Abstract
The amount of extra-pair paternity in socially monogamous bird species varies from 0% to 76% extra-pair offspring. The causes of this remarkable interspecific variation are largely unknown, although intraspecific analyses suggest that females seek extra-pair matings to improve the genetic quality of their offspring. If this is a general explanation for the occurrence of extra-pair matings, then proportionally more females should seek to modify the paternity of their clutch when there is more variation among males in their genetic quality. Here we test this prediction in birds and show that interspecific variation in the proportion of extra-pair offspring is positively related to the proportion of polymorphic loci as measured by protein electrophoresis, even when controlling for potentially confounding variables. Genetic variability was also assessed, for sister pairs of species and populations differing significantly in extra-pair paternity, by using random priming, which provides an estimate of genome-wide diversity. We found that genetic diversity was higher in the populations with a higher level of extra-pair paternity. These results suggest that the amount of genetic variability in a population may be an important factor influencing mating patterns.
Understanding the factors, ecological or otherwise, that determine the prevalent mating system of a population is central to evolutionary biology because it is the mating system that ultimately determines patterns of gene transmission across generations. However, despite considerable research, we have made little progress in identifying the main factors involved. This lack of progress may be partly the result of previous categorizations of mating systems that were too broad to be meaningful. For example, in birds 90% of species were, until very recently, categorized as monogamous, but the use of genetic markers has revealed that in many species social monogamy does not result in genetic monogamy. Females can seek extra-pair copulations from males other than their social mate, leading to surprising levels of polygamy.
Levels of extra-pair paternity vary enormously among bird species, with some species showing low or nonexistent extra-pair paternity (1–3), whereas other species have levels of extra-pair offspring exceeding 35% (4–8). Understanding this variation provides us with an opportunity to understand which factors may be generally important in promoting polygamy.
There have been few attempts to explain this interspecific variation (see refs. 9 and 10 for recent reviews). Some authors have suggested that factors such as the density of breeding pairs (for example, when birds breed colonially) or breeding synchrony may be important in promoting high levels of extra-pair paternity (11, 12). However, a recent comparative study controlling for similarity among species caused by common descent revealed that breeding density, coloniality, or breeding synchrony could not account for any interspecific variation in extra-pair paternity (13).
Levels of extra-pair paternity are a population-level description of events that occur at the level of the individual. To explain why levels of extra-pair paternity are particularly high, we need to explain when it will pay a higher proportion of the females in a population to modify the paternity of their clutch. Intraspecific analyses of the patterns of extra-pair behavior have suggested that the main benefit females gain from seeking extra-pair matings is an improvement in the genetic quality of their offspring (refs. 14 and 15; review in ref. 10). Females do not gain any obvious direct benefits from seeking an extra-pair partner and tend to prefer the most extravagantly ornamented, colored, or displaying males as an extra-pair sire (10). Moreover, extra-pair paternity is particularly common in species in which the most ornamented males provide the least amount of parental care (16). If female choice of indirect fitness benefits is a general explanation for the occurrence of extra-pair paternity, we can predict that more females will seek to modify the paternity of their clutch when there are larger differences between the genetic quality of potential fathers. With low variation in male genetic quality, it would not pay females to be choosy and seek extra-pair sires (17). The aim of this paper is to test this prediction.
We investigate in two ways whether populations with a lower level of extra-pair paternity also have lower genetic variability. First, we ask whether the proportion of polymorphic loci, measured by protein gel electrophoresis, is a predictor of the level of extra-pair paternity among species by using data extracted from the literature. Second, we compare the genome-wide genetic variation, measured with random priming [random amplified polymorphic DNA (RAPD) techniques], of sister species or populations known to differ significantly in levels of extra-pair paternity.
MATERIALS AND METHODS
Allozyme Data.
Genetic variation is an attribute that cannot be measured exhaustively, but variation in proteins provides an estimate of variation in the structural genes of an organism. Gel electrophoresis has been used to study protein variation in a wide array of species, and we searched the literature by using standard abstract and reference books, such as Zoological Record and Biological Abstracts and also references cited in papers to find 432 different samples of allozyme variation in birds.
