Abstract
Developing protein therapeutics has posed challenges due to short circulating times and toxicities. Recent advances using poly(ethylene) glycol (PEG) conjugation have improved their performance. A PEG-conjugated hemoglobin (Hb), Hemospan, is in clinical trials as an oxygen therapeutic. Solutions of PEG-hemoglobin with two (P5K2) or six to seven strands of 5-kD PEG (P5K6) were studied by small-angle x-ray scattering. PEGylation elongates the dimensions (Hb < P5K2 < P5K6) and leaves the tertiary hemoglobin structure unchanged but compacts its quaternary structure. The major part of the PEG chains visualized by ab initio reconstruction protrudes away from hemoglobin, whereas the rest interacts with the protein. PEGylation introduces intermolecular repulsion, increasing with conjugated PEG amount. These results demonstrate how PEG surface shielding and intermolecular repulsion may prolong intravascular retention and lack of reactivity of PEG-Hb, possibly by inhibiting binding to the macrophage CD163 hemoglobin-scavenger receptor. The proposed methodology for assessment of low-resolution structures and interactions is a powerful means for rational design of PEGylated therapeutic agents.
INTRODUCTION
Poly(ethylene) glycol (PEG) is an inert, synthetic polymer increasingly being used to modify drugs and proteins to enhance stability and retention time and lower toxicity (1,2). For example, covalent attachments of PEG to the surfaces of acetylcholinesterase or interferon-β-1b increase their circulatory residency times (3,4). In parallel, PEG-conjugated hemoglobin (Hb) is being explored for use as a hemoglobin-based oxygen carrier (HBOC). Hemospan is human oxyHb conjugated to an average of six to seven PEGs (5-kD) per Hb tetramer. This product, also referred to in the literature as MP4, is formulated at ∼4 g/dl and has been the subject of biochemical (5–8) and physiologic investigations that have shown it to be a safe and effective oxygen-transport agent in animal models of hemodilution (9,10) and hemorrhage (11,12). Hemospan has successfully completed Phase I (13) and Phase II clinical trials (14), with Phase III trials underway in Europe.
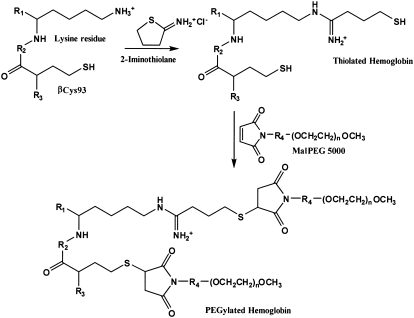
Despite ongoing development of several therapeutic PEG-conjugated proteins, including Hb, the effect of the PEG polymer on the three-dimensional (3D) structure of proteins is poorly understood. The relatively large, highly flexible PEG-polymer conjugates impede crystallization and structural determination by protein crystallography. Thus, considering the substantial amount of biochemical and structural data on Hb, the corresponding PEG-conjugated Hb (PEG-Hb) provides an excellent model to study the general architecture of the PEG envelope and its impact on the 3D structure of the conjugated protein. Accordingly, in this study, we present small angle x-ray scattering (SAXS) solution structures of Hb conjugated to 5-kD maleimide-activated PEG (MalPEG) with either two (P5K2) or an average of six to seven (P5K6) PEGs attached per Hb tetramer. In the case of P5K2, the MalPEGs are conjugated at the two free thiols of Cys-β93, whereas in P5K6, additional MalPEGs are conjugated after reaction at surface-exposed lysines with 2-iminothiolane (Fig. 1).
FIGURE 1.
Schematic representation of the reaction to produce P5K6. In addition to the specific reaction of the two β-subunit Cys-93 residues to produce P5K2, an additional four to five lysine surface residues are modified by thiolation, followed by the maleimide-PEGylation reaction.
MATERIALS AND METHODS
Sample preparation
Hb and P5K6 were prepared as described previously for Hb and P5K6, respectively (5). Another PEG-Hb derivative, P5K2, with two PEGs attached per tetramer was prepared by the same method described for P5K6, except that no thiolation was performed before the reaction with PEG; in this case, the maleimide-PEG reacts only at the two Cys-β93 residues. Reagents and enzymes of the highest purity available were obtained from Sigma Chemicals (St. Louis, MO). Maleimide-activated PEG was provided by NOF (Tokyo, Japan).
SAXS analysis
All samples were equilibrated in Ringer's lactate buffer. Synchrotron radiation scattering data were collected on the X33 beamline of the European Molecular Biology Laboratory, Hamburg Outstation (DORIS III storage ring, DESY) (15). The sample-detector distance was 2.5 m with the wavelength λ = 1.5 Å covering the momentum transfer range 0.01 < s < 0.5 Å−1 (here, s = 4π sin(θ)/λ where 2θ is the scattering angle). Solutions of free and PEGylated Hbs were measured on a MAR345 image plate detector for at least four solute concentrations, each ranging from 2.5 to 31.5 mg/ml. The exposure time was 3 min; to check for radiation damage, two successive 2-min exposures were compared, and no radiation-induced effects were observed. The data were processed and extrapolated to zero-solute concentration, following standard procedures using the program package PRIMUS (16). The forward scattering I(0) and the radius of gyration Rg were calculated by Guinier plot (17). The distance distribution functions and the maximum particle dimension Dmax were evaluated using the program GNOM (18). The molecular masses (MM) of the solutes were estimated by comparison of the I(0) values with that from a reference solution of bovine serum albumin. For the PEG-containing constructs, this calibration was corrected by the difference in the partial specific volumes of Hb (taken as 0.74 cm3/g (19)) and PEG (taken as 0.83 cm3/g (20)). The volumes occupied by the hydrated particles in solution were computed using the Porod invariant (21). The scattering from the atomic models of Hb was calculated using the program CRYSOL (22).
