Abstract
The localization of proteins to late-Golgi membranes (TGN) of Saccharomyces cerevisiae is conferred by targeting motifs containing aromatic residues in the cytosolic domains of these proteins. These signals could act by directing retrieval from a post-Golgi compartment or by preventing exit from the TGN. To investigate the mechanism of localization of yeast TGN proteins, we used the heterologous protein A-ALP (consisting of the cytosolic domain of dipeptidyl aminopeptidase A [DPAP A] fused to the transmembrane and luminal domains of the vacuolar protein alkaline phosphatase [ALP]), which localizes to the yeast TGN. Insertion of the aromatic residue–based TGN localization motif (FXFXD) of DPAP A into the cytosolic domain of ALP results in a protein that resides in the TGN. We demonstrate that the FXFXD motif confers Golgi localization through retrieval from a post-Golgi compartment by detecting a post-Golgi processed form of this protein in the TGN. We present an assay that uncouples retrieval-mediated Golgi localization from static retention-based localization, allowing measurement of the rate at which proteins exit the yeast TGN. We also demonstrate that the cytosolic domain of DPAP A contains additional information, separate from the retrieval motif, that slows exit from the TGN. We propose a model for DPAP A localization that involves two distinct mechanisms: one in which the FXFXD motif directs retrieval from a post-Golgi compartment, and a second that slows the rate at which DPAP A exits the TGN.
The secretory pathway of eukaryotic cells is composed of a series of membrane-bound organelles, each with its own unique complement of components. Transport of components between these organelles is achieved by means of vesicular transport, which results in a flow of membrane traffic throughout the pathway (Rothman, 1994). To maintain its identity, an organelle must ensure that its resident proteins do not get swept with those proteins passing through en route to other destinations in the cell. To achieve this localization, resident proteins can either be retrieved once they have exited the organelle, or they may be prevented from ever leaving that organelle in the first place (Pelham, 1993). These two mechanisms need not be mutually exclusive, since efficient localization of a protein to an organelle could arise through a combination of both mechanisms.
Both luminal and membrane proteins are localized to the ER through continuous retrieval from a post-ER compartment after recognition of specific localization signals (Lewis and Pelham, 1992; Townsley et al., 1993). In addition, it seems that the selection of cargo for entry into vesicles budding from the ER also plays an important role in the retention of proteins in this organelle (Schekman and Orci, 1996). The mechanism for localization of proteins to the Golgi apparatus is less clearly understood, but it is likely that both modes of localization are at work. Studies on glycosyltransferases have shown that the transmembrane domains of these molecules are essential for Golgi localization in both yeast and mammalian cells (Munro, 1991; Nilsson et al., 1991; Machamer et al., 1993; Chapman and Munro, 1994; Graham and Krasnov, 1995; Lussier et al., 1995). Current theories favor models in which the transmembrane domains prevent entry of resident proteins into transport vesicles. This may be achieved either by the formation of aggregates too large to enter transport vesicles (Swift and Machamer, 1991; Nilsson et al., 1993) or by exclusion of proteins from vesicles because of the length of their membrane spanning domain (Bretscher and Munro, 1993). However, localization of the yeast glycosyltransferase, Och1p, which resides in the cis-Golgi, requires retrieval from a more distal Golgi compartment (Harris and Waters, 1996).
Other Golgi membrane proteins residing in the TGN achieve their localization through more dynamic means. TGN38, furin, and the mannose-6-phosphate receptor are localized to the TGN of mammalian cells by virtue of aromatic residue containing signals in their cytosolic tails (Bos et al., 1993; Wong and Hong, 1993; Humphrey et al., 1992; Canfield et al., 1991; Takahashi et al., 1995; Voorhees et al., 1995; Schafer et al., 1995). These proteins are localized through a similar mechanism to that in operation at the ER: they continuously exit the TGN and are later retrieved from endosomal compartments (Kornfeld, 1992).
Although the morphology of the last definable subcompartment of the Golgi of Saccharomyces cerevisiae has not been described at the ultrastructural level, it is taken as being functionally equivalent to the TGN of mammalian cells (Graham and Emr, 1991; Wilcox and Fuller, 1991; Wilsbach and Payne, 1993; Nothwehr et al., 1995). The yeast TGN is defined as the compartment where proteins destined for the cell surface are sorted from those destined for delivery to the vacuole, and contains the three processing proteinases involved in the maturation of the mating pheromone α-factor (Kex2p, Kex1p, and Ste13p; also called dipeptidyl aminopeptidase A [DPAP A]1; Bryant and Boyd, 1993; Nothwehr et al., 1993), as well as the vacuolar protein sorting receptor (Vps10p; Marcusson et al., 1994; Cereghino et al., 1995; Cooper and Stevens, 1996). The cytosolic tail of each of these four integral membrane proteins is required for localization in the yeast TGN (Roberts et al., 1992; Wilcox et al., 1992; Cooper and Bussey, 1992; Cereghino et al., 1995; Cooper and Stevens, 1996), and more specifically aromatic residues have been shown to be essential for the proper localization of DPAP A, Kex2p, and Vps10p (Wilcox et al., 1992; Nothwehr et al., 1993; Cereghino et al., 1995; Cooper and Stevens, 1996).
The cytosolic tail of DPAP A is sufficient to localize the transmembrane and luminal domains of the vacuolar membrane protein alkaline phosphatase (ALP) in Golgi membranes (Nothwehr et al., 1993). The resulting fusion protein, A-ALP, has served as a model TGN marker protein and has been used to show that a 10-residue segment (containing the motif FXFXD) within the cytosolic domain of DPAP A is critical for the protein's localization to Golgi membranes (Nothwehr et al., 1993). Insertion of this motif into the cytosolic domain of ALP (RS-ALP; retention sequence ALP) results in a protein that colocalizes with Kex2p in the yeast TGN (Nothwehr et al., 1993). Mutagenesis of the FXFXD motif leads to delivery of the protein to the vacuole (Nothwehr et al., 1993), an observation that contributed to the formulation of a model of how proteins are retained in the S. cerevisiae TGN (Nothwehr and Stevens, 1994; Nothwehr et al., 1995). In this model, proteins continually leave the Golgi and enter a late endosomal/prevacuolar compartment (PVC), from which they are retrieved back to the Golgi apparatus after recognition of the FXFXD motif.
Evidence in support of the above model comes from the observation that the exaggerated PVC that accumulates in class E vps mutants contains proteins normally resident to the TGN (Vps10p, Kex2p, DPAP A, and A-ALP), as well as vacuolar proteins and endocytosed proteins (Raymond et al., 1992; Piper et al., 1995). Vps27p has been shown to be involved in controlling traffic through the PVC, and it functions to allow Vps10p to return to the TGN from the PVC (Piper et al., 1995). These data support a model in which proteins resident to the yeast TGN enter the PVC, from where they are efficiently retrieved back to the Golgi apparatus.
To date, little direct evidence exists to discriminate between this sort of retrieval mechanism and a static retention mechanism where proteins would be excluded from vesicles leaving Golgi membranes and, consequently, have a longer residency period in the TGN.
The mechanistic role of the aromatic localization signals identified in TGN proteins of S. cerevisiae is not known. To determine the mechanism by which information within the cytosolic tail of DPAP A confers Golgi localization, we devised an assay to uncouple “retrieval”-mediated Golgi localization from “static retention”–mediated Golgi localization by analyzing the trafficking of a series of A-ALP mutants in vps27Δ cells. Here, we report that the aromatic residue based motif FXFXD of the DPAPA tail mediates retrieval from the PVC. We also report the discovery of a second signal near the NH2 terminus of the DPAP A tail that acts independently of the retrieval mechanism to slow the rate of departure of this protein from the TGN.
Materials and Methods
Materials
Enzymes used in DNA manipulations were purchased from New England Biolabs (Beverly, MA), Boehringer Mannheim Biochemicals (Indianapolis, IN), Bethesda Research Laboratories (Gaithersburg, MD), or United States Biochemical Corp. (Cleveland, OH). Goat α-rabbit and goat α-mouse HRP-conjugated antibodies, as well as the ECL kit used for the development of immunoblots, were purchased from Amersham Corp. (Arlington Heights, IL). Secondary antibodies used for indirect immunofluorescence (all cross-species absorbed) were purchased from Jackson Immunoresearch Laboratories Inc. (West Grove, PA). mAbs specific for Vph1p (10D7-A7-B2) and ALP (1D3-A10) were obtained from Molecular Probes, Inc. (Eugene, OR). Fixed Staphylococcus aureus cells (Ig sorb) were obtained from The Enzyme Center (Malden, MA). 35S-Express label was from New England Nuclear (Boston, MA), and oxalyticase was from Enzogenetics (Corvallis, OR). All other reagents were purchased from Sigma Chemical Co. (St. Louis, MO).
Strains, Media, and Microbiological Techniques
The yeast strains used in this study are listed in Table I. Strains were constructed by standard genetic techniques and grown in rich media (1% yeast extract, 1% peptone, 2% dextrose; YEPD) or standard minimal medium (SD) with appropriate supplements as described by Sherman et al. (1986). Strain NBY60 was constructed by transforming SNY36-9A with the vps27Δ::LEU2 disruption cassette (BamHI/EcoRI fragment) from pKJH2, and by screening Leu+ transformants first for the secretion of carboxypeptidase Y (CPY) and subsequently for loss of production of Vps27p, as determined by immunoblot analysis of cell lysates. The PHO8 gene of MY1885 was disrupted to give rise to NBY67 by transforming pSN111 linearized with SalI, selecting for Ura+ colonies, and then selecting for Ura− loop outs on media containing 5-fluoroorotic acid. Immunoblot analysis was used to identify Ura− loop outs lacking Pho8p. Yeast strains NBY68 and NBY69 were constructed by integrating the VPS10-10* allele into the VPS10 locus of SNY36-9A and NBY60, respectively, as described by Cooper and Stevens (1996).
