Abstract
The amphibian Xenopus provides a model organism for investigating microRNA expression during vertebrate embryogenesis and development. Searching available Xenopus genome databases using known human pre-miRNAs as query sequences, more than 300 genes encoding 142 Xenopus tropicalis miRNAs were identified. Analysis of Xenopus tropicalis miRNA genes revealed a predominate positioning within introns of protein-coding and nonprotein-coding RNA Pol II-transcribed genes. MiRNA genes were also located in pre-mRNA exons and positioned intergenically between known protein-coding genes. Many miRNA species were found in multiple locations and in more than one genomic context. MiRNA genes were also clustered throughout the genome, indicating the potential for the cotranscription and coordinate expression of miRNAs located in a given cluster. Northern blot analysis confirmed the expression of many identified miRNAs in both X. tropicalis and X. laevis. Comparison of X. tropicalis and X. laevis blots revealed comparable expression profiles, although several miRNAs exhibited species-specific expression in different tissues. More detailed analysis revealed that for some miRNAs, the tissue-specific expression profile of the pri-miRNA precursor was distinctly different from that of the mature miRNA profile. Differential miRNA precursor processing in both the nucleus and cytoplasm was implicated in the observed tissue-specific differences. These observations indicated that post-transcriptional processing plays an important role in regulating miRNA expression in the amphibian Xenopus.
MicroRNAs are small noncoding RNAs of between 19 and 23 nucleotides in length that play fundamental roles in regulating gene expression in both plants and animals (Carrington and Ambros 2003; Bartel 2004; Valencia-Sanchez et al. 2006). MiRNAs base pair with messenger RNA and the established RNA:RNA duplex promotes mRNA cleavage and/or inhibition of translation, thus down-regulating the synthesis of the encoded protein. Estimates of miRNAs in a given organism number in the hundreds with each miRNA targeting its own set of 1–50 messenger RNAs (John et al. 2004; Bentwich 2005). It is now estimated that ∼30% of human protein-coding genes are post-transcriptionally regulated by miRNAs (Lewis et al. 2005; Rajewsky 2006).
MiRNAs are well conserved in evolution, particularly in animals, suggesting that they play conserved functions in different organisms (Pasquinelli et al. 2000; Chen and Rajewsky 2007). They are often expressed in a cell-specific manner or during different stages of development, thus suggesting regulatory roles in cellular differentiation and organism development (Ason et al. 2006; Sood et al. 2006). More recently, some miRNAs have also been implicated in human cancer, heart failure, and neurodegenerative diseases (Bilen et al. 2006; Kloosterman and Plasterk 2006; van Rooij et al. 2006).
Analysis of miRNA genes has revealed a variety of genomic organizations (Lai et al. 2003; Rodriguez et al. 2004; Griffiths-Jones et al. 2006). A large fraction of animal miRNAs are encoded within introns of protein-coding genes, most often on the coding strand of the pre-mRNA. A few are located with pre-mRNA exons, while others are positioned within intergenic regions. Most pri-miRNAs are transcribed by RNA Pol II and subsequently polyadenylated (Cai et al. 2004; Lee et al. 2004) before cleavage in the nucleus by the RNase III endonuclease Drosha, which is found in the microprocessor complex (Seitz and Zamore 2006). The pre-miRNA is then exported to the cytoplasm using the exportin 5 pathway (Lund et al. 2004). It is further cleaved by the endonuclease Dicer and the resulting ∼22-nt miRNA/miRNA* duplex is incorporated into the RNA-induced silencing or RISC complex (Tang 2005).
A number of approaches have been used to define miRNAs in various organisms. The cloning and sequencing of total small RNAs of appropriate size from isolated tissues or whole organisms has led to the identification of many miRNA species (for review, see Berezikov et al. 2006). Alternatively, computational approaches such as miRscan and miRseeker (Lai et al. 2003; Lim et al. 2003a, b) have identified miRNAs using either homology searches or finding potential RNA hairpins that exhibit those features characteristic of the double-stranded stem–loop structures of the folded pre-miRNA (Rodriguez et al. 2004; Weber 2005). A distinct advantage of computational searching is the ability to identify miRNAs that are often only expressed in specific tissues or stages of development or at low-copy number that may be missed by cloning and sequencing approaches.
The objective of this study was to computationally identify Xenopus miRNA genes, characterize their genomic organization, and then examine their expression in adult frog tissues. We present here 142 Xenopus miRNAs homologs. Like other vertebrates, ∼75% of these miRNAs are located within pre-mRNA introns. Analysis of miRNA expression in both Xenopus tropicalis and Xenopus laevis revealed the differential expression of some miRNAs in different adult tissues, suggesting significant post-transcriptional regulation of miRNA expression. These initial investigations also help to establish Xenopus as a model amphibian for further investigations of miRNA biogenesis and function during development.
Results
Computational identification of Xenopus microRNAs
Human pre-miRNA sequences were obtained from the microRNA database (miRBase: Giffiths-Jones et al. 2006) of the Sanger Institute and used as queries to search for Xenopus miRNA homologs using available X. tropicalis databases. Although still under construction, the X. tropicalis genome is most complete, consisting of DNA scaffolds assembled from overlapping contig sequences (Klein et al. 2002). Three different X. tropicalis databases were used (JGI, Ensembl, and the UCSC X. tropicalis genome server) as well as available EST databases (TIGR, Sanger, and the NCBI databases) (Smalheiser 2003; Li et al. 2006). The homology search was carried out simultaneously using all of the genomic and EST databases (see Supplemental Fig. 1 for the search strategy).
