Abstract
Advances in the adaptation of optical spectroscopy to monitor photo-induced or enzyme catalyzed reactions in the crystalline state have enabled x-ray crystal structures to be accurately linked with spectroscopically defined intermediates. This in turn has led to a deeper understanding of the role protein structural changes play in function. The integration of optical spectroscopy with x-ray crystallography is growing, and now extends beyond linking crystal structure to reaction intermediate. Recent examples of this synergy include applications in protein crystallization, x-ray data acquisition, radiation damage and acquisition of phase information important for structure determination.
Introduction
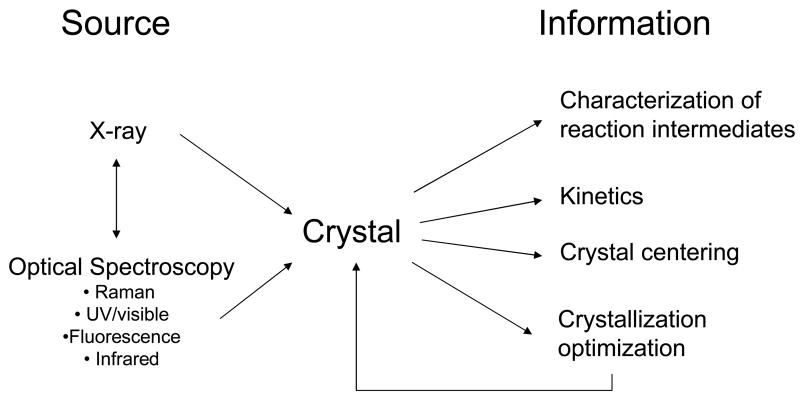
The last two years have brought about significant progress in the development of single crystal microspectrophotometers, and associated methodologies [1•,2]. As these become more widely available, they are being used to find conditions that allow reaction intermediates to accumulate in the crystal for x-ray structure determination, thus challenging the dogma that protein crystals are static entities [3• •,4• •]. Advances in methodology and equipment manufacturing have enabled the incorporation of optical spectroscopy and crystallography into common platforms, thereby increasing our knowledge of how complex biological systems behave in the crystalline state (Figure 1).
Figure 1.
Synergy between single crystal optical spectroscopy and x-ray crystallography.
The most common use of optical spectroscopy in crystallography is to confirm the identity of reaction intermediates trapped within a crystal, and thus directly link a structure to its position along the reaction co-ordinate [5–13• •,14–16•,17•,18,19]. Since spectroscopic techniques can be used to complement each other, researchers are beginning to combine different types of optical spectroscopies to assist in the identification of species contained within the crystal [13• •,14]. As the number and variety of crystal structures of trapped intermediates has increased, interest has also grown in how the crystalline state affects reaction kinetics. Obtaining kinetic data from the reaction occurring in the crystal [17•–19] enables comparison with solution measurements [20–26]. Not surprisingly reaction steps that depend on conformational change often slow down within the crystal, and this can enable key intermediates to accumulate and be trapped by rapid freezing. In the case of photosensitive or redox centers, the need to verify that the crystals still contain the desired intermediate during x-ray exposure and crystallographic data acquisition, has become an important requirement. X-rays are highly oxidizing, and they generate photoelectrons from solvent within the crystals. Therefore, different kinds of microspectrophotometers are being incorporated into x-ray beamlines for direct monitoring of x-ray induced changes [2,14,15,27,28•–31•,32,33].
Recently optical spectroscopy has been incorporated into other areas of crystallography, such as in the automation of protein crystallization and x-ray data acquisition [27,34], assessment of protein damage due to intense x-ray exposure [31•] and acquisition of phase information for structure determination [28•].
This review focuses on recent developments in single crystal optical spectroscopy (Raman, infrared, fluorescent and UV/visible) that complement and enhance x-ray crystallography of protein and DNA.
Kinetics and structures of trapped reaction intermediates
Optical spectroscopy of protein crystals is most often used to confirm the presence of a reaction intermediate for structure determination. Research has focused on redox or light-sensitive proteins where there is a wealth of solution spectroscopic studies to compare to the data generated from the crystalline state. Most times a single spectroscopy technique is sufficient to detect the intermediate [5–12,15,16•,17•–19]. A recent and elegant example involves the use of Raman spectroscopy coupled to a crystal structure of superoxide reductase (SOR) [16•]. SOR is a bacterial non-heme mononuclear iron enzyme that catalyzes the transformation of toxic superoxide anion radical (O2•−) into hydrogen peroxide through a one-electron reduction pathway. Electron transfer is expected to be the first step in the SOR catalytic mechanism to form iron(III)-peroxo, which is then singly protonated to form an iron(III)-hydroperoxo, followed by a second protonation to give the product hydrogen peroxide, which dissociates from the enzyme. Crystals of the mutant enzyme E114A, known to stabilize the iron(III)-peroxo state, were treated with hydrogen peroxide. Raman spectroscopy demonstrated the presence of isotope-sensitive bands at ~567 cm−1 and ~838 cm−1, which fit within the expected range for iron(III)-peroxo species. Surprisingly there were three distinct end-on iron(III)-peroxo conformations within the same asymmetric unit, suggesting to the authors possible sources for the two protons required for product formation. As this study demonstrates, Raman spectroscopy is a particularly powerful technique for identifying small ligands bound to proteins, such as dioxygen species that are key players in aerobic biology.
