Abstract
It is widely accepted that newly arisen duplicate gene pairs experience an altered selective regime that is often manifested as an increase in the rate of protein sequence evolution. Many details about the nature of the rate acceleration remain unknown, however, including its typical magnitude and duration, and whether it applies to both gene copies or just one. We provide initial answers to these questions by comparing the rate of protein sequence evolution among eight yeast species, between a large set of duplicate gene pairs that were created by a whole-genome duplication (WGD) and a set of genes that were returned to single-copy after this event. Importantly, we use a new method that takes into account the tendency for slowly evolving genes to be retained preferentially in duplicate. We show that, on average, proteins encoded by duplicate gene pairs evolved at least three times faster immediately after the WGD than single-copy genes to which they behave identically in non-WGD lineages. Although the high rate in duplicated genes subsequently declined rapidly, it has not yet returned to the typical rate for single-copy genes. In addition, we show that although duplicate gene pairs often have highly asymmetric rates of evolution, even the slower members of pairs show evidence of a burst of protein sequence evolution immediately after duplication. We discuss the contribution of neofunctionalization to duplicate gene preservation and propose that a form of subfunctionalization mediated by coding region activity-reducing mutations is likely to have played an important role.
Theory indicates that one of three fates awaits all newly created duplicate gene pairs (Force et al. 1999; Lynch et al. 2001): nonfunctionalization (one copy is disabled and eventually lost, restoring the ancestral genotype and phenotype), subfunctionalization (the ancestral gene functions are partitioned between the two duplicates, thus restoring the ancestral phenotype but altering the genotype), or neofunctionalization (retention of both gene copies confers an advantage, so both genotype and phenotype are altered). In the event that one member of a pair becomes nonfunctionalized, the selective constraints that operated on the single ancestral gene are presumed to be inherited by the remaining functional duplicate. Indeed, unless the retained gene copy resides at a different genomic location than the ancestral gene (in which case reproductive isolation may emerge between lineages) (Lynch and Force 2000a; Scannell et al. 2006), the net effect of nonfunctionalization is likely to be the restoration of the preduplication status quo. In contrast, if a gene pair is either subfunctionalized or neofunctionalized, then both members will be maintained by selection, but the presence of (partial) redundancy may result in one or both genes experiencing an altered selective regime relative to the ancestral single-copy state. Thus, gene duplication may initiate a period of altered molecular evolution and duplicate preservation may result in this being prolonged. However, because the vast majority of new genes originate by gene duplication, the distinction between duplicated and single-copy genes is essentially a semantic one, and it is apparent that the evolutionary dynamics of a gene formed by duplication must eventually change into the dynamics of a single-copy gene.
Several authors have reported that duplicate genes exhibit an elevated rate of protein sequence evolution (Lynch and Conery 2000; Nembaware et al. 2002; Jordan et al. 2004), and this has been interpreted to mean that both members of a pair are subject to weaker purifying selection than single-copy genes (Kondrashov et al. 2002). However, it has also been observed that duplicated genes may exhibit asymmetric protein sequence evolution (i.e., the pair consists of a “slow” gene copy and a “fast” gene copy) (Van de Peer et al. 2001; Conant and Wagner 2003; Zhang et al. 2003; Brunet et al. 2006), and this asymmetry has been taken as support for the Ohno model of evolution after gene duplication (Kellis et al. 2004), which hypothesizes that one member of a pair (the “slow” copy) maintains the ancestral rate of evolution (and the ancestral role), while the “fast” copy may evolve to optimize a novel beneficial function (Ohno 1970). It is worth pointing out, however, that the observations themselves are not mutually exclusive. For instance, it is possible that young and old duplicated pairs are subject to different selection pressures, and that age differences between data sets have contributed to different conclusions. Moreover, in either case, substitutions are presumed to be accepted for the same underlying reason: The presence of a redundant gene copy complements any loss of the ancestral function (due either to a loss-of-function mutation or to the gain of an alternative function) in its paralogous partner. An important corollary of this idea is that as duplicates accumulate substitutions, they become progressively less able to complement one another (Gu et al. 2003) and at some point must fail to do so completely. Surprisingly, few investigators have studied when this loss of complementation may occur (Lynch and Conery 2000). In this study we examine the rate of protein sequence evolution in multiple time intervals after gene duplication and attempt to clarify previous observations by focusing on three distinct aspects of duplicate gene-pair evolution: the magnitude of the increase in the rate of protein sequence evolution exhibited by duplicate genes; the symmetry of this effect (whether it is exhibited equally by both copies); and the duration of the effect (how soon after gene duplication the rate of protein sequence evolution returns to the preduplication level). We do this by comparing rates of protein sequence evolution in 85 loci that were retained in duplicate and 808 loci that were returned to single-copy after the yeast whole-genome duplication (WGD) (Wolfe and Shields 1997; Dietrich et al. 2004; Kellis et al. 2004).
Our approach differs in a number of ways from that of previous investigators. First, we have chosen to study only genes for which either single-copy orthologs or double-copy co-orthologs are available in eight yeast species, four of which diverged after a WGD in their common ancestor (post-WGD yeasts; Saccharomyces cerevisiae, Saccharomyces bayanus, Candiuda glabrata, and Saccharomyces castellii) and four of which diverged from this lineage prior to the WGD (Kluyveromyces waltii, Kluyveromyces lactis, Ashbya gossypii, and Candida albicans; we refer to the first three as non-WGD yeasts and use C. albicans as an outgroup). More specifically, our set of single-copy loci consists of genes that are single-copy and orthologous in the three non-WGD yeasts and that are also currently single-copy and orthologous in all four post-WGD yeasts. In contrast, although the genes in our double-copy data set also possess only a single ortholog in each of the non-WGD yeasts, they have been retained in duplicate in all four post-WGD yeasts. Our motivation for requiring that all genes in our data sets have single-copy orthologs in multiple non-WGD species is discussed below, but the motivation for studying gene pairs that are retained in duplicate in multiple post-WGD yeasts is simple: It allows us to study the same gene pairs in successive time intervals after gene duplication.
The second major difference between our approach and previous studies is that we use concatenated alignments to study the group properties of duplicates and single-copy genes. We estimate the average increase, after the WGD, in the rate of protein sequence evolution in double-copy sequences on different branches of the phylogenetic tree. Although concatenating alignments in this manner prevents us from identifying individual gene pairs that exhibit particularly asymmetric protein sequence evolution or that are evolving very rapidly (Byrne and Wolfe 2006), it increases our power to identify general evolutionary trends associated with gene duplication. In this regard, our experimental design is similar to the study by Lynch and Conery (2000), in which data from a large number of pairs were fitted to an evolutionary model in order to make inferences about the evolution of the “average” or “ideal” gene pair.
Finally, we use a method we developed recently (Scannell et al. 2006) to correct for the fact that genes that are retained in duplicate do not comprise a random sample of the genome but are, on average, more slowly evolving (prior to duplication) than genes that are not retained in duplicate (Davis and Petrov 2004). This bias can lead to a scenario where an interspecies comparison of the rates of protein sequence evolution between sets of orthologous genes that either have paralogs or do not have paralogs can fail to detect a true increase in the rate of protein sequence evolution in the former set. It is likely that this effect has been a significant source of error in previous studies (Davis and Petrov 2004) and, by correcting for it, we show that although the rate of protein sequence evolution in duplicated genes in modern S. cerevisiae has declined significantly from its high immediately after the WGD, it still has not returned to the preduplication rate for at least one member of most gene pairs.
Results
Assessing the effect of gene duplication on protein sequence evolution
To assess the impact of gene duplication on the rate of protein sequence evolution, we first assembled two super-alignments, called A1 and A2. A1 is a concatenation of the aligned protein sequences of genes in our single-copy data set (324,540 columns from 808 loci that are single-copy in all seven ingroup species), and A2 is a concatenation of the aligned protein sequences in our double-copy data set (33,720 columns from 85 loci that are double-copy in the four post-WGD species and single-copy in the three non-WGD species). We then used the procedure described in Scannell et al. (2006) to mitigate rate biases between sequences in the super-alignments A1 and A2 due to the preferential retention of slowly evolving genes in duplicate (Davis and Petrov 2004). Briefly, this procedure pairs each column in A2 with a “control” column in A1 that contains exactly the same amino acid residues in some of the non-WGD species (usually three species; see below), and so can be considered to be following a similar evolutionary trajectory in the non-WGD species. We then use only these matched columns to assemble two new super-alignments, A1′ and A2′. Because there are about 10 times more columns in A1 than A2, it is possible to find a matching column in A1 for almost every column in A2.