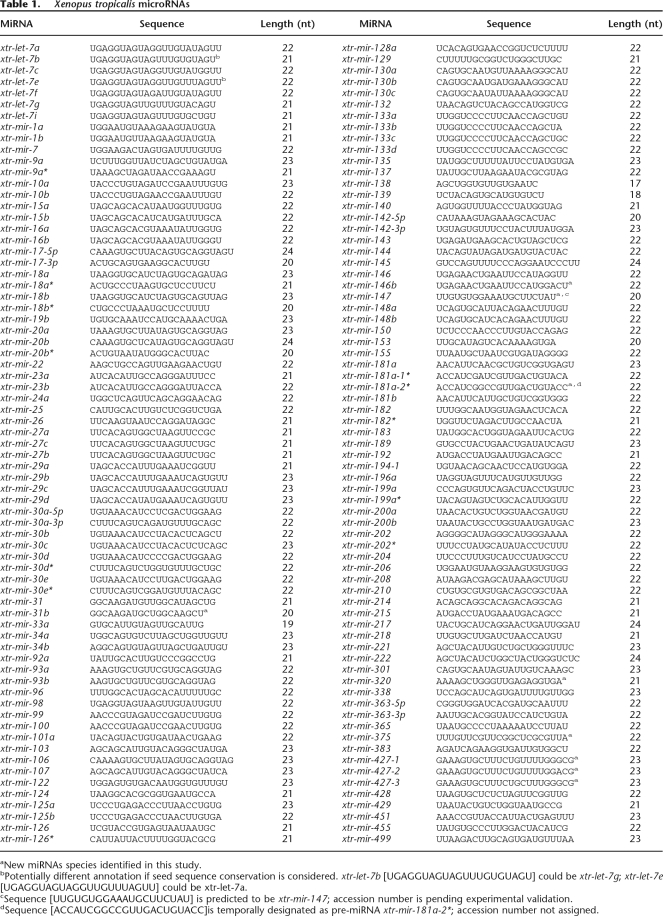
A total of 142 putative X. tropicalis miRNAs were identified in our computational search (Table 1; Supplemental Table 1). Included in these 142 species are 28 of the 29 homologs previously shown to be differentially expressed in X. laevis during embryogenesis (Watanabe et al. 2005). During the course of this investigation, many of our identified miRNAs also appeared on the Sanger microRNA registry (http://microrna.sanger.ac.uk/cgi-bin/sequences/mirna_summary.pl?org=xtr).
Table 1.
Xenopus tropicalis microRNAs
aNew miRNAs species identified in this study.
bPotentially different annotation if seed sequence conservation is considered. xtr-let-7b [UGAGGUAGUAGUUUGUGUAGU] could be xtr-let-7g; xtr-let-7e [UGAGGUAGUAGGUUGUUUAGUU] could be xtr-let-7a.
cSequence [UUGUGUGGAAAUGCUUCUAU] is predicted to be xtr-mir-147; accession number is pending experimental validation.
dSequence [ACCAUCGGCCGUUGACUGUACC]is temporally designated as pre-miRNA xtr-mir-181a-2*; accession number not assigned.
Each of the identified Xenopus miRNAs is able to fold into the hairpin–loop structure characteristic of a folded pre-miRNA. Folding analysis was first accomplished using Mfold (Zuker 2003) and the resulting secondary structures manually adjusted using more recently established criteria for pre-miRNA identification as detailed in the Methods and Supplemental Figure 1. (The folding of those X. tropicalis miRNAs not exhibiting 100% miRNA similarity with its human homolog are shown folded as pre-miRNAs in Supplemental Table 1.) These computational approaches were most productive in identifying miRNAs for X. tropicalis. However, some X. laevis miRNAs were also identified in our search and they are designated in Supplemental Tables 2 and 3.
Xenopus miRNA-coding sequences are predominantly located within introns of RNA Pol II-transcribed host genes
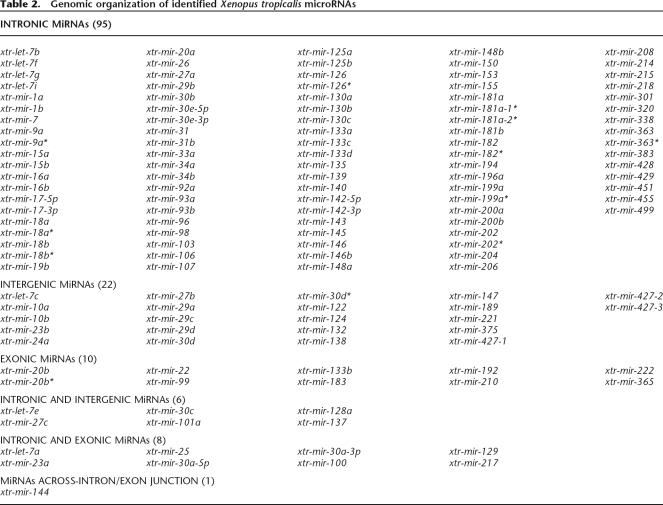
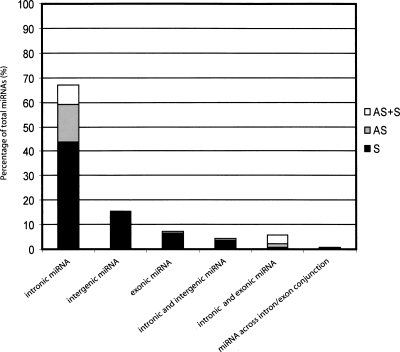
The available X. tropicalis genome databases enabled a determination of miRNA genomic context for all 142 X. tropicalis species (Supplemental Table 2). Approximately 90% of the miRNAs are found exclusively in a single genomic context even if a given miRNA is present in the genome in multiple copies. The predominant context is miRNA positioning within introns (77%) of RNA Pol II-transcribed genes (Table 2; Fig. 1). A few are located in pre-mRNA exons (∼7%) with slightly more located within intergenic regions (∼15%).
Table 2.
Genomic organization of identified Xenopus tropicalis microRNAs
Figure 1.
