Abstract
Embryo liver morphogenesis takes place after gastrulation and starts with a ventral foregut evagination that reacts to factor signaling from both cardiac mesoderm and septum transversum mesenchyme. Current knowledge of the progenitor stem cell populations involved in this early embryo liver development is scarce. We describe here a population of 11-day postcoitus c-Kitlow(CD45/TER119)– liver progenitors that selectively expressed hepatospecific genes and proteins in vivo, was self-maintained in vitro by long-term proliferation, and simultaneously differentiated into functional hepatocytes and bile duct cells. Purified c-Kitlow(CD45/TER119)– liver cells cocultured with cell-depleted fetal liver fragments engrafted and repopulated the hepatic cell compartments of the latter organoids, suggesting that they may include the embryonic stem cells responsible for liver development.
Introduction
The liver is the central organ of nutrient digestion and processing, where most of the individual’s metabolism occurs. In adult life, it is a quiescent organ, but when subjected to pathological aggressions, it responds with neoproliferation of mature hepatocytes and subsequently of the other liver cells to the point of complete organ regeneration (1–3). When the proliferation capacity of mature cells is exhausted or inhibited, however, immature progenitors expand in order to generate new functional cells. Adult liver progenitors are located in the periportal region, deriving from terminal biliary ductules (canals of Hering). They give rise to highly proliferating, transient oval cells that subsequently differentiate into mature hepatocytes and bile duct cells (4–6). Few stem cell– or oval cell–specific markers are available, and some are also shared by hematopoietic progenitors (CD34, Thy1, Sca1, c-Kit, CD45), suggesting a developmental relationship between the two cell compartments (7–10). Attempting to recapitulate liver development and regeneration, experimental protocols have been designed in which mature cells and progenitors from intra- and extrahepatic sources were subjected to hepatic-promoting conditions. These assays revealed the inductive influences of nonparenchymal liver cells, ECM and cell adhesion receptors, soluble factors, cell-density conditions, and vasculoendothelial cells over the differentiation of progenitors in order to create a functional liver (11–14). There is a consensus that highly proliferating embryo fetal liver progenitors are more efficient than those derived from adult sources in high-level, long-term liver regeneration (15–17). We focused our attention on embryonic progenitors of the early postgastrulation mouse liver. The embryo liver starts as an endodermal evagination of the ventral foregut endoderm (7.5–8.5 days postcoitus, or dpc) in response to signaling derived from the cardiac mesoderm (FGF-1, -2, and –3) and the septum transversum mesenchyme (bone morphogenetic protein-4) (18–21). Very early on (9–10 dpc), hematopoietic progenitors engraft in the liver, where they expand and differentiate during the second half of gestation and also contribute to the maturation of liver epithelial compartments (22–25). Around birth, the hepatic cells stop cycling and reach full maturity, while the hematopoietic niches are exhausted and their cells exported to the bone marrow.
The genetic mechanisms of liver morphogenesis have been well characterized (22–24), but limited information exists about the hepatospecific progenitors involved, thus impeding their purification and study of potentials for differentiation and regeneration. A stem cell/progenitor phenotype would also contribute to elucidating the differentiation pathways of liver-regenerating cells from various lineages and sources (embryonic stem cells, bone marrow and cord blood cells, fetal and adult hepatocytes, biliary cells, oval cells, etc.). During studies of liver lymphohematopoiesis, we detected two cell populations that expressed high and low levels of c-Kit, respectively, but no other hematopoietic marker, such as CD45 (26) or the erythroid-specific TER119 (27). These cell populations did not generate any lymphohematopoetic cell lineage in ad hoc assays. We speculated about the possibility that they represented progenitors for the epithelial compartment of the liver (28). Upon purification, these c-Kitlow(CD45/TER119)– cells and, to a lesser extent, the c-Kithigh(CD45/TER119)– cells expressed hepatospecific genes. When established in hepatic-promoting cultures, only the c-Kitlow(CD45/TER119)– cell population became established in long-term cultures, were self-maintained by proliferation after successive replatings, and differentiated into hepatocytes and cholangiocytes. Finally, the purified c-Kitlow(CD45/TER119)– cells engrafted into 2′-deoxyguanosine–treated (2′deo-treated) embryo livers, where they repopulated the hepatic cell lineages. The findings support that 11-dpc liver c-Kitlow(CD45/TER119)– cells contain the earliest-described hepatic progenitors, also displaying features of liver-repopulating stem cells.
Methods
Mice and cells.
BALB/c, C57BL/6, and C57BL/6.GFP transgenic mice (29) were maintained under pathogen-free conditions. Day 0 of gestation was taken to be detection of a vaginal plug after overnight mating. Embryos were microdissected under a surgical stereoscopic microscope (Nikon SMZ-1; Nikon Corp., Tokyo, Japan), and contaminating blood was washed out by embryo exsanguination and passages through PBS with 2% FCS and heparin (Rovi Pharmaceutical Labs, Madrid, Spain). Liver cell suspensions were prepared by mechanical dissociation, and cell viability (>95%) was measured by trypan blue dye exclusion.
FACS analysis and cell purification.
Cell suspensions from pooled embryo livers of the same gestational age were first incubated with Fc Block (PharMingen, BD Biosciences, San Diego, California, USA) for 10 minutes. After washing, the cells were incubated with biotinylated, FITC-conjugated, or purified mAb. Biotinylated mAb were revealed by phycoerythrin-streptavidin. Cyanine 5–conjugated mouse anti-rat Ig mAb (clone MAR18.5) and biotinylated rat anti-mouse Igκ mAb (clone 187.1) revealed purified rat and mouse mAb, respectively. Cells were analyzed in a FACScalibur equipped with the CellQuest System (Immunocytometry Systems, BD Biosciences, San Jose, California, USA). Cell debris and dead cells were discarded by light parameters and by propidium iodide exclusion. The mAb-specific signals were defined against the background of irrelevant, isotype-matched mAb. Cells were purified in a FACS Vantage (Immunocytometry Systems, BD Biosciences) under sterile conditions. The sorting windows of intermixing cell populations were established over the most separated fluorescence channels; the intermediate cells were uncollected. Reanalysis in the FACScalibur revealed degrees of purity of more than 97%. The mAb’s used were biotinylated anti-CD45.2 and TER119, FITC anti–c-Kit, and purified anti-mouse CD34, anti-CD49f, and anti-CD29 (PharMingen) and mouse anti–c-Met (Upstate Biotechnology Inc., Lake Placid, New York, USA). Purified rat anti-mouse AA4.1 (fetal hematopoietic stem cell antigen), Sca-1, Thy-1, ICAM-1, and MHC-I, 187.1, and MAR18.5 mAb’s were obtained from hybridoma supernatants by affinity chromatography (Pharmacia Biotech AB, Uppsala, Sweden).
RT-PCR analyses.