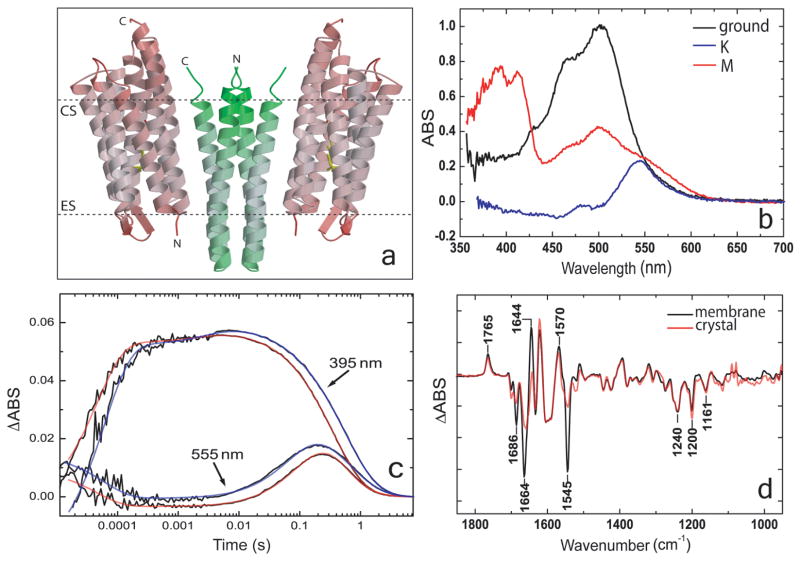
As more single crystal spectroscopy instruments become available, studies are beginning to combine two complementary spectroscopic techniques to investigate reaction intermediates observed in crystals [13• •,14,21,22,24,26]. The power of using multiple spectroscopies is illustrated by a study to understand structural changes that accompany the photocycle of microbial phototaxis receptor sensory rhodopsin II (NpSRII) protein [13• •]. NpSRII is a membrane protein found in the haloarchaeon Natronomonas pharaonis that upon light exposure triggers a signal through its transducer protein NpHtrII to induce photophobic movement in the bacterium. The physiological protein complex, observed both in membrane preparations and crystals, consists of two molecules of NpSRII and two molecules of NpHtrII. The two crystal structures in the study correspond to the K and late M (M2) states of the NpSRII photocycle. Formation of the K intermediate is the initial step following excitation by light, and late M is the signaling state that is translated to the receptor. UV/visible and Fourier-transform infrared (FTIR) spectroscopies were key to identifying the intermediates trapped in the crystals, and for comparison between the photocycle kinetics in the crystals to that in membrane preparations (Figure 2).
Figure 2.
Crystal structure of the NpSRII / NpHtrII complex and its spectroscopic characterization. a, Crystal structure of the complex in the ground state consisting of two molecules of NpSRII (red) with two molecules of NpHtrII (green). The ribbon diagram of the side view of the complex in B-factor coloring: light red and green indicate less mobile; dark red and green indicate mobile. ES, extracellular side; CS, cytoplasmic side. The dotted lines confine the hydrophobic core of the protein. b, Low-temperature (100 K) absorbance spectrum of the complex in crystals: ground state (black), trapped M state (red) and a difference spectrum of trapped K state minus ground state (blue). c, Photocycle kinetics of the complex in crystals and of the complex reconstituted in polar lipids from purple membranes of Halobacterium salinarum in 3 M sodium phosphate pH 5.6 at 25 °C. Traces of transient absorbance difference of the complex at 390 and 555 nm show the time evolution of the M and O states. Raw data are shown in black, and exponential fits are shown in red and blue for the complex in liposomes and in crystals, respectively. d, Difference FTIR spectra of illuminated minus non-illuminated complex in the crystal (red) and reconstituted in liposomes (black) measured at room temperature. Figure and legend reprinted with permission from [13••].
In addition, researchers are now revisiting previously published crystal structures of reaction intermediates. Undertaking kinetic studies in crystals, monitored by spectroscopic techniques, several studies have been able to clarify how the crystal structures fit with solution data [20,23,25,35].
