Abstract
The C57BL/6J strain is one of the most widely used animal models for biomedical research, and individual mice within the strain are often assumed to be genetically identical after more than 70 yr of inbreeding. Using a single nucleotide polymorphism (SNP) genotyping panel, we assessed if copy number variations (CNVs) could be detected within the C57BL/6J strain by comparing relative allele frequencies in first generation (F1) progeny of C57BL/6J mice. Sequencing, quantitative PCR, breeding, and array comparative genomic hybridization (CGH) together confirmed the presence of two CNVs. Both CNVs span genes encoded on chromosome 19, and quantitative RT-PCR demonstrated that they result in altered expression of the insulin-degrading enzyme (Ide) and fibroblast growth factor binding protein 3 (Fgfbp3) genes. Analysis of 39 different C57BL/6J breeders revealed that 64% of mice from the Jackson Laboratory colony were heterozygous for the CNV spanning Ide. Homozygotes with and without the duplication were present in concordance with Hardy-Weinberg equilibrium (13% and 23%, respectively), and analysis of archived samples from the C57BL/6J colony suggests that the duplication has rapidly reached a high frequency in the colony since 1994. The identification of two CNVs in the small portion of the genome screened demonstrates that individual mice of highly inbred strains are not isogenic and suggests other CNVs may be segregating within C57BL/6J as well as other carefully maintained inbred strains. These differences can influence interpretations of physiological, biomedical, and behavioral experiments and can be exploited to model CNVs apparent in the human genome.
The recent identification of extensive genomic structural variation among normal, healthy individuals has highlighted the importance of such diversity in human evolution and variation (Eichler 2001; Iafrate et al. 2004; Sebat et al. 2004; Sharp et al. 2006a). Structural variation can take many forms including inversions, deletions, and duplications of DNA segments ranging in size from single-base pairs to large rearrangements affecting several Mb of DNA. Specifically, copy number variations (CNVs) have been defined as genomic alterations involving duplication or deletion of >1 kb of DNA (Freeman et al. 2006). CNVs can result in gene copy number differences and recently have been implicated in a diverse group of human diseases including nervous system disorders (Lee and Lupski 2006), mental retardation (Sharp et al. 2006b), and cancer (La Starza et al. 2007).
The ability to model these and other human diseases in mouse is extremely important in a broad range of biomedical fields and is made possible in part by the existence of well characterized inbred mouse strains whose importance as an experimental system has been well documented (Paigen 1995; Rubin and Barsh 1996; Hamilton and Frankel 2001). One of the oldest used inbred control strains, C57BL/6J, is derived from the C57BL line first bred in 1921 by Dr. C.C. Little, founder of the Jackson Laboratory (JAX Notes 1989). While many lines of C57BL/6 are maintained throughout the world, one of the most widely used is the C57BL/6J line that has been carefully inbred at the Jackson Laboratory for over seventy years through more than 200 generations of brother-sister mating. Because of its widespread use in medical genetics and basic science, C57BL/6J was selected as the basis of the mouse genome sequence build (Mouse Genome Sequencing Consortium 2002) and is also one of the strains chosen to create a collection of knockout lines for the entire genome in association with the NIH mouse genome knockout project (KOMP) (The International Mouse Knockout Consortium 2007). Additionally, C57BL/6J is a common strain used for ethyl nitrosourea (ENU) mutagenesis projects as well as in large-scale efforts to map quantitative trait loci (QTLs) with designer mice including chromosome substitution, congenic, consomic, and recombinant inbred lines (www.nmf.org; Kile et al. 2003; Wang et al. 2003; Churchill et al. 2004; Singer et al. 2004; Peters et al. 2007).
It is clear that different inbred strains have polymorphisms in genomic copy number (Li et al. 2004; Snijders et al. 2005; Graubert et al. 2007), and, within substrains, specific genetic differences are expected (Bailey 1982) and have been identified (Specht and Schoepfer 2001). However, the presence of possible CNVs within carefully maintained inbred stocks such as C57BL/6J has not been investigated. To assess the extent of CNV within inbred strains, we utilized an Illumina single nucleotide polymorphism (SNP) genotyping platform to measure the ratio of parental alleles in first generation (F1) progeny of C57BL/6J and BALB/cJ mice. After screening a small fraction of the murine genome, we identified two CNVs on chromosome 19 within the C57BL/6J strain. Sequencing, quantitative PCR, and array comparative genomic hybridization (CGH) confirmed the CNVs and showed that they significantly alter expression of the insulin-degrading enzyme (Ide) and fibroblast growth factor binding protein 3 (Fgfbp3) genes. Our findings suggest that even the most carefully maintained inbred strains carry genetic differences that could have widespread implications for gene expression and phenotype studies.
Results
Identification of CNVs within C57BL/6J
An Illumina SNP genotyping platform was used to measure the relative ratio of parental alleles in the first generation (F1) progeny of two inbred strains, C57BL/6J and BALB/cJ. In a genome scan of 804 SNPs, a candidate CNV was identified at one SNP on chromosome 19 (refSNP ID rs30920120) where DNA samples from 158 F1 animals did not show identical heterozygosity but, instead, fell into two discrete genotype classes that differed in their signal intensity ratio. Some heterozygotes appeared to carry a single copy of the C57BL/6J allele (B6/BALB) while others appeared to carry a duplicated copy of the C57BL/6J allele (DupB6/BALB) (Fig. 1A).
Figure 1.
Three techniques support a CNV in C57BL/6J × BALB/cJ first generation offspring. (A) An Illumina platform SNP genotyping assay reveals two classes of heterozygote F1 animals that differ in their ratio of C57BL/6J allele intensity compared to BALB/cJ allele intensity at SNP rs30920120. (B) A real-time PCR assay was used to confirm the two classes of heterozygote F1 animals detected in the SNP genotyping assay shown in panel A. (C) Sequencing of SNP rs30920120 confirms an altered parental ratio in two representative individuals from each of the two heterozygote F1 genotype classes.