Total RNA was extracted, and cDNA was prepared by using 1 μg oligo-(dT) as primer and avian myeloblastosis virus reverse transcriptase (Promega Corp., Madison, Wisconsin, USA). The RT products (one-tenth of each reaction) were amplified with 1 U Taq DNA polymerase (Sigma-Aldrich, St. Louis, Missouri, USA). After an initial denaturation (95°C, 5 minutes), PCR cycles were performed (95°C, 1 minute; annealing temperature, 1 minute; 72°C, 1 minute) followed by an extension step (72°C, 10 minutes). PCR cycles were repeated 30 times for α-fetoprotein (AFP), albumin (ALB), and β-actin; 35 times for c-Kit; and 40 times for the rest of the PCRs, except for oncostatin M receptor (OSMR) and tyrosin aminotransferase (TAT), which received 70 cycles by using 2 μl of their first PCR products (40 cycles) as starting material in a second round of 30 cycles. The intron-spanning oligonucleotides used were as follows: 5′-TCAACAGACCAGACAGTGAGCT and 5′-AAAGGGTGGTCGAAGGCGA (α1-antitrypsin [AAT]); 5′-TCACACCCGCTTCCCTCATCCT and 5′-CATCCTGCAGACACTCCAG (AFP); 5′-AGAAGACACCCTGATTACTCT and 5′-TCGAGAAGCAGGTGTCCTTGT (ALB); 5′-TTCTTGGCTATGGAATCCTGT and 5′-CTAGAAGCACTTGCGGTGCACG (β-actin); 5′-AGGCATCTTCGTGCACGAGCA and 5′-TATTCACAGAGAATTTGGCAGCCA (c-Kit); 5′-GGAAGAGGGCATTTTGGCTG and 5′-AGCACACCGAAGGACCACAC (c-Met); 5′-AACCCATTGTGAGGCGAGAGG and 5′-TACTCATTACACTAGTTGGTC (glucose-6-phosphatase [G6P]); 5′-ACAAACAACAGTAGGGTGGA and 5′-AGCAGGAACAATGACACCAA (HGF); 5′-TCTGAAGGTGCCAACCTCAATTCA and 5′-CTCGACCAGGACCAGCAGCTG (hepatocyte nuclear factor–4α [HNF-4α]); 5′-TCACTCCGTACACCAGCGCT and 5′-TGTTAGGTGGAGCCAATAGT (OSMR, first round) (30); 5′-TCAGCATCATTGTCTGCTACT and 5′-TGTTAGGTGGAGCCAATAGT (OSMR, second round); 5′-AAGACCTTCAATCCCATCCGAGC and 5′-TCATGAATGAGGATCCAGCCCAA (TAT first round); 5′-AGCCCAATCCGAACAAGACCG and 5′-AGCCAGCGCTTGGCCAGCCC (TAT second round); 5′-ATCCACAAGCTCCTGACAG and 5′-ACTGCTTTGGCAAGATCCTGG (transthyretin [TTR]); and 5′-CTGGTGGACGAGGCTAT and 5′-CACAGACTGCATGAGGTTC (vinculin). The annealing temperatures were 63°C (TAT), 60°C AAT, c-Kit, HNF, HNF-4α, and TTR), 55°C (AFP, ALB, β-actin, c-Met, G6P, OSMR, TAT, and vinculin), and 52°C (HGF). Adult and 14-dpc livers and ST2 stromal cell cDNAs were positive and negative controls. Genomic DNA amplifications were either negative or yielded fragments of larger size than those from cDNA. The amplification products were separated electrophoretically on 2% agarose gels, transferred to Zeta-Probe membranes (Bio-Rad Laboratories Inc., Hercules, California, USA), and hybridized at 52°C with the 32P-labeled oligonucleotides: 5′-AAGAACCATTATCAGGCAGA (AAT), 5′-TTGGGATAGCTTCCACGT (AFP), 5′-TGCTACGGCACAGTGCTTGCT (ALB), 5′-AACACAGTGTTGTCTGGTGGT (β-actin), 5′-ACATTCAAGCAGGTTGTCCA (c-Kit), 5′-AGTTTGTCCACAGAGACTTAG (c-Met), 5′-AGTGTGAATGCTGGAAACTGA (G6P), 5′-TGAAGGCTCAGACTTGGTT (HGF), 5′-GAACCAGTGTCGTTACTGCA (HNF-4α), 5′-ACTGCATTCCAGATGTCCTT (OSMR), 5′-GGGAACCTGCCTACAGACC (TAT), 5′-TCCTGGGCTGAGTCTCTCAAATTC (TTR), and 5′-TCTGCCTGAGGGTGAAGT (vinculin). Amplification products were quantified from nonsaturated PCRs (registering less than 40 cycles) in a Fujibas-1000 detector (Fuji Photo Film Co., Tokyo, Japan) with the TINA program (Raytest GmbH, Stauhenhardt, Germany). Transcript levels represented the value of PCR-amplified cDNAs relative to one of the β-actin transcripts performed simultaneously in separated tubes for each sample.
High- and low-density cell cultures.
Liver cell populations were plated onto 50 μg/ml collagen I–coated (Discovery Labware, BD Biosciences, Bedford, Massachusetts, USA) 96-well plates, or collagen I–coated chamber slides (Nalge Nunc International, Naperville, Illinois, USA) (2 × 105 to 4 × 105 cells/cm2), or onto Terasaki microwell plates (twofold dilutions from 240–30 cells/cm2, 100–150 wells/group) and maintained for 8–12 days at 37°C, 5% CO2. The medium was DMEM (BioWhittaker Europe, Verviers, Belgium) supplemented with 10% heat-inactivated FCS, 100 U/ml penicillin-streptomycin (BioWhittaker Europe), 2 mM L-glutamine (Gibco BRL; Invitrogen Life Technologies, Paisley, Scotland), 1 mM pyruvate (Sigma-Aldrich), 50 μM 2-mercaptoethanol (Sigma-Aldrich), nonessential amino acids (BioWhittaker Europe), 10 ng/ml EGF (PreproTech Inc., Rocky Hill, New Jersey, USA), 1 μg/ml human insulin (Novo Nordisk A/S, Bagsvaerd, Denmark), and 10–7 M dexamethasone (Sigma-Aldrich) renewed every 4 days. The soluble factors were as follows: human HGF (40 ng/ml; Discovery Labware, BD Biosciences), murine OSM (10 ng/ml; Sigma-Aldrich), stem cell factor (SCF) (20 ng/ml; R&D Systems Europe Ltd., Abingdon, United Kingdom), FGF-1 (50 ng/ml, supplemented with 50 ng/ml heparin sulfate; Sigma-Aldrich), and their combinations. Clonable precursor frequencies were calculated as described (31).
Immunofluorescence assays.
Slides prepared with ex vivo purified cells after centrifugation in a Cytospin-2 (32.2 g, 5 minutes; Shandon Southern Products Ltd., Runcorn, United Kingdom) and with cells cultured on collagen I–coated slide chambers were fixed and permeabilized with methanol/acetone (95:5) (–20°C, 20 minutes). They were then incubated with the primary Ab: mouse anti-cytokeratin 8 (anti-CK8) (clone 35bH11; DAKO A/S, Glostrup, Denmark), mouse anti-CK18 (clone LE61; DAKO A/S), mouse anti-CK19 (clone RCK 108; DAKO A/S), and rabbit anti-mouse ALB (Nordic Immunology, Tilburg, The Netherlands) in a moist chamber (37°C, 1 hour). Slides were then incubated with either rhodamine-labeled goat anti-mouse IgG (Southern Biotechnology Associates, Birmingham, Alabama, USA) or Alexa 488–conjugated goat anti-rabbit IgG (37°C, 1 hour; Molecular Probes Inc., Eugene, Oregon, USA). The frequency of Ki-67+, S-phase cycling, cells per population was established by three-color immunofluorescence along the following steps: (a) incubation with NH4Cl (50 mM in PBS, pH 8; room temperature (rt), 10 minutes), blocking with 10% mouse serum (37°C, 30 minutes), and washing with PBS; (b) incubation with anti-CK19 mAb (37°C, 90 minutes) and washing with PBS; (c) antigen retrieval with citrate buffer (0.01 M, pH 6.7; 7 minutes in a microwave oven at 70 W, 100°C) and washing with PBS; (d) incubation with rabbit anti–Ki-67 Ab (4°C, overnight; Novocastra Laboratories Ltd., Newcastle upon Tyne, United Kingdom); (e) incubation with Alexa 647–conjugated goat anti-rabbit IgG (37°C, 90 minutes; Molecular Probes Inc.) to reveal the anti–Ki-67 Ab; and (f) staining with rhodamine-conjugated goat anti-mouse IgG to reveal the anti-CK19 mAb and FITC-conjugated rabbit anti-mouse ALB (37°C, 90 minutes; Nordic Immunology). Nuclei were counterstained with 10 mM 4′ DAPI (Molecular Probes Inc.) in glycerol (rt, 10 minutes), and cell preparations were mounted with Fluorsave (rt, 30 minutes; Calbiochem, Darmstadt, Germany). These samples were examined under a Leitz DMRD microscope (Leica Microsystems AG, Wetzlar, Germany).
The GFP+ cells of the chimeric organotypic cultures were detected with a rabbit anti-GFP Ab (37°C, 90 minutes; Molecular Probes Inc.) revealed with Alexa 647–conjugated goat anti-rabbit Ab. This treatment blocked GFP fluorescence and identified the GFP+ cells at 647 nm, allowing the simultaneous use of other FITC-conjugated Ab’s. The samples were subsequently incubated with mouse anti-CK19 mAb (4°C, overnight) and revealed with rhodamine-conjugated goat anti-mouse IgG and with FITC-conjugated rabbit anti-mouse ALB (37°C, 90 minutes). The frequencies of cycling Ki-67+/GFP+ donor cells of the organotypic cultures were detected with the rabbit anti–Ki-67 Ab as described above, maintaining the original green fluorescence of GFP+ donor cells. The slides were mounted with Fluorsave and examined as above and under a confocal microscope (Radiance 2500; Bio-Rad Laboratories Inc.). All of the images were processed using Adobe Photoshop 6.0 (Adobe Systems Inc., San Jose, California, USA). Quantitative cell data in photomicrographs were obtained by three independent observers.