Low-resolution 3D models of Hb, P5K2, and P5K6 were computed ab initio by the program GASBOR (23), which represents a protein by an assembly of dummy residues (DRs) within the search volume with diameter Dmax to fit the experimental scattering data Iexp(s) by minimizing discrepancy:
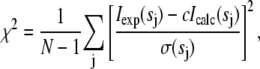 |
(1) |
where N is the number of experimental points, c is a scaling factor, and Icalc(sj) and σ(sj) are the calculated intensity and the experimental error at the momentum transfer sj, respectively. GASBOR was originally designed for proteins, but it also can be applicable for the PEG chains. The density of PEG5000 is ∼1.2 g/cm3 (20), i.e., its electron density (∼0.4 e/Å3) is not significantly different from that of Hb (∼0.43 e/Å3). The models of Hb were built from 574 residues, as in its sequence. For P5K2 and P5K6, 664 and 842 residues were used, respectively, to account for the extra MM (10 and 30 kDa, respectively) of PEG chains in these constructs. The overall shapes of the PEGylated models were not sensitive to ±5% alterations of the number of residues in the model. The shapes of the DR models were aligned to the crystal structure of the most recently refined model of oxyHb (Protein Data Bank (PDB) entry code 2DN1) using the program Supcomb (24).
In an alternative approach, an ab initio model highlighting Hb and PEG moieties was generated by a multiphase bead modeling program MONSA (25,26). This program describes the model by an assembly of small beads densely packed with packing radius 2.5 Å inside a spherical search volume with Dmax = 130 Å as for P5K6. The beads may belong either to solvent or to any of three phases (structural components): i), Hb, ii), two PEG chains in P5K2; iii), the remaining PEG chains in P5K6. This model accounts for the difference between the contrasts of Hb and PEG (the latter is taken to be 0.80 of the former). Starting from a random assignment of the phases, simulated annealing was employed to find which bead belongs to which phase by simultaneous fitting of the three scattering curves from Hb, P5K2, and P5K6. Before the MONSA fitting, appropriate constants were subtracted from each data point of these scattering curves to force the s−4 decay of the intensity at higher angles, following the Porod's law for homogeneous particles (21) as required for the bead modeling assuming uniform density inside each phase. Subject to this uniformity requirement, a limited portion of the scattering data up to s = 0.37 Å−1 (resolution of ∼17 Å) was accounted for in MONSA analysis.
Both GASBOR and MONSA were run without symmetry constraints and using P222 symmetry restrictions, and these runs yielded similarly elongated models for the PEGylated Hb. In symmetric runs, the PEG chains protruded toward both sides of Hb (see Figs. 4 and 5), whereas in asymmetric runs models could be also obtained where the PEG chains protruded toward one side of Hb (data not shown). The asymmetric models showed otherwise similar arrangement of PEG chains with respect to Hb, and this ambiguity does not influence the conclusions of the article. Given the isotropic environment of Hb in solution, symmetric models presented in Figs. 4 and 5 appear to be much more plausible (although the presence of asymmetric shapes or a mixture of symmetric and asymmetric shapes cannot be excluded based in the SAXS data).
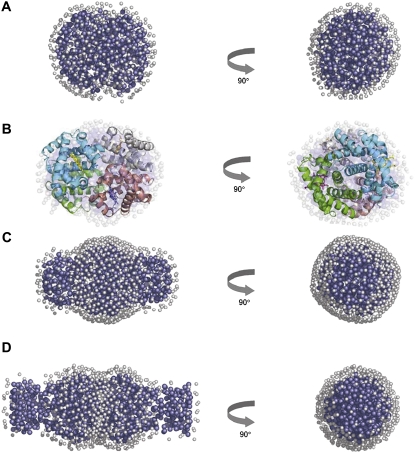
FIGURE 4.
Representative GASBOR models of Hb (A), P5K2 (C), and P5K6 (D). Dark blue beads show the protein and/or PEG dummy residues, light gray beads represent the dummy waters imitating the solvation shell. Structural alignment of the Hb model (shown as a transparent surface) to the crystal structure of oxyHb (shown as a ribbon model) (B).
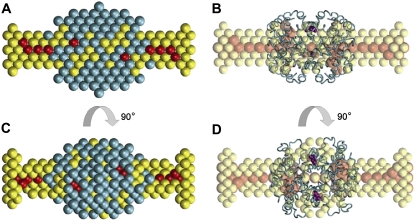
FIGURE 5.
Low-resolution multiphase model of PEGylated Hb constructed ab initio using the program MONSA. Cyan beads belong to Hb, red beads belong to the two PEG chains in P5K2, and yellow beads are the rest of the four to five PEG chains in P5K6; bead radius, 2.5 Å (A and C). In the right panel (B and D), the Cα chains in oxyHb (PDB entry 2DN1) were positioned by Supcomb (24) to best match the beads depicting the Hb volume (the latter are not displayed for clarity). The Cys-β93 residues are shown in the crystal structure in purple. Also in panels B and D, the red beads were made larger to better see them; in panels A and C, they are of equal size and thus masked by yellow beads. The bottom view is rotated counterclockwise by 90° around the vertical axis.
