Abstract
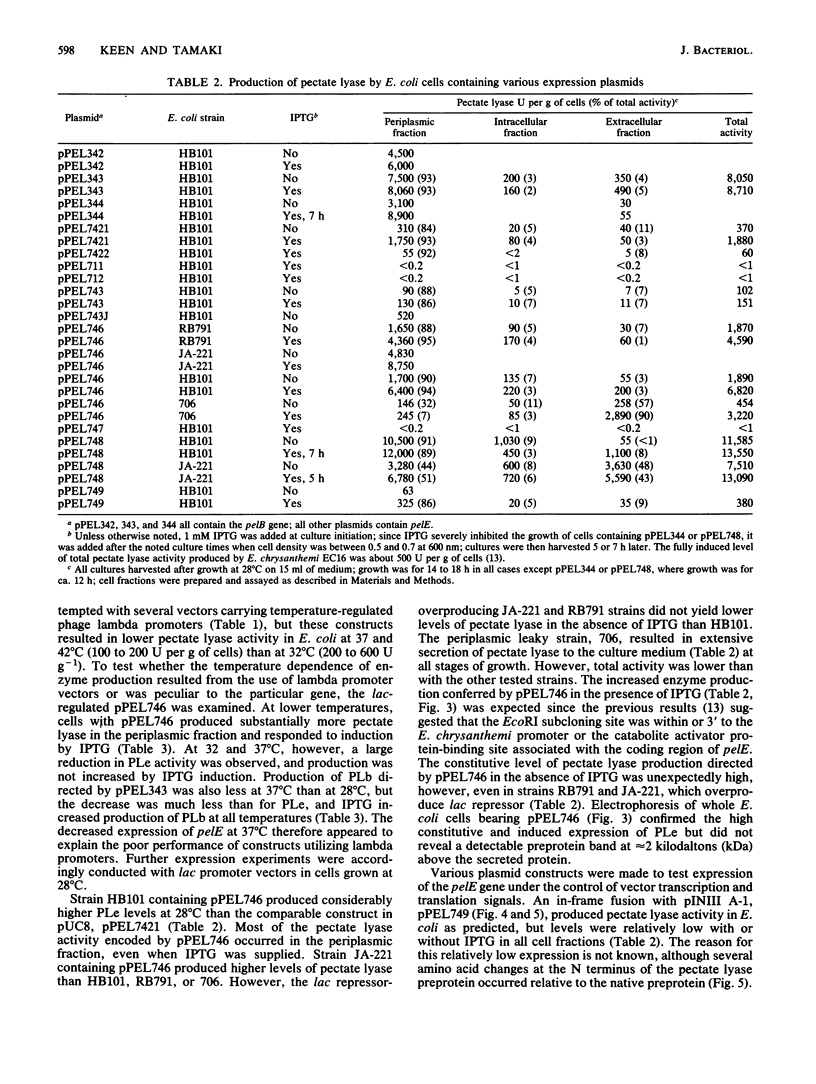
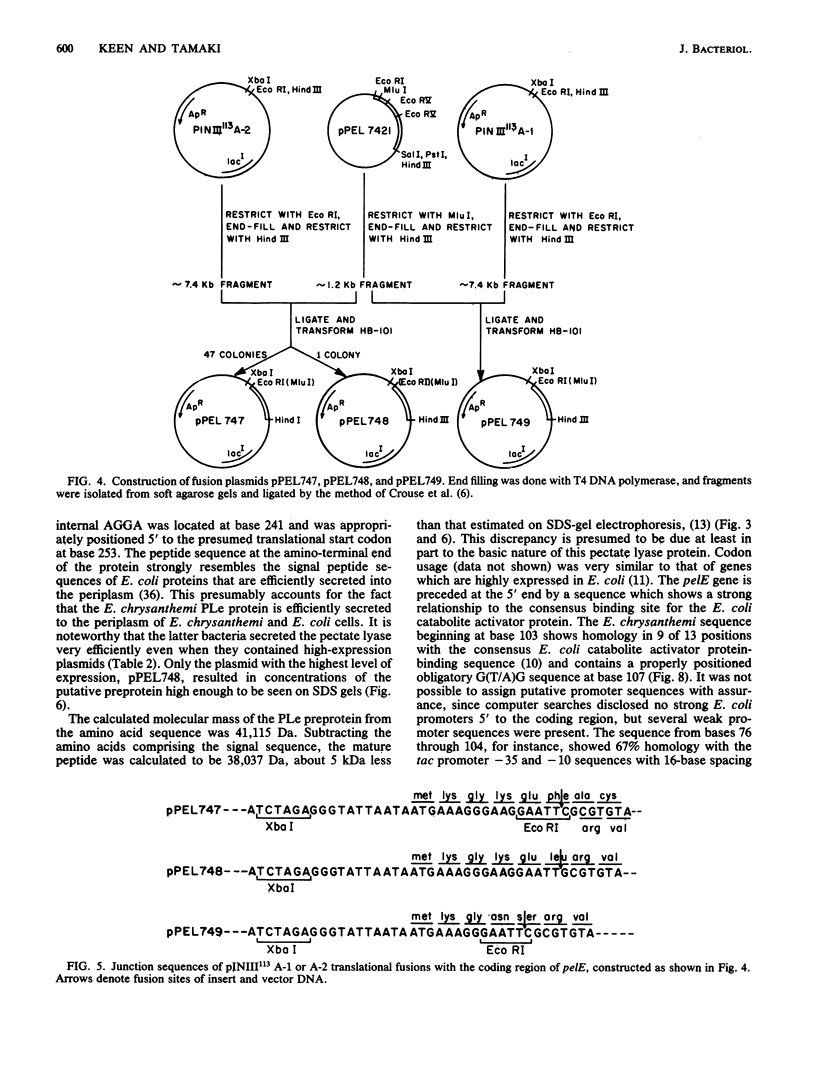
The pelB and pelE genes from Erwinia chrysanthemi EC16, which encode different pectate lyase enzymes, were sequenced and expressed at a high level in Escherichia coli. The genes possessed little similarity to each other in 5' signal regions, signal peptide sequences, coding sequences, or 3' noncoding regions. Both genes contained their own promoters as well as sequences 3' to the coding regions with considerable secondary structure which may function as rho-independent transcriptional termination signals. High-level expression plasmids were constructed with both genes, which led to 20% or more of E. coli cellular protein. The pectate lyases were secreted efficiently to the periplasm and, to a lesser extent, the culture medium. The mature proteins in E. coli periplasmic fractions were obtained in milligram amounts and high purity with a single-column affinity purification method. E. coli cells which produced high amounts of the pelE protein macerated potato tuber tissue as efficiently as E. chrysanthemi EC16 cells but cells producing high amounts of the pelB protein were less effective. Thus, the pelE gene product is an important pathogenicity factor which solely enables E. coli to cause a soft-rot disease on potato tuber tissue under laboratory conditions.
