Abstract
The finishing of the Human Genome Project largely completed the detailing of human euchromatic sequences; however, the most highly repetitive regions of the genome still could not be assembled. The 12 gene clusters producing the structural RNA components of the ribosome are critically important for cellular viability, yet fall into this unassembled region of the Human Genome Project. To determine the extent of human variation in ribosomal RNA gene content (rDNA) and patterns of rDNA cluster inheritance, we have determined the physical lengths of the rDNA clusters in peripheral blood white cells of healthy human volunteers. The cluster lengths exhibit striking variability between and within human individuals, ranging from 50 kb to >6 Mb, manifest essentially complete heterozygosity, and provide each person with their own unique rDNA electrophoretic karyotype. Analysis of these rDNA fingerprints in multigenerational human families demonstrates that the rDNA clusters are subject to meiotic rearrangement at a frequency >10% per cluster, per meiosis. With this high intrinsic recombinational instability, the rDNA clusters may serve as a unique paradigm of potential human genomic plasticity.
Each human ribosome contains four RNA molecules (rRNA) that play indispensable roles in all protein translation. In order to produce sufficient rRNA for the highly abundant ribosomes, the genes encoding the rRNA are represented genomically in multiple copies. The 5S rRNA molecule (OMIM 180420) is produced from a cluster of repeated 2.2-kb genes (Little and Braaten 1989; Sorensen and Frederiksen 1991) on maternal and paternal Chromosome 1q42 (Sorensen et al. 1991). Similarly, the remaining three rRNA molecules—18S, 5.8S, and 28S—are produced by post-transcriptional processing of a 45S precursor transcript expressed from clusters of repeated 43-kb genes (Gonzalez and Sylvester 1995) on maternal and paternal 13p12, 14p12, 15p12, 21p12, and 22p12 (OMIM 180450–180454) (Henderson et al. 1972). Overall, the rDNA clusters consist of repeated individual rRNA genes abutting each other in a largely head-to-tail orientation, devoid of intervening non-rDNA sequences (Little and Braaten 1989; Sorensen and Frederiksen 1991; Shiels et al. 1997; Schofer et al. 1998; Caburet et al. 2005).
The repetitive, clustered nature of the rDNA was noted in early quantitative hybridization studies (Schmickel and Knoller 1977) and led to the hypothesis that, “the number of rRNA genes, although a polymorphic character, is an inheritable attribute of [a] given nucleolar organizer” (Guanti and Petrinelli 1974). This prediction was supported by tracking patterns of silver-stained acrocentric chromosomes in human families (Markovic et al. 1978; Taylor and Martin-DeLeon 1981), although the stain was noted to be a function of both rDNA quantity and transcriptional activity (Miller et al. 1976). Through analysis of sequence polymorphisms within individual rDNA repeat units, it was determined that there was strong recombination activity both intra- and interchromosomally within the rDNA, leading to concerted evolution of rDNA sequences and clusters (Arnheim et al. 1980; Worton et al. 1988; Kuick et al. 1996; Gonzalez and Sylvester 2001). Nevertheless, direct biochemical evidence of meiotic rDNA cluster recombination and quantitation of rDNA within individual rDNA clusters has proven elusive. We present here the determination of individual rDNA cluster lengths, the corresponding degree of person-to-person variability, and patterns of meiotic rDNA cluster inheritance and recombination.
Results
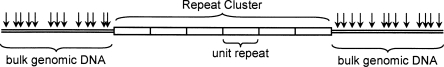
Our experimental approach is shown in Figure 1. Very high molecular weight genomic DNA is prepared from human peripheral blood (Birren and Lai 1993), and intact rDNA clusters are released from the bulk genomic DNA by restriction digestion. Since the individual rRNA genes are very highly self-similar (Sorensen et al. 1990; Gonzalez and Sylvester 2001), liberating intact rDNA gene clusters is feasible, as a restriction enzyme without a recognition site in a single rDNA gene will typically lack a recognition site throughout the entire cluster. The intact clusters are separated by size in a pulsed field gel and detected by hybridization with either a 5S or 45S rDNA-specific radiolabeled probe.
Figure 1.
Experimental Strategy. Release of intact rDNA clusters from bulk genomic DNA. (Open rectangles) Individual rDNA repeat units; (double line) non-rDNA genomic DNA; (arrows) hypothetical restriction sites.
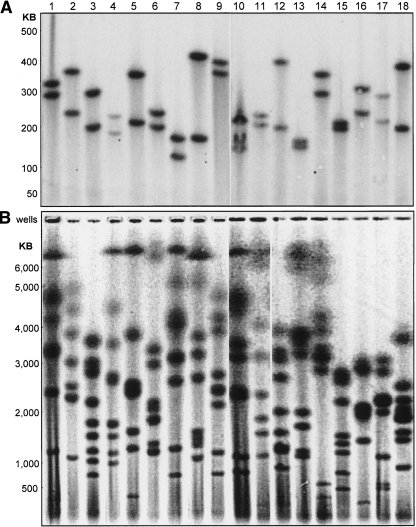
5S rDNA clusters (Fig. 2A) isolated from anonymous human blood donors demonstrate tremendous allelic diversity, with most of the donors showing two distinct 5S rDNA cluster lengths that clearly indicate differentially sized 5S rDNA clusters between maternal and paternal 1q42. Indeed, we have yet to observe a person homozygous for 5S rDNA cluster lengths. Occasionally, as seen in donor #10, we resolve three 5S rDNA bands; the presence of an extra band is enzyme-specific, and we infer that a chance polymorphism in one of the 5S repeats has coincidentally created this specific restriction site within the gene cluster.