Organotypic chimeric liver cultures.
Cocultures of purified 11-dpc liver progenitor cells and cell-depleted 12-dpc liver fragments were modified from Jenkinson et al. (32). Three-millimeter sections of 12-dpc C57BL/6 livers were treated with 2′deo (1.35 mmol/l; Sigma-Aldrich) for 5 days to remove hematopoietic and hepatic cycling cells. Liver fragments were then washed and cocultured with 1 × 103 to 5 × 103 11-dpc purified liver cells from C57BL/6.GFP mice in hanging drops in Terasaki plates. After 48 hours, the liver fragments were transferred to nucleopore filters, layered over gel foam sponges, and cultured for 3–4 weeks. The resulting chimeric liver organoids were frozen in Jung tissue-freezing medium (Leica Microsystems AG). Liver sections (10 μm) were obtained in a cryostat (Leica Microsystems AG), mounted on Superfrost microscope slides (Fisher Scientific Co., Pittsburgh, Pennsylvania, USA), and fixed in acetone (4°C, 10 minutes).
Transmission electron microscopy.
The cells were cultured on collagen I–coated Thermanox sheets (Electron Microscopy Science, Fort Washington, Pennsylvania, USA) in 1-cm2 wells. They were washed in 0.1 M, pH 7.4, cacodylate buffer (Sigma-Aldrich), fixed in 2% glutaraldehyde/2% tannic acid/cacodylate buffer (rt, 1 hour), and incubated with 1% OsO4/H2O (rt, 1 hour). The samples were dehydrated with ethanol, stained with 2% uranyl acetate/ethanol, and embedded in Epoxy resin. After polymerization (60°C, 48 hours), ultrathin sections (70–80 nm) were counterstained with uranyl acetate and lead citrate and examined under a JEOL-1010 electron microscope (JEOL Ltd., Tokyo, Japan).
Periodic acid-Schiff staining.
Cells grown on collagen I–coated chamber slides were oxidized in 1% periodic acid (5 minutes), rinsed in dH2O (three times), treated with Schiff’s reagent (15 minutes), rinsed again in dH2O (5–10 minutes), stained with Mayer’s hematoxylin (1 minute), and finally rinsed in dH2O.
Statistical analysis.
The GraphPad Prism 3.0 program (GraphPad Software Inc., San Diego, California, USA) was used to calculate mean plus or minus SEM. Data were compared by the Student t test.
Results
Cell populations of 11- to 13-dpc mouse embryo livers.
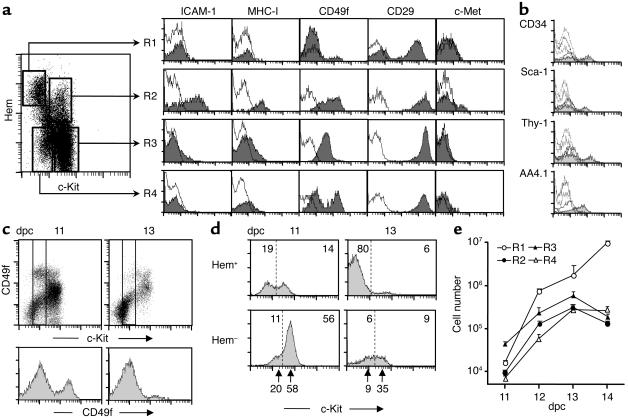
Embryo fetal liver contains lymphohematopoietic progenitors characterized by the CD45 and TER119 antigens. Double staining of 11-dpc liver cells with anti-mAb (TER119 + CD45) (hematopoietic (Hem), in Figure 1) and anti–c-Kit mAb revealed four cell populations: (R1) c-Kit–(CD45/TER119)+ (20%), (R2) c-Kit+(CD45/TER119)+ (15%), (R3) c-Kithigh(CD45/TER119)– (55%), and (R4) c-Kitlow(CD45/TER119)– (10%). ICAM-1 and MHC class I antigens (33) were seen in most R2 (hematopoietic) cells, but only in few R3 cells and not at all in R1 or R4 cells. The β1-integrin chain (CD29) was highly expressed in all liver cells, while α6-integrin chain expression (CD49f) differed between the liver populations: R1 cells were negative, R2 and R3 cells had strong signals, and R4 cells displayed two peaks of low and high CD49f levels. The HGF receptor c-Met was detected in a small fraction of R2 cells, and low levels were present in the R3 and R4 cell subsets (Figure 1a). Only the R2 liver cells had other lymphohematopoietic-specific antigens, such as CD34, Sca-1, Thy-1, and AA4.1 (Figure 1b). Since integrin levels change during liver ontogeny (34), we compared the CD49f expressions of 11- and 13-dpc livers: a depletion of c-KitlowCD49fhigh cells was observed in the 13-dpc livers (Figure 1c). The c-Kit peaks of R3 and R4 in Hem– liver cells were also reduced in 13-dpc compared with 11-dpc livers (Figure 1d, bottom arrows). The absolute numbers of liver cell populations (see total liver cells per gestational day in ref. 35) varied during midgestation ontogeny, with a marked increase in R1 cells (mostly erythrocytes), while R2 and R3 cells increased up to 13 dpc and decreased afterward; R4 cells also increased, reaching a plateau at 13 to 14 dpc (Figure 1, d and e).
Figure 1.
Liver cell populations of mouse midgestation embryo. (a) Three-color FACS analyses of 11-dpc liver cells. A mixture of biotinylated anti-CD45 and anti-TER119 mAb versus FITC anti-c-Kit mAb defined the four populations of the dot plot. The histograms represented antigen expressions of each population detected with specific mAb’s (gray) and irrelevant, isotype-matched mAb controls (white). (b) Histograms for hematopoietic-specific antigens of R2 (gray) and R1, R3, and R4 cell populations (white). (c) CD49f levels (α6-integrin) in 11- and 13-dpc total liver cells (upper plots) and in c-Kitlow cells (bottom histograms) defined by the two vertical lines in the plots. (d) The 11- and 13-dpc liver c-Kit expressions. The numbers inside the histograms are the cell frequencies of each population separated by vertical dotted lines, and the bottom arrows point to the c-Kit immunofluorescence peaks (n = 7 independent analyses). (e) Absolute numbers of liver cell populations during midgestation (log scale). The data were calculated with the FACS percentages and total recovered liver cell numbers per day (35). Hem, hematopoietic.
Hepatospecific gene expression and cytoplasmic ALB in embryo liver cell populations.
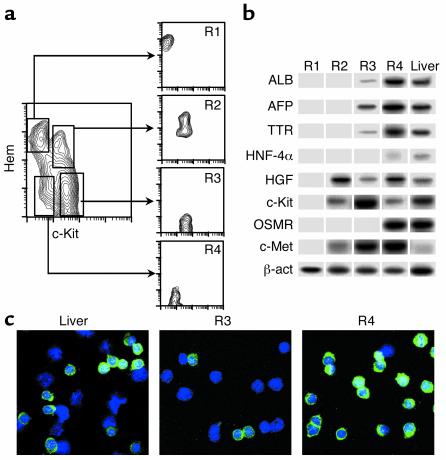
The 11-dpc liver cell populations were purified by FACS (Figure 2a), and hepatospecific gene expressions were tested in them by RT-PCR. The selected genes coded for transcription factors (HNF-4α), secreted proteins (ALB, AFP, TTR), growth factors (HGF), and receptors (c-Kit, OSMR, c-Met) (Figure 2b). R1 erythroid cells did not express any of these genes. R2 c-Kit+(CD45/TER119)+ cells expressed the HGF, c-Met, and c-Kit genes. R3 c-Kithigh(CD45/TER119)– cells showed high transcript levels of c-Kit and c-Met, intermediate of AFP, and lower than ALB, TTR, and HGF genes. Finally, R4 c-Kitlow(CD45/TER119)– cells had moderate HNF-4α and c-Kit transcript titers, the latter being one-fifth of those of R3 cells, in agreement with FACS data. All the other hepatic gene transcripts (ALB, AFP, TTR, HGF, OSMR, and c-Met) were highly expressed in R4 cells. The ratios of expressed transcripts between R3 and R4 populations (10% for ALB and TTR, 40% for AFP; six independent samples) make it unlikely that these findings in R3 were due to R4 cell contaminants. The putative hepatic lineage of R3 and R4 liver cells was also supported by the detection of cytoplasmic ALB (Figure 2c): R3 cells were predominantly ALB negative, although over 10% of them exhibited dull staining. By contrast, most R4 cells represented a homogeneous population of cytoplasmic ALB+ small cells.