Table I.
Yeast Strains Used in This Study
| Strain | Genotype | Reference | ||
|---|---|---|---|---|
| SNY36-9A | MATa ura3-52 leu2-3, 112 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ::ADE2 | Nothwehr et al., 1995 | ||
| SNY63 | MATα ura3-52 leu2-3, 112 his3-Δ200 trp1-Δ901 lys2-801 pho8Δ::LEU2 pep4-3 | Nothwehr et al., 1996 | ||
| NBY60 | MATa ura3-52 leu2-3, 112 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ::ADE2 vps27Δ::LEU2 | This study | ||
| MY1885 | MATa vps27-59 (vpl23-5) pep4-3 leu2-3,112 ura3-52 his4-519 gal2 | Raymond et al., 1992 | ||
| NBY67 | MATa vps27-59 (vpl23-5) pep4-3 leu2-3,112 ura3-52 his4-519 gal2 pho8-ΔX | This study | ||
| NBY68 | MATa vps10-10*ura3-52 leu2-3,112 his3-Δ200 trp1Δ901 suc2-Δ9 pho8D::ADE2 | This study | ||
| NBY69 | MATa vps10-10*ura3-52 leu2-3,112 his3-Δ200 trp1Δ901 suc2-Δ9 pho8Δ::ADE2 vps27Δ::LEU2 | This study | ||
| RPY82 | MATα pep4-3 leu2-3, 112 ura3-52 his4-519 gal2 vps27-123(ts) pho8Δ::LEU2 | Tom Stevens |
Plasmid Construction
This plasmids used in this study are listed in Table II. Deletions within the cytosolic domain of A-ALP were constructed by oligonucleotide-directed mutagenesis of pCJR71 according to the method of Kunkel et al. (1987). To incorporate these deletions into the A-ALP fusion protein, EagI-BglII fragments from derivatives of pCJR71 carrying the appropriate deletions were subcloned into the EagI-BglII sites of pSN55. The following deletions were made: pNB81 (Δ2-11), pNB82 (Δ2-21), pNB83 (Δ2-31), and pNB84 (Δ2-41), where the deletion endpoints are indicated (e.g., Δ2-11 is missing amino acid residues 2–11).
Table II.
Plasmids Used in This Study
| Plasmid | Description | Reference | ||
|---|---|---|---|---|
| pSN55 | CEN plasmid encoding A-ALP | Nothwehr et al., 1993 | ||
| pSN97 | CEN plasmid encoding RS-ALP | Nothwehr et al., 1993 | ||
| pSN100 | CEN plasmid encoding (F/A)A-ALP | Nothwehr et al., 1993 | ||
| pSN111 | pho8-ΔX allele in pRS306 | Nothwehr et al., 1993 | ||
| pKJH2 | vps27Δ::LEU2 in pUC19 | Raymond et al., 1992 | ||
| pHY5 | CEN plasmid harboring VPS27 under GAL1 promotor control | Tom Stevens | ||
| pCJR71 | 650bp EagI-PstI STE13 fragment in pKS+ | Roberts et al., 1992 | ||
| pSN25 | CEN plasmid encoding A-ALP with residues 2–51 deleted | Nothwehr et al., 1993 | ||
| pSN27 | CEN plasmid encoding A-ALP with residues 68–106 deleted | Nothwehr et al., 1993 | ||
| pNB81 | CEN plasmid encoding A-ALP with residues 2–11 deleted | This study | ||
| pNB82 | CEN plasmid encoding A-ALP with residues 2–21 deleted | This study | ||
| pNB83 | CEN plasmid encoding A-ALP with residues 2–31 deleted | This study | ||
| pNB84 | CEN plasmid encoding A-ALP with residues 2–41 deleted | This study |
Radiolabeling and Immunoprecipitation
35Met labeling and immunoprecipitation of ALP fusion proteins, Vps10prelated proteins, and CPY were performed as described previously (Piper et al., 1994; Nothwehr et al., 1995; Cooper and Stevens, 1996). Briefly, yeast cultures were grown overnight in selective synthetic media without methionine to OD600 = 1. Cells were harvested and resuspended in fresh media to the same OD600. Cells were pulse labeled for 10 min with 100 mCi 35S-Express label/0.5 OD600, followed by the addition of unlabeled methionine and cysteine, both to 50 μg/ml. At specified times, samples were removed and treated by the addition of sodium azide to 10 mM at 4°C. Vps10p, CPY, ALP, and related proteins were immunoprecipitated from protein extracts of these cells using polyclonal antibodies specific for these proteins, as described previously (Piper et al., 1994; Nothwehr et al., 1995; Cooper and Stevens, 1996). Half-times of processing of ALP- and Vps10p-related proteins were determined as described previously (Nothwehr et al., 1993; Cooper and Stevens, 1996) using a Radioanalytic Imaging System (AMBIS, Inc., San Diego, CA), and linear regression analysis plotting percentage total protein was processed as a function of time.
Cell Fractionation
Fractionation studies were carried out essentially as described using the differential sedimentation of subcellular membranes followed by equilibrium gradient analysis (Becherer et al., 1996). In brief, cells were grown in 100 ml SD medium lacking uracil to an OD600 = 1.0. Sodium azide was added to a final concentration of 10 mM before the cells were harvested and resuspended in 10 ml 200 mM Tris-HCl, pH 8.0, 20 mM EDTA, 1% β-mercaptoethanol. After incubation at 30°C for 10 min, cells were harvested and resuspended in 1.2 M sorbitol, 50 mM KPO4, pH 7.3, 1 mM MgCl2 (spheroplast buffer) containing 200 μg/ml zymolyase. Cells were incubated for 1 h at 30°C and washed twice with spheroplast buffer. Spheroplasts were lysed in 1 ml 50 mM Tris-HCl, pH 7.5, 0.2 M sorbitol, 1 mM EDTA (lysis buffer) containing added proteinase inhibitors (PMSF [1 mM], leupeptin [1 μg/ml], antipain [1 μg/ml], chymostatin [1 μg/ml], and pepstatin [1 μg/ml]) using 10 strokes in a Dounce homogenizer (this and subsequent steps were all performed at 4°C). Cell debris and unlysed cells were removed by centrifugation at 500 g for 5 min. Centrifugation at 13,000 g for 10 min yielded pellet (P13) and supernatant (S13) fractions. The S13 fraction was further separated by centrifugation at 100,000 g for 30 min to yield a membrane pellet fraction. The 100,000 g membrane pellet was resuspended in 2 ml cold lysis buffer and loaded on the top of a 20– 50% sucrose step gradient. After centrifugation at 170,000 g for 18 h, 14 × 1–ml fractions were collected from the top of the gradient, and the proteins from these fractions were precipitated using TCA. Equal percentages of each fraction were subjected to immunoblot analysis after SDS-PAGE.
Immunofluorescence Microscopy
The preparation of fixed spheroplasted yeast cells, attachment to microscope slides, and costaining of ALP fusion proteins using mAb 1D3-A10 (Molecular Probes) and Vph1p using affinity-purified polyclonal antibodies were carried out as described previously (Nothwehr et al., 1996). Essentially, fixed spheroplasts attached to slides were incubated with the following solutions, followed by extensive washing with 5 mg/ml BSA in PBS after each step: (a) PBS-BSA containing a 1:3 dilution of mouse anti-ALP mAb 1D3-A10 (Molecular Probes) and a 1:20 dilution of affinity-purified rabbit anti-Vph1p polyclonal antibody; (b) 1:200 dilution of a biotin-conjugated goat anti–mouse IgG (heavy and light chains [H + L]); and (c) 1:200 dilution of both FITC-conjugated streptavidin and rhodamine-conjugated goat anti–rabbit IgG (H + L). Staining using affinity-purified antibodies specific for Vps10p was also carried out as described previously (Cooper and Stevens, 1996), using a 1:200 dilution of the affinity-purified polyclonal antibody, followed by a 1:200 dilution of a biotin-conjugated goat anti– mouse IgG (H + L) and, subsequently, streptavidin-conjugated FITC.
Temperature-sensitive Experiments
To assess the morphological redistribution of ALP fusion proteins and Vps10p in cells harboring a temperature-sensitive allele of VPS27, cells were grown in YEPD at 22°C to OD600 = 1, at which time cycloheximide was added to a final concentration of 100 μg/ml, and an aliquot of these cells was fixed and prepared for immunofluorescence microscopy. To follow the fate of these proteins upon loss of Vps27p function, the remainder of the cells were warmed rapidly to 37°C, and the culture was moved to a 37°C water bath. Samples were removed at various times and fixed in preparation for immunofluorescence microscopy.
