copies of pNeo1–5 constructs in stably transfected cell lines. DNA was purified from transfected HeLa cells, dot-blotted, and hybridized either with a probe for the Neor gene or the control GAPDH gene. The quantification was carried out using the standard concentrations of plasmid DNA mixed with nontransfected HeLa DNA as described in Methods. (D) Analysis of expression of Neor gene in stably transfected cell lines. Total RNA was purified from transfected HeLa cells, separated by gel electrophoresis, blotted, and hybridized either with a probe for the Neor gene or the control GAPDH gene. The gene expression was quantified using the ImageQuant software (Fuji).
These results appeared somewhat surprising in the light of the generally accepted notion that S/MARs augment the level of transcription of integrated transgenic constructs by insulating them from a generally unfavorable chromatin environment (Bode et al. 2000). In the present situation, FR-MAR seemed to act differently since it did not protect the construct from an inhibitory environment (compare pNeo04 and pNeo05) but rather blocked the activity of adjacent enhancers. It is known that the S/MAR activities are highly context-dependent (Schubeler et al. 1997). While boundary functions are mostly provided by extended S/MAR elements (Kalos and Fournier 1995), short elements show a spectrum of activities depending on the system The 368-bp FR-MAR may simply be too short to provide boundary functions.
A similar situation could well occur in FSHD myoblasts. Indeed, we have previously shown that, while FR-MAR is associated with the nuclear matrix in normal myoblasts, in myoblasts from FSHD patients, its counterpart present on the deleted chromosome 4q is delocalized from the nuclear matrix.
FR-MAR containing constructs are associated with the nuclear matrix
As shown previously in HeLa cells and human primary myoblasts, native FR-MARs associate with the nuclear matrix in vivo (Petrov et al. 2006). It thus appeared important to explore whether this property was conserved for FR/MARs present in the constructs used here to generate stable transfectants. To this purpose, we used the in vivo nuclear matrix mapping assay we have previously described (Ioudinkova et al. 2005). Nuclei were isolated from individual clones, treated with DNase I and high-salt extracted as described in Methods. The resulting nuclear matrices contain DNA sequences preferentially associated with the nuclear matrix and protected from DNase I digestion by nuclear matrix-associated proteins. This nuclear matrix-associated DNA was used for hybridization with short 29–31-mer oligonucleotides specifically designed to hybridize with FR-MAR, with a DNA segment lacking any affinity to the nuclear matrix (Petrov et al. 2006) and with a well-characterized structural S/MAR in the MYC gene locus (Gromova et al. 1995; Girard-Reydet et al. 2004) used here as a positive control.
In this assay, we evaluated the frequency of matrix attachment of the FR-MAR present in pNeo01 and pNeo04 constructions. As seen in Figure 4 and as expected, no association to the matrix was observed when the negative control oligonucleotide was used. In contrast, the positive MYC probe was found bound to the matrix in all transfectants. Similarly, the FR-MAR probe was found consistently bound to the matrix. To exclude that this FR-MAR association to the matrix was due to the native (genomic) FR-MAR, a DNA fragment adjacent to the FR-MAR in the constructs was used as a probe (pNeo). Similar to the adjacent FR-MAR, this probe associated with the matrix, thus confirming that the FR-MAR in the constructs behaves as does its genomic counterpart.
Figure 4.
The FR-MARs from the transgenic constructs are associated with the nuclear matrix in stable HeLa cell transfectants. The nuclear matrix-associated DNA isolated from transfected HeLa cells generated using the pNeo01and pNeo04 constructs was hybridized with an oligonucleotide mini-array consisting of Control, a 4q35 DNA fragment that does not associate with the nuclear matrix; Myc, a constitutive S/MAR from the upstream region of the human MYC gene used as a positive control; FR-MAR; and pNeo, a fragment adjacent to FR-MAR in the pNeo plasmid with no homology with human DNA. Association of the c-myc S/MAR to the nuclear matrix was considered as 100%.
Overall, the data we have obtained can be summarized as schematized in Figure 5. In normal cells (Fig. 5, top panel), the FR-MAR that lies between the centromeric FRG1/FRG2 genes and the telomeric array of D4Z4 repeats is attached to the nuclear matrix, physically separating and functionally protecting the FRG genes from the regulatory activity of the strong transcriptional enhancer present in each D4Z4 repeat unit. In contrast, in deleted myoblasts from FSHD patients (Fig. 5, bottom panel), the FR-MAR is dissociated from the nuclear matrix because of the chromosomal deletion on its telomeric side (Petrov et al. 2006). Transcription of FRG1/FRG2 genes is accordingly activated by the enhancer elements present in the remaining repeats. Whether and how this phenomenon could play a role in the genesis of FSHD will require further studies.