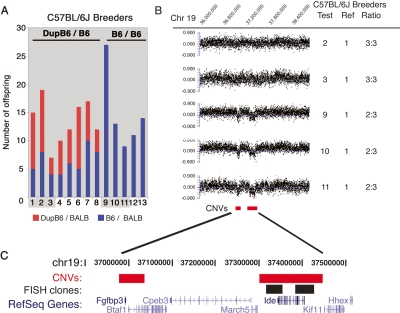
To further investigate a possible copy number variation at this locus we developed a real-time PCR assay to quantitatively determine the relative ratio of the two parental alleles in heterozygous samples. Genotyping of samples included in the genome scan using this independent technique confirmed the presence of two genotype classes of heterozygote F1 animals (Fig. 1B). Additionally, sequencing across SNP rs30920120 in representative samples from each F1 genotype class revealed altered peak height ratios consistent with the ratios we observed on the Illumina platform and real-time PCR assays (Fig. 1C). Because the initial population of heterozygote F1 animals screened on the Illumina platform included progeny that were bred as part of an ENU screen, we could not rule out that ENU treatment was introducing a high frequency of de novo duplications or deletions. Thus, we bred additional mice to confirm the presence of a CNV in a non-ENU-treated, wild-type colony. In total, 166 heterozygote F1 progeny from the wild-type colony were genotyped at rs30920120 using our real-time PCR SNP genotyping assay. These progeny were bred from C57BL/6J and BALB/cJ inbred mice obtained directly from the Jackson Laboratory. We observed both classes of F1 animals in crosses set up in reciprocal directions with respect to maternal and paternal strain, and the two classes did not segregate with gender of the F1 offspring. However, we observed that not every breeding pair produced both genotype classes. A subset of C57BL/6J breeders produced both B6/BALB and DupB6/BALB F1 offspring in a 50:50 ratio whereas other breeders produced offspring only of a single genotype class (Fig. 2A). The genotype of the F1 offspring produced by each C57BL/6J breeder was consistent when rotated through multiple BALB/cJ breeders. However, the reciprocal was not true. Individual BALB/cJ breeders produced different classes of F1 offspring that could vary from litter to litter as C57BL/6J breeders were rotated. For example, a single BALB/cJ breeder produced 11 B6/BALB offspring with one C57BL/6J mating partner and then in subsequent litters the same BALB/cJ breeder produced a mix of offspring (six B6/BALB and six DupB6/BALB) with a second C57BL/6J mating partner. This result suggested that the C57BL/6J breeders were introducing the variability we observed in the F1 offspring and led us to hypothesize that a CNV was present within the C57BL/6J inbred colony and some individual C57BL/6J mice were heterozygous with respect to their copy number at this SNP.
Figure 2.
Breeding and CGH confirm CNV between individual C57BL/6J breeders. (A) The number of F1 offspring in each genotype class is shown for 13 different C57BL/6J breeders. Eight breeders (1–8) are heterozygous at the locus (DupB6/B6) and transmitted an extra copy of the Ide locus to ∼50% of their offspring. Five breeders (breeders 9–13) are presumed homozygous for the locus (B6/B6) and did not transmit a duplicated copy of the locus. Shading highlights C57BL/6J breeders where copy number was confirmed by CGH analysis. (B) CGH detects CNVs in the C57BL/6J strain. Data are shown for hybridizations of five different C57BL/6J breeders each compared to a sixth C57BL/6J breeder as a reference sample. The normalized log2 ratio is plotted for each probe within the portion of chromosome 19 indicated. Test and reference sample numbers correspond to the animal numbers for individual C57BL/6J breeders shown in panel A. For each pair of samples the copy number ratio is given to indicate if hybridizations revealed an equal genomic copy number (3:3) or a reduced copy number (2:3) between the test and reference sample. (C) The location of the CNV and fosmid clones used for FISH are shown relative to RefSeq genes in the region.
To confirm the presence of a CNV in our individual C57BL/6J breeders, we carried out array CGH analysis using DNA samples from 14 animals within our pedigree. The array CGH confirmed that the source of the CNV was indeed differences among individual C57BL/6J breeders (Fig. 2B). Analysis of two BALB/cJ animals, one that had produced only B6/BALB offspring and one that had produced a mix of B6/BALB and DupB6/BALB offspring, showed that both BALB/cJ breeders had the same genomic copy number in the region of chromosome 19 flanking SNP rs30920120. However, among seven different C57BL/6J animals included in the CGH analysis, three carried an extra copy of this region. In all three cases those animals had produced both F1 genotype classes in their offspring. Four C57BL/6J animals showed no increased copy number in the CGH analysis, indicating they were homozygous for a single copy of the locus and consistent with breeding results where all four produced a single genotype (B6/BALB) in their progeny. The CGH results pinpointed the boundaries of the C57BL/6J CNV to an ∼112-kb region of chromosome 19 (NCBI Build 36, chromosome 19 37.303–37.415 Mb) including the entire insulin-degrading enzyme locus (Ide) and the first exon of Kif11 (Fig. 2C). CGH analysis also identified a second CNV of ∼60 kb (NCBI Build 36, chromosome 19 36.977–37.037) containing fibroblast growth factor binding protein 3 (Fgfbp3) and 5′ exons of Btaf1 RNA polymerase II (Btaf1) (Fig. 2C).
Linkage analysis
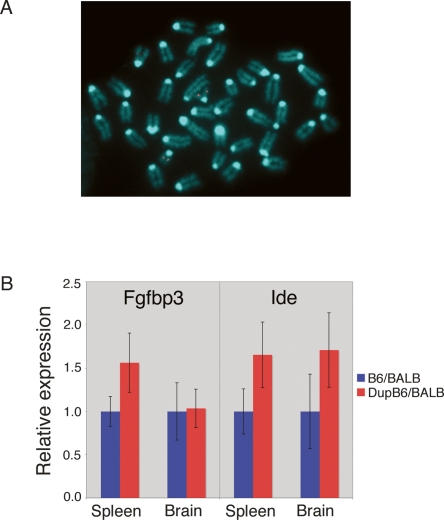
Our CGH results were consistent with individual C57BL/6J mice having a variable copy number at the Ide locus. However, we did not know if the extra copy of the Ide locus observed in some animals was located in close physical proximity to the known chromosome 19 location or in an unlinked region of the genome. Two methods confirm that the multiple copies of Ide in DupB6/BALB animals are tightly linked on chromosome 19. First, two fosmid clones (G135P64894C2/WI-1411L21 and G135P68275D8/WI-807I9), each spanning ∼40 kb of the 112 kb affected region identified by CGH, were used for fluorescent in situ hybridization (FISH). Metaphase chromosome spreads of an F1 animal from the DupB6/BALB group revealed signal at a single location on chromosome 19 (Fig. 3A), consistent with the multiple copies of this locus being within several Mb of each other. Also, in genotyping of 27 heterozygote offspring from a (DupB6/BALB F1 × BALB/cJ) cross, the multiple C57BL/6J copies of the Ide locus co-segregated, providing additional evidence that all copies of the Ide locus reside in close proximity on chromosome 19.