Figure 2.
Normal human rDNA cluster length variability. Eighteen healthy anonymous human donors are shown. (A) 5S rDNA gene clusters. (B) 45S rDNA gene clusters. Cluster lengths are based on chromosome size standards from Schizosaccharomyces pombe, Hansenula wingei, and lambda phage concatemers.
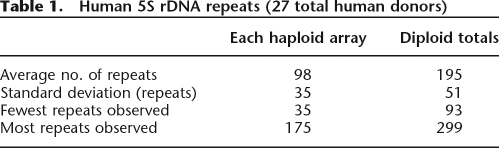
The Human Genome Project (build 36.2) (International Human Genome Sequencing Consortium 2004) shows 17.2 5S rDNA repeats on 1q42 with an overall sequence conservation of 99.6% (Clamp et al. 2004). The Genome Project and other sources (Little and Braaten 1989; Sorensen and Frederiksen 1991; Timofeeva et al. 1993) give a unit 5S rDNA repeat length of 2236 bp. Using our 5S rDNA cluster length data and the lengths of the unit 5S rDNA repeat and non-rDNA flanking sequences (International Human Genome Sequencing Consortium 2004), we directly calculated the number of human 5S rDNA repeats (Table 1) in a total of 27 human donors (18 shown in Fig. 2A and nine additional unrelated individuals from Fig. 3A). Our average 5S rDNA cluster length of 98 repeats is consistent with earlier hybridization studies (Sorensen and Frederiksen 1991); however, the Human Genome Project’s total of 17.2 repeats is >2.3 standard deviations outside this value, likely as a result of the difficulty the Genome Project has in correctly assembling very highly self-similar sequences (She et al. 2004).
Table 1.
Human 5S rDNA repeats (27 total human donors)
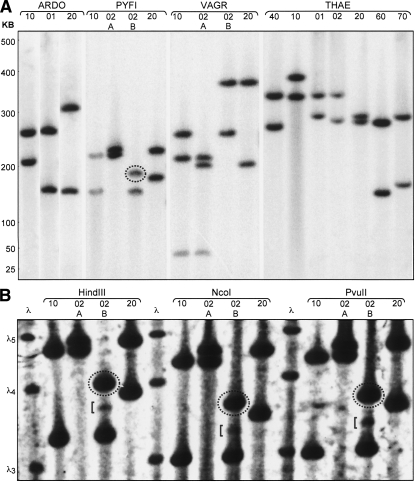
Figure 3.
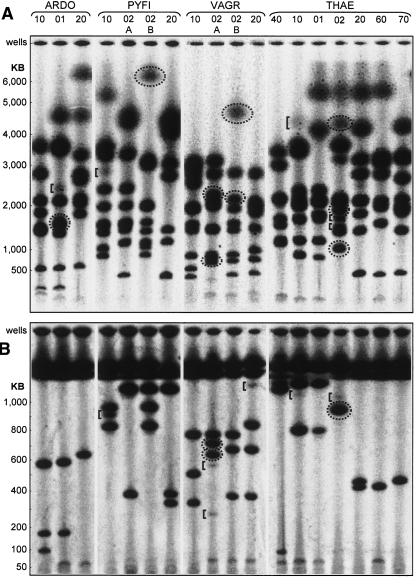
(A) Genomic architecture and patterns of human inheritance of 5S ribosomal DNA gene clusters. Four families, “ARDO,” “PYFI,” “VAGR,” and “THAE” are shown. Relationship codes: (01) daughter, (02) son, (10) mother, (20) father, (40) maternal grandmother, (60) paternal grandmother, (70) paternal grandfather. Siblings of the same sex and generation are additionally designated “A,” “B,” in descending birth order. (Dotted oval) A band not represented in either parent. rDNA cluster lengths are measured in kilobases and calibrated using lambda phage concatemers. (B) PYFI family 5S rDNA structure at high resolution and contrast. Restriction enzymes used to liberate 5S gene clusters from bulk genomic DNA are shown above sets of family samples. (Dotted ovals) Bands not represented in either parent. (Square brackets) Bands of nonstoichiometric signal intensity. (Left) Lambda phage concatemer size references: λ3 = 145,506 bp, λ4 = 194,008 bp, λ5 = 242,510 bp.