RESULTS
Overall parameters from SAXS
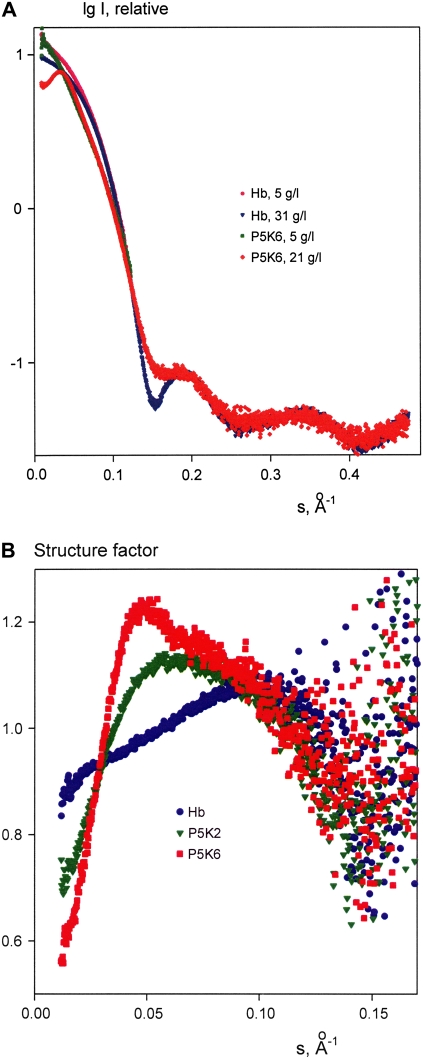
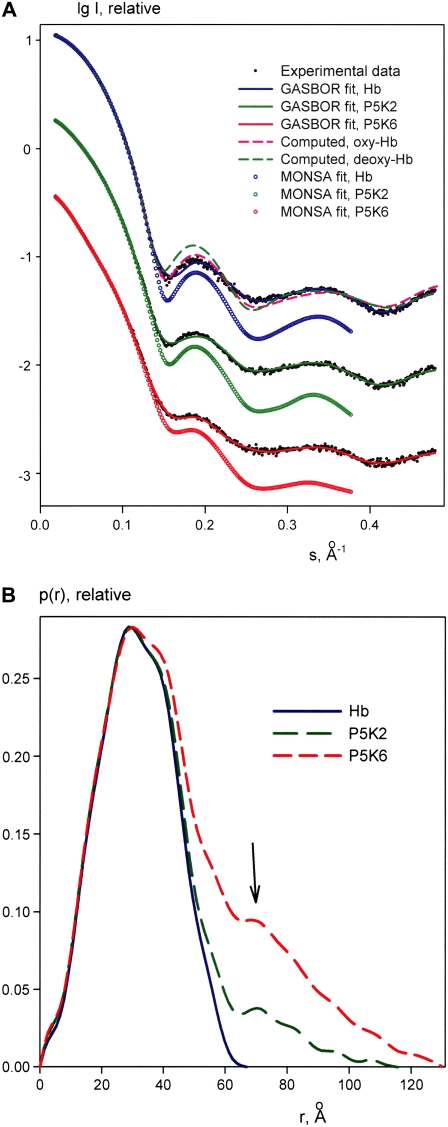
The processed scattering patterns from PEGylated Hb reveal strong repulsive interactions between particles in solution, which manifest as interference peaks at small angles in solutions at higher solute concentration. This is illustrated in Fig. 2 A, comparing Hb and P5K6 data recorded at low and high solute concentrations. The structure factors of Hb, P5K2, and P5K6 for the highest measured concentrations (computed as ratios to the scattering at the lowest concentration) are presented in Fig. 2 B. Although the highest concentration of native Hb (31.5 mg/ml) exceeded those of P5K2 and P5K6 (25 and 21 mg/ml, respectively), the interference effect of Hb is relatively small compared to the PEGylated samples. The average distance between Hb molecules computed from the maximum in the structure factor is ∼75 Å, which is close to the diameter of the crystallographic model of oxyHb (70 Å; PDB entry code 2DN1). The maxima in the structure factors of P5K2 and especially P5K6 are much more pronounced and reveal larger interparticle separations (∼100 and 140 Å, respectively). The strong repulsive interactions thus can be attributed unambiguously to the attached PEG chains.
FIGURE 2.
Scattered intensities of Hb at 5 g/l (pink) and 31 g/l (blue) and of P5K6 at 5 g/l (green) and 21 g/l (red) (A). Structure factors of Hb (blue), P5K2 (green), and P5K6 (red) (B).
Further analysis of the structure of the Hb constructs was performed using the data extrapolated to infinite dilution to exclude the interference effects (Fig. 3 A). The scattering pattern from Hb yields a radius of gyration Rg = 23.7 ± 0.3 Å, in agreement with the value calculated from the crystallographic model of oxyHb (23.5 Å). Moreover, the scattering computed from the latter model fits the experimental data well with χ = 1.4, whereas the fit from deoxyHb (PDB entry code 2DN2) is worse (χ = 1.8; the fits are displayed as dashed lines in Fig. 3 A). This indicates that the PEGylated Hb core adopts the oxy, R-state conformation under our experimental conditions. As expected, the scattering patterns from P5K2 and P5K6 yield larger Rg values (28.3 ± 0.5 Å and 35.6 ± 0.6 Å, respectively) and cannot be fitted by the computed scattering from the oxyHb model (χ = 3.9 and 5.0, respectively). Further, the computed volumes of the hydrated particles are (1.05 ± 0.05) × 105 Å3 for Hb, (1.20 ± 0.05) × 105 Å3 for P5K2, and (1.50 ± 0.10) × 105 Å3 for P5K6, reflecting the volume increase due to the PEG chains. The contribution of the latter becomes even more evident in the distance distribution functions displayed in Fig. 3 B. The maximum particle diameter Dmax = 70 ± 5 Å of Hb coincides with that of the crystal structure of oxyHb (27), whereas P5K2 and P5K6 yield Dmax = 115 ± 5 Å and Dmax = 130 ± 10 Å, respectively. For interatomic distances below 50 Å, which largely reflect the internal structure of Hb, the three p(r) functions nearly coincide, indicating that the protein structure remains intact upon PEGylation. This is further corroborated by the fact that the three experimental scattering patterns from native and PEGylated Hb neatly overlap at higher angles (scattering vectors s > 0.2 Å, i.e., resolution better than 30 Å; see Fig. 2 A) and remain similar to the computed scattering from the atomic model of oxyHb in this angular range. The p(r) functions of the two PEGylated samples display decaying tails due to the PEG contribution. Interestingly, both functions of P5K2 and P5K6 reveal a peak at the interatomic distance ∼75 Å (indicated by an arrow in Fig. 3 B), just 5 Å beyond the diameter of Hb, which indicates that a portion of PEG chains is bound on the surface of Hb. However, a significant amount of PEG chains moves away from the surface, leading to a noticeable increase in the particle size upon PEGylation.