Dealing with adversity
The increased x-ray flux and improved focusing at synchrotron beamlines has enabled protein structures to be determined to high resolution from increasingly small crystals. However, this can lead to increased radiation damage and/or photoreduction of functionally important redox centers in proteins [36]. General radiation damage of protein side-chains, such as decarboxylation of acidic side-chains and oxidation of disulfide bonds, occurs on a relatively slow timescale, and although evident in final electron density maps, it can be tolerated if not in a functionally important part of the protein [37,38]. To counter this, radioprotection using free-radical scavengers has been suggested as a possible means of reducing the rate of radiation damage [32,39•]. A systematic study of radiation damage to thiols and disulfide bonds has been recently carried out, monitored by an on-line UV/visible microspectrophotometer, that suggests ascorbate, 2,2,6,6-tetramethyl-4-piperidone and reduced dithiothreitol might be useful radioprotectants [39•]. X-ray damage to brominated DNA crystals has recently been followed using on-line Raman spectroscopy during synchrotron x-ray data collection [33].
In the case of redox centers, their very nature tends to lead to rapid photoreduction by electrons generated from solvent oxidation [40], giving final electron density dominated by the reduced cofactor [31•]. As these cofactors are at the heart of protein function, this is a serious issue. To counter this problem, researchers now generate composite datasets, either through translation of a single crystal during x-ray exposure or from multiple crystals. If the reduction can be followed spectroscopically, the ideal solution is to simultaneously collect x-ray and spectroscopic data from multiple crystals exposed for a short amount of time to the x-ray beam [29,30,33]. This approach directly tracks the chromophore reduction in each individual crystal, which may vary in thickness and shape leading to different dose rates. A cutoff, either based on reduction rate or maximum x-ray dose, can be selected for each crystal. Only the data prior to the cutoff is then used in the creation of the composite dataset [29–31•]. However, this is complicated by the anisotropic nature of single crystals, and it is often impossible to track redox changes accurately using static on-line microspectrophotometer optics with a rotating crystal [31•]. If reduction is not rapid, the crystal can be rotated back to the starting position for the acquisition of optical spectra at regular intervals during x-ray data collection. However, in the case of rapid reduction of redox centers this is not possible, as once the process reaches a critical point it continues in the absence of x-rays [31•]. Recently, a general strategy has been presented that allows these variables to be accounted for, and has enabled the effective monitoring of the reduction of three separate oxidized species within the same crystal [31•]. An on-line UV/visible microspectrophotometer with static optics was used on the same beamline where experimental x-ray data were collected (Figure 3). Multiple crystals were exposed to x-rays in dummy data collections that exactly duplicated the experimental data collection, only the crystal was not rotated. Although over a 2-fold difference was observed in the time taken, for example, to reach 20% reduction of the Cu(II) ion contained within each crystal, this enabled the selection of a conservative time cutoff for all crystals that was less than the shortest time observed. Using multiple crystals to define an allowable time gave confidence that no oxidized species would be at less than 80% occupancy at any point during x-ray data collection. Radioprotectants may also help in reducing redox center reduction, with nicotinamide, and to a lesser extent sodium nitrate, being found to decrease the rate of heme reduction in myoglobin crystals [32].
Figure 3.
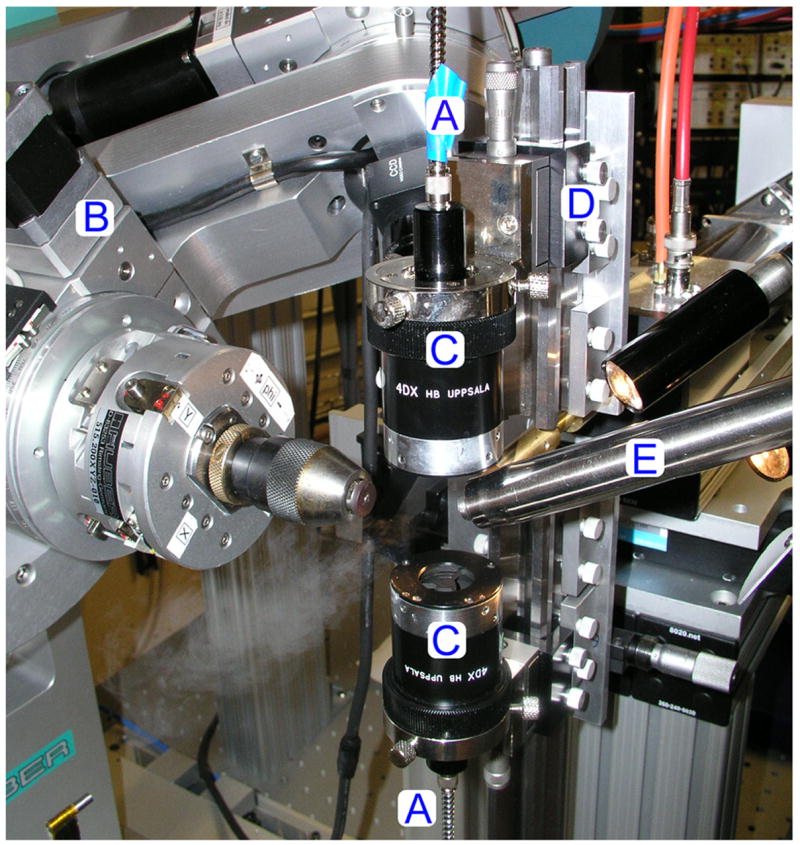
The 4DX Systems microspectrophotometer optics mounted on-line at BioCARS beamline 14-BM-C. A, fiber-optic light guides. B,κ-diffractometer. C, 4DX focusing optics. D, custom mount for 4DX optics. E, cryostream. Figure and legend reprinted with permission from [31•].
