Summary
Background
In pre-mitotic plant cells, the future division plane is predicted by a cortical ring of microtubules and F-actin called the preprophase band (PPB). The PPB persists throughout prophase, but is disassembled upon nuclear envelope break down as the mitotic spindle forms. Following nuclear division, a cytokinetic phragmoplast forms between the daughter nuclei, and expands laterally to attach the new cell wall at the former PPB site. A variety of observations suggest that expanding phragmoplasts are actively guided to the former PPB site, but little is known about how plant cells “remember” this site after PPB disassembly.
Results
In pre-mitotic plant cells, Arabidopsis TANGLED fused to YFP (AtTAN::YFP) colocalizes at the future division plane with PPBs. Strikingly, cortical AtTAN::YFP rings persist after PPB disassembly, marking the division plane throughout mitosis and cytokinesis. The AtTAN::YFP ring is relatively broad during preprophase/prophase and mitosis, narrows to become a sharper, more punctate ring during cytokinesis, and then rapidly disassembles upon completion of cytokinesis. The initial recruitment of AtTAN::YFP to the division plane requires microtubules and the kinesins POK1 and POK2, but subsequent maintenance of AtTAN::YFP rings appears to be microtubule-independent. Consistent with the localization data, analysis of Arabidopsis tan mutants shows that AtTAN plays a role in guidance of expanding phragmoplasts to the former PPB site.
Conclusions
AtTAN is implicated as a component of a cortical guidance cue that remains behind when the PPB is disassembled and directs the expanding phragmoplast to the former PPB site during cytokinesis.
Introduction
In plants, where cells are embedded in a matrix of wall material and do not migrate, relative cell positions are permanently established when daughter cells are formed at cytokinesis. Consequently, proper orientation of division planes during development is critical for the cellular organization of plant tissues. Unlike most other eukaryotic cells, the division planes of plant cells are established prior to mitosis. During S or G2 phase of the cell cycle, most plant cells form a cortical ring of microtubules and F-actin called the preprophase band (PPB) at the future division plane as the premitotic nucleus migrates into this plane [1]. The PPB persists throughout prophase, but is disassembled upon nuclear envelope break down as the mitotic spindle forms [1,2]. During cytokinesis, a new cell wall (cell plate) is initiated through the action of the phragmoplast, another F-actin- and microtubule-based structure. The disk-shaped phragmoplast is assembled between daughter nuclei and expands laterally to complete cytokinesis, guiding cell plate attachment at the site formerly occupied by the PPB [3,4].
The observation that PPBs accurately predict the future division plane in a wide variety of plant cell types strongly suggests that the PPB plays a key role in division plane establishment [1,5]. Further supporting this idea, genetic [6] or pharmacological [7] disruption of PPBs causes cells to divide in aberrant orientations. A variety of observations have indicated that some type of cue is present at the former location of the PPB that guides phragmoplast/cell plate expansion to this site [8]. For example, if a spindle is mechanically displaced from the plane defined by the former PPB, phragmoplasts often migrate back to the former PPB site as they expand [9,10]. Furthermore, in some cell types, spindles normally rotate to an oblique orientation during mitosis. When this occurs, the phragmoplast is initially oriented obliquely, but rotates as cytokinesis proceeds so that the cell plate attaches at the former PPB site [11,12]. Thus, the PPB has long been thought to function during prophase to establish a “cortical division site” that guides the expanding phragmoplast during cytokinesis [5,8,13].
Identification of the molecular features of cortical division site has been a longstanding challenge. During mitosis and cytokinesis, the former PPB site is “negatively marked” by local depletion of cortical F-actin [14-16] and a kinesin, KCA1 [7]. The depletion of these two proteins has been proposed to play an important role in the maintenance or function of the cortical division site. A novel microtubule-associated protein, AIR9, colocalizes with PPBs but disappears from the cortex upon PPB disassembly, later reappearing at the cortical division site when the cell plate inserts there [17]. Similarly, two other proteins are localized at the cortical division site or adjacent cell wall just as the cell plate attaches there: RSH, a hydroxyproline-rich glycoprotein [18], and T-PLATE, a protein with domains similar to those of vesicular coat proteins [19]. Thus, AIR9, RSH and T-PLATE are all implicated in cell plate attachment at the cortical division site and/or cell plate maturation. However, proteins that are recruited to the division site by the PPB and remain localized there after PPB disassembly have not been identified.
Previous studies established that the tangled (tan) gene plays an important role in the spatial control of cytokinesis in maize [20]. In tan mutants, cells in all tissues divide in aberrant orientations due to the frequent failure of phragmoplasts to be guided to former PPB sites [12]. The highly basic maize TAN protein binds to microtubules in vitro, but is not closely related to other proteins of known function [21]. Proteins recognized by anti-TAN antibodies were shown by immunofluorescence microscopy to associate with PPBs, spindles, and phragmoplasts in dividing cells and to be present in the cytoplasm throughout the cell cycle [21]. However, these antibodies were not completely specific for TAN [21], apparently also recognizing TAN-related proteins that are likely to be the products of expressed, tan-related genes that were recently identified via EST/cDNA sequencing (e.g., TIGR Gene Indexes TC311793, TC290411, DR791368). Therefore, the intracellular localization of maize TAN has not been definitively determined.
To advance our understanding of TAN localization and function, we investigated the Arabidopsis TANGLED homolog (AtTAN). Here we show that AtTAN::YFP is recruited in a microtubule and kinesin-dependent manner to the cortical division site, where it colocalizes with PPBs and persists at the division site after PPB disassembly, positively marking this site throughout the remainder of the cell cycle. We also present genetic evidence that AtTAN plays a role in phragmoplast guidance. Thus, AtTAN is implicated as a component of the cortical division site that preserves the memory of the PPB throughout mitosis and cytokinesis.
Results
Arabidopsis TANGLED::YFP localizes as a ring at the cortical division site throughout mitosis and cytokinesis
A BLAST search of the Arabidopsis genome revealed a single gene that encodes a protein significantly similar to maize TAN. The full length coding sequence of Arabidopsis TAN (AtTAN; At3g05330) was determined by sequencing of a cDNA clone and RT-PCR products. Like maize TAN (pI 12.6), AtTAN is highly basic, with a pI of 12.3. AtTAN (53 kD) is 29% larger than maize TAN (41 kD) due mainly to internal sequence duplications near the carboxy terminus of the Arabidopsis protein that are not found in maize TAN or its other dicot homologs (Supplementary Figure 1). AtTAN is 35% identical overall to maize TAN; its amino terminal half (245 amino acids) is 48% identical to the corresponding region of maize TAN (Supplementary Figure 1). Analysis of an AtTAN promoter::β-glucuronidase reporter together with Northern blot analysis demonstrated that AtTAN expression correlates with cell division (Supplementary Figure 2), as previously shown for maize tan [21].
To investigate the localization of AtTAN protein, we fused yellow fluorescent protein (YFP) to its carboxy terminus and expressed the fusion protein from either the native promoter or a constitutive CaMV 35S promoter in transgenic plants. In both cases, AtTAN::YFP localized in well-defined, peripheral rings found in a subset of root tip cells. Two distinct classes of rings were observed: broad, diffuse rings (Figure 1A), and sharper, denser rings (Figure 1B). When expressed from the CaMV 35S promoter in transgenic Arabidopsis, maize TAN fused at its carboxy terminus to GFP localized similarly, also forming either broad (Figure 1C) or sharp (Figure 1D) rings in a subset of root tip cells.