Figure 3.
Multiple copies of chromosome 19 CNVs are tightly linked and result in altered gene expression. (A) FISH using a fosmid clone from the region hybridized to metaphase spreads from a DupB6/BALB F1 animal. Signal observed in the four chromatids of two homologous chromosomes corresponds to a single location on mouse chromosome 19. (B) Fgfbp3 and Ide expression are increased in animals carrying gene duplications. Expression of Fgfbp3 is significantly (P < 0.001) increased in spleen, but not affected in brain. Expression of Ide is significantly increased in both spleen (P < 0.001) and brain (P < 0.01). The graph depicts the mean±one standard deviation for data from four to six replicates from each of seven to eight animals in each group.
CNVs alter expression of duplicated genes
Since the entire coding region of Ide and Fgfbp3 are included in the two CNVs identified, we used real-time RT-PCR to investigate if their expression differed between B6/BALB and DupB6/BALB F1 animals. Indeed, DupB6/BALB F1 animals showed significantly increased expression of Ide in spleen and brain (1.7-fold increase) and of Fgfbp3 expression in spleen (1.6-fold increase; Fig. 3B). Interestingly, Fgfbp3 was expressed but not altered in brain, suggesting that a brain-specific regulatory element may be altered in the extra copy of the gene. We also looked at expression of four other genes in the regions that either overlap the breakpoints or lie between the CNVs. We did not detect any significant expression differences in brain or spleen between the genotype classes for these other four genes (Btaf1, March5, Cpeb3, Kif11), indicating that either the regulation of these genes is not affected by the rearrangement, expression was measured in an inappropriate tissue, or feedback mechanisms exist to correct for any changes in copy number.
High frequency in C57BL/6J stock
The CNV spanning Ide is present at a high frequency within the C57BL/6J stock and was stable through at least three generations of breeding in our colony. Using DNA from archived crosses involving C57BL/6J and BALB/cJ, we found animals heterozygous for the CNV in four independent shipments received from Jackson Laboratories between April 2005 and November 2006. In total, we genotyped F1s from 39 different C57BL/6J breeders, each having litter sizes large enough to allow us to make assumptions about their genotype status by testing for significance (P < 0.05) if a single genotype class was observed rather than the 50:50 ratio of B6/BALB:DupB6/BALB F1s expected from a C57BL/6J breeder heterozygous for the duplication (B6/DupB6). In the sample of 39 breeders, 25 (64%) produced F1 progeny of both genotype classes consistent with heterozygosity for the CNV spanning Ide. Additionally, we observed nine breeders (23%) that only produced B6/BALB F1 offspring, suggesting they were homozygous for a single copy of the Ide locus (B6/B6), and five breeders (13%) that produced only DupB6/BALB F1 as expected from homozygotes carrying a duplicated copy of the locus (DupB6/DupB6). The observed frequency of apparent homozygotes within the C57BL/6J animals we used in this study is in concordance with Hardy–Weinberg equilibrium. To further investigate if there would be selection against homozygotes within the colony, we intercrossed two F1 animals carrying duplications of the C57BL/6J Ide locus (DupB6/BALB × DupB6/BALB). The genotypes of 26 progeny (19% DupB6/DupB6, 46% DupB6/BALB, 35% BALB/BALB) from this intercross did not differ significantly from the expected frequency (25%, 50%, 25%), providing additional support that homozygotes for a duplicated copy of Ide are viable and there is no significant selection against them in the population. Additionally, our CGH analysis included analysis of two animals homozygous for a single copy of the Ide locus and two animals homozygous for a duplicated copy of the Ide locus, thus confirming the presence of viable homozygotes of each class. We also backcrossed and intercrossed F1 mice with and without the duplication, and in each case the parental chromosome was stably transmitted and no de novo rearrangements or reversions were observed.
The concordance with Hardy–Weinberg equilibrium could suggest that this genomic rearrangement occurred a long time ago in the C57BL/6J colony. To address when this rearrangement arose, we analyzed samples from a closely related strain, C57BL/10, which was separated from C57BL/6 sometime prior to 1937 (Festing 1998) as well as samples from the C57BL/6J colony prior to 1994. Ten individual C57BL/10J animals were bred to BALB/cJ, and F1 progeny were genotyped using the realtime PCR assay used in our C57BL/6J F1 progeny. No variation in copy number was observed for any F1 offspring of the 10 breeders. These results support that all 10 C57BL/10J breeders were homozygous for a single copy of the Ide locus. Additionally, the status of the C57BL/6J colony prior to 1994 was assessed by genotyping samples from the BSS and BSB mouse interspecific backcross DNA panels (Rowe et al. 1994). This community mapping resource was bred prior to publication in 1994 and variation within the backcross progeny would reflect any variation present in the parental C57BL/6J breeders. For this analysis we genotyped the 188 N2 animals by measuring peak heights for a simple sequence length polymorphism (SSLP) marker (D19Mit88) within the CNV spanning Ide to quantitate the ratio of the C57BL/6J allele compared to the SPRET/Ei allele in each backcross sample. No variation in the ratio of C57BL/6J to SPRET/Ei was observed in any of the heterozygote N2 samples, and our observed ratios were consistent with all C57BL/6J breeders used to establish this cross being homozygous for a single copy of the region. We conclude that the CNV we observe spanning the Ide locus arose after 1994, however, because of the small number of breeders used to establish the backcross panels, we cannot eliminate the possibility that the CNV was present at a low frequency in the population at that time.
Discussion
To identify possible CNVs within inbred mouse strains, we measured the relative ratio of parental alleles in the F1 progeny of two inbred strains, C57BL/6J and BALB/cJ. A CNV was identified on chromosome 19 and verified by independent techniques including quantitative PCR and sequencing. Subsequent pedigree analysis and array CGH identified a second CNV on chromosome 19 and confirmed that the two CNVs are present within the highly inbred C57BL/6J colony and alter expression of the Ide and Fgfbp3 genes.