Genomic distribution of identified Xenopus tropicalis microRNAs. Graphical representation of the X. tropicalis miRNA’s genomic organization. For intronic miRNAs, the solid sector of the bar indicates the proportion of miRNA genes positioned on the coding strand (S), the gray sector indicates the proportion of miRNA genes located on the noncoding strand (AS), and the white sector indicates miRNA genes found on both coding and noncoding strands (AS + S).
The X. tropicalis intronic miRNAs can be grouped into three types. Some miRNAs are located within introns of known protein-coding genes. Others are encoded in putative introns, predicted to be within hypothetical protein-coding sequences (designated in Supplemental Table 2 by scaffold or protein ID numbers). MicroRNAs were also identified within designated introns of “non-spliced EST” sequences (Mott 1997) as listed in the UCSC and Ensembl genome server data bases (see Supplemental Table 3). The transcription of miRNA precursors for those encoded in known protein-coding sequences and those identified in the EST database is clear. Transcription of miRNAs encoded in hypothetical putative protein-coding sequences may be less certain. However, a majority of these are also found in the EST database, indicating that they are also transcribed (Supplemental Table 3). Comparison of the Xenopus intronic miRNAs with their human homologs revealed strong conservation in genomic location (see Supplemental Table 4).
Xenopus miRNA genes are clustered in the genome
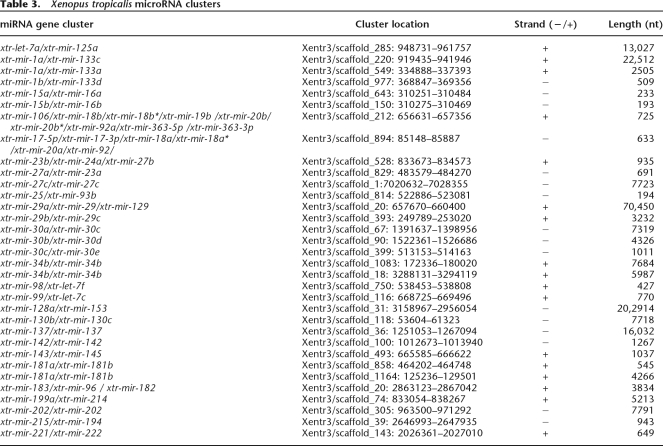
Analysis of miRNA gene location within the X. tropicalis genome frequently revealed multiple miRNAs clustered on a given DNA scaffold. Almost 50% of the 142 identified X. tropicalis miRNA species were observed to occur at least once in a miRNA cluster (Table 3). For many of the miRNAs appearing in a cluster, additional copies of the same miRNA are often found elsewhere as a single, nonclustered miRNA-coding sequence. A given cluster could be a simple repeat of a single miRNA species (e.g., xtr-mir-137/xtr-mir-137 cluster [Xentr3/scaffold 36] and xtr-mir-202/xtr-mir-202 [Xentr3/scaffold 305]). However, the vast majority of clusters possessed different miRNAs. Interestingly, several clusters were found to be repeated in the Xenopus genome. Repeated clusters could be identical in miRNA composition, such as the xtr-mir-34b/xtr-mir-34b cluster found on two different DNA scaffolds (Xentr3/scaffolds 194, 350, and 366). Also observed was a X. tropicalis miRNA xtr-mir-17/xtr-mir-92 cluster that is designated the “oncogene cluster” in human (He et al. 2005). This cluster possesses six miRNAs (xtr-mir-17-5p/xtr-mir-17-3p/xtr-mir-18a/xtr-mir-18a*/xtr-mir-20a/xtr-mir-92; Xentr3/scaffold 894).
Table 3.
Xenopus tropicalis microRNA clusters
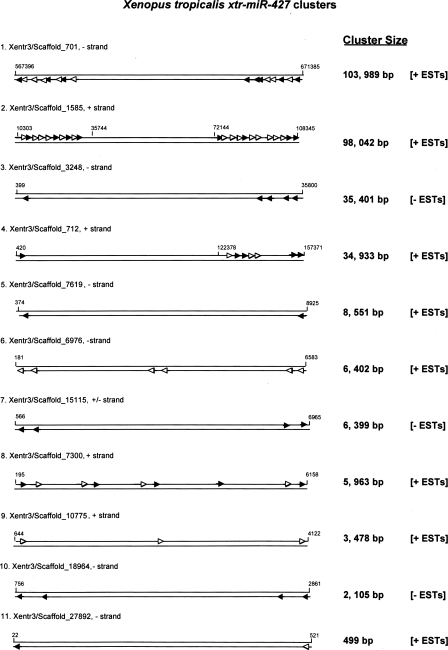
A particularly striking example of miRNA clustering was the Xenopus xtr-mir-427 coding sequences. We have identified ∼80 xtr-mir-427 genes in the X. tropicalis genome (Supplemental Table 2), many of them clustered on specific DNA scaffolds as shown in Figure 2. The length of DNA scaffold containing xtr-mir-427 miRNAs varied greatly from only several hundred to more than 100,000 bp. The close proximity of some clustered miRNAs suggests cotranscription in a single pri-miRNA transcript, whereas distantly positioned miRNAs may suggest otherwise. Strikingly, analysis of available X. tropicalis EST data indicates that all but two indicated clusters could well be transcribed as a single, common precursor transcript (designated as +ESTs). Even more striking was the observation that some of the miRNAs within a cluster were absent from the EST (open triangles), indicating differential processing of the pri-miRNA transcript.
Figure 2.
Xenopus tropicalis mir-427 clusters. Schematic presentation of identified X. tropicalis mir-427 clusters. Specific scaffolds are designated with scaffold boundary nucleotides indicated at each terminus. The largest to smallest DNA scaffolds are presented from top to bottom with the scaffold total length in nucleotides indicated at the side. Arrows indicate individual xtr-mir-427 genes. For some clusters, the open triangles indicate absent miRNAs in transcribed ESTs. The top strand of each scaffold is the +DNA strand and the bottom strand is the –DNA strand. +ESTs indicate evidence for cotranscription of each cluster (Supplemental Table 3).