Figure 1.
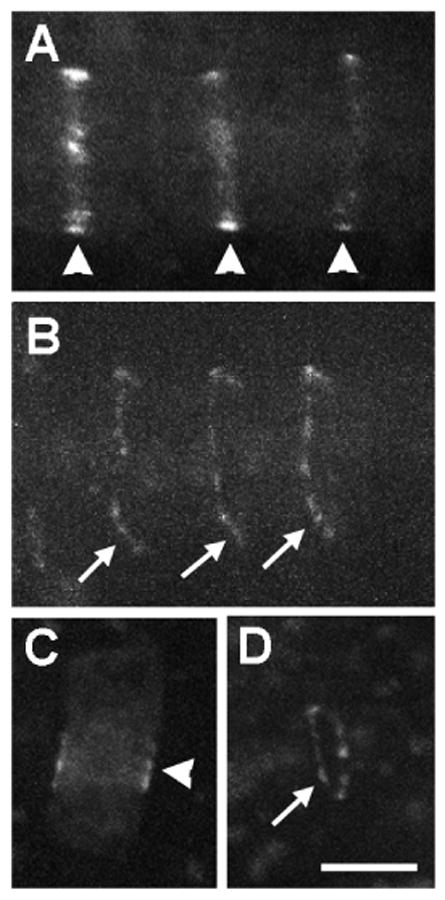
Arabidopsis and maize TAN fluorescent fusion proteins form rings in a subset of root tip cells in transgenic Arabidopsis. Arrowheads point to broad, diffuse rings found in cells with a single nucleus, and arrows point to sharper, denser, more punctate rings found in cells with divided nuclei. A and B, Localization of Arabidopsis TAN::YFP expressed from its native promoter (1.34 kb of genomic sequence upstream of the ATG). C and D, Localization of maize TAN::GFP expressed from the CaMV 35S promoter. Scale bar = 10 μm.
To clarify how AtTAN::YFP rings relate to mitotic microtubule arrays, plants expressing AtTAN::YFP were transformed with a 35S promoter::CFP::α-tubulin (CFP::TUA1) construct [22]. In root tip cells with PPBs, AtTAN::YFP formed a diffuse ring that colocalized precisely with the PPB (n>60; Figure 2A-D). In cells with mitotic spindles, AtTAN::YFP was localized at the future division plane as a diffuse ring of the same width as a PPB-associated ring (n>35; Figure 2E-H). In cells with phragmoplasts, AtTAN::YFP rings were sharper and denser than those in cells with PPBs or spindles, and precisely circumscribed the midplane of the phragmoplast (n>100; Figure 2I-P). These sharp rings were relatively continuous in cells at early stages of cytokinesis (Figure 2I-L), but appeared distinctly punctate at later stages (Figure 2M-P). Analysis of AtTAN::YFP and CFP::TUA1 was focused mainly on root tips where most cell divisions are transverse to the long axis of the root, but we observed similar results in tissues where other patterns of division occur, such as newly emerged cotyledons and leaf primordia (e.g. Figure 5A).
Figure 2.
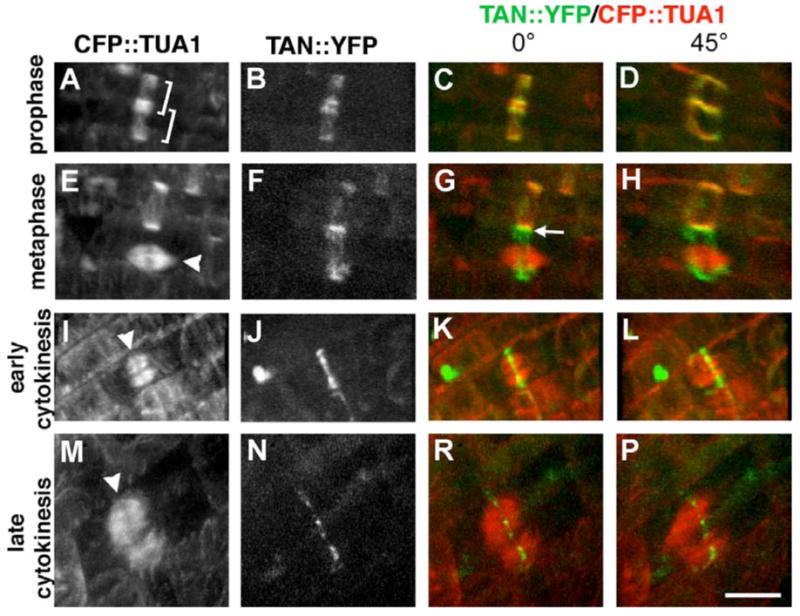
Arabidopsis TAN::YFP localizes as a ring at the division site throughout mitosis and cytokinesis in root tip cells. A-P, Dual localization of 35S-AtTAN::YFP (monochrome in second column, green in third and fourth column) and 35S-CFP::TUA1 (monochrome in first column, red in third and fourth columns) in cells at the indicated cell cycle stages. Brackets in A indicate a pair of adjacent PPBs. Arrowheads point to a metaphase spindle in E, and to phragmoplasts in I and M. Arrow in G points to the junction between a spindle-associated AtTAN::YFP ring and the adjacent PPB. 45° rotations in D, H, and L show that AtTAN::YFP forms a complete ring encircling the cell. Scale bar = 10 μm.
Figure 5.
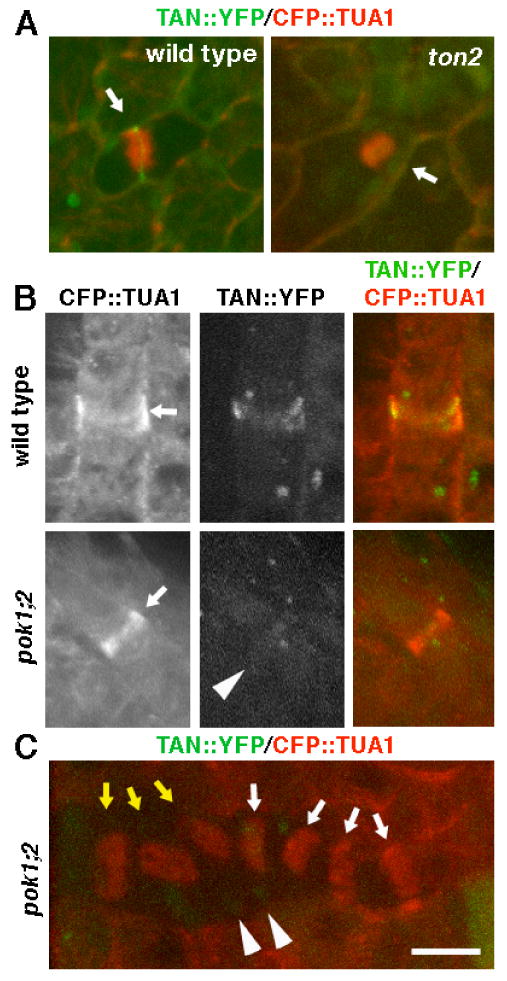
TON2 and POK1/POK2 are required for localization of AtTAN::YFP at the division site. A, Arrows point to cells with phragmoplasts in plants expressing AtTAN::YFP (green) and CFP::TUA1 (red) that are wild-type (left) or ton2 mutant (right). The wild-type cell has a well defined AtTAN::YFP ring and the ton2 mutant cell does not. B, Arrows point to a PPB in a wild-type plant (top) and a pok1;2 double mutant plant (bottom) expressing AtTAN::YFP (monochrome in second column, green in third column) and CFP::TUA1 (monochrome in first column, red in third column). The wild-type cell has a well-defined AtTAN::YFP ring, while the pok1;2 double mutant cell has only a very faint accumulation of AtTAN::YFP (arrowhead). C, pok1;2 double mutant cells with spindles (yellow arrows) and phragmoplasts (white arrows) labeled with CFP::TUA1 (red) lack well defined AtTAN::YFP rings (green). Arrowheads point to faint local accumulations of AtTAN::YFP (compare to wild-type AtTAN::YFP ring in A). Scale bar = 10μm.