The identification of the CNV spanning Ide in the small portion of the genome we initially screened for allelic ratios suggests that other CNVs are likely to be present within C57BL/6J as well as other carefully maintained inbred strains. The Illumina SNP genotyping technology we utilized for a small-scale screen of 804 informative SNPs distributed across the genome has the potential to detect CNVs of any length that span the SNPs utilized, and thus we can assume that we might have detected the majority of very large CNVs. However, little is known about the average size of potential CNVs within the mouse genome. Until recently, studies were biased toward the identification of the largest CNVs because of the limited resolution of available techniques for detection. More recently, a comprehensive study in mouse utilized array CGH with an average oligo spacing of ∼5 kb to identify 80 likely CNVs after screening animals from 20 different strains. The authors identified an average number of 22 CNVs per strain with an average size of 271.5 kb (Graubert et al. 2007). However, recent data using the higher resolution platforms available for human studies suggest that the average CNV size may be much smaller. For example, the median size of all CNVs in the Database of Genomic Variants was reported in 2006 as 18 kb, despite an average size of 118 kb (Freeman et al. 2006), consistent with the majority of human CNVs being relatively small. Additionally, analysis of more than 1000 CNVs identified in human using SNP-based methods, rather than array CGH, found the majority were <10 kb (Eichler 2006). If the majority of CNVs in mouse are similar in size to what is being observed in human, then the SNP panel we used with an average spacing of >3 Mb would only be sufficient to detect a very small fraction of existing CNVs. Full characterization of the extent of intra-strain CNVs will require additional work taking advantage of the latest generation of high-density array CGH technologies.
To limit genetic drift in the Jackson Laboratory colonies, C57BL/6J along with other strains are rederived from a frozen embryo stock every five generations (Taft et al. 2006). The high frequency of heterozygosity we observed for these CNVs in the C57BL/6J colony (64%) suggests that they may exist within the frozen stock and will persist in the population independent of any possible selection. Alternatively, the CNVs could have originated in a single animal after cryopreservation and quickly reached a high frequency in the population because of a population bottleneck created during the recovery of the cryopreserved strain. If this is the case, additional intra-strain CNVs could rapidly introduce unexpected variation into these carefully maintained stocks. Collectively, CNVs like the two we report could account for some of the phenotypic variability reported within strains and previously attributed to environmental, epigenetic, or stochastic differences (Crabbe et al. 1999; de Fourmestraux et al. 2004) and could also alter interpretations of large-scale efforts to collect phenotypic data from inbred strains (Bogue et al. 2007). Possible intra-strain CNVs also need to be considered when interpreting efforts to identify copy number differences among different strains. Indeed, the intra-strain CNV we report spanning Ide has been previously published as a strain-specific difference between C57BL/6J and BALB/cJ (Lakshmi et al. 2006). Additional effort to identify and characterize existing intra-strain CNVs within carefully maintained stocks could increase the diversity of available inbred strains and strengthen their potential to model human variation, especially in light of recent work highlighting the importance of CNVs in human disease (Freeman et al. 2006; Lee and Lupski 2006; Sharp et al. 2006b).
The CNVs we identified cause duplication of the Ide and Fgfbp3 genes in a high proportion of C57BL/6J individuals and result in increased gene expression. The implications of these findings are important considering that Ide expression differences in engineered mice have been shown to have significant effects on disease phenotypes (Farris et al. 2003; Leissring et al. 2003) and that variants near the IDE locus have been associated with risk for human type 2 diabetes (T2D) (Scott et al. 2007) and Alzheimer disease (AD) (Mueller et al. 2007; Vepsäläinen et al. 2007). Mice carrying subtle differences in Ide expression will be useful for understanding the specific role, if any, of IDE in the pathogenesis of T2D and AD since studies on the association of IDE with disease have been inconclusive (Qiu and Folstein 2006). It is crucial that the status of the Ide CNV in various C57BL/6 colonies be determined since both T2D and AD are often modeled in C57BL/6J mice, whose gene copy number differences identified here could directly influence the exact phenotypes under investigation.
Given the extensive use of C57BL/6J in research, knowledge of the precise content of its genome is extremely important. Over 90% of the mouse genome sequence is thought to be complete with NCBI Build 37.1; however, the extent of intra-strain variation is not likely to be represented by such efforts. With the increasing availability of techniques to rapidly and inexpensively characterize intra-strain variation, it is important for the research community to begin to identify existing CNVs in the most widely used strains where genetic drift has been limited by cryopreservation programs. This effort would allow the affects of CNVs in humans to be modeled on controlled genetic backgrounds. Additionally, if intra-strain CNVs are widespread within inbred colonies, they may complicate large scale efforts in mouse to study complex diseases using recombinant inbred and congenic strains, which may indeed carry polymorphisms at loci outside the selected regions. While we cannot limit de novo events that will surely introduce minor variability into inbred strains, documentation of CNVs that are fixed in existing populations will provide details that will ultimately extend the utility of the genomes of mouse inbred strains.
Methods
Genome scan
Genotyping services were provided by the Center for Inherited Disease Research (CIDR, Baltimore, MD) using a commercially available medium density mouse linkage panel (Illumina) to genotype 804 autosomal SNPs polymorphic between C57BL/6J and BALB/cJ on 158 first generation offspring (F1s). Data were analyzed using BeadStudio software (Illumina) and Excel (Microsoft). A single SNP (gnf19.035.019 corresponding to refSNP ID rs30920120) showed evidence of CNV.
Mouse husbandry
BALB/cJ and C57BL/6J inbred mouse strains were purchased from The Jackson Laboratories (Bar Harbor, ME). All other mice were bred and housed in an NHGRI animal facility according to NIH guidelines.
Real-time PCR
Taqman MGB probes (Applied Biosystems) were designed across SNP rs30920120 specific for the C57BL/6J allele (Fam-ACGTGTTAGGTCGTTGG) and the BALB/cJ allele (Vic-ACGTGTTAGGTGGTTGG). PCR reactions were carried out in 1× GenAmp Fast PCR Master Mix, 900 nM each primer (AGGGAAC TGAACTGACTGTACTCA and CAAATATCACAAAGAAATTG GAAAGGTATAAATATCCC), and 200 nM each allele-specific probe. Cycling conditions were 20 sec at 95°C followed by 40 cycles of 92°C for 3 sec, and 60°C for 30 sec. Relative quantitation of the SNP locus was determined in an end-point assay.
CGH
Genomic DNA samples were prepared from tail or liver samples using a PUREGENE DNA purification kit (Gentra Systems, Inc.) according to the manufacturer’s instructions. In some cases DNA was repurified using a Qiagen DNeasy Tissue Kit (Qiagen) and if necessary concentrated using Microcon YM-100 centrifugal filter devices (Millipore). Labeling and hybridization was carried out by a commercial array CGH service (Nimblegen Systems). An oligonucleotide array (Nimblegen #B4351–08–01) of 38,500 probes of 50–75 bp with an average spacing of 656 bp was used covering a portion of mouse chromosome 17 and all of chromosomes 18, 19, X, and Y. Analysis was not done on the data from the X and Y chromosome because of low probe density. The autosomal probes contained on this array represented ∼7% of the NCBI Build 36 mouse genome. Cy3 and Cy5 signals were normalized to one another (Workman et al. 2002), and DNA segmentation analysis (Olshen et al. 2004) was carried out after window averaging. DNA samples from 14 animals within our pedigree were used in various combinations and all hybridization results revealed consistent breakpoints.