45S rDNA cluster lengths from the same anonymous donors (Fig. 2B) also exhibit striking person-to-person variability. Consistent with 45S clusters on the five pairs of human acrocentric chromosomes, our pulsed field gel analysis revealed eight to 10 bands per person. We assume that individuals showing fewer than 10 bands have yet additional bands >10 Mb that are incapable of entering a pulsed field gel (Birren and Lai 1993), although complete loss of rDNA sequences on specific acrocentric chromosomes is an alternative possibility. The 45S rDNA cluster size variation is consistent with our 5S rDNA cluster data: the 45S rDNA clusters display distinct and independent size variation on parentally homologous chromosomes. The 45S rDNA clusters shown here range in length from ∼70 kb to >6 Mb. For a unit 45S rDNA repeat size of 43 kb (Gonzalez and Sylvester 1995), these data indicate cluster sizes from a single 45S rDNA unit alone up to >140 repeats. While the lack of resolution above 6 Mb precludes an exact determination of human total diploid 45S rDNA content, our results are consistent with the upper range of earlier hybridization experiments of ∼600 copies per diploid human cell (Schmickel 1973). In contrast to the 5S rDNA clusters on 1q42, there are currently no data from the Human Genome Project concerning 45S rDNA cluster lengths.
Beyond variation in cluster length between individuals, we observed other intriguing features of 5S and 45S rDNA architecture, including evidence of significant meiotic rearrangement. In our analysis of the 5S rDNA clusters from four different human families (Fig. 3A), it can be seen that the paternal 5S rDNA cluster of PYFI-02B was inherited with a meiotic rearrangement. We verified the presence of this de novo 5S rDNA cluster length, relative to the 5S clusters in PYFI-02B’s parents PYFI-10 and PYFI-20, with a panel of three different restriction enzymes (Fig. 3B). Commercial short tandem repeat (STR) testing verified that PYFI-20 is, indeed, the biological father of PYFI-02B (see Supplemental Material).
The higher resolution of the 5S rDNA clusters in Figure 3B also revealed an additional faint band in PYFI-02B of intermediate length at ∼2% of the intensity of the two major 5S rDNA cluster bands for this individual. The intensity of the bands is directly proportional to both the cluster length and the percentage of cells in the sample harboring a particular 5S rDNA cluster in these experiments. Therefore, we interpret this fainter band to be the product of a somatic recombination event occurring in the hematopoietic stem cell population during the embryogenesis of PYFI-02B, making the peripheral blood of PYFI-02B genetically mosaic for the 5S rDNA clusters. Given that three distinct restriction enzymes yielded the same banding pattern, we can rule out the possibility of partial digestion as a source of this band.
The 45S rDNA clusters display an even larger degree of both meiotic rearrangement and somatic mosaicism (Fig. 4A). There are clear examples of inherited rearrangement in five of the offspring (ARDO-01, PYFI-02B, VAGR-02A, VAGR-02B, and THAE-02), as indicated by the bands enclosed in dotted circles. For example, VAGR-02B possesses a cluster of 4.6 Mb that is 1.6 Mb larger than the largest cluster from either his mother (VAGR-10) or his father (VAGR-20). A similar expansion is observed in the 6-Mb cluster of PYFI-02B that is at least 0.5 Mb larger than the largest rDNA cluster in either of his parents. Since clusters of repeated genes are potentially susceptible to recombinational erosion by single-strand annealing (Stark et al. 2004), the ability to extend the cluster lengths through meiotic recombination may be evolutionarily important for maintaining a steady-state level of rRNA genes.
Figure 4.
(A) Genomic architecture and patterns of human inheritance of 45S ribosomal DNA gene clusters. Four families, “ARDO,” “PYFI,” “VAGR,” and “THAE” are shown. Relationship codes: (01) daughter, (02) son, (10) mother, (20) father, (40) maternal grandmother, (60) paternal grandmother, (70) paternal grandfather. Siblings of the same sex and generation are additionally designated “A,” “B” in descending birth order. (Dotted ovals) Bands not represented in either parent. (Square brackets) Bands of nonstoichiometric signal intensity. Cluster lengths are calibrated on chromosomal size standards from S. pombe, H. wingei, and S. cerevisiae and lambda phage concatemers. (B) High resolution. Note resolution of two bands at 600–700 kb in VAGR-02A with respect to the unresolved doublet in this size range in panel A.
A high-resolution gel for the 45S rDNA clusters <1 Mb in size (Fig. 4B) provides a superior view of this meiotic rearrangement. VAGR-02A has clusters of 625 kb and 715 kb (dotted circles) that are not found in either his mother (VAGR-10) or his father (VAGR-20). Likewise, the 950-kb cluster found in THAE-02 is not found in either of his parents (THAE-10 and THAE-20). Collectively, we observe 11 clear examples of 5S or 45S rDNA cluster meiotic restructuring in five of the eight parent–child trios analyzed.
Similar to the 5S rDNA cluster analysis of PYFI-02B (Fig. 3B), minor bands arising from 45S rDNA cluster somatic mosaicism are also apparent (Fig. 4B). PYFI-10 shows the strongest minor banding with a 910-kb length cluster at ∼25% of the intensity of her major 45S rDNA bands in that size range. The VAGR-02A, VAGR-20, THAE-10, and THAE-02 lanes all possess minor bands ranging from 5% to 10% the intensity of their respective major banding patterns. It is possible that the minor band in THAE-10 at 4.3 Mb (Fig. 4A) represents a 45S rDNA mosaicism that exists in some fraction of her germline, such that her son THAE-02 inherited this cluster in Mendelian fashion from her. In total, we can detect minor-intensity mosaic banding patterns in about one-third of the individuals tested.