Expression of X. tropicalis and X. laevis miRNAs
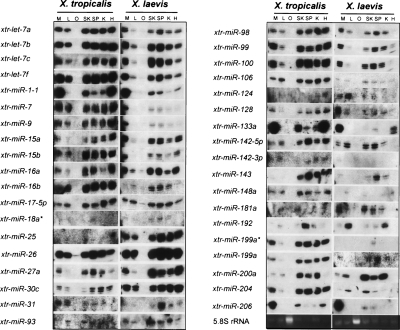
Total RNA was prepared from X. tropicalis and X. laevis adult tissues and miRNA expression assessed using Northern blot analysis. Of the 63 miRNAs assessed, 47 species or 77% (designated in Supplemental Table 1) were observed in one or more of the tissue-specific RNA preparations (38 of these are presented in Fig. 3). Those miRNAs tested but not detected may be expressed in other tissues not examined, expressed during specific stages of embryogenesis, or expressed at such low levels as to make detection by Northern blot analysis difficult. Most of the observed miRNAs were expressed in almost all of the adult tissues assayed. The lack of miRNA expression in oocytes is consistent with miRNA roles in regulating gene expression during embryogenesis and organism development.
Figure 3.
Northern blot validation of computationally identified Xenopus tropicalis and Xenopus laevis miRNAs. Total RNA was isolated from X. tropicalis and X. laevis oocytes and adult tissues, resolved on denaturing polyacrylamide gels, electroblotted to nylon membranes, and hybridized with DNA oligonucleotides complementary to computationally identified Xenopus miRNAs. Specific Xenopus miRNAs sequences probed with complementary DNA oligonucleotides are indicated at left. Xenopus tissues extracted for RNA analysis are indicated at the top. (M) Muscle; (L) liver; (O) oocyte; (SK) skin; (SP) spleen; (K) kidney; (H) heart.
The expression patterns were comparable for most of the miRNAs in both X. tropicalis and X. laevis tissues. However, apparent quantitative differences in the expression patterns of some miRNAs were observed between the two different Xenopus species. These miRNAs included xtr-mir-98, xtr-mir-133a, xtr-mir-142-5p, and xtr-mir-143 (Fig. 3). Differences in expression patterns could reflect species-specific differences in transcription or differential pri-mRNA processing. Some clustered miRNAs exhibited comparable expression patterns (e.g., xtr-mir-99/xtr-let-7c cluster), consistent with cotranscription in a common pri-miRNA as previously suggested (Lagos-Quintana et al. 2001; Baskerville and Bartel 2005). However, this was not true for all miRNA clusters (see below).
Evidence for the regulated, tissue-specific expression of X. laevis miRNAs
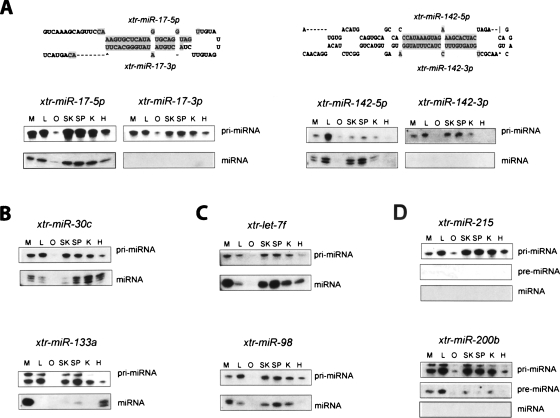
More detailed examination of selected X. laevis miRNAs indicated their post-transcriptional regulation. Northern blot analysis of the xtr-mir-17-5p/xtr-mir-17-3p and the xtr-mir-142-5p/xtr-miR142-3p miRNAs transcribed in common pre-miRNAs revealed that while both pri-miRNA precursors were transcribed in all adult frog tissues, only xtr-mir-17-5p and xtr-mir-142-5p were observed as mature miRNAs (Fig. 4A). This suggested differential processing of the pre-miRNA in the cytoplasm to produce the miRNA found on the 5′ strand, but not on the 3′ “passenger strand”. Northern blot analysis of both xtr-mir-30c and xtr-mir-133a revealed different expression patterns of the pri-miRNA precursor and the mature miRNA (Fig. 4B). The distinct tissue-specific expression patterns of precursor and mature miRNAs suggested differential pri- or pre-miRNA processing and/or differential stabilization of the mature miRNA. The observation of two pri-miRNA precursors may also suggest the transcription of xtr-mir-133a pri-miRNA from different promoters or the differential processing of the pri-miRNA transcript.
Figure 4.
Differential expression of Xenopus laevis miRNAs and pri-miRNAs. Total RNA was isolated from X. laevis oocytes and adult tissues were resolved on denaturing polyacrylamide gels, electroblotted to nylon membranes, and hybridized with DNA oligonucleotides complementary to computationally identified Xenopus miRNAs. Specific Xenopus miRNAs sequences probed are indicated at the top and detected miRNA transcripts indicated at right. Xenopus tissues extracted for RNA analysis are indicated at the top. (M) muscle; (L) liver; (O) oocyte; (SK) skin; (SP) spleen; (K) kidney; (H) heart. (A) Differential expression of Xenopus laevis miRNAs located on the 5′ and 3′ arms of the same pre-miRNA. The folded structure of these pre-miRNAs is illustrated. (B) Tissue-specific expression of Xenopus laevis miRNAs. (C) Tissue-specific expression of Xenopus laevis miRNAs clustered on the same pre-miRNA transcript. (D) Differential expression of transcribed Xenopus laevis pri-miRNA precursors at the level of pre-miRNA and/or miRNA processing.