Figure 5.
Loop domain organization and transcriptional control in the 4q35 region. (Top) In the presence of an intact D4Z4 array, the FR-MAR is attached to the nuclear matrix, protecting the genes in one chromatin loop from the enhancing activity of D4Z4 located in the next loop. (Bottom) The D4Z4 repeat array is reduced in size, delocalizing FR-MAR from the matrix, resulting in the FRG1/FRG2 genes being present in the same loop as and up-regulated by the remaining D4Z4 repeats.
The mechanism of dissociation of the FR-MAR from the nuclear matrix in FSHD cells remains unknown; one of the hypotheses is based on the observation that a partial loss of D4Z4 repeat array in FSHD patients is linked to hypomethylation of the D4Z4 array and adjacent sequences (van Overveld et al. 2003; de Greef et al. 2007). It is known that some proteins that mediate specific association of DNA with the nuclear matrix, for example, MECP2 (Stratling and Yu 1999; Horike et al. 2005), only interact with the methylated DNA. Thus, interaction with the hypomethylated FR-MAR may be lost in FSHD patients. We are currently exploring this hypothesis and searching for the protein factor(s) that are involved in interaction with the FR-MAR in normal cells and myoblasts from the FSHD patients. Whether and how this phenomenon could play a role in the genesis of FSHD will require further studies.
Methods
Cell lines
C2C12 and HeLa cell line was purchased from American Type Culture Collection and grown in the Dulbecco’s Modified Eagle Medium (Invitrogen) supplemented with 10% fetal calf serum. RMS cells were a kind gift of Dr. S. Leibowitz.
Vectors and cloning
A series of pGL3 vectors (Promega) was used for transient transfection studies. The pGL3-Promoter vector contains an SV40 promoter upstream of the luciferase gene. The D4Z4 repeat containing putative enhancer/silencer elements was inserted either upstream or downstream of the promoter-luc+ transcriptional unit. The pGL3-Control vector contains SV40 promoter and enhancer sequences, resulting in strong expression of the reporter gene in many types of mammalian cells. It served as a positive control in the experiments on identification of a putative enhancer within the D4Z4.
A series of pNEO vectors was used for stable transfection studies. The pNeo02 vector was obtained by inserting the neomycin resistance cassette from the pIRES vector (Clontech), in place of the Luciferase unit in the pGL3-Pro-170 vector. This vector contains the 170-bp D4Z4 minimal enhancer upstream of the Luciferase promoter. The vectors were digested by BamHI and HindIII.
The D4Z4 repeat and the FR-MAR were obtained from the pGEM42 plasmid containing two D4Z4 repeats and the flanking sequences (Gabriels et al. 1999). The pGEM42 plasmid, cloned into a pGEM vector (Promega), was digested by HindIII. The 1263-bp fragment containing the FR-MAR was inserted into the pNeo02 vector, between the 170-bp D4Z4 minimal enhancer and the Neomycine cassette. This new vector was named pNeo01.
The FR-MAR was excised from the 1263-bp fragment by HincII digestion and religated to construct the pNeo03 vector. pNeo04 and pNeo05 were obtained by excising the 170-bp D4Z4 minimal enhancer by MluI digestion from the pNeo01 and pNeo03 vectors, respectively.
Transient transfection and reporter gene assays
The effect of the D4Z4 repeat on expression of the reporter gene was evaluated by transient transfection of the constructs into primary human myoblasts, murine myoblast cell line C2C12, HeLa, and RMS cells followed by estimation of the luciferase activity in transfected cells. Transfection was carried out using the JET PEI reagent (Qbiogene) according to the manufacturer’s instructions.
The constructs (with the firefly luciferase gene) were cotransfected with an internal control (with a Renilla luciferase gene) in each case. Dual-Luciferase Reporter Assay System (Promega) was used for the reporter gene expression assay. In this assay, the activities of firefly (construct) and Renilla (control) luciferases are measured sequentially from a single sample using the Top count NXT fluorimeter (Hewlett-Packard).
Stable transfection
Stable transfection was carried out using the JET PEI reagent (Qbiogene) according to the manufacturer’s instructions. The resistant clones were selected in the culture medium supplemented with G418 (1.2 μg/mL). The effect of the FR-MAR on expression of the reporter gene was also evaluated in stable clones by Northern blots.