A positive outcome of this process is that photoreduction can sometimes be used to generate and solve the structure of a reduced species. Kühnel et al. used photoreduction to their advantage in chloroperoxidase crystals [29]. Chloroperoxidase is a heme enzyme, and is unusual in that it contains many features associated with the enzymatic activity of cytochromes P450, such as a proximal Cys ligand to heme. Cytochromes P450 are the major players in drug metabolism, carcinogen activation, and biosynthesis of physiologically important molecules, such as steroids, fat-soluble vitamins and fatty acids. Chloroperoxidase has been found to be a crucial model system for understanding cytochrome P450 reaction intermediates. The researchers first trapped compound III within the crystals. However, when the chloroperoxidase crystals were undergoing x-ray data collection at the synchrotron, UV/visible spectra from an online microspectrophotometer revealed that compound III was reduced first to a peroxo species, and then rapidly to compound 0. These researchers were thus able to obtain the structure of compound 0 through the construction of a high x-ray dose dataset.
Innovative uses of optical spectroscopy in crystallography
There are two emerging areas in crystallography where optical spectroscopy is being put to good use. The first is in the identification of protein crystals within crystallization trials. Noda et al. employed an automated confocal Raman spectroscopy microscope to screen hanging drop vapor diffusion protein crystallization trays for emerging crystals [34]. This microscope analyzed trays with high spatial resolution so that it was able to distinguish between real protein crystals and pseudo-protein crystals. The second area involves finding the location of protein crystals once they have been mounted for x-ray data collection. This is commonly done by eye using a video camera and visible light. With the increasing use of very small crystals for x-ray data collection at synchrotrons, and the advent of microdiffractometers and microfocus beamlines [41], one of the current challenges is locating the position of the crystal within a much larger loop. Vernede et al. have used fluorescence spectroscopy to aid in locating protein crystals that were either too small or too embedded in mother liquor to be easily observed with the naked eye [27]. Using UV light (266 nm) to excite the aromatic residues within the protein to fluoresce, they were able to clearly locate glowing crystals within loops (Figure 4).
Figure 4.
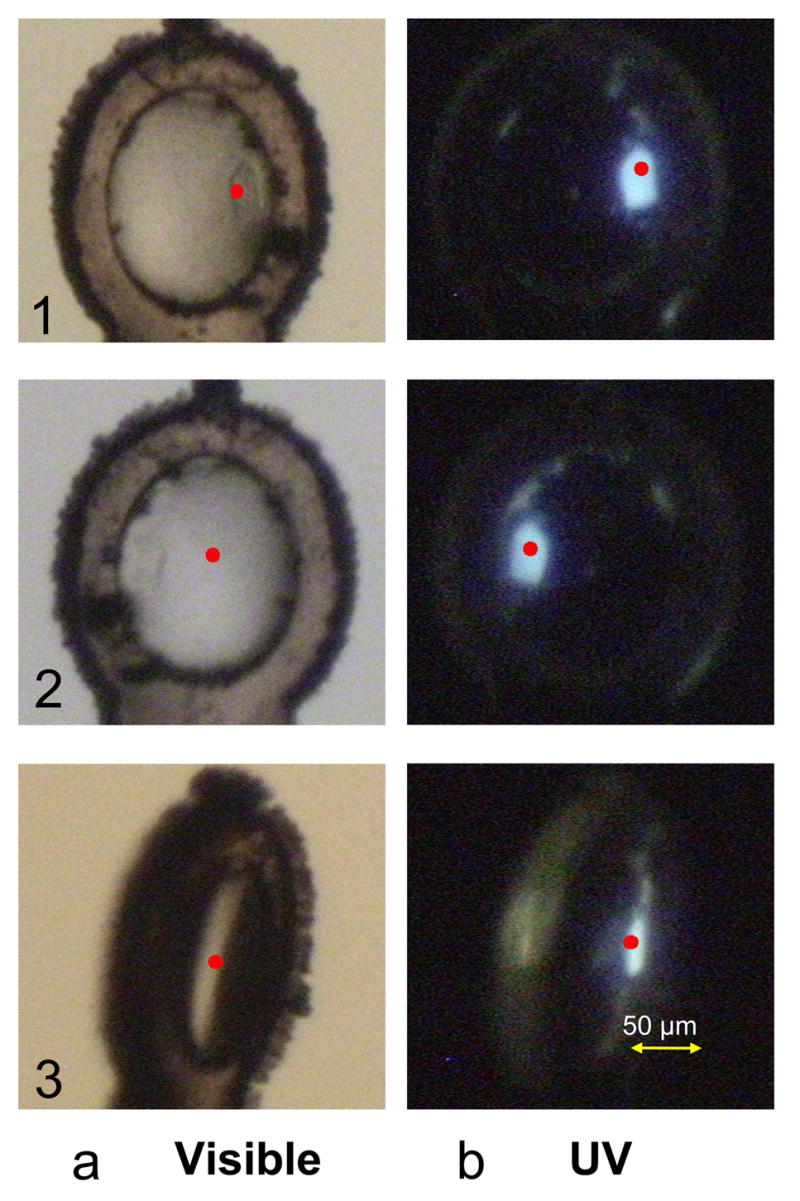
Fluorescence images recorded with the standard setup. A crystal of cephamycinase 2 (~20 μm in thickness) is shown in three different orientations in visible light, a or UV light, b. Red points show the crystal center as detected by the C3D software. The crystal is hardly detectable in visible light, so C3D fails to identify it correctly in orientations 2 and 3. In contrast, the crystal is easily identified under UV-laser illumination by both the user and the software, whatever the loop orientation. Figure and legend reprinted with permission from [27].