Both xtr-mir-98 and xtr-let-7f are clustered on a 427-nt DNA fragment (Xentr3_750) and can reasonably be assumed to be cotranscribed as a common pri-miRNA precursor transcript. The size of the pri-miRNA transcript observed is consistent with the cotranscription of these three miRNAs. Notably, while the patterns of xtr-let-7f and xtr-mir-98 pri-miRNA expression are identical, the patterns of tissue-specific expression for the two mature miRNAs are clearly different (Fig. 4C). Particularly prominent is the presence of xtr-let-7f miRNA in heart tissue, but the absence of xtr-mir-98 in the same tissue. This suggested the differential processing of the pri-miRNA precursor or the pre-miRNA intermediate to produce xtr-let-7f but not xtr-mir-98.
Finally, both mature xtr-mir-215 and xtr-mir-220b were not observed in initial Northern blot analysis. However, additional examination revealed the presence of pri-miRNA primary transcripts for both miRNAs in all adult tissues and the presence of xtr-mir-200b pre-miRNA in some adult tissues (Fig. 4D). These observations suggested the lack of xtr-mir-215 pri-miRNA primary transcript processing to produce either pre-miRNAs or mature miRNAs in all tissues and the differential processing of xtr-pre-mir-200b in some tissues, along with the possible disruption of pre-miRNA transport and/or cytoplasmic Dicer processing in all tissues.
Discussion
A computational approach using human pre-miRNA query sequences has identified more than 300 miRNA genes encoding some 142 Xenopus miRNA species. Xenopus miRNA genes are predominantly located within introns and highly clustered in the genome. Using Northern blot analysis as well as the EST databases, a substantial proportion of the identified Xenopus miRNAs have been verified as expressed RNA transcripts. The shared expression patterns of almost all X. tropicalis and X. laevis miRNAs in the various adult tissues are consistent with highly conserved miRNA function in vertebrates. However, a few probed miRNAs exhibited tissue-specific differences in these two Xenopus species, suggesting perhaps species-specific miRNA expression. Most striking however, was the observation of post-transcriptional regulation of miRNA expression at both nuclear and cytoplasmic processing steps.
Xenopus miRNAs exhibit several genomic contexts, but their predominant positioning within pre-mRNA introns is similar to other vertebrates, including human, where it has been estimated that 70% of the miRNAs are intronic (Rodriguez et al. 2004). The intronic miRNAs are located within many known protein-coding host genes, but also found within putative, unidentified Pol II transcripts and within EST sequences. Notably, many of the Xenopus intronic miRNAs are encoded within the same host gene as their human homolog, extending their conservation beyond simply the pre-miRNA-coding sequence (Supplemental Table 4). In addition, this conservation also extends to clusters of intronic miRNAs conserved in both human and Xenopus. Conserved miRNA clusters within the same host gene suggest the possible coordinated expression of both host pre-mRNA and included miRNAs as previously suggested (Ying and Lin 2006).
Like other vertebrates, miRNA clustering is a prominent feature in the Xenopus genome. Approximately half of the identified Xenopus miRNAs are found in clusters in varying combinations. The clustering of Xenopus miRNA genes could also have important implications for the coordinated, tissue-specific expression of clustered miRNAs during development. Certainly, pri-miRNAs containing clustered miRNAs have been reported (Lau et al. 2001; Cai et al. 2004) and some Xenopus RNA transcripts found in the EST databases support clustered miRNA cotranscription in a common pri-miRNA. Some clustered Xenopus miRNAs are positioned on lengthy DNA fragments, raising the question as to the maximum size of pri-miRNA transcripts. Bartel and coworkers (Baskerville and Bartel 2005) have suggested pri-miRNAs as large as 50 kb, and some of the transcripts found in the Xenopus EST database support this proposition. Comparison of human miRNA clusters (Altuvia et al. 2005) with the Xenopus miRNA clusters has revealed many of the same clustered miRNAs, yet miRNA clustering in human and Xenopus is not entirely conserved, possibly suggesting differences in coordinated miRNA transcription in these two vertebrates.
Of particular importance will be defining promoters for pri-miRNA transcription of intergenic, intronic, and clustered miRNAs. Analysis of miRNA synthesis has revealed that the majority of pri-miRNAs are transcribed by Pol II polymerase and possess 5′ caps and 3′ poly (A) tails (Cai et al. 2004; Lee et al. 2004), although recent work has shown that some pri-miRNAs are transcribed by RNA Pol III (Borchert et al. 2006). The predominance of intronic miRNAs has implied cotranscription with the host pre-mRNAs and pre-miRNA processing concurrent with host pre-mRNA splicing (Cai et al. 2004; for review, see Kim and Nam 2006). However, differing pre-mRNA/pri-miRNA expression profiles with respect to their mature, fully processed miRNA may suggest miRNA-specific promoters within the host gene (Thomson et al. 2007). Alternatively, regulated alternative pre-mRNA splicing coupled with intronic miRNA processing could explain the different expression profiles observed. Clearly, for those miRNAs positioned with a host gene intron but on the noncoding strand, pri-miRNA-specific promoters will be required.
Further examination of transcribed Xenopus miRNA sequences in the EST databases may suggest an even more complex picture of miRNA transcription and processing. Those miRNAs occurring in the EST databases are indicated in Supplemental Tables 2 and 3. These previously designated “spliced” miRNAs are flanked by upstream and downstream transcript corresponding to the genomic sequences flanking the pre-miRNA sequence. However, far more intriguing is the occurrence of so-called “non-spliced” RNA transcripts (Mott 1997). These RNA transcripts consist of transcribed upstream and downstream genomic sequence, but lack the pre-miRNA sequence. These RNAs suggest that during transcription, a processing event has excised the pre-miRNA sequence. Fully, more than 70% of the Xenopus miRNAs identified in this study are found in these nonspliced EST transcripts, consistent with cotranscriptional processing of pre-miRNA sequences. For the intronic miRNAs, this may suggest a complex biogenesis pathway distinct from pre-mRNA transcriptional events.