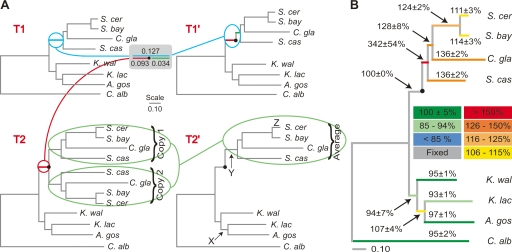
We then performed maximum likelihood branch-length evaluation on A1′ and A2′ using the established phylogenetic relationships among the yeast species represented in these super-alignments (see Methods; Scannell et al. 2006). Tree T1 is derived from the super-alignment A1′ and tree T2 is derived from the super-alignment A2′ (Fig. 1A, left). We then modify the topologies of T1 and T2 to produce a final pair of trees, T1′ and T2′, with a single topology (Fig. 1A, right). In the case of T2, we simply average the lengths of all the duplicated branches between the clades labeled “Copy 1” and “Copy 2” (Fig. 1A, bottom) and collapse one of the redundant clades. In the case of T1, we partition the branch on which the WGD occurred into pre- and post-duplication branches as described (Fig. 1A, top; Scannell et al. 2006). Because T1′ and T2′ have identical topologies (Fig. 1A, right) we can estimate the rate of protein sequence evolution on T2′ relative to T1′ by comparing branch lengths between them. For convenience, we report the length of each branch on T2′ as a percentage of the length of the corresponding branch on T1′ in all subsequent analyses. In addition, because we are only interested in this scaled value (i.e., the rate of protein sequence evolution of double-copy sequences relative to appropriate single-copy control sequences) and not the actual length of the branches on either T1′ or T2′, we will refer to this percentage simply as the “relative rate of protein sequence evolution.”
Figure 1.
Measuring the increase in the rate of protein sequence evolution after gene duplication. (A) Construction of a pair of topologically identical trees, T1′ and T2′ (right), from a tree derived from single-copy sequences only (T1; obtained from super-alignment A1′) and a tree derived from single- and double-copy sequences (T2; obtained from super-alignment A2′). The tree T1′ was derived from the tree T1 by partitioning the branch between the divergence of the non-WGD yeasts and the divergence of S. castellii from the S. cerevisiae lineage into pre- and post-duplication segments (light-blue line and gray box). As in Scannell et al. (2006), we assumed that the length of the preduplication branch on T1 is the same as that on T2 (red line). The tree T2′ was derived from the tree T2 by averaging the lengths of all duplicated branches between the post-WGD clades labeled “Copy 1” and “Copy 2” (light-green ovals). A filled circle (●) indicates the inferred point of duplicate gene divergence. The branches labeled X, Y, and Z on T2′ are referred to in the text. (B) Tree showing the length of branches on T2′ as a percentage of the length of the corresponding branches on T1′, which is a measure of the rate of evolution of double-copy sequences relative to single-copy sequences. Percentages (± one standard deviation) are averages from 100 bootstrap replicates (see Methods). The branch lengths drawn are the averages on T1′ from the same 100 bootstrap replicates. Branches are colored according to the arbitrary scale shown.
In Supplemental Figure 1, we show that our column-matching procedure can substantially reduce the effect of the bias noted by Davis and Petrov (2004) that slowly evolving genes are more likely to be retained as duplicates. We first confirm their result for our data set. In the non-WGD species K. waltii, K. lactis, and A. gossypii, the average rate of protein sequence evolution of genes that were retained in duplicate in post-WGD species is only 78%–80% of the average rate of those that were not retained in duplicate (Supplemental Fig. 1A). That is, the median evolutionary rate in the non-WGD species for genes in set A2 is about 20% lower than for those in set A1, even though the distinction between sets A2 and A1 concerns whether or not they are duplicated in a different group of species. We then demonstrate that the column-matching procedure can reduce this rate bias. We performed column-matching in three ways, by matching columns in A1 to those in A2 on the basis of having identical amino acid residues in two, three, or four non-WGD species (the three pre-WGD species noted above and Saccharomyces kluyveri) (see Methods). As the number of matched species increases, the median relative rate of protein sequence evolution in non-WGD species in A2′ relative to A1′ increases from 92% to 94% to 97% (Supplemental Fig. 1B–D), indicating that Davis and Petrov’s bias is being eliminated. We note that this is not a trivial consequence of the site-pairing procedure (which causes the non-WGD sequences to be identical in A1′ and A2′), because the branch lengths in the post-WGD clade change by a similar amount (Supplemental Fig. 1, cf. A and D; the median changes in the relative rate of protein sequence evolution in the non-WGD and post-WGD clades are 18% and 19%, respectively). Column-matching with four non-WGD species is the most effective method, but because of the more demanding match requirement (identical residues in four non-WGD species), it is not possible to find matches for 6.3% of the columns in A2 (Supplemental Fig. 1D). For the remainder of this study, we therefore chose to use super-alignments made by column-matching with three non-WGD species (K. lactis, K. waltii, and A. gossypii), because this criterion allows matching of almost all columns in A2 (99.7%) and the amelioration of the rate bias is only slightly less than when four non-WGD yeasts are used (Supplemental Fig. 1C). In addition, for convenience, we will refer to corresponding sequences in alignments to which the column-matching procedure has been applied (e.g., the K. lactis derived sequences from both A1′ and A2′) as “equivalent” sequences because, in lineages that have not undergone duplication, such sequence pairs exhibit approximately equal rates of protein sequence evolution.
Elevated rate of protein sequence evolution in double-copy sequences relative to equivalent single-copy sequences
After controlling for the Davis and Petrov effect as described above, we find that the relative rate of sequence evolution of proteins in the A2′ set is greater than the expected 100% in all branches descended from the WGD (median 128%; range 111%–342%; Fig. 1B) but very close to this value for all other branches (median 95%; range 93%–107%). The observation that all of the branches in the post-WGD clade are significantly longer than expected indicates that double-copy sequences experience a considerable increase in the rate of protein sequence evolution relative to equivalent single-copy sequences. As we discuss in more detail below, this appears to be true for duplicates derived from the WGD even in modern S. cerevisiae (the relative rate of protein sequence evolution on the terminal S. cerevisiae branch is 111 ± 3%) and appears to be especially true on the earliest branch after duplication (342 ± 54%). We also note that the change in the relative rate of protein sequence evolution on successive branches after the WGD in Figure 1B declines monotonically on successive branches from the WGD to modern S. cerevisiae (342% > 128% > 124% > 111%), which is consistent with a progressive restoration of purifying selection after gene duplication and conforms precisely to the expectation under the model outlined previously.
We performed three control experiments to confirm our observations. First, we considered the possibility that the column-matching procedure we used might artificially inflate the estimated relative rate of protein sequence evolution among double-copy sequences (although Supplemental Fig. 1A strongly suggests that this is not the case). To test this, we replaced A2 with an equal number of randomly sampled columns from A1 and carried out all other steps as previously. As expected for a negative control, we detected no acceleration on any branch (Supplemental Fig. 2A). Second, we considered that spurious matches between columns in A1 and A2 based on rare combinations of amino acids in K. lactis, K. waltii, and A. gossypii might cause us to overestimate the relative rate of protein sequence evolution in double-copy sequences. We therefore excluded all sites from A1′ and A2′ that possessed an amino acid combination in K. lactis, K. waltii, and A. gossypii that was observed fewer than five times in either A1 or A2. Excluding these sites causes our estimates of the relative rate of protein sequence evolution in double-copy sequences to be slightly increased for the post-WGD clade, and probably slightly improved for the non-WGD clade (Supplemental Fig. 2B), but ultimately supports the same conclusions as Figure 1B. Third, to exclude the possibility that the differing numbers of sequences in A1 and A2 made a comparison between trees derived from these super-alignments inappropriate, or that the tree-processing steps introduced an error of some kind, we removed all of the sequences from one of the duplicate clades from A2 (e.g., the sequences corresponding to “Copy 2” in Fig. 1A, bottom, left) and repeated all other previous steps. Again, the results were not significantly affected (Supplemental Fig. 2C), and we conclude that sequences of retained duplicate gene pairs evolve faster at the protein sequence level than equivalent single-copy sequences.