FIGURE 3.
Processed experimental scattering data (from top to bottom: Hb, P5K2, and P5K6), and the scattering calculated from the models. Solid lines and open circles are the fits by ab initio models from GASBOR and MONSA, respectively; dashed lines are scattering patterns computed from the crystallographic models of oxyHb or deoxyHb. The curves are displaced along the abscissa axis for clarity (A). Distance distribution functions of Hb (blue), P5K2 (green), and P5K6 (red); the arrow marks the maximum appearing in the PEG-containing constructs at r = 75 Å (B).
Ab initio structure determination
The low-resolution structures of Hb, P5K2, and P5K6 were further elucidated ab initio using the program GASBOR. First, the shape of Hb was reconstructed in several independent runs, without symmetry or by imposing P2 and P222 symmetry restrictions. All these reconstructions yielded models that neatly fit the experimental scattering from Hb (χ = 1.05; Fig. 3 A), which are also consistent with the crystallographic structure of oxyHb (see the overlap of the typical GASBOR model in P222 symmetry with the PDB entry 2DN1 in Fig. 4, A and B). The models for P5K2 and P5K6 (fits with χ = 1.2 and 1.3, respectively; Fig. 3 A) resulted in elongated structures containing a core with the shape similar to that of oxyHb with two protrusions at opposite poles, apparently representing the PEG chains (see Fig. 4, C and D, for models generated in P222 symmetry). Of great interest from this analysis, the core of PEG-Hb appears to be more compact compared to that of unmodified Hb.
An alternative model of the PEGylated Hb was built using the multiphase ab initio program MONSA by simultaneous fitting of the three available data sets by a bead model showing the three structural components (Hb, Hb plus two PEG chains as in P5K2, and an additional four chains as in P5K6). The typical model constructed using P222 symmetry yields very good fits to the scattering patterns after subtraction of the appropriate Porod constants (as described in Materials and Methods) with χ =1.2, 1.3, and 1.5 from Hb, P5K2, and P5K6, respectively. The MONSA fits are displayed in Fig. 3 A (note that due to the constant subtraction, the computed patterns go below the raw experimental data at higher angles). The model is shown in Fig. 5, A and C, where the colored beads represent volumes occupied by the different structural components. The shape of Hb (cyan beads in panels A and C) is adequately reconstructed, with results similar to those using GASBOR. In Fig. 5, B and D, the Cα chains in oxyHb (PDB entry 2DN1) are positioned to best match the Hb part of the model. The PEG chains display elongated configurations, and the overall MONSA-generated shapes are compatible with those from GASBOR (see Fig. 4). Note that in the MONSA model, the PEG chains of P5K2 appear to protrude to extremities of the model, such that the apparent maximum size of the P5K2 model nearly matches that of P5K6. The few beads of the P5K2 chains in the MONSA model making up the larger distances however make little contribution to the computed scattering, and their presence may simply reflect the flexibility of the PEG chains in P5K2. The experimental value Dmax for P5K2 (115 Å) is clearly smaller than that of P5K6 (130 Å), in agreement with the GASBOR-generated models. Overall, the multiphase bead MONSA modeling yields a lower resolution than GASBOR but distinguishes between Hb and PEG chains and provides direct evidence that PEG is found on the periphery of the Hb volume, perhaps interacting with surface residues. As seen from Fig. 5, B and D, some PEG beads also penetrate into the Hb-occupied volume. Given the low resolution of the model, it is of course impossible to discuss details of localization of these PEG chains, but it appears conceivable that they may occupy the internal cavities between the Hb subunits.
DISCUSSION
In this study, we present SAXS solution structures for Hb and Hb conjugated with two or six to seven copies of 5-kD PEG. This is the first example of a 3D structural determination of a PEG-conjugated hemoglobin, albeit at a low resolution. The most important findings from this study are: i), PEG conjugation does not induce any gross distortion of the tertiary structure of Hb (at this resolution), but its quaternary structure appears compacted; ii), part of the PEG chains enters into cavities between the Hb subunits, part remains near the protein surface, and the rest protrudes away from the surface; iii), PEGylation changes the spherical Hb molecule into an elongated structure with either two or six PEGs conjugated; iv), PEG conjugation leads to repulsion between the PEGylated Hb molecules caused by the PEG layer that surrounds the protein Hb, which is manifested by a marked concentration effect in the SAXS data.
The experimental scattering from Hb is well fitted by the calculated curve from the crystal structure of oxyHb, whereas the deoxyHb crystal structure yields a worse fit. Both P5K2 and P5K6 are conjugated at Cys-β93 adjacent to the proximal His-β92, which is the only residue in the β-subunits of Hb directly coordinated to the heme iron. It seems likely that the attachment of a 5-kD PEG causes local, minor structural distortions of the β-subunit heme pocket that are not detectable at this resolution (∼12 Å). This is supported by studies of heme loss kinetics that are 5 times higher for the β-subunit of P5K6 compared to nonconjugated Hb, whereas the α-subunits show similar kinetics to Hb (8). Consistent with this interpretation, 1H NMR studies of a PEG-conjugated Hb, which is similar in chemical structure to the P5K6 studied here, showed a shift in the proximal histidine of the β- but not α-subunits (28). Moreover, residues in the vicinity of Cys-β93 are located in a conformationally plastic region that is directly linked to the allosteric properties of Hb (29,30).