Discussion
Collectively, we observed one occurrence of 5S and 10 occurrences of 45S rDNA gene cluster meiotic restructuring in five of eight parent–child trios analyzed, for a total of 11 clearly identifiable events. Given a potential total of 96 different rDNA clusters (eight children, each with two 5S and up to 10 45S rDNA clusters), this yields an average recombination rate of 11% per generation per gene cluster (95% confidence interval: 6.4%–20%) (Agresti and Coull 1998). This rapid meiotic restructuring likely accounts for the present-day interperson variability in both rDNA quantity and genomic organization. We cannot currently ascertain the relative degree to which this diversity is generated by recombination between rDNA clusters on nonhomologous chromosomes, parental homologs, or sister chromatids, although familial analysis of flow-sorted acrocentric chromosomes may be able to answer this question.
Molecular combing experiments indicate that the rDNA clusters largely consist of repeats oriented in a head-to-tail configuration, but also with a substantial fraction of individual repeats within any given rDNA cluster in the inverted or palindromic orientation (Caburet et al. 2005). Intermolecular recombination between such inverted rDNA repeats would generate a dicentric chromosomal translocation product linked by an rDNA cluster, reminiscent of the Robertsonian translocations between the various acrocentric chromosomes frequently observed in the human population. Actual Robertsonian translocations, however, almost invariably have fusion breakpoints not in the rDNA, but rather in satellite III DNA proximal to the centromere, with resulting complete loss of rDNA sequences (Page et al. 1996; Sullivan et al. 1996), and this is also true for dup(21q) chromosomes that account for 2% of Down syndrome patients (Antonarakis et al. 1990). Although there are scattered examples (Boyd et al. 2005), why rDNA recombination-mediated acrocentric chromosome fusions are generally not observed is unclear. Conceivably, the greater distance between centromeres in such an rDNA acrocentric fusion, relative to the smaller intercentromeric distance of satellite III fused Robertsonians, would allow both centromeres to retain activity and lead to fusion chromosome loss, whereas inactivation of one of the two Robertsonian centromeres allows stable chromosomal transmission (Page and Shaffer 1998).
The human 5S and 45S rDNA gene clusters are clearly capable of very rapid copy number evolution. Although there may be considerable flexibility in rRNA expression at the transcriptional level via epigenetic control (Kawasaki et al. 1992; Mayer et al. 2006), for long-term maintenance of population equilibrium rDNA levels, there must be underlying evolutionary pressures constraining the total rDNA copy number between specific minimum and maximum values. It is not intuitively obvious what the phenotype of an rDNA deficiency would be. Chicken strains in which rDNA levels are reduced to 56% of wild-type levels are viable (Delany et al. 1995); however, a reduction to 45% of wild-type rDNA levels results in teratogenic embryonic failure (Delany et al. 1994). We suggest that in humans, a partial rDNA insufficiency may manifest as bone marrow failure, similar (Ellis and Massey 2006) to the way in which ribosomal protein haploinsufficiency can cause Diamond-Blackfan anemia (Draptchinskaia et al. 1999; Gazda et al. 2006).
We anticipate that further study of the rDNA gene clusters, which are fundamentally required for cellular metabolism yet are highly variable in copy number and exhibit significant instability, will provide further insight into the role of copy number variation in human genomic architecture and stability.
Methods
High molecular weight genomic DNA was prepared in agarose from peripheral blood via ammonium chloride erythrocyte lysis (generally according to methods described in Birren and Lai 1993) with a final concentration of 1 × 107 white blood cells per milliliter of 0.8% low-melting temperature agarose. Agarose/DNA slices of ∼15 μL volume were digested with 50 units of indicated restriction enzymes (New England Biolabs) overnight at 37°C and subsequently sealed with low melting temperature agarose into gels for electrophoresis. All electrophoresis was performed using a CHEF Mapper (Bio-Rad) system. Gels were subsequently processed according to Lueders and Fewell (1994) and Ehtesham and Hasnain (1991) with minor modifications.
Hybridization probes
5S rDNA
Human genomic DNA was PCR-amplified using primers 5′-GGGCTCGAGGACAAAAGTAGCGCGAGGTC and 5′-GGGT CTAGACAGCCACCGGGAAAACAG, yielding a 489-bp fragment.
45S rDNA
Human genomic DNA was PCR-amplified using primers 5′-GGGCTCGAGATTTGGGACGTCAGCTTCTG and 5′-GGGTCT AGAGTGCTCCCTTCCTCTGTGAG, yielding a 532-bp fragment.
PCR products were subsequently digested with XhoI and XbaI, subcloned individually into pBluescript II SK− (Stratagene), and verified by DNA sequencing. XhoI/XbaI fragments from these plasmids were 32P radiolabeled using a Prime-It II kit (Stratagene).
Pulsed field gels
The conditions in Figure 2A were genomic DNA digested with EcoRV; 1% Pulse Field Certified (PFC) agarose (Bio-Rad); 0.5× TBE (44.5 mM Tris, 44.5 mM borate, 1 mM EDTA at pH 8.0); 14°C; 6 V/cm; 120° included angle; 3–90-sec field switching with linear ramp; and 24 h of total electrophoresis.