Determination of the number of integrated pNeo constructs
Determination of the number of integrated pNeo constructs was carried out essentially as described in http://www.med.umich.edu/tamc/std.pdf. One microgram of genomic DNA purified from stably transfected HeLa cell clones was dot-blotted and hybridized either with a probe for the Neor gene or the control GAPDH gene. Standard samples containing the amounts of plasmid corresponding to 2, 5, and 10 copies per genome were generated by mixing 1.29 pg, 3.24 pg, and 6.48 pg of plasmid DNA with 1 μg of genomic DNA from nontransfected HeLa cells. All experiments were carried out in triplicate. The quantification was carried out using the ImageQuant software (Fuji).
Nuclei and nuclear matrices
Nuclei were isolated from stably transfected HeLa cells essentially as described in Gasser and Vassetzky (1998).
Nuclear matrices were prepared by treatment of the isolated nuclei with NaCl as follows: Digestion buffer (100 mM NaCl, 25 mM KCl, 10 mM Tris-HCl at pH 7.5, 0.25 mM spermidine) was added to 105 nuclei to a final volume of 400 μl. DNase I was added to a final concentration of 100 μg/mL, and the samples were digested for 2 h at 4°C, followed by the addition of CuCl2 to a final concentration of 1 mM for 10 min at 4°C. The nuclei were then extracted by addition of one volume of a buffer containing 4 M NaCl, 20 mM EDTA, and 40 mM Tris-HCl at pH 7.5. The resulting nuclear matrices were spun in a microfuge at 2000g for 10 min at 4°C and then washed three times with a buffer containing 2 M NaCl, 10 mM EDTA, and 20 mM Tris-HCl at pH 7.5.
Nuclear matrices were digested with proteinase K and extracted with phenol-chloroform. The obtained nuclear matrix-associated DNA was treated with RNase A and either radioactively labeled using the Ready-to-Go kit (APBiotech) or labeled with DIG using DIG-High Prime Kit (Roche) and used as a probe for hybridization.
Acknowledgments
We thank Dr. A. Belayev for her kind gift of the pGEM42 plasmid. We thank Ms. Ingrid Lenglart for her help in some experiments. The research has been supported by grants from the Association Française contre les Myopathies (AFM) to Y.S.V. and D.L. A.P. was supported by a post-doctoral fellowship from the Fondation pour la Recherche Médicale. I.P. was supported by a post-doctoral fellowship from AFM.
Footnotes
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6620908
References
- Bode J., Benham C., Knopp A., Mielke C., Benham C., Knopp A., Mielke C., Knopp A., Mielke C., Mielke C. Transcriptional augmentation: Modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements) Crit. Rev. Eukaryot. Gene Expr. 2000;10:73–90. [PubMed] [Google Scholar]
- Cockerill P.N., Garrard W.T., Garrard W.T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- de Greef J.C., Wohlgemuth M., Chan O.A., Hansson K.B., Smeets D., Frants R.R., Weemaes C.M., Padberg G.W., van der Maarel S.M., Wohlgemuth M., Chan O.A., Hansson K.B., Smeets D., Frants R.R., Weemaes C.M., Padberg G.W., van der Maarel S.M., Chan O.A., Hansson K.B., Smeets D., Frants R.R., Weemaes C.M., Padberg G.W., van der Maarel S.M., Hansson K.B., Smeets D., Frants R.R., Weemaes C.M., Padberg G.W., van der Maarel S.M., Smeets D., Frants R.R., Weemaes C.M., Padberg G.W., van der Maarel S.M., Frants R.R., Weemaes C.M., Padberg G.W., van der Maarel S.M., Weemaes C.M., Padberg G.W., van der Maarel S.M., Padberg G.W., van der Maarel S.M., van der Maarel S.M. Hypomethylation is restricted to the D4Z4 repeat array in phenotypic FSHD. Neurology. 2007;69:1018–1026. doi: 10.1212/01.wnl.0000271391.44352.fe. [DOI] [PubMed] [Google Scholar]