Induction of Vps27p
To observe the fate of various ALP fusion proteins after their accumulation in the PVC as a result of a loss of Vps27p function, vps27 mutant cells harboring VPS27 under control of the GAL1 promoter (+) or empty plasmid (−) were grown to OD600 = 1 in synthetic media containing 2% raffinose. Galactose was added to a final concentration of 2%, and the cultures were split in half. One half was treated immediately (− galactose) and the other half was returned to 30°C for 90 min (+ galactose). At both time points, samples were prepared for immunofluorescence microscopy, and whole-cell extracts were prepared to monitor the induction of Vps27p production by immunoblot analysis. 0.5 OD units were analyzed for CPY secretion after pulse-chase labeling with [35S]methionine.
Results
A Late-Golgi Membrane Protein Is Retrieved From a Post-Golgi Compartment
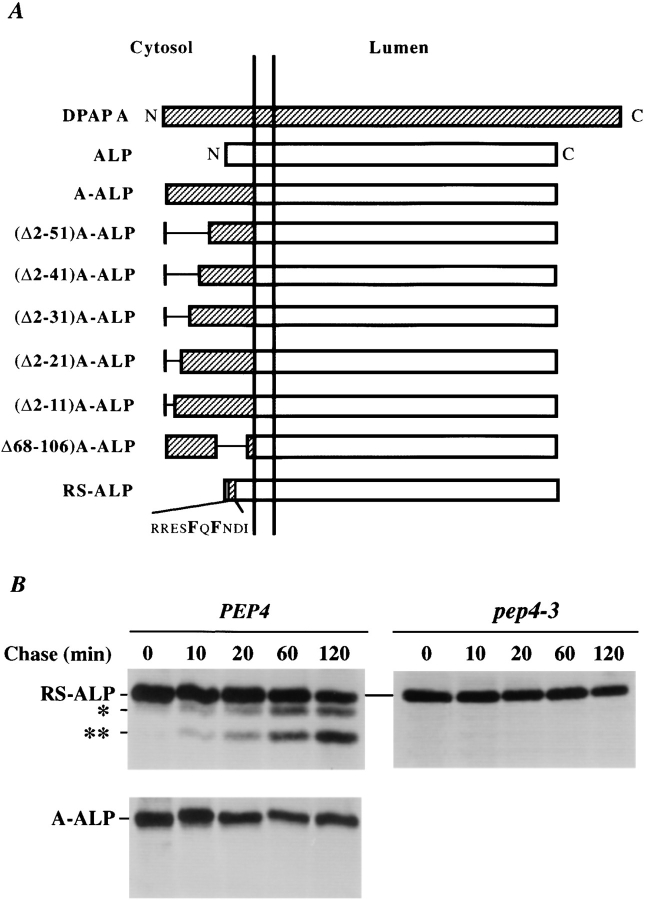
The cytosolic domain of DPAP A is necessary and sufficient to retain the transmembrane and luminal domains of the vacuolar protein ALP in the last definable Golgi subcompartment in yeast (Nothwehr et al., 1993), which is functionally equivalent to the TGN of mammalian cells (Franzusoff et al., 1991; Redding et al., 1991; Graham and Emr, 1991). Mutational analysis identified the aromatic residue–containing motif, FXFXD, as being essential for the localization of the fusion protein A-ALP (consisting of the cytosolic domain of DPAP A fused to the transmembrane and luminal domains of ALP) to the yeast TGN. Forms of the A-ALP protein lacking this signal (such as (F/A)A-ALP, in which the two phenylalanine residues have been mutated to alanines) fail to be retained in the Golgi complex and are delivered to the vacuole (Roberts et al., 1992; Wilcox et al., 1992; Cooper and Bussey, 1992). Delivery of proteins containing the luminal domain of ALP to the vacuole can be monitored by detecting a vacuolar protease-dependent (PEP4-dependent) cleavage, which results in the production of a lower molecular weight form of the protein (Klionsky and Emr, 1989; Nothwehr et al., 1993).
Whereas ALP is delivered to the vacuole and processed with a half-time of <10 min (Klionsky and Emr, 1989), A-ALP is localized to the TGN and does not undergo any significant processing (Nothwehr et al., 1993). Insertion of the FXFXD motif into the cytosolic domain of ALP causes the protein RS-ALP to be retained in the TGN, as determined by indirect immunofluorescence (Nothwehr et al., 1993). Like A-ALP, RS-ALP has been shown to colocalize with the TGN protein Kex2p (Nothwehr et al., 1993), but RS-ALP undergoes PEP4-dependent proteolytic cleavage with a half-time of 90 min in wild-type cells, whereas A-ALP does not undergo proteolysis, even after chase times of up to 2 h (Fig. 1 B). These data indicate that even though RS-ALP localizes to the TGN in the steady state, it becomes exposed to vacuolar proteases in a post-Golgi compartment.
Figure 1.
(A) Schematic illustration of modifications made within the cytosolic tail of DPAP A in the context of A-ALP. Deleted regions are indicated by a single line. (B) Immunoprecipitation of A-ALP from wild-type cells and RS-ALP from wild-type and pep4-3 cells. SNY36-9A (PEP4) cells harboring pSN55 (A-ALP) or pSN97 (RS-ALP) and SNY63 (pep4-3) cells harboring pSN97 (RS-ALP) were labeled with [35S]methionine for 10 min and chased by adding unlabeled methionine and cysteine each to a final concentration of 50 μg/ml. At the indicated times, the cells were spheroplasted and extracts were immunoprecipitated with a polyclonal antibody against ALP, followed by SDS-PAGE and fluorography. The product of the initial PEP4-dependent proteolysis is indicated by a single asterisk, with a double asterisk indicating a further breakdown product of this primary product. This breakdown pattern differs from that seen for A-ALP (which has only one PEP4-dependent breakdown product, e.g., Fig. 6), but clearly both of the lower bands shown here are produced in a PEP4-dependent manner.
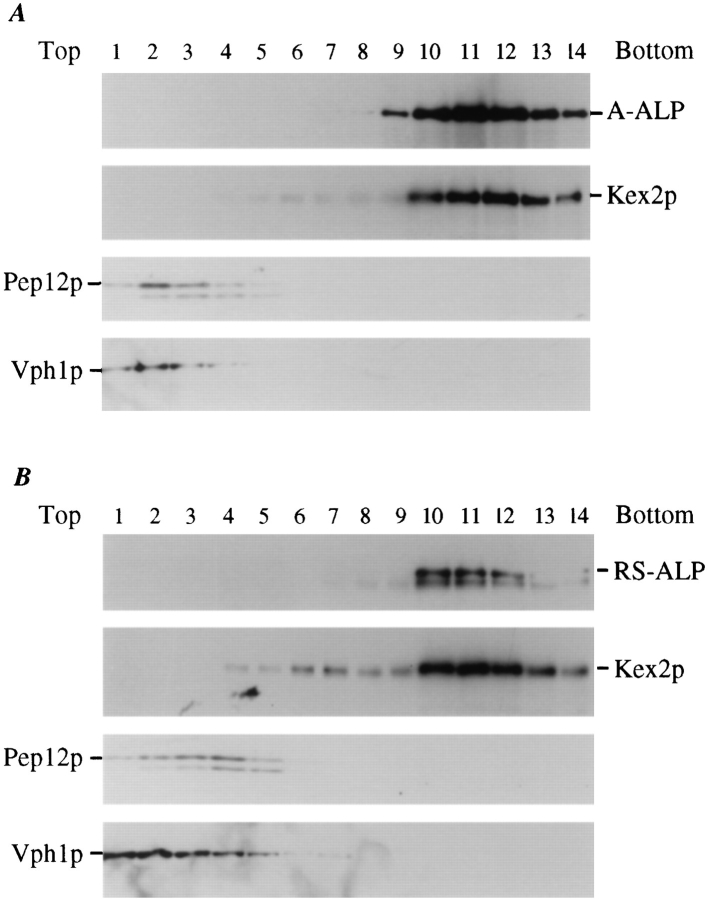
If RS-ALP is being retrieved back to the TGN from a vacuolar protease–containing, post-Golgi compartment (i.e., the PVC), then proteolytically processed RS-ALP should fractionate with TGN membranes. To test this possibility, we separated yeast membranes by sucrose density gradient centrifugation. Membranes from cell lysates were sedimented at 13,000 g to obtain a membrane pellet containing ER, vacuolar, mitochondrial, and plasma membranes (Paravicini et al., 1992; Piper et al., 1994). The resulting supernatant was subjected to centrifugation at 100,000 g, and the resulting membrane pellet (containing ∼10% of the total cellular Vph1p and 80–90% of the total cellular Pep12p, Kex2p, and A-ALP or RS-ALP) was resuspended and loaded on a 20–50% sucrose gradient. In this gradient, the residual vacuolar membranes, defined by Vph1p (Manolson et al., 1992; Piper et al., 1994), and prevacuolar membranes, defined by Pep12p (Becherer et al., 1996; Piper, R.C., and T.H. Stevens, unpublished results), were found at the top of the gradient (fractions 1–4, Fig. 2 A). By contrast, TGN membrane proteins (A-ALP and Kex2p) fractionated near the bottom of the gradient. Both the processed and unprocessed forms of RS-ALP also cofractionated with Kex2p in the TGN membrane fractions (Fig. 2 B). These data indicate that RS-ALP has reached a post-Golgi compartment containing activated vacuolar proteases, and has been retrieved back to the TGN.
Figure 2.