Figure 2.
Hepatospecific gene expressions and ALB production in purified 11-dpc liver cell populations. (a) Liver cell populations were purified in a FACS Vantage and reanalyzed in a FACScalibur. (b) The mRNA from purified cells was prepared and RT-PCRs were performed for the genes displayed in the figure in each purified cell subset. β-actin (β-act) gene expression and 11-dpc liver unseparated cells were used as controls for mRNA content per sample and positive gene transcripts, respectively. The OSMR gene required a nested PCR, and its signals were not quantitative (see Methods). A representative experiment for all the gene expressions (of a total of six independent ones) is shown. (c) 11-dpc liver and purified R3 and R4 cells were spun on slides and stained for cytoplasmic ALB (green signals). Total cells per sample were evaluated by DAPI (blue). ×63 amplification.
In vitro growth and differentiation of liver cells in response to HGF and OSM.
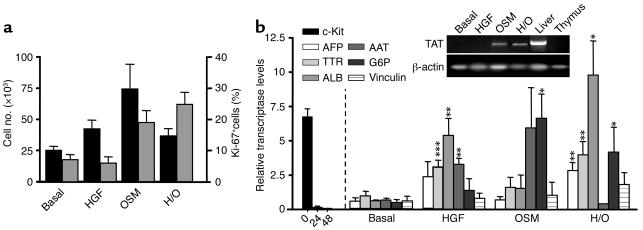
In preliminary cultures, mixed Hem+ (R1 + R2) and Hem– (R3 + R4) 11-dpc liver cell populations were purified and seeded in hepatic-promoting cultures (105 cells/well, 8–12 days) due to the low cell numbers of early embryo livers. All the c-Kit+/– (CD45/TER119)+ Hem+ cells and more than two-thirds of the c-Kit+ (CD45/TER119)– Hem– cells disappeared during the first culture days (not shown). The early mortality of Hem– cells was accounted for by the predominant c-Kithigh R3 cell population (see below). Later, 15 × 103 to 20 × 103 cells/well survived and slightly expanded under basal conditions (1.5-fold to twofold). Supplementing with HGF and/or OSM multiplied cell recoveries by a factor of 3–4 (Figure 3a, black columns). Ten to twenty percent of cells of basal and HGF-supplemented cultures were Ki-67+ cycling cells, and OSM increased these frequencies up to 50–60%. Cultured cells progressively evolved from the ex vivo round cells with big nuclei to flattened and adherent epithelioid cells, which organized into two-dimensional and globular cell aggregates (data not shown). The cells of the primary cultures were successively replated and remained viable in long-term cultures (up to 3 months, at the time of this writing). The hepatic differentiation of the cultured cells was tested by their gene expressions in RT-PCRs (c-Kit, AFP, TTR, ALB, AAT, TAT, G6P, and vinculin genes). The small cell numbers required for RT-PCRs allowed us to analyze separately both R3 and R4 cells. The first observation was that purified R3 cells didn’t survive in the hepatic cultures, which explains the early cell losses of Hem– cell cultures. Second, the ex vivo c-Kit expression of R4 cells was rapidly downregulated after 24–48 hours in vitro. HGF markedly stimulated the differentiation of c-Kitlow(CD45/TER119)– R4 cells, significantly upregulating the expressions of TTR, ALB, and AAT. OSM alone provided a weaker differentiation stimulus, although it selectively activated the expression of G6P and TAT genes, which are upregulated at the perinatal stages of liver maturation (36). Both HGF and OSM cooperated in differentiation. Finally, the bile-specific vinculin gene was also expressed in these cultures (Figure 3b).
Figure 3.
(a) Cell recoveries (left axis; black bars) and proliferative rates (right axis of Ki-67+ cells; gray bars) of 11-dpc liver c-Kit+(CD45/TER119)– (R3 + R4) progenitors established in hepatospecific cell cultures (H/O, HGF + OSM). (b) Gene expression titers (relative to those of β-actin) in cultured 11-dpc c-Kitlow(CD45+TER119)– R4 cells for 8–12 days. The c-Kit expressions disappeared after 48-hour culture. The RT-PCR results for TAT gene, obtained in a nonquantitative assay, are shown in the upper right inset. Adult liver and thymus were positive and negative controls, respectively. Mean ± SEM of five to six replicates per gene and experimental group. P values (asterisks) are from Student two-tailed unpaired t test. *P < 0.05; **P < 0.01; ***P < 0.001, by comparing the results of factor supplemented with those of basal cultures.
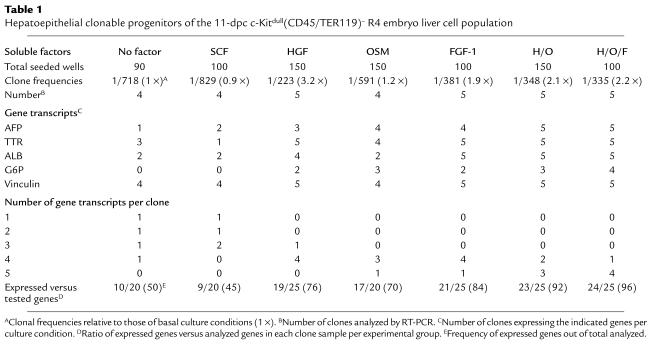
Colony-forming progenitors of the c-Kitlow(CD45/TER119)– cell population.
The 11-dpc R1–R4 liver cell populations were seeded in low-density cultures to calculate the frequencies of clonable progenitors generating colonies (>50 cells/colony). The colony differentiation was evaluated by the gene transcripts (AFP, TTR, ALB, G6P, and vinculin) detected in significant clone samples per experimental group. As seen in high-density cultures, the purified 11-dpc R1-R3 liver cell populations didn’t generate any colony (only two clones in 360 wells of R3 cells, probably due to R4 cell contaminants; not shown). In contrast, purified c-Kitlow(CD45/TER119)– R4 cells generated different colony numbers per group (Table 1). The basal frequencies were 1 in 718, similar to those of SCF- and OSM-enriched cultures (0.9- and 1.2-fold, respectively). The addition of either HGF or FGF-1, however, rescued more clonable progenitors (more than threefold and 1.9-fold, respectively), as did the HGF and OSM (H/O) and HGF, OSM, and FGF-1 (H/O/F) factor combinations. Clone differentiation was markedly promoted by HGF, as in the high cell-density cultures, and by FGF-1. OSM, which also upregulated hepatospecific transcripts, reached lower titers per gene than HGF (not shown). The factor combinations induced the strongest differentiation stimuli: more than 90% of the tested hepatic genes were activated (three to four positive out of five analyzed transcripts per clone). When either the ratios of expressed versus analyzed transcripts per clone and of total expressed genes per clone sample and condition were calculated, the intensity of differentiation stimuli was distributed in the following order: H/O/F > H/O > FGF-1 > HGF > OSM > no factor > SCF.
Table 1.
Hepatoepithelial clonable progenitors of the 11-dpc c-Kitdull(CD45/TER119)– R4 embryo liver cell population
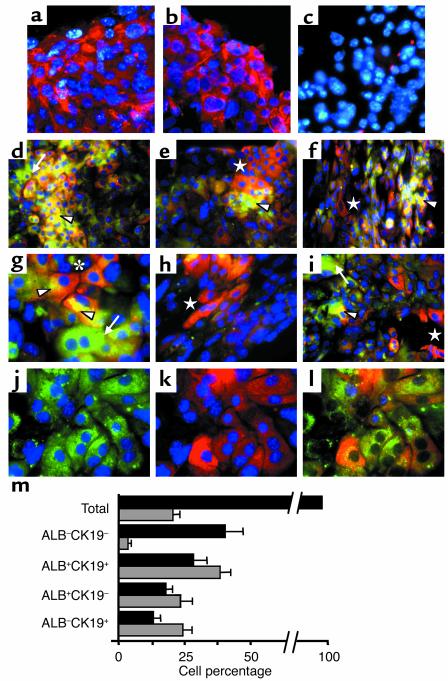
In vitro c-Kitlow(CD45/TER119)– 11-dpc liver cell progenitors self-proliferate and differentiate into mature hepatocytes and bile duct cells. The RT-PCR results suggested that hepatic differentiation programs were activated in the cultured c-Kitlow(CD45/TER119)– R4 embryo cells. To establish whether mature hepatocytes and bile duct cells were generated, we undertook immunofluorescence analyses in the H/O cultures. The hepatospecific CK8 and CK18 cytokeratins were expressed by many of the cultured cells (Figure 4, a–c). Two-color ALB/CK19 analyses revealed four cell types in the cultures: ALB–CK19– (40.2% of total cultured cells), ALB+CK19+ (28.1%), ALB+CK19– hepatocytes (17.8%), and ALB–CK19+ cholangiocytes (13%) (Figure 4, d–h, and m). Cell subsets aggregated in clusters, with ALB+CK19+ cells frequently located centrally, with hepatocytes and bile duct cells in peripheral segregated areas (Figure 4, d–f). Frequently, the ALB–CK19+ biliary cells appeared as ductlike elongated tracts (Figure 4, f and h). Intracellular distribution of hepatic proteins was not always uniform in ALB+CK19+ cells, with CK19 accumulating in cell-to-cell contact areas and ALB enrichment on the opposite cytoplasm (Figure 4g). Many binucleated cells, most of them hepatocytes, were present. Views from isolated green and red channels, with nuclei in blue (DAPI), are shown (Figure 4, j and k). We also evaluated the proliferation rates of each cell subset by measuring their frequencies of S-phase, Ki-67+ cells (an example is shown in Figure 4j). Twenty percent of all cells were Ki-67+, but were distributed in different proportions per cell subset: 3.5% of ALB–CK19– cells, 38.3% of ALB+CK19+ cells, 23.3% of ALB+CK19– cells, and 24.3% of ALB–CK19+ cells were Ki-67+ (Figure 4m, gray bars). These findings show that embryo c-Kitlow(CD45/TER119)– progenitors differentiate in vitro up to mature hepatic stages, while maintaining high proliferation rates.