Full text
PDF



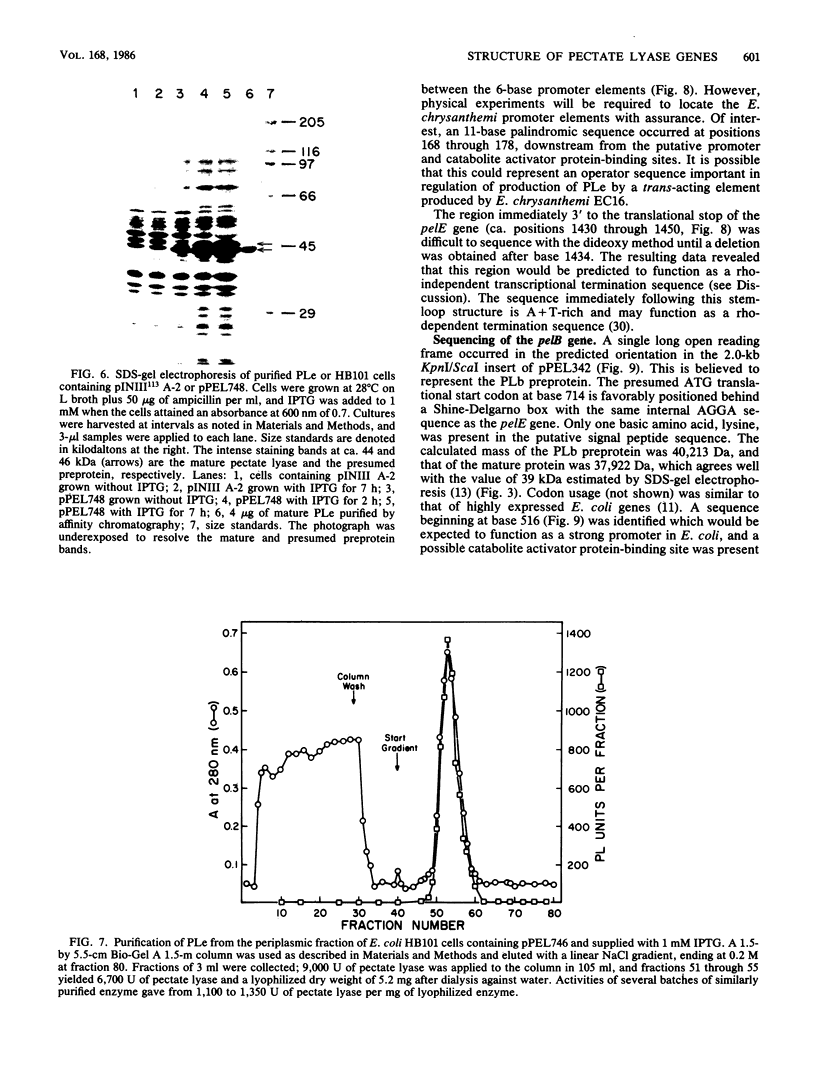
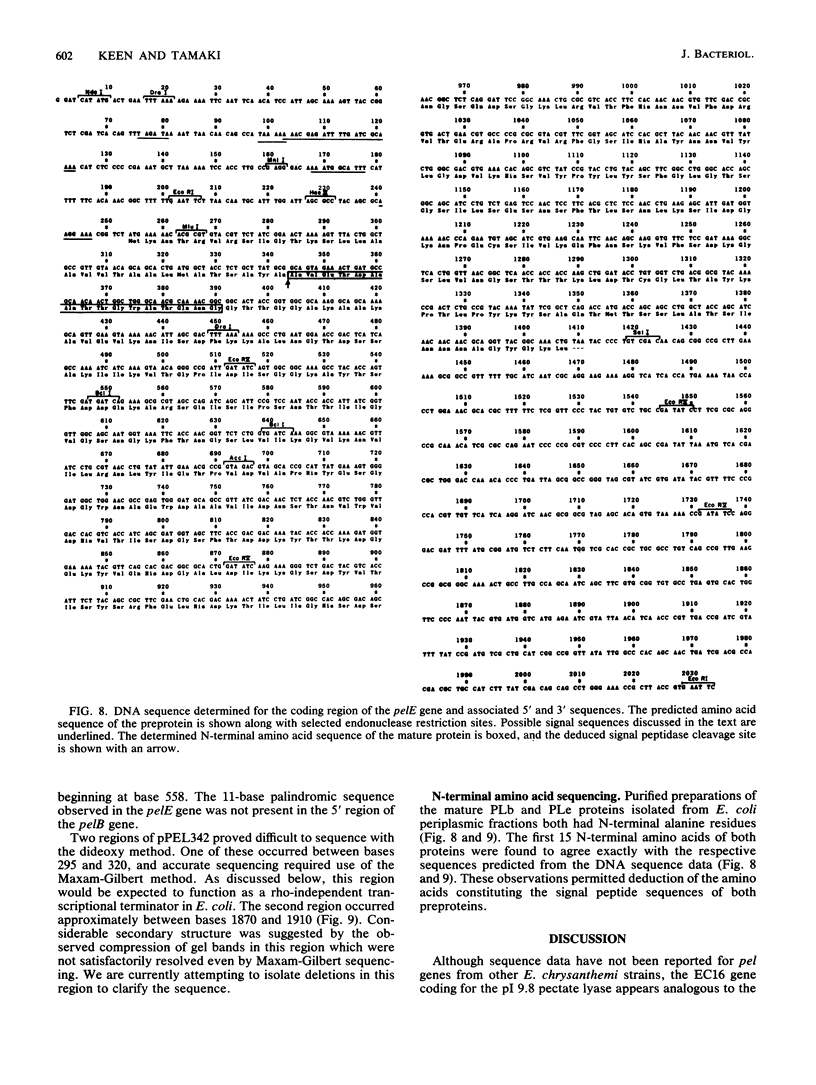
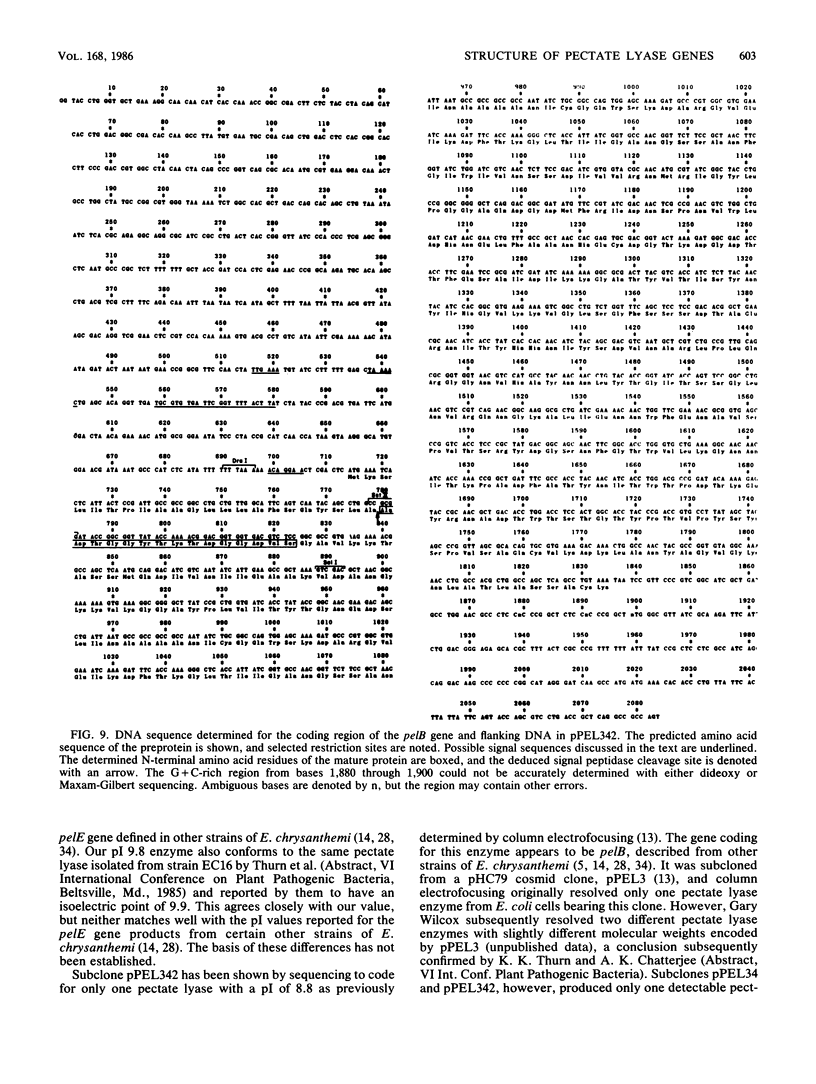
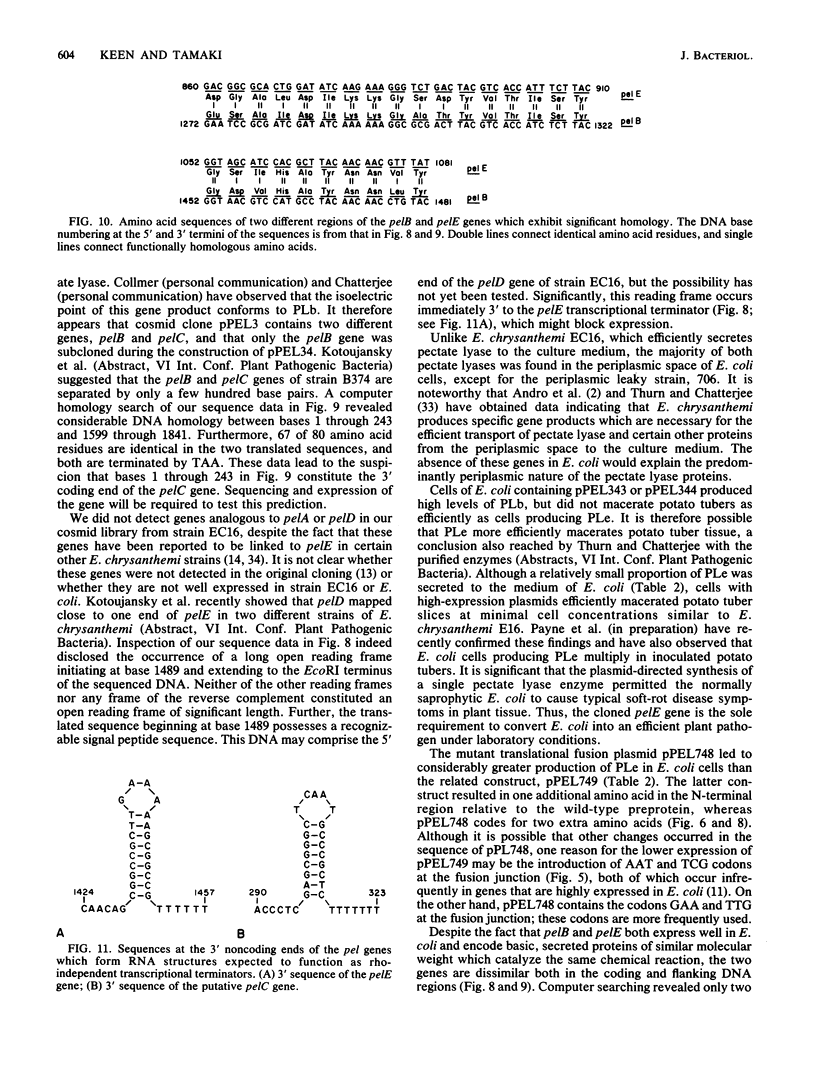


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann E., Bröker M., Wurm F. Expression of Herpes simplex virus type 1 glycoprotein C antigens in Escherichia coli. Gene. 1984 Dec;32(1-2):203–215. doi: 10.1016/0378-1119(84)90048-9. [DOI] [PubMed] [Google Scholar]
- Andro T., Chambost J. P., Kotoujansky A., Cattaneo J., Bertheau Y., Barras F., Van Gijsegem F., Coleno A. Mutants of Erwinia chrysanthemi defective in secretion of pectinase and cellulase. J Bacteriol. 1984 Dec;160(3):1199–1203. doi: 10.1128/jb.160.3.1199-1203.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biville F., Guiso N. Evidence for the presence of cAMP, cAMP receptor and transcription termination factor rho in different gram-negative bacteria. J Gen Microbiol. 1985 Nov;131(11):2953–2960. doi: 10.1099/00221287-131-11-2953. [DOI] [PubMed] [Google Scholar]
- Collmer A., Schoedel C., Roeder D. L., Ried J. L., Rissler J. F. Molecular cloning in Escherichia coli of Erwinia chrysanthemi genes encoding multiple forms of pectate lyase. J Bacteriol. 