Double-copy sequences experience a burst of protein sequence evolution immediately after duplication
As expected, the greatest increase in the relative rate of protein sequence evolution among double-copy sequences is observed immediately after the WGD. On the branch between the WGD and the divergence of S. castellii from the S. cerevisiae lineage, we estimate that double-copy sequences evolved on average at 342 ± 54% the rate of equivalent single-copy sequences (Fig. 1B), and this is probably a lower-bound estimate for several reasons. One is that we averaged the relative rate of protein sequence evolution between the two duplicate clades and if (as we show below) the increase in the relative rate of sequence evolution is usually experienced primarily by one member of each duplicate pair, the increase in some gene copies could be up to twice that shown in Figure 1B. Such a value would be similar to the 10-fold average increase in the nonsynonymous substitution rate detected by Lynch and Conery (2000). In addition, we did not attempt to remove duplicated pairs that are undergoing gene conversion from our data set, except for those encoding cytosolic ribosomal proteins (see Methods). Since gene conversion will cause us to underestimate the lengths of branches on T2 only (Fig. 1A, bottom, left), it is possible that it has depressed our estimates of the rate increase in double-copy sequences. Finally, we note that all the sequences in A1 must have been duplicated for at least a short period of time after the WGD (Scannell et al. 2006), so it is possible that they also experienced a brief increase in the rate of protein sequence evolution. If this is the case, then comparing branch lengths between T1′ and T2′ will tend to underestimate the increase in the rate of protein sequence evolution attributable to gene duplication.
The branch from the WGD to the first speciation event accounts for ∼10% of the time from the WGD to the present (Scannell et al. 2006), so the increase in the relative rate of protein sequence evolution we observe on that branch is the average value over a reasonably long period of time (roughly 10 Myr). This suggests that the increase may have been more modest toward the end of this branch and potentially much greater immediately after the WGD. We used the genome sequence of K. polysporus (Scannell et al. 2007a) to investigate this possibility further. Because K. polysporus diverged from the S. cerevisiae lineage on the branch between the WGD and the divergence of S. castellii, it should allow us to partition the branch immediately after the WGD into two segments. On the branch immediately after the WGD, we expect the estimated relative rate of protein sequence evolution to be greater than 342 ± 54%, and on the other we expect it to be less. Surprisingly, however, when we applied our method to super-alignments that included K. polysporus sequences, A1Kpol and A2Kpol (similar to A1 and A2 above, but including sequences from K. polysporus; see Methods), we were unable to estimate reliably the length of the branch between the WGD and the divergence of K. polysporus on tree T1′ (this is done by comparison to T2′; see Fig. 1A). In 33 of 100 pseudo-replicates, we obtained a very short branch length (mean 0.009 amino acid substitutions per site) and consequently estimated the rate of protein sequence evolution in double-copy sequences immediately after the WGD to be >1000% of the single-copy rate in many cases. However, the remaining 67 pseudo-replicates indicated a short negative branch, and the average of all 100 pseudo-replicates was not distinguishable from zero (−0.003 ± 0.011 amino acid substitutions per site). Although this is nominally consistent with our previous conclusion that K. polysporus and S. cerevisiae diverged very soon after the WGD (Scannell et al. 2007a), additional data (not shown) indicate that two sources of error may be contributing to underestimation of the length of the branch between the WGD and this divergence event. For example, it is possible that gene-conversion events between duplicate pairs that occurred prior to the divergence of the K. polysporus and S. cerevisiae lineages are causing the WGD to appear to occur at a later time on tree T2 than was actually the case. In addition, we have previously shown that it is very difficult to determine whether genes in K. polysporus are orthologs or paralogs (created by the WGD) of their closest homologs in the other sequenced post-WGD yeast species (Scannell et al. 2007a). If some of the single-copy K. polysporus sequences in A1Kpol are paralogs rather than orthologs of the sequences from the other post-WGD species in A1Kpol, then we will infer that K. polysporus diverged from these species earlier than was actually the case. The combination of these two sources of error (gene conversion in T2 and cryptic paralogs in T1) will cause us to underestimate the length of the branch between the WGD and the divergence of K. polysporus on T1′ (Fig. 1A, top). We are therefore currently unable to confirm that the relative rate of protein sequence evolution on the branch between the WGD and the divergence of K. polysporus is >342 ± 54%. However, we were able to estimate that the relative rate of protein sequence evolution on the branch between the divergence of K. polysporus and S. castellii is 252 ± 37%, which is consistent with the pattern of a sudden increase in the rate of protein sequence evolution after WGD followed by a slowdown.
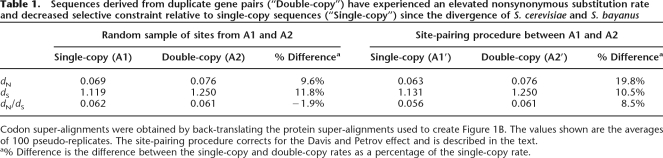
An elevated rate of protein sequence evolution persists in double-copy sequences for a long time after duplication
The rate of protein sequence evolution in double-copy sequences on the terminal S. cerevisiae branch is higher than for equivalent single-copy sequences (111 ± 3%), suggesting that duplicate pairs still experience a more permissive selective regime due to the presence of a partially redundant gene copy. Because it is surprising that this effect is still observed so long after the WGD (on the order of 100 Myr) (Wolfe and Shields 1997; Friedman and Hughes 2001), we verified this result by performing a codon-based analysis of selective constraint between orthologous sequences from S. cerevisiae and S. bayanus that are either derived from single-copy or double-copy sequences (see Methods). The divergence time between this pair of species is ∼15% of the age of the WGD (Scannell et al. 2006). The nonsynonymous substitution rate is significantly (19.8%) higher between orthologs that are members of duplicate pairs than between orthologs that are single-copy genes (Table 1). The former are also ∼10% less constrained (as inferred from the dN/dS ratio) and we note that this effect is only observed if the biased retention of slowly evolving sequences in duplicate identified by Davis and Petrov (2004) is corrected for (compare the “% Difference” in dN/dS values between columns labeled “Site-matched” and “Random sample”). Table 1 also confirms that the column-matching procedure operates by selecting a subset of sites from the super-alignment A1 that are evolving more slowly than average, but does not otherwise affect the data (compare the dN and dS values between single-copy loci for the columns labeled “Site-matched” and “Random sample”). Most importantly, however, it is clear that the altered molecular evolution of duplicated gene pairs can persist for a very long period of time after the initial duplication event.
Table 1.
Sequences derived from duplicate gene pairs (“Double-copy”) have experienced an elevated nonsynonymous substitution rate and decreased selective constraint relative to single-copy sequences (“Single-copy”) since the divergence of S. cerevisiae and S. bayanus
Codon super-alignments were obtained by back-translating the protein super-alignments used to create Figure 1B. The values shown are the averages of 100 pseudo-replicates. The site-pairing procedure corrects for the Davis and Petrov effect and is described in the text.
a% Difference is the difference between the single-copy and double-copy rates as a percentage of the single-copy rate.
Double-copy sequences evolve asymmetrically at the protein sequence level
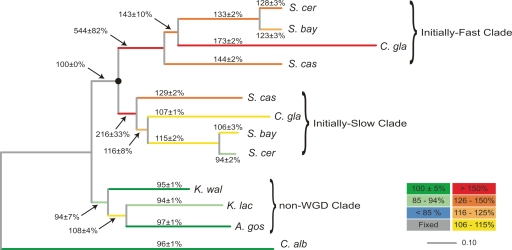
To examine the possibility of asymmetric rates of protein sequence evolution between members of duplicated pairs, we performed maximum-likelihood branch-length evaluation individually on each of the 85 double-copy loci we collected (Methods). We designated the duplicate clades (e.g., the clades labeled “Copy 1” and “Copy 2” in Fig. 1A, bottom, left) as either “initially fast–” or “initially slow–” evolving based on the relative lengths of only the first branches after the WGD (i.e., the branches from the WGD event to the divergence of S. cerevisiae and S. castellii). We then assembled a new super-alignment, A2asym, by concatenating sequences from all the “initially fast” clades together. Using A2asym and A1, we repeated all the steps performed to create Figure 1B except the final averaging step between the two duplicate clades (Fig. 1A, bottom). Instead, we compared the lengths of branches in the “initially fast” and “initially slow” clades on the tree reconstructed from A2asym separately to the equivalent branches on the tree reconstructed from single-copy loci. Branches in the “initially fast” and “initially slow” clades exhibit radically different rates of protein sequence evolution (Fig. 2). Between the WGD and the present, sequences on the “initially fast” S. cerevisiae lineage have evolved on average at 140% of the rate of those on the “initially slow” lineage. Importantly, although we treat these data as a measure of the asymmetry of protein sequence evolution, we note that the method by which we constructed A2asym (on the basis of the initial branch after the WGD only) represents an implicit test of the hypothesis that duplicated sequences evolve asymmetrically. If this hypothesis is false, then no difference in the relative rate of protein sequence evolution between “initially fast” and “initially slow” clades should be observed on any branch other than the first branch after the WGD (which we forced to be asymmetric). In fact, the distinction is apparent on every pair of branches (Fig. 2; range 15%–67%) and all differences are greater than the sum of the standard deviations on the relevant branches. Thus, within gene pairs, the copy that evolved faster immediately after the WGD tended to keep evolving faster in later time periods too.