FISH
Fosmid clones from the WI1 library (G135P64894C2/WI-1411L21 and G135P68275D8/WI-807I9) were obtained from BACPAC Resources (Children’s Hospital and Research Center, Oakland, CA). DNA was prepared using a Qiagen Large Construct Kit (Qiagen). Metaphase spreads were prepared from blood collected by retro-orbital bleeds. Labeling and hybridization was performed using standard techniques as previously described (Dutra et al. 1996).
Gene expression
Animals from each heterozygote F1 genotype group were used. For each tissue, two independent RNA samples were prepared using Trizol Reagent (Invitrogen Corporation) according to the manufacturer’s instructions. First strand cDNA was prepared using Superscript II (Invitrogen). Relative gene expression assays were done using Taqman assays commercially available from ABI for each gene. Gene expression was normalized to GAPDH in multiplex reactions, and fold expression in DupB6/BALB F1s relative to B6/BALB F1s was calculated using the Comparative CT Method. Seven to eight animals from each group were used to quantitate gene expression in the DupB6/BALB animals relative to the B6/BALB animals. Expression was normalized to GAPDH, and four to six replicates for each animal were averaged. The mean of the two groups was compared using an unpaired t-test performed with QuickCalcs free online software (www.graphpad.com; GraphPad Software). We did not detect any significant differences in gene expression for four other genes in the region that either overlap the breakpoints of the rearrangement or lie between the two CNVs (Btaf1, March5, Cpeb3, Kif11).
Acknowledgments
Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number N01-HG-65403. We thank the NHGRI cytogenetics core for FISH services and Deanna Church at NCBI for genome sequence analysis. SSLP genotyping was performed by Ursula Harper and MaryPat Jones through the NHGRI Genomics Core. The Jackson Laboratory kindly provided archived DNA from the BSS/BSB backcross panels. This research was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (USA). Thank you to Julie Segre, Leslie Biesecker, Pamela Schwartzberg, Eric Green, Lawrence Brody, and members of the Pavan Laboratory for helpful comments and discussions.
Footnotes
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6927808
References
- Bailey D.W. How pure are inbred strains of mice? Immunol. Today. 1982;3:210–214. doi: 10.1016/0167-5699(82)90093-7. [DOI] [PubMed] [Google Scholar]
- Bogue M.A., Grubb S.C., Maddatu T.P., Bult C.J., Grubb S.C., Maddatu T.P., Bult C.J., Maddatu T.P., Bult C.J., Bult C.J. Mouse Phenome Database (MPD) Nucleic Acids Res. 2007;35:D643–D649. doi: 10.1093/nar/gkl1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G.A., Airey D.C., Allayee H., Angel J.M., Attie A.D., Beatty J., Beavis W.D., Belknap J.K., Bennett B., Berrettini W., Airey D.C., Allayee H., Angel J.M., Attie A.D., Beatty J., Beavis W.D., Belknap J.K., Bennett B., Berrettini W., Allayee H., Angel J.M., Attie A.D., Beatty J., Beavis W.D., Belknap J.K., Bennett B., Berrettini W., Angel J.M., Attie A.D., Beatty J., Beavis W.D., Belknap J.K., Bennett B., Berrettini W., Attie A.D., Beatty J., Beavis W.D., Belknap J.K., Bennett B., Berrettini W., Beatty J., Beavis W.D., Belknap J.K., Bennett B., Berrettini W., Beavis W.D., Belknap J.K., Bennett B., Berrettini W., Belknap J.K., Bennett B., Berrettini W., Bennett B., Berrettini W., Berrettini W., et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Crabbe J.C., Wahlsten D., Dudek B.C., Wahlsten D., Dudek B.C., Dudek B.C. Genetics of mouse behavior: Interactions with laboratory environment. Science. 1999;284:1670–1672. doi: 10.1126/science.284.5420.1670. [DOI] [PubMed] [Google Scholar]
- de Fourmestraux V., Neubauer H., Poussin C., Farmer P., Falquet L., Burcelin R., Delorenzi M., Thorens B., Neubauer H., Poussin C., Farmer P., Falquet L., Burcelin R., Delorenzi M., Thorens B., Poussin C., Farmer P., Falquet L., Burcelin R., Delorenzi M., Thorens B., Farmer P., Falquet L., Burcelin R., Delorenzi M., Thorens B., Falquet L., Burcelin R., Delorenzi M., Thorens B., Burcelin R., Delorenzi M., Thorens B., Delorenzi M., Thorens B., Thorens B. Transcript profiling suggests that differential metabolic adaptation of mice to a high fat diet is associated with changes in liver to muscle lipid fluxes. J. Biol. Chem. 2004;279:50743–50753. doi: 10.1074/jbc.M408014200. [DOI] [PubMed] [Google Scholar]
- Dutra A.S., Mignot E., Puck J.M., Mignot E., Puck J.M., Puck J.M. Gene localization and syntenic mapping by FISH in the dog. Cytogenet. Cell Genet. 1996;74:113–117. doi: 10.1159/000134395. [DOI] [PubMed] [Google Scholar]
- Eichler E.E. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 2001;17:661–669. doi: 10.1016/s0168-9525(01)02492-1. [DOI] [PubMed] [Google Scholar]
- Eichler E.E. Widening the spectrum of human genetic variation. Nat. Genet. 2006;38:9–11. doi: 10.1038/ng0106-9. [DOI] [PubMed] [Google Scholar]