1985 Mar;161(3):913–920. doi: 10.1128/jb.161.3.913-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse G. F., Frischauf A., Lehrach H. An integrated and simplified approach to cloning into plasmids and single-stranded phages. Methods Enzymol. 1983;101:78–89. doi: 10.1016/0076-6879(83)01006-x. [DOI] [PubMed] [Google Scholar]
- Davis K. R., Lyon G. D., Darvill A. G., Albersheim P. Host-Pathogen Interactions : XXV. Endopolygalacturonic Acid Lyase from Erwinia carotovora Elicits Phytoalexin Accumulation by Releasing Plant Cell Wall Fragments. Plant Physiol. 1984 Jan;74(1):52–60. doi: 10.1104/pp.74.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison J., Heusterspreute M., Merchez M., Brunel F. Vectors with restriction-site banks. I. pJRD158, a 3903-bp plasmid containing 28 unique cloning sites. Gene. 1984 Jun;28(3):311–318. doi: 10.1016/0378-1119(84)90148-3. [DOI] [PubMed] [Google Scholar]
- Dean C., Elzen P., Tamaki S., Dunsmuir P., Bedbrook J. Differential expression of the eight genes of the petunia ribulose bisphosphate carboxylase small subunit multi-gene family. EMBO J. 1985 Dec 1;4(12):3055–3061. doi: 10.1002/j.1460-2075.1985.tb04045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijsegem F., Toussaint A., Schoonejans E. In vivo cloning of the pectate lyase and cellulase genes of Erwinia chrysanthemi. EMBO J. 1985 