Electrophoretic data give the frequency of electromorphs that are assumed to correspond to one allele. Two measures of genetic variation can be deduced from allozyme data, PL and H. PL is the measure of loci found to be polymorphic in any one sample (a locus is considered to be polymorphic when the frequency of the most common allele is no greater than a certain value, usually 0.99 or 0.95). The range of PL for the bird species studied varied from 0% to 71.4% [mean (SE) = 19.1% (0.7)]. Estimates of heterozygosity (H) reflect the mean number of heterozygous loci per individual. Both heterozygosity and the proportion of polymorphic loci are generally expected to covary. We did not find evidence for such an interspecific correlation in our data (Pearson’s r = 0.04; n = 203 species of birds). When based on a few loci, estimates of heterozygosity provide poor estimates of genome-wide genetic variation (18–21). Therefore, we used only the proportion of polymorphic loci. Most estimates of PL were based on a large number of loci surveyed. Any samples originating from small introduced populations or island populations were excluded from the analyses. Similarly, studies without information on sample sizes were excluded from the analyses. Summed sample sizes for the estimates of the number of polymorphic loci in the original sources were used to correct the analyses for any bias caused by a different number of individuals being sampled.
We obtained estimates of the frequency of extra-pair paternity by using an extensive survey of the literature, relying entirely on studies based on molecular methods and enzyme polymorphism (the latter only being used when the estimates of extra-pair paternity had been corrected for the probability of exclusion of sires).
The proportion of polymorphic loci and the frequency of extra-pair paternity were estimated in multiple studies for 26 and 17 species, respectively. From these multiple studies, we calculated the repeatability of these parameters (22). The repeatability of the proportion of polymorphic loci was 0.55 and statistically significant (F = 3.61; df = 25, 27; P = 0.0008). The repeatability of extra-pair paternity was also high at 0.68 and statistically highly significant (F = 7.00; df = 16, 31; P < 0.001). These high repeatabilities imply that estimates of both parameters are consistent across studies of the same species despite any methodological and/or environmental differences and that a single estimate for a species would provide a reliable species-specific value. If more than a single estimate of the proportion of polymorphic loci and frequency of extra-pair paternity was available, we used the mean value of the available estimates in the analyses because the mean value would be closer to the true species-specific mean than any randomly chosen single estimate. The fact that the allozyme data do not derive from the same populations as those surveyed for extra-pair paternity makes any test of association conservative because spatial or temporal variability would tend to eliminate any correlation between genetic variation and paternity.
As a measure of sexual dichromatism, we used the difference between mean male and female color score in the visual spectrum made by three independent scorers (23). Such scores are highly repeatable among scorers, and they correlated well with extra-pair paternity in two other studies (23, 24), implying that scores estimate important features of color signals related to sexual selection. Information on body mass was obtained from Brough (25). The data set is reproduced as supplemental data to this article on the PNAS website (www.pnas.org).
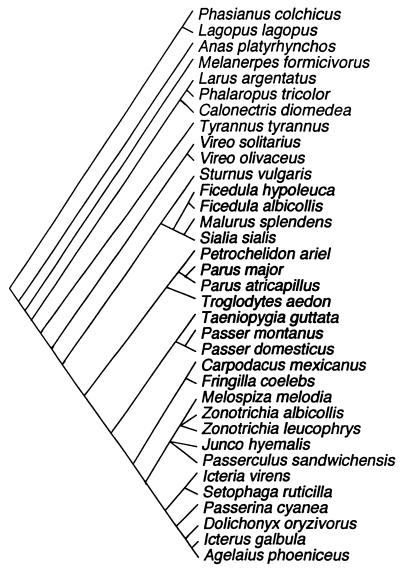
Because species cannot be considered statistically independent due to similarities arising from common descent (26), we corrected for this problem by investigating the relationship between extra-pair paternity and polymorphic loci by using standardized contrasts (or differences) between taxa. We adopted the software CAIC (27) to calculate standardized differences between taxa for the two variables of interest and for the potentially confounding variables. Here we present the results based on a model of gradual evolution assuming that branch lengths are related to the number of species in a clade, but the results based on a model of punctuated evolution (with all branch lengths being equal) gave qualitatively similar results. Information on phylogenetic relationships among taxa was obtained from Sibley and Ahlquist (28). Although this study has been severely criticized (29–31), several parts of the phylogeny have been confirmed with independent data sets and stringent phylogenetic analysis (review in ref. 32). The clade Melospiza melodia, Zonotrichia albicollis, and Zonotrichia leucophrys was unresolved and treated as a polytomy, and the same was true for Junco hyemalis and Passerculus sandwichensis, which was considered to constitute a polytomy nearer the root of the same branch. The phylogeny is shown in Fig. 1.
Figure 1.
A phylogeny of the species included in the allozyme study, mainly based on Sibley and Ahlquist (28).
Before the analyses, we made a number of transformations of variables to meet the assumptions of normal frequency distributions. The proportion of extra-pair paternity and the proportion of polymorphic loci were square root-arcsine-transformed, and sample size and body mass were log10-transformed, whereas sexual dichromatism was untransformed.