One area of crystallography where there is always room for ingenuity is in the calculation of an initial phase set for structure determination. Lately, the use of x-ray induced radiation damage for phasing has been increasing, as it provides the ability to collect x-ray data from a single crystal without the need for the introduction of heavy atoms [42,43]. However, it does require access to intense x-ray sources, in other words synchrotrons. One new direction is the use of UV-induced radiation damage, which has been used to obtain phases for multiple protein crystals, including lysozyme, trypsin, and ribonuclease A [28•]. The use of an on-line UV/visible microspectrophotometer was key to aligning the x-ray beam, spindle and microspectrophotometer optics, so that one of the microspectrophotometer optic cables could be used to deliver light to the crystal from a 266 nm UV laser. As this method does not require an intense x-ray source to induce damage, it may prove useful for phasing on a home source.
Improvements in single crystal microspectrophotometers
A new fluorescence microspectrophotometer, specifically designed for kinetic studies on protein crystals in combination with x-ray crystallography, can be used easily on or off x-ray beamlines [2]. One of the most appealing features of this setup is that it is built for 0o fluorescence detection, rather than the more normal 90o configuration. It uses a single objective, and is thus much more compact. The authors introduced a fluorophore into the switch region of GTPase H-Ras p21, a key protein in intracellular signaling that is linked to the development of a variety of cancer tumors. Upon GTP hydrolysis, the protein undergoes a conformational change involving the switch region. This conformational change can occur within the crystal lattice without loss of diffraction. Using a caged GTP, the reaction was initiated by photolysis. The conformational change was followed directly in the hanging drop crystallization tray, as well as in a crystal mounted in a loop in a vapor saturated gas stream, by combining the microspectrofluorimeter with a stereomicroscope. X-ray radiolysis, often used to decage substrates, was also investigated, and the system proved to be highly sensitive, with observed fluorescence changes suggesting that more was occurring in the crystals than simply the decaging event. The ability to combine this microspectrofluorimeter with minimal or no modification to stereomicroscopes and a variety of beamlines, promises to make this instrument a useful and adaptable tool within the single crystal microspectrophotometer arsenal.
Conclusions
Improvements in integrating optical spectroscopic methods with crystallographic techniques have increased our ability to follow chemical reactions in crystallized proteins, and trap reaction intermediates for x-ray crystal structure solution. These results have led to a profound understanding of complex enzyme and photoreceptor mechanisms. However, the synergy now extends beyond this. Other examples of optical spectroscopic applications within x-ray crystallography are now emerging, including the identification of crystalline material in crystallization experiments, in locating microcrystals mounted within loops for x-ray data collection, and following radiation damage in protein and DNA crystals, including its use in providing initial phases for structure determination. The future appears bright for the continuing integration of single crystal optical spectroscopy with x-ray crystallography.