Finally, analysis of Xenopus miRNA expression has revealed unanticipated expression profiles for some Xenopus pri-miRNAs and their processed, mature miRNAs. Differences in tissue-specific expression between the two Xenopus species were surprising, yet other laboratories have reported similar observations (Ason et al. 2006; Sood et al. 2006). Unfortunately, Northern blot analysis is not sufficiently quantitative to make definitive statements about miRNA amounts, and more detailed analysis awaits quantitative PCR investigation. Evidence has recently been presented indicating differential pri-miRNA processing of some clustered miRNAs (Samols et al. 2005; Obernosterer et al. 2006) and our observations also indicated the differential processing of the miR-98/let-7f/let-7a-1 cluster. However, most striking were the distinctly different, tissue-specific expression patterns of some Xenopus pri-miRNAs with respect to their processed mature miRNAs. Similar observations have been made by Hammond and coworkers (Thomson et al. 2007) as well as Martinez and coworkers (Obernosterer et al. 2006). Collectively, these observations suggest significant post-transcriptional regulation in the miRNA biogenesis pathway at various processing steps in both the nucleus and cytoplasm. The consequences of such regulation could have a large impact on the ultimate expression of a specific miRNA species. Future experiments concerning miRNA transcription and post-transcriptional processing are likely to reveal a complex and highly regulated pathway for miRNA expression.
Methods
Computational identification of Xenopus microRNAs
Human pre-miRNA sequences were retrieved from miRbase∷sequences (http://microrna.sanger.ac.uk/sequences/) and used as query sequences against available X. tropicalis genomic databases (Xentr3-Oct. 2004 assembly and the Xentr4-Aug. 2005 assembly with most of our reported data obtained from the 2004 assembly and designated as Xentr3). This computational approach is summarized in Supplemental Figure 1 and consisted of homology searching, alignment and annotation, and pre-miRNA secondary structural analysis. The X. tropicalis genome database is presently under construction (Klein et al. 2002) and consists of sequenced DNA fragments or “scaffolds” and overlapping “contig” sequences. The three genomic databases utilized were the JGI X. tropicalis genome server (two servers on the JGI Web site: http://genome.jgi-psf.org/Xentr3/Xentr3.home.html and http://genome.jgi-psf.org/Xentr4/Xentr4.home.html), Ensembl X.tropicalis (http://www.ensembl.org/Xenopus_tropicalis/blastview), and the UCSC X. tropicalis genomic server (http://www.genome.ucsc.edu/cgi-bin/hgBlat). The human pre-miRNA query sequences were used in either BLAT or BLASTN algorithms for searching. An alternative search path utilized the available EST databases: TIGR’s Xenopus EST database (http://tigrblast.tigr.org/tgi/ with options for X. laevis and X. tropicalis) and the Sanger Xenopus EST project (http://www.sanger.ac.uk/cgi-bin/blast/submitblast/x_tropicalis). Homology searching was carried out in all genomic and EST databases with initial analysis revealing that pre-miRNA sequences were more useful than the shorter miRNA sequences. Preliminary results provided sequence alignments as well as the candidate Xenopus miRNA genomic locations with possible host genes.
Candidate Xenopus miRNAs were further analyzed for their folded structures with specific pre-miRNA secondary structural features being used as filters in the verification process. Identified Xenopus sequences (60–120 nt long) were folded using the Mfold program (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/) and manually adjusted according to specific criteria based upon recent analysis and definition of conserved or preferred pre-miRNA structures. These features included (1) the length of the pre-miRNA core region between 60 and 80 nt (Ambros et al. 2003), (2) a pre-miRNA hairpin stem is ≥22 nt (deduced from the listed microRNAs in the microRNA registry database), (3) a loop size of ≥10 nt (Zeng et al. 2005), (4) hairpin bulges within the base-paired stem of not more than 6 nt (Ambros et al. 2003; Weber 2005), (5) an initial preferred nucleotide of U (Lau et al. 2001), (6) well-conserved miRNA seed sequences or nucleotides 2–7 (Lewis et al. 2004), (7) total G:U base pairs within the hairpin stem of <5 nt (Bonnet et al. 2004), and (8) similar locations of the mature human and Xenopus microRNAs on the same hairpin arm.
Northern blot analysis
Total RNA of adult female X. tropicalis and X. laevis frogs was prepared from various harvested tissues by extraction using TRIzol reagent according to the manufacturer’s protocols (Invitrogen). A total of 10 μg of total tissue RNA was resolved on 12% denaturing polyacrylamide gels containing 7 M urea and then electroblotted onto Genescreen Nylon membrane (NEN) in TBE buffer. Northern blots were dried and then incubated overnight in 10 mL of ULTRAhyb-Oligo (Ambion) containing 5′-32P radio-labeled oligonucleotides (21–23 nt) complementary to the predicted microRNA sequences. Prehybridization and hybridization procedures were performed at 37°C as previously detailed (Liu et al. 2005). Nylon membranes were washed three times for 10 min at room temperature in 2X SSC containing 0.5% SDS and then once for 15 min at 42°C in 2X SSC containing 0.5% SDS (Bentwich 2005). Both phosphorimaging and autoradiography were used to reveal hybridized microRNAs, pri-miRNAs, and pre-miRNAs.