The contrasts were analyzed by forcing a regression of the dependent variable (extra-pair paternity) on the independent variable (polymorphic loci) through the origin (27). The effects of potentially confounding variables were controlled by using the same procedure with multiple linear regression analysis.
RAPD Data.
A survey of the literature allowed us to identify several pairs of populations differing in their frequency of extra-pair paternity both within species and between closely related species (at least within families). The genetic variability of these populations was then assessed from individuals originating from the same populations for which extra-pair paternity had been estimated. We obtained DNA or blood samples from ≈20 individuals for 7 sister pairs of populations. The names of the species and populations studied are given in Table 2. For each pair, the populations differed significantly in their level of extra-pair paternity (Fisher exact test, P < 0.05), except for pair 2 for which P = 0.055.
Table 2.
Level of extra-pair paternity (EPP) and genetic variability measured with RAPD techniques
| Species | Ni | Nl | Primers UBC | EPP | Ref. | Pol | Ho |
|---|---|---|---|---|---|---|---|
| 1. Calonectis diomedea | 15 | 22 | 219(4)/229(2)/230(3)/243(5)/244(5)/283(3) | 0 | 38 | 0.36 | 0.070 (19) |
| 1. Puffinus tenuirostris | 15 | 22 | 219(5)/229(2)/230(6)/243(3)/244(3)/283(3) | 0.13 | 39 | 0.50 | 0.079 (20) |
| 2. Erithacus rubecula | 18 | 17 | 215(2)/229(2)/230(5)/232(3)/238(3)/239(2) | 0.04 | ∗ | 0.59 | 0.171 (15) |
| 2. Luscinia s. svecica | 18 | 24 | 215(2)/229(4)/230(4)/232(5)/238(5)/239(4) | 0.20 | 40 | 0.79 | 0.198 (23) |
| 3. Acrocephalus vaughani taiti | 16 | 20 | 219(5)/226(2)/231(5)/238(4)/239(4) | 0–0.07 | 41 | 0.30 | 0.015 (16) |
| 3. A. paludicola | 16 | 20 | 219(4)/226(3)/231(5)/238(4)/239(4) | 0.36 | 42 | 0.40 | 0.094 (20) |
| 4. Carpodacus mexicanus | 20 | 24 | 215(5)/228(3)/229(4)/238(4)/239(5)/283(3) | 0.08 | 43 | 0.67 | 0.186 (21) |
| 4. Emberiza schoeniclus | 16 | 29 | 215(4)/228(6)/229(5)/238(3)/239(4)/283(7) | 0.55 | 8 | 0.76 | 0.173 (24) |
| 5. Cardinalis cardinalis | 17 | 18 | 219(3)/226(3)/228(5)/237(2)/283(5) | 0.13 | 44 | 0.72 | 0.154 (16) |
| 5. Icterus galbula | 19 | 19 | 219(4)/226(5)/228(4)/237(2)/283(4) | 0.37 | † | 0.79 | 0.195 (19) |
| 6. Phylloscopus trochilus 1 | 18 | 18 | 215(4)/219(5)/228(2)/232(4)/238(3) | 0.00 | 1 | 0.78 | 0.121 (14) |
| 6. P. trochilus 2 | 18 | 17 | 215(4)/219(4)/228(3)/232(3)/238(3) | 0.33 | 45 | 0.82 | 0.228 (16) |
| 7. Agelaius phoenicus 1 | 19 | 17 | 226(4)/228(4)/229(4)/237(2)/283(3) | 0.25 | 46 | 0.82 | 0.199 (15) |
| 7. A. phoenicus 2 | 19 | 16 | 226(2)/228(4)/229(3)/237(3)/283(4) | 0.35 | 47 | 0.75 | 0.194 (15) |
Pol is the ratio of polymorphic loci, and Ho is the gene diversity. The number of loci considered to estimate the gene diversity according to Linch and Milligan (34), is given between parenthesis. Ni is the number of individuals analysed and Nl is the total number of bands considered. The number of bands for each primer UBC (University of British Columbia, set #3) is given in parentheis.