Acknowledgments
TMR would like to thank Bryan Johnson for helpful discussions. We acknowledge financial support from the National Institutes of Health (GM-66569).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- •1.Pearson AR, Mozzarelli A, Rossi GL. Microspectrophotometry for structural enzymology. Curr Opin Struct Biol. 2004;14:656–662. doi: 10.1016/j.sbi.2004.10.007. Excellent beginners review on single crystal microspectrophotometry. [DOI] [PubMed] [Google Scholar]
- 2.Klink BU, Goody RS, Scheidig AJ. A newly designed microspectrofluorometer for kinetic studies on protein crystals in combination with x-ray diffraction. Biophys J. 2006;91:981–992. doi: 10.1529/biophysj.105.078931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••3.Bourgeois D, Royant A. Advances in kinetic protein crystallography. Curr Opin Struct Biol. 2005;15:538–547. doi: 10.1016/j.sbi.2005.08.002. An inclusive review focused on the mechanisms used by researchers to analyze kinetic crystallographic experiments. [DOI] [PubMed] [Google Scholar]
- ••4.Carey PR. Raman crystallography and other biochemical applications of Raman microscopy. Annu Rev Phys Chem. 2006;57:527–554. doi: 10.1146/annurev.physchem.57.032905.104521. A comprehensive review that highlights different ways single crystal Raman spectroscopy has been used in the study of biological systems. [DOI] [PubMed] [Google Scholar]
- 5.Enami N, Yoshimura K, Murakami M, Okumura H, Ihara K, Kouyama T. Crystal structures of archaerhodopsin-1 and -2: Common structural motif in archaeal light-driven proton pumps. J Mol Biol. 2006;358:675–685. doi: 10.1016/j.jmb.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 6.Padayatti PS, Sheri A, Totir MA, Helfand MS, Carey MP, Anderson VE, Carey PR, Bethel CR, Bonomo RA, Buynak JD, et al. Rational design of a beta-lactamase inhibitor achieved via stabilization of the trans-enamine intermediate: 1.28 A crystal structure of wt SHV-1 complex with a penam sulfone. J Am Chem Soc. 2006;128:13235–13242. doi: 10.1021/ja063715w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamichi H, Okada T. Local peptide movement in the photoreaction intermediate of rhodopsin. Proc Natl Acad Sci U S A. 2006;103:12729–12734. doi: 10.1073/pnas.0601765103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlsson GH, Nicholls P, Svistunenko D, Berglund GI, Hajdu J. Complexes of horseradish peroxidase with formate, acetate, and carbon monoxide. Biochemistry. 2005;44:635–642. doi: 10.1021/bi0483211. [DOI] [PubMed] [Google Scholar]
- 9.Echalier A, Goodhew CF, Pettigrew GW, Fulop V. Activation and catalysis of the di-heme cytochrome c peroxidase from Paracoccus pantotrophus. Structure. 2006;14:107–117. doi: 10.1016/j.str.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 10.Sukumar N, Chen ZW, Ferrari D, Merli A, Rossi GL, Bellamy HD, Chistoserdov A, Davidson VL, Mathews FS. Crystal structure of an electron transfer complex between aromatic amine dehydrogenase and azurin from Alcaligenes faecalis. Biochemistry. 2006;45:13500–13510. doi: 10.1021/bi0612972. [DOI] [PubMed] [Google Scholar]
- 11.Roujeinikova A, Scrutton NS, Leys D. Atomic level insight into the oxidative half-reaction of aromatic amine dehydrogenase. J Biol Chem. 2006;281:40264–40272. doi: 10.1074/jbc.M605559200. [DOI] [PubMed] [Google Scholar]
- 12.Key J, Moffat K. Crystal structures of deoxy and CO-bound bjFixLH reveal details of ligand recognition and signaling. Biochemistry. 2005;44:4627–4635. doi: 10.1021/bi047942r. [DOI] [PubMed] [Google Scholar]
- ••13.Moukhametzianov R, Klare JP, Efremov R, Baeken C, Goppner A, Labahn J, Engelhard M, Buldt G, Gordeliy VI. Development of the signal in sensory rhodopsin and its transfer to the cognate transducer. Nature. 2006;440:115–119. doi: 10.1038/nature04520. UV/visible and FTIR spectroscopies confirmed the trapping of two intermediates in phoborhodopsin crystals, and enabled comparison of the photocycle kinetics in crystals to that in membrane preparations. [DOI] [PubMed] [Google Scholar]
- 14.Katona G, Snijder A, Gourdon P, Andreasson U, Hansson O, Andreasson LE, Neutze R. Conformational regulation of charge recombination reactions in a photosynthetic bacterial reaction center. Nat Struct Mol Biol. 2005;12:630–631. doi: 10.1038/nsmb948. [DOI] [PubMed] [Google Scholar]