Time lapse observations were made to investigate the timing of changes in AtTAN::YFP localization in single cells as they progress through the cell cycle. As illustrated in Figure 3A-E for the cell shown at time zero (metaphase) in Figure 2E-H, broad AtTAN::YFP rings began to narrow as soon as a phragmoplast was initiated (Figure 3C), and sharpened further during the course of phragmoplast expansion to the cell periphery (n=5; Figure 3D-E). Time lapse imaging of cells at later stages of cytokinesis, when phragmoplasts had already expanded to the cell periphery, showed that AtTAN::YFP rings persisted through the completion of cell plate insertion and then disintegrated (n=11). For example, in the cell shown in Figure 3F-I, an expanded phragmoplast initially appeared in an arc across the upper half of the cell, having previously reached the plasma membrane and disassembled in the lower half of the cell; the punctate AtTAN::YFP ring was clearly visible as bright spots around the entire cell equator (Figure 3F). As cytokinesis proceeded (Figure 3G-I), disintegration of AtTAN::YFP puncta followed disassembly of the phragmoplast from the corresponding position at the cell periphery by 10-20 minutes. Low levels of 35S promoter-driven AtTAN::YFP were observed in the cytoplasm of interphase cells, but native promoter-driven AtTAN::YFP was only detectable in cells with cortical rings (data not shown), suggesting cell cycle-regulated accumulation of TAN protein and/or TAN mRNA.
Figure 3.
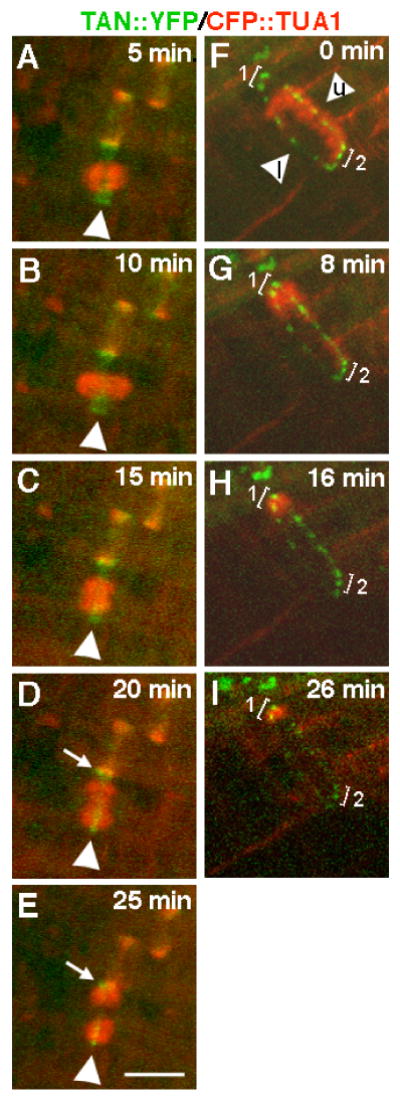
Timelapse analysis of changes in the appearance of AtTAN::YFP rings (green) during cell division with CFP::TUA1 shown in red. A-F, The metaphase cell shown at time zero in Figure 2E-H was subsequently imaged every five minutes as indicated. Arrowheads point to an AtTAN::YFP ring in a cell with a metaphase spindle (A), an elongated spindle (B), a newly initiated phragmoplast (D), an expanded phragmoplast (D), and a more expanded phragmoplast (E). Arrows in D and E point to the edge of a broad, PPB-associated AtTAN::YFP ring in the adjacent cell. F-I, A cell completing cytokinesis was imaged every 8-10 minutes as indicated. In F, a highly punctate, but complete, AtTAN::YFP ring encircles a cell with a phragmoplast that has expanded to the upper face of the cell (arrowhead marked “u”) and has already disassembled at the lower face (arrowhead marked “l”; note that AtTAN::YFP puncta in bracketed areas 1 and 2 are equally bright at time zero). Subsequently, the phragmoplast completes its expansion into the corner marked by bracket 1 while disassembling elsewhere. By 8 minutes (G), the AtTAN::YFP ring has already begun to disintegrate at the lower face of the cell and by 26 minutes (I), has also disintegrated at the upper face and at the area marked by bracket 2 while persisting at the corner marked with bracket 1, where the phragmoplast has not yet disassembled. Scale bar = 10μm.
In summary, AtTAN::YFP forms cortical rings that, together with PPBs, demarcate the future division plane in pre-mitotic cells. Unlike PPBs, however, AtTAN::YFP rings persist at the cortical division site throughout mitosis. The initially broad AtTAN::YFP rings seen in cells with PPBs and spindles narrow down during cytokinesis to become sharper, more punctate rings, which disintegrate shortly after cell plate insertion.
Initial recruitment of Arabidopsis TAN::YFP to the division site requires microtubules
To investigate the role of the PPB in AtTAN::YFP localization at the division site, inhibitor studies were carried out to determine whether AtTAN::YFP localization requires microtubules. 5 μM oryzalin, a microtubule destabilizing drug, was applied to cells that had formed PPBs and these cells were subsequently observed at 5 minute intervals for 10-20 minutes. As shown in Figure 4A, the addition of 5 μM oryzalin caused disassembly of the PPB microtubules within 5 minutes of drug addition, but failed to alter AtTAN::YFP localization during the observation period (n=13). Therefore, short-term maintenance of broad AtTAN::YFP rings that have already formed appears to be largely microtubule-independent.
Figure 4.