- Gabellini D., Green M.R., Tupler R., Green M.R., Tupler R., Tupler R. Inappropriate gene activation in FSHD: A repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- Gabellini D., D’Antona G., Moggio M., Prelle A., Zecca C., Adami R., Angeletti B., Ciscato P., Pellegrino M.A., Bottinelli R., D’Antona G., Moggio M., Prelle A., Zecca C., Adami R., Angeletti B., Ciscato P., Pellegrino M.A., Bottinelli R., Moggio M., Prelle A., Zecca C., Adami R., Angeletti B., Ciscato P., Pellegrino M.A., Bottinelli R., Prelle A., Zecca C., Adami R., Angeletti B., Ciscato P., Pellegrino M.A., Bottinelli R., Zecca C., Adami R., Angeletti B., Ciscato P., Pellegrino M.A., Bottinelli R., Adami R., Angeletti B., Ciscato P., Pellegrino M.A., Bottinelli R., Angeletti B., Ciscato P., Pellegrino M.A., Bottinelli R., Ciscato P., Pellegrino M.A., Bottinelli R., Pellegrino M.A., Bottinelli R., Bottinelli R., et al. Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature. 2005;439:973–977. doi: 10.1038/nature04422. [DOI] [PubMed] [Google Scholar]
- Gabriels J., Beckers M.C., Ding H., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., Beckers M.C., Ding H., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., Ding H., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., Frants R.R., Hewitt J.E., Collen D., Hewitt J.E., Collen D., Collen D., et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- Gasser S.M., Vassetzky Y.S., Vassetzky Y.S. Analysis of nuclear scaffold attachment regions. In: Gould H., editor. Chromatin: A Practical Approach. Oxford University Press; Oxford, UK: 1998. pp. 111–124. [Google Scholar]
- Gaszner M., Felsenfeld G., Felsenfeld G. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Girard-Reydet C., Gregoire D., Vassetzky Y., Mechali M., Gregoire D., Vassetzky Y., Mechali M., Vassetzky Y., Mechali M., Mechali M. DNA replication initiates at domains overlapping with nuclear matrix attachment regions in the xenopus and mouse c-myc promoter. Gene. 2004;332:129–138. doi: 10.1016/j.gene.2004.02.031. [DOI] [PubMed] [Google Scholar]
- Gromova I.I., Thomsen B., Razin S.V., Thomsen B., Razin S.V., Razin S.V. Different topoisomerase II antitumor drugs direct similar specific long-range fragmentation of an amplified c-MYC gene locus in living cells and in high-salt-extracted nuclei. Proc. Natl. Acad. Sci. 1995;92:102–106. doi: 10.1073/pnas.92.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horike S., Cai S., Miyano M., Cheng J.F., Kohwi-Shigematsu T., Cai S., Miyano M., Cheng J.F., Kohwi-Shigematsu T., Miyano M., Cheng J.F., Kohwi-Shigematsu T., Cheng J.F., Kohwi-Shigematsu T., Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- Ioudinkova E., Petrov A., Razin S.V., Vassetzky Y.S., Petrov A., Razin S.V., Vassetzky Y.S., Razin S.V., Vassetzky Y.S., Vassetzky Y.S. Mapping long-range chromatin organization within the chicken α-globin gene domain using oligonucleotide DNA arrays. Genomics. 2005;85:143–151. doi: 10.1016/j.ygeno.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Kalos M., Fournier R.E., Fournier R.E. Position-independent transgene expression mediated by boundary elements from the apolipoprotein B chromatin domain. Mol. Cell. Biol. 1995;15:198–207. doi: 10.1128/mcb.15.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoudj-Chenivesse D., Carnac G., Bisbal C., Hugon G., Bouillot S., Desnuelle C., Vassetzky Y., Fernandez A., Carnac G., Bisbal C., Hugon G., Bouillot S., Desnuelle C., Vassetzky Y., Fernandez A., Bisbal C., Hugon G., Bouillot S., Desnuelle C., Vassetzky Y., Fernandez A., Hugon G., Bouillot S., Desnuelle C., Vassetzky Y., Fernandez A., Bouillot S., Desnuelle C., Vassetzky Y., Fernandez A., Desnuelle C., Vassetzky Y., Fernandez A., Vassetzky Y., Fernandez A., Fernandez A. Increased levels of adenine nucleotide translocator 1 protein and response to oxidative stress are early events in facioscapulohumeral muscular dystrophy muscle. J. Mol. Med. 2005;83:216–224. doi: 10.1007/s00109-004-0583-7. [DOI] [PubMed] [Google Scholar]