Sucrose gradient fractionation of A-ALP/RS-ALP containing membranes. High speed pellet membrane fractions (100,000 g) were prepared from 100 OD units of SNY36-9A cells harboring either pSN55 (A-ALP) or pSN97 (RS-ALP). These membranes were fractionated by a 20–50% sucrose density step gradient. After centrifugation, 14 × 1–ml fractions were collected from the top of the gradient, and the proteins were precipitated by the addition of TCA. Equal percentages of each fraction were subjected to immunoblot analysis after SDS-PAGE to detect the following proteins: A-ALP/RS-ALP using an anti-ALP mAb (1D3-A10); Kex2p was used as a marker protein for TGN membranes and detected using affinity-purified polyclonal antibodies; anti-Vph1p mAb 10D7-A7-B2 was used to detect Vph1p as a marker protein for vacuolar membranes; and Pep12p, a marker protein for membranes of the prevacuolar compartment, was detected using affinity-purified polyclonal antibodies.
The FXFXD Motif Is Necessary and Sufficient for Retrieval from the Prevacuolar Compartment
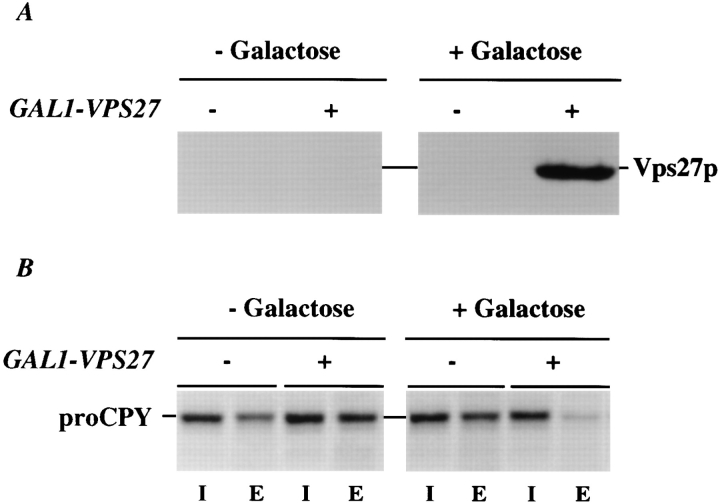
To investigate retrieval from the PVC in greater detail, we turned to yeast cells harboring mutations in the VPS27 gene. VPS27 is required for the movement of traffic out of the PVC, both back to the TGN and onto the vacuole (Raymond et al., 1992; Piper et al., 1995). vps27 mutant cells accumulate an exaggerated form of the PVC (the class E compartment) containing endocytosed proteins, as well as TGN membrane proteins and proteins en route to the vacuole, such as activated vacuolar proteases (Fig. 3; Raymond et al., 1992; Piper et al., 1995). We have followed the fate of A-ALP, (F/A)A-ALP, and RS-ALP in a vps27 mutant before and after induction of VPS27 under control of the GAL1 promoter. Immunoblot analysis (Fig. 4 A) revealed that vps27 mutant cells carrying the GAL1VPS27 plasmid (pHY5) produced Vps27p only after the addition of galactose. Pulse-chase immunoprecipitation of the vacuolar protease CPY demonstrated that secretion of CPY was suppressed in vps27 mutant cells now producing Vps27p (Fig. 4 B), indicating that the CPY sorting receptor (Vps10p) had regained the ability to cycle between the PVC and the TGN.
Figure 3.
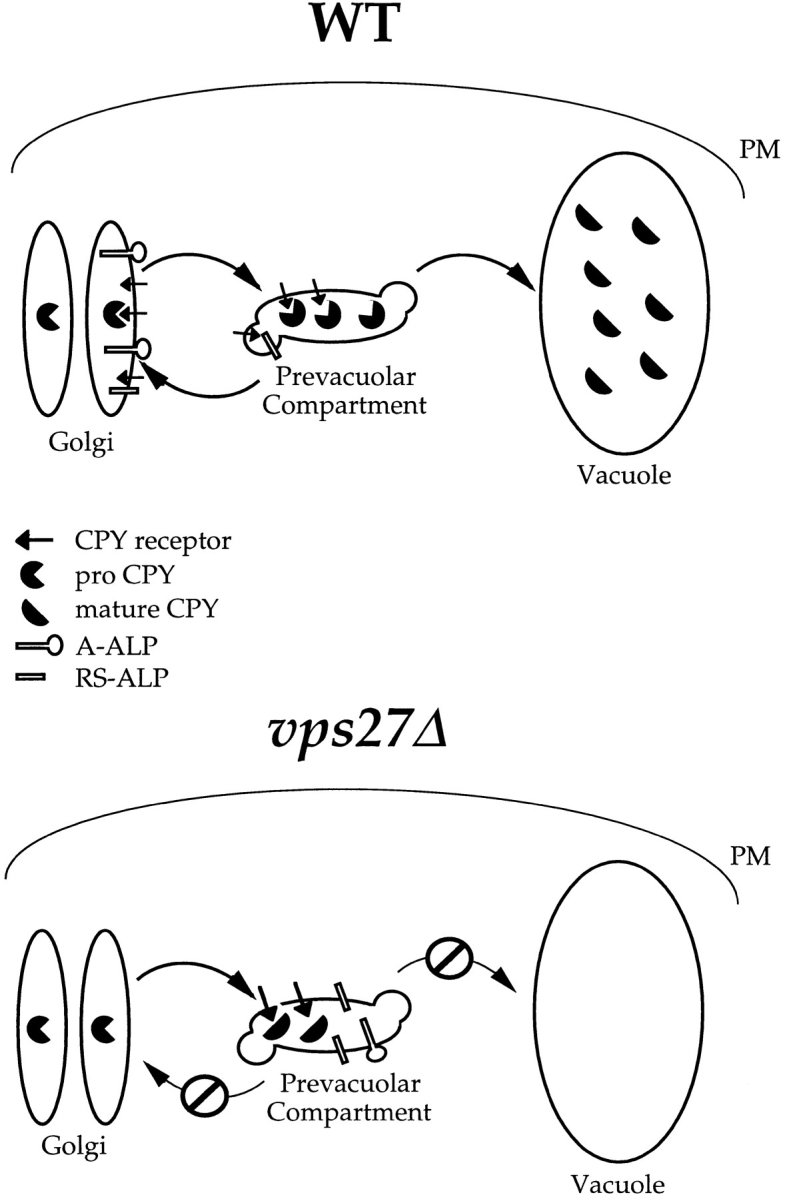
Model of Vps27p function in the retention of yeast TGN membrane proteins. TGN membrane proteins such as Vps10p, A-ALP, and RS-ALP continuously leave Golgi membranes and enter the PVC, from which they are retrieved via a mechanism that involves the recognition of an aromatic residue– containing motif. This retrieval requires Vps27p, and in vps27Δ cells, TGN membrane proteins accumulate in an exaggerated form of the PVC, the class E compartment (Raymond et al., 1992; Piper et al., 1995). The class E compartment is proteolytically active, and the rate at which proteolytically sensitive proteins are processed in vps27Δ cells can be used to measure the rate at which they enter this compartment. This is the basis for the assay to measure the leaving rates of A-ALP– and Vps10p–related proteins from the TGN. WT, wild type.
Figure 4.
Induction of synthesis of Vps27p in vps27 cells. NBY67 (vps27) cells harboring either pSN55 (A-ALP), pSN100 ((F/A)AALP) or pSN97 (RS-ALP), and pHY5 (VPS27 under GAL1 control) (+) or empty vector (−) were grown in synthetic media containing 2% raffinose. At time t = 0, galactose was added to a final concentration of 2%, and cells were allowed to grow for an additional 90 min. At times t = 0 (− galactose) and t = 90 min (+ galactose), samples were removed and treated in the following ways (also see Fig. 5): (A) Whole-cell extracts were prepared from 10 OD units of NBY67 cells harboring pSN97 and pHY5 or empty vector to detect the presence of Vps27p using immunoblot analysis (similar results were obtained using the same cells harboring either pSN55 or pSN100 in conjunction with pHY5; data not shown); and (B) 0.5 OD units of cells were radiolabeled to determine whether the cells were secreting CPY.
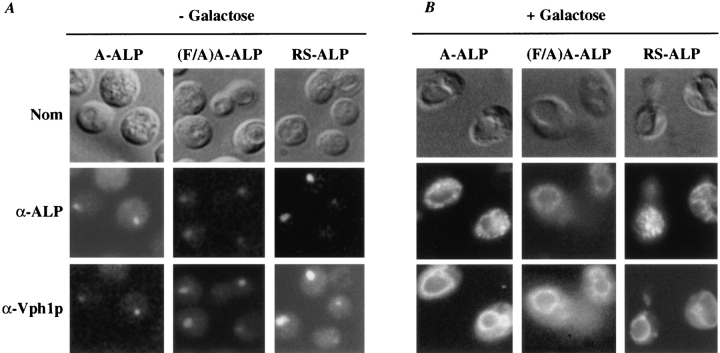
We performed indirect immunofluorescence to determine the fate of A-ALP, RS-ALP, and the retrieval-defective form of A-ALP, (F/A)A-ALP in vps27 mutant cells after restoration of Vps27p function. All three proteins localized to the exaggerated PVC in vps27 mutant cells (Fig. 5 A), indicating that they are transported to the PVC, but either fail to be retrieved to the TGN (A-ALP and RSALP) or fail to be transported to the vacuole ((F/A)AALP). Approximately 90 min after induction of Vps27p synthesis by addition of galactose (Fig. 5 B), A-ALP and RS-ALP redistributed to a punctate pattern as commonly observed for Golgi proteins in wild-type cells (Franzusoff et al., 1991; Redding et al., 1991; Roberts et al., 1992; Wilcox et al., 1992; Nothwehr et al., 1993). In the same cells in which A-ALP and RS-ALP redistributed to the TGN, the vacuolar membrane protein Vph1p redistributed from the class E compartment to the vacuole (Fig. 5 B). By contrast to A-ALP and RS-ALP, 90 min after induction of Vps27p synthesis, (F/A)A-ALP colocalized to the vacuole membrane with Vph1p (Fig. 5 B). Finally, in vps27 cells that did not contain VPS27 under GAL1 control, A-ALP and Vph1p remained in the class E compartment after the addition of galactose (data not shown). Taken together, these data show that the FXFXD motif found in the DPAP A cytosolic domain is both necessary and sufficient for retrieval from the PVC to the TGN.