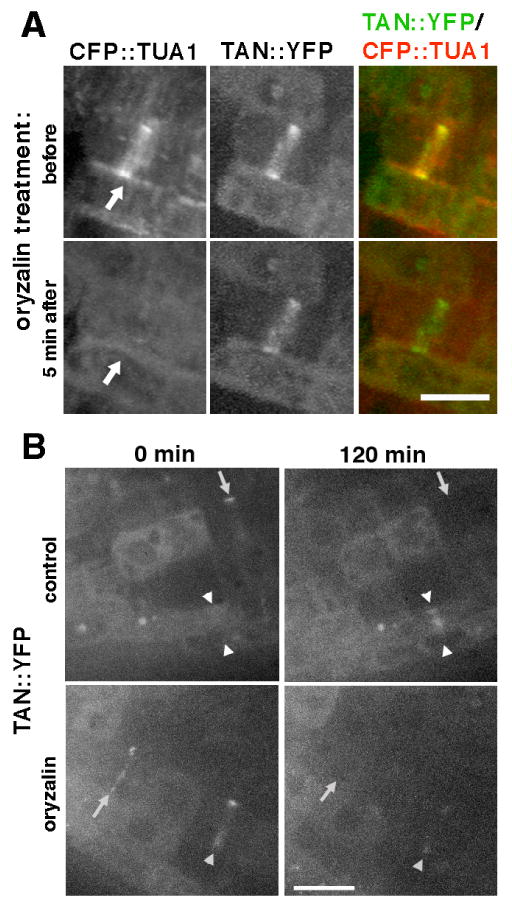
Analysis of the role of microtubules in AtTAN::YFP recruitment and retention at the division site (Z-projections of confocal stacks). A, CFP::TUA1 (monochrome in first column, red in third column) and AtTAN::YFP (monochrome in second column, green in third column) in a cell with a PPB (arrows) before (top) and 5 minutes after (bottom) perfusion of 5 μM oryzalin under the coverslip. The AtTAN::YFP ring persists after disassembly of PPB microtubules. B, AtTAN::YFP rings (monochrome only) observed in root tips treated with 5 μM oryzalin (bottom) or no oryzalin (top). Root tips were examined at 0 minutes (left) and again at 120 minutes (right). Comparison of 0 minute and 120 minute images for the same roots revealed rings that disappeared (arrows) or faded (single arrowhead) during the observation period in the presence or absence of oryzalin, but new rings (i.e., absent at 0 minutes but present at 120 minutes; double arrowheads) formed only in the absence of oryzalin. Scale bar = 10 μm.
Further experiments were conducted to determine whether the initial recruitment of AtTAN::YFP to the division site requires microtubules. Seedlings were mounted on cover slips in the presence or absence of 5 μM oryzalin. Root tips were examined immediately, and again 120 minutes later, for the presence of AtTAN::YFP rings. Time zero and 120 minute images of the same root tips were directly compared to distinguish new rings (those present at 120 minutes but absent at time zero) from older rings. In the absence of oryzalin, 16 of 24 broad rings present at 120 minutes were new (Table 1). Consistent with our earlier finding that AtTAN::YFP rings sharpen late in the cell cycle and disintegrate shortly after completion of cytokinesis, no sharp rings present at time zero remained after 120 minutes in the absence of oryzalin: 12 of 16 were new (Table 1) and the remaining 4 arose from rings that were broad at time zero. Extending the results of the short-term oryzalin treatment experiments described earlier, 11 of 25 broad rings present at time zero were still present after 120 minutes in oryzalin (e.g. single arrowhead, Figure 4B), but no new broad rings formed in the presence of oryzalin (Table 1). As in control roots, all sharp rings present at time zero disappeared during 120 minutes of oryzalin treatment, and no new sharp rings formed (Table 1). Thus, new AtTAN::YFP rings consistently formed in the absence but not in the presence of oryzalin, suggesting that microtubules are required for the initial recruitment of AtTAN::YFP to the division site.
Table 1.
Summary of results for all AtTAN::YFP rings observed in 10 roots of each genotype treated and analyzed as described in the legend to Figure 4B. Broad rings are like those seen in cells with PPBs and spindles (e.g., Figure 2B-D and F-H); sharp rings are like those seen in cells with phragmoplasts (e.g., Figure 2J-L and N-P). Pairs of corresponding control vs. oryzalin values that are significantly different at p<0.001 or p<0.05 (determined via Mann-Whitney U test analysis of results for individual roots, which were combined to obtain the cumulative values shown in the table) are marked with * or †.
| 0 min | 120 min | new | ||
|---|---|---|---|---|
| control | broad | 32 | 24† | 16* |
| sharp | 39 | 16* | 12* | |
| total | 71 | 40* | 28* | |
| oryzalin | broad | 25 | 11† | 0* |
| sharp | 38 | 0* | 0* | |
| total | 63 | 11* | 0* |
p<0.05
p<0.001
Additional experiments were carried out to investigate whether the lack of AtTAN::YFP ring formation in the presence of oryzalin could be due simply to inhibition of cell cycle initiation by oryzalin. Seedlings were treated as before for two hours with or without 5 μM oryzalin, then fixed and stained for nucleic acids to identify cells with condensed chromosomes, indicating that they were in mitosis (Supplementary Figure 3). The frequency of cells with condensed chromosomes was higher in oryzalin treated roots (13.4/root, n=10 roots) than in control roots (3.6/root, n=10 roots), as expected if cells continue to enter the cell cycle during the oryzalin treatment, but are unable to progress through mitosis due to lack of microtubule function. Indeed, microtubule depolymerizing drugs are commonly used as a tool to enrich for mitotic cells because of their ability to block the cell cycle at prometaphase [e.g. 23]. Thus, the failure of AtTAN::YFP rings to form in oryzalin does not appear to be due to inhibition of cell cycle initiation by the drug treatment, supporting the conclusion that initial recruitment of AtTAN::YFP to the division plane is microtubule-dependent. However, inhibition of cell cycle progression through metaphase by oryzalin precludes any conclusions from these experiments regarding the possible role of microtubules in maintaining existing AtTAN::YFP rings after prophase or in sharpening of AtTAN::YFP rings during cytokinesis.
Taking a complementary approach to further investigate the role of PPB microtubules in recruitment of AtTAN to the division plane, we also expressed AtTAN::YFP and CFP::TUA1 in tonneau 2 (ton2) mutants, which fail to form PPBs but do form spindles and phragmoplasts (6,24). AtTAN::YFP rings were observed in 12/12 cells with spindles and 10 /13 cells with phragmoplasts in YFP+ wild-type siblings of ton2 mutants, but were not observed in YFP+ ton2 mutant cells with phragmoplasts (n=10) or spindles (n=9; Figure 5A).
In summary, a combination of pharmacological and genetic experiments strongly supports the conclusion that the initial recruitment of AtTAN::YFP to the cortical division site depends on microtubules, but that microtubules are not required for subsequent retention of broad AtTAN::YFP rings at the cortex once they have formed.
Localization of Arabidopsis TAN::YFP to the division site requires the kinesins POK1 and POK2
We previously described a pair of closely related Arabidopsis kinesins that are required for the spatial control of cytokinesis, POK1 and POK2 [25]. Similar to tan mutants of maize, a high proportion of pok1;2 double mutant phragmoplasts fail to be guided to former PPB sites (25). The COOH terminus of POK1 interacts with AtTAN in the yeast two hybrid system, suggesting that POK1 might interact directly with AtTAN in vivo [25]. To test the hypothesis that these kinesins function in the localization of AtTAN to the division plane, we introduced AtTAN::YFP and CFP::TUA1 into pok1;2 double mutants. In 9 of 11 YFP+ pok1;2 mutant cells with PPBs, AtTAN::YFP rings were either faint or absent (Figure 5B), while the remaining 2 had well-defined AtTAN::YFP rings. None of the 29 YFP+ pok1;2 mutant cells observed with spindles (n=12) or phragmoplasts (n=17) had well-defined AtTAN::YFP rings, although faint, diffuse AtTAN::YFP accumulations were sometimes observed (Figure 5C). By contrast, all cells with PPBs (n=11; Figure 5B), spindles (n=12) and phragmoplasts (n=11) in YFP+ wild-type siblings of pok1;2 mutants had well-defined AtTAN::YFP rings. Therefore, POK1 and POK2 in combination are required for efficient localization of AtTAN::YFP at the division site.