Figure 4.
In vitro differentiation of 11-dpc c-Kitlow(CD45/TER119)– R4 cells into distinct hepatic cell populations. The immunofluorescence pictures were obtained in H/O-supplemented cultures (12 days). (a–c) Expression of CK8 and CK18 integrins and control samples stained with primary irrelevant Ab’s. The blue dots are DAPI nuclear signals. (d–i) Two-color ALB/CK19 pictures showing the general topography (d–f) and detailed views (g and i) of the different cell types: ALB–CK19– (only DAPI+, blue), ALB+CK19+ (yellow and orange, arrowheads), ALB+CK19– (green, arrows), and ALB–CK19+ (red, stars). (d–f) ALB+CK19+ cells accumulate in clusters, while ALB+CK19– hepatocytes and ALB–CK19+ cholangiocytes are located in different peripheral areas. (g) Nonuniform intracellular distribution of both CK19 (close to cell-cell contact areas) and ALB (opposite cytoplasm, arrowheads) in two interacting ALB+CK19+ cells (star). (e and h) Cholangiocytes distributed in elongated tracts resembling ducts. (j and k) Green and red channels (with DAPI, blue) of a representative field, and (l) three-color ALB/CK19/Ki-67 staining (blue nuclei correspond to Ki-67+ cells) in the same field. Amplification was ×63 for a–c, g–h, and j; ×40 for i; and ×20 for d–f photomicrographs. (m) Distribution of hepatic cell subsets in the cultures (black bars) and frequencies of S-phase Ki-67+ cells per subset (gray bars) are shown in the bottom graph. The data are means ± SEM of cells counted by three independent observers on 25 different images (approximately 2,500 total cells).
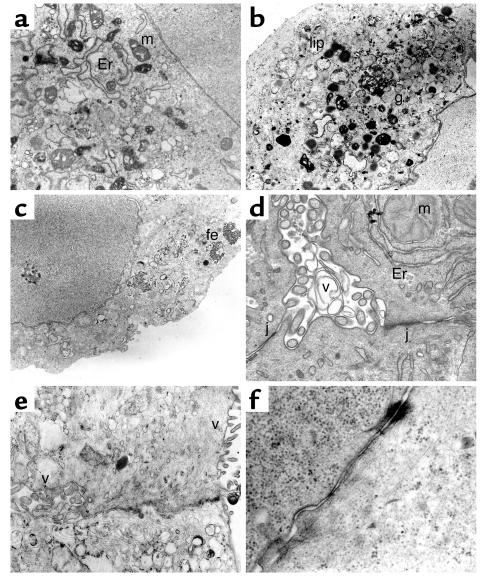
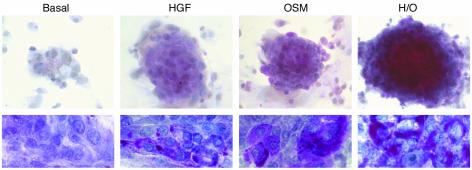
Transmission electron microscopy (TEM) analyses revealed polygonal cells enriched in organelles such as ribosomes, smooth endoplasmic reticulum, and mitochondria, the latter two frequently clustered in complexes (Figure 5, a, b, and d). Lipid vacuoles and granules suggestive of glycogen were detected (Figure 5b). Interestingly, ferritin deposits existed only in the OSM-supplemented cultures (Figure 5c), suggesting that this cytokine might control ferritin expression, directly and/or through IL-6 (37, 38). Small extracellular spaces were suggestive of biliary canaliculi and bile ducts, both enclosed by tight junctions (Figure 5, d and e). In evolved cultures, complexes of intermediate filaments and desmosomes were seen (Figure 5f). The endothelial ductal cells had well-developed microvilli (Figure 5, d and e). Biliary development was absent in 11-dpc liver (refs. 39, 40, and data not shown), suggesting that in vitro differentiation stimuli were stronger than those present in vivo in early liver ontogeny. Glycogen-specific periodic acid-Schiff (PAS) staining confirmed the presence of intracellular glycogen. As seen in Figure 6 for globular aggregates and cell monolayers, the amount of glycogen per cell was higher under H/O than in basal cultures. The greatest increases were due to the addition of OSM, which also induced bigger globular cell clusters.
Figure 5.
TEM analyses of hepatic cells differentiating in vitro. The selected views were obtained in H/O cultures of 11-dpc c-Kitlow(CD45/TER119)– R4 cells, (b and e) except those that were from R3 + R4 cell cultures. (a and b) Cytoplasms enriched in organelles and deposits such as mitochondria, smooth endoplasmic reticulum, lipidic, and glycogen accumuli. (c) Ferritin deposits. (d and e) Examples of a small bile duct and a biliary canaliculus with microvilli and closing tight junctions. (f) Endosome linked to intermediate filaments. Er, endoplasmic reticulum; m, mitochondria; g, glycogen deposits; lip, lipid vacuoles; fe, ferritin; v, microvilli; j, tight junction. The original magnifications were between ×8,000 and ×12,000.
Figure 6.
Intracellular glycogen detection by PAS staining. The upper photographs (×10 magnification) correspond to globular aggregates of 11-dpc R3 + R4 cell cultures, while the bottom pictures (×100 magnification) were obtained in cell monolayers from purified R4 cell cultures, both established as indicated above.
C-Kitlow(CD45/TER119)– liver progenitors engrafted and repopulated cell-depleted 12-dpc liver organoids.
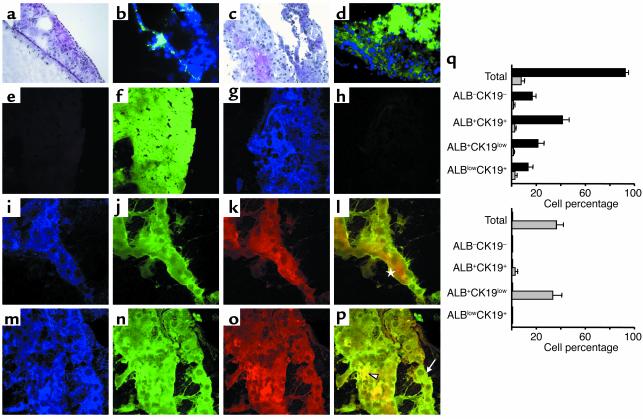
To analyze the developmental potential of the embryo liver progenitors, we designed an organotypic coculture consisting of 12-dpc C57BL/6 liver fragments depleted of cycling cells with 2′-deo and then seeded with c-Kitlow(CD45/TER119)– R4 cells purified from C57BL/6.GFP embryo livers. Livers treated only with control 2′-deo were reduced to small pieces of disrupted stromal and ECM components with a few GFP– cells (Figure 7, a and b, and bottom right gray bars). In contrast, the livers seeded with c-Kitlow(CD45/TER119)– R4 cells showed increased size and cellularity, predominantly of GFP+ cells (Figure 7, c and d). The liver-engrafted GFP+ R4 liver cell population (90% of total liver cells) gave rise to different hepatic cell types: ALB–CK19– (16%) and ALB+CK19+ (41%), as well as the partially matured ALB+CK19low (21%) and ALBlowCK19+ (13%) (Figure 7, e–m, photographs, bottom left columns). Rare mature cells of either ALB+CK19– or ALB–CK19+ phenotype were seen. Among the remaining GFP– cells (10%) of these liver organoids, most were ALB–CK19– cells (8.5% DAPI+ cells). Approximately 20% of the GFP+ cells were cycling (Ki-67+), although we could not quantify the proliferation rates in each GFP+ cell subset. Livers seeded with R3 cells included only a minor population of ALB+CK19+ GFP+ cells (not shown). We conclude that the embryonic c-Kitlow(CD45/TER119)– liver progenitors not only self-proliferate and differentiate in hepatic cultures, but also repopulate the hepatic compartments of cell-deficient livers.