J. Tobias and I. R. Hartley, personal communication.
D. Ritchardson, personal communication.
The DNA concentration of each sample was checked with a Hoeffer DyNA Quant 200 Fluorometer, If necessary, a dilution was performed to get a working solution of 20 ng of DNA/μl. PCR mixtures (12.5 μl final volume) contained 20 ng of template DNA; 100 μM each of dATP, dGTP, dCTP, and dTTP; 0.2 μM 10-base random primer; 3 mM MgCl2, 1× Taq Buffer IV; and 0.5 unit Taq DNA polymerase (Advanced Biotechnologies). Each reaction was overlaid with a drop of mineral oil to prevent evaporation. Amplifications were performed in a Perkin–Elmer DNA Thermal Cycler by using the following parameters: 1 min at 94°C followed by 40 cycles each of 1 min at 94°C, 1 min at 36°C, and 2 min at 72°C. After the final cycle, samples were incubated for 5 min at 72°C and then held at 4°C before analysis. Amplification products were loaded into a 1.4% agarose gel in a 1× TBE buffer, electrophoretically separated at 80 V for 6–8 h (according to the primer), stained with ethidium bromide, and photographed over a UV light. PCR reactions for the individuals of the two populations to be compared were performed simultaneously, when possible, to avoid any artefactual variation as a result of differences in the concentration of PCR solutions or in the temperature of the thermal cycler (33). PCR products from a population were run on the same gel to facilitate subsequent reading. We performed all PCR reactions twice to test for the repeatability of amplifications. The gels were read blind with respect to the level of extra-pair paternity.
We screened approximately 20 primers (University of British Colombia Biotechnology Laboratory, set #3) by using four individuals for each species to retain five to six primers for subsequent analysis (the sequences of the primers used are available on request). An effort was made to use the same primers for each species pair (see Table 2). The primers were selected for each pair to fill the following criteria: (i) bands were clearly identifiable, (ii) the pattern of most bands was repeatable, and (iii) at least one band was polymorphic for the species analyzed. For each of the selected primers, bands were scored only if the same banding pattern was observed in two PCR reactions of similar amplification strength.
Fragments for each polymorphic locus were scored as present (1) or absent (0). To estimate the amount of genetic variability for each population, we calculated (i) the proportion of polymorphic markers for each population (Pol) and (ii) the gene diversity (Ho). We assumed that each band represents a Mendelian locus with two alleles in Hardy–Weinberg equilibrium, the visible dominant allele and the null recessive one, and that the alleles of different loci do not migrate to the same position on the gel. The allelic frequency of the null allele can then be estimated as qi = √fi, where fi is the frequency of individuals not showing the band i. The gene diversity was then calculated according to Lynch and Milligan (34). To get an unbiased estimate of gene diversity, we used the loci for which the null phenotype frequency was below 3/N as advised by Lynch and Milligan (34). To avoid missing data, only individuals for which the genotypes could be determined for all loci without ambiguity were used. This is the reason why occasionally the number of individuals used is not the same for both sister groups.
Identified pairs were subsequently compared by using a pairwise comparative method, which automatically controls for confounding variables because closely related species generally are very similar in ecology and evolutionary history because of their mainly common evolutionary past (35–37). We used a paired t test to investigate differences in genetic variability in relation to frequency of extra-pair paternity.
RESULTS
Allozyme Data.
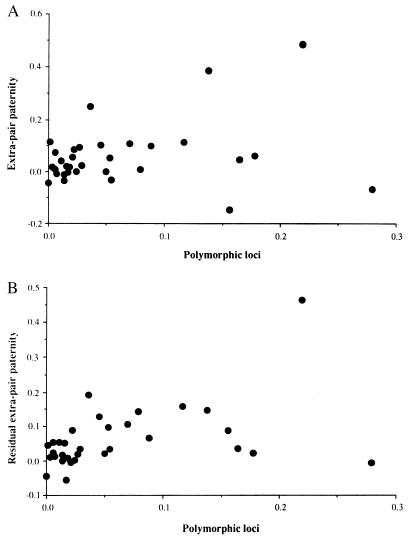
We found a positive correlation between the proportion of polymorphic loci and extra-pair paternity. Species with low levels of polymorphic loci also had low levels of extra-pair paternity. We do not provide statistics for this relationship because data points are inherently statistically dependent and thus cannot be used for formal statistical analyses. It is possible that this simple correlation is confounded by phylogeny if several closely related species have similar amounts of polymorphic loci and levels of extra-pair paternity. To remove such potential effects, a contrast analysis was used to compare the levels of extra-pair paternity and polymorphic loci among species. Levels of polymorphic loci and extra-pair paternity were significantly positively related, explaining 22% of the variance (Fig. 2a; F = 8.90; df = 1, 31; r2 = 0.22; P = 0.0055).
Figure 2.