Modification of Cys-β93 residues with N-ethyl maleimide alone causes a loss in cooperativity, and this effect, along with compaction of the quaternary structure, may explain the apparent loss in cooperativity in Hemospan (5). Quaternary movement could be restricted, giving rise to only partial transition between R and T states or an intermediate quaternary structure. Such an effect has been considered previously to interpret the kinetics of O2 binding to Hemospan (7).
Given that the wide-angle portions of scattering from Hb and PEG-Hb coincide, significant tertiary/secondary structure changes due to PEGylation can be excluded, and our results thus point to a more compact quaternary structure of the PEGylated protein. This can be explained by a partial dehydration of the intersubunit interfaces caused by the PEG chains, which are, rather unexpectedly, partially located in the interior of the PEG-Hbs. The quaternary structure of oxyHb creates a central cavity between the four subunits of hemoglobin. The cavity can accommodate at least 80 water molecules as shown in the high-resolution crystal structure of oxyHb (27). Accordingly, the volume is sufficient for PEG to enter the intersubunit interface and displace water molecules located in the interior of the tetramer. Moreover, PEG is known to cause dehydration and is often used as a cosolvent for protein crystallization. In conjunction with displacement of water molecules located in the central cavity, a general dehydration may contribute to the compact quaternary structure of PEG-Hbs. This conclusion is consistent with molecular dynamic simulations that have shown a correlation between extent of PEGylation and PEG-Hb molecular volume, with a corresponding displacement of water molecules from the Hb structure (31).
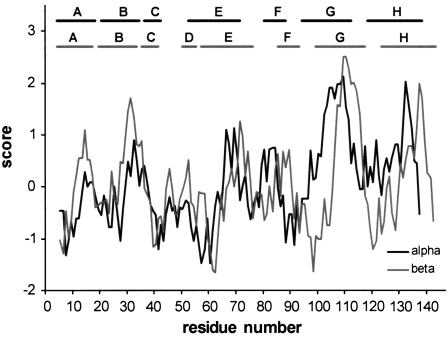
As the major part of PEG chains is located outside Hb, our results agree with earlier studies showing that PEG is preferentially excluded from proteins by steric exclusion (32). Moreover, but in competition with exclusion forces, PEG has been shown to interact with nonpolar protein surface residues (32), and interaction of 5-kD PEG has been reported for hydrophobic clusters on the surface of cytochrome c (33). The hydrophobicity score for Hb was calculated according to Kyte and Doolittle (34) (Fig. 6), showing residue numbers aligned with helical segments. A region of particular interest in possible PEG-Hb interactions is over the relatively hydrophobic G-H helices at the α1β1 packing contacts between dimers. Our models display surface interactions of PEG with Hb, although the resolution is insufficient to define the Hb residues involved or their polarity. Analysis of SAXS spectra recorded at a relatively high protein concentration shows a clear concentration effect for both P5K2 and P5K6, whereas this effect is nearly absent in nonconjugated Hb. As might be expected, the concentration effect is more pronounced for P5K6 than for P5K2, suggesting that the introduction of additional PEG chains results in a better shielding of the Hb molecule, also in agreement with molecular dynamics calculations of theoretical PEG-conjugated Hbs (31).
FIGURE 6.
Hydrophobicity plot for Hb (34). The hydrophobicity score is shown for residue numbers aligned with subunit helical segments.
Intuitively, one would expect the PEG-Hbs to have the same shape as the nonPEGylated Hb, but here we show that the shape of a PEGylated protein is not defined by that of the core protein. The nonspherical solution shape of the PEG-Hbs was not anticipated and provides new information by which to calculate PEG-Hb diffusion constants using the Stokes-Einstein equation. We have reported simulations of oxygen transport by cell-free Hbs, including PEG-Hb, where Hb molecular diffusion was shown to be one critical factor in overall oxygen transport (35). Although the conclusion holds, that study used the simplifying assumption that the Hb molecules were spherical. Since the SAXS solution structure contradicts this assumption, correct molecular dimensions can now be employed to provide more accurate simulations.
Further, although both P5K2 and P5K6 have similar shapes and overall dimensions, the difference in the repulsive effect of the particles in solution provides a highly critical property that may explain some of the positive effects of PEGylated Hbs in vivo. The generality of these observations can be attributed to the attached PEG chains and suggests that the concentration effect is a general consequence of PEG conjugation. Accordingly, in vivo, one may hypothesize that this effect not only prevents intermolecular interactions between adjacent PEG-conjugated proteins but also between conjugated and nonconjugated proteins or even cellular structures.
Several general benefits, such as increased intravascular retention time, decreased immunogenicity, and improved solubility, are frequently associated with the attachment of a PEG polymer to a protein. For instance, attaching a 40-kDa branched PEG polymer to interferon-β-1b markedly improved its circulating half-life from 1.1 h for the nonconjugated protein to 9.4 h for the PEG-conjugated drug; moreover, attachment of PEG dramatically decreased the immunogenicity of the protein and the tendency for aggregation (4). The ability of PEG-Hb to exclude other proteins or cells within its excluded volume is particularly critical in the design of oxygen therapeutics. Hemoglobin-based products are being optimized to extend circulating time, while minimizing or eliminating toxicity. This result is consistent with the extended circulatory half-life of Hemospan, which is ∼20 h in the absence of hemoglobinuria (14), compared to the very rapid renal clearance of unmodified, cell-free Hb (36). Prolongation of Hemospan circulation time may arise if it does not bind to the macrophage CD163 receptor, an Hb scavenger that shows progressively decreasing binding of polymerized Hbs with increasing size (37). Studies with Hemospan in this system are underway. In addition, repulsion of PEG-Hb from other immune cells may be critical to avoid inflammatory responses. Prevailing theories about cell-free hemoglobin-induced vasoconstriction are that the Hb molecules come into close approximation with endothelium (38) or extravasate across the endothelium (39), either of which would make NO scavenging more efficient, thus leading to an increase in vascular tone and negative side effects of hypertension and decreased perfusion (40). It remains speculative at this point, but the repulsive nature of the PEG modification may reduce its interaction with the surface glycocalyx of endothelial cells; thus the repulsive effect provides an explanation for the lessened vasoactivity with Hemospan (10).