- Farris W., Mansourian S., Chang Y., Lindsley L., Eckman E.A., Frosch M.P., Eckman C.B., Tanzi R.E., Selkoe D.J., Guenette S., Mansourian S., Chang Y., Lindsley L., Eckman E.A., Frosch M.P., Eckman C.B., Tanzi R.E., Selkoe D.J., Guenette S., Chang Y., Lindsley L., Eckman E.A., Frosch M.P., Eckman C.B., Tanzi R.E., Selkoe D.J., Guenette S., Lindsley L., Eckman E.A., Frosch M.P., Eckman C.B., Tanzi R.E., Selkoe D.J., Guenette S., Eckman E.A., Frosch M.P., Eckman C.B., Tanzi R.E., Selkoe D.J., Guenette S., Frosch M.P., Eckman C.B., Tanzi R.E., Selkoe D.J., Guenette S., Eckman C.B., Tanzi R.E., Selkoe D.J., Guenette S., Tanzi R.E., Selkoe D.J., Guenette S., Selkoe D.J., Guenette S., Guenette S. Insulin-degrading enzyme regulates the levels of insulin, amyloid β-protein, and the β-amyloid precursor protein intracellular domain in vivo. Proc. Natl. Acad. Sci. 2003;100:4162–4167. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festing M.F.W. 1998. Inbred strains of mice. http://www.informatics.jax.org/external/festing/mouse/STRAINS.shtml. [Google Scholar]
- Freeman J.L., Perry G.H., Feuk L., Redon R., McCarroll S.A., Altshuler D.M., Aburatani H., Jones K.W., Tyler-Smith C., Hurles M.E., Perry G.H., Feuk L., Redon R., McCarroll S.A., Altshuler D.M., Aburatani H., Jones K.W., Tyler-Smith C., Hurles M.E., Feuk L., Redon R., McCarroll S.A., Altshuler D.M., Aburatani H., Jones K.W., Tyler-Smith C., Hurles M.E., Redon R., McCarroll S.A., Altshuler D.M., Aburatani H., Jones K.W., Tyler-Smith C., Hurles M.E., McCarroll S.A., Altshuler D.M., Aburatani H., Jones K.W., Tyler-Smith C., Hurles M.E., Altshuler D.M., Aburatani H., Jones K.W., Tyler-Smith C., Hurles M.E., Aburatani H., Jones K.W., Tyler-Smith C., Hurles M.E., Jones K.W., Tyler-Smith C., Hurles M.E., Tyler-Smith C., Hurles M.E., Hurles M.E. Copy number variation: New insights in genome diversity. Genome Res. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- Graubert T.A., Cahan P., Edwin D., Selzer R.R., Richmond T.A., Eis P.S., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., Cahan P., Edwin D., Selzer R.R., Richmond T.A., Eis P.S., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., Edwin D., Selzer R.R., Richmond T.A., Eis P.S., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., Selzer R.R., Richmond T.A., Eis P.S., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., Richmond T.A., Eis P.S., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., Eis P.S., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., Li X., McLeod H.L., Cheverud J.M., McLeod H.L., Cheverud J.M., Cheverud J.M., et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B.A., Frankel W.N., Frankel W.N. Of mice and genome sequence. Cell. 2001;107:13–16. doi: 10.1016/s0092-8674(01)00514-1. [DOI] [PubMed] [Google Scholar]
- Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Lee C., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Lee C., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Lee C., Listewnik M.L., Donahoe P.K., Qi Y., Lee C., Donahoe P.K., Qi Y., Lee C., Qi Y., Lee C., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- International Mouse Knockout Consortium A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- JAX Notes Profile: C57BL/6J. JAX Notes. 1989;438 http://jaxmice.jax.org/library/notes/438b.html. [Google Scholar]
- Kile B.T., Hentges K.E., Clark A.T., Nakamura H., Salinger A.P., Liu B., Box N., Stockton D.W., Johnson R.L., Behringer R.R., Hentges K.E., Clark A.T., Nakamura H., Salinger A.P., Liu B., Box N., Stockton D.W., Johnson R.L., Behringer R.R., Clark A.T., Nakamura H., Salinger A.P., Liu B., Box N., Stockton D.W., Johnson R.L., Behringer R.R., Nakamura H., Salinger A.P., Liu B., Box N., Stockton D.W., Johnson R.L., Behringer R.R., Salinger A.P., Liu B., Box N., Stockton D.W., Johnson R.L., Behringer R.R., Liu B., Box N., Stockton D.W., Johnson R.L., Behringer R.R., Box N., Stockton D.W., Johnson R.L., Behringer R.R., Stockton D.W., Johnson R.L., Behringer R.R., Johnson R.L., Behringer R.R., Behringer R.R., et al. Functional genetic analysis of mouse chromosome 11. Nature. 2003;425:81–86. doi: 10.1038/nature01865. [DOI] [PubMed] [Google Scholar]
- La Starza R., Crescenzi B., Pierini V., Romoli S., Gorello P., Brandimarte L., Matteucci C., Kropp M.G., Barba G., Martelli M.F., Crescenzi B., Pierini V., Romoli S., Gorello P., Brandimarte L., Matteucci C., Kropp M.G., Barba G., Martelli M.F., Pierini V., Romoli S., Gorello P., Brandimarte L., Matteucci C., Kropp M.G., Barba G., Martelli M.F., Romoli S., Gorello P., Brandimarte L., Matteucci C., Kropp M.G., Barba G., Martelli M.F., Gorello P., Brandimarte L., Matteucci C., Kropp M.G., Barba G., Martelli M.F., Brandimarte L., Matteucci C., Kropp M.G., Barba G., Martelli M.F., Matteucci C., Kropp M.G., Barba G., Martelli M.F., Kropp M.G., Barba G., Martelli M.F., Barba G., Martelli M.F., Martelli M.F., et al. A common 93-kb duplicated DNA sequence at 1q21.2 in acute lymphoblastic leukemia and Burkitt lymphoma. Cancer Genet. Cytogenet. 2007;175:73–76. doi: 10.1016/j.cancergencyto.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Lakshmi B., Hall I.M., Egan C., Alexander J., Leotta A., Healy J., Zender L., Spector M.S., Xue W., Lowe S.W., Hall I.M., Egan C., Alexander J., Leotta A., Healy J., Zender L., Spector M.S., Xue W., Lowe S.W., Egan C., Alexander J., Leotta A., Healy J., Zender L., Spector M.S., Xue W., Lowe S.W., Alexander J., Leotta A., Healy J., Zender L., Spector M.S., Xue W., Lowe S.W., Leotta A., Healy J., Zender L., Spector M.S., Xue W., Lowe S.W., Healy J., Zender L., Spector M.S., Xue W., Lowe S.W., Zender L., Spector M.S., Xue W., Lowe S.W., Spector M.S., Xue W., Lowe S.W., Xue W., Lowe S.W., Lowe S.W., et al. Mouse genomic representational oligonucleotide microarray analysis: Detection of copy number variations in normal and tumor specimens. Proc. Natl. Acad. Sci. 2006;103:11234–11239. doi: 10.1073/pnas.0602984103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.A., Lupski J.R., Lupski J.R. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron. 2006;52:103–121. doi: 10.1016/j.neuron.2006.09.027. [DOI] [PubMed] [Google Scholar]