Figure 7.
Chimeric organotypic cultures of purified 11-dpc c-Kitlow(CD45/TER119)– R4 cells obtained from C57BL/6.GFP mouse embryos, cocultured with 2′deo-treated, 12-dpc liver fragments from C57BL/6 mice were analyzed after 3–5 weeks, as described in Methods. H&E and direct GFP views of cell depleted-only (a and b) and R4 cell–seeded (c and d) liver organoids. GFP+ cells are in green and GFP– cells show only the blue signal from DAPI (b and d) (×40). (e–p) R4 cell–seeded livers. (e and f) Livers stained with secondary Alexa 647 goat anti-rabbit and rhodamine, goat anti-mouse Ab’s. Merged views of blue and red channels (e) and green channel (f) (×40). (g and h) Livers stained with anti-GFP mAb and revealed with Alexa 647–conjugated mAb. Blue GFP signals (g) and no signals in the merged green and red channels (h) (×60). (i–p) Two representative areas of the liver organoids subjected to GFP/ALB/CK19 staining: GFP+ blue (i and m), ALB+ green (j and m), and CK19+ red channels (k and o), and overlayed green/red signals (l and p) (×60). Arrowhead, ALB+CK19+ cells; arrow, ALB+CK19low cells; star, ALBlowCK19+ cells. (q) Frequencies of both GFP+ (black bars) and GFP– (gray bars) hepatic cell subsets detected in R4 cell–seeded livers (top) and in 2′deo-treated livers (bottom). The results shown are the mean frequencies ± SEM, related to the GFP+ + GFP– total cells recovered from five R4-seeded liver fragments. Cells in 15 different photomicrographs were counted by three independent observers (3,000 total cells).
Discussion
Unlike many somatic systems, which depend on stem cells to replace the losses from cell turnover and pathological damage (41), liver regeneration is initially due to mature, quiescent cells. These cells restart proliferation, maintaining it until normal organ size is achieved (1), or may reconstitute several livers through sequential cell transfers (2, 3). Immature progenitors reconstitute the liver when mature cells become exhausted or their function is blocked (4–6, 42). All these cells are subjected to inducing stimuli of liver microenvironments. The apparently confusing multiplicity of cells and signals giving rise to a liver demands (a) progenitor-specific cell phenotypes defining cell populations and their commitment events and (b) model systems that, while including as many variables as possible, can be subjected to controlled manipulations. The liver progenitors can be divided into embryonic and adult groups: the reconstitution capacity of cells from embryonic sources is much greater than that of adult cells (15–17, 43). The recipient liver also plays an active role in modulating progenitor potentials through intrinsic cell proliferation rates and by the engraftment of progenitors inside lobular compartments (44, 45), two conditions that are difficult to reproduce in culture. We investigated the initial stages of liver development and characterized a 11-dpc liver cell population of c-Kitlow(CD45/TER119)– cells that expressed hepatospecific gene transcripts, had intracytoplasmic ALB, and were the only liver cells to survive in long-term hepatic cultures, where they proliferated and differentiated into hepatocytes and biliary cells. HGF and FGF-1 rescued greater numbers of clonable progenitors from c-Kitlow(CD45/TER119)– cells than did those of basal cultures, and they provided strong differentiation stimuli. OSM collaborated with the previous factors in differentiation and, by itself, activated traits of mature liver stages (G6P and TAT genes and glycogen production). These embryonic cells are reminiscent of those recently described by Suzuki et al. in 13.5-dpc livers (46, 47), although theirs were considered to be c-Kit–. This divergence with our c-Kit+ progenitors may be related to the fact that these authors checked only gene expression in cultured cells and not in ex vivo purified cells. We observed that in vivo c-Kit expression was rapidly shut off in vitro. Also, the c-Kit levels of 11-dpc cells were reduced at 13 dpc, and, finally, the culture conditions differed between the two studies. The developmental potentials of the other subset of c-Kithigh(CD45/TER119)– liver cells remain to be elucidated. They might represent a mixed population, perhaps including “more immature” hepatic progenitors evolving from c-Kithigh to c-Kitlow cells, as happens in other cell lineages. The c-Kit expression of our embryonic progenitors reproduces that of adult oval cells (9, 48). Other hematopoietic markers, however, were undetected in the embryonic c-Kitlow(CD45/TER119)– cells (7, 8, 10). Specific traits such as Dlk (49), low ASPG-R (50), indocyanine green uptake (51), PRAJA-1 (52), etc., have been ascribed to fetal liver multipotent progenitors that are characterized as highly proliferating cells. This latter cell behavior is characteristic of transient stages where population expansion occurs, but not of bona fide stem cells that are usually quiescent. Recently, cells from extrahepatic sources have been induced to generate mature hepatic cells. Of these, embryonic stem cells (51, 53, 54), hematopoietic progenitors from bone marrow and cord blood (55–58), and the so-called multipotent adult progenitor cells (59) are the best studied. Although interlineage transdifferentiation mechanisms may be involved in these phenomena, cell fusions with recipient cells are the cause of others (60, 61).
Somatic stem cells maintain their population size throughout the life span, simultaneously generating cell progenies committed to differentiation pathways through asymmetric cell divisions. Their experimental proof consists of the capacity for long-term regeneration of mature cells in primary and secondary recipients. The 11-dpc c-Kitlow(CD45/TER119)– liver cells gave rise to four populations in vitro: ALB–CK19–, ALB+CK19+, ALB+CK19– hepatocytes, and ALB–CK19+ bile duct cells. Significant fractions of S-phase cycling cells (20–30%) were present in these cell subsets, except for the ALB–CK19– cell population, which was more quiescent (3% S-phase cells). The presence of proliferating cells and their persistence after replating suggest the presence of stem cells in the cultures. The intracellular labeling, however, did not allow isolation of viable cell subsets in order to study their potential in secondary replatings. The ALB–CK19– cell subset could be the best candidate to include hepatic stem cells, as suggested by Suzuki et al. (47): they did not express differentiation traits and were highly quiescent. Ex vivo 11-dpc c-Kitlow(CD45/TER119)– cells, however, already expressed intracytoplasmic ALB. Either a tiny ALB– cell subset escaped detection and preferentially expanded in vitro, or the ALB–CK19– cultured cells were the result of dedifferentiation events (62, 63). Stem cell vital tracers are required to resolve this issue. Organ repopulations are approached by in vivo adoptive cell transfers, where the final result is analyzed, but the intermediate pathways and mechanisms are missed. We designed an alternative model of chimeric liver organoids by coculturing GFP+ embryo liver progenitors and 2′deo-treated fetal livers from congenic GFP– mice. The 11-dpc c-Kitlow(CD45/CK19)– cells selectively engrafted the cell-depleted liver fragments and expanded their size and cellularity while proliferating and differentiating into hepatic cell populations. The mature stages were less represented in these organotypic livers than in conventional cell cultures, likely recapitulating better the early liver ontogeny, where differentiation is slower than that forced in the cultures. We conclude that the embryo liver c-Kitlow(CD45/TER119)– cells contained long-term proliferating precursors that gave rise to the hepatic cell lineages. They reconstituted cell-deficient fetal livers in organotypic conditions, suggesting the presence of stem cells. As shown with other organotypic protocols (11, 40, 52), this new chimeric model may reproduce the in vivo liver regeneration by progenitors in a more accessible situation. Furthermore, the chimeric set-up allows segregation of the specific roles of both purified cell subsets and environmental signals in a highly controlled manner.