Figure 2.
Asymmetric protein sequence evolution is initiated very soon after gene duplication and persists in modern duplicates. The tree was reconstructed from A2asym and shows branch lengths expressed as percentages of the length of the corresponding branches on a tree reconstructed from equivalent single-copy sequences (see text for details). Branch lengths are the averages of 100 pseudo-replicates and the coloring scheme is the same as in Figure 1.
To formulate an explicit test, we calculated the sum of the differences in relative rates of protein sequence evolution between the “initially fast” and “initially slow” clades for all pairs of duplicate branches excluding the first branches after the WGD, and compared the value obtained from the tree derived from A2asym to the distribution of values obtained from 100 randomized data sets (i.e., clades were not partitioned into “initially fast” and “initially slow” clades prior to concatenation). Using this measure of asymmetry, the tree derived from A2asym was more asymmetric than all but one of the randomized data sets, suggesting a nominal significance of P < 0.02. We confirmed this conclusion by comparing the number of substitutions observed on the terminal S. cerevisiae (or S. bayanus) branches with that expected, assuming equal rates of protein sequence evolution. Despite the fact that these branches are separated from the WGD—and the branches that we used to partition the data into “initially fast” and “initially slow” clades—by tens of millions of years, we find that 37% more substitutions accumulated on the terminal S. cerevisiae branch in the “initially fast” clade than on the terminal branch in the “initially slow” clade (χ2 goodness-of-fit test, P < 1 × 10−11; absolute branch-lengths 0.078 and 0.057 substitutions per site, respectively).
Although the analysis of substitutions on the terminal S. cerevisiae branches implies that our observation of asymmetric protein sequence evolution is not conditional on the inclusion of C. glabrata, it is clear from Figure 2 that C. glabrata contributes significantly to our measure of asymmetry. The difference in the relative rates of protein sequence evolution is 67% for the terminal C. glabrata branches, whereas for all other pairs of branches that contribute to our measure the contribution is in the range of from 15% to 34%. By systematically excluding one or more post-WGD species from the alignments A1 and A2asym, we show in Supplemental Table 1 that the observation of asymmetric evolution is not conditional on the inclusion of C. glabrata or any other post-WGD lineage (P < 0.02 in all cases). Similarly, we considered the possibility that the asymmetry we observe is attributable to the influence of just a small number of highly asymmetric gene pairs and is not a property shared by most double-copy loci. To address this, we generated 100 bootstrap replicates of A2asym by sampling 50% of the double-copy loci without replacement from our data set of 85 loci. Following partitioning into “initially fast” and “initially slow” clades, the mean asymmetry of these 100 data sets was significantly greater than that of 100 randomized data sets (P < 1 × 10−15 by Wilcoxon rank–sum test; Supplemental Table 2). This confirms that a substantial fraction of double-copy loci contribute to asymmetric protein sequence evolution.
Three other features of Figure 2 are notable. First, the relative rate of protein sequence evolution on the first branch after the WGD is significantly >100% in both the “initially fast” and “initially slow” clades. The relative rate on this branch in the “initially fast” clade is greater than five times the expected (single-copy) rate. More surprisingly, the relative rate in the “initially slow” clade is 216 ± 33%, more than twice the expected value. This strongly suggests that both members of duplicated pairs experience a burst of protein sequence evolution after gene duplication. This result is unlikely to be an artifact of the method we used to estimate the rate on this branch (Fig. 1B, top), because even if we assume that the WGD occurred immediately after the divergence of the non-WGD yeasts (i.e., reducing the red branch in Fig. 1A to zero, which minimizes the estimated increase in the rate of sequence evolution on the first branch after the WGD), we find that the first branch after the WGD in the “initially slow” clade is 127 ± 5% of the length of the equivalent branch on T1′. In addition, the terminal S. castellii branch in the “initially slow” clade also shows significant acceleration (Fig. 2; 129 ± 2%).
Second, although both the “initially fast” and “initially slow” duplicate clades experience a rapid decline in the rate of protein sequence evolution (Fig. 2), the levels to which they fall are very different. The terminal branches in the “initially fast” clade are still evolving much faster than expected (123%–173%), but in the “initially slow” clade the rate increase attributable to the presence of a paralog has virtually disappeared on all branches after the divergence of S. castellii (94%–116%). Again, these observations are unlikely to be artifactual, since our comparison of dN and dS between S. bayanus and S. cerevisiae (see above) supports a real difference in rates between single-copy and double-copy sequences, and alignment error is expected to be negligible at this distance. This observation is discussed in more detail in the Discussion section.
Finally, we infer that the rapid emergence of “fast” and “slow” members of gene pairs represents a decisive and largely irreversible evolutionary change, because our partitioning of genes into “initially fast” and “initially slow” copies based on the rate on the first branch after WGD is a remarkably accurate predictor of the rates of evolution on all subsequent branches (Fig. 2). In independent work, our laboratory has described this evolutionary pattern as “consistent asymmetry” (Byrne and Wolfe 2006) and possible bases for this pattern are considered there and in the Discussion section.
The pattern of amino acid substitution does not differ between double-copy and single-copy sequences
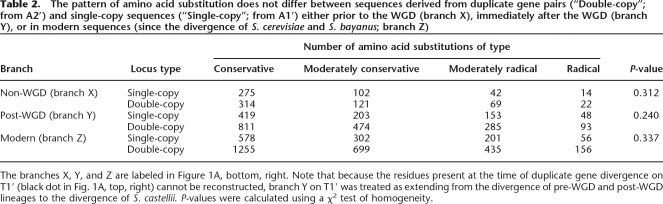
Because gene duplication is often associated with evolutionary innovation, we considered the possibility that the mode as well as the tempo of protein sequence evolution may be affected by gene duplication. We therefore compared the pattern of amino acid substitutions occurring in T2′ and T1′ on three different branches (labeled X, Y, and Z in Fig. 1A, bottom, right). We chose these branches because they are short (minimizing the number of sites that have sustained multiple substitutions), they have similar lengths (so results can be compared between branches), and because branches Y (immediately after the WGD) and Z (the branch from the divergence of S. bayanus to modern S. cerevisiae) are of particular interest. We used maximum-likelihood to reconstruct internal nodes in the trees and inferred substitutions by parsimony. We classified substitutions on a spectrum from “Conservative” to “Radical” using the Universal Evolutionary Index (Tang et al. 2004), an empirically derived index specifying the relative frequencies of amino acid-changing single nucleotide substitutions (see Methods). We did not detect any difference in the proportions of substitutions of different types between corresponding branches on T2′ and T1′ (Table 2). We obtained similar results (data not shown) when substitutions were classified using the “Grantham Matrix” (Li et al. 1985), which is based on physico-chemical properties of amino acids (Grantham 1974). Because we have sufficient statistical power to detect even a small departure from expected values, we conclude that gene duplication does not lead to a disproportionate increase in certain types of amino acid substitutions, but results in a general increase in the rate of protein sequence evolution. This is consistent with results suggesting that neither positive selection (which is likely to have contributed to asymmetric evolution of duplicate pairs after the WGD) (Fares et al. 2006) nor gene duplication per se is associated with altered patterns of amino acid substitution (Seoighe et al. 2003; Conant et al. 2006; Hanada et al. 2006).
Table 2.