The relationship between extra-pair paternity and polymorphic loci across birds by using contrasts (a) and residual contrasts (b) (after controlling for the effects of sexual dichromatism, body mass, and sample size) as independent observations.
The proportion of extra-pair paternity may not depend on the proportion of polymorphic loci; instead, the proportion of polymorphic loci may be related to some unknown third variable that also predicts levels of extra-pair paternity. Although it is possible that some ecological factor could be involved, this does seem unlikely given that the only good correlate of the variation in the proportion of extra-pair paternity is sexual dichromatism (11, 23, 24). It has been suggested that the proportion of polymorphic loci might increase with a larger sample of individuals surveyed. We investigated this effect by including sample size (the number of individuals surveyed) as an independent variable. Variation in extra-pair paternity may be related to body size because small passerines tend to have more frequent extra-pair paternity than large nonpasserines (11, 23). A multiple regression analysis, incorporating sexual dichromatism, sample size, body size, and the interaction between sexual dichromatism and the proportion of polymorphic loci as independent variables, explained more than 85% of the variance in the data set (Table 1). Extra-pair paternity was still significantly positively related to the proportion of polymorphic loci (Fig. 2b). Extra-pair paternity was positively related to sexual dichromatism (Table 1), as in previous studies (23, 24) and negatively related to body size. The interaction term between sexual dichromatism and the proportion of polymorphic loci was negative and highly significant (Table 1), implying that genetic variation was more important in less dichromatic species.
Table 1.
A linear multiple regression analysis of contrasts relating extra-pair paternity (square root-arcsine-transformed) to the proportion of polymorphic loci (square root-arcsine-transformed), sexual dichromatism, body mass (log-transformed), and sample size (log-transformed)
| Variable | Coefficient | SE | t | P |
|---|---|---|---|---|
| Polymorphic loci | 1.952 | 0.203 | 9.61 | <0.0001 |
| Dichromatism | 0.255 | 0.027 | 9.29 | <0.0001 |
| Body mass | −0.400 | 0.054 | −7.38 | <0.0001 |
| Sample size | 0.090 | 0.021 | 4.27 | 0.0002 |
| Polymorphic loci ∗ dichromatism | −0.587 | 0.092 | −6.41 | <0.0001 |
The regression model has the following statistics F = 31.01; d.f. = 5, 26; r2 = 0.85; P < 0.0001. The model includes the only interaction term that explained a significant amount of the variation.
RAPD Data.
The genetic variability indices ranged from 0.30 to 0.82 for the ratio of polymorphic loci (Pol) and from 0.015 to 0.228 for the gene diversity (Ho) (Table 2). A significant positive correlation was observed between the number of individuals analyzed per population and the genetic variability indices (product-moment correlation, r = 0.68, n = 14, P < 0.01 for Pol and r = 0.77, n = 14, P < 0.01 for Ho). However, the effect of sample size on genetic diversity could not bias our results because the number of individuals compared is the same for five out of seven sister pairs and very similar in the two remaining species (Table 2).
The level of extra-pair paternity for our samples ranged from 0% to 55% (Table 2). Populations with a lower level of extra-pair paternity had a lower amount of genetic variability for six out of seven pairs for Pol and five out of seven for Ho (Table 2). This trend was close to significance at the 5% level (t6 = 2.35, P = 0.057 for Pol (arcsine-transformed data) and t6 = 2.09, P = 0.082 for Ho). The low number of pairs decreases the power of our tests. Unfortunately, obtaining DNA samples of individuals from the same population for which the level of extra-pair paternity had also been estimated was a necessary but strong constraint and reduced the number of pairs available to study.
Combined Probability Test.
We have used two different sets of data (allozyme data and RAPD data) to test the same scientific hypothesis that extra-pair paternity depends on levels of polymorphic loci and it is therefore valid to combine the probability values for both of these tests (ref. 48, pp. 794–797). If we use the P value from the univariate test for the allozyme data to calculate a combined probability, we find that χ2 = 15.74, df = 4, and P < 0.005. If instead we use the P value from Table 1 for the allozyme data (after controlling for the confounding variables), we find χ2 = 28.31, df = 4, and P < 0.0001. Thus, this combined test leads to an overall rejection of the null hypothesis that the level of extra pair paternity is unrelated to genetic variation.
DISCUSSION
The results presented here show that variation in levels of polymorphic loci are positively related to species levels of extra-pair paternity. This trend is to be expected under indirect benefit models of female choice, which predict that if there is little variation in genetic quality among males, then the benefits of gaining an extra-pair mating do not outweigh any costs to females. The results therefore support indirect benefit models of female choice as well as point to the importance of the population genetic structure for understanding variation in levels of extra-pair paternity.