Acknowledgments
This work has been supported by grants from Comunidad Autónoma de Madrid (08.3/0009/1997), Ministerio de Ciencia y Tecnologia (PM99-0104), and Instituto de Salud Carlos III (01/34-02/34), Spain. S. Minguet, I. Cortegano, P. Gonzalo, D. Melero, and B. de Andrés received fellowships from MCyT, ISCIII, and the Ramón y Cajal Program. The Centro de Biología Molecular Severo Ochoa has institutional support from the Fundación Ramón Areces. We thank M. Rejas and F. Manzarbeitia for help in the analysis of TEM data, C. Sánchez and V. Azcoitia for expert suggestions about immunofluorescence and confocal microscopy, and P. Mason for editorial assistance. GFP mice were generously provided by M. Okabe (Osaka University, Osaka, Japan) (see ref. 29).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: day(s) postcoitus (dpc); 2′-deoxyguanosine (2′deo); α-fetoprotein (AFP); albumin (ALB); oncostatin M receptor (OSMR); tyrosin aminotransferase (TAT); α1-antitrypsin (AAT); glucose-6-phosphatase (G6P); hepatocyte nuclear factor (HNF); transthyretin (TTR); stem cell factor (SCF); cytokeratin (CK); room temperature (rt); transmission electron microscopy (TEM); periodic acid-Schiff (PAS); hematopoietic (Hem); HGF and OSM (H/O); HGF, OSM, and FGF-1 (H/O/F).
References
- 1.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 2.Overturf K, al-Dhalimy M, Ou CN, Finegold M, Gromp M. Serial transplantation reveals the stem-cell-like regenerative potential of adult mouse hepatocytes. Am. J. Pathol. 1997;151:1273–1280. [PMC free article] [PubMed] [Google Scholar]
- 3.Overturf K, et al. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat. Genet. 1996;12:266–273. doi: 10.1038/ng0396-266. [DOI] [PubMed] [Google Scholar]
- 4.Shiojiri N, Lemire JM, Fausto N. Cell lineages and oval cell progenitors in rat liver development. Cancer Res. 1991;51:2611–2620. [PubMed] [Google Scholar]
- 5.Fausto N, Lemire JM, Shiojiri N. Cell lineages in hepatic development and the identification of progenitor cells in normal and injured liver. Proc. Soc. Exp. Biol. Med. 1993;204:237–241. doi: 10.3181/00379727-204-43659. [DOI] [PubMed] [Google Scholar]
- 6.Theise ND, et al. The canals of Hering and hepatic stem cells in humans. Hepatology. 1999;30:1425–1433. doi: 10.1002/hep.510300614. [DOI] [PubMed] [Google Scholar]
- 7.Petersen BE, Goff JP, Greenberger JS, Michalopoulos GK. Hepatic oval cells express the hematopoietic stem cell marker Thy-1 in the rat. Hepatology. 1998;27:433–445. doi: 10.1002/hep.510270218. [DOI] [PubMed] [Google Scholar]
- 8.Omori N, et al. Partial cloning of rat CD34 cDNA and expression during stem cell-dependent liver regeneration in the adult rat. Hepatology. 1997;26:720–727. doi: 10.1002/hep.510260325. [DOI] [PubMed] [Google Scholar]
- 9.Fujio K, Evarts RP, Hu Z, Marsden ER, Thorgeirsson SS. Expression of stem cell factor and its receptor, c-kit, during liver regeneration from putative stem cells in adult rat. Lab. Invest. 1994;70:511–516. [PubMed] [Google Scholar]
- 10.Petersen BE, et al. Mouse A6-positive hepatic oval cells also express several hematopoietic stem cell markers. Hepatology. 2003;37:632–640. doi: 10.1053/jhep.2003.50104. [DOI] [PubMed] [Google Scholar]
- 11.Michalopoulos GK, et al. Morphogenetic events in mixed cultures of rat hepatocytes and nonparenchymal cells maintained in biological matrices in the presence of hepatocyte growth factor and epidermal growth factor. Hepatology. 1999;29:90–100. doi: 10.1002/hep.510290149. [DOI] [PubMed] [Google Scholar]
- 12.Mitaka T, Sato F, Mizuguchi T, Yokono T, Mochizuki Y. Reconstruction of hepatic organoid by rat small hepatocytes and hepatic nonparenchymal cells. Hepatology. 1999;29:111–125. doi: 10.1002/hep.510290103. [DOI] [PubMed] [Google Scholar]
- 13.Matsumoto K, Yoshitomi H, Rossant J, Zaret KS. Liver organogenesis promoted by endothelial cells prior to vascular function. Science. 2001;294:559–566. doi: 10.1126/science.1063889. [DOI] [PubMed] [Google Scholar]
- 14.Nagai H, et al. Differentiation of liver epithelial (stem-like) cells into hepatocytes induced by coculture with hepatic stellate cells. Biochem. Biophys. Res. Commun. 2002;293:1420–1425. doi: 10.1016/S0006-291X(02)00406-0. [DOI] [PubMed] [Google Scholar]
- 15.Sandhu JS, Petkov PM, Dabeva MD, Shafritz DA. Stem cell properties and repopulation of the rat liver by fetal liver epithelial progenitor cells. Am. J. Pathol. 2001;159:1323–1334. doi: 10.1016/S0002-9440(10)62519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dabeva MD, et al. Proliferation and differentiation of fetal liver epithelial progenitor cells after transplantation into adult rat liver. Am. J. Pathol. 2000;156:2017–2031. doi: 10.1016/S0002-9440(10)65074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malhi H, Irani AN, Gagandeep S, Gupta S. Isolation of human progenitor liver epithelial cells with extensive replication capacity and differentiation into mature hepatocytes. J. Cell Sci. 2002;115:2679–2688. doi: 10.1242/jcs.115.13.2679. [DOI] [PubMed] [Google Scholar]
- 18.Le Douarin NM. An experimental analysis of liver development. Med. Biol. 1975;53:427–455. [PubMed] [Google Scholar]
- 19.Houssaint E. Differentiation of the mouse hepatic primordium. I. An analysis of tissue interactions in hepatocyte differentiation. Cell Differ. 1980;9:269–279. doi: 10.1016/0045-6039(80)90026-3. [DOI] [PubMed] [Google Scholar]
- 20.Jung J, Zheng M, Goldfarb M, Zaret KS. Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science. 1999;284:1998–2003. doi: 10.1126/science.284.5422.1998. [DOI] [PubMed] [Google Scholar]
- 21.Zaret KS. Regulatory phases of early liver development: paradigms of organogenesis. Nat. Rev. Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 22.Johnson GR, Moore MA. Role of stem cell migration in initiation of mouse foetal liver haemopoiesis. Nature. 1975;258:726–728. doi: 10.1038/258726a0. [DOI] [PubMed] [Google Scholar]
- 23.Houssaint E. Differentiation of the mouse hepatic primordium. II. Extrinsic origin of the haemopoietic cell line. Cell Differ. 1981;10:243–252. doi: 10.1016/0045-6039(81)90007-5. [DOI] [PubMed] [Google Scholar]
- 24.Godin IE, Garcia-Porrero JA, Coutinho A, Dieterlen-Lievre F, Marcos MA. Para-aortic splanchnopleura from early mouse embryos contains B1a cell progenitors. Nature. 1993;364:67–70. doi: 10.1038/364067a0. [DOI] [PubMed] [Google Scholar]
- 25.Kinoshita T, Miyajima A. Cytokine regulation of liver development. . Biochim. Biophys. Acta. 2002; 1592:303–312. doi: 10.1016/s0167-4889(02)00323-3. [DOI] [PubMed] [Google Scholar]
- 26.Kawamoto H, Ikawa T, Ohmura K, Fujimoto S, Katsura Y. T cell progenitors emerge earlier than B cell progenitors in the murine fetal liver. Immunity. 2000;12:441–450. doi: 10.1016/s1074-7613(00)80196-x. [DOI] [PubMed] [Google Scholar]
- 27.Kina T, et al. The monoclonal antibody TER-119 recognizes a molecule associated with glycophorin A and specifically marks the late stages of murine erythroid lineage. Br. J. Haematol. 2000;109:280–287. doi: 10.1046/j.1365-2141.2000.02037.x. [DOI] [PubMed] [Google Scholar]
- 28.Minguet, S., et al. 1999. A population of liver stem cells in early mouse liver organogenesis. In Liver development, gene regulation and disease. C. Ciliberto and M. Tripodi, editors. Istituto de Ricerche di Biologia Molecolare P. Angeletti and Universitá degli Studi di Roma “La Sapienza.” Rome, Italy. 82. (Abstr.)