The conditions in Figures 2B and 4A were genomic DNA digested with EcoRV; 1% SeaKem Gold agarose (Cambrex); 1× TAE (40 mM Tris, 40 mM acetate, 1 mM EDTA at pH 8.0); 14°C; 2 V/cm; 106° included angle; 5–40-min field switching with linear ramp; and 92 h of total electrophoresis.
The conditions in Figure 3A were genomic DNA digested with HindIII; 1% PFC agarose (Bio-Rad); 0.5× TBE; 14°C; 6 V/cm; 120° included angle; 10–40-sec field switching with linear ramp; and 20 h of total electrophoresis.
The conditions in Figure 3B were 1% PFC agarose; 0.5× TBE; 14°C; 6 V/cm; sequence of electric field vectors relative to 0° as “forward”: −60°, 180°, +60°, −60°, +60°, 180°, −60°, +60°; field durations: ±60° field vectors: 10 sec; 180° field vectors: 5 sec, 42 h of total electrophoresis. Pulse field parameters were adapted from Clark et al. (1988).
The conditions in Figure 4B were as in Figure 2A, except we used a “ramp factor” (Bio-Rad) of 0.357.
Gel post-electrophoresis processing and hybridization
Gels were equilibrated to a final concentration of 0.5% glycerol in water, dried at 65°C, and subsequently rehydrated with two exchanges of distilled water. Rehydrated gels were denatured with 0.4 N NaOH/0.8 M NaCl followed by neutralization in 0.5 M Tris (pH 8.0)/0.8 M NaCl. Gels were pre-hybridized in 2× SSC (300 mM NaCl, 30 mM Na-citrate), 7% SDS, and 0.5% casein at 65°C and then hybridized to radiolabeled probe sequences overnight at 65°C in fresh pre-hybridization solution. Gels were rinsed at 65°C with 2× SSC and 0.1% SDS for 30 min and then again with fresh solution for 60 min. Gels were then rinsed twice at 65°C with 0.5× SSC and 0.1% SDS for 2 h per rinse. Gels were finally rinsed briefly with 2× SSC at room temperature and imaged using a PhosphorImager system (Molecular Dynamics).
Commercial STR allele testing
Dried blood spotted on FTA Micro Cards (Whatman) was analyzed by The Genetic Testing Laboratories, Inc. (GTL), Las Cruces, NM. We use simplified identifying labels for the PYFI family in the main text. The labels used for the samples submitted for allelic testing shown in the Supplemental material are as follows: PYFI-10 (mother): PYFI-10-MA, PYFI-02A (elder son): PYFI-02-GA, PYFI-02B (younger son): PYFI-02-TY, PYFI-20 (father): PYFI-20-BR.
Human subjects
All human subjects protocols were reviewed and approved by the Institutional Review Board of the University of Kentucky.
Acknowledgments
We thank our human families and the Kentucky Blood Center for providing blood. Funding for these studies was provided by the Markey Cancer Center and aided by an Institutional Research Grant from the American Cancer Society (A.J.P.). D.M.S. performed the experiments in Figures 2A and 3. M.W.K. helped prepare genomic DNA from blood. H.H.P. recruited and consented human subjects. A.J.P. performed the experiments in Figures 2B and 4. D.M.S. and A.J.P. wrote the paper. All authors discussed the results and commented on the manuscript.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6858507
References
- Agresti A., Coull B.A., Coull B.A. Approximate is better than “exact” for interval estimation of binomial proportions. Am. Stat. 1998;2:119–125. [Google Scholar]
- Antonarakis S.E., Adelsberger P.A., Petersen M.B., Binkert F., Schinzel A.A., Adelsberger P.A., Petersen M.B., Binkert F., Schinzel A.A., Petersen M.B., Binkert F., Schinzel A.A., Binkert F., Schinzel A.A., Schinzel A.A. Analysis of DNA polymorphisms suggests that most de novo dup(21q) chromosomes in patients with Down syndrome are isochromosomes and not translocations. Am. J. Hum. Genet. 1990;47:968–972. [PMC free article] [PubMed] [Google Scholar]
- Arnheim N., Krystal M., Schmickel R., Wilson G., Ryder O., Zimmer E., Krystal M., Schmickel R., Wilson G., Ryder O., Zimmer E., Schmickel R., Wilson G., Ryder O., Zimmer E., Wilson G., Ryder O., Zimmer E., Ryder O., Zimmer E., Zimmer E. Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proc. Natl. Acad. Sci. 1980;77:7323–7327. doi: 10.1073/pnas.77.12.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birren B., Lai E., Lai E., editors. Pulsed field gel electrophoresis—A practical guide. Academic Press; San Diego, CA: 1993. [Google Scholar]