The pattern of amino acid substitution does not differ between sequences derived from duplicate gene pairs (“Double-copy”; from A2′) and single-copy sequences (“Single-copy”; from A1′) either prior to the WGD (branch X), immediately after the WGD (branch Y), or in modern sequences (since the divergence of S. cerevisiae and S. bayanus; branch Z)
The branches X, Y, and Z are labeled in Figure 1A, bottom, right. Note that because the residues present at the time of duplicate gene divergence on T1′ (black dot in Fig. 1A, top, right) cannot be reconstructed, branch Y on T1′ was treated as extending from the divergence of pre-WGD and post-WGD lineages to the divergence of S. castellii. P-values were calculated using a χ2 test of homogeneity.
Discussion
In this study we examined the relative rates of protein sequence evolution of double-copy genes in successive time intervals after the yeast WGD by comparing the lengths of equivalent branches between trees drawn from double-copy and single-copy sequences (Fig. 1). Because the time between speciation events is fixed, any differences in branch lengths compared in this way must be due to differences in substitution rates. Moreover, because of the large size of our data set (Rokas et al. 2003) and provided that no systematic biases exist (Phillips et al. 2004), any observed rate differences can be attributed to gene duplication. This approach is conceptually similar to that taken by Halligan and Keightley (2006), who compared the rate of nucleotide substitution between putatively neutrally evolving intronic sites and promoter regions and concluded that the rate of evolution in promoters is constrained by purifying selection. In our case we sought to identify a sample of single-copy sites that were under a level of constraint similar to that experienced by double-copy sequences prior to the WGD. We showed empirically that matching sites between A1 and A2 that were following similar evolutionary trajectories in non-WGD yeasts is an effective way to do this (Supplemental Fig. 1).
The column-matching procedure we used here could be improved if more data were available. We used sequences from the same three non-WGD species both to pair sites between A1 and A2 and subsequently to evaluate the efficacy of the procedure, whereas it would be desirable to use different sets of taxa for these two purposes. More generally, the site-matching procedure as implemented here is an ad hoc approximation of a principled method. By pairing sites with identical combinations of amino acids we sought to pair sites that, on average, are evolving at similar rates in species that have not undergone the WGD. If genome sequences from more non-WGD species were available, however, it should be possible to assign sites in A1 and A2 to rate classes by maximum likelihood and then derive A1′ and A2′ by simply sampling the appropriate number of sites from each rate class.
The observation that both the “initially fast” and “initially slow” duplicate clades experienced a burst of protein sequence evolution after the WGD (Fig. 2) is the most striking result of this study. Previous work has typically focused either on identifying cases of asymmetric protein sequence evolution (Van de Peer et al. 2001; Conant and Wagner 2003; Zhang et al. 2003; Brunet et al. 2006) or on testing whether gene duplication leads to an increase in the rate of protein sequence evolution (Lynch and Conery 2000; Nembaware et al. 2002; Jordan et al. 2004) and has not attempted to quantify the relative contributions of the two processes. Indeed, it was often unclear whether observation of the latter effect was a consequence of failure to control for the former. Our data clarify these issues and we estimate that the “initially slow” clade evolved at twice the expected rate (216 ± 33%) on the first branch after the WGD, while the “initially fast” clade evolved at more than twice this rate again (544 ± 82%).
One possible explanation for the dramatic change in the pattern of molecular evolution after gene duplication is that it is a consequence of the mechanism of duplicate gene preservation (Kellis et al. 2004). While this view has been criticized because the long-term molecular evolution of double-copy genes does not necessarily reflect the (potentially subtle) molecular events that led to their initial preservation (Lynch and Katju 2004), we believe that it has merit in the present case. Specifically, although our analysis of the relative rates on the very short branch between the WGD and the divergence of K. polysporus is incomplete, the data are clearly compatible with our other results and indicate that the rate acceleration stems from the earliest timepoints after the WGD. On this basis, we suggest that any account of the preservation of genes in duplicate after the WGD must incorporate the dramatic (though asymmetric) increase in the rate of molecular evolution of both gene copies after duplication. Ultimately, only three explanations are possible: The presence of redundant gene copies either permitted the accumulation of previously forbidden deleterious mutations, permitted the accumulation of previously forbidden beneficial mutations (i.e., permitted an adaptive valley to be crossed) (Poelwijk et al. 2007), or a combination of both (Piatigorsky and Wistow 1991; Hughes 1994). Although we recognize that the last possibility may be the most likely, we initially consider only the first two. Of these, we believe that the rapid accumulation of slightly deleterious mutations due to the presence of a (partially) complementing paralog is much more likely than an explosive accumulation of beneficial substitutions by both duplicates. Indeed, it is not hard to imagine that immediately after the WGD, many previously deleterious mutations could be fixed. Moreover, if each member of a duplicate pair were to fix activity-reducing mutations, then neither copy could immediately be lost and more such mutations might accumulate. For the remainder of the Discussion we refer to the process of duplicate preservation due to partial loss-of-function mutations in coding regions as ARMs (activity-reducing mutations) (Stoltzfus 1999; but see also Dermitzakis and Clark 2001; Postlethwait et al. 2004) because our results pertain only to protein sequences. We point out, however, that ARMs are conceptually identical to quantitative subfunctionalization as proposed by Lynch and coworkers (Force et al. 1999; Lynch and Force 2000b).
An important consequence of ARMs is that genes will be retained in duplicate in the population for longer than would otherwise have been the case (Stoltzfus 1999; Lynch and Force 2000b). This is because the segregation of partial loss-of-function alleles can slow the dynamics of gene loss, while the fixation of a partial loss-of-function allele by one duplicate will prevent fixation of a null allele by its paralog. Note, however, that even in the latter case, the subsequent fixation of suppressor mutations at the first locus can result in a genotype that will again permit the fixation of a null allele at the second locus. Thus, ARMs do not preclude the eventual loss of one of the duplicates or a role for other mechanisms of duplicate preservation, but may function primarily to retard the return of double-copy loci to a single-copy state. This points to a key difference between ARMs (quantitative subfunctionalization) and “classical” subfunctionalization. Whereas the latter describes the loss of discrete subfunctions that may be very difficult to regain (e.g., a protein domain), the former proposes a quantitative decrease in function that is much more likely to be reversible (e.g., an enzyme whose activity has been compromised by ARMs may sustain a gain-of-function mutation that restores the ancestral level of activity). As discussed below, ARMs may thus be unusually qualified to facilitate additional evolutionary changes. In recent work, we have proposed that selection for increased gene dosage (quantitative neofunctionalization) (Scannell et al. 2007b) may have similar consequences (Conant and Wolfe 2007).
Because duplicate genes that are subject to ARMs are retained in the genome for longer, they have an increased probability of sustaining mutations that confer novel beneficial functions (Force et al. 1999; Stoltzfus 1999). Indeed, we have previously argued that neofunctionalization occurred at many loci after the WGD (Byrne and Wolfe 2006) because we showed that if one post-WGD species had lost one member of a duplicate pair, it was always the copy that was faster evolving in the other species (see, for example, REG1/REG2 in Byrne and Wolfe 2006). We observed that this is incompatible with classical subfunctionalization on the shared post-WGD branch. This observation is important because, of the genes that have been retained in duplicate in S. cerevisiae, 50% are not duplicated in K. polysporus (Scannell et al. 2007a) and 25% are not duplicated in S. castellii (Scannell et al. 2006), so they cannot have been preserved by classical subfunctionalization prior to the divergence of these species. A possibility is that some of the duplicates were preserved by classical subfunctionalization after the divergence of S. cerevisiae from the other lineage. This could occur if a locus was preserved in duplicate (perhaps due to ARMs) up to the time of divergence and was subsequently resolved independently in the two daughter lineages (Scannell et al. 2006). This appears to have occurred to the SNF12/RSC6 duplicate pair. S. cerevisiae SNF12 and RSC6 have been shown experimentally to perform nonoverlapping subsets of the functions performed by the single S. kluyveri ortholog (van Hoof 2005), yet only a single SNF12/RSC6 gene has been retained in S. castellii and K. polysporus (at the SNF12 locus; as can be seen using the Yeast Gene Order Browser) (Byrne and Wolfe 2005). These lineages must therefore have fixed null alleles at the SNF12 locus after their divergence from the S. cervisaie lineage. Although no well-supported examples of neofunctionalization after the WGD are known, by increasing the opportunity for novel beneficial functions to evolve, it is almost certain that ARMs have also facilitated the preservation of genes in duplicate by neofunctionalization. In summary, we propose that three outcomes are possible for a pair of duplicates initially maintained in duplicate by ARMs: The pair may be permanently preserved either by neofunctionalization or classical subfunctionalization; one of the copies may sustain a mutation that restores the ancestral level of function and allows the second copy to be lost; or both copies are preserved indefinitely if no suppressor, subfunctionalizing, or neofunctionalizing mutations are fixed.