Arabidopsis TANGLED plays a role in phragmoplast guidance in dividing root tip cells
Maize TAN is required for guidance of phragmoplasts to former PPB sites [12]. Phenotypes resulting from mutations in the Arabidopsis TAN gene were examined to determine whether AtTAN also has this function. Plants homozygous for each of three atn alleles with insertions near the beginning of the gene (tan-csh, tan-mad and tan-riken; see Supplementary Data for additional characterization) appeared morphologically normal but exhibited cell pattern defects in root tips, indicative of aberrantly oriented cell divisions (Figure 6A-D). Analysis of microtubule arrays in root tips tan-csh mutants revealed a defect in phragmoplast guidance similar to, but weaker than, that observed previously in tan mutants of maize [12] and pok1;2 double mutants of Arabidopsis [25]. In both wild-type and tan-csh mutant root tip cells, 100% of PPBs were oriented transversely or longitudinally to the cell's long axis (wild-type n=56, tan-csh n=51; Figure 6E,F). In wild-type root tips, 100% of late phragmoplasts (those spanning the full width of the mother cell) were also oriented within 15° of the transverse or londitudinal cell axis (n=54; Figure 6G). By contrast, 9 of 79 late phragmoplasts observed in tan-csh mutant root tips (11%) were tilted ≥15° from the transverse or longitudinal axis of the mother cell (Figure 6H). Thus, in tan-csh mutant root tips, a significant (p < 0.01 by χ2 test) proportion of phragmoplasts were not properly guided to former PPB sites, demonstrating that TAN plays a role in phragmoplast guidance in Arabidopsis roots. The low frequency of aberrantly oriented divisions observed in Arabidopsis tan mutant roots may indicate the existence of a TAN-independent mechanism acting together with TAN to guide phragmoplast expansion in root cells, or may be due to the presence of residual/ truncated TAN proteins that could potentially accumulate in all three mutants analyzed (see Supplementary Data). The aberrant division pattern observed in Arabidopsis tan mutant roots did not alter root growth rate or diameter (Supplementary Data), consistent with earlier findings for maize tan mutants where misoriented divisions in leaves have no impact on leaf shape [20].
Figure 6.
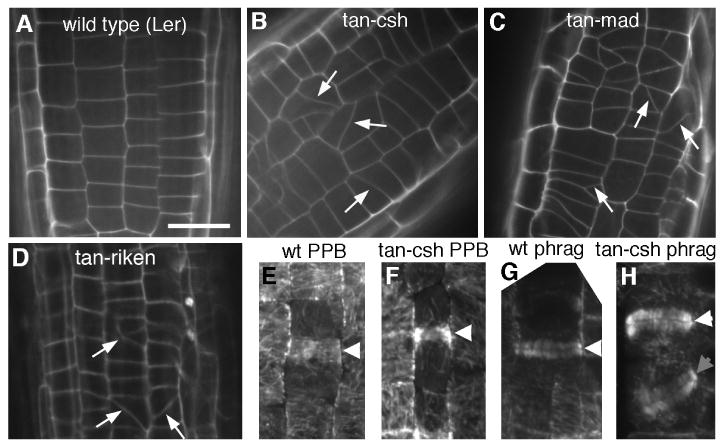
Genetic analysis of AtTAN function. A-D, Root tip of 5-6 day old wild-type (A), tan-csh homozygote (B), tan-mad homozygote (C), and tan-riken homozygote (D). Arrows indicate oblique cell walls indicative of recent, misoriented cell divisions. E-H, White arrowheads point to normal, transversely oriented PPBs (E,F) and phragmoplasts (G,H) observed in wild-type (E,G) and tan-csh mutant root tips (F,H). Gray arrowhead points to oblique phragmoplast observed in tan-csh mutant root tip (H). Scale bar = 10μm.
Discussion
A long-standing hypothesis [5,8,13] holds that the PPB functions to direct the assembly of a cortical division site, which preserves the memory of the division plane following PPB disassembly and interacts with the phragmoplast during cytokinesis to guide cell plate insertion at the former PPB site. Our findings for the localization and function of Arabidopsis TANGLED (AtTAN) fulfill several predictions of this hypothesis. Genetic analysis showed, as previously demonstrated for maize TAN, that AtTAN plays a role in guidance of phragmoplasts to former PPB sites in dividing root cells. AtTAN::YFP colocalizes with PPBs in preprophase/prophase cells and remains localized as a cortical ring after PPB disassembly, positively marking the former PPB site throughout mitosis and cytokinesis. Consistent with the hypothesis that the PPB directs the formation of AtTAN::YFP rings, pharmacological and genetic studies showed that the initial recruitment of AtTAN::YFP to the cortical division site requires microtubules and also requires the kinesins POK1 and POK2. Since most kinesins function as microtubule-based motors, these findings suggest that AtTAN may be a cargo for POK1/2, and that POK1/2 function may be at least partly responsible for microtubule-dependent localization of AtTAN::YFP to the division plane. Thus, POK1/2 may help to tether AtTAN to cortical PPB microtubules during recruitment of AtTAN to the division site, or these kinesins may function to deliver AtTAN to the division site along other microtubules, such as those linking the nuclear surface to the PPB [26-28].
The persistence of broad AtTAN::YFP rings following depolymerization of existing PPB microtubules with oryzalin, and following spontaneous PPB disassembly later in the cell cycle in non drug-treated cells, indicates that once AtTAN::YFP has been recruited to the division site it is retained there in a microtubule-independent manner. Since AtTAN lacks a predicted transmembrane domain, this finding suggests that it may be anchored at the division site through a direct or indirect binding interaction with an unknown transmembrane protein with very limited mobility at the division site. Recent work suggesting that PPB microtubules accomplish their function in marking of the division plane long before the PPB normally disassembles [29] is consistent with the view that microtubule-dependent recruitment of AtTAN to the division site is an important feature of division plane establishment.
Interestingly, as cells make the transition from mitosis to cytokinesis, AtTAN::YFP narrows from an initially broad, diffuse ring to a sharper, denser ring that precisely marks the division plane during cytokinesis. This sharpening process strikingly resembles that observed for the initially broad cortical ring of Mid1p that forecasts the division plane in fission yeast, which becomes a sharper ring at the onset of cytokinesis [30,31]. Mid1p is an anillin-like protein that is unrelated in sequence to TANGLED, and functions to recruit several other proteins that bring about the assembly and contraction of an F-actin based cytokinetic ring. Coalescence of the Mid1p ring has been proposed to be driven by an acto-myosin dependent mechanism [31], but actin is unlikely to be involved in narrowing of the AtTAN ring since cortical F-actin is locally depleted at the cortical division site of plant cells. Thus, in spite of the resemblance of AtTAN to Mid1p in some respects, the function of the AtTAN ring, and the mechanism of its narrowing at the onset of cytokinesis, are most likely quite different. Sharpening of AtTAN rings may refine the definition of the division plane in preparation for cell plate insertion, but further work will be required to establish the functional significance of ring sharpening and to identify the mechanism by which it occurs.