Mar;4(3):787–792. doi: 10.1002/j.1460-2075.1985.tb03698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982 Nov 25;10(22):7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen N. T., Dahlbeck D., Staskawicz B., Belser W. Molecular cloning of pectate lyase genes from Erwinia chrysanthemi and their expression in Escherichia coli. J Bacteriol. 1984 Sep;159(3):825–831. doi: 10.1128/jb.159.3.825-831.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotoujansky A., Diolez A., Boccara M., Bertheau Y., Andro T., Coleno A. Molecular cloning of Erwinia chrysanthemi pectinase and cellulase structural genes. EMBO J. 1985 Mar;4(3):781–785. doi: 10.1002/j.1460-2075.1985.tb03697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazzaroni J. C., Portalier R. C. Genetic and biochemical characterization of periplasmic-leaky mutants of Escherichia coli K-12. J Bacteriol. 1981 Mar;145(3):1351–1358. doi: 10.1128/jb.145.3.1351-1358.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg E. M. Plasmid prefix designations registered by the Plasmid Reference Center 1977-1985. Plasmid. 1986 Jan;15(1):57–92. doi: 10.1016/0147-619x(86)90014-4. [DOI] [PubMed] [Google Scholar]
- Lei S. P., Lin H. C., Heffernan L., Wilcox G. Cloning of the pectate lyase genes from Erwinia carotovora and their expression in Escherichia coli. Gene. 1985;35(1-2):63–70. doi: 10.1016/0378-1119(85)90158-1. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Morrison D. A. Transformation in Escherichia coli: cryogenic preservation of competent cells. J Bacteriol. 1977 Oct;132(1):349–351. doi: 10.1128/jb.132.1.349-351.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwankwo D. O., Guterman S. K. Purification of RNA polymerase and transcription-termination factor Rho from Erwinia carotovora. Eur J Biochem. 1985 Jan 15;146(2):383–389. doi: 10.1111/j.1432-1033.1985.tb08664.x. [DOI] [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A convenient and adaptable package of computer programs for DNA and protein sequence management, analysis and homology determination. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):643–655. doi: 10.1093/nar/12.1part2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C. A vector that uses phage signals for efficient synthesis of proteins in Escherichia coli. J Mol Appl Genet. 1983;2(1):1–10. [PubMed] [Google Scholar]
- Remaut E., Tsao H., Fiers W. Improved plasmid vectors with a thermoinducible expression and temperature-regulated runaway replication. Gene. 1983 Apr;22(1):103–113. doi: 10.1016/0378-1119(83)90069-0. [DOI] [PubMed] [Google Scholar]