This study is based on two assumptions. The first is that the level of extra-pair paternity reflects the proportion of females that would benefit from modifying the genetic quality of some of their offspring. The possibility that females can control paternity is now clearly recognized because females choose extra-pair partners by accepting or rejecting extra-pair copulations or by actively seeking them (10, 11, 47). There are, however, other factors affecting the population level of extra-pair paternity, factors such as the costs to females of seeking extra-pair copulations and constraints to female choice (9). For example, mate guarding or repetitive copulation by the male partner could prevent females from achieving extra-pair copulations. Moreover, environmental factors determining density or breeding synchrony could affect the availability of potential partners (11). However, all of these other possible factors influencing the level of extra-pair paternity would tend to reduce the chance of detecting the predicted relationship.
The second assumption is that our measures of genetic variability are representative of the general amount of genetic variability and also of any variability in male fitness. We used two different measures of genetic variability. For the allozyme data, there is no a priori reason to suppose that the proportion of polymorphic loci of the analyzed allozymes differs from allozymes in general and may therefore provide an estimate of genome-wide genetic variability. Several studies have shown that variation in proteins as measured by gel electrophoresis can have fitness consequences and be under weak selection (49, 50). For the RAPD data, the primers used are randomly designed and should not bind to a specific region of the genome, and will therefore reflect genome-wide genetic variability. Unfortunately, there are very few empirical studies that allow us to determine whether this variation could possibly reflect fitness variation. There is some suggestion that the within-population structure for RAPD markers is not significantly different from quantitative traits where the latter are expected to be under selection (51). It is possible that our assumption is invalid and that our measures of genetic variation do not reflect variation in fitness but merely reflect neutral variation. However, then it is hard to interpret the relationships that we report here and to explain why females would be more likely to modify the paternity of their clutch when there is more neutral variation.
It has been claimed that populations at equilibrium should be deprived of variation in heritable fitness and this historically is one of the main objections to the “good-genes” hypothesis for the maintenance of female choice. However, empirical estimates have revealed that the heritability of fitness traits is not zero (52, 53). Moreover, theoretical models of the maintenance of female preferences allow the maintenance of substantial genetic variability through two main mechanisms: mutation-selection balance (54, 55) and frequency-dependent selection, especially induced by antagonistic coevolution between host and parasite (56). Other factors could maintain genetic variability across the genome and cause differences between species. For example, the frequency of recombination, the intensity of migration, the prevalence of genetic drift, and the probability of population bottle-necks (57).
Nevo et al. (58) argued that genetic variation should be low in populations or species in which a few males gain most matings because under these circumstances the effective population size is small. In species in which the proportion of extra-pair paternity is high, the variance in male mating success is high (review in ref. 10). Extra-pair paternity gives rise to almost a doubling of the relative variance in male reproductive success in a sample of eight studies (10). We can conclude that any factor that enhances the frequency of extra-pair paternity also tends to enhance the variance in male reproductive success (10). Surprisingly, and contrary to expectations from effective population size, we found that in species with a high frequency of extra-pair paternity, the genetic variance measured as the proportion of polymorphic allozyme loci was also high, and this was the case even when controlling for a number of potentially confounding variables.
Are there any other potential benefits to multiple mating that make predictions about how the level of extra-pair paternity might relate to genetic variation? One possibility is that females may be modifying paternity to avoid the expression of lethal or deleterious genes that can occur with inbreeding (59). This possibility does make predictions about the frequency of extra-pair copulations in relation to the overall genetic diversity in populations but in the opposite direction to that expected from genetic quality benefits. In populations with lower genetic diversity, the risks of inbreeding depression are higher, and under these circumstances, females might be expected to be more likely to seek extra-pair matings if they were avoiding inbreeding (60).
A previous study of genetic variation and mating systems revealed no significant difference in heterozygosity between groups of fish and birds classified as monogamous, polygamous, or social breeders (58). However, sample sizes were small and other confounding factors were not considered; moreover, the discovery of extra-pair paternity in so-called monogamous species raises doubts about such a broad categorization of mating systems based on the number of social mates per male.
In spite of the potential variation in the environmental and behavioral characteristics of the different populations studied, the relationships observed here provide evidence that the genetic variability of the population can predict levels of polygamy. Previous workers who have looked for ecological correlates of mating systems may have largely failed if the key variable determining the mating system is underlying genetic variation. Genetic variation of a population can be influenced by a variety of ecological factors, but it may also reflect factors such as history and gene flow. The absence of any relationships between the ecology of a species and its social mating system is then not particularly surprising.