- Lunt P.W., Harper P.S., Harper P.S. Genetic counselling in facioscapulohumeral muscular dystrophy. J. Med. Genet. 1991;28:655–664. doi: 10.1136/jmg.28.10.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M.E., Laemmli U.K., Mirault M.E., Laemmli U.K., Laemmli U.K. Organization of the higher-order chromatin loop: Specific DNA attachment sites on nuclear scaffold. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Petrov A.P., Laoudj D., Vassetzky Y.S., Laoudj D., Vassetzky Y.S., Vassetzky Y.S. Genetics and epigenetics of progressive facioscapulohumeral (Landouzy-Dejerine) muscular dystrophy. Genetika. 2003;39:147–151. [PubMed] [Google Scholar]
- Petrov A., Pirozhkova I., Laoudj D., Carnac G., Lipinski M., Vassetzky Y.S., Pirozhkova I., Laoudj D., Carnac G., Lipinski M., Vassetzky Y.S., Laoudj D., Carnac G., Lipinski M., Vassetzky Y.S., Carnac G., Lipinski M., Vassetzky Y.S., Lipinski M., Vassetzky Y.S., Vassetzky Y.S. Chromatin loop domain organization within the 4q35 locus in facioscapulohumeral dystrophy patients versus normal human myoblasts. Proc. Natl. Acad. Sci. 2006;103:6982–6987. doi: 10.1073/pnas.0511235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkers T., Deidda G., van Koningsbruggen S., van Geel M., Lemmers R.J., van Deutekom J.C., Figlewicz D., Hewitt J.E., Padberg G.W., Frants R.R., Deidda G., van Koningsbruggen S., van Geel M., Lemmers R.J., van Deutekom J.C., Figlewicz D., Hewitt J.E., Padberg G.W., Frants R.R., van Koningsbruggen S., van Geel M., Lemmers R.J., van Deutekom J.C., Figlewicz D., Hewitt J.E., Padberg G.W., Frants R.R., van Geel M., Lemmers R.J., van Deutekom J.C., Figlewicz D., Hewitt J.E., Padberg G.W., Frants R.R., Lemmers R.J., van Deutekom J.C., Figlewicz D., Hewitt J.E., Padberg G.W., Frants R.R., van Deutekom J.C., Figlewicz D., Hewitt J.E., Padberg G.W., Frants R.R., Figlewicz D., Hewitt J.E., Padberg G.W., Frants R.R., Hewitt J.E., Padberg G.W., Frants R.R., Padberg G.W., Frants R.R., Frants R.R., et al. FRG2, an FSHD candidate gene, is transcriptionally upregulated in differentiating primary myoblast cultures of FSHD patients. J. Med. Genet. 2004;41:826–836. doi: 10.1136/jmg.2004.019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D., Mielke C., Bode J., Mielke C., Bode J., Bode J. Excision of an integrated provirus by the action of FLP recombinase. In Vitro Cell. Dev. Biol. 1997;33:825–830. doi: 10.1007/s11626-997-0163-6. [DOI] [PubMed] [Google Scholar]
- Stratling W.H., Yu F., Yu F. Origin and roles of nuclear matrix proteins. Specific functions of the MAR-binding protein MeCP2/ARBP. Crit. Rev. Eukaryot. Gene Expr. 1999;9:311–318. doi: 10.1615/critreveukargeneexpr.v9.i3-4.150. [DOI] [PubMed] [Google Scholar]
- Tupler R., Perini G., Pellegrino M.A., Green M.R., Perini G., Pellegrino M.A., Green M.R., Pellegrino M.A., Green M.R., Green M.R. Profound misregulation of muscle-specific gene expression in facioscapulohumeral muscular dystrophy. Proc. Natl. Acad. Sci. 1999;96:12650–12654. doi: 10.1073/pnas.96.22.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maarel S.M., Frants R.R., Padberg G.W., Frants R.R., Padberg G.W., Padberg G.W.2006Facioscapulohumeral muscular dystrophy Biochim Biophys Acta 1772 :186–194. [DOI] [PubMed] [Google Scholar]