Together, our findings suggest that AtTAN is a component of the guidance cue that has been proposed to remain behind when the PPB is disassembled, and to function during cytokinesis to direct the expanding phragmoplast to the former PPB site. How might AtTAN contribute to phragmoplast guidance? Like maize TAN [21], AtTAN is distantly related to the basic domains of vertebrate adenomatous polyposis coli (APC) proteins. Interestingly, in a variety of dividing cell types in Drosophila, APC homologs that are localized to specific cortical domains appear to interact with EB1 at the plus ends of astral microtubules to orient the mitotic spindle [32-34]. Spindle position then dictates the division plane by directing the site of contractile ring assembly. Recent work has revealed the presence of EB1 at the plus ends of microtubules radiating from the phragmoplast/daughter nuclei to the cell cortex in dividing plant cells [27,28]. Like APC, AtTAN might interact with microtubule plus ends via EB1 to help guide the expanding phragmoplast toward the division site. However, the EB1-binding domain of APC is not present in maize or Arabidopsis TAN, and no obvious similarities to this domain are found in any Arabidopsis proteins. Thus, such a mechanism would require a cryptic EB1 binding domain in AtTAN, or the involvement of one or more as yet unidentified proteins linking AtTAN to EB1. Identification of the mechanism by which AtTAN at the division site interacts with the expanding phragmoplast during cytokinesis is an important goal for future studies.
Experimental Procedures
Primers Used
ATLP: 5′-GATCCGTTACGAAAGTGAACACCTTTATC-3′
ATRP: 5′-ATCTCTTAGGAACCAAAACCGGACGCTGT-3′
ATUP: 5′-TTGGGTTAATCTAGTGAGAA-3′
ATIP: 5′-TAGGTGAGGAAGCAGGAAAC-3′
JL202 (T-DNA border): 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′
Ds5-4: 5′-TACGATAACGGTCGGTACGG-3′
Ds3-4: 5′-CCGTCCCGCAAGTTAAATATG-3′
ATN-6: 5′-GCTTTGGTGAAACCCTGTTGG3′
ATN-7: 5′-GCCAAGTTACTGCCATTTCTCG-3′
ATN-20: 5′-CGAGCTCGGTAGAGTTGAACCAGATGCTCCAG-3′
ATATG: 5′-ATGGTTGCAAGAACCCCACAG-3′
PITA: 5′-GAAGTTTCCTGCTTCCTCACC-3′
NUBQ5: 5′-GGTGCTAAGAAGAGGAAGAAT-3′
CUBQ5: 5′-CTCCTTCTTTCTGGTAAACGT-3′
Growth conditions
For localization studies and examination of Arabidopsis tan mutant phenotypes at seedling stages, seedlings were grown on plates containing 1X Murashige and Skoog basal medium (MP Biomedical), 0.05% MES and 1% agarose or agar. For growth to reproductive maturity, plants were grown on soil. Plates and pots were incubated at 20-22°C on a 16 hours light/8 hours dark cycle.
Construction of full length AtTAN cDNA clone
The entire Arabidopsis TAN coding region was amplified from genomic DNA using ATLP + ATRP and used to probe 106 plaques from a flower bud cDNA library (Ler background; a gift from Detlef Weigel, Max Planck Institute, Tübingen). A 1.2 kb cDNA was isolated, which was not full length. Arabidopsis flower bud RNA was used to amplify, via RT-PCR with primers ATUP + ATIP, a 500 base fragment from the 5′ end of AtTAN, which was ligated into the cDNA clone using an internal BsaMI site to create a full length AtTAN cDNA.
Creation of AtTAN::YFP and maize TAN::GFP transgenic plants
To express a C-terminal AtTAN::YFP fusion protein from the CaMV 35S promoter, the full length AtTAN cDNA (above) minus the stop codon was cloned in frame with YFP in the vector pEZRK-LNY, which confers kanamycin resistance in plants (http://deepgreen.stanford.edu/cell%20imaging%20site%20/html/vectors.html). To express a C-terminal maize TAN::GFP fusion protein from the CaMV 35S promoter, the full length maize TAN cDNA [21] minus the stop codon was cloned in frame with GFP in the vector pEZT-CL, which confers BASTA resistance in plants (http://deepgreen.stanford.edu/cell%20imaging%20site%20/html/vectors.html). To express AtTAN::YFP from its native promoter, primers ATN-7 + ATN-20 were used to amplify a genomic fragment comprising 1.3 kb of sequence upstream of the start codon together with most of the AtTAN coding region (including all 3 introns). The CaMV 35S promoter and most of the AtTAN cDNA were removed from the pEZRK-based AtTAN::YFP construct (above) on a SacI/Bsu361 fragment, and replaced with a SacI/Bsu361 fragment from the AtTAN genomic PCR product. After sequencing to ensure there were no errors in the coding regions of these constructs, they were transformed into Arabidopsis via Agrobacterium-mediated transformation as described previously [35]. YFP/GFP rings were observed via confocal microscopy in several T2 lines for all three constructs. One of the lines expressing CaMV 35S::AtTAN::YFP was transformed with a CaMV 35S-driven CFP::α-tubulin (CFP::TUA1) construct conferring BASTA resistance21 to generate plants co-expressing AtTAN::YFP and CFP::TUA1.
Localization of AtTAN::YFP and CFP::TUA1
AtTAN::YFP and CFP::TUA1 were examined in root tips of 4-5 day old seedlings mounted on glass coverslips in 1X MS liquid media or water using a spinning disk confocal microscope system described previously [22]. YFP was excited with 488 or 514 nm and was viewed with a 525/50 or a 570/65 nm bandpass emission filter, respectively. CFP was visualized with 442 nm excitation and a 480/40 bandpass emission filter. Image integration times varied from 400 to 800 msec. with a Roper Cascade 512b camera using on-chip gain and reading off at 5 Mhz. Z-series were captured with a 63X 1.2 NA water immersion objective at 0.5mm intervals. 2D projections and 3D reconstructions of Z-stacks were produced with either MetaMorph v5.0r7 (Universal Imaging) or ImageJ vs. 1.34s (http://rsb.info.nih.gov/ij/). Further image processing and red-green color merges were carried out using NIH ImageJ vs. 1.34s or Adobe Photoshop v8.0 (Adobe Systems). Only linear adjustments to pixel values were applied. For brightest point projections at angles other than zero, linear interpolation was used between Z planes.
Oryzalin treatments
4-5 day old seedlings expressing AtTAN::YFP and CFP::TUA1 were treated with oryzalin in two different ways. For short term treatments, seedlings were mounted in water and imaged as described above to identify cells with microtubule PPBs/AtTAN::YFP rings. Water was then exchanged with 5 μM oryzalin and cells that had PPBs prior to oryzalin treatment were reanalyzed for YFP and CFP fluorescence at 5 min intervals for 10-20 minutes. For long-term treatments, seedlings were mounted on cover slips coated with 1X MS, 1% agar, and 0.025% methanol with or without 5 μM oryzalin. Root tips were imaged immediately for YFP and CFP fluorescence as described above. After two hours incubation in a humidified box inside a growth chamber, root tips were imaged again for YFP and CFP fluorescence. Images were analyzed to identify all AtTAN::YFP rings present before and after treatment. To analyze mitotic activity at the end of the treatment period, seedlings were fixed (3:1 methanol/acetic acid) and stained for nucleic acids with 1 μM 4′-6-diamidino-2-phenylindole (DAPI) or 10 μg/ml propidium iodide (PI). PI was imaged on the spinning disk confocal microscope system described earlier with 488 nm excitation and a 570-650 nm bandpass emission filter. DAPI was visualized on a Nikon microscope equipped for epifluorescence imaging using a standard DAPI filter cube and a SPOT RT camera (Diagnostics, Inc.) for image acquisition. Images were analyzed to identify cells with condensed chromosomes.