- Leissring M.A., Farris W., Chang A.Y., Walsh D.M., Wu X., Sun X., Frosch M.P., Selkoe D.J., Farris W., Chang A.Y., Walsh D.M., Wu X., Sun X., Frosch M.P., Selkoe D.J., Chang A.Y., Walsh D.M., Wu X., Sun X., Frosch M.P., Selkoe D.J., Walsh D.M., Wu X., Sun X., Frosch M.P., Selkoe D.J., Wu X., Sun X., Frosch M.P., Selkoe D.J., Sun X., Frosch M.P., Selkoe D.J., Frosch M.P., Selkoe D.J., Selkoe D.J. Enhanced proteolysis of β-amyloid in APP transgenic mice prevents plaque formation, secondary pathology, and premature death. Neuron. 2003;40:1087–1093. doi: 10.1016/s0896-6273(03)00787-6. [DOI] [PubMed] [Google Scholar]
- Li J., Jiang T., Mao J., Balmain A., Peterson L., Harris C., Rao P.H., Havlak P., Gibbs R., Cai W.W., Jiang T., Mao J., Balmain A., Peterson L., Harris C., Rao P.H., Havlak P., Gibbs R., Cai W.W., Mao J., Balmain A., Peterson L., Harris C., Rao P.H., Havlak P., Gibbs R., Cai W.W., Balmain A., Peterson L., Harris C., Rao P.H., Havlak P., Gibbs R., Cai W.W., Peterson L., Harris C., Rao P.H., Havlak P., Gibbs R., Cai W.W., Harris C., Rao P.H., Havlak P., Gibbs R., Cai W.W., Rao P.H., Havlak P., Gibbs R., Cai W.W., Havlak P., Gibbs R., Cai W.W., Gibbs R., Cai W.W., Cai W.W. Genomic segmental polymorphisms in inbred mouse strains. Nat. Genet. 2004;36:952–954. doi: 10.1038/ng1417. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium 2002Initial sequencing and comparative analysis of the mouse genome Nature 420520 -562 [DOI] [PubMed] [Google Scholar]
- Mueller J.C., Riemenschneider M., Schoepfer-Wendels A., Gohlke H., Konta L., Friedrich P., Illig T., Laws S., Förstl H., Kurz A., Riemenschneider M., Schoepfer-Wendels A., Gohlke H., Konta L., Friedrich P., Illig T., Laws S., Förstl H., Kurz A., Schoepfer-Wendels A., Gohlke H., Konta L., Friedrich P., Illig T., Laws S., Förstl H., Kurz A., Gohlke H., Konta L., Friedrich P., Illig T., Laws S., Förstl H., Kurz A., Konta L., Friedrich P., Illig T., Laws S., Förstl H., Kurz A., Friedrich P., Illig T., Laws S., Förstl H., Kurz A., Illig T., Laws S., Förstl H., Kurz A., Laws S., Förstl H., Kurz A., Förstl H., Kurz A., Kurz A. Weak independent association signals between IDE polymorphisms, Alzheimer’s disease, and cognitive measures. Neurobiol. Aging. 2007;28:727–734. doi: 10.1016/j.neurobiolaging.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Olshen A.B., Venkatraman E.S., Lucito R., Wigler M., Venkatraman E.S., Lucito R., Wigler M., Lucito R., Wigler M., Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Paigen K. A miracle enough: The power of mice. Nat. Med. 1995;1:215–220. doi: 10.1038/nm0395-215. [DOI] [PubMed] [Google Scholar]
- Peters L.L., Robledo R.F., Bult C.J., Churchill G.A., Paigen B.J., Svenson K.L., Robledo R.F., Bult C.J., Churchill G.A., Paigen B.J., Svenson K.L., Bult C.J., Churchill G.A., Paigen B.J., Svenson K.L., Churchill G.A., Paigen B.J., Svenson K.L., Paigen B.J., Svenson K.L., Svenson K.L. The mouse as a model for human biology: A resource guide for complex trait analysis. Nat. Rev. Genet. 2007;8:58–69. doi: 10.1038/nrg2025. [DOI] [PubMed] [Google Scholar]
- Qiu W.Q., Folstein M.F., Folstein M.F. Insulin, insulin-degrading enzyme and amyloid-beta peptide in Alzheimer’s disease: Review and hypothesis. Neurobiol. Aging. 2006;27:190–198. doi: 10.1016/j.neurobiolaging.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Rowe L.B., Nadeau J.H., Turner R., Frankel W.N., Letts V.A., Eppig J.T., Ko M.S.H., Thurston S.J., Birkenmeier E.H., Nadeau J.H., Turner R., Frankel W.N., Letts V.A., Eppig J.T., Ko M.S.H., Thurston S.J., Birkenmeier E.H., Turner R., Frankel W.N., Letts V.A., Eppig J.T., Ko M.S.H., Thurston S.J., Birkenmeier E.H., Frankel W.N., Letts V.A., Eppig J.T., Ko M.S.H., Thurston S.J., Birkenmeier E.H., Letts V.A., Eppig J.T., Ko M.S.H., Thurston S.J., Birkenmeier E.H., Eppig J.T., Ko M.S.H., Thurston S.J., Birkenmeier E.H., Ko M.S.H., Thurston S.J., Birkenmeier E.H., Thurston S.J., Birkenmeier E.H., Birkenmeier E.H. Maps from two interspecific backcross DNA panels available as a community genetic mapping resource. Mamm. Genome. 1994;5:253–274. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]
- Rubin E.M., Barsh G.S., Barsh G.S. Biological insights through genomics: Mouse to man. J. Clin. Invest. 1996;97:275–280. doi: 10.1172/JCI118413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L.J., Mohlke K.L., Bonnycastle L.L., Willer C.J., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., Mohlke K.L., Bonnycastle L.L., Willer C.J., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., Bonnycastle L.L., Willer C.J., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., Willer C.J., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., Stringham H.M., Chines P.S., Jackson A.U., Chines P.S., Jackson A.U., Jackson A.U., et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Månér S., Massa H., Walker M., Chi M., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Månér S., Massa H., Walker M., Chi M., Troge J., Alexander J., Young J., Lundin P., Månér S., Massa H., Walker M., Chi M., Alexander J., Young J., Lundin P., Månér S., Massa H., Walker M., Chi M., Young J., Lundin P., Månér S., Massa H., Walker M., Chi M., Lundin P., Månér S., Massa H., Walker M., Chi M., Månér S., Massa H., Walker M., Chi M., Massa H., Walker M., Chi M., Walker M., Chi M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Sharp A.J., Cheng Z., Eichler E.E., Cheng Z., Eichler E.E., Eichler E.E. Structural variation of the human genome. Annu. Rev. Genomics Hum. Genet. 2006a;7:407–442. doi: 10.1146/annurev.genom.7.080505.115618. [DOI] [PubMed] [Google Scholar]