- van Deutekom J.C., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R., van Ommen G.J., Hofker M.H., Frants R.R., Hofker M.H., Frants R.R., Frants R.R. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- van Geel M., Heather L.J., Lyle R., Hewitt J.E., Frants R.R., de Jong P.J., Heather L.J., Lyle R., Hewitt J.E., Frants R.R., de Jong P.J., Lyle R., Hewitt J.E., Frants R.R., de Jong P.J., Hewitt J.E., Frants R.R., de Jong P.J., Frants R.R., de Jong P.J., de Jong P.J. The FSHD region on human chromosome 4q35 contains potential coding regions among pseudogenes and a high density of repeat elements. Genomics. 1999;61:55–65. doi: 10.1006/geno.1999.5942. [DOI] [PubMed] [Google Scholar]
- van Koningsbruggen S., Straasheijm K.R., Sterrenburg E., de Graaf N., Dauwerse H.G., Frants R.R., van der Maarel S.M., Straasheijm K.R., Sterrenburg E., de Graaf N., Dauwerse H.G., Frants R.R., van der Maarel S.M., Sterrenburg E., de Graaf N., Dauwerse H.G., Frants R.R., van der Maarel S.M., de Graaf N., Dauwerse H.G., Frants R.R., van der Maarel S.M., Dauwerse H.G., Frants R.R., van der Maarel S.M., Frants R.R., van der Maarel S.M., van der Maarel S.M. FRG1P-mediated aggregation of proteins involved in pre-mRNA processing. Chromosoma. 2006;116:53–64. doi: 10.1007/s00412-006-0083-3. [DOI] [PubMed] [Google Scholar]
- van Overveld P.G., Lemmers R.J., Sandkuijl L.A., Enthoven L., Winokur S.T., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M., Lemmers R.J., Sandkuijl L.A., Enthoven L., Winokur S.T., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M., Sandkuijl L.A., Enthoven L., Winokur S.T., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M., Enthoven L., Winokur S.T., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M., Winokur S.T., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M., van Ommen G.J., Frants R.R., van der Maarel S.M., Frants R.R., van der Maarel S.M., van der Maarel S.M. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- Vassetzky Y., Lemaitre J.M., Mechali M., Lemaitre J.M., Mechali M., Mechali M. Specification of chromatin domains and regulation of replication and transcription during development. Crit. Rev. Eukaryot. Gene Expr. 2000;10:31–38. [PubMed] [Google Scholar]
- Winokur S.T., Chen Y.W., Masny P.S., Martin J.H., Ehmsen J.T., Tapscott S.J., Van Der Maarel S.M., Hayashi Y., Flanigan K.M., Chen Y.W., Masny P.S., Martin J.H., Ehmsen J.T., Tapscott S.J., Van Der Maarel S.M., Hayashi Y., Flanigan K.M., Masny P.S., Martin J.H., Ehmsen J.T., Tapscott S.J., Van Der Maarel S.M., Hayashi Y., Flanigan K.M., Martin J.H., Ehmsen J.T., Tapscott S.J., Van Der Maarel S.M., Hayashi Y., Flanigan K.M., Ehmsen J.T., Tapscott S.J., Van Der Maarel S.M., Hayashi Y., Flanigan K.M., Tapscott S.J., Van Der Maarel S.M., Hayashi Y., Flanigan K.M., Van Der Maarel S.M., Hayashi Y., Flanigan K.M., Hayashi Y., Flanigan K.M., Flanigan K.M. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum. Mol. Genet. 2003;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth M., Lemmers R.J., Van Der Kooi E.L., Van Der Wielen M.J., Van Overveld P.G., Dauwerse H., Bakker E., Frants R.R., Padberg G.W., Van Der Maarel S.M., Lemmers R.J., Van Der Kooi E.L., Van Der Wielen M.J., Van Overveld P.G., Dauwerse H., Bakker E., Frants R.R., Padberg G.W., Van Der Maarel S.M., Van Der Kooi E.L., Van Der Wielen M.J., Van Overveld P.G., Dauwerse H., Bakker E., Frants R.R., Padberg G.W., Van Der Maarel S.M., Van Der Wielen M.J., Van Overveld P.G., Dauwerse H., Bakker E., Frants R.R., Padberg G.W., Van Der Maarel S.M., Van Overveld P.G., Dauwerse H., Bakker E., Frants R.R., Padberg G.W., Van Der Maarel S.M., Dauwerse H., Bakker E., Frants R.R., Padberg G.W., Van Der Maarel S.M., Bakker E., Frants R.R., Padberg G.W., Van Der Maarel S.M., Frants R.R., Padberg G.W., Van Der Maarel S.M., Padberg G.W., Van Der Maarel S.M., Van Der Maarel S.M. Possible phenotypic dosage effect in patients compound heterozygous for FSHD-sized 4q35 alleles. Neurology. 2003;61:909–913. doi: 10.1212/wnl.61.7.909. [DOI] [PubMed] [Google Scholar]