- 15.Nienhaus K, Ostermann A, Nienhaus GU, Parak FG, Schmidt M. Ligand migration and protein fluctuations in myoglobin mutant L29W. Biochemistry. 2005;44:5095–5105. doi: 10.1021/bi047513t. [DOI] [PubMed] [Google Scholar]
- •16.Katona G, Carpentier P, Niviere V, Amara P, Adam V, Ohana J, Tsanov N, Bourgeois D. Raman-assisted crystallography reveals end-on peroxide intermediates in a nonheme iron enzyme. Science. 2007;316:449–453. doi: 10.1126/science.1138885. Raman spectroscopy confirmed the entrapment of iron(III)-peroxo intermediates in superoxide reductase crystals, which displayed different conformations / interactions within the crystallographic asymmetric unit. [DOI] [PubMed] [Google Scholar]
- •17.Rajagopal S, Anderson S, Srajer V, Schmidt M, Pahl R, Moffat K. A structural pathway for signaling in the E46Q mutant of photoactive yellow protein. Structure. 2005;13:55–63. doi: 10.1016/j.str.2004.10.016. Laue data supplied structural images of the signaling event in the E46Q mutant of PYP. UV/visible spectroscopy allowed the identification of five different intermediates, whose structures were used to propose a mechanism for signaling. [DOI] [PubMed] [Google Scholar]
- 18.Padayatti PS, Helfand MS, Totir MA, Carey MP, Carey PR, Bonomo RA, van den Akker F. High resolution crystal structures of the trans-enamine intermediates formed by sulbactam and clavulanic acid and E166A SHV-1 β-lactamase. J Biol Chem. 2005;280:34900–34907. doi: 10.1074/jbc.M505333200. [DOI] [PubMed] [Google Scholar]
- 19.Totir MA, Padayatti PS, Helfand MS, Carey MP, Bonomo RA, Carey PR, van den Akker F. Effect of the inhibitor-resistant M69V substitution on the structures and populations of trans-enamine β-lactamase intermediates. Biochemistry. 2006;45:11895–11904. doi: 10.1021/bi060990m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gall A, Gardiner AT, Cogdell RJ, Robert B. Carotenoid stoichiometry in the LH2 crystal: no spectral evidence for the presence of the second molecule in the alpha/beta-apoprotein dimer. FEBS Lett. 2006;580:3841–3844. doi: 10.1016/j.febslet.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Pascal AA, Liu Z, Broess K, van Oort B, van Amerongen H, Wang C, Horton P, Robert B, Chang W, Ruban A. Molecular basis of photoprotection and control of photosynthetic light-harvesting. Nature. 2005;436:134–137. doi: 10.1038/nature03795. [DOI] [PubMed] [Google Scholar]
- 22.Sanii LS, Schill AW, Moran CE, El-Sayed MA. The protonation-deprotonation kinetics of the protonated Schiff base in bicelle bacteriorhodopsin crystals. Biophys J. 2005;89:444–451. doi: 10.1529/biophysj.105.059675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeremenko S, van Stokkum IH, Moffat K, Hellingwerf KJ. Influence of the crystalline state on photoinduced dynamics of photoactive yellow protein studied by ultraviolet-visible transient absorption spectroscopy. Biophys J. 2006;90:4224–4235. doi: 10.1529/biophysj.105.074765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanii LS, El-Sayed MA. Partial dehydration of the retinal binding pocket and proof for photochemical deprotonation of the retinal Schiff base in bicelle bacteriorhodopsin crystals. Photochem Photobiol. 2005;81:1356–1360. doi: 10.1562/2005-03-09-RA-458. [DOI] [PubMed] [Google Scholar]
- 25.Baxter RH, Krausz E, Norris JR. Photoactivation of the photosynthetic reaction center of Blastochloris viridis in the crystalline state. J Phys Chem B. 2006;110:1026–1032. doi: 10.1021/jp053697p. [DOI] [PubMed] [Google Scholar]
- 26.Efremov R, Gordeliy VI, Heberle J, Buldt G. Time-resolved microspectroscopy on a single crystal of bacteriorhodopsin reveals lattice-induced differences in the photocycle kinetics. Biophys J. 2006;91:1441–1451. doi: 10.1529/biophysj.106.083345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vernede X, Lavault B, Ohana J, Nurizzo D, Joly J, Jacquamet L, Felisaz F, Cipriani F, Bourgeois D. UV laser-excited fluorescence as a tool for the visualization of protein crystals mounted in loops. Acta Crystallogr D Biol Crystallogr. 2006;62 :253–261. doi: 10.1107/S0907444905041429. [DOI] [PubMed] [Google Scholar]
- •28.Nanao MH, Ravelli RB. Phasing macromolecular structures with UV-induced structural changes. Structure. 2006;14:791–800. doi: 10.1016/j.str.2006.02.007. A proposed phasing method that promises to be more effective than using x-ray damage, as it introduces more protein specific changes without the need for a synchrotron. [DOI] [PubMed] [Google Scholar]