Generation of ton2 and pok1;2 mutants expressing AtTAN::YFP and CFP::TUA1
Plants expressing 35S-AtTAN::YFP and CFP::TUA1 were crossed to ton2-13/+ plants [24] (a gift from Martine Pastuglia, Centre de Versailles), and to both pok1-2;2-2 and pok1-1;2-1 double mutants [25]. In wild-type siblings of ton2 mutant or pok1;2 double mutant F2 progeny, diffuse cytoplasmic YFP fluorescence in interphase cells correlated reliably with the presence of YFP rings in mitotic cells. Thus, diffuse cytoplasmic YFP fluorescence allowed us to positively identify ton2 and pok1;2 mutant individuals expressing AtTAN::YFP even though AtTAN::YFP rings were faint or absent in these plants. YFP + shoot tissue (ton2 and wild-type siblings) or root tips (pok1;2 and wild-type siblings) were scanned for cells with CFP::TUA1-labeled phragmoplasts (all genotypes), spindles (all genotypes), and PPBs (pok1;2 double mutants and wild-type siblings only). Cells with these microtubule arrays were then analyzed for localization of AtTAN::YFP.
Identification and analysis of tan insertion mutants
The tan-mad allele (Ws background) was identified in collaboration with the Arabidopsis Knockout Facility at the University of Wisconsin via a PCR-based approach [36]. PCR genotyping was carried out using primers JL202 + ATRP to amplify the tan-mad mutant allele, and ATRP + ATLP to amplify the wild-type allele. A BLAST search of the Cold Spring Harbor Arabidopsis Genetrap [37] database at http://genetrap.cshl.org/ led to identification of tan-csh (stock CSHL_GT6634; Ler background). PCR genotyping was carried out using Ds5-4 + ATRP or ATIP to amplify the tan-csh mutant allele and ATLP + ATRP to amplify the wild-type allele. A keyword search on At3g05330 of the RIKEN insertion database [38,39] at http://rarge.gsc.riken.jp/dsmutant/keyword.pl resulted in identification of the tan-riken allele (line 13-0229-1, Nossen background). PCR genotyping was carried out using ATN-7 + Ds3-4 to amplify the tan-riken mutant allele and ATLP + ATRP to amplify the wild-type allele. For all three alleles, progeny of multiple homozygous mutants, their homozygous wild-type siblings, and wild-type plants of the same genetic background were examined. Root tips from 4-6 day old seedlings, and unexpanded hypocotyls and petioles from 3 day old seedlings, were stained with 10 μg/ml propidium iodide to fluorescently label cell walls, and imaged via confocal microscopy as previously described [40]. Unexpanded pedicels were examined by scanning electron microscopy as previously described [40]. Immunofluorescent labeling and confocal microscopy analysis of root tip microtubules were carried out as described previously [25].
Northern blots and RT-PCR
RNA was extracted using TRIzol reagent (Invitrogen). For Northern blots poly A+ mRNA was enriched with the PolyATract mRNA isolation kit (Promega). Northern blots were performed as described previously (41) using the full length AtTAN cDNA (described earlier) as a probe. Blots were stripped and reprobed with a β tubulin fragment [42] (a gift from Martin Yanofsky, UCSD) to demonstrate equal loading. For RT-PCR, reverse transcriptase reactions were performed with the RETROscript kit (Ambion). PCR amplification of AtTAN cDNA was then carried out with primers ATUP + ATN-6 or ATATG for reactions spanning the insertions in tan-csh, tan-riken and tan-mad, and ATN-6 + PITA for reactions 3′ of these insertions. As a control, a fragment of the UBIQUITIN5 gene was amplified with primers NUBQ5 and CUBQ5.
GUS staining
GUS activity was visualized in plants containing the tan-csh allele by staining with 125 μg/ml X-Gluc as described previously [43] except that tissue was incubated for 3-5 days at 37° C.
Supplementary Material
Acknowledgments
We thank the Cold Spring Harbor Laboratory Arabidopsis Genetrap Project for tan-csh, the Arabidopsis Knockout Facility at University of Wisconsin for tan-mad, RIKEN BRC for tan-riken, Martine Pastuglia for ton2, Detlef Weigel for the flower bud cDNA library, Suzanne Gerttula, Janine Peroutka, Cole Iliff, and Annette Ho for help with experimental work, and members of the Smith laboratory for helpful discussions and comments on the manuscript. This work was supported by NIH grant GM53137 to L.G.S. and by the Carnegie Institution to D.W.E.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mineyuki Y. The preprophase band of microtubules: its function as a cytokinetic apparatus in higher plants. International Review of Cytology. 1999;187:1–49. [Google Scholar]
- 2.Dixit R, Cyr RJ. Spatio-temporal relationship between nuclear envelope breakdown and preprophase band disappearance in cultured tobacco cells. Protoplasma. 2002;219:116–121. doi: 10.1007/s007090200012. [DOI] [PubMed] [Google Scholar]
- 3.Staehelin LA, Hepler PK. Cytokinesis in higher plants. Cell. 1996;84:821–824. doi: 10.1016/s0092-8674(00)81060-0. [DOI] [PubMed] [Google Scholar]
- 4.Jürgens G. Cytokinesis in higher plants. Annu Rev Plant Biol. 2005;56:281–283. doi: 10.1146/annurev.arplant.55.031903.141636. [DOI] [PubMed] [Google Scholar]
- 5.Smith LG. Plant cell division: Building walls in the right places. Nature Rev Molec Cell Biol. 2001;10:33–39. doi: 10.1038/35048050. [DOI] [PubMed] [Google Scholar]
- 6.Traas J, Bellini C, Nacry P, Kronenberger J, Bouchez D, Caboche M. Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature. 1995;375:676–677. [Google Scholar]
- 7.Vanstraelen M, Van Damme D, De Rycke R, Mylle E, Inze D, Geelen D. Cell cycle-dependent targeting of a kinesin at the plasma membrane demarcates the division site in plant cells. Curr Biol. 2006;16:308–314. doi: 10.1016/j.cub.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Gunning BES. The cytokinetic apparatus: its development and spatial regulation. In: Lloyd CW, editor. The Cytoskeleton in Plant Growth and Development. London: Academic Press; 1982. pp. 229–292. [Google Scholar]
- 9.Ota T. The role of cytoplasm in cytokinesis of plant cells. Cytologia. 1961;26:428–447. [Google Scholar]
- 10.Gunning BS, Wick SM. Preprophase bands, phragmoplasts, and spatial control of cytokinesis. J Cell Sci. 1985;2:S157–S179. doi: 10.1242/jcs.1985.supplement_2.9. [DOI] [PubMed] [Google Scholar]
- 11.Palevitz BA. Division plane determination in guard mother cells of Allium: Video time-lapse analysis of nuclear movements and phragmoplast rotation in the cortex. Dev Biol. 1986;117:644–654. [Google Scholar]