Figure 5.
Redistribution of A-ALP, (F/A)A-ALP, and RS-ALP upon induction of synthesis of Vps27p in vps27 cells. NBY67 (vps27) cells harboring either pSN55 (A-ALP), pSN100 ((F/A)A-ALP), or pSN97 (RS-ALP) and pHY5 (VPS27 under GAL1 control) (+) or empty vector (−) were grown in synthetic media containing 2% raffinose. At time t = 0, galactose was added to 2%, and cells were allowed to grow for an additional 90 min. At times (A) t = 0 (− galactose) and (B) t = 90 min (+ galactose) (also see Fig. 4), cells were fixed and prepared for immunofluorescence using anti-ALP antibodies in combination with biotin-linked secondary antibody and streptavidin-conjugated FITC, as well as anti-Vph1p antibodies in combination with rhodamine-conjugated goat anti–rabbit IgG (H + L).
The Cytosolic Domain of DPAP A Slows Exit From the TGN
We were interested to further investigate the observation that whereas both A-ALP and RS-ALP localize to the TGN, only RS-ALP undergoes measurable proteolytic processing in wild-type yeast cells. One possibility, suggested by the observation that (F/A)A-ALP is processed with a half-time of 60 min while ALP is processed with a half-time of <10 min (Nothwehr et al., 1993; Klionsky and Emr, 1989), is that the cytosolic tail of DPAP A contains a second signal, not present in RS-ALP, that slows its exit from the TGN. To test this hypothesis, we designed an assay to estimate the rate of exit of membrane proteins from the TGN. The assay is based on measuring the rate of proteolytic processing of TGN proteins in vps27 mutant cells, since these proteins accumulate in the class E compartment and this compartment contains activated vacuolar proteases (Raymond et al., 1992; Piper et al., 1995). Therefore, we predict that proteins that exit the TGN slowly will exhibit long half-times for proteolytic processing in vps27 mutant cells whether or not they are competent to be retrieved to the TGN. By contrast, proteins that exit rapidly from the TGN would be rapidly proteolytically processed in vps27 mutant cells independently of their potential to be retrieved.
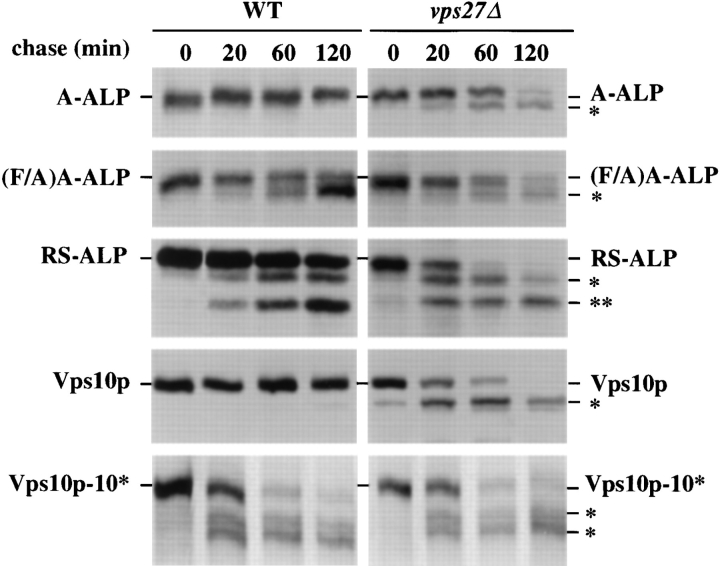
Pulse-chase immunoprecipitations (Fig. 6 and Table III) revealed that whereas A-ALP was very stable in wild-type cells, this protein became proteolytically processed with a half-time of 70 min in vps27 mutant cells. By contrast, retrieval-defective A-ALP, (F/A)A-ALP, was proteolytically processed with a half-time of ∼60 min in both wildtype and vps27 mutant cells. RS-ALP was proteolytically processed with half-times of 90 min in wild-type cells and 15 min in vps27 cells, suggesting that RS-ALP exits rapidly from the TGN and relies solely on retrieval for Golgi localization. These data indicate that the DPAP A cytosolic domain indeed slows exit from the TGN, and that A-ALP may avoid proteolytic processing in the PVC of wild-type cells by only rarely exiting the TGN. A slow rate of TGN exit would be consistent with the function of DPAP A, which is to proteolytically process the α-factor polyprotein in the TGN (Sprague and Thorner, 1992).
Figure 6.
Kinetics of processing of A-ALP, (F/A)A-ALP, RSALP, Vps10p, and Vps10p-10* (the recycling defective form of Vps10p) in wild-type and vps27Δ cells. SNY36-9A (wild-type) and NBY60 (vps27Δ) cells harboring pSN55 (A-ALP), pSN100 ((F/A)A-ALP) or pSN97 (RS-ALP), as well as NBY68 (vps1010*) and NBY69 (vps10-10* vps27Δ) cells were labeled with [35S]methionine for 10 min and chased by adding unlabeled methionine and cysteine, each to a final concentration of 50 μg/ml. At the indicated times, the cells were spheroplasted, and the extracts were immunoprecipitated with polyclonal antibodies against ALP and/or Vps10p, followed by SDS-PAGE and fluorography. The products of PEP4-dependent proteolysis are indicated using asterisks, as described in Fig. 1.
Table III.
Processing and Immunolocalization of Membrane Proteins
| Half-time of processing‡ | Localization by IIF§ | |||||
|---|---|---|---|---|---|---|
| WT | vps27Δ | |||||
| min | ||||||
| A-ALP | >180 | 70 | Golgi | |||
| (F/A)-ALP | 60 | 60 | Vacuolar | |||
| RS-ALP | 90 | 15 | Golgi | |||
| (Δ2-51)A-ALP | 85 | 15 | Golgi | |||
| (Δ2-41)A-ALP | 95 | 20 | Golgi | |||
| (Δ2-31)A-ALP | 100 | 25 | Golgi | |||
| (Δ2-21)A-ALP | 85 | 15 | Golgi | |||
| (Δ2-11)A-ALP | 90 | 15 | Golgi | |||
| (Δ68-106)A-ALP | 65 | 60 | Vacuolar | |||
| Vps10p | >180 | 20 | Golgi | |||
| Vps10p-10* | 20 | 20 | Vacuolar | |||
The kinetics of processing of the proteins listed in Table III were determined in SNY36-9A (wild-type) and NBY60 (vps27Δ) cells harboring the appropriate plasmid or producing Vps10p-10*. In brief, cells were labeled with [35S]methionine for 10 min and chased for various times by adding methionine and cysteine each to a final concentration of 50 μg/ml. Cells were spheroplasted and extracts were immunoprecipitated with a polyclonal antibody against ALP and/or Vps10p, followed by SDS-PAGE and fluorography. Half-times of processing were determined using linear regression analysis, as described in Materials and Methods.
The localization of each of the proteins was also determined in SNY36-9A (wildtype) cells by indirect immunofluorescence (IIF) using affiniity-purified polyclonal antibodies specific for ALP.
To extend the analysis of the exit rates of TGN membrane proteins, we compared the rates of proteolytic processing of the CPY sorting receptor, Vps10p, in wild-type and vps27 mutant cells. To gain further insight into Vps10p, we also determined processing rates for the recycling-defective form of Vps10p (Vps10p-10*), which lacks the cytosolic domain but otherwise binds CPY precursor (proCPY) normally (Cereghino et al., 1995; Cooper and Stevens, 1996). Whereas Vps10p was very stable in wildtype cells, Vps10p-10* was cleaved by vacuolar proteases with a half-time of ∼20 min (Fig. 6 and Table III). Interestingly, both Vps10p and Vps10p-10* were proteolyzed with a half-time of ∼20 min in vps27 mutant cells, suggesting that Vps10p exits the TGN rapidly, independently of the presence of the recycling/retrieval signal in the cytosolic domain. Rapid exit from the TGN by Vps10p is consistent with its function as the CPY sorting receptor, binding proCPY in the TGN and releasing it in the PVC before recycling back for more rounds of vacuolar hydrolase sorting.