- Sharp A.J., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Stewart H., Price S.M., Blair E., Hennekam R.C., Price S.M., Blair E., Hennekam R.C., Blair E., Hennekam R.C., Hennekam R.C., et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 2006b;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- Singer J.B., Hill A.E., Burrage L.C., Olszens K.R., Song J., Justice M., O’Brien W.E., Conti D.V., Witte J.S., Lander E.S., Hill A.E., Burrage L.C., Olszens K.R., Song J., Justice M., O’Brien W.E., Conti D.V., Witte J.S., Lander E.S., Burrage L.C., Olszens K.R., Song J., Justice M., O’Brien W.E., Conti D.V., Witte J.S., Lander E.S., Olszens K.R., Song J., Justice M., O’Brien W.E., Conti D.V., Witte J.S., Lander E.S., Song J., Justice M., O’Brien W.E., Conti D.V., Witte J.S., Lander E.S., Justice M., O’Brien W.E., Conti D.V., Witte J.S., Lander E.S., O’Brien W.E., Conti D.V., Witte J.S., Lander E.S., Conti D.V., Witte J.S., Lander E.S., Witte J.S., Lander E.S., Lander E.S., et al. Genetic dissection of complex traits with chromosome substitution strains of mice. Science. 2004;304:445–448. doi: 10.1126/science.1093139. [DOI] [PubMed] [Google Scholar]
- Snijders A.M., Nowak N.J., Huey B., Fridlyand J., Law S., Conroy J., Tokuyasu T., Demir K., Chiu R., Mao J.H., Nowak N.J., Huey B., Fridlyand J., Law S., Conroy J., Tokuyasu T., Demir K., Chiu R., Mao J.H., Huey B., Fridlyand J., Law S., Conroy J., Tokuyasu T., Demir K., Chiu R., Mao J.H., Fridlyand J., Law S., Conroy J., Tokuyasu T., Demir K., Chiu R., Mao J.H., Law S., Conroy J., Tokuyasu T., Demir K., Chiu R., Mao J.H., Conroy J., Tokuyasu T., Demir K., Chiu R., Mao J.H., Tokuyasu T., Demir K., Chiu R., Mao J.H., Demir K., Chiu R., Mao J.H., Chiu R., Mao J.H., Mao J.H., et al. Mapping segmental and sequence variations among laboratory mice using BAC array CGH. Genome Res. 2005;15:302–311. doi: 10.1101/gr.2902505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht C.G., Schoepfer R., Schoepfer R. Deletion of the alpha-synuclein locus in a subpopulation of C57BL/6J inbred mice. BMC Neurosci. 2001;2:11. doi: 10.1186/1471-2202-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft R.A., Davisson M., Wiles M.V., Davisson M., Wiles M.V., Wiles M.V. Know thy mouse. Trends Genet. 2006;22:649–653. doi: 10.1016/j.tig.2006.09.010. [DOI] [PubMed] [Google Scholar]
- The International Mouse Knockout Consortium 2007A mouse for all reasons Cell 1289 -13 [DOI] [PubMed] [Google Scholar]
- Vepsäläinen S., Parkinson M., Helisalmi S., Mannermaa A., Soininen H., Tanzi R., Bertram L., Hiltunen M., Parkinson M., Helisalmi S., Mannermaa A., Soininen H., Tanzi R., Bertram L., Hiltunen M., Helisalmi S., Mannermaa A., Soininen H., Tanzi R., Bertram L., Hiltunen M., Mannermaa A., Soininen H., Tanzi R., Bertram L., Hiltunen M., Soininen H., Tanzi R., Bertram L., Hiltunen M., Tanzi R., Bertram L., Hiltunen M., Bertram L., Hiltunen M., Hiltunen M. Insulin degrading enzyme is genetically associated with Alzheimer’s disease in the Finnish population. J. Med. Genet. 2007;44:606–608. doi: 10.1136/jmg.2006.048470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Le Roy I., Nicodeme E., Li R., Wagner R., Petros C., Churchill G.A., Harris S., Darvasi A., Kirilovsky J., Le Roy I., Nicodeme E., Li R., Wagner R., Petros C., Churchill G.A., Harris S., Darvasi A., Kirilovsky J., Nicodeme E., Li R., Wagner R., Petros C., Churchill G.A., Harris S., Darvasi A., Kirilovsky J., Li R., Wagner R., Petros C., Churchill G.A., Harris S., Darvasi A., Kirilovsky J., Wagner R., Petros C., Churchill G.A., Harris S., Darvasi A., Kirilovsky J., Petros C., Churchill G.A., Harris S., Darvasi A., Kirilovsky J., Churchill G.A., Harris S., Darvasi A., Kirilovsky J., Harris S., Darvasi A., Kirilovsky J., Darvasi A., Kirilovsky J., Kirilovsky J., et al. Using advanced intercross lines for high-resolution mapping of HDL cholesterol quantitative trait loci. Genome Res. 2003;13:1654–1664. doi: 10.1101/gr.1185803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman C., Jensen L.J., Jarmer H., Berka R., Gautier L., Nielser H.B., Nielsen C., Brunak S., Knudsen S., Jensen L.J., Jarmer H., Berka R., Gautier L., Nielser H.B., Nielsen C., Brunak S., Knudsen S., Jarmer H., Berka R., Gautier L., Nielser H.B., Nielsen C., Brunak S., Knudsen S., Berka R., Gautier L., Nielser H.B., Nielsen C., Brunak S., Knudsen S., Gautier L., Nielser H.B., Nielsen C., Brunak S., Knudsen S., Nielser H.B., Nielsen C., Brunak S., Knudsen S., Nielsen C., Brunak S., Knudsen S., Brunak S., Knudsen S., Knudsen S. Genome Biol. Vol. 3. 2002. A new nonlinear normalization method for reducing variability in DNA microarray experiments. [DOI] [PMC free article] [PubMed] [Google Scholar]