- Boyd L.J., Livingston J.S., Brown M.G., Lawce H.J., Gilhooly J.T., Wildin R.S., Linck L.M., Magenis R.E., Pillers D.A., Livingston J.S., Brown M.G., Lawce H.J., Gilhooly J.T., Wildin R.S., Linck L.M., Magenis R.E., Pillers D.A., Brown M.G., Lawce H.J., Gilhooly J.T., Wildin R.S., Linck L.M., Magenis R.E., Pillers D.A., Lawce H.J., Gilhooly J.T., Wildin R.S., Linck L.M., Magenis R.E., Pillers D.A., Gilhooly J.T., Wildin R.S., Linck L.M., Magenis R.E., Pillers D.A., Wildin R.S., Linck L.M., Magenis R.E., Pillers D.A., Linck L.M., Magenis R.E., Pillers D.A., Magenis R.E., Pillers D.A., Pillers D.A. Meiotic exchange event within the stalk region of an inverted chromosome 22 results in a recombinant chromosome with duplication of the distal long arm. Am. J. Med. Genet. A. 2005;138:355–360. doi: 10.1002/ajmg.a.30895. [DOI] [PubMed] [Google Scholar]
- Caburet S., Conti C., Schurra C., Lebofsky R., Edelstein S.J., Bensimon A., Conti C., Schurra C., Lebofsky R., Edelstein S.J., Bensimon A., Schurra C., Lebofsky R., Edelstein S.J., Bensimon A., Lebofsky R., Edelstein S.J., Bensimon A., Edelstein S.J., Bensimon A., Bensimon A. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res. 2005;15:1079–1085. doi: 10.1101/gr.3970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M., Cuff J., Searle S.M., Barton G.J., Cuff J., Searle S.M., Barton G.J., Searle S.M., Barton G.J., Barton G.J. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Clark S.M., Lai E., Birren B.W., Hood L., Lai E., Birren B.W., Hood L., Birren B.W., Hood L., Hood L. A novel instrument for separating large DNA molecules with pulsed homogeneous electric fields. Science. 1988;241:1203–1205. doi: 10.1126/science.3045968. [DOI] [PubMed] [Google Scholar]
- Delany M.E., Muscarella D.E., Bloom S.E., Muscarella D.E., Bloom S.E., Bloom S.E. Effects of rRNA gene copy number and nucleolar variation on early development: Inhibition of gastrulation in rDNA-deficient chick embryos. J. Hered. 1994;85:211–217. doi: 10.1093/oxfordjournals.jhered.a111437. [DOI] [PubMed] [Google Scholar]
- Delany M.E., Taylor R.L., Bloom S.E., Taylor R.L., Bloom S.E., Bloom S.E. Teratogenic development in chicken embryos associated with a major deletion in the rRNA gene cluster. Dev. Growth Differ. 1995;37:403–412. doi: 10.1046/j.1440-169X.1995.t01-3-00007.x. [DOI] [PubMed] [Google Scholar]
- Draptchinskaia N., Gustavsson P., Andersson B., Pettersson M., Willig T.N., Dianzani I., Ball S., Tchernia G., Klar J., Matsson H., Gustavsson P., Andersson B., Pettersson M., Willig T.N., Dianzani I., Ball S., Tchernia G., Klar J., Matsson H., Andersson B., Pettersson M., Willig T.N., Dianzani I., Ball S., Tchernia G., Klar J., Matsson H., Pettersson M., Willig T.N., Dianzani I., Ball S., Tchernia G., Klar J., Matsson H., Willig T.N., Dianzani I., Ball S., Tchernia G., Klar J., Matsson H., Dianzani I., Ball S., Tchernia G., Klar J., Matsson H., Ball S., Tchernia G., Klar J., Matsson H., Tchernia G., Klar J., Matsson H., Klar J., Matsson H., Matsson H., et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- Ehtesham N.Z., Hasnain S.E., Hasnain S.E. Direct in-gel hybridization, without blotting, using nick-translated cloned DNA probe. Biotechniques. 1991;11:718–722. [PubMed] [Google Scholar]
- Ellis S.R., Massey A.T., Massey A.T. Diamond Blackfan anemia: A paradigm for a ribosome-based disease. Med. Hypotheses. 2006;66:643–648. doi: 10.1016/j.mehy.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Gazda H.T., Grabowska A., Merida-Long L.B., Latawiec E., Schneider H.E., Lipton J.M., Vlachos A., Atsidaftos E., Ball S.E., Orfali K.A., Grabowska A., Merida-Long L.B., Latawiec E., Schneider H.E., Lipton J.M., Vlachos A., Atsidaftos E., Ball S.E., Orfali K.A., Merida-Long L.B., Latawiec E., Schneider H.E., Lipton J.M., Vlachos A., Atsidaftos E., Ball S.E., Orfali K.A., Latawiec E., Schneider H.E., Lipton J.M., Vlachos A., Atsidaftos E., Ball S.E., Orfali K.A., Schneider H.E., Lipton J.M., Vlachos A., Atsidaftos E., Ball S.E., Orfali K.A., Lipton J.M., Vlachos A., Atsidaftos E., Ball S.E., Orfali K.A., Vlachos A., Atsidaftos E., Ball S.E., Orfali K.A., Atsidaftos E., Ball S.E., Orfali K.A., Ball S.E., Orfali K.A., Orfali K.A., et al. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am. J. Hum. Genet. 2006;79:1110–1118. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez I.L., Sylvester J.E., Sylvester J.E. Complete sequence of the 43-kb human ribosomal DNA repeat: Analysis of the intergenic spacer. Genomics. 1995;27:320–328. doi: 10.1006/geno.1995.1049. [DOI] [PubMed] [Google Scholar]