An important implication of our proposal is that loci that are double-copy in multiple post-WGD species may not have been preserved by the same mechanism in all lineages. For example, in one lineage, ARMs may culminate in a complete partitioning of ancestral subfunctions (van Hoof 2005; Hickman and Rusche 2007), while in the other lineage, the increased mutational opportunity afforded by ARMs may facilitate the emergence of a novel beneficial allele at one locus. If this occurs, there may also be positive selection for the restoration of the ancestral level of function at the paralogous locus. This is consistent with the observation that substitutions favored by positive selection occurred at both fast- and slow-evolving members of duplicate gene pairs (Fares et al. 2006) and suggests that a combination of neutral and directed evolution contributed to the burst of protein sequence evolution experienced by both members of duplicate pairs after the WGD. The conclusion that loci may have been preserved in duplicate independently in different species has both theoretical and practical implications. First, by causing (sub)functions to be located at different chromosomal locations in different lineages, it may contribute to reproductive isolation between species (Lynch and Force 2000a) in a manner similar to the reciprocal loss of ancestrally duplicated genes (Scannell et al. 2006). Second, it reinforces the view that experimentation in multiple yeast species will ultimately be required to determine why particular gene pairs have been retained in duplicate (van Hoof 2005; Hickman and Rusche 2007).
Are ARMs consistent with the asymmetric protein sequence evolution seen in Figure 2? The answer to this may reside in the manner in which substitutions are accumulated after gene duplication. During the initial burst of substitution that followed the WGD, it is likely that the fraction of acceptable mutations declined rapidly as the ability of duplicates to perform the ancestral function was eroded: Every time one copy fixed a partial loss-of-function mutation, the other copy was committed to supplying more of the required function and was consequently unable to fix mutations that would have been acceptable on a different genetic background (Weinreich et al. 2006). Importantly, due to the stochastic nature of protein sequence evolution, this is likely to be an inherently asymmetric process, making an unequal division of labor between duplicates a much more likely outcome than both duplicates declining to half the ancestral level of activity. Thus, the existence of fast and slow clades in Figure 2 may primarily be a reflection of the existence of “minor” and “major” members of gene pairs, respectively (though, as noted above, directional selection is also likely to contribute). Under this view, a natural interpretation of the persistent asymmetry observed in modern sequences (Fig. 2; Table 1) is that the process of accumulation of slightly deleterious mutations is incomplete and that duplicate pairs retain redundant capacity (Harrison et al. 2007). An alternative interpretation is that the increase in the rate of sequence evolution attributable to the presence of a paralog no longer operates, but that “minor” and “major” gene copies are now expressed at different levels. In this scenario, the persistent difference in evolutionary rates may be due to the dependence of protein sequence evolution on factors such as selection for translational efficiency (Drummond et al. 2005, 2006; Kim and Yi 2006). Although we do not consider this issue resolved, we prefer the former explanation because it accords with our previous observation that the process of gene loss that was begun by the WGD may not yet be fully complete (Scannell et al. 2007a). Moreover, the continuing slowdown seen in both the “initially fast” and “initially slow” clades (Fig. 2) suggests that their rates have not yet reached equilibrium (as would be indicated by successive branches showing similar rates). We conclude that the altered selective regime experienced by double-copy sequences begins immediately after duplication and can persist for tens of millions of years.
Methods
Generation of super-alignments
We used the Yeast Gene Order Browser (YGOB) (Byrne and Wolfe 2005) to identify loci at which both duplicates derived from the WGD have been retained in the four post-WGD species S. cerevisiae, S. bayanus, C. glabrata, and S. castellii, and for which the orthology/paralogy relationships between duplicates in different species are known (double-copy loci) (Scannell et al. 2006). We also assembled a set of loci at which only single-copy syntenic orthologs have been retained in the same four post-WGD species (single-copy loci). We discarded any loci for which syntenic orthologs in the non-WGD species K. waltii, K. lactis, and A. gossypii were unavailable in YGOB, as well as any loci for which we could not identify an ortholog in C. albicans using the reciprocal-best-hit BLAST methodology between C. albicans and K. lactis. We also discarded any loci that code for cytosolic ribosomal proteins. Coding sequences for all genes were obtained from the Web site of the consortium that sequenced the relevant genome (Kellis et al. 2003, 2004; Dietrich et al. 2004; Dujon et al. 2004) except for S. castellii (Cliften et al. 2003), which we have previously reannotated (wolfe.gen.tcd.ie/ygob/scas/; Scannell et al. 2006), S. cerevisiae (www.yeastgenome.org), and C. albicans (www.candidagenome.org). We translated and aligned the sequences at each locus, removed gapped sites, and discarded alignments shorter than 50-amino-acid sites in length. Finally, we generated super-alignments by concatenating all of the alignments derived from the remaining single-copy (808) or double-copy (85) loci as appropriate. The resulting super-alignments, which we refer to as A1 and A2, consist of 324,540 and 33,720 amino acid sites (columns), respectively.
Generation of pseudo-replicates and confidence estimates
In the main text we review a sampling procedure (which we have previously described; Scannell et al. 2006) to select sub-alignments from A1 and A2 (called A1′ and A2′) that we use for phylogenetic reconstruction (see “Phylogenetics,” below). Unless otherwise stated, however, we always performed the sampling procedure 100 times and generated 100 pairs of pseudo-replicate super-alignments ([A1′1, A2′1]. . .[A1′100, A2′100]). All subsequent steps were then performed separately on each pseudo-replicate pair ([A1′n, A2′n]), and we report the average results with the associated standard deviations. Prior to generating each pseudo-replicate pair, we also randomized the relationships between duplicate clades from different loci in A2. Consider two alignments, each of which consists of two duplicate clades (DC1 and DC2), which in turn consist of four orthologs each. Because all of the sequences in DC1 (or DC2) at each locus are orthologs, but there is no relationship between sequences in DC1 (or DC2) at different loci, it is possible to concatenate the sequences in DC1 from locus one with the sequences from either DC1 or DC2 from locus two.
Phylogenetics
We determined the topology of the species tree for the yeasts used in this study by removing the C. glabrata sequence from the super-alignment A1 and generating 100 pseudo-replicate alignments (30,000 sites each) from the remaining sequences. C. glabrata was omitted because although its phylogenetic relationship to the other species is known with certainty from gene loss and other data, phylogenetic inferences based on C. glabrata sequence data have been shown to be unreliable (Scannell et al. 2006). We then used the WAG+G(8)+I+F model (as implemented in Tree-Puzzle) (Schmidt et al. 2002) to determine the maximum-likelihood topology for each bootstrap replicate and obtained a consensus topology, which is supported by all 100 pseudo-replicates. Since this topology (modified to include C. glabrata) (Fig. 1A) recapitulates the putative relationships between these yeast species (Scannell et al. 2006), it was imposed for all subsequent analyses: All parameters (branch-lengths, gamma rate classes, etc.) other than the topology were optimized for all trees derived from the super-alignments A1′ and A2′. The imposed topology was modified as necessary to accommodate the existence of duplicate gene pairs in A2′, and the WAG+G(8)+I+F model as implemented in Tree-Puzzle was used for all figures in the main text. The model WAG+G(8)+F (no invariant sites) was used for all Supplemental figures because it significantly reduces the required computation time.
S. kluyveri and K. polysporus analyses
We generated super-alignments including the non-WGD species S. kluyveri exactly as described above, in “Generation of super-alignments,” but with the additional requirement that orthologous S. kluyveri genes could be identified at each locus using the reciprocal-best-hit BLAST methodology between S. kluyveri and K. lactis proteins. We obtained 793 single-copy and 81 double-copy loci and the resulting super-alignments, A1Sklu and A2Sklu, consist of 307,374 and 29,918 aligned sites, respectively. The phylogenetic relationship between S. kluyveri and the other species was determined using the super-alignment A1Sklu and the procedure described above, in “Phylogenetics.” For analyses involving K. polysporus, we used 11 alignments of double-copy loci and 59 alignments of single-copy loci from Scannell et al. (2007a). These were concatenated (as described above, in “Generation of super-alignments”) to produce A1Kpol and A2Kpol, which consist of 23,157 and 4904 aligned sites, respectively.