Supplementary Material
Acknowledgments
We are very grateful to J. Austin, I. R. Hartley, J. T. Lifjeld, R. Montgomerie, D. Ritchardson, K. Schulze-Hagen, D. Westneat, J. Wetton, and M. Wink for kindly providing us with DNA samples; to T. Burke for advice, laboratory space, and help with the molecular analyses; and to P. J. Cordero, I. R. Hartley, J. T. Lifjeld. M. Magrath, E. S. Morton, D. Ritchardson, J. N. M. Smith, J. Tobias, and D. F. Westneat who kindly provided unpublished information on paternity. A.P.M. was supported by a grant from the Danish Natural Science Research Council, and the RAPD work was supported by a Human Capital and Mobility Scientific and Technical Collaboration Network grant from the European Union.
ABBREVIATION
- RAPD
random amplified polymorphic DNA
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Gyllensten U B, Jacobsson S, Temrin H. Nature (London) 1990;343:168–170. doi: 10.1038/343168a0. [DOI] [PubMed] [Google Scholar]
- 2.Hunter F M, Burke T, Watts S E. Anim Behav. 1992;44:149–156. [Google Scholar]
- 3.Hartley I R, Shepherd M, Robson T, Burke T. Behav Ecol. 1993;4:310–317. [Google Scholar]
- 4.Westneat D F. Anim Behav. 1987;35:865–876. [Google Scholar]
- 5.Westneat D F. Behav Ecol Sociobiol. 1990;27:67–76. [Google Scholar]
- 6.Sherman P W, Morton M L. Behav Ecol Sociobiol. 1988;22:413–420. [Google Scholar]
- 7.Lifjeld J T, Dunn P O, Robertson R J, Boag P T. Anim Behav. 1993;45:213–229. [Google Scholar]
- 8.Dixon A, Ross D, O’Malley S L C, Burke T. Nature (London) 1994;371:698–700. [Google Scholar]
- 9.Petrie M, Kempenaers B. Trends Ecol Evol. 1998;13:52–58. doi: 10.1016/s0169-5347(97)01232-9. [DOI] [PubMed] [Google Scholar]
- 10.Møller A P. In: Sperm Competition and Sexual Selection. Birkhead T R, Møller A P, editors. London: Academic; 1998. [Google Scholar]
- 11.Birkhead T R, Møller A P. Sperm Competition in Birds. London: Academic; 1992. [Google Scholar]
- 12.Stutchbury B J, Morton E S. Behaviour. 1995;132:675–690. [Google Scholar]
- 13.Westneat D F, Sherman P W. Behav Ecol Sociobiol. 1997;41:205–215. [Google Scholar]
- 14.Kempenaers B, Verheyen G R, Broeck M v d, Burke T, Broeckhoven C V, Dhondt A A. Nature (London) 1992;357:494–496. [Google Scholar]
- 15.Hasselquist D, Bensch S, von Schantz T. Nature (London) 1996;381:229–232. [Google Scholar]
- 16.Møller, A. P. & Thornhill, R. (1998) Anim. Behav., in press. [DOI] [PubMed]
- 17.Petrie M, Lipsitch M. Proc R Soc London B. 1994;256:275–280. [Google Scholar]
- 18.Mitton J B, Pierce B A. Genetics. 1980;95:1043–1054. doi: 10.1093/genetics/95.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakraborty R. Genetics. 1981;98:461–466. doi: 10.1093/genetics/98.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitton J B. Genetica. 1993;89:47–65. [Google Scholar]
- 21.Mitton J B. Acta Theriol Suppl. 1995;3:33–54. [Google Scholar]
- 22.Falconer D S, Mackay T F C. Introduction to Quantitative Genetics. 4th Ed. New York: Longman; 1996. [Google Scholar]
- 23.Møller A P, Birkhead T R. Evolution. 1994;48:1089–1100. doi: 10.1111/j.1558-5646.1994.tb05296.x. [DOI] [PubMed] [Google Scholar]
- 24.Møller A P. Proc R Soc London B. 1997;264:561–566. [Google Scholar]
- 25.Brough T. Average Weights of Birds. Worplesdon Laboratory, Guilford, U.K.: Aviation Bird Unit; 1983. [Google Scholar]