- Gonzalez I.L., Sylvester J.E., Sylvester J.E. Human rDNA: Evolutionary patterns within the genes and tandem arrays derived from multiple chromosomes. Genomics. 2001;73:255–263. doi: 10.1006/geno.2001.6540. [DOI] [PubMed] [Google Scholar]
- Guanti G., Petrinelli P., Petrinelli P. rDNA and acrocentric chromosomes in man. Cell Differ. 1974;2:319–324. doi: 10.1016/0045-6039(74)90010-4. [DOI] [PubMed] [Google Scholar]
- Henderson A.S., Warburton D., Atwood K.C., Warburton D., Atwood K.C., Atwood K.C. Location of ribosomal DNA in the human chromosome complement. Proc. Natl. Acad. Sci. 1972;69:3394–3398. doi: 10.1073/pnas.69.11.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Kawasaki K., Minoshima S., Kudoh J., Fukuyama R., Shimizu N., Minoshima S., Kudoh J., Fukuyama R., Shimizu N., Kudoh J., Fukuyama R., Shimizu N., Fukuyama R., Shimizu N., Shimizu N. Methylation status of ribosomal RNA gene clusters in the flow-sorted human acrocentric chromosomes. Mamm. Genome. 1992;3:173–178. doi: 10.1007/BF00352463. [DOI] [PubMed] [Google Scholar]
- Kuick R., Asakawa J., Neel J.V., Kodaira M., Satoh C., Thoraval D., Gonzalez I.L., Hanash S.M., Asakawa J., Neel J.V., Kodaira M., Satoh C., Thoraval D., Gonzalez I.L., Hanash S.M., Neel J.V., Kodaira M., Satoh C., Thoraval D., Gonzalez I.L., Hanash S.M., Kodaira M., Satoh C., Thoraval D., Gonzalez I.L., Hanash S.M., Satoh C., Thoraval D., Gonzalez I.L., Hanash S.M., Thoraval D., Gonzalez I.L., Hanash S.M., Gonzalez I.L., Hanash S.M., Hanash S.M. Studies of the inheritance of human ribosomal DNA variants detected in two-dimensional separations of genomic restriction fragments. Genetics. 1996;144:307– 316. doi: 10.1093/genetics/144.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little R.D., Braaten D.C., Braaten D.C. Genomic organization of human 5 S rDNA and sequence of one tandem repeat. Genomics. 1989;4:376–383. doi: 10.1016/0888-7543(89)90345-5. [DOI] [PubMed] [Google Scholar]
- Lueders K.K., Fewell J.W., Fewell J.W. Hybridization of DNA in dried gels provides increased sensitivity compared with hybridization to blots. Biotechniques. 1994;16:66–67. [PubMed] [Google Scholar]
- Markovic V.D., Worton R.G., Berg J.M., Worton R.G., Berg J.M., Berg J.M. Evidence for the inheritance of silver-stained nucleolus organizer regions. Hum. Genet. 1978;41:181–187. doi: 10.1007/BF00273100. [DOI] [PubMed] [Google Scholar]
- Mayer C., Schmitz K.M., Li J., Grummt I., Santoro R., Schmitz K.M., Li J., Grummt I., Santoro R., Li J., Grummt I., Santoro R., Grummt I., Santoro R., Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Miller D.A., Dev V.G., Tantravahi R., Miller O.J., Dev V.G., Tantravahi R., Miller O.J., Tantravahi R., Miller O.J., Miller O.J. Suppression of human nucleolus organizer activity in mouse–human somatic hybrid cells. Exp. Cell Res. 1976;101:235–243. doi: 10.1016/0014-4827(76)90373-6. [DOI] [PubMed] [Google Scholar]
- Page S.L., Shaffer L.G., Shaffer L.G. Chromosome stability is maintained by short intercentromeric distance in functionally dicentric human Robertsonian translocations. Chromosome Res. 1998;6:115–122. doi: 10.1023/a:1009286929145. [DOI] [PubMed] [Google Scholar]
- Page S.L., Shin J.C., Han J.Y., Choo K.H., Shaffer L.G., Shin J.C., Han J.Y., Choo K.H., Shaffer L.G., Han J.Y., Choo K.H., Shaffer L.G., Choo K.H., Shaffer L.G., Shaffer L.G. Breakpoint diversity illustrates distinct mechanisms for Robertsonian translocation formation. Hum. Mol. Genet. 1996;5:1279–1288. doi: 10.1093/hmg/5.9.1279. [DOI] [PubMed] [Google Scholar]
- Schmickel R.D. Quantitation of human ribosomal DNA: Hybridization of human DNA with ribosomal RNA for quantitation and fractionation. Pediatr. Res. 1973;7:5–12. doi: 10.1203/00006450-197301000-00002. [DOI] [PubMed] [Google Scholar]
- Schmickel R.D., Knoller M., Knoller M. Characterization and localization of the human genes for ribosomal ribonucleic acid. Pediatr. Res. 1977;11:929–935. doi: 10.1203/00006450-197708000-00015. [DOI] [PubMed] [Google Scholar]