- 29.Kuhnel K, Derat E, Terner J, Shaik S, Schlichting I. Structure and quantum chemical characterization of chloroperoxidase compound 0, a common reaction intermediate of diverse heme enzymes. Proc Natl Acad Sci U S A. 2007;104:99–104. doi: 10.1073/pnas.0606285103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubnovitsky AP, Ravelli RB, Popov AN, Papageorgiou AC. Strain relief at the active site of phosphoserine aminotransferase induced by radiation damage. Protein Sci. 2005;14:1498–1507. doi: 10.1110/ps.051397905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •31.Pearson AR, Pahl R, Kovaleva EG, Davidson VL, Wilmot CM. Tracking X-ray- derived redox changes in crystals of a methylamine dehydrogenase/amicyanin complex using single-crystal UV/Vis microspectrophotometry. J Synchrotron Radiat. 2007;14:92–98. doi: 10.1107/S0909049506051259. The authors devised a general composite data collection strategy that tracked reduction at multiple redox centers to ensure the resulting structures represented the true oxidized intermediates. [DOI] [PubMed] [Google Scholar]
- 32.Beitlich T, Kuhnel K, Schulze-Briese C, Shoeman RL, Schlichting I. Cryoradiolytic reduction of crystalline heme proteins: analysis by UV-Vis spectroscopy and X-ray crystallography. J Synchrotron Radiat. 2007;14:11–23. doi: 10.1107/S0909049506049806. [DOI] [PubMed] [Google Scholar]
- 33.McGeehan JE, Carpentier P, Royant A, Bourgeois D, Ravelli RB. X-ray radiation- induced damage in DNA monitored by online Raman. J Synchrotron Radiat. 2007;14:99–108. doi: 10.1107/S0909049506043251. [DOI] [PubMed] [Google Scholar]
- 34.Noda K, Sato H, Watanabe S, Yokoyama S, Tashiro H. Efficient characterization for protein crystals using confocal Raman spectroscopy. Appl Spectrosc. 2007;61:11–18. doi: 10.1366/000370207779701514. [DOI] [PubMed] [Google Scholar]
- 35.Portuondo-Campa E, Schenkl S, Dolder M, Chergui M, Landau EM, Haacke S. Absorption spectroscopy of three-dimensional bacteriorhodopsin crystals at cryogenic temperatures: effects of altered hydration. Acta Crystallogr D Biol Crystallogr. 2006;62:368–374. doi: 10.1107/S0907444906001399. [DOI] [PubMed] [Google Scholar]
- 36.Garman EF, McSweeney SM. Progress in research into radiation damage in cryo-cooled macromolecular crystals. Journal of Synchrotron Radiation. 2007;14:1–3. doi: 10.1107/S0909049506053015. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu N, Hirata K, Hasegawa K, Ueno G, Yamamoto M. Dose dependence of radiation damage for protein crystals studied at various X-ray energies. Journal of Synchrotron Radiation. 2007;14:4–10. doi: 10.1107/S0909049506049296. [DOI] [PubMed] [Google Scholar]
- 38.Fioravanti E, Vellieux FMD, Amara P, Madern D, Weik M. Specific radiation damage to acidic residues and its relation to their chemical and structural environment. Journal of Synchrotron Radiation. 2007;14:84–91. doi: 10.1107/S0909049506038623. [DOI] [PubMed] [Google Scholar]
- •39.Southworth-Davies RJ, Garman EF. Radioprotectant screening for cryocrystallography. Journal of Synchrotron Radiation. 2007;14:73–83. doi: 10.1107/S0909049506044177. Very interesting study analyzing the use of ascorbate, 2,2,6,6-tetramethyl-4-piperidone and reduced dithiothreitol as radioprotectants for thiols and disulfide bond radiation damage, monitored by an on-line UV/vis microspectrophotometer. [DOI] [PubMed] [Google Scholar]
- 40.Garman EF, Owen RL. Cryocooling and radiation damage in macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2006;62:32–47. doi: 10.1107/S0907444905034207. [DOI] [PubMed] [Google Scholar]
- 41.Perrakis A, Cipriani F, Castagna JC, Claustre L, Burghammer M, Riekel C, Cusack S. Protein microcrystals and the design of a microdiffractometer: current experience and plans at EMBL and ESRF/ID13. Acta Crystallogr D Biol Crystallogr. 1999;55:1765–1770. doi: 10.1107/s0907444999009348. [DOI] [PubMed] [Google Scholar]
- 42.Nanao MH, Sheldrick GM, Ravelli RB. Improving radiation-damage substructures for RIP. Acta Crystallogr D Biol Crystallogr. 2005;61:1227–1237. doi: 10.1107/S0907444905019360. [DOI] [PubMed] [Google Scholar]
- 43.Schiltz M, Bricogne G. Modelling and refining site-specific radiation damage in SAD/MAD phasing. Journal of Synchrotron Radiation. 2007;14:34–42. doi: 10.1107/S0909049506038970. [DOI] [PubMed] [Google Scholar]