These new insights from this SAXS analysis can now be used to design PEG-Hbs for maximal or optimal repulsion effects and particle size and PEG conformations. Intermolecular repulsive forces increase substantially with the total mass of conjugated PEGs with the same polymer length (10 vs. ∼30 kD for P5K2 versus P5K6), an effect that was not predicted by changes in molecular shape and/or dimensions. Thus, solution conformations of the conjugated PEGs, which includes its interaction with Hb, other PEG polymers, and surrounding water layers, must play a large role in the repulsion effect.
Effective PEG lengths are defined by the Flory dimension, RF, which is determined from the effective length of an oxyethylene unit, a = 3.5 Å (41), and the number of units per polymer, N (42–44):
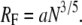 |
The relative dimensions of RF and the distance between PEG grafting sites, DG, on a surface determine the secondary structure of the grafted PEG polymer: for DG > RF, the PEG polymer is able to fold in on itself over the grafted surface, giving a “mushroom” conformation; for DG < RF, the PEGs become extended due to steric interactions between PEG chains to form a “brush” conformation; and at DG ≈ RF, the PEGs are in a “mushroom-to-brush” transition phase (45). For 5-kD PEG, N = 113 and RF ∼ 60 Å. Since the half-maximal circumference DG on the Hb surface from one Cys-β93 to the other on the symmetrically opposite side of Hb is ∼110 Å, DG > RF, thus allowing a mushroomed conformation of the grafted PEG polymers in P5K2. This is reflected in the PEG features from the SAXS structure of P5K2, showing an overall ellipsoidal shape with Dmax = 115 Å. Calculation of the maximal diameter for P5K2 using the Flory dimension for 5-kD PEG gives a molecule with Dmax ∼ 190 Å (i.e., Hb diameter = 70 Å + PEGs, 2 × 60 Å), thus providing an estimation for the mushroom conformation of the PEG chains on P5K2 that fold to ∼60% of their full Flory dimensions.
The more highly PEG-coordinated P5K6 molecule contains the two specific attachment sites (Cys-β93) and an additional four to five more PEG grafted sites. The SAXS solution structure implies that the PEG chains of P5K6 have greater steric interaction to form an even more elongated shape compared to P5K2 (Fig. 4). Using an approximation that the PEG chains are evenly distributed over the surface of Hb, DG is decreased to ∼55 Å, and DG ≈ RF, thus suggesting a more extended PEG conformation but still in a mushroom-to-brush transition. Using the maximal Flory dimension compared to the measured Dmax = 130 Å, the 5-kD PEG chains on P5K6 appear to fold to ∼70% of their full Flory dimension, consistent with an increase in PEG extension in P5K6 compared to P5K2 by steric constraints.
By direct determination of particle size by SAXS combined with Flory analysis, we have predicted that P5K6 is in a mushroom-brush transition and thus was not designed to reach its maximal particle size when the conjugated PEGs are fully extended. New PEG-Hb molecules can be engineered for larger particles, such that DG < RF either by: 1), creating a more highly conjugated Hb with more PEG attachment sites, thus decreasing DG, or 2), increasing PEG length, thus increasing RF. Doubling the size of PEG to 10 kD (i.e., P10Kx, where x is the number of conjugation sites), increases RF to ∼90 Å, which we predict would provide a mushroom conformation in P10K2 but would approach a fuller brush conformation in P10K6. It is, however, unknown how extending PEG polymer lengths alters steric interactions during the PEG-Hb chemical conjugation reaction, and so new designs warrant experimental practice with reaction ratios in conjunction with SAXS studies to verify the mushroom-to-brush transition point.
SUMMARY
In summary, we have demonstrated that SAXS in conjunction with ab initio structure determination methods is a technique suitable for structural studies of PEGylated proteins. Using this approach, we can confirm that the overall tertiary structure of Hb remains intact upon conjugation with two or six to seven PEG chains, but its quaternary structure appears to be compacted, presumably due to the dehydration of the intersubunit interfaces. The reconstructed shapes of Hb, P5K2, and P5K6 illustrate that the PEG chains interact with the core Hb molecule and also protrude to form either a dual mushroom (P5K2) or a dual mushroom-to-brush (P5K6) conformation. SAXS measurements reveal two properties critical for the pharmacodynamic development of PEG-Hbs as oxygen therapeutics: 1), The repulsion effect growing with the degree of PEGylation opens the way to use SAXS for optimization of PEG lengths and sites of attachment to the protein by maximizing the concentration effect. 2), The transition point between PEG mushroom and brush conformations is critical in determining overall particle size and shape. A brush conformation would lead to larger molecular dimensions, which in turn will affect the diffusion behavior of the particle and perhaps its rate of clearance.
Acknowledgments
This work was supported by grant R01 HL 076163 from the National Institutes of Health, National Heart, Lung, and Blood Institute.