- Schofer C., Weipoltshammer K., Almeder M., Wachtler F., Weipoltshammer K., Almeder M., Wachtler F., Almeder M., Wachtler F., Wachtler F. Arrangement of individual human ribosomal DNA fragments on stretched DNA fibers. Histochem. Cell Biol. 1998;110:201–205. doi: 10.1007/s004180050282. [DOI] [PubMed] [Google Scholar]
- She X., Jiang Z., Clark R.A., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Jiang Z., Clark R.A., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Clark R.A., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Sutton G., Halpern A.L., Eichler E.E., Halpern A.L., Eichler E.E., Eichler E.E. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004;431:927–930. doi: 10.1038/nature03062. [DOI] [PubMed] [Google Scholar]
- Shiels C., Coutelle C., Huxley C., Coutelle C., Huxley C., Huxley C. Analysis of ribosomal and alphoid repetitive DNA by fiber-FISH. Cytogenet. Cell Genet. 1997;76:20–22. doi: 10.1159/000134504. [DOI] [PubMed] [Google Scholar]
- Sorensen P.D., Frederiksen S., Frederiksen S. Characterization of human 5S rRNA genes. Nucleic Acids Res. 1991;19:4147–4151. doi: 10.1093/nar/19.15.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen P.D., Simonsen H., Frederiksen S., Simonsen H., Frederiksen S., Frederiksen S. Nucleotide sequence of a human 5S rRNA gene. Nucleic Acids Res. 1990;18:3060. doi: 10.1093/nar/18.10.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen P.D., Lomholt B., Frederiksen S., Tommerup N., Lomholt B., Frederiksen S., Tommerup N., Frederiksen S., Tommerup N., Tommerup N. Fine mapping of human 5S rRNA genes to chromosome 1q42.11–q42.13. Cytogenet. Cell Genet. 1991;57:26–29. doi: 10.1159/000133107. [DOI] [PubMed] [Google Scholar]
- Stark J.M., Pierce A.J., Oh J., Pastink A., Jasin M., Pierce A.J., Oh J., Pastink A., Jasin M., Oh J., Pastink A., Jasin M., Pastink A., Jasin M., Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B.A., Jenkins L.S., Karson E.M., Leana-Cox J., Schwartz S., Jenkins L.S., Karson E.M., Leana-Cox J., Schwartz S., Karson E.M., Leana-Cox J., Schwartz S., Leana-Cox J., Schwartz S., Schwartz S. Evidence for structural heterogeneity from molecular cytogenetic analysis of dicentric Robertsonian translocations. Am. J. Hum. Genet. 1996;59:167–175. [PMC free article] [PubMed] [Google Scholar]
- Taylor E.F., Martin-DeLeon P.A., Martin-DeLeon P.A. Familial silver staining patterns of human nucleolus organizer regions (NORs) Am. J. Hum. Genet. 1981;33:67–76. [PMC free article] [PubMed] [Google Scholar]
- Timofeeva M., Kost M.V., Tikhomirova T.P., Sarafanov A.G., Elbert B.L., Alimov A.A., Kvitsiniia S.L., Beritashvili D.R., Zelenin A.V., Kost M.V., Tikhomirova T.P., Sarafanov A.G., Elbert B.L., Alimov A.A., Kvitsiniia S.L., Beritashvili D.R., Zelenin A.V., Tikhomirova T.P., Sarafanov A.G., Elbert B.L., Alimov A.A., Kvitsiniia S.L., Beritashvili D.R., Zelenin A.V., Sarafanov A.G., Elbert B.L., Alimov A.A., Kvitsiniia S.L., Beritashvili D.R., Zelenin A.V., Elbert B.L., Alimov A.A., Kvitsiniia S.L., Beritashvili D.R., Zelenin A.V., Alimov A.A., Kvitsiniia S.L., Beritashvili D.R., Zelenin A.V., Kvitsiniia S.L., Beritashvili D.R., Zelenin A.V., Beritashvili D.R., Zelenin A.V., Zelenin A.V. Organization of a 5S ribosomal RNA gene cluster in the human genome. Mol. Biol. 1993;27:861–868. [PubMed] [Google Scholar]
- Worton R.G., Sutherland J., Sylvester J.E., Willard H.F., Bodrug S., Dube I., Duff C., Kean V., Ray P.N., Schmickel R.D., Sutherland J., Sylvester J.E., Willard H.F., Bodrug S., Dube I., Duff C., Kean V., Ray P.N., Schmickel R.D., Sylvester J.E., Willard H.F., Bodrug S., Dube I., Duff C., Kean V., Ray P.N., Schmickel R.D., Willard H.F., Bodrug S., Dube I., Duff C., Kean V., Ray P.N., Schmickel R.D., Bodrug S., Dube I., Duff C., Kean V., Ray P.N., Schmickel R.D., Dube I., Duff C., Kean V., Ray P.N., Schmickel R.D., Duff C., Kean V., Ray P.N., Schmickel R.D., Kean V., Ray P.N., Schmickel R.D., Ray P.N., Schmickel R.D., Schmickel R.D. Human ribosomal RNA genes: Orientation of the tandem array and conservation of the 5′ end. Science. 1988;239:64–68. doi: 10.1126/science.3336775. [DOI] [PubMed] [Google Scholar]