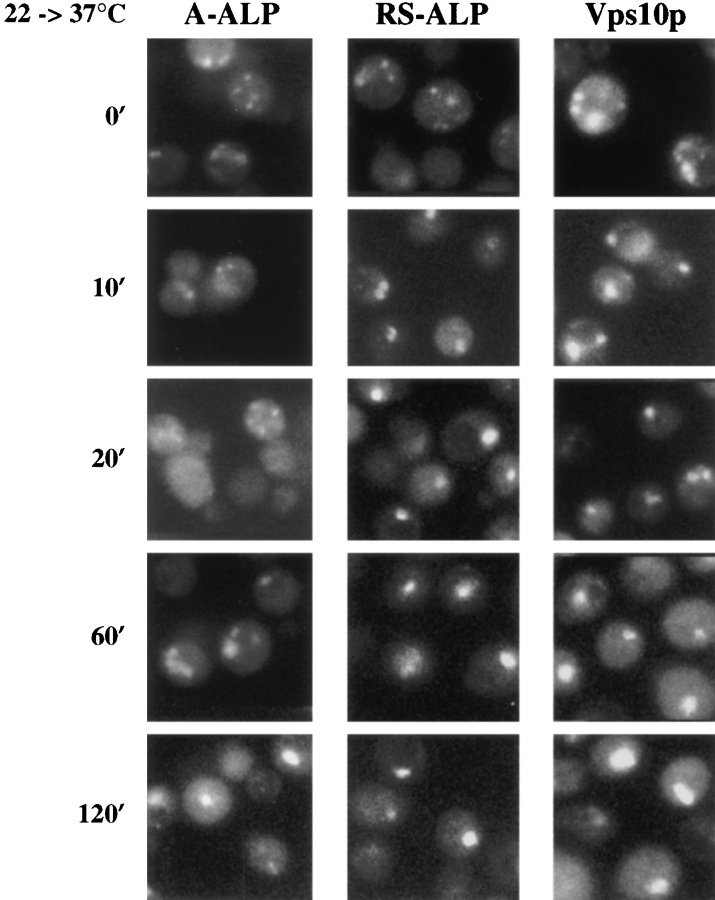
To further address the rate of protein exit from the TGN, we followed the transfer of three membrane proteins from the TGN to the PVC (class E compartment) in vps27 mutant cells by indirect immunofluorescence. Yeast cells carrying a temperature-sensitive allele of VPS27 accumulate Vps10p rapidly into the PVC upon shift to 37°C, and this protein redistributes to Golgi membranes once cells are returned to the permissive temperature (Piper et al., 1995). The rapid onset of the transport defect in vps27-ts cells allowed us to monitor the rate at which A-ALP, RSALP, and Vps10p enter the exaggerated PVC. When vps27-ts cells were maintained at 22°C, indirect immunofluorescence revealed (Fig. 7) that A-ALP, RS-ALP, and Vps10p exhibited staining patterns typical of Golgi membrane proteins (5–10 dispersed dots) in a vast majority of the cells (>90%). 10 min after shifting vps27-ts cells to 37°C, both RS-ALP and Vps10p had redistributed to the exaggerated PVC, yet A-ALP staining appeared Golgilike as long as 60–120 min after the temperature shift. These immunofluorescence data support the conclusion that exit from the TGN is the rate-limiting step for A-ALP processing in vps27 mutant cells. Therefore, by both pulsechase and indirect immunofluorescence analyses, A-ALP has been shown to exit the TGN more slowly than RSALP and Vps10p.
Figure 7.
Redistribution of A-ALP, RS-ALP, and Vps10p in vps27-ts cells. RPY82 (vps27-ts) cells harboring either pSN55 (A-ALP) or pSN97 (RS-ALP) were grown in minimal media at 22°C, and then shifted to 37°C. Aliquots were removed and fixed at the times indicated. Fixed cells were spheroplasted and labeled with anti-ALP mAbs or affinity-purified anti-Vps10p polyclonal antibodies in combination with biotin-linked secondary antibody and streptavidin-conjugated FITC.
The Signal to Slow DPAP A Exit from the TGN Resides Near the NH2 Terminus
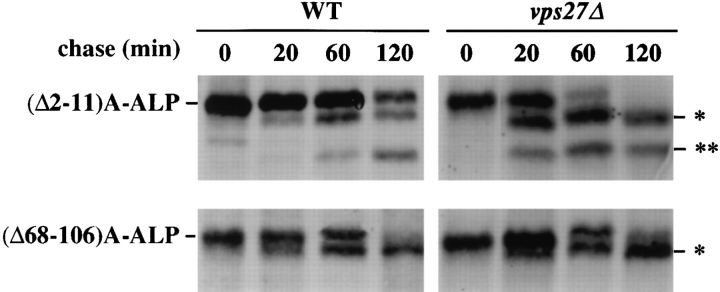
The differences in the rate of exit from the TGN between A-ALP and RS-ALP/Vps10p could either arise through information contained within the tails of ALP and Vps10p, specifying fast exit of the latter two proteins, or it could arise through information contained within the tail of DPAP A that acts to slow the exit of A-ALP. To determine whether tail sequences are responsible for slowing the exit of A-ALP from the TGN, a series of deletions was made in the 118–amino acid cytosolic tail of Ste13p in the context of A-ALP (Fig. 1 A). The proteolytic processing rates of a number of mutated A-ALP fusion proteins were determined in both wild-type and vps27 mutant cells (Table III and Fig. 1). The A-ALP protein lacking the first 50 amino acids of the DPAP A cytosolic domain ((Δ2-51)AALP), which localizes to the TGN (Nothwehr et al., 1993; our unpublished results), was found to behave like RSALP. (Δ2-51)A-ALP was processed with a half-time of 85 min in wild-type cells and ∼15 min in vps27 cells (Table III), indicating that it exits the Golgi rapidly and achieves its Golgi localization through retrieval alone. By contrast, (Δ68-106)A-ALP behaved just like (F/A)A-ALP in that it localized to vacuolar membranes (Nothwehr et al., 1993; our unpublished results) and was processed with halftimes of ∼60–65 min in both wild-type and vps27 mutant cells (Fig. 8 and Table III) presumably since it contains information to slow exit from the TGN. These data indicate that the signal to slow TGN exit resides in the first 50 amino acids of the DPAP A cytosolic domain.
Figure 8.
Kinetics of processing of (Δ2-11)A-ALP and (Δ68106)A-ALP in wild-type and vps27Δ cells. SNY36-9A (wild-type) and NBY60 (vps27Δ) cells harboring either pNB81 ((Δ2-11)AALP) or pSN27 ((Δ68-106)A-ALP) were labeled with [35S]methionine for 10 min and chased by adding unlabeled methionine and cysteine, each to a final concentration of 50 μg/ml. At the indicated times, the cells were spheroplasted, and the extracts were immunoprecipitated with a polyclonal antibody against ALP, followed by SDS-PAGE and fluorography. The products of PEP4-dependent proteolysis are indicated using asterisks, as described in Fig. 1.
In an experiment similar to that described above, vps27-ts cells producing a version of DPAP A lacking residues 2–11 accumulated the protein in the class E compartment after 10–15 min at the restrictive temperature, whereas those producing the full-length protein took longer (∼60 min) to accumulate Ste13p in the class E compartment (data not shown). These data indicate that amino acid residues 2–11 are important for slowing the exit of full-length, wild-type DPAP A from the TGN.
Analysis of a set of deletions lacking various portions of the cytosolic tail of DPAP A revealed that A-ALP missing only amino acid residues 2–11 exited the TGN with a rate of 15–20 min (Fig. 8). These data indicate that the DPAP A TGN retention signal can be mutationally separated into two components, an NH2-terminal signal for slowing exit from the TGN, and the FXFXD motif (residues 85–89 of the cytosolic domain), which directs retrieval back to the TGN from the PVC.
Discussion
Eukaryotic cells face the challenge of maintaining the integrity of secretory organelles despite a continuous and dynamic flow of membranes and proteins through the pathway. The data presented in this paper demonstrate that the yeast S. cerevisiae uses two different modes of retention to ensure localization of DPAP A to the TGN. There are two separable signals for Golgi localization of DPAP A found in the cytosolic domain. The first is the well-characterized motif containing aromatic amino acid residues (FXFXD in DPAP A), which functions in retrieval from the prevacuolar compartment (PVC) back to the TGN. A novel assay for measuring the rate of membrane protein exit from the TGN revealed the second signal, which serves to slow exit from the TGN. These two retention signals serve to efficiently localize DPAP A to the yeast TGN.
The Aromatic Residue Signal Contained in the DPAP A Cytosolic Domain Specifies Retrieval from the PVC
While Kex2p, DPAP A, and Vps10p have been shown to contain Phe and/or Tyr residues in their cytosolic domains that are required for Golgi localization (Wilcox et al., 1992; Nothwehr et al., 1993; Cereghino et al., 1995; Cooper and Stevens, 1996), the evidence that these localization motifs direct retrieval from a post-Golgi compartment has remained indirect. However, subcellular fractionation of membranes from wild-type yeast cells expressing RS-ALP (ALP containing FXFXD) revealed that the vacuolar protease-activated form of RS-ALP cofractionated with the TGN membrane protein Kex2p. These data demonstrate that the FXFXD retention motif of DPAP A functions as a retrieval signal, and that RS-ALP was exposed to active vacuolar proteases in a post-Golgi compartment and then retrieved back to the TGN. Further evidence for the involvement of the FXFXD motif in retrieval from the PVC comes from our data that this motif is required to specify redistribution of A-ALP from the class E compartment (exaggerated PVC) to Golgi membranes after induction of Vps27p synthesis, an experiment that also indicates that the Golgi localization of A-ALP involves retrieval from the PVC.