Calculation of synonymous and nonsynonymous substitution rates
We calculated the average dN and dS between orthologous single-copy S. cerevisiae and S. bayanus sequences by removing all sequences other than those from S. cerevisiae and S. bayanus from A1 either before or after performing the site-pairing procedure to correct for the over-representation of slow-evolving genes in double-copy (see Results). We then replaced each amino acid with the codon that encodes it and used yn00 in the PAML package to estimate synonymous and nonsynonymous distances between the two nucleotide sequences. The procedure to estimate the average dN and dS between orthologous double-copy S. cerevisiae and S. bayanus sequences was identical except that duplicated sequences from each species were concatenated to produce a single pairwise nucleotide super-alignment (201,708 nucleotides in length; the nucleotide super-alignment derived from single-copy sequences is 100,854 nucleotides in length) prior to using yn00 to estimate synonymous and nonsynonymous distances.
Partitioning duplicates into fast-evolving and slow-evolving copies
We performed maximum-likelihood branch-length estimation individually for each of the 85 double-copy loci in our data set as described in the “Phylogenetics” section, but with four, rather than eight, gamma rate classes. We then compared the lengths of the branches between the nodes corresponding to the WGD event and the divergence of S. castellii (i.e., the first branches after the WGD) and considered the longer branch to be at the base of the “initially fast” evolving clade and the shorter branch to be at the base of the “initially slow” evolving clade. We assembled a super-alignment, A2asym, from the alignments of the remaining loci by ensuring that the fast-evolving clades were always concatenated together.
Comparison of substitution patterns between single-copy and double-copy sequences
We performed a joint reconstruction of the sequences at internal nodes of a randomly chosen pair of super-alignments A1′ and A2′ (see “Generation of pseudo-replicates and confidence estimates”) using Fastml and the model WAG+G(8) (Pupko et al. 2002). We then used parsimony to infer the substitutions between the nodes at which the marginal probability of the most likely amino acid was at least twice the probability of the next most likely one and not less than 0.25. Finally, we classified all substitutions as “Conservative,” “Moderately Conservative,” “Moderately Radical,” and “Radical” using either the Universal Evolutionary Index (Tang et al. 2004) or the Grantham Matrix (Grantham 1974; Li et al. 1985) and compared the proportions of substitutions of each type between equivalent branches on T1′ and T2′ using a χ2 test of homogeneity. Comparisons were made between three pairs of branches (X, Y, and Z) (see main text; Fig. 1A, bottom, right).
Acknowledgments
We are grateful to Mark Johnston for allowing us to use unpublished S. kluyveri sequences and to Aoife McLysaght and Cathal Seoighe for comments. We thank the members of the Wolfe lab for helpful discussions. This project was supported by Science Foundation Ireland.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6341207
References
- Brunet F.G., Crollius H.R., Paris M., Aury J.M., Gibert P., Jaillon O., Laudet V., Robinson-Rechavi M., Crollius H.R., Paris M., Aury J.M., Gibert P., Jaillon O., Laudet V., Robinson-Rechavi M., Paris M., Aury J.M., Gibert P., Jaillon O., Laudet V., Robinson-Rechavi M., Aury J.M., Gibert P., Jaillon O., Laudet V., Robinson-Rechavi M., Gibert P., Jaillon O., Laudet V., Robinson-Rechavi M., Jaillon O., Laudet V., Robinson-Rechavi M., Laudet V., Robinson-Rechavi M., Robinson-Rechavi M. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol. Biol. Evol. 2006;23:1808–1816. doi: 10.1093/molbev/msl049. [DOI] [PubMed] [Google Scholar]
- Byrne K.P., Wolfe K.H., Wolfe K.H. The Yeast Gene Order Browser: Combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne K.P., Wolfe K.H., Wolfe K.H. Consistent patterns of rate asymmetry and gene loss indicate widespread neofunctionalization of yeast genes after whole-genome duplication. Genetics. 2006;175:1341–1350. doi: 10.1534/genetics.106.066951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Majors J., Waterston R., Cohen B.A., Johnston M., Waterston R., Cohen B.A., Johnston M., Cohen B.A., Johnston M., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Conant G.C., Wagner A., Wagner A. Asymmetric sequence divergence of duplicate genes. Genome Res. 2003;13:2052–2058. doi: 10.1101/gr.1252603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant G.C., Wolfe K.H., Wolfe K.H. Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol. Syst. Biol. 2007;3:129. doi: 10.1038/msb4100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant G.C., Wagner G.P., Stadler P.F., Wagner G.P., Stadler P.F., Stadler P.F. Modeling amino acid substitution patterns in orthologous and paralogous genes. Mol. Phylogenet. Evol. 2006;42:298–307. doi: 10.1016/j.ympev.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Davis J.C., Petrov D.A., Petrov D.A. Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2004;2:E55. doi: 10.1371/journal.pbio.0020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis E.T., Clark A.G., Clark A.G. Differential selection after duplication in mammalian developmental genes. Mol. Biol. Evol. 2001;18:557–562. doi: 10.1093/oxfordjournals.molbev.a003835. [DOI] [PubMed] [Google Scholar]
- Dietrich F.S., Voegeli S., Brachat S., Lerch A., Gates K., Steiner S., Mohr C., Pohlmann R., Luedi P., Choi S., Voegeli S., Brachat S., Lerch A., Gates K., Steiner S., Mohr C., Pohlmann R., Luedi P., Choi S., Brachat S., Lerch A., Gates K., Steiner S., Mohr C., Pohlmann R., Luedi P., Choi S., Lerch A., Gates K., Steiner S., Mohr C., Pohlmann R., Luedi P., Choi S., Gates K., Steiner S., Mohr C., Pohlmann R., Luedi P., Choi S., Steiner S., Mohr C., Pohlmann R., Luedi P., Choi S., Mohr C., Pohlmann R., Luedi P., Choi S., Pohlmann R., Luedi P., Choi S., Luedi P., Choi S., Choi S., et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Drummond D.A., Bloom J.D., Adami C., Wilke C.O., Arnold F.H., Bloom J.D., Adami C., Wilke C.O., Arnold F.H., Adami C., Wilke C.O., Arnold F.H., Wilke C.O., Arnold F.H., Arnold F.H. Why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D.A., Raval A., Wilke C.O., Raval A., Wilke C.O., Wilke C.O. A single determinant dominates the rate of yeast protein evolution. Mol. Biol. Evol. 2006;23:327–337. doi: 10.1093/molbev/msj038. [DOI] [PubMed] [Google Scholar]
- Dujon B., Sherman D., Fischer G., Durrens P., Casaregola S., Lafontaine I., de Montigny J., Marck C., Neuvéglise C., Talla E., Sherman D., Fischer G., Durrens P., Casaregola S., Lafontaine I., de Montigny J., Marck C., Neuvéglise C., Talla E., Fischer G., Durrens P., Casaregola S., Lafontaine I., de Montigny J., Marck C., Neuvéglise C., Talla E., Durrens P., Casaregola S., Lafontaine I., de Montigny J., Marck C., Neuvéglise C., Talla E., Casaregola S., Lafontaine I., de Montigny J., Marck C., Neuvéglise C., Talla E., Lafontaine I., de Montigny J., Marck C., Neuvéglise C., Talla E., de Montigny J., Marck C., Neuvéglise C., Talla E., Marck C., Neuvéglise C., Talla E., Neuvéglise C., Talla E., Talla E., et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Fares M.A., Byrne K.P., Wolfe K.H., Byrne K.P., Wolfe K.H., Wolfe K.H. Rate asymmetry after genome duplication causes substantial long-branch attraction artifacts in the phylogeny of Saccharomyces species. Mol. Biol. Evol. 2006;23:245–253. doi: 10.1093/molbev/msj027. [DOI] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J., Pickett F.B., Amores A., Yan Y.L., Postlethwait J., Amores A., Yan Y.L., Postlethwait J., Yan Y.L., Postlethwait J., Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R., Hughes A.L., Hughes A.L. Gene duplication and the structure of eukaryotic genomes. Genome Res. 2001;11:373–381. doi: 10.1101/gr.155801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- Gu Z., Steinmetz L.M., Gu X., Scharfe C., Davis R.W., Li W.H., Steinmetz L.M., Gu X., Scharfe C., Davis R.W., Li W.H., Gu X., Scharfe C., Davis R.W., Li W.H., Scharfe C., Davis R.W., Li W.H., Davis R.W., Li W.H., Li W.H. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- Halligan D.L., Keightley P.D., Keightley P.D. Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res. 2006;16:875–884. doi: 10.1101/gr.5022906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Gojobori T., Li W.H., Gojobori T., Li W.H., Li W.H. Radical amino acid change versus positive selection in the evolution of viral envelope proteins. Gene. 2006;385:83–88. doi: 10.1016/j.gene.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Harrison R., Papp B., Pal C., Oliver S.G., Delneri D., Papp B., Pal C., Oliver S.G., Delneri D., Pal C., Oliver S.G., Delneri D., Oliver S.G., Delneri D., Delneri D. Plasticity of genetic interactions in metabolic networks of yeast. Proc. Natl. Acad. Sci. 2007;104:2307–2312. doi: 10.1073/pnas.0607153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman M.A., Rusche L.N., Rusche L.N. Substitution as a mechanism for genetic robustness: The Duplicated deacetylases Hst1p and Sir2p in Saccharomyces cerevisiae. PLoS Genet. 2007;3:e126. doi: 10.1371/journal.pgen.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A.L. The evolution of functionally novel proteins after gene duplication. Proc. R. Soc. Lond. B. Biol. Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- Jordan I.K., Wolf Y.I., Koonin E.V., Wolf Y.I., Koonin E.V., Koonin E.V. Duplicated genes evolve slower than singletons despite the initial rate increase. BMC Evol. Biol. 2004;4:22. doi: 10.1186/1471-2148-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S., Patterson N., Endrizzi M., Birren B., Lander E.S., Endrizzi M., Birren B., Lander E.S., Birren B., Lander E.S., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kellis M., Birren B.W., Lander E.S., Birren B.W., Lander E.S., Lander E.S. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Yi S.V., Yi S.V. Correlated asymmetry of sequence and functional divergence between duplicate proteins of Saccharomyces cerevisiae. Mol. Biol. Evol. 2006;23:1068–1075. doi: 10.1093/molbev/msj115. [DOI] [PubMed] [Google Scholar]
- Kondrashov F.A., Rogozin I.B., Wolf Y.I., Koonin E.V., Rogozin I.B., Wolf Y.I., Koonin E.V., Wolf Y.I., Koonin E.V., Koonin E.V.2002. Selection in the evolution of gene duplicationsGenome Biol. 3research0008 10.1186/gb-2002-3-2-research0008 [DOI] [Google Scholar]
- Li W.H., Wu C.I., Luo C.C., Wu C.I., Luo C.C., Luo C.C. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol. Biol. Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- Lynch M., Conery J.S., Conery J.S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Lynch M., Force A., Force A. The origin of interspecies genomic incompatibility via gene duplication. Am. Nat. 2000a;156:590–605. doi: 10.1086/316992. [DOI] [PubMed] [Google Scholar]
- Lynch M., Force A., Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000b;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Katju V., Katju V. The altered evolutionary trajectories of gene duplicates. Trends Genet. 2004;20:544–549. doi: 10.1016/j.tig.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Lynch M., O’Hely M., Walsh B., Force A., O’Hely M., Walsh B., Force A., Walsh B., Force A., Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159:1789–1804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nembaware V., Crum K., Kelso J., Seoighe C., Crum K., Kelso J., Seoighe C., Kelso J., Seoighe C., Seoighe C. Impact of the presence of paralogs on sequence divergence in a set of mouse-human orthologs. Genome Res. 2002;12:1370–1376. doi: 10.1101/gr.270902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. George Allen and Unwin; London, UK: 1970. [Google Scholar]
- Phillips M.J., Delsuc F., Penny D., Delsuc F., Penny D., Penny D. Genome-scale phylogeny and the detection of systematic biases. Mol. Biol. Evol. 2004;21:1455–1458. doi: 10.1093/molbev/msh137. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J., Wistow G., Wistow G. The recruitment of crystallins: New functions precede gene duplication. Science. 1991;252:1078–1079. doi: 10.1126/science.252.5009.1078. [DOI] [PubMed] [Google Scholar]
- Poelwijk F.J., Kiviet D.J., Weinreich D.M., Tans S.J., Kiviet D.J., Weinreich D.M., Tans S.J., Weinreich D.M., Tans S.J., Tans S.J. Empirical fitness landscapes reveal accessible evolutionary paths. Nature. 2007;445:383–386. doi: 10.1038/nature05451. [DOI] [PubMed] [Google Scholar]
- Postlethwait J., Amores A., Cresko W., Singer A., Yan Y.L., Amores A., Cresko W., Singer A., Yan Y.L., Cresko W., Singer A., Yan Y.L., Singer A., Yan Y.L., Yan Y.L. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Pupko T., Pe’er I., Hasegawa M., Graur D., Friedman N., Pe’er I., Hasegawa M., Graur D., Friedman N., Hasegawa M., Graur D., Friedman N., Graur D., Friedman N., Friedman N. A branch-and-bound algorithm for the inference of ancestral amino-acid sequences when the replacement rate varies among sites: Application to the evolution of five gene families. Bioinformatics. 2002;18:1116–1123. doi: 10.1093/bioinformatics/18.8.1116. [DOI] [PubMed] [Google Scholar]
- Rokas A., Williams B.L., King N., Carroll S.B., Williams B.L., King N., Carroll S.B., King N., Carroll S.B., Carroll S.B. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- Scannell D.R., Byrne K.P., Gordon J.L., Wong S., Wolfe K.H., Byrne K.P., Gordon J.L., Wong S., Wolfe K.H., Gordon J.L., Wong S., Wolfe K.H., Wong S., Wolfe K.H., Wolfe K.H. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440:341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
- Scannell D.R., Frank A.C., Conant G.C., Byrne K.P., Woolfit M., Wolfe K.H., Frank A.C., Conant G.C., Byrne K.P., Woolfit M., Wolfe K.H., Conant G.C., Byrne K.P., Woolfit M., Wolfe K.H., Byrne K.P., Woolfit M., Wolfe K.H., Woolfit M., Wolfe K.H., Wolfe K.H. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc. Natl. Acad. Sci. 2007a;104:8397–8402. doi: 10.1073/pnas.0608218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell D.R., Butler G., Wolfe K.H., Butler G., Wolfe K.H., Wolfe K.H. Yeast genome evolution-the origin of the species. Yeast. 2007b doi: 10.1002/yea.1515. [DOI] [PubMed] [Google Scholar]
- Schmidt H.A., Strimmer K., Vingron M., von Haeseler A., Strimmer K., Vingron M., von Haeseler A., Vingron M., von Haeseler A., von Haeseler A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Seoighe C., Johnston C.R., Shields D.C., Johnston C.R., Shields D.C., Shields D.C. Significantly different patterns of amino acid replacement after gene duplication as compared to after speciation. Mol. Biol. Evol. 2003;20:484–490. doi: 10.1093/molbev/msg059. [DOI] [PubMed] [Google Scholar]
- Stoltzfus A. On the possibility of constructive neutral evolution. J. Mol. Evol. 1999;49:169–181. doi: 10.1007/pl00006540. [DOI] [PubMed] [Google Scholar]
- Tang H., Wyckoff G.J., Lu J., Wu C.I., Wyckoff G.J., Lu J., Wu C.I., Lu J., Wu C.I., Wu C.I. A universal evolutionary index for amino acid changes. Mol. Biol. Evol. 2004;21:1548–1556. doi: 10.1093/molbev/msh158. [DOI] [PubMed] [Google Scholar]
- de Van Peer Y., Taylor J.S., Braasch I., Meyer A., Taylor J.S., Braasch I., Meyer A., Braasch I., Meyer A., Meyer A. The ghost of selection past: Rates of evolution and functional divergence of anciently duplicated genes. J. Mol. Evol. 2001;53:436–446. doi: 10.1007/s002390010233. [DOI] [PubMed] [Google Scholar]
- van Hoof A. Conserved functions of yeast genes support the Duplication, Degeneration and Complementation model for gene duplication. Genetics. 2005;171:1455–1461. doi: 10.1534/genetics.105.044057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich D.M., Delaney N.F., Depristo M.A., Hartl D.L., Delaney N.F., Depristo M.A., Hartl D.L., Depristo M.A., Hartl D.L., Hartl D.L. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- Wolfe K.H., Shields D.C., Shields D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Zhang P., Gu Z., Li W.H., Gu Z., Li W.H., Li W.H. Different evolutionary patterns between young duplicate genes in the human genome. Genome Biol. 2003;4:R56. doi: 10.1186/gb-2003-4-9-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]