K.V., A.M., D.B., and R.W. are employees of Sangart Inc. K.V., A.M., D.B., and R.W. hold stock or stock options in the company. R.W. is President, Chief Executive Officer, and Chairman of the Board of Sangart.
Editor: Jill Trewhella.
References
- 1.Veronese, F. M., and J. Harris. 2002. Introduction and overview of peptide and protein pegylation. Adv. Drug Deliv. Rev. 54:453–456. [DOI] [PubMed] [Google Scholar]
- 2.Morar, A. S. 2006. PEGylation of proteins: a structural approach. BioPharm. Internat. 34–48.
- 3.Kronman, C., O. Cohen, L. Raveh, O. Mazor, A. Ordentlich, and A. Shafferman. 2006. Polyethylene-glycol conjugated recombinant human acetylcholinesterase serves as an efficacious bioscavenger against soman intoxication. Toxicology. 233:40–46. [DOI] [PubMed] [Google Scholar]
- 4.Basu, A., K. Yang, M. Wang, S. Liu, R. Chintala, T. Palm, H. Zhao, P. Peng, D. Wu, Z. Zhang, J. Hua, M.-C. Hsieh, et al. 2006. Structure-function engineering of interferon-β-1b for improving stability, solubility, potency, immunogenicity, and pharmacokinetic properties by site-selective mono-PEGylation. Bioconjug. Chem. 17:618–630. [DOI] [PubMed] [Google Scholar]
- 5.Vandegriff, K. D., A. Malavalli, J. Wooldridge, J. Lohman, and R. M. Winslow. 2003. MP4, a new nonvasoactive PEG-Hb conjugate. Transfusion. 43:509–516. [DOI] [PubMed] [Google Scholar]
- 6.Manjula, B. N., A. Tsai, R. Upadhya, K. Perumalsamy, P. K. Smith, A. Malavalli, K. D. Vandegriff, R. M. Winslow, M. Intaglietta, M. Prabhakaran, J. M. Friedman, and A. S. Acharya. 2003. Site-specific PEGylation of hemoglobin at Cys-93(β): correlation between the colligative properties of the PEGylated protein and the length of the conjugated chain. Bioconjug. Chem. 14:464–472. [DOI] [PubMed] [Google Scholar]
- 7.Vandegriff, K. D., A. Bellelli, M. Samaja, A. Malavalli, M. Brunori, and R. M. Winslow. 2004. Kinetics of NO and O2 binding to a maleimide poly(ethylene glycol)-conjugated human haemoglobin. Biochem. J. 382:183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vandegriff, K. D., A. Malavalli, C. Mann, E. Jiang, J. Lohman, M. A. Young, M. Samaja, and R. M. Winslow. 2006. Oxidation and haeme loss kinetics of poly(ethylene gycol)-conjugated hemoglobin (MP4): dissociation between in vitro and in vivo oxidation rates. Biochem. J. 399:463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winslow, R. M., J. Lohman, A. Malavalli, and K. D. Vandegriff. 2004. Comparison of PEG-modified albumin and hemoglobin in extreme hemodilution in the rat. J. Appl. Physiol. 97:1527–1534. [DOI] [PubMed] [Google Scholar]
- 10.Tsai, A. G., K. D. Vandegriff, M. Intaglietta, and R. M. Winslow. 2003. Targeted O2 delivery by low-P50 hemoglobin: a new basis for O2 therapeutics. Am. J. Physiol. 285:H1411–H1419. [DOI] [PubMed] [Google Scholar]
- 11.Drobin, D., B. T. Kjellström, E. Malm, A. Malavalli, J. Lohman, K. D. Vandegriff, M. A. Young, and R. M. Winslow. 2004. Hemodynamic response and oxygen transport in pigs resuscitated with maleimide-polyethylene glycol-modified hemoglobin (MP4). J. Appl. Physiol. 96:1843–1853. [DOI] [PubMed] [Google Scholar]
- 12.Young, M. A., L. Riddez, B. T. Kjellström, J. Bursell, F. Winslow, J. Lohman, and R. M. Winslow. 2005. MalPEG-hemoglobin (MP4) improves hemodynamics, acid-base status, and survival after uncontrolled hemorrhage in anesthetized swine. Crit. Care Med. 33:1794–1804. [DOI] [PubMed] [Google Scholar]
- 13.Björkholm, M., B. Fagrell, R. Przybelski, N. Winslow, M. Young, and R. M. Winslow. 2005. A phase I single blind clinical trial of a new oxygen transport agent (MP4), human hemoglobin modified with maleimide-activated polyethylene glycol. Haematologica. 90:505–515. [PubMed] [Google Scholar]
- 14.Olofsson, C., T. Ahl, T. Johansson, S. Larsson, P. Nellgård, S. Ponzer, B. Fagrell, R. Przybelski, P. Keipert, N. Winslow, and R. M. Winslow. 2006. A multi-center clinical study of the safety and activity of maleimide-polyethylene glycol hemoglobin (Hemospan®) in patients undergoing major orthopedic surgery. Anesthesiology. 105:1153–1163. [DOI] [PubMed] [Google Scholar]
- 15.Roessle, M. W., R. Klaering, S. Ristau, B. Robrahn, D. Jahn, T. Gehrmann, P. V. Konarev, A. Round, S. Fiedler, C. Hermes, and D. I. Svergun. 2007. Upgrade of the small angle X-ray scattering Beamline X33 at the EMBL Hamburg. J. Appl. Cryst. 40:s190–s194. [Google Scholar]
- 16.Konarev, P. V., V. V. Vokov, A. V. Sokolova, M. H. J. Koch, and D. I. Svergun. 2003. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 36:1277–1282. [Google Scholar]
- 17.Guinier, A. 1939. La diffraction des rayons X aux très petit angles: application à l'étude de phénomènes ultramicroscopique. Ann. Phys. 12:161–167. [Google Scholar]
- 18.Svergun, D. I. 1992. Determination of the regularization parameter in indirect-transform methods using perceptual data. J. Appl. Cryst. 25:495–503. [Google Scholar]
- 19.Mylonas, E., and D. I. Svergun. 2007. Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering. J. Appl. Cryst. 40:s245–s249. [Google Scholar]
- 20.Mallinckrodt Chemicals. 2006. Material Safety Data Sheet: Polyethylene Glycol. http://www.jtbaker.com/msds/englishhtml/p5029.htm. [Online].