- 12.Cleary AL, Smith LG. The tangled1 gene is required for spatial control of cytoskeletal arrays associated with cell division during maize leaf development. Plant Cell. 1998;10:1875–1888. doi: 10.1105/tpc.10.11.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pickett-Heaps JD, Northcote DH. Organization of microtubules and endoplasmic reticulum during mitosis and cytokinesis in wheat meristems. J Cell Sci. 1966;1:109–120. doi: 10.1242/jcs.1.1.109. [DOI] [PubMed] [Google Scholar]
- 14.Cleary AL, Gunning BES, Wasteneys GO, Hepler PK. Microtubule and F-actin dynamics at the division site in living Tradescantia stamen hair cells. J Cell Sci. 1992;103:977–988. [Google Scholar]
- 15.Liu B, Palevitz BA. Organization of cortical microfilaments in dividing root cells. Cell Motil Cytoskel. 1992;23:252–264. [Google Scholar]
- 16.Hoshino H, Yoneda A, Kumagai F, Hasezawa S. Roles of actin-depleted zone and preprophase band in determining the division site of higher-plant cells, a tobacco BY-2 cell line expressing GFP-tubulin. Protoplasma. 2003;222:157–165. doi: 10.1007/s00709-003-0012-8. [DOI] [PubMed] [Google Scholar]
- 17.Buschmann H, Chan J, Sanchez-Pulido L, Andrade-Navarro MA, Doonan JH, Lloyd CW. Microtubule-associated AIR9 recognizes the cortical division site at preprophase and cell-plate insertion. Curr Biol. 2006;16:1938–1943. doi: 10.1016/j.cub.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 18.Hall Q, Cannon MC. The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell. 2002;14:1161–1172. doi: 10.1105/tpc.010477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Damme D, Coutuer S, De Rycke R, Bouget FY, Inzé D, Geelen D. Somatic cytokinesis and pollen maturation in Arabidopsis depend on TPLATE, which has domains similar to coat proteins. Plant Cell. 2006;18:3502–3518. doi: 10.1105/tpc.106.040923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith LG, Hake SC, Sylvester AW. The tangled1 mutation alters cell division orientations throughout maize leaf development without altering leaf shape. Development. 1996;122:481–489. doi: 10.1242/dev.122.2.481. [DOI] [PubMed] [Google Scholar]
- 21.Smith LG, Gerttula SM, Han S, Levy J. TANGLED1: a microtubule binding protein required for the spatial control of cytokinesis in maize. J Cell Biol. 2001;152:231–236. doi: 10.1083/jcb.152.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- 23.Samuels AL, Giddings TH, Jr, Staehelin LA. Cytokinesis in tobacco BY-2 and root tip cells: a new model of cell plate formation in higher plants. J Cell Biol. 1995;130:1345–1357. doi: 10.1083/jcb.130.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camilleri C, Azimzadeh J, Pastuglia M, Bellini C, Granjean O, Bouchez D. The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell. 2002;14:833–845. doi: 10.1105/tpc.010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller S, Han S, Smith LG. Two kinesins are involved in the spatial control of cytokinesis in Arabidopsis. Curr Biol. 2006;16:888–894. doi: 10.1016/j.cub.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 26.Wick SM. Immunofluorescence microscopy of tubulin and microtubule arrays in plant cells. III. Transition between mitotic/cytokinetic and interphase microtubule arrays. Cell Biol Int Rep. 1985;9:357–371. doi: 10.1016/0309-1651(85)90031-1. [DOI] [PubMed] [Google Scholar]
- 27.Dhonukshe P, Mathur J, Hulskamp M, Gadella TWJ. Microtubule plus-ends reveal essential links between intracellular polarization and localized modulation of endocytosis during division-plane establishment in plant cells. BMC Biol. 2005;3:11. doi: 10.1186/1741-7007-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan J, Calder G, Fox S, Lloyd C. Localization of the microtubule end binding protein EB1 reveals alternative pathways of spindle development in Arabidopsis suspension cells. Plant Cell. 2005;17:1737–1748. doi: 10.1105/tpc.105.032615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcus AI, Dixit R, Cyr RJ. Narrowing of the preprophase microtubule band is not required for cell division plane determination in cultured plant cells. Protoplasma. 2005;226:169–174. doi: 10.1007/s00709-005-0119-1. [DOI] [PubMed] [Google Scholar]
- 30.Wu JQ, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- 31.Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu B, Roegiers F, Jan LY, Jan YN. Adherens junctions inhibit asymmetric division in the Drosophila epithelium. Nature. 2001;409:522–525. doi: 10.1038/35054077. [DOI] [PubMed] [Google Scholar]
- 33.McCartney BM, McEwen DG, Grevengoed E, Maddox P, Bejsovec A, Peifer M. Drosophila APC2 and Armadillo participate in tethering mitotic spindles to cortical actin. Nat Cell Biol. 2001;3:933–938. doi: 10.1038/ncb1001-933. [DOI] [PubMed] [Google Scholar]
- 34.Yamashita YM, Jones DL, Fuller MT. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–1550. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- 35.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 36.Krysan PJ, Young JC, Sussman MR. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell. 1999;11:2283–2290. doi: 10.1105/tpc.11.12.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martienssen RA. Functional genomics: probing plant gene function and expression with transposons. Proc Natl Acad Sci USA. 1998;95:2021–2026. doi: 10.1073/pnas.95.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito T, Motohashi R, Kuromori T, Mizukado S, Sakurai T, Kanahara H, Seki M, Shinozaki K. A new resource of locally transposed Dissociation elements for screening gene-knockout lines in silico on the Arabidopsis genome. Plant Physiol. 2002;129:1695–1699. doi: 10.1104/pp.002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuromori T, Hirayama T, Kiyosue Y, Takabe H, Mizukado S, Sakurai T, Akiyama K, Kamiya A, Ito T, Shinozaki K. A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 2004;37:897–905. doi: 10.1111/j.1365.313x.2004.02009.x. [DOI] [PubMed] [Google Scholar]
- 40.Djakovic SN, Dyachok J, Burke MP, Frank MJ, Smith LG. BRICK1/HSPC300 acts with SCAR and the ARP2/3 complex to regulate epidermal cell shape in Arabidopsis. Development. 2006;133:1091–1100. doi: 10.1242/dev.02280. [DOI] [PubMed] [Google Scholar]
- 41.Luehrsen KB. Northern blotting. In: Walbot V, Freeling M, editors. The Maize Handbook. New York: Springer-Verlag; 1994. pp. 572–574. [Google Scholar]
- 42.Marks MD, West J, Weeks DP. The relatively large beta-tubulin gene family of Arabidopsis contains a member with an unusual transcribed 5′ non-coding sequence. Plant Mol Biol. 1987;10:91–104. doi: 10.1007/BF00016147. [DOI] [PubMed] [Google Scholar]
- 43.Meister RJ, Kotow LM, Gasser CS. SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments. Development. 2002;129:4281–4289. doi: 10.1242/dev.129.18.4281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