- 26.Harvey P H, Pagel M. The Comparative Method in Evolutionary Biology. Oxford: Oxford Univ. Press; 1991. [Google Scholar]
- 27.Purvis A, Rambaut A. Comput Appl Biosci. 1995;11:247–251. doi: 10.1093/bioinformatics/11.3.247. [DOI] [PubMed] [Google Scholar]
- 28.Sibley C G, Ahlquist J E. Phylogeny and Classification of Birds: A Study in Molecular Evolution. New Haven, CT: Yale Univ. Press; 1990. [Google Scholar]
- 29.Krajewski C. Auk. 1991;108:987–990. [Google Scholar]
- 30.O’Hara R J. Auk. 1991;108:990–994. [Google Scholar]
- 31.Raikow R J. Auk. 1991;108:985–987. [Google Scholar]
- 32.Sibley C G. J Avian Biol. 1995;25:87–92. [Google Scholar]
- 33.Ellsworth D L, Rittenhouse K D, Honeycutt R L. BioTechniques. 1993;14:214–217. [PubMed] [Google Scholar]
- 34.Lynch M, Milligan B G. Mol Ecol. 1994;3:91–99. doi: 10.1111/j.1365-294x.1994.tb00109.x. [DOI] [PubMed] [Google Scholar]
- 35.Birkhead T R, Atkin L, Møller A P. Behaviour. 1987;101:101–138. [Google Scholar]
- 36.Read A F. Am Nat. 1991;138:434–459. [Google Scholar]
- 37.Møller A P, Birkhead T R. Am Nat. 1992;139:644–656. [Google Scholar]
- 38.Swatschek I, Ristow D, Wink M. Mol Ecol. 1994;3:259–262. [Google Scholar]
- 39.Austin J J, Parkin D T. Mol Ecol. 1996;5:145–150. [Google Scholar]
- 40.Krokene C, Anthonisen K, Lifjeld J T, Amundsen T. Anim Behav. 1996;52:405–417. [Google Scholar]
- 41.Brooke M d L, Hartley I R. Auk. 1995;112:77–86. [Google Scholar]
- 42.Schulze-Hagen K, Swatschek I, Dyrcz A, Wink M. J Ornithol. 1993;134:145–154. [Google Scholar]
- 43.Hill G E, Montgomerie R, Roeder R, Boag P. Behav Ecol Sociobiol. 1994;35:193–199. [Google Scholar]
- 44.Ritchison G, Klatt P H, Westneat D F. Condor. 1994;96:1055–1063. [Google Scholar]
- 45.Bjørnstad G, Lifjeld J T. J Avian Biol. 1997;28:319–324. [Google Scholar]
- 46.Westneat D F. Behav Ecol. 1993;4:49–60. [Google Scholar]
- 47.Gray E M. Behav Ecol Sociobiol. 1996;38:267–278. [Google Scholar]
- 48.Sokal R R, Rohlf F J. Biometry. 3rd Ed. New York: Freeman; 1995. [Google Scholar]
- 49.Mitton J B. Selection in Natural Populations. Oxford: Oxford Univ. Press; 1997. [Google Scholar]
- 50.David P, Jarne P. Genetics. 1997;146:335–344. doi: 10.1093/genetics/146.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonnin I, Prosperi J M, Olivieri I. Genetics. 1996;143:1795–1805. doi: 10.1093/genetics/143.4.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mousseau J A, Roff D A. Heredity. 1987;59:181–198. doi: 10.1038/hdy.1987.113. [DOI] [PubMed] [Google Scholar]
- 53.Houle D, Hoffmaster D K, Assimacopoulus S, Charlesworth B. Nature (London) 1992;359:58–60. doi: 10.1038/359058a0. [DOI] [PubMed] [Google Scholar]
- 54.Pomiankowski A. Oxford Surv Evol Biol. 1988;5:136–184. [Google Scholar]
- 55.Iwasa Y, Pomiankowski A, Nee S. Evolution. 1991;45:1431–1442. doi: 10.1111/j.1558-5646.1991.tb02646.x. [DOI] [PubMed] [Google Scholar]
- 56.Pomiankowski A. Proc R Soc London B. 1987;231:123–146. [Google Scholar]
- 57.Hartl D L, Clark A G. Principles of Population Genetics. 2nd Ed. Sunderland, MA: Sinauer; 1989. [Google Scholar]
- 58.Nevo E, Beiles A, Ben-Shlomo R. In: Lecture Notes in Biomathematics. Mani G S, editor. Vol. 53. Berlin: Springer; 1984. pp. 13–213. [Google Scholar]
- 59.Brown J L. Behav Ecol. 1997;8:60–65. [Google Scholar]
- 60.Petrie M, Kempenaers B. Trends Ecol Evol. 1998;13:280–281. doi: 10.1016/s0169-5347(98)01379-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.