- 21.Porod, G. 1982. General theory. In Small-Angle X-ray Scattering. O. Glatter and O. Kratky, editors. Academic Press, London, UK. 17–51.
- 22.Svergun, D. I., C. Barberato, and M. H. J. Koch. 1995. CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 28:768–773. [Google Scholar]
- 23.Svergun, D. I., M. V. Petoukhov, and M. H. J. Koch. 2001. Determination of domain structure of proteins from x-ray solution scattering. Biophys. J. 80:2946–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozin, M. B., and D. I. Svergun. 2001. Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 34:33–41. [Google Scholar]
- 25.Svergun, D. I. 1999. Restoring low-resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76:2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petoukhov, M. V., and D. I. Svergun. 2006. Joint use of small-angle X-ray and neutron scattering to study biological macromolecules in solution. Eur. Biophys. J. 35:567–576. [DOI] [PubMed] [Google Scholar]
- 27.Park, S. Y., T. Yokoyama, N. Shibayama, Y. Shiro, and J. R. Tame. 2006. 1.25 Å resolution crystal structures of human haemoglobin in the oxy, deoxy and carbonmonoxy forms. J. Mol. Biol. 360:690–701. [DOI] [PubMed] [Google Scholar]
- 28.Manjula, B. N., A. G. Tsai, M. Intaglietta, C.-H. Tsai, C. Ho, P. K. Smith, K. Perumalsamy, N. D. Kanika, J. M. Friedman, and S. A. Acharya. 2005. Conjugation of multiple copies of polyethylene glycol to hemoglobin facilitated through thiolation: influence on hemoglobin structure and function. Protein J. 24:133–146. [DOI] [PubMed] [Google Scholar]
- 29.Perutz, M. F. 1970. Stereochemistry of cooperative effects in haemoglobin. Nature. 228:726–734. [DOI] [PubMed] [Google Scholar]
- 30.Perutz, M. F., G. Fermi, B. Luisi, B. Shaanan, and R. C. Liddington. 1987. Stereochemistry of cooperative mechanisms in hemoglobin. Acc. Chem. Res. 20:309–321. [DOI] [PubMed] [Google Scholar]
- 31.Prabhakaran, M., B. N. Manjula, and S. A. Acharya. 2006. Molecular modeling studies of surface decoration of hemoglobin by maleimide PEG. Artif. Cells Blood Substit. Immobil. Biotechnol. 34:381–393. [DOI] [PubMed] [Google Scholar]
- 32.Bhat, R., and S. Timasheff. 1992. Steric exclusion is the principal source of the preferential hydration of proteins in the presence of polyethylene glycols. Protein Sci. 1:1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Arellano, H., B. Valderrama, G. Saab-Rincon, and R. Vazquez-Duhalt. 2002. High temperature biocatalysis by chemically modified cytochrome c. Bioconjug. Chem. 13:1336–1344. [DOI] [PubMed] [Google Scholar]
- 34.Kyte, J., and R. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132. [DOI] [PubMed] [Google Scholar]
- 35.Cole, R. H., K. D. Vandegriff, A. J. Szeri, O. Savas, D. A. Baker, and R. M. Winslow. 2007. A quantitative framework for the design of acellular hemoglobins as blood substitutes: implications of dynamic flow conditions. Biophys. Chem. 128:63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winslow, R. M. 1991. Hemoglobin-Based Red Cell Substitutes. Johns Hopkins University Press, Baltimore, MD.
- 37.Schaer, D. J., C. A. Schaer, P. W. Buehler, R. A. Boykins, G. Schoedon, A. I. Alayash, and A. Schaffner. 2006. CD163 is the macrophage scavenger receptor for native and chemically modified hemoglobins in the absence of haptoglobin. Blood. 107:373–380. [DOI] [PubMed] [Google Scholar]
- 38.Lancaster, J. R. 1994. Simulation of the diffusion and reaction of endogenously produced nitric oxide. Biochemistry. 91:8137–8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bucci, E., B. Matheson, H. Kwansa, and R. Koehler. 2006. Development of non-extravasating hemoglobin-based oxygen carriers. In Blood Substitutes. R.Winslow, editor. Elsevier, London, UK. 488–97.
- 40.Winslow, R. M. 2007. Red cell substitutes. Semin. Hematol. 44:51–59. [DOI] [PubMed] [Google Scholar]
- 41.Kenworthy, A. K., D. Hristova, D. Needham, and T. J. McIntosh. 1995. Range and magnitude of the steric pressure between bilayers containing phospholipids with covalently attached poly(ethylene glycol). Biophys. J. 68:1921–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Gennes, P. G. 1976. Scaling theory of polymer adsorption. J. Phys. 37:1445–1452. [Google Scholar]
- 43.Alexander, S. 1977. Adsorption of chain molecules with a polar head. A scaling description. J. Phys. 38:983–987. [Google Scholar]
- 44.Alexander, S. 1977. Polymer adsorption on small spheres. A scaling approach. J. Phys. 38:977–981. [Google Scholar]
- 45.de Gennes, P. G. 1987. Polymers at an interface; a simplified view. Adv. Colloid Interface Sci. 27:189–209. [Google Scholar]