Having established that localization of A-ALP directed through aromatic amino acid residues involves retrieval from the PVC, we used vps27 mutant cells (which are blocked in traffic from the PVC back to the Golgi apparatus) to measure the rate at which proteins exit the TGN and enter the proteolytically active and exaggerated form of the PVC (Raymond et al., 1992; Piper et al., 1995). As described below, this assay successfully uncouples retrieval-mediated localization from static retention–based Golgi localization by blocking the retrieval mechanism. Comparison of protein processing half-times in wild-type and vps27 mutant cells allows us to determine whether a particular protein is localized to Golgi membranes through retrieval from the PVC, retarded exit from the TGN, or a combination of both mechanisms. For example, when the signal that directs retrieval from the PVC is defective, there is no difference between the half-time of processing in wild-type and vps27 mutant cells. In contrast to this, when a signal that acts to slow the exit of a protein from the TGN is defective, increased processing in vps27 mutant cells is observed.
There Are Two Signals for TGN Localization of DPAP A
Consistent with earlier work (Nothwehr et al., 1993), mutations in the FXFXD retrieval motif in the cytosolic domain of DPAP A were found to result in vacuolar delivery of the A-ALP fusion protein with a half-time of ∼60 min. To determine the molecular basis for the half-time of delivery of (F/A)A-ALP to the vacuole (∼60 min) being so much slower than for ALP (∼5–10 min), we investigated whether there was additional localization information in the DPAP A cytosolic domain. vps27 mutant cells, in which TGN membrane proteins accumulate in a proteolytically active class E compartment (exaggerated PVC; Piper et al., 1995), were used to estimate whether the various A-ALP fusion proteins were all delivered to the PVC at the same rate. Based on proteolytic processing of the lumenal (COOH-terminal) propeptide of ALP, these studies revealed that A-ALP and (F/A)A-ALP were delivered to the PVC with a half-time of ∼60 min, while RS-ALP and (Δ2-51)A-ALP exhibited half-times of ∼15–20 min. These data indicate that there is information in the NH2-terminal 50 amino acids of the DPAP A cytosolic domain that confers the slower rate of delivery of the A-ALP fusion proteins to the PVC, and that this rate is independent of the presence or absence of the aromatic amino acid–based retrieval motif located elsewhere (FXFXD, amino acids 85–89) in the cytosolic domain. Since all our data indicate that the A-ALP fusion proteins are delivered rapidly to the Golgi (A-ALP receives Golgi modifications with similar kinetics to ALP; Bryant, N.J., and T.H. Stevens, unpublished results), the slow rate of transport of A-ALP (and (F/A) A-ALP) to the PVC reflects a slow rate of exit from the TGN.
Vps10p, the CPY sorting receptor, is a yeast TGN membrane protein that was found to be delivered rapidly to the PVC (half-time of ∼15–20 min). As with A-ALP, the rate of Vps10p delivery to the PVC was independent of the presence or absence (Vps10p-10*) of the Tyr-based retrieval motif. Therefore, Vps10p exits the Golgi at a rapid rate and achieves Golgi localization through retrieval, whereas DPAP A contains both retrieval and static retention signals. Such a result is consistent with the biological functions of the two proteins. DPAP A is involved in the processing of the mating pheromone α-factor precursor as this polyprotein passes through the Golgi apparatus before its packaging into secretory vesicles (Sprague and Thorner, 1992). By contrast, Vps10p must continuously cycle between the Golgi and the PVC, binding proCPY in the Golgi and releasing it in the PVC before recycling back to the Golgi to bind more ligand (Cereghino et al., 1995; Cooper and Stevens, 1996). It is thus easy to rationalize the rapid TGN exit rate of Vps10p and the rather slow rate of DPAP A. Consistent with this interpretation are the observations that overexpression of Vps10p leads to processing of A-ALP in wild-type cells, yet the rate of processing of A-ALP in vps27Δ cells is unaffected by overexpression of Vps10p (Bryant, N.J., and T.H. Stevens, unpublished results). These observations suggest that Vps10p and A-ALP compete for the retrieval machinery but not for entry into TGN-derived vesicles bound for the PVC.
DPAP A NH2 Terminus Specifies Static Golgi Retention
Deletion of the first 50 amino acids of the DPAP A cytosolic domain did not affect the localization of the resulting (Δ2-51)A-ALP fusion protein (Nothwehr et al., 1993). Aanalysis of the rate of delivery of (Δ2-51)A-ALP to the PVC in vps27 mutant cells, however, revealed that this protein rapidly exited the TGN just like RS-ALP, containing only the FXFXD retrieval motif. Conversely, deletion of residues 68–106 of the DPAP A cytosolic domain produced a retention-defective form of A-ALP, (Δ68-106)AALP (localized to the vacuole membrane), which was delivered to the PVC slowly (half-time of ∼60 min) in vps27 mutant cells. These results indicate that the first 50 amino acid residues of the DPAP A cytosolic domain are necessary for slowing the TGN exit rate, and that the first 68 are sufficient to slow the rate.
The region important to slow the rate of A-ALP from the TGN was further narrowed by construction and analysis of a set of deletions, each smaller by 10 amino acids. The smallest deletion, removing only residues 2–11 of the DPAP A cytosolic domain, still eliminated the static mode of retention. Thus, the first 11 amino acid residues of DPAP A are required for static retention. Deletion of these same amino acid residues from wild-type DPAP A caused the protein to accumulate rapidly in the class E compartment of vps27-ts cells (data not shown), indicating that this static retention motif is important in the context of the full-length protein. Additional studies will be required to test whether this region is sufficient for static retention.
The presence of the static retention signal in the cytosolic domain of DPAP A helps explain why both A-ALP and RS-ALP are localized to the TGN, yet only RS-ALP gets proteolytically processed by vacuolar proteases in a postGolgi compartment. While we cannot as yet rule out the possibility that RS-ALP is retrieved from the vacuole itself, it is most likely that both RS-ALP and A-ALP are transported to and retrieved from the same PVC, and that this compartment contains low levels of activated vacuolar proteases en route to the vacuole. Evidence for low levels of activated vacuolar proteases in the yeast late-endosomal/ prevacuolar compartment comes from the work of Schimmoller and Riezman, 1993, indicating that α-factor endocytosed into late endosomes encounters proteases. There is abundant evidence from mammalian cells that hydrolases are distributed throughout the endocytic pathway (Blum et al., 1991). If RS-ALP and A-ALP are in fact retrieved from the same PVC, then the failure to detect proteolytic processing of A-ALP must reflect its very slow exit rate from the TGN, thus reducing the time A-ALP spends in the PVC compared to RS-ALP. If RS-ALP is being retrieved from the same PVC as Vps10p, then the differential rate of proteolysis between these two proteins in wild-type cells may reflect a differential susceptibility to the low levels of proteases in the PVC.
Models for Localization of TGN Membrane Proteins in Yeast
The data presented in this paper indicate that DPAP A requires both static retention and retrieval for efficient localization to the yeast TGN. It remains to be determined whether the two other TGN proteins that have been identified (Kex1p and Kex2p) also contain signals to slow their exit from the TGN. We propose that the NH2 terminus of DPAP A serves to keep this protein anchored in the TGN membrane, and that this protein only occasionally exits the Golgi apparatus (half-time of ∼60 min). Molecules of DPAP A that enter the PVC are efficiently recognized by virtue of the FXFXD motif and are recycled back to the TGN. Some proteins, such as the CPY sorting receptor Vps10p, are only retained in the TGN by retrieval because they must continually cycle between the TGN and PVC to fulfill their function in the cell.
The identification of separable signals for retention of DPAP A in the TGN enables us to determine whether the recently identified grd mutants (Golgi retention defective; Nothwehr et al., 1996) are defective in the static retention of DPAP A, or defective in its retrieval from a post-Golgi compartment. We are currently investigating whether the Grd proteins are involved in recognition of either the static or retrieval signals in the cytosolic domain of DPAP A.
Acknowledgments
We thank Rob Piper and Antony Cooper for useful discussions about this work and for providing Pep12p and Vps10p antibodies. Laurie Graham is thanked for affinity-purified anti-Vph1p antibody and for critical reading of the manuscript. We also thank Jason Brickner and Bob Fuller for providing anti-Kex2p serum, as well as members of the Stevens lab and Greg Flynn for reading the manuscript.
This work was supported by a grant from the National Institutes of Health (38006) to T.H. Stevens.