Acknowledgments
We gratefully acknowledge support from the College of Agriculture and Life Sciences at North Carolina State University, Raleigh, NC.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6539108
References
- Altuvia Y., Landgraf P., Lithwick G., Elefant N., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Landgraf P., Lithwick G., Elefant N., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Lithwick G., Elefant N., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Elefant N., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Brownstein M.J., Tuschl T., Margalit H., Tuschl T., Margalit H., Margalit H. Clustering and conservation patterns of human microRNAs. Nucl. Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V., Bartel B., Bartel D.P., Burge C., Carrington J., Chen X., Dreyfuss G., Eddy S., Griffiths-Jones S., Marshall M., Bartel B., Bartel D.P., Burge C., Carrington J., Chen X., Dreyfuss G., Eddy S., Griffiths-Jones S., Marshall M., Bartel D.P., Burge C., Carrington J., Chen X., Dreyfuss G., Eddy S., Griffiths-Jones S., Marshall M., Burge C., Carrington J., Chen X., Dreyfuss G., Eddy S., Griffiths-Jones S., Marshall M., Carrington J., Chen X., Dreyfuss G., Eddy S., Griffiths-Jones S., Marshall M., Chen X., Dreyfuss G., Eddy S., Griffiths-Jones S., Marshall M., Dreyfuss G., Eddy S., Griffiths-Jones S., Marshall M., Eddy S., Griffiths-Jones S., Marshall M., Griffiths-Jones S., Marshall M., Marshall M., et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ason B., Darnell D., Wittbrodt B., Berezikov E., Kloosterman W., Wittbrodt J., Antin P., Plasterk R., Darnell D., Wittbrodt B., Berezikov E., Kloosterman W., Wittbrodt J., Antin P., Plasterk R., Wittbrodt B., Berezikov E., Kloosterman W., Wittbrodt J., Antin P., Plasterk R., Berezikov E., Kloosterman W., Wittbrodt J., Antin P., Plasterk R., Kloosterman W., Wittbrodt J., Antin P., Plasterk R., Wittbrodt J., Antin P., Plasterk R., Antin P., Plasterk R., Plasterk R. Differences in vertebrate microRNA expression. Proc. Natl. Acad. Sci. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S., Bartel D., Bartel D. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bentwich I. Prediction and validation of microRNAs and their targets. FEBS Lett. 2005;579:5904–5910. doi: 10.1016/j.febslet.2005.09.040. [DOI] [PubMed] [Google Scholar]
- Berezikov E., Cuppen E., Plasterk R., Cuppen E., Plasterk R., Plasterk R. Approaches to microRNA discovery. Nat. Genet. 2006;38:S2–S7. doi: 10.1038/ng1794. (Suppl) [DOI] [PubMed] [Google Scholar]
- Bilen J., Liu N., Bonini N., Liu N., Bonini N., Bonini N. A new role for microRNA pathways: Modulation of degeneration induced by pathogenic human disease proteins. Cell Cycle. 2006;5:2835–2838. doi: 10.4161/cc.5.24.3579. [DOI] [PubMed] [Google Scholar]
- Bonnet E., Wuyts J., Rouze P., de Van Peer Y., Wuyts J., Rouze P., de Van Peer Y., Rouze P., de Van Peer Y., de Van Peer Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. 2004;101:11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert G., Lanier W., Davidson B., Lanier W., Davidson B., Davidson B. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- Cai X., Hagedorn C., Cullen B., Hagedorn C., Cullen B., Cullen B. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington J., Ambros V., Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Chen K., Rajewsky N., Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R., van Dongen S., Bateman A., Enright A., Grocock R., van Dongen S., Bateman A., Enright A., van Dongen S., Bateman A., Enright A., Bateman A., Enright A., Enright A. miRBase: microRNA sequences, targets and gene nomenclature. Nucl. Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Thomson J., Hemann M., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S., Hannon G., Thomson J., Hemann M., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S., Hannon G., Hemann M., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S., Hannon G., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S., Hannon G., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S., Hannon G., Goodson S., Powers S., Cordon-Cardo C., Lowe S., Hannon G., Powers S., Cordon-Cardo C., Lowe S., Hannon G., Cordon-Cardo C., Lowe S., Hannon G., Lowe S., Hannon G., Hannon G., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B., Enright A., Aravin A., Tuschl T., Sander C., Marks D., Enright A., Aravin A., Tuschl T., Sander C., Marks D., Aravin A., Tuschl T., Sander C., Marks D., Tuschl T., Sander C., Marks D., Sander C., Marks D., Marks D. Human microRNA targets. Erratum. PLoS Biol. PLoS Biol. 2004;2005;23:e363. e264]. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., Nam J., Nam J. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Klein S., Strausberg R., Wagner L., Pontius J., Clifton S., Richardson P., Strausberg R., Wagner L., Pontius J., Clifton S., Richardson P., Wagner L., Pontius J., Clifton S., Richardson P., Pontius J., Clifton S., Richardson P., Clifton S., Richardson P., Richardson P. Genetic and genomic tools for Xenopus research: The NIH Xenopus initiative. Dev. Dyn. 2002;225:384–391. doi: 10.1002/dvdy.10174. [DOI] [PubMed] [Google Scholar]