- Reverchon S., Hugouvieux-Cotte-Pattat N., Robert-Baudouy J. Cloning of genes encoding pectolytic enzymes from a genomic library of the phytopathogenic bacterium, Erwinia chrysanthemi. Gene. 1985;35(1-2):121–130. doi: 10.1016/0378-1119(85)90164-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Ho Y. S., Shatzman A. The use of pKc30 and its derivatives for controlled expression of genes. Methods Enzymol. 1983;101:123–138. doi: 10.1016/0076-6879(83)01009-5. [DOI] [PubMed] [Google Scholar]
- Thurn K. K., Chatterjee A. K. Single-site chromosomal Tn5 insertions affect the export of pectolytic and cellulolytic enzymes in Erwinia chrysanthemi EC16. Appl Environ Microbiol. 1985 Oct;50(4):894–898. doi: 10.1128/aem.50.4.894-898.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witholt B., Boekhout M., Brock M., Kingma J., Heerikhuizen H. V., Leij L. D. An efficient and reproducible procedure for the formation of spheroplasts from variously grown Escherichia coli. Anal Biochem. 1976 Jul;74(1):160–170. doi: 10.1016/0003-2697(76)90320-1. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zink R. T., Chatterjee A. K. Cloning and expression in Escherichia coli of pectinase genes of Erwinia carotovora subsp. carotovora. Appl Environ Microbiol. 1985 Mar;49(3):714–717. doi: 10.1128/aem.49.3.714-717.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]