Abbreviations used in this paper
- ALP
alkaline phosphatase
- CPY
carboxypeptidase Y
- DPAP A
dipeptidyl aminopeptidase A
- E
extracellular
- I
intracellular
- proCPY
CPY precursor
- PM
plasma membrane
- PVC
prevacuolar compartment
- RS-ALP
retention sequence ALP
Footnotes
Address all correspondence to Tom H. Stevens, Institute of Molecular Biology, University of Oregon, Eugene, OR 97403-1229. Tel.: (541) 3465884. Fax: (541) 346-4854. e-mail: stevens@molbio.uoregon.edu
References
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum JS, Fiani ML, Stahl PD. Localization of cathepsin D in endosomes: characterization and biological importance. Adv Exp Med Biol. 1991;306:281–287. doi: 10.1007/978-1-4684-6012-4_34. [DOI] [PubMed] [Google Scholar]
- Bos K, Wraight C, Stanley KK. TGN38 is maintained in the transGolgi network by a tyrosine-containing motif in the cytoplasmic domain. EMBO (Eur Mol Biol Organ) J. 1993;12:2219–2228. doi: 10.1002/j.1460-2075.1993.tb05870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher MS, Munro S. Cholesterol and the Golgi apparatus. Science (Wash DC) 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- Bryant NJ, Boyd A. Immunoisolation of Kex2p-containing organelles from yeast demonstrates colocalisation of three processing proteinases to a single Golgi compartment. J Cell Sci. 1993;106:815–822. doi: 10.1242/jcs.106.3.815. [DOI] [PubMed] [Google Scholar]
- Canfield WM, Johnson RD, Ye RD, Gregory W, Kornfeld S. Localization of the signal for rapid internalization of the bovine cation-independent mannose 6-phosphate/insulin-like growth factor-II receptor to amino acids 24-29 of the cytoplasmic tail. J Biol Chem. 1991;266:5682–5688. [PubMed] [Google Scholar]
- Cereghino JL, Marcusson EG, Emr SD. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPSgene products regulate receptor stability, function, and localization. Mol Biol Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RE, Munro S. The functioning of the yeast Golgi apparatus requires an ER protein encoded by ANP1, a member of a new family of genes affecting the secretory pathway. EMBO (Eur Mol Biol Organ) J. 1994;13:4896–4907. doi: 10.1002/j.1460-2075.1994.tb06817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AA, Bussey H. Yeast Kex1p is a Golgi-associated membrane protein: deletions in a cytoplasmic targeting domain result in mislocalization to the vacuolar membrane. J Cell Biol. 1992;119:1459–1468. doi: 10.1083/jcb.119.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AA, Stevens TH. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzusoff A, Redding K, Crosby J, Fuller RS, Schekman R. Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J Cell Biol. 1991;112:27–37. doi: 10.1083/jcb.112.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Emr SD. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18(NSF) mutant. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Krasnov VA. Sorting of yeast alpha 1,3 mannosyltransferase is mediated by a lumenal domain interaction, and a transmembrane domain signal that can confer clathrin-dependent Golgi localization to a secreted protein. Mol Biol Cell. 1995;6:809–824. doi: 10.1091/mbc.6.7.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S, Waters MG. Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J Cell Biol. 1996;132:985–988. doi: 10.1083/jcb.132.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey JS, Peters PJ, Yuan LC, Bonifacino JS. Localization of TGN38 to the trans-Golgi network: involvement of a cytoplasmic tyrosine-containing sequence. J Cell Biol. 1992;120:1123–1135. doi: 10.1083/jcb.120.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO (Eur Mol Biol Organ) J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld S. Structure and function of the mannose-6-phosphate/insulin growth factor-II receptors. Annu Rev Biochem. 1992;61:307–330. doi: 10.1146/annurev.bi.61.070192.001515. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Pelham HR. Ligand-induced redistribution of a human KDEL receptor from the Golgi complex to the endoplasmic reticulum. Cell. 1992;68:353–364. doi: 10.1016/0092-8674(92)90476-s. [DOI] [PubMed] [Google Scholar]
- Lussier M, Sdicu M, Ketela T, Bussey H. Localization and targeting of the Saccharomyces cerevisiaeKre2p/Mnt1p alpha 1,2-mannosyltransferase to a medial-Golgi compartment. J Cell Biol. 1995;131:913–927. doi: 10.1083/jcb.131.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer CE, Grim MG, Esquela A, Chung SW, Rolls M, Ryan K, Swift AM. Retention of a cis-Golgi protein requires polar residues on one face of a predicted alpha-helix in the transmembrane domain. Mol Biol Cell. 1993;4:695–704. doi: 10.1091/mbc.4.7.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolson MF, Proteau D, Preston RA, Stenbit A, Roberts BT, Hoyt MA, Preuss D, Mulholland J, Botstein D, Jones EW. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H(+)-ATPase. J Biol Chem. 1992;267:14294–14303. [PubMed] [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast carboxypeptidase Y is encoded by the VPS10gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Munro S. Sequences within and adjacent to the transmembrane segment of alpha-2,6-sialyltransferase specify Golgi retention. EMBO (Eur Mol Biol Organ) J. 1991;10:3577–3588. doi: 10.1002/j.1460-2075.1991.tb04924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Lucocq JM, Mackay D, Warren G. The membrane spanning domain of beta-1,4-galactosyltransferase specifies transGolgi localization. EMBO (Eur Mol Biol Organ) J. 1991;10:3567–3575. doi: 10.1002/j.1460-2075.1991.tb04923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Slusarewicz P, Hoe MH, Warren G. Kin recognition. A model for the retention of Golgi enzymes. FEBS Lett. 1993;330:1–4. doi: 10.1016/0014-5793(93)80906-b. [DOI] [PubMed] [Google Scholar]
- Nothwehr SF, Stevens TH. Sorting of membrane proteins in the yeast secretory pathway. J Biol Chem. 1994;269:10185–10188. [PubMed] [Google Scholar]
- Nothwehr SF, Roberts CJ, Stevens TH. Membrane protein retention in the yeast Golgi apparatus: dipeptidyl aminopeptidase A is retained by a cytoplasmic signal containing aromatic residues. J Cell Biol. 1993;121:1197–1209. doi: 10.1083/jcb.121.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Conibear E, Stevens TH. Golgi and vacuolar membrane proteins reach the vacuole in vps1mutant yeast cells via the plasma membrane. J Cell Biol. 1995;129:35–46. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Bryant NJ, Stevens TH. The newly identified yeast GRDgenes are required for retention of late-Golgi membrane proteins. Mol Cell Biol. 1996;16:2700–2707. doi: 10.1128/mcb.16.6.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paravicini G, Horazdovsky BF, Emr SD. Alternative pathways for the sorting of soluble vacuolar proteins in yeast: a vps35null mutant missorts and secretes only a subset of vacuolar hydrolases. Mol Biol Cell. 1992;3:415–427. doi: 10.1091/mbc.3.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HR, Munro S. Sorting of membrane proteins in the secretory pathway. Cell. 1993;75:603–605. doi: 10.1016/0092-8674(93)90479-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Yang H, Stevens TH. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. . J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Whitters EA, Stevens TH. Yeast Vps45p is a Sec1plike protein required for the consumption of vacuole-targeted, post-Golgi transport vesicles. Eur J Cell Biol. 1994;65:305–318. [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vpsmutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K, Holcomb C, Fuller RS. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. . J Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Nothwehr SF, Stevens TH. Membrane protein sorting in the yeast secretory pathway: evidence that the vacuole may be the default compartment. J Cell Biol. 1992;119:69–83. doi: 10.1083/jcb.119.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE. Mechanisms of intracellular protein transport. Nature (Lond) 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Schafer W, Stroh A, Berghofer S, Seiler J, Vey M, Kruse ML, Kern HF, Klenk HD, Garten W. Two independent targeting signals in the cytoplasmic domain determine trans-Golgi network localization and endosomal trafficking of the proprotein convertase furin. EMBO (Eur Mol Biol Organ) J. 1995;14:2424–2435. doi: 10.1002/j.1460-2075.1995.tb07240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R, Orci L. Coat proteins and vesicle budding. Science (Wash DC) 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- Schimmoller F, Riezman H. Involvement of Ypt7p, a small GTPase, in traffic from late endosome to the vacuole in yeast. J Cell Sci. 1993;106:823–830. doi: 10.1242/jcs.106.3.823. [DOI] [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 61–64.
- Sprague, G.F., and J. Thorner. 1992. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 657–744.
- Swift AM, Machamer CE. A Golgi retention signal in a membrane-spanning domain of coronavirus E1 protein. J Cell Biol. 1991;115:19–30. doi: 10.1083/jcb.115.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Nakagawa T, Banno T, Watanabe T, Murakami K, Nakayama K. Localization of furin to the trans-Golgi network and recycling from the cell surface involves Ser and Tyr residues within the cytoplasmic domain. J Biol Chem. 1995;270:28397–28401. doi: 10.1074/jbc.270.47.28397. [DOI] [PubMed] [Google Scholar]
- Townsley FM, Wilson DW, Pelham HR. Mutational analysis of the human KDEL receptor: distinct structural requirements for Golgi retention, ligand binding and retrograde transport. EMBO (Eur Mol Biol Organ) J. 1993;12:2821–2829. doi: 10.1002/j.1460-2075.1993.tb05943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees P, Deignan E, van Donselaar E, Humphrey J, Marks MS, Peters PJ, Bonifacino JS. An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO (Eur Mol Biol Organ) J. 1995;14:4961–4975. doi: 10.1002/j.1460-2075.1995.tb00179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox CA, Fuller RS. Posttranslational processing of the prohormone-cleaving Kex2 protease in the Saccharomyces cerevisiaesecretory pathway. J Cell Biol. 1991;115:297–307. doi: 10.1083/jcb.115.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox CA, Redding K, Wright R, Fuller RS. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992;3:1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsbach K, Payne G. Dynamic retention of TGN membrane proteins in Saccharomyces cerevisiae. . Trends Cell Biol. 1993;3:426–432. doi: 10.1016/0962-8924(93)90031-u. [DOI] [PubMed] [Google Scholar]
- Wong SH, Hong W. The SXYQRL sequence in the cytoplasmic domain of TGN38 plays a major role in transGolgi network localization. J Biol Chem. 1993;268:22853–22862. [PubMed] [Google Scholar]