- 29.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. Green mice as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 30.Schaper F, et al. Hepatocyte growth factor/scatter factor (HGF/SF) signals via the STAT3/APRF transcription factor in human hepatoma cells and hepatocytes. FEBS Lett. 1997;405:99–103. doi: 10.1016/s0014-5793(97)00167-1. [DOI] [PubMed] [Google Scholar]
- 31.Martínez MJ, et al. Long-lived polyclonal B-cell lines derived from midgestation mouse embryo lymphohematopoietic progenitors reconstitute adult immunodeficient mice. Blood. 2001;98:1862–1871. doi: 10.1182/blood.v98.6.1862. [DOI] [PubMed] [Google Scholar]
- 32.Jenkinson EJ, Franchi LL, Kingston R, Owen JJT. Effect of deoxyguanosin on lymphopoiesis in the developing thymus rudiment in vitro: application in the production of chimeric thymus rudiments. Eur. J. Immunol. 1982;12:583–587. doi: 10.1002/eji.1830120710. [DOI] [PubMed] [Google Scholar]
- 33.Kubota H, Reid LM. Clonogenic hepatoblasts, common precursors for hepatocytic and biliary lineages, are lacking classical major histocompatibility complex class I antigen. Proc. Natl. Acad. Sci. U. S. A. 2000;97:12132–12137. doi: 10.1073/pnas.97.22.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Couvelard A, et al. Expression of integrins during liver organogenesis in humans. Hepatology. 1998;26:839–847. doi: 10.1002/hep.510270328. [DOI] [PubMed] [Google Scholar]
- 35.Marcos MA, et al. Antigenic phenotype and gene expression pattern of lymphohemopoietic progenitors during early mouse ontogeny. J. Immunol. 1997;158:2627–2637. [PubMed] [Google Scholar]
- 36.Pan CJ, Lei KJ, Chen H, Ward JM, Chou JY. Ontogeny of the murine glucose-6-phosphatase system. Arch. Biochem. Biophys. 1998;358:17–24. doi: 10.1006/abbi.1998.0849. [DOI] [PubMed] [Google Scholar]
- 37.Kobune M, Kohgo Y, Kato J, Miyazaki E, Niitsu Y. Interleukin-6 enhances hepatic transferrin uptake and ferritin expression in rats. Hepatology. 1994;19:1468–1475. [PubMed] [Google Scholar]
- 38.Brown TJ, Rowe JM, Liu JW, Shoyab M. Regulation of IL-6 expression by oncostatin M. J. Immunol. 1991;147:2175–2180. [PubMed] [Google Scholar]
- 39.Shiojiri N. The origin of intrahepatic bile duct cells in the mouse. J. Embryol. Exp. Morphol. 1984;79:25–39. [PubMed] [Google Scholar]
- 40.Shiojiri N, Mizuno T. Differentiation of functional hepatocytes and biliary epithelial cells from immature hepatocytes of the fetal mouse in vitro. Anat. Embryol. (Berl.). 1993;187:221–229. doi: 10.1007/BF00195759. [DOI] [PubMed] [Google Scholar]
- 41.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 42.Fausto N, Campbell JS. The role of hepatocytes and oval cells in liver regeneration and repopulation. Mech. Dev. 2003;120:117–130. doi: 10.1016/s0925-4773(02)00338-6. [DOI] [PubMed] [Google Scholar]
- 43.Cantz T, et al. Quantitative gene expression analysis reveals transition of fetal liver progenitor cells to mature hepatocytes after transplantation in uPA/RAG-2 mice. Am. J. Pathol. 2003;162:37–45. doi: 10.1016/S0002-9440(10)63796-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Notenboom RGE, de Boer PAJ, Moorman AFM, Lamers WH. The establishment of the hepatic architecture is a prerequisite for the development of a lobular patterns of gene expression. Development. 1996;122:321–332. doi: 10.1242/dev.122.1.321. [DOI] [PubMed] [Google Scholar]
- 45.Gupta S, et al. Position-specific gene expression in the liver lobule is directed by the microenvironment and not by the previous cell differentiation state. J. Biol. Chem. 1999;274:2157–2165. doi: 10.1074/jbc.274.4.2157. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki A, et al. Flow-cytometric separation and enrichment of hepatic progenitor cells in the developing mouse liver. Hepatology. 2000;32:1230–1239. doi: 10.1053/jhep.2000.20349. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki A, et al. Clonal identification and characterization of self-renewing pluripotent stem cells in the developing liver. J. Cell Biol. 2002;156:173–184. doi: 10.1083/jcb.200108066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsusaka S, et al. Role of c-kit receptor tyrosine kinase in development of oval cells in the rat 2-acetylaminofluorene/partial hepatectomy model. Hepatology. 1999;29:670–676. doi: 10.1002/hep.510290304. [DOI] [PubMed] [Google Scholar]
- 49.Tanimizu N, Nishikawa M, Saito H, Tsujimura T, Miyajima A. Isolation of hepatoblasts based on the expression of Dlk/Pref-1. J. Cell Science. 2003;116:1775–1786. doi: 10.1242/jcs.00388. [DOI] [PubMed] [Google Scholar]
- 50.Ise H, Sugihara N, Negishi N, Nikaido T, Akaike T. Low asialoglycoprotein receptor expression as markers for highly proliferative potential hepatocytes. Biochem. Biophys. Res. Commun. 2001;285:172–182. doi: 10.1006/bbrc.2001.5139. [DOI] [PubMed] [Google Scholar]
- 51.Yamada T, et al. In vitro differentiation of embryonic stem cells into hepatocyte-like cells identified by cellular uptake of indocyanine green. Stem Cells. 2002;20:146–154. doi: 10.1634/stemcells.20-2-146. [DOI] [PubMed] [Google Scholar]
- 52.Monga SP, et al. Expansion of hepatic and hematopoietic stem cells utilizing mouse embryonic liver explants. Cell Transplant. 2001;10:81–99. [PubMed] [Google Scholar]
- 53.Jones EA, Tosh D, Wilson DI, Lindsay S, Forrester LM. Hepatic differentiation of murine embryonic stem cells. Exp. Cell Res. 2002;272:15–22. doi: 10.1006/excr.2001.5396. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto H, et al. Differentiation of embryonic stem cells into hepatocytes: biological functions and therapeutic application. Hepatology. 2003;37:983–993. doi: 10.1053/jhep.2003.50202. [DOI] [PubMed] [Google Scholar]
- 55.Petersen BE, et al. Bone marrow as a potential source of hepatic oval cells. Science. 1999;284:1168–1170. doi: 10.1126/science.284.5417.1168. [DOI] [PubMed] [Google Scholar]
- 56.Lagasse E, et al. Purified hematopoietic stem cells can differentiate into hepatocytes in vivo. Nat. Med. 2000;6:1229–1234. doi: 10.1038/81326. [DOI] [PubMed] [Google Scholar]
- 57.Avital I, et al. Isolation, characterization, and transplantation of bone marrow-derived hepatocyte stem cells. Biochem. Biophys. Res. Commun. 2001;288:156–164. doi: 10.1006/bbrc.2001.5712. [DOI] [PubMed] [Google Scholar]
- 58.Kakinuma S, et al. Human umbilical cord blood as a source of transplantable hepatic progenitor cells. Stem Cells. 2003;21:217–227. doi: 10.1634/stemcells.21-2-217. [DOI] [PubMed] [Google Scholar]
- 59.Schwartz RE, et al. Multipotent adult progenitor cells from bone marrow differentiate into functional hepatocyte-like cells. J. Clin. Invest. 2002;109:1291–1302. doi:10.1172/JCI200215182. doi: 10.1172/JCI15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X, et al. Cell fusion is the principal source of bone-marrow-derived hepatocytes. Nature. 2003;422:897–901. doi: 10.1038/nature01531. [DOI] [PubMed] [Google Scholar]
- 61.Vassilopoulos G, Wang PR, Russell DW. Transplanted bone marrow regenerates liver by cell fusion. Nature. 2003;42:901–904. doi: 10.1038/nature01539. [DOI] [PubMed] [Google Scholar]
- 62.Runge D, Runge DM, Bowen WC, Locker J, Michalopoulos GK. Matrix induced re-differentiation of cultured rat hepatocytes and changes of CCAAT/enhancer binding proteins. Biol. Chem. 1997;378:873–881. doi: 10.1515/bchm.1997.378.8.873. [DOI] [PubMed] [Google Scholar]
- 63.Nishikawa Y, Tokusashi Y, Kadohama T, Nishimori H, Ogawa K. Hepatocytic cells form bile duct-like structures within a three-dimensional collagen gel matrix. Exp. Cell Res. 1996;223:357–371. doi: 10.1006/excr.1996.0091. [DOI] [PubMed] [Google Scholar]