- Kloosterman W., Plasterk R., Plasterk R. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T., Rauhut R., Lendeckel W., Tuschl T., Lendeckel W., Tuschl T., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lai E., Tomancak P., Williams R., Rubin G., Tomancak P., Williams R., Rubin G., Williams R., Rubin G., Rubin G. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N., Lim L., Weinstein E., Bartel D., Lim L., Weinstein E., Bartel D., Weinstein E., Bartel D., Bartel D. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee Y., Kim M., Han J., Yeom K., Lee S., Baek S., Kim V., Kim M., Han J., Yeom K., Lee S., Baek S., Kim V., Han J., Yeom K., Lee S., Baek S., Kim V., Yeom K., Lee S., Baek S., Kim V., Lee S., Baek S., Kim V., Baek S., Kim V., Kim V. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B., Burge C., Bartel D., Burge C., Bartel D., Bartel D. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li S., Pan C., Lin W., Pan C., Lin W., Lin W. Bioinformatic discovery of microRNA precursors from human ESTs and introns. BMC Genomics. 2006;7:164. doi: 10.1186/1471-2164-7-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L., Lau N., Weinstein E., Abdelhakim A., Yekta S., Rhoades M., Burge C., Bartel D., Lau N., Weinstein E., Abdelhakim A., Yekta S., Rhoades M., Burge C., Bartel D., Weinstein E., Abdelhakim A., Yekta S., Rhoades M., Burge C., Bartel D., Abdelhakim A., Yekta S., Rhoades M., Burge C., Bartel D., Yekta S., Rhoades M., Burge C., Bartel D., Rhoades M., Burge C., Bartel D., Burge C., Bartel D., Bartel D. The microRNAs of Caenorhabditis elegans. Genes & Dev. 2003a;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L., Glasner M., Yekta S., Burge C., Bartel D., Glasner M., Yekta S., Burge C., Bartel D., Yekta S., Burge C., Bartel D., Burge C., Bartel D., Bartel D. Vertebrate microRNA genes. Science. 2003b;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Liu S., Sun Y.H., Shi R., Clark C., Li L., Chiang V.L., Sun Y.H., Shi R., Clark C., Li L., Chiang V.L., Shi R., Clark C., Li L., Chiang V.L., Clark C., Li L., Chiang V.L., Li L., Chiang V.L., Chiang V.L. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell. 2005;17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Guttinger S., Calado A., Dahlberg J., Kutay U., Guttinger S., Calado A., Dahlberg J., Kutay U., Calado A., Dahlberg J., Kutay U., Dahlberg J., Kutay U., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Mott R. EST_GENOME: A program to align spliced DNA sequences to unspliced genomic DNA. Comput. Appl. Biosci. 1997;13:477–488. doi: 10.1093/bioinformatics/13.4.477. [DOI] [PubMed] [Google Scholar]
- Obernosterer G., Leuschner P., Alenius M., Martinez J., Leuschner P., Alenius M., Martinez J., Alenius M., Martinez J., Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A., Reinhart B., Slack F., Martindale M., Kuroda M., Maller B., Hayward D., Ball E., Degnan B., Muller P., Reinhart B., Slack F., Martindale M., Kuroda M., Maller B., Hayward D., Ball E., Degnan B., Muller P., Slack F., Martindale M., Kuroda M., Maller B., Hayward D., Ball E., Degnan B., Muller P., Martindale M., Kuroda M., Maller B., Hayward D., Ball E., Degnan B., Muller P., Kuroda M., Maller B., Hayward D., Ball E., Degnan B., Muller P., Maller B., Hayward D., Ball E., Degnan B., Muller P., Hayward D., Ball E., Degnan B., Muller P., Ball E., Degnan B., Muller P., Degnan B., Muller P., Muller P., et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Rajewsky N. MicroRNA target predictions in animals. Nat. Genet. 2006;38:S8–S13. doi: 10.1038/ng1798. (Suppl) [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Griffiths-Jones S., Ashurst J., Bradley A., Griffiths-Jones S., Ashurst J., Bradley A., Ashurst J., Bradley A., Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samols M., Hu J., Skalsky R., Renne R., Hu J., Skalsky R., Renne R., Skalsky R., Renne R., Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 2005;79:9301–9315. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H., Zamore P., Zamore P. Rethinking the microprocessor. Cell. 2006;125:827–829. doi: 10.1016/j.cell.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Smalheiser N. EST analyses predict the existence of a population of chimeric microRNA precursor-mRNA transcripts expressed in normal human and mouse tissues. Genome Biol. 2003;4:403. doi: 10.1186/gb-2003-4-7-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood P., Krek A., Zavolan M., Macino G., Rajewsky N., Krek A., Zavolan M., Macino G., Rajewsky N., Zavolan M., Macino G., Rajewsky N., Macino G., Rajewsky N., Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G. siRNA and miRNA: An insight into RISCs. Trends Biochem. Sci. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Thomson J., Newman M., Parker J., Morin-Kensicki E., Wright T., Hammond S., Newman M., Parker J., Morin-Kensicki E., Wright T., Hammond S., Parker J., Morin-Kensicki E., Wright T., Hammond S., Morin-Kensicki E., Wright T., Hammond S., Wright T., Hammond S., Hammond S. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes & Dev. 2007;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez M., Liu J., Hannon G., Parker R., Liu J., Hannon G., Parker R., Hannon G., Parker R., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes & Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- van Rooij E., Sutherland L., Liu N., Williams A., McAnally J., Gerard R., Richardson J., Olson E., Sutherland L., Liu N., Williams A., McAnally J., Gerard R., Richardson J., Olson E., Liu N., Williams A., McAnally J., Gerard R., Richardson J., Olson E., Williams A., McAnally J., Gerard R., Richardson J., Olson E., McAnally J., Gerard R., Richardson J., Olson E., Gerard R., Richardson J., Olson E., Richardson J., Olson E., Olson E. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Takeda A., Mise K., Okuno T., Suzuki T., Minami N., Imai H., Takeda A., Mise K., Okuno T., Suzuki T., Minami N., Imai H., Mise K., Okuno T., Suzuki T., Minami N., Imai H., Okuno T., Suzuki T., Minami N., Imai H., Suzuki T., Minami N., Imai H., Minami N., Imai H., Imai H. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 2005;579:318–324. doi: 10.1016/j.febslet.2004.11.067. [DOI] [PubMed] [Google Scholar]
- Weber M. New human and mouse microRNA genes found by homology search. FEBS J. 2005;272:59–73. doi: 10.1111/j.1432-1033.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- Ying S., Lin S., Lin S. Current perspectives in intronic microRNAs (miRNAs) J. Biomed. Sci. 2006;13:5–15. doi: 10.1007/s11373-005-9036-8. [DOI] [PubMed] [Google Scholar]
- Zeng Y., Yi R., Cullen B., Yi R., Cullen B., Cullen B